Abstract
Background
Established bacterial diagnostic techniques for orthopaedic-related infections rely on a combination of imperfect tests that often can lead to negative culture results. Spectroscopy is a tool that potentially could aid in rapid detection and differentiation of bacteria in implant-associated infections.
Questions/purposes
We asked: (1) Can principal component analysis explain variation in spectral curves for biofilm obtained from Staphylococcus aureus, Staphylococcus epidermidis, and Pseudomonas aeruginosa? (2) What is the accuracy of Fourier transformed-near infrared (FT-NIR)/multivariate data analysis in identifying the specific species associated with biofilm?
Methods
Three clinical isolates, S aureus, S epidermidis, and P aeruginosa were cultured to create biofilm on surgical grade stainless steel. At least 52 samples were analyzed per group using a FT-NIR spectrometer. Multivariate and principal component analyses were performed on the spectral data to allow for modeling and identification of the bacterial species.
Results
Spectral analysis was able to correctly identify 86% (37/43) of S aureus, 89% (16/18) of S epidermidis, and 70% (28/40) of P aeruginosa samples with minimal error. Overall, models developed using spectral data preprocessed using a combination of standard normal variant and first-derivative transformations performed much better than models developed with the raw spectral data in discriminating between the three classes of bacteria because of its low Type 1 error and large intermodel distinction.
Conclusions
The use of spectroscopic methods to identify and classify bacterial biofilms on orthopaedic implant material is possible and improves with advanced modeling that can be obtained rapidly with little error. The sensitivity for identification was 97% for S aureus (95% CI, 88-99%), 100% for S epidermidis (95% CI, 95–100%), and 77% for P aeruginosa (95% CI, 65–86%). The specificity of the S aureus was 86% (95% CI, 3–93%), S epidermidis was 89% (95% CI, 67–97%), and P aeruginosa was 70% (95% CI, 55–82%).
Clinical Relevance
This technique of spectral data acquisition and advanced modeling should continue to be explored as a method for bacterial biofilm identification. A spectral databank of bacterial and potentially contaminating tissues should be acquired initially through an in vivo animal model and quickly transition to explanted devices and the clinical arena.
Introduction
Some bacteria live in surface-associated communities referred to as biofilms. By volume, biofilms are composed of approximately 5% bacteria and 95% extracellular polymeric substance secreted by bacteria and composed of lipids, proteins, and polysaccharides [11]. These biofilms provide an environment in which bacteria communicate through various signaling pathways such as quorum sensing or detachment, take on differing forms of virulence, and establish a tolerance to typical antimicrobial therapy [5, 9, 22]. Biofilms are one of the reasons orthopaedic implant-associated infections are so difficult to treat [20]. Diagnosis of implant-related infections is often difficult. Established bacterial diagnostic techniques include joint aspiration with cell count and culture and intraoperative tissue culture after explantation for guidance of antibiotic therapy, but false-negative test results are common [15], raising the suspicion that patients with a diagnosis of aseptic failure of prostheses may in fact have undiagnosed infections [17, 23]. Newer microscopic, molecular, and spectroscopic techniques have been used for detection of bacteria on metal implants ex vivo which requires explantation, ultrasonication of the explant, and destruction of biofilm and explant [5, 13]. These methods often aid in bacterial diagnosis that could guide surgical treatment and antibiotic therapy. Despite decreasing processing times (traditional culture, 48 to 120 hours; PCR, 6 hours; immunofluorescent microscopy, 3 hours), these methods remain unavailable intraoperatively and require implant removal [4].
A quick method that would allow for rapid detection and characterization of bacterial biofilms at the time of surgery and without implant removal or destruction as is needed with other techniques such as sonication, has been proposed with the use of Fourier-transformed near infrared (FT-NIR) spectroscopy [3, 16]. Infrared and Raman vibrational spectroscopy are capable of collecting detailed chemical, biochemical, and physical information regarding biological systems from properties of chemical bonds in a short time (seconds). Traditional spectral data have been analyzed by assignment of specific peaks to a single property (specific chemical markers) using a univariate approach. This analysis is difficult for complex biological systems that are best described using multiple factors that often are closely related to each other. The average single colony bacillus, Escherichia coli, contains 2000 to 4500 different types of small molecules, comprised of amino acids, carbohydrates, and fatty acids. Since the same groups are present in other bacteria, it is difficult to use assignment of specific functional groups as a basis for classification [14]. The use of two spectroscopic methods, infrared (IR) and Raman are complimentary vibrational spectroscopic techniques and, when coupled to fiber optic probes, have emerged as potential tools for biological and diagnostic medical applications [3, 16]. One advantage of near infrared (NIR, 700 nm–2500 nm) and Raman spectroscopy is that they can easily be coupled via the existing optical technology, whereas the better known mid-infrared ([MIR], 2500 nm–10,000 nm) requires optical halide fibers. Near infrared is capable of greater penetration (mm) of materials compared with MIR (micron) allowing for better characterization. Although NIR spectra are broad with no visible spectral differences, it is the primary tool used in characterization of biological and nonbiological materials.
Two methods to evaluate and differentiate complex biological systems such as biofilms are multivariate data analysis and principal component analysis; these methods allow all the properties of test material and spectra to be analyzed at the same time [6]. Principal component analysis visually shows the variance between and within objects being analyzed.
Currently the diagnostic capabilities of implant-associated infections are limited to imperfect tests. This can lead to culture negative infections of 20% or more of the time. The purpose of our study was to show the use of FT-NIR coupled with multivariate data analysis to characterize commonly occurring implant-associated bacterial biofilms adherent to a common orthopaedic implant material for real-time analysis that may improve our capabilities to identify and treat implant-related infections. Specifically, we asked: (1) Can principal component analysis differentiate the spectral curves for biofilm obtained from Staphylococcus aureua, Staphylococcus epidermidis, and Pseudomonas aeruginosa? (2) What is the accuracy of FT-NIR/multivariate data analysis in identifying the specific species associated with biofilm?
Materials and Methods
Bacteria Preparation
Three clinical isolates of S aureus, S epidermidis, and P aeruginosa were identified and characterized by our institution’s microbiology department. One colony of each type of bacteria isolate was placed in a separate vial containing 20 cc of Brain Heart Infusion broth (Fisher Scientific, Pittsburgh, PA, USA) and then incubated at 37° C for 18 hours to create an inoculum. These three separate preparations were diluted to a standard optical density with absorbance of 1.0 at 600 nm wavelength using an ultraviolet spectrometer. The bacterial concentrations at that standard optical density were calculated as an average of 5.3 × 108 colony forming units/mL.
Reactor Setup and Sample Preparation
Small discs were cut from stainless steel 316 L (1 cm diameter, 0.004 cm thickness) (Small Parts, Miami Lakes, FL, USA; wet jet cut and polished by Wilson Works, Morgantown, WV, USA) and were secured in the CDC biofilm reactor. The biofilm reactor is a rugged system that incorporates eight polypropylene rods in a circular orientation inside a 1-L container; in the center of the aligned rods is a baffled stir bar [8] (Fig. 1). The back of each disc, which faces away from the interior of the reactor, was marked before assembly. Each disc was sonicated for 2 minutes at 42 kHz (Crest Ultrasonics, Springfield, NJ, USA) and then washed with 70% alcohol. Three discs were secured in the eight columns for a total of 24 discs per reactor. The reactor was assembled completely before autoclaving it for 15 minutes at 132° C. After allowing the reactor to cool to room temperature, 450 mL of phosphate buffered saline (Fisher Scientific, Waltham, MA, USA) was placed in the reactor with 45 mL of Brain Heart Infusion broth. Five milliliters from the inoculums prepared as above were added to the solution in the reactor. The reactor was placed under a sterile hood on a stirring hotplate set to 100 rpm and 37° C using a temperature probe to ensure a stable temperature during incubation. The reactor operated under batch conditions for 72 hours before removal of samples for spectroscopic analysis. A total of 96 samples (discs) were cultured for S aureus, 80 for S epidermidis, and 52 for P aeruginosa. Sample groups were limited owing to technical difficulties, machine error, and gross contamination. Batches of samples were obtained throughout the course of a year. Fifteen sample discs were incubated in a similar fashion without the presence of bacteria to provide a control group.
Fig. 1.

The biofilm reactor is a device that incorporates eight polypropylene rods in a circular orientation inside a 1-L container; in the center of the aligned rods is a baffled stir bar.
Development of NIR Calibration
Using sterile technique, each column was removed under the hood and washed five times in 50-mL tubes of phosphate buffered saline for 10 seconds each to remove any planktonic bacteria. Each disc was removed individually, placed in small sterile plastic Petri dishes, and underwent immediate spectroscopic analysis using the Bruker Matrix FT-NIR spectrometer (Billerica, MA, USA). This unit was equipped with a fiberoptic sampling probe for solids and liquids (IN263E) operating in the diffuse reflectance mode 120,004 cm−1 to 4000 cm−1 (833 nm–2500 nm). The probe was aligned such that its collection optic tip was perpendicular to the surface of the sample disc for each scan at a distance of approximately 1.5 mm from the sample. Each specimen was scanned 15 times and averaged to a single spectrum producing one spectrum for each disc. The 15 scans were completed in less than 30 seconds.
The total sample set was 228 spectral specimens consisting of 96 samples of S aureus, 80 samples of S epidermidis, and 52 samples of P aeruginosa. Spectral specimens were divided into calibration and testing specimen sets, respectively. A priori determination of the percentage of total samples to be assigned to the calibration and validation set was determined. Fifty-six percent of the total samples of each species were randomly assigned to the calibration set. The remaining 44% of each species was assigned to their respective test set. The calibration data set (127 samples) consisted of 53, 34, and 40 samples that were randomly selected from S aureus, S epidermidis, and P aeruginosa, respectively. Before development of calibration models, all spectra data were standardized using auto scaling, and principal component analysis was performed on the specimens to observe any clustering or separation in the specimen set. Separate calibration models were developed for each of the bacteria species using the whole FT-NIR spectra range (833 nm–2500 nm). The raw spectra data and three other preprocessed spectra variations were used to develop models for each of the three species. These preprocessing methods include First derivative Savitzky-Golay preprocessing (15-point smoothing second polynomial order), standard normal variate, and a combination of standard normal variate and first derivative. To prevent underfitting or overfitting of the calibration models, the Unscrambler® X software (CAMO Software Inc, Woodbridge, NJ, USA; http://www.camo.com/rt/Products/Unscrambler/unscrambler.html) for multivariate data analysis was used to determine the optimal number of principal components on the explained and residual variance plots of each model.
Spectroscopic Data Acquisition
The validation set (101 samples) consisted of 43, 40, and 18 samples of S aureus, S epidermidis, and P aeruginosa, respectively. These samples were evaluated in a fashion similar to the calibration set.
Spectroscopic Data Analysis
Multivariate data analysis of raw spectral data was performed using The Unscrambler® 9.7 software. The raw spectra and spectra preprocessed using standard normal variant, first derivative, and combination of standard normal variant and first-derivative preprocessed spectra were used for development of calibrations to identify possible improvements in interpretation of the spectra, integrity, and applicability of the calibration models.
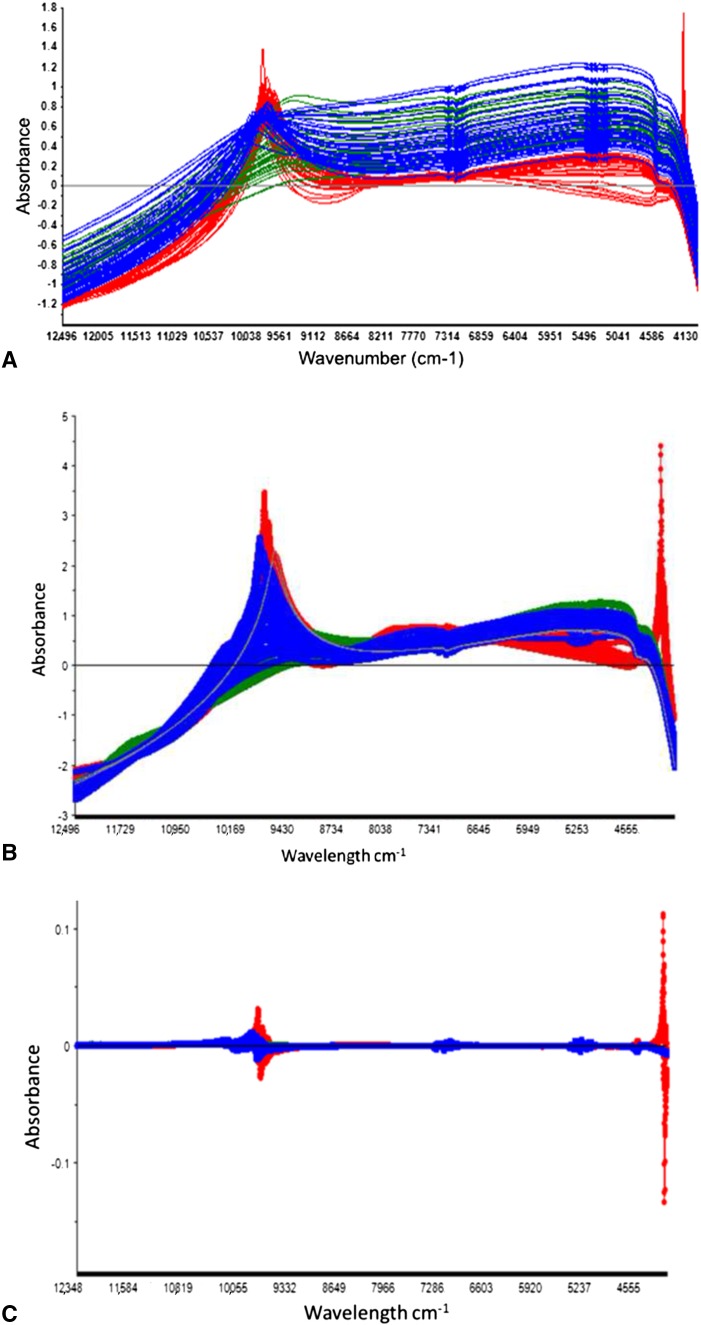
The NIR raw spectra, standard normal variant, first derivative, and combination of standard normal variant and first-derivative preprocessed spectra for all three bacterial species were used for development of calibrations (Fig. 2). The raw spectra of P aeruginosa appeared different from the other two groups, with higher absorption at the region 10,000 to 9245 cm−1 (Fig. 2A). The first derivative, standard normal variant, and standard normal variant + first-derivative preprocessed spectra displayed baseline differences along the NIR spectra region, but no clear visual differentiation according to bacteria group was seen because maximum peaks in the NIR spectra region 10,000 to 9245 cm−1 exhibited broad absorbance ranges (Fig. 2B–D).
Fig. 2A–D.
The raw spectra of the three classes of bacteria we used for development of calibrations are shown. (A) P aeruginosa (red) has higher absorption at 10,000 to 9245 cm−1. (B) First-derivative preprocessed spectra show broad absorbance ranges and separation is difficult owing to lack of distinct peaks. (C) Standard normal variant preprocessed data show improved distinction between the P aeruginosa and the gram-positive species. (D) Standard normal variant + first-derivative preprocessed data have increasingly distinct characteristics to the gram-positive species. Red = P aeruginosa. Blue = S aureus. Green = S epidermidis.
After development of calibration models for each class of bacteria, the models were tested on validation samples that were not included in model development according to the soft independent modeling of class analogies classification method [5] at the 5% significance level. Before using the models to predict class membership for new samples, the model specificity was evaluated to check whether the classes overlapped or were sufficiently distinct from each other. Specific tools in The Unscrambler® software setup, such as model distance and modeling power, were available for this purpose. The model distance plot showed the distances between different models; a distance larger than three was an indication that the models were different and good for class separation. The model power plot showed the contribution of variables to the model; variables with modeling power close to one were important for the model while variables less than 0.3 were of little importance for the model.
Before testing new samples, the specificities of the calibration models were evaluated to ensure that model classes did not overlap or were sufficiently distant from each other. The model distance for all models developed using raw spectral data and other spectral data preprocessed using standard normal variant, first derivative, and standard normal variant + first-derivative models was greater than three when compared with models for S aureus.
The 15 control sample discs showed an absence of spectral readings on all discs. These control discs were evaluated for colony forming units and showed no bacterial growth.
Determining Bacterial Count and Presence of Biofilm
After spectroscopic data acquisition, samples of discs were placed in 1 mL of phosphate buffered saline in a 15-mL tube which was sonicated at 42 kHz for 60 seconds to remove biofilm from the disc into solution. Each tube then was vortexed for 30 seconds to break up biofilm clusters in the phosphate buffered saline [1]. Using serial log dilutions and agar plating, viable colony forming units were counted after incubation at 37° C for 48 hours. These samples were used for species identification by the clinical microbiologist (JGT) to confirm the presence of bacteria and absence of contamination on the discs used for spectroscopic analysis. We found that all discs produced colony forming units that were concordant with inoculant.
Results
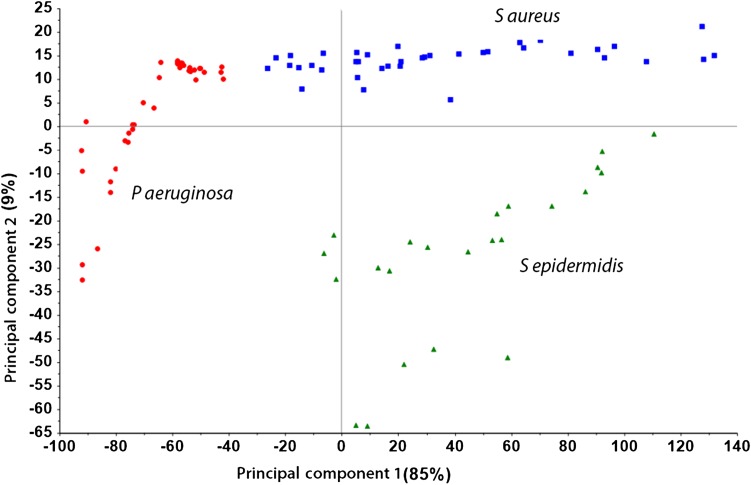
Principal component analysis was able to transform the correlated variables (which the intensities at different wavelengths are) to small uncorrelated variables (latent variables). The first principal component explained 85% of variation in spectral data and the first four principal components explained almost 98% of the variation observed. Using the optimum number of principal components and the total explained variance (%) in spectra data for principal component analysis, models were developed for each bacterial class using raw spectra (S aureus 1, S epidermidis 2, P aeruginosa 4), standard normal variant (S aureus 3, S epidermidis 3, P aeruginosa 4), first-derivative (S aureus 6, S epidermidis 5, P aeruginosa 6), and a combination of standard normal variant + first-derivative (S aureus 4, S epidermidis 5, P aeruginosa 5) preprocessing methods (Table 1). Overall, models developed using spectral data preprocessed using a combination of standard normal variant and first-derivative transformation performed much better than models developed with the raw spectral data in discriminating between the three classes of bacteria because of its low Type 1 error and large intermodel distance. Model distance is a measurement of model accuracy. Principal component analysis of all raw spectral data clustered spectra into groups for which the first two principal components are best seen visually (Fig. 3).
Table 1.
Optimum number of principal components and explained variance
| Preprocessing method | S aureus | S epidermidis | P aeruginosa |
|---|---|---|---|
| Raw spectra | |||
| Number of principal components | 1 | 2 | 4 |
| Explained variance | 99% | 99% | 99% |
| Model distance | 1 | 33 | 16 |
| First derivative | |||
| Number of principal components | 6 | 5 | 6 |
| Explained variance | 99% | 98% | 97% |
| Model distance | 1 | 87 | 77 |
| Single normal variant | |||
| Number of principal Components | 3 | 3 | 4 |
| Explained variance | 99% | 99% | 98% |
| Model distance | 1 | 107 | 42 |
| Single normal variant + first derivative | |||
| Number of principal components | 4 | 5 | 5 |
| Explained variance | 98% | 97% | 98% |
| Model distance | 1 | 268 | 92 |
Fig. 3.
This plot is an example of the first principal component against the second principal component of S aureus, S epidermidis, and P aeruginosa (Blue = S aureus, Green = S epidermidis, Red = P aeruginosa). Principal component analysis was able to transform the correlated variables (the intensities at different wavelengths) to small uncorrelated variables (latent variables). The first principal component explained 85% of variation in spectral data.
In a principal component space, a plot of score vectors (score plot) visually reveals the variance between and within objects being analyzed; in other words, how objects being analyzed are related to each other is captured in this principal component space. The higher the score between two objects the greater the chances are that the two objects are different from each other.
The first principal component captures the most important variance between the three bacterial species (83%) (Fig. 3). The majority of the S aureus and S epidermidis samples were clustered on the positive axis of the first principal component whereas P aeruginosa samples mostly were clustered on the negative side.
The second principal component captures only 9% of the variability in the three class of bacteria. The second score vector seems to capture the difference between the S aureus and S epidermidis samples.
Given the clustering we observed, we believe the three sample classes are different from each other based on their spectra characteristics. The S aureus and S epidermidis sample groups may share some similar characteristics since the majority are on the same side of the principal components. The similarity in spectra characteristics between S aureus and S epidermidis may be a result of their chemical or biological structure.
Accuracy of FT-NIR/Multivariate Data Analysis
Calibration models developed using raw spectral analysis were able to correctly identify 86% of S aureus, 89% of S epidermidis, and 70% of P aeruginosa. The computational analysis used in the soft independent modeling by class analogy classification and the test that quantified the risk of Type I error associated to the models ranged from 11% to 30%. Sensitivities associated with the models ranged from 77% to 100% with sensitivity reaching 100% for all calculations using the standard normal variant + first-derivative model (Table 2). Overall, spectra preprocessing using a combination of a single normal variant + first derivative appears best to improve the performance of the classification models in separation of the three bacterial species when evaluating via the modeling distance (Table 1), although there was reduction in the accuracies of identification of individual species but the Type II error was low (Table 2).
Table 2.
Members identified in each class
| Measured values | S aureus (n = 43) | S epidermidis (n = 18) | P aeruginosa (n = 40) |
|---|---|---|---|
| Adherent bacteria (colony forming units/cm2) | 5 × 103 ± 3 × 103 | 5.8 × 104 ± 3.9 × 104 | 1.66 × 105 ± 1.03 × 105 |
| Raw spectra | |||
| Discs identified | 37 | 16 | 28 |
| Sensitivity | 97% (88%–99%) | 100% (95%–100%) | 77% (65%–86%) |
| Specificity | 86% (73%–93%) | 89% (67%–97%) | 70% (55%–82%) |
| First derivative | |||
| Discs identified | 36 | 16 | 25 |
| Sensitivity | 100% (93%–100%) | 83% (74%–90%) | 100% (94%–100%) |
| Specificity | 84% (68%–92%) | 89% (67%–97%) | 63% (47%–78%) |
| Single normal variant | |||
| Discs identified | 38 | 15 | 29 |
| Sensitivity | 93% (84%–97%) | 95% (88%–98%) | 93% (83%–98%) |
| Specificity | 88% (75%–95%) | 83% (60%–94%) | 73% (57%–85%) |
| Single normal variant + first derivative | |||
| Discs identified | 32 | 17 | 26 |
| Sensitivity | 100% (93%–100%) | 100% (95%–100%) | 100% (94%–100%) |
| Specificity | 74% (60%–85%) | 94% (74%–99%) | 65% (50%–78%) |
Discussion
The ability to diagnose and identify causative organisms in orthopaedic infections continues to cause physicians difficulty. We found that principal component analysis can explain the variation in spectral curves for biofilm obtained from S aureus, S epidermidis, and P aeruginosa using FT-NIR. This analysis also provides a sensitivity that increases with advanced modeling. We used the soft independent modeling by class analogy method to develop calibration models to classify and identify three common bacterial species identified in infected prostheses (Gram negative and Gram positive) that are known to establish biofilms on common implant materials. Validation samples were compared with the class models and assigned to classes according to their proximity to the calibration (training) samples. Soft independent modeling by class analogy also allows additional class groups to be added to the model without rebuilding the classification models [7].
The concerns for FT-NIR assessment of bacterial presence and speciation are the limited sample area and time required for multiple sampling [3]. Other concerns for this technique are the heterogeneity in one strain and across strains [18]. Our evaluation of the spectral data collected and analyzed through multivariate data and principal component analyses showed improvement in recognition with decreased errors with continued modeling including combined standard normal variant and first derivative. The continued evaluation of different bacterial strains and implant surfaces will allow for continued evolution of this potentially clinically useful application. All of the bacteria for this study were grown from the same clinical isolate that has the potential to affect the results with other strains; however, the study was performed during a prolonged course with many different runs of the biofilm reactor thus strengthening our results. Additional studies to develop models for classification are needed to create a database of infecting organisms and consideration for contaminants such as surrounding tissues. Although use of the technology requires new instrumentation, the low cost and portability of NIR machines would make them easily obtainable. The most challenging aspect would be in developing and maintaining the calibration. Rapid detection and inexpensiveness of the technology would make the translation of this technology easier compared with PCR and immunoassays, which can be labor intensive and time consuming.
Principal component analysis explains variation in spectral curves for biofilm obtained from S aureus, S epidermidis, and P aeruginosa. This is an indication that separation of the spectral data is possible. Principal component analysis gives the overview of the spectra data and reduces the dimensionality of the data using a linear combination of original data to generate new latent variables that are orthogonal and uncorrelated to each other. Wold et al. [24] described principal component analysis as a process that distills the information from the original data variables into a lower number of variables (latent variables) that maximizes the variance in the data set. The FT-NIR also produced no spectral readings of the steel discs without the presence of bacteria, confirming that the spectral readings are correlated with the presence of biofilm.
Without processing of the data, biofilm of the studied species were identified using FT-NIR after establishing a calibration set, or databank. By further increasing this databank this technique potentially could identify many other types of bacteria localized as adherent to implants. Using the calibration models developed from the raw spectral analysis, the sample biofilm sets were correctly identified in 86% of S aureus samples, 89% of S epidermidis, and 70% of P aeruginosa. We identified bacterial species grown as biofilms on an orthopaedic implant material using FT-NIR. Specific peaks of importance on the bacterial spectra using different ranges of spectroscopy were identified in previous studies [12, 18, 20]. The full NIR spectrum was used in our model development because a study that used biofilm-based materials showed that the region between 1000 nm to 2500 nm (combination of first overtone, second overtone, and combination band region) of the NIR hold the most distinct biological spectral information [19]. Spectral data preprocessed or transformed using standard normal variant, first derivative, and standard normal variant + first derivative were used to develop calibration models with good prediction accuracy and acceptable error. Overall, spectral preprocessing using combined modeling of standard normal variant + first derivative appeared to best improve the performance of the classification models by improved modeling distance and separation of the three bacterial species with the best reduction in Type II error. In this modeling situation the Type 1 error rejects a specimen that belongs to the classification group and the Type II error accepts a specimen as a member when the specimen does not belong to the classification group. In a proposed translational application we hope to identify not only the presence or absence of bacteria but also to identify a particular strain thus increasing the importance of improving Type II error. Previous studies have used spectroscopy to analyze biologic specimens [2, 10, 12, 18, 21]. Ngo-Thi et al. [18] reported the ability of conventional FT-NIR spectroscopy in the MIR region and multivariate data analysis to correctly classify different nonbiofilm-associated bacteria in groups. Karadenizli et al. [12] used FT-IR and multivariate data analysis to classify biofilm-associated extracellular polymeric substance separated from S epidermis and S aureus correctly 80% and 75% of the time, respectively [12]. Ojeda et al. [21] used reflectance FT-IR (MIR region) to characterize Aquabacterium commune on stainless steel discs containing biofilm on intact samples. Reflectance spectroscopy provided a similar spectrum compared with conventional techniques in a nondestructive manner [21]. Janbu et al. [10] showed that micro-FT-IR spectroscopy using stepwise canonical discriminant analysis could correctly identify strains of Listeria 93% of the time. Bosch et al. [2] characterized Bordetella pertussis using spectroscopy and showed that planktonic bacteria have different spectra from its biofilm formation secondary to the spectroscopic absorbance of the glycocalyx or extracellular polymeric substances.
A rapid method to diagnose bacterial biofilm infections real-time would be beneficial to practicing orthopaedic surgeons. A real-time analysis would have the potential to allow surgeons to make decisions intraoperatively regarding the need for revision surgery and the choice of local antibiotics or systemic antibiotics tailored to the specific organism responsible for the infection. Future applications also could include diagnostic applications via arthroscopy or aspiration. The translation of this potential diagnostic technique will be confounded by the presence of nonbacterial cells, conditioning films, and other materials. Future studies will need to focus on the sensitivity of the spectroscopy using an infected implant in vivo model. With this model we will explore the ability to limit the potential interference caused by surrounding tissues, materials, and cells. This could be accomplished by establishing a dataset from which specific signals could be subtracted to improve a signal to noise ratio. We then can begin to explore our abilities to identify biofilm on explanted materials and then transition to clinical evaluations in the operative setting. Continued development of the diagnosis and characterization of implant-related biofilms is imperative to improve treatment of implant-related infections.
Acknowledgments
We thank Suzanne Danley BS (Department of Orthopaedics, West Virginia University) for critical review of this manuscript, Therwa Hamza PhD (School of Dentistry, University of Maryland, Baltimore, MD) for assistance with sample preparation and data collection, and John G. Thomas PhD (Department of Clinical Microbiology, West Virginia University) for providing clinical bacterial isolates.
Footnotes
The institution of one or more of the authors (JET) has received, during the study period, funding from the Orthopaedic Research and Education Foundation, and The American Association of Hip and Knee Surgeons with funding provided by Zimmer, Inc.
All ICMJE Conflict of Interest Forms for authors and Clinical Orthopaedics and Related Research ® editors and board members are on file with the publication and can be viewed on request.
Each author certifies that his or her institution approved or waived approval for the reporting of this investigation and that all investigations were conducted in conformity with ethical principles of research.
The work was performed at West Virginia University, Morgantown, WV.
References
- 1.Barth E, Myrvik QM, Wagner W, Gristina AG. In vitro and in vivo comparative colonization of Staphylococcus aureus and Staphylococcus epidermidis on orthopaedic implant materials. Biomaterials. 1989;10:325–328. doi: 10.1016/0142-9612(89)90073-2. [DOI] [PubMed] [Google Scholar]
- 2.Bosch A, Serra D, Prieto C, Schmitt J, Naumann D, Yantorno O. Characterization of Bordetella pertussis growing as biofilm by chemical analysis and FT-IR spectroscopy. Appl Microbiol Biotechnol. 2006;71:736–747. doi: 10.1007/s00253-005-0202-8. [DOI] [PubMed] [Google Scholar]
- 3.Burgula Y, Khali D, Kim S, Krishnan SS, Cousin MA, Gore JP, Reuhs BL, Mauer LJ. Review of mid-infrared Fourier transform-infrared spectroscopy applications for bacterial detection. J Rapid Methods Automation Microbiol. 2007;15:146–175. doi: 10.1111/j.1745-4581.2007.00078.x. [DOI] [Google Scholar]
- 4.Campoccia D, Speziale P, An YH, Del Pozo JL, Ceresa L, Pegreffi F, Montanaro L, Arciola CR. Innovative methods of rapid bacterial quantification and applicability in diagnostics and in implant materials assessment. Int J Artif Organs. 2007;30:842–851. doi: 10.1177/039139880703000914. [DOI] [PubMed] [Google Scholar]
- 5.Donlan RM. New approaches for the characterization of prosthetic joint biofilms. Clin Orthop Relat Res. 2005;437:12–19. doi: 10.1097/01.blo.0000175120.66051.29. [DOI] [PubMed] [Google Scholar]
- 6.Dunteman GH. Principal Components Analysis (Quantitative Applications in the Social Sciences) Newbury Park, CA: Sage Publications Inc; 1989. [Google Scholar]
- 7.Esbensen KH, Guyot D, Westad F, Houmoller LP. Multivariate Data Analysis: In Practice: An Introduction to Multivariate Data Analysis and Experimental Design. 5. Oslo, Norway: CAMO Software AS; 2002. [Google Scholar]
- 8.Goeres DM, Loetterle LR, Hamilton MA, Murga R, Kirby DW, Donlan RM. Statistical assessment of a laboratory method for growing biofilms. Microbiology. 2005;151:757–762. doi: 10.1099/mic.0.27709-0. [DOI] [PubMed] [Google Scholar]
- 9.Gristina AG, Oga M, Webb LX, Hobgood CD. Adherent bacterial colonization in the pathogenesis of osteomyelitis. Science. 1985;228:990–993. doi: 10.1126/science.4001933. [DOI] [PubMed] [Google Scholar]
- 10.Janbu AO, Moretro T, Bertrand D, Kohler A. FT-IR microspectroscopy: a promising method for the rapid identification of Listeria species. FEMS Microbiol Lett. 2008;278:164–170. doi: 10.1111/j.1574-6968.2007.00995.x. [DOI] [PubMed] [Google Scholar]
- 11.Jass J, Surman S, Walker JT, editors. Medical Biofilms: Detection, Prevention and Control. Chichester, England: John Wiley and Sons; 2003. [Google Scholar]
- 12.Karadenizli A, Kolayli F, Ergen K. A novel application of Fourier-transformed infrared spectroscopy: classification of slime from staphylococci. Biofouling. 2007;23:63–71. doi: 10.1080/08927010601143524. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi N, Bauer TW, Tuohy MJ, Fujishiro T, Procop GW. Brief ultrasonication improves detection of biofilm-formative bacteria around a metal implant. Clin Orthop Relat Res. 2007;457:210–213. doi: 10.1097/BLO.0b013e3180312042. [DOI] [PubMed] [Google Scholar]
- 14.Morgan S, Fox A, Rogers J, Watt B. Identification of chemical markers for microbial differentiation and detection by gas chromatography-mass spectometry. In: Nelson W, editor. Modern Techniques for Rapid Microbiological Analysis. New York, NY: VCH Publishers Inc; 1991. pp. 1–49. [Google Scholar]
- 15.Mortazavi SM, Vegari D, Ho A, Zmistowski B, Parvizi J. Two-stage exchange arthroplasty for infected total knee arthroplasty: predictors of failure. Clin Orthop Relat Res. 2011;469:3049–3054. doi: 10.1007/s11999-011-2030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naumann D. Vibrational spectroscopy in microbiology and medical diagnostics. In: Lasch P, Kniepp J, editors. Biomedical Vibrational Spectroscopy. Hoboken, NJ: Wiley-Interscience; 2008. pp. 1–8. [Google Scholar]
- 17.Nelson CL, McLaren AC, McLaren SG, Johnson JW, Smeltzer MS. Is aseptic loosening truly aseptic? Clin Orthop Relat Res. 2005;437:25–30. doi: 10.1097/01.blo.0000175715.68624.3d. [DOI] [PubMed] [Google Scholar]
- 18.Ngo-Thi NA, Kirschner C, Naumann D. Characterization and identification of microorganisms by FT-IR microspectrometery. J Mol Struct. 2003;661–662:371–380. doi: 10.1016/j.molstruc.2003.08.012. [DOI] [Google Scholar]
- 19.Nkansah K, Dawson-Andoh B, Slahor J. Rapid characterization of biomass using near infrared spectroscopy coupled with multivariate data analysis: Part 1. Yellow-poplar (Liriodendron tulipifera L.) Bioresour Technol. 2010;101:4570–4576. doi: 10.1016/j.biortech.2009.12.046. [DOI] [PubMed] [Google Scholar]
- 20.System NNIS. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2003, issued August 2003. Am J Infect Control. 2003;31:481–498. doi: 10.1016/j.ajic.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Ojeda JJ, Romero-Gonzalez ME, Banwart SA. Analysis of bacteria on steel surfaces using reflectance micro-Fourier transform infrared spectroscopy. Anal Chem. 2009;81:6467–6473. doi: 10.1021/ac900841c. [DOI] [PubMed] [Google Scholar]
- 22.Trampuz A, Osmon DR, Hanssen AD, Steckelberg JM, Patel R. Molecular and antibiofilm approaches to prosthetic joint infection. Clin Orthop Relat Res. 2003;414:69–88. doi: 10.1097/01.blo.0000087324.60612.93. [DOI] [PubMed] [Google Scholar]
- 23.Tunney MM, Patrick S, Curran MD, Ramage G, Hanna D, Nixon JR, Gorman SP, Davis RI, Anderson N. Detection of prosthetic hip infection at revision arthroplasty by immunofluorescence microscopy and PCR amplification of the bacterial 16S rRNA gene. J Clin Microbiol. 1999;37:3281–3290. doi: 10.1128/jcm.37.10.3281-3290.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wold S, Esbensen K, Geladi P. Principal component analysis. Chemometr Intell Lab Syst. 1987;2:37–52. doi: 10.1016/0169-7439(87)80084-9. [DOI] [Google Scholar]