Abstract
Werner syndrome (WS) is characterized by the premature onset of several age-associated pathologies. The protein defective in WS patients (WRN) is a helicase/exonuclease involved in DNA repair, replication, telomere maintenance, and transcription. Here, we present the results of a large-scale proteome analysis to determine protein partners of WRN. We expressed fluorescent tagged-WRN (eYFP-WRN) in human 293 embryonic kidney cells and detected interacting proteins by co-immunoprecipitation from cell extract. We identified by mass spectrometry 220 nuclear proteins that complexed with WRN. This number was reduced to 40 when broad-spectrum nucleases were added to the lysate. We consider these 40 proteins as directly interacting with WRN. Some of these proteins have previously been shown to interact with WRN, whereas most are new partners. Among the top 15 hits, we find the new interactors TMPO, HNRNPU, RPS3, RALY, RPS9 DDX21, and HNRNPM. These proteins are likely important components in understanding the function of WRN in preventing premature aging and deserve further investigation. We have confirmed endogenous WRN interaction with endogenous RPS3, a ribosomal protein with endonuclease activities involved in oxidative DNA damage recognition. Our results suggest that the use of nucleases during cell lysis severely restricts interacting protein partners and thus enhances specificity.
Keywords: LC-MS/MS, proteomics, Werner syndrome, ribosomal protein, nuclease treatments
Graphical abstract
INTRODUCTION
Werner Syndrome (WS) is an autosomal recessive disorder that displays many of the clinical symptoms of normal aging at an early age. From their second decade of life onward, WS patients develop pathologies that prematurely resemble many traits of normal aging such as osteoporosis, ocular cataracts, graying and loss of hair, diabetes mellitus, arteriosclerosis, and cancer.1–4 Death generally occurs in the fifth decade of life from heart demise or cancer. Accumulating evidence indicates that WRN protein is a suppressor of illegitimate recombination. Indeed, WS cells are characterized by the presence of deletions and variegated chromosomal translocations.5,6 Processes such as DNA replication or transcription generate regions of single-stranded DNA, which may inadvertently provide a substrate for the initiation of recombination. Various mechanisms have evolved to ensure that recombination does not occur promiscuously during these processes and the WRN protein may be part of one such mechanism. Thus, one potential role for WRN would be to actually monitor recombinational repair of double-strand breaks at sites of DNA replication like it has been demonstrated for the RecQ DNA helicase orthologue in Escherichia coli.7 During the process of recombination, nonhomologous regions of DNA could inadvertently be used as templates for repair. WRN protein will not inhibit the initiation of recombination but will dissociate abnormal recombination intermediates.8–10 Accordingly, purified WRN protein has an affinity for DNA fork structures such as those observed during DNA recombination.11 Furthermore, WRN can migrate Holliday junctions (a recombination intermediate).8 Thus, a mutation in WRN may lead to an increased frequency of illegitimate recombination during the repair of breaks at transcriptional sites or DNA replication forks, creating small deletions or variegated chromosomal translocations. Consequently, it is likely that repetitive sequences will show multiple recombinational errors affecting the length of such structures during DNA replication in WS cells. Incidentally, there is evidence indicating accelerated shortening and lengthening of telomeres in serially passaged human WS cultures12,13 and in murine Wrn helicase mutant cell lines.14,15 This telomeric instability in WS cells is believed to be due to illegitimate recombination between telomeres of different chromosomes.16 It is known that an increase in chromosome instability may be associated with loss of telomeric repeats.17 In addition to homologous recombination, WRN is involved with the KU complex and DNA-PK in nonhomologous end joining reactions.18,19 Finally, it also interacts with RPA and PARP-1.20,21 Thus, WRN is likely to be involved in several DNA repair pathways. In the absence of a functional WRN protein, accumulation of deletions and translocations could potentially inactivate tumor suppressor genes or activate oncogenes, accelerating tumor formation and/or aggressiveness. Such mutations could also potentially inactivate genes involved in normal cellular metabolism inducing senescence and accelerated aging. In addition to defects in DNA replication and repair, defects in transcription have also been observed in WS cell lines implicating WRN in some aspects of transcription as well. Analysis of the expression profile of primary human fibroblast cell lines derived from young donors, old donors, and WS patients have indicated that transcription alterations in WS are strikingly similar to those in normal cells established from old individuals.22 Furthermore, we recently determined that microarray analyses on short-term knock down of WRN protein was sufficient to induce an expression profile resembling that of fibroblasts derived from old individuals.23
Despite advances in understanding how the WRN exonuclease/helicase enzyme contributes to DNA replication and repair, precisely how the absence of functional WRN protein leads to the numerous pathologies characteristic of Werner syndrome, some of which might be independent of genome maintenance defects (e.g., dyslipidemia, diabetes and cataracts), is still unknown. In this study, we conducted the first large-scale proteomics investigation to identify new candidates for WRN binding that would give us clues on biological processes altered by WRN in cells. As WRN is a nucleic acid binding protein, two different cell extract approaches were used, one using Benzonase and RNase A during lysis and one without nuclease pretreatments before immunoprecipitation of a tagged WRN construct in HEK 293 cells. Mass spectrometry analysis of the co-immunoprecipitate in the absence of nucleases revealed 220 nuclear proteins potentially interacting with WRN peptide. In nucleic acids depleted extracts, however, only 40 nuclear proteins were identified as direct interactants of the WRN protein in HEK 293 cells.
MATERIALS AND METHODS
Cell Line and Expression Vectors
Human 293 embryonic kidney cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (250 IU/mL), and streptomycin (250 µL/mL) at 37 °C in atmosphere of 5% CO2. The eYFP control vector was purchased from Clontech (Mountain View, CA). The eYFP-WRN construct is described elsewhere.24
eYFP-WRN Immunoprecipitation
Human 293 embryonic kidney cells were seeded onto nine 150 mm cell-culture dishes and grown up to 80–90% confluency. Transfections were performed with the Effectene transfection kit from Qiagen, Inc. (Mississauga, ON). All further steps were performed 24 h after the transfections on ice or at 4 °C. Two PBS washes were carried out prior to the extraction (cell scraping) with 2 mL/plates of lysis buffer [40 mM HEPES pH 7.5, 120 mM NaCl, 0.3% CHAPS, 1 mM EDTA, Complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN)]. When indicated, Benzonase (25 U/mL) and RNase A (100 µg/mL) (EMD, Gibbstown, NJ) were added in the buffer. Cells were kept on ice for 15 min and genlty lysed for another 15 min on a rotating device. Cell extracts were centrifuged for 5 min at 3000g to remove cellular debris and unsoluble material. Immunoprecipitation experiments were performed using Dynabeads magnetic beads covalently coupled with Protein G (Invitrogen, Burlington, ON). The Dynabeads were washed two times with 1 mL of 0.1 M sodium acetate buffer, pH 5.0, coated with 10–15 µg of mouse monoclonal anti-GFP antibody, which also recognizes eYFP protein (Roche Applied Science, Indianapolis, IN). The antibody-coupled Dynabeads were incubated for 1 h at room temperature with 1 mL of PBS containing 1% (w/v) BSA (Sigma-Aldrich, Oakville, ON) to block nonspecific antibody binding sites. The beads were finally washed three times with 1 mL of lysis buffer and added to the protein extract for 4 h incubation with gentle agitation in a cold room. (Protein extracts had been precleared with empty Dynabeads for 45 min in a cold room before adding the anti-GFP coupled beads.) Samples were then washed three times with 2 vol of lysis buffer for 5 min. Protein complexes were eluted using 150 µL of 2× Laemmli sample buffer containing 5% β-mercaptoethanol and boiled for 5 min in a water bath. Proteins were resolved using 4–12% Criterion XT Bis-Tris gradient gel (Bio-Rad, Mississauga, Canada) and stained with Sypro Ruby (Bio-Rad, Mississauga, ON) according to the manufacturer’s instructions. Images were acquired using the Geliance CCD-based imaging system (Perkin-Elmer, Shelton, CT).
Exonuclease and Helicase Activities of Immunoprecipitated eYFP-WRN
One 150-mm Petri dish of HEK 293 cells (1.8 × 108 cells) was transfected with 2 µg of the eYFP or the eYFP-WRN plasmids with the Effectene transfection kit from Qiagen, Inc. The next day, eYFP and eYFP-WRN transfected cells were lysed in 1 mL of a stringent buffer [50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1.0% NP-40, 0.1% SDS, 0.5% Sodium deoxycholate, Complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN)]. eYFP and eYFP-WRN were immunoprecipitated with 2 µg of an antibody against YFP and magnetic beads as described above. Immunoprecipitation was carried out for 2 h in a cold room. Beads containing the immune complexes were washed once with 1 mL of a buffer containing 20 mM Tris-HCl pH 8.0, 0.5 M NaCl, 1 mM EDTA, 0.5 mM DTT, 0,5% NP-40, 25% Glycerol, 0.2 mM PMSF. Beads were then washed twice with 2 mL of a buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 25% Glycerol, 0.5% NP-40, 0.05% Sodium deoxycholate, 0.005% SDS. Finally, beads were resuspended in 15 µL of buffer (25 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 1 mM DTT, 0.05% NP-40, 25% glycerol). Resuspended beads containing the immune complexes (∼0.1 µg/µL of antibody) were diluted as indicated in the figures in assay reaction buffer (40 mM Tris-HCl pH 7.4, 4 mM MgCl2, 0.1 mg/mL BSA, 5 mM DTT, and 100 nM of splayed arms labeled on one DNA strand25). The reaction was incubated for 20 min at 37 °C, and stopped with one-fifth volume of Stop buffer (40% glycerol, 50 mM EDTA, 2% SDS, 3% xylene cyanol and 3% bromophenol blue). Reaction samples were loaded on a 6% PAGE/TBE 1× for autoradiography.
LC-MS/MS Analysis
SDS-PAGE protein lanes corresponding to anti-eYFP immunoprecipitated extracts were cut into 33 gel slices per lane using a disposable lane picker (The Gel Company, CA). Gel slices were deposited into 96-well plates. In-gel protein digest was performed on a MassPrep liquid handling station (Waters, Mississauga, Canada) according to the manufacturer’s specifications and using sequencing-grade modified trypsin (Promega, Madison, WI). Peptide extracts were dried out using a SpeedVac.
Peptide extracts were separated by online reversed-phase (RP) nanoscale capillary LC (nanoLC) and analyzed by electrospray MS (ES MS/MS). The experiments were performed on a Thermo Surveyor MS pump connected to a LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA) equipped with a nanoelectrospray ion source (Thermo Electron, San Jose, CA). Peptide separation took place within a PicoFrit column BioBasic C18, 10 cm × 0.075 mm internal diameter (New Objective, Woburn, MA) with a linear gradient from 2% to 50% solvent B (acetonitrile, 0.1% formic acid) in 30 min, at 200 nL/min. Mass spectra were acquired using data-dependent acquisition mode (Xcalibure software, version 2.0). Each full-scan mass spectrum (400–2000 m/z) was followed by collision-induced dissociation of the seven most intense ions. The dynamic exclusion function was enabled (30 s exclusion), and the relative collisional fragmentation energy was set to 35%.
Interpretation of Tandem MS Spectra
All MS/MS samples were analyzed using Mascot (Matrix Science, London, U.K.; version 2.3). Mascot was set up to search against human Uniref_100 protein database (Homo sapiens: Taxon 9606, 100 683 entries) assuming a digestion with trypsin. Fragment and parent ion mass tolerance were, respectively, of 0.5 and 2.0 Da. Iodoacetamide derivative of cysteine was specified as a fixed modification and oxidation of methionine was specified as variable modification. Two missed cleavages were allowed. Peak list was generated by Mascot Deamon with Extract_MSN.exe.
Criteria for Protein Identification
Scaffold (version 03_00_02; Proteome Software, Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at >95.0% probability as specified by the Peptide Prophet algorithm.26 Protein identifications were accepted if they could be established at >95.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm.27 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Using these stringent identification parameters, 15 013 spectra were annotated (646 protein identifications with a false discovery rate of 0.1% calculated from Scaffold’s statistical analysis). Scaffold 3 analysis files (IP-WRN.sf3 and IP-WRN+ Benzonase.sf3) are provided as Supporting Information. These files can be accessed with the free viewer available in Proteome Software, Inc. Web site (http://www.proteomesoftware.com). These files contain all the spectral information, including the accession number for each protein sequence, Mascot scores, protein sequence coverage, statistical probability modeling, and spectral counting.
Immunoblotting
Protein extracts eluted from Dynabeads were separated on SDS-PAGE and then transferred onto 0.2 µm nitrocellulose membrane (Bio-Rad). After incubating 1 h with blocking solution (PBS-T containing 5% nonfat milk), the membrane was probed overnight at 4 °C with either a mouse monoclonal antibody against KI-67 antigen (clone MIB-1; Dako, Denmark), a mouse monoclonal antibody against nucleophosmin (B-23) (clone FC82291; Sigma, St. Louis, MO), a mouse monoclonal antibody against MSH2 (clone FE11; Oncogene Research Products, Boston, MA), a mouse monoclonal antibody against Lamin B1 (clone P3×63-Ag.653; US Biologicals, Swampscott, MA), a mouse monoclonal antibody against HNRPK (D-6; Santa Cruz Biotechnology, Santa Cruz, CA), a rabbit polyclonal antibody against HNRPC1/C2 (Santa Cruz Biotechnology, Santa Cruz, CA), a rabbit polyclonal antibody against MSH3 (Santa Cruz Biotechnology, Santa Cruz, CA), a rabbit polyclonal antibody against XRCC1 (Santa Cruz Biotechnology, Santa Cruz, CA), a rabbit polyclonal antibody against POLR2B (Novus Biologicals, Littleton, CO), or a rabbit polyclonal antibody against WRN (Santa Cruz Biotechnology, Santa Cruz, CA). The antibody against GFP (or eYFP) was purchased from BD Biosciences (Palo Alto, CA). After washing with PBS-T, species-specific horseradish peroxidase-conjugated secondary antibody was added for 2 h at room temperature. Signals were detected with Western Lightning Chemiluminescence reagent plus kit (GE Healthcare Limited, Piscataway, NJ). Immunoprecipitation of the endogenous WRN was performed with a goat polyclonal antibody from Santa Cruz Biotechnology (antibody C-19; Santa Cruz, CA).
Bioinformatic Tool for Protein Network Analysis
The PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System is a unique resource that classifies genes by their functions, using published scientific experimental evidence and evolutionary relationships to predict function even in the absence of direct experimental evidence. Proteins are classified by expert biologists into families and subfamilies of shared function, which are then categorized by molecular function and biological process ontology terms (www.pantherdb.org). Enrichments for specific biological functions were considered significant with a Benjamini value smaller than 0.05. The Benjamini value corresponds to an adjusted p-value using the Benjamini–Hochberg method to correct for multiple hypotheses tested during gene enrichment analyses.
Evaluation of Protein Domain and Family Distribution
The Pfam (http://pfam.sanger.ac.uk/) domains were obtained by parsing the ‘swisspfam’ (version Pfam 22.0, July 2007, 9318 families) database using in-house Ruby scripts (version 1.8.6), which output Microsoft Excel-compatible spreadsheets suitable for further analysis. Data sets were generated from all the proteins identified by LC–MS/MS from the eYFP-WRN immunoprecipitation experiments (without nucleases).
Live Cell Imaging and Laser Microirradiation
Live cell imaging combined with laser microirradiation was carried out as described previously.28 Briefly, eYFP-WRN was expressed in HeLa cells by overnight transfection with Effectene reagent (Qiagen). Cells were placed in medium containing 1 µg/mL Hoechst 33258 for 30 min. The medium was then removed and replaced with fresh one. A 37 °C preheated stage was used during the acquisition period. Fluorescence in a living cell was monitored using a Zeiss LSM510 NLO laser-scanning confocal microscope. DNA damage was generated in a defined area of the nucleus by microirradiation with a 750-nm femtosecond pulsed titanium-sapphire laser. Time-lapse images were acquired and fluorescence intensities within the microirradiated nuclear region were quantitated. Background and photobleaching corrections were applied to each data sets as described.
RESULTS
Identification of WRN-Interacting Proteins by Mass Spectrometry
To identify proteins that interact with the human WRN gene product, we transfected human 293 embryonic kidney cells with an eYFP-WRN expression construct. We used a tagged version of the WRN protein because one potential limitation in using anti-WRN antibodies is the interference of the antibodies with WRN protein complex formation in cells. Furthermore, the WRN epitopes recognized by the antibodies maybe embedded in protein complexes and unreachable, impeding the identification of important interactors in our analyses. The advantage of using the eYFP-WRN chimera is the presence of good commercial antibodies against eYFP for large-scale immunoprecipitation and the behavior of the eYFP-WRN protein, which is similar to the endogenous WRN protein in cell culture. 29,30 Indeed, eYFP-WRN proteins were mainly localized to nucleoli in HEK 293 cells (Figure 1A) and translocated with very fast kinetics to regions of DNA breaks generated by laser-induced DNA damage micro-irradiation (Figure 1B). In addition, we analyzed the enzymatic activities of immunoprecipitated eYFP-WRN on a radioactive forked DNA structure (Figure 1C). As a control, we transfected 293 embryonic kidney cells with the eYFP expression vector. The immunoprecipitate was stringently washed with buffers containing high salt concentration, sodium deoxycholate and SDS. As indicated in Figure 1C,D, there was no helicase or exonuclease activities associated with the eYFP immunoprecipitate. In contrast, the eYFP-WRN immunoprecipitate showed both activities on the forked DNA substrate indicating that the catalytic sites of the eYFP-WRN construct are functional. Such controls ensured that the eYFP-tagged WRN protein co-localized to the same compartment as the endogenous protein and that the fusion protein retained the biochemical functions of the endogenous WRN protein.
Figure 1.
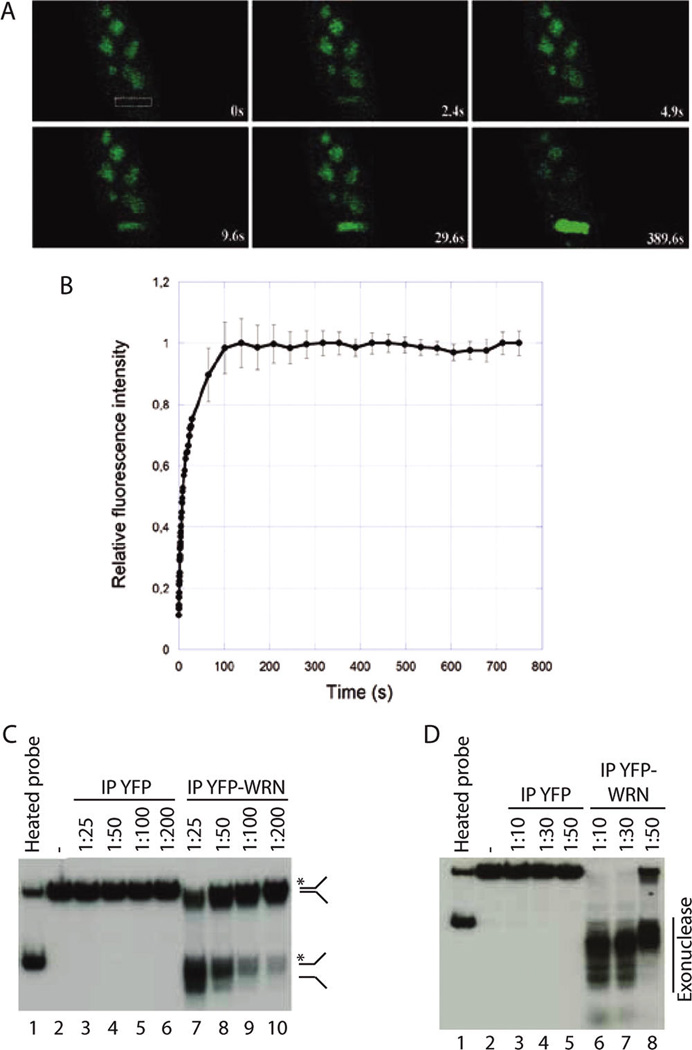
Cellular and enzymatic activities of the eYFP-WRN protein. (A) Time course localization of the eYFP-WRN protein in a representative HEK 293 cell by live imaging after microlaser beam application. The path of the laser is indicated by a rectangle box in the image at 0 s. Magnification is 40×. (B) Recruitment kinetics of eYFP-WRN shown by the fluorescence intensity to sites of DNA damage after microlaser beam application. (C) Helicase activity of the eYFP-WRN protein. Immunoprecipitated eYFP (lanes 3–6) or YFP-WRN (lanes 7–10) were diluted in reaction buffer 1/25, 1/50, 1/100, and 1/200, as indicated. The position of the displaced strand is indicated on the right. Autoradiogram represents a 6 h exposition. Lane 1, heated splayed arm substrate; lane 2, no protein. (D) Exonuclease activity of the eYFP-WRN protein. Immunoprecipitated eYFP (lanes 3–5) or YFP-WRN (lanes 6–8) were diluted in reaction buffer 1/10, 1/30, and 1/50 to better see the exonuclease activity. Nuclease fragments are indicated on the right. Lane 1, heated splayed arm substrate; lane 2, no protein. Autoradiogram represents 18 h of exposition.
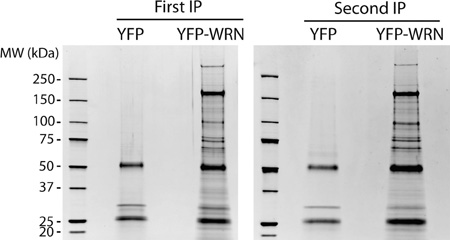
For the mass spectrometry analyses, immunoprecipitation assays were carried out under rather mild detergent and ionic strength that allowed efficient isolation of intact protein complexes (see Materials and Methods). The immunoprecipitated proteins were resolved by SDS-PAGE and stained with Sypro Ruby. A first eYFP-WRN immunoprecipitation experiment was performed in a context where nucleases were omitted in the lysis buffer to get potentially the whole WRN interactome. In addition, the entire protein load resolved by SDS-PAGE was extracted for a complete coverage of the co-immunprecipitated proteins rather than limited to high-abundance protein bands. Protein tracks were thus cut into several gel slices for trypsin digestion followed by liquid chromatography tandem mass spectrometry (LC–MS/MS). MS/MS spectra were assigned to peptides using Mascot search engine and further validated using Scaffold’s program statistical analysis. Assignments were grouped according to corresponding proteins and the probability of correct protein assignment was computed. Scaffold reports were generated to include all spectral information. This information is displayed in the supplementary Scaffold files (IP-WRN.sf3 and IP-WRN+Benzonase.sf3) and can be modulated according to appropriate filters criteria. Readers can refer to these files using the free Scaffold viewer easily accessible on the Proteome Software Web site. Scaffold uses proven statistical algorithms (PeptideProphet and ProteinProphet) to calculate the probability that proteins are actually in the biological samples. The data from the Mascot search engine is mapped on a histogram demarcated by discriminating scores. In addition, Bayesian statistics were used to determine the probability that a match is correct at each discriminating score. Detailed results help detect false positives by allowing the reader to focus on a protein and examine the peptide and spectra evidence supporting the identification. For this study, we applied stringent probability thresholds (>95% confidence for peptide and proteins annotations supported by at least two unique peptides). These criteria result in an estimated protein false discovery rate (Prot FDR) of only 0.1% as calculated by Scaffold. Supplementary Table S1 gives a list of all the proteins identified by LC–MS/MS. A protein interacting with WRN was considered positive if at least two unique peptides for the same protein were identified. Proteins identified in control HEK 293 immunoprecipitates (expressing eYFP alone) were considered artifacts and removed from the final list of potential eYFP-WRN-interacting proteins. The Supplementary Table S2a gives a list of total proteins identified without eYFP interacting proteins. The Supplementary Table S2b gives a list of all the identified proteins in the eYFP-WRN immunprecipitate with at least two identified unique peptides. Since WRN is exclusively a nuclear protein, we finally restricted the remaining of our study to a list containing exclusively nuclear proteins or proteins known to shuttle to the nucleus. Supplementary Table S3 gives a list of 220 nuclear proteins co-immunoprecipitating with the eYFP-WRN construct.
Our nuclear interactome data set was compared to six public protein interaction databases that store both potential and confirmed WRN-interacting proteins. Queried databases included the Biomolecular Interaction Network Database (BIND), the Database of Interacting Proteins (DIP), the Human Protein Reference Database (HPRD), the Online Predicted Human Interaction Database (OPHID), the Human Protein-Protein Interaction Prediction (PIPs), and the Molecular Interactions Database (MINT). Out of the 49 proteins predicted or confirmed to interact with WRN (Table 1), 21 of them are present in our list from Supplementary Table S3. Comparison of the proteomics data set with interactions reported in the literature revealed a high success rate in detection of known complexes (see Discussion). However, some reported WRN-interacting proteins are yet to be validated using proteomics approaches. Clearly, the composition of the WRN interactome is highly subject to modulation in the context of DNA damage. Importantly, transfected HEK 293 cells used in this study were not stressed by genotoxic agents known to cause decompartmentalization of WRN from the nucleoli to the nucleoplasm and its accumulation into DNA damage foci. It is also likely that transient interaction of WRN might occur with components of the DNA damage repair machinery and might have been missed in this study. Finally, it is unknown whether WRN interacts with the same partners in different cell types an issue that warrants further investigation.
Table 1.
List of Proteins Previously Identified or Predicted in the Literature to Interact with WRN
| protein name | description | mass spectrometry |
|---|---|---|
| ABL | c-Ableson tyrosine kinase | |
| ATR | Serine-protein kinase ATM-related | Identified in our analysis |
| ATM | Serine-protein kinase ATM | |
| BLM | Bloom syndrome protein | Identified in our analysis |
| BRCA1 | Breast Cancer susceptibility Region 1 | |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (p14ARF) | |
| DDX17 | Probable ATP-dependent RNA helicase DDX17 | Identified in our analysis |
| DDX3X | ATP-dependent RNA helicase DDX3X | Identified in our analysis |
| DDX5 | Probable ATP-dependent RNA helicase DDX5 | Identified in our analysis |
| PRKDC | DNA dependent protein kinase catalytic subunit | Identified in our analysis |
| DPOE2 | DNA polymerase epsilon subunit 2 | |
| EXO1 | Exonuclease 1 | |
| FEN1 | Flap structure-specific endonuclease 1 | |
| H2AX | Histone H2AX | |
| KU complex | KU70 and KU86 proteins | Identified in our analysis |
| LIG1 | Ligase I, DNA, ATP-dependent | |
| MCM2 | Minichromosome maintenance complex component 2 | |
| MRN complex | MRE11, NBS1, and RAD50 proteins | Identified in our analysis |
| MSH2 | DNA mismatch repair protein MSH2 | Identified in our analysis |
| MSH3 | DNA mismatch repair protein MSH3 | Identified in our analysis |
| MSH6 | DNA mismatch repair protein MSH6 | |
| NEIL1 | DNA glycosylase (nei endonuclease VIII-like 1) | |
| PARP-1 | Poly (ADP-ribose) polymerase-1 | Identified in our analysis |
| PARP-2 | Poly (ADP-ribose) polymerase-2 | |
| PCNA | Proliferating cell nuclear antigen | Identified in our analysis |
| POLB | DNA polymerase beta subunit | |
| POLD1 | DNA polymerase delta 1 subunit | |
| POLR1C | DNA-directed RNA polymerases I subunit RPAC1 | |
| POT1 | Protection of telomere homologue 1 | |
| PRKDC | DNA dependent protein kinase catalytic subunit | Identified in our analysis |
| RAD51 | DNA repair protein RAD51 | |
| RAD52 | DNA repair protein RAD52 | |
| RAD54 | DNA repair protein RAD54 | |
| RECQL | ATP-dependent DNA helicase Q1 | |
| RFC1 | Replication factor C (activator 1) 1, 145 kDa | Identified in our analysis |
| RPA1 | Replication protein A 70 kDa DNA-binding subunit | Identified in our analysis |
| RPA2 | Replication protein A2, 32 kDa | Identified in our analysis |
| SUMO1 | SMT3 suppressor of mif two 3 homologue 1 (S. cerevisiae) | |
| TP53 | Tumour Protein p53 | Identified in our analysis |
| TRF1 | Telomeric repeat binding factor 1 | |
| TRF2 | Telomeric repeat binding factor 2 | |
| UBC9 | SUMO-conjugating enzyme | |
| VCP | Transitional endoplasmic reticulum ATPase | |
| WRNIP1 | Werner helicase-interacting protein 1 (WHIP) | |
| YBX1 | Y box 1 transcription factor | Identified in our analysis |
Identification of WRN-Interacting Proteins in DNA and RNA Depleted Cell Extracts by Mass Spectrometry Analysis
A Pfam classification of the 220 nuclear proteins identified in eYFP-WRN immunoprecipitation extracts indicated that the largest families of proteins associated with WRN were helicases and proteins with RNA binding motifs (Supplementary Table S4). One potential caveat with this type of analysis, however, is the co-immunoprecipitation of proteins that form complexes with WRN via nucleic acid molecules without any direct protein–protein interaction. To exclude this possibility, additional immunoprecipitation experiments were performed in cells extracts depleted of nucleic acids. All DNA and RNA molecules were extensively digested with Benzonase, a highly active nuclease degrading all forms of DNA and RNA even at 4 °C. Extracts were also supplemented with RNase A. As before, eYFP immunoprecipitates were used to assess unspecific binding. The immunoprecipitation and mass spectrometry analysis were duplicated in two independent experiments. Figure 2A shows the proteins in the anti-eYFP immunoprecipitates resolved by SDS-PAGE and stained with Sypro Ruby. Staining of the gels indicates that the immunoprecipitations (performed on two different weeks) were technically highly reproducible (compare the left and right panels in Figure 2A). On average, the ratio of eYFP-WRN protein to endogenous WRN in transfected cells was approximately 5:1 based on Western blot analyses with an antibody against WRN protein (Figure 2B). Finally, we examined the efficiency of the nucleases in the buffer during the extraction and immunoprecipitation steps. Figure 2C indicates that nucleic acids were totally degraded in the presence of both Benzonase and RNases A.
Figure 2.
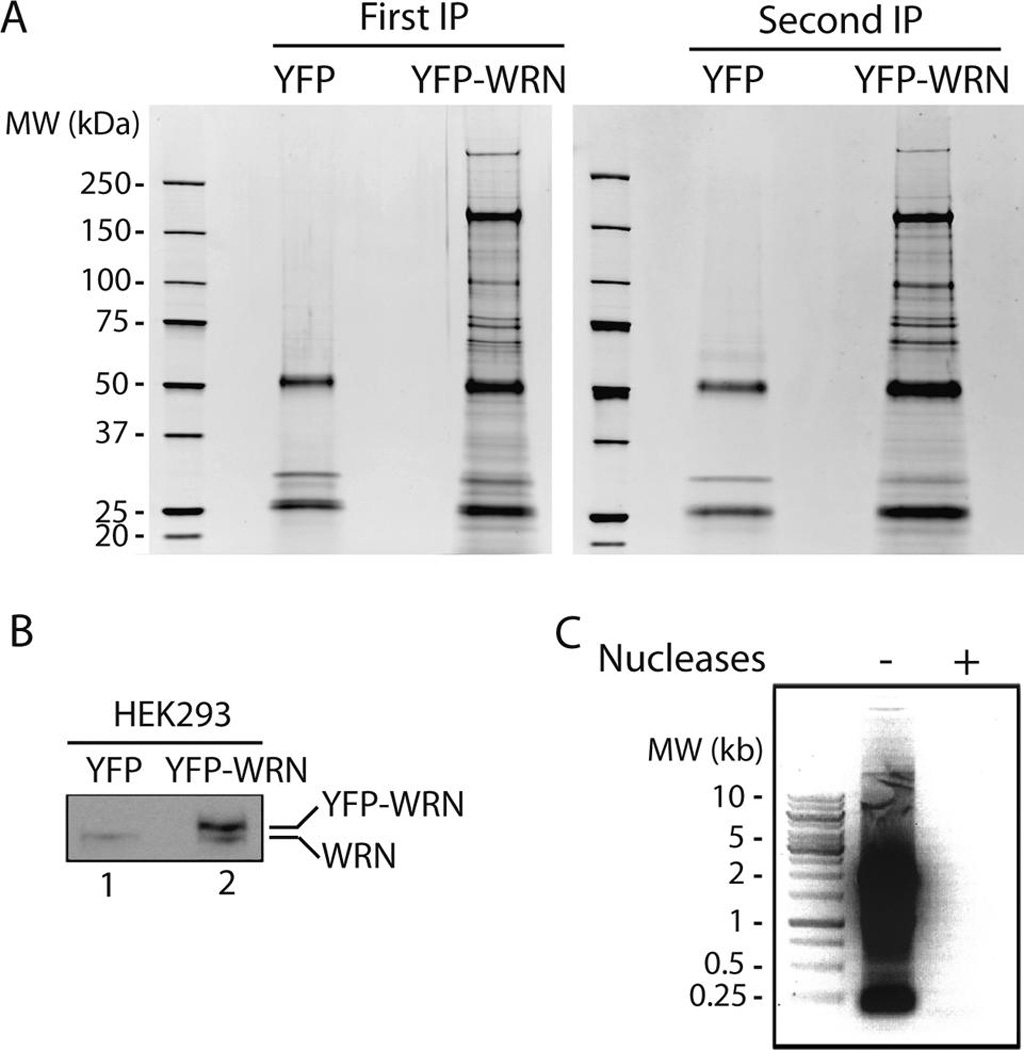
Co-immunoprecipitation of proteins from cells transfected with eYFP or eYFP-WRN constructs. (A) SYPRO Ruby-staining pattern of the co-immunoprecipitated proteins from transfected cells with the anti-YFP antibody. The immunoprecipitation was performed a second time (panel on the right) to show the reproducibility of the experiment. All immunoprecipitations were performed 24 h after the transfection reactions. Molecular-mass sizes are indicated in kilodaltons (kDa). (B) Levels of eYFP-WRN in transfected cells. HEK 293 cells were transfected with eYFP or eYFP-WRN contructs. Twenty-four hours later, whole cell lysates were analyzed on 6% SDS-PAGE followed by Western blotting with an antibody against WRN protein. The positions of the EYFP-WRN and of the endogenous WRN proteins are indicated on the right. (C) Agarose gel showing the presence of nucleic acids without or with Benzonase and RNase A in the lysis buffer prior to the immunoprecipitation step. An aliquot (representing one 150-mm Petri dish of HEK 293 cells) was treated with phenol-chloroform and the nucleic acids were precipitated in ethanol. The whole precipitate were analyzed on a 1% agarose gel.
Protein tracks were cut into gel slices for trypsin digestion followed by liquid chromatography tandem mass spectrometry (LC–MS/MS) as described in the preceding section. A protein interacting with the WRN enzyme was considered positive if at least two unique peptides for that same protein were identified. Proteins identified in control HEK 293 immunoprecipitates (expressing eYFP alone) were considered artifacts and removed from the final list of potential eYFP-WRN-interacting proteins. Supplementary Table 2c gives the complete lists of proteins co-immunoprecipitated with the eYFP-WRN in the presence of nucleases. From these, we parsed a list of nuclear proteins as described in the preceding section. Table 2 lists 40 nuclear proteins co-immunoprecipitating with the eYFP-WRN construct from the two consolidated data sets. Supplementary Table S5 gives more details on the origin of the proteins identified by mass spectrometry for each analysis. The most abundant co-immunoprecipitated proteins (identified with more than 10 unique peptides) were very similar whether cell extracts were treated with or without nucleases (Supplementary Table S5). Proteins such as DNA-PK, KU70, KU80, DHX9, RFA1, DNA ligase 3 or PARP-1 were among the proteins identified with the highest peptide coverage in both intact and nucleic acids depleted extracts. Since these interactions are not strictly tethered by DNA, it is likely that these proteins interact with WRN to form a stable complex which carry out the basic genome maintenance activities reported for WRN. Most interesting were also the identification of proteins such as the lamina-associated polypeptide 2-alpha (LAP2A) or the 40S ribosomal protein S3 (RPS3) in the same high abundance range as the proteins aforementioned, suggesting that these proteins might also be involved in strong binding to WRN. Finally, nuclease treatment could have also facilitated the release of some proteins tightly bound to DNA/ RNA, allowing new unidentified transient interactions with WRN. Examples of such proteins include Histone 1 (H1e), THOC1, PELP1, BAT1, DDX1, and Histone H2A type 1-B/E proteins (see Supplementary Table S5 for a more detailed description of these proteins).
Table 2.
List of WRN-Interacting Proteins Identified by Mass Spectrometry Using Benzonase and RNAse A in the Lysis Buffer
| number of unique peptides |
|||
|---|---|---|---|
| gene name | First IP | Second IP | description |
| WRN | 70 | 77 | Werner syndrome gene product |
| PRKDC | 62 | 53 | DNA-dependent protein kinase catalytic subunit |
| TMPO | 29 | 29 | Thymopoietin (lamina-associated polypeptide 2) |
| XRCC5 | 21 | 17 | KU86 ATP-dependent DNA helicase 2 subunit 2 |
| HNRNPU | 16 | 28 | Heterogeneous nuclear ribonucleoprotein U |
| XRCC6 | 15 | 18 | KU70 ATP-dependent DNA helicase 2 subunit 1 |
| DHX9 | 13 | 13 | DHX9 ATP-dependent RNA helicase A (DNA helicase II) |
| RPS3 | 9 | 14 | 40S ribosomal protein S3 |
| RALY | 9 | 7 | Autoantigenic hnRNP-associated with lethal yellow |
| RPA1 | 7 | 11 | Replication protein A 70 kDa DNA-binding subunit |
| RPS9 | 7 | 11 | 40S ribosomal protein S9 |
| LIG3 | 7 | 6 | DNA ligase 3 |
| DDX21 | 6 | 3 | Nucleolar RNA helicase 2 |
| HNRNPM | 6 | 5 | Heterogeneous nuclear ribonucleoprotein M |
| PARP-1 | 5 | 7 | Poly [ADP-ribose] polymerase 1 |
| RFC4 | 5 | 2 | Replication factor C subunit 4 |
| RPS3A | 5 | 4 | 40S ribosomal protein S3A (v-fos effector protein 1) |
| VRK3 | 4 | 7 | Vaccinia related kinase 3 |
| HIST1H1E | 4 | 7 | Histone 1, H1e |
| HIST1H4A | 4 | 6 | Histone H4 |
| DDX3X | 4 | 5 | DDX3X ATP-dependent RNA helicase |
| THOC1 | 4 | 2 | THO complex 1 (nuclear matrix protein p84) |
| RPL5 | 4 | 3 | 60S ribosomal protein L5 |
| TRIM28 | 3 | 4 | Tripartite motif-containing 28 |
| PELP1 | 3 | 4 | Proline, glutamate and leucine rich protein 1 |
| RBMX | 3 | 3 | HnRNP G, N-terminally processed |
| BAT1 | 3 | 2 | HLA-B associated transcript 1 |
| TOP1 | 3 | 2 | DNA topoisomerase 1 |
| RPA2 | 3 | 2 | Replication protein A 32 kDa subunit |
| CCT8 | 3 | 2 | T-complex protein 1 subunit theta |
| HNRNPUL2 | 2 | 4 | HnRNP U-like protein 2 |
| TFAM | 2 | 3 | Transcription factor A, mitochondrial |
| HIST1H2BK | 2 | 3 | Histone H2B type 1-K |
| RPL11 | 2 | 2 | 60S ribosomal protein L11 |
| SUPT16 | 2 | 2 | Facilitates chromatin remodeling subunit SPT16 |
| DDX1 | 2 | 2 | DDX1 ATP-dependent RNA helicase |
| HSPA5 | 2 | 2 | Heat shock 70 kDa protein 5 (glucose-regulated) |
| SSRP1 | 2 | 2 | Structure specific recognition protein 1 |
| HERC2 | 2 | 2 | Hect domain-containing protein 2 (E3 ubiquitin ligase) |
| SFRS1 | 2 | 2 | Splicing factor, arginine/serine-rich 1 |
| HIST1H2AB | 2 | 2 | Histone H2A type 1-B/E |
Supplementary Table S5 (bottom part of the Table) also indicates that 34 additional proteins were identified with high statistical confidence but these identifications rely on at least two unique peptides in one of the immunoprecipitation experiments (Ex: NPM1, RAD50). These proteins may represent the limit of detection for our mass spectrometry analysis.
Validation of the Proteomic Analyses by Western Analysis of Specific WRN Interactants in Untreated and Nuclease-Treated Immunoprecipitations
To confirm several new WRN-interacting proteins, we transfected HEK 293 cells with the eYFP and the eYFP-WRN expression vectors for co-immunoprecipitation experiments with an anti-eYFP antibody. The immunoprecipitates (without nuclease treatments) were analyzed by Western blotting. Western analyses were performed with antibodies against POLR2A, POLR2B, HNRPK, MSH2, XRCC1, Lamin B1, and nucleophosmin (B-23 or NPM1). As indicated in Figure 3, all the antibodies tested confirmed their co-immunoprecipitation with the eYFP-WRN construct but not with the eYFP control. When nucleases were used in the lysis buffer during the course of the immunoprecipitation, however, we could only detect significant amount of nucleophosmin in the eYFP-WRN immunoprecipitate over the eYFP immunoprecipitation background level (Figure 3, right panel) confirming the limit of detection of our mass spectrometry study. These results suggest that the interactions of POLR2A, POLR2B, HNRPK, MSH2, XRCC1, and Lamin B1 proteins in WRN-containing multiprotein complexes are highly dependent on the presence of nucleic acids.
Figure 3.
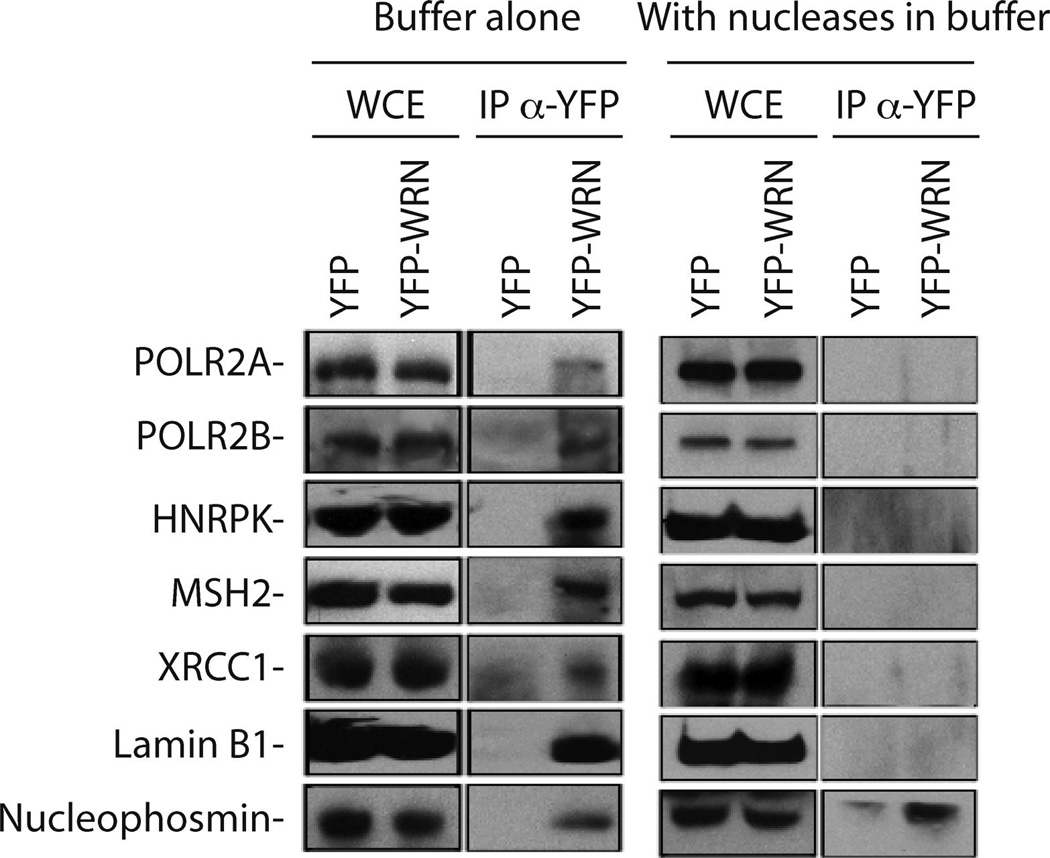
Co-immunoprecipitation of selected proteins with the eYFP-WRN construct. Human 293 embryonic kidney cells were transfected with an eYFP or a eYFP-WRN expression vector. Whole cell extracts (WCE) were immunoprecipitated with an anti-eYFP antibody (IP α-YFP). The immunoprecipitate was analyzed by immunoblotting with antibodies against POLR2A, POLR2B, HNRPK, MSH2, XRCC1 Lamin B1, and nucleophosmin. Panels on the left represent the immunoprecipitation performed in the absence of nuclease. Panels on the right represent the immunoprecipitation performed in the presence of Benzonase and RNase A in the lysis buffer.
Validation of RPS3 Interactions with Endogenous WRN
A good concordance between nuclease and non-nuclease treated immunoprecipitations was obtained for the top hits (see Supplementary Table S5). More importantly, in the nuclease treated immunoprecipitation, the top 15 hits are very plausible direct interactors of WRN as many of them were previously shown to bind WRN protein directly. These include DNA-PK (PRKDC),31,32 XRCC5 (KU86) and XRCC6 (KU70),18,19 DHX9 (DNA helicase II),33 RPA1,20,34 LIG3 (DNA ligase III),35 and PARP-1.14,21 Out of the 40 proteins listed in Table 2, 30 of them are potentially new direct interactors of WRN protein. Among the top 15 hits, we find the new interactors TMPO, HNRNPU, RPS3, RALY, RPS9 DDX21, and HNRNPM. We focused our attention on RPS3, which is a ribosomal protein with endonuclease activities that is involved in oxidative DNA damage recognition.36–38 Increased oxidative stress has been described in WS patients39 and increase oxidative DNA damage has recently been described in mice with a deletion of part of the Wrn helicase domain.40,41 Furthermore, there is evidence that WRN stimulates repair of oxidative DNA base damage.42 We first validated the presence of this protein in the nuclease-treated eYFP-WRN immunoprecipitate. As indicated in Figure 4A, RPS3 was present in the eYFP-WRN immunoprecipitate and absent in the eYFP immunoprecipitate. We next examined whether we could co-immunoprecipitate these proteins with endogenous WRN protein in untransfected HEK 293 cells. As indicated in Figure 4B, RPS3 was co-immunoprecipitated with the endogenous WRN protein while a control IgG gave no signal. We also performed the reverse immunoprecipitation with an antibody against RPS3. As indicated in Figure 4B, WRN was co-immunoprecipitated with the endogenous RPS3. All immunoprecipitations were performed in the presence of Benzonase and RNase A. We also performed immunoprecipitation experiments on HEK 293 cells treated with different concentrations of hydrogen peroxide for 1 to 2 h. We did not detect an increase in the amount of WRN/RPS3 complex in the immunoprecipitates (data not shown). Additional stress conditions with different DNA damaging agents are warranted to determine the function of the WRN/RPS3 complex in cells.
Figure 4.
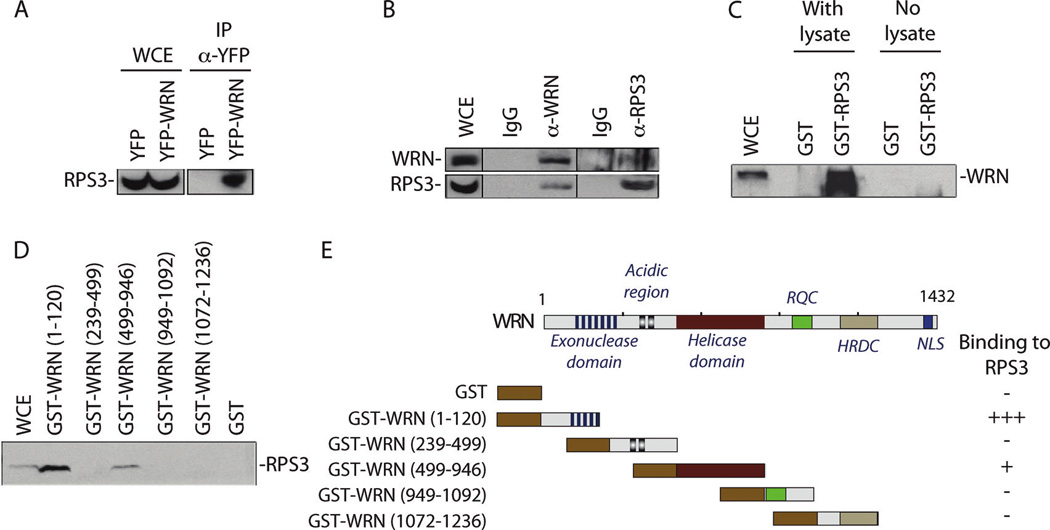
Interaction of RPS3 with the WRN protein. (A) Co-precipitation of RPS3 with the eYFP-WRN protein in HEK 293 cells. (WCE represents the whole cell extract.) (B) Co-immunoprecipitation of human RPS3 protein with endogenous WRN protein. Approximately 2 mg of proteins from HEK 293 cells was immunoprecipitated with antibodies against the C-terminus region of human WRN protein H300 from Santa Cruz Biotechnology. Control antibodies were of the same IgG species. The immunoprecipitates were analyzed by Western blotting with the anti-WRN antibody (WRN; top panel) and an antibody against RPS3. Proteins were revealed with an ECL kit. The anti-WRN antibody for the immunoblot is from Novus Biologicals. (C) Interaction of RPS3 with different domains of WRN in whole cell extract. Immunoblot against RPS3 protein bound to different GST-WRN affinity Sepharose beads. HEK 293 whole cell extracts were incubated with 50 µg of the GST-WRN fragments or GST linked glutathione-sepharose beads overnight. Proteins bound to the affinity beads were analyzed by SDS-PAGE with antibodies against RPS3. (D) Interaction of WRN with RPS3 in whole cell extract. Immunoblot against WRN protein bound to GST-RPS3 but not GST affinity Sepharose beads. HEK 293 whole cell extracts were incubated with 50 µg of the GST-RPS3 or GST linked glutathione-sepharose beads overnight. Proteins bound to the affinity beads were analyzed by SDS-PAGE with antibodies against WRN. (E) Schematic representation of different WRN fragments that were used in the WRN affinity chromatography experiments. Each domain of the WRN protein is indicated on the full WRN protein figure. The amino acid residues of the WRN fragments used in this study are indicated on the left of each construct. Binding of RPS3 is indicated on the right by the “+” sign. The “−” sign indicates no binding detected.
We next determined whether WRN could bind directly to a GST-RPS3 chimeric protein in vitro. As indicated in Figure 4C, WRN interacted directly to a GST-RPS3 construct but not to sepharose beads containing GST alone. Finally, we determined which region of WRN is required for RPS3 interaction. As indicated in Figure 4D, GST pull-down assays showed that RPS3 interacted strongly to a region of the WRN protein containing part of the exonuclease domain (amino acids 1–120) and more weakly to part of the helicase domain of WRN (amino acids 499–946) (summarized in Figure 4E). Overall, these experiments suggest that RPS3 interacts directly to WRN protein in cells.
DISCUSSION
The greatest advantage of a proteome-wide study like the one presented here lies in the acceleration of the pace at which WRN-binding candidates are discovered compared with traditional approaches. By combining large-scale LC-MS/MS identification of immunoprecipitated eYFP-WRN-associated proteins and bioinformatics-based predictions, this study represents the first large-scale proteomic identification of WRN-binding proteins and provides insights into the pathways that can be modulated by WRN protein. Although these candidates will require additional validation, their disclosure opens up considerable opportunity for new hypothesis-driven experiments.
In this study, two approaches have been used to elucidate WRN’s intermolecular interactions. The first approach was focused on a more global, systems-wide analysis of WRN complexes composition. While this approach is more likely to capture low affinity and transient interactions, it cannot rule out recruitment via nucleic acid tethering. Therefore, we adopted a second approach that minimizes physical interactions between WRN protein and nucleic acids. Many laboratories add ethidium bromide or nucleases to cell extraction procedures before the immunoprecipitation step. These strategies have been used on a case-by-case level to confirm that protein–protein interactions are not due to the presence of nucleic acids only. Here, we use this concept for the first time at a larger scale, with a complete proteomic analysis on human WRN. We did not use ethidium bromide in this study because there is evidence that such chemical intercalates into the minor groove of the DNA leading to a nucleic acid structure recognized by specific DNA binding proteins. For example, a recent study indicated that p53 has a higher affinity for DNA treated with various concentrations of ethidium bromide.43 We thus opted for broad-spectrum nucleases in our analyses. To our surprise, more than 80% of the WRN interactors were removed with the addition of Benzonase and RNase A prior to immunoprecipitation and mass spectrometry. The 40 co-immunoprecipitated proteins in the presence of nucleases are likely forming stable and direct protein–protein interaction complexes that persisted during all the purification steps. Our strategy is thus amenable to proteomics analysis of other nucleic acid binding proteins and serves as a cautionary note in identifying protein partners of nucleic acid binding proteins.
The major interactors of WRN identified in this study were the proteins of the nonhomologous end joining repair system (DNA-PKc and the KU70/86 complex). A role for WRN in end joining reaction has been established through other studies,44 showing functional interactions between WRN and key factors in end joining. We did not detect all the proteins that have been experimentally shown to interact or to co-immunoprecipitate with the WRN protein in our system. For example, EXO1, RAD51, RAD52, and BRCA1 (for DNA recombination), APE1 and FEN1 (base excision repair)24,45–49 were absent in our list (Table 1). This may be due to the fact that we used 293 embryonic kidney cells in unstressed conditions. Nevertheless, we did detect proteins involved in these repair pathways such as RAD50, SFPQ, and NONO for recombination50 (when nucleases were omitted in the lysis buffers) or PARP-1, and DNA ligase 3 for base excision repair (even with the presence of nucleases in the lysis buffer). We also identified MSH2 and MSH3 (when no nucleases were added in the lysis buffer), which are involved in both mismatch repair and recombination pathways51 confirming a previous report.52 Several sister chromatid cohesion proteins, such as SMC1A, SMC3, and NIPBL, involved in DNA repair with BRCA1 were identified in the eYFP-WRN immunoprecipitate as well but only when nucleases were omitted in our lysis buffer.53–56 Finally, the ATR serine/threonine protein kinase involved in sensing double-strand breaks was also associated with the WRN protein based on our preliminary list. Similarly, we did not detect any DNA polymerase.57,58 We did identify, however, several accessory proteins required for the activities of these polymerases such as several RFCs and RPAs, DHX9, and topoisomerase I. These accessory proteins are likely to be direct interactors as they were present even in nucleic acids depleted extracts.
Interestingly, our results indicated that WRN also co-immunoprecipitated the POLR2A and POLR2B subunits of the RNA polymerase II transcriptional machinery. These results complement a study conducted a decade ago showing that purified WRN protein increases the transcription efficiency of a plasmid template bearing an RNA polymerase II-specific promoter in an in vitro assay.59 It is also in agreement with a report describing the importance of WRN protein in the transcription and thus the replication of the HIV-1 retrovirus.60 With nucleases in the buffers, however, both subunits of the RNA polymerase II machinery (POLR2A and POLR2B) were absent from the eYFP-WRN immunoprecipitates. It is possible that the WRN protein facilitates transcription by remodeling the chromatin structure or by simply changing the topology of the DNA61 in close proximity to some RNA polymerase subunits during transcription of specific genes without direct contact. Alternatively, WRN may interact directly to transcription factors during transcription which themselves interact directly with the RNA polymerase machinery. Several transcription factors or cofactors are in the new list of WRN interactors even in the presence of nucleases in the immunoprecipitates (SUPT16H, SSRP1, TFAM, PELP1). Validation and the significance of these potential interactions are warranted.
Except for RALY, the top 15 proteins associated with WRN were found in both nuclease treated and untreated immunoprecipitation experiments. Several of these interactions have been confirmed in the literature. We are thus confident that the mass spectrometry analyses on the eYFP-WRN immunoprecipitation containing Benzonase and RNase A have identified direct interactors of the WRN protein. Several proteins have never been reported as potential WRN interactors, although some may possess biologic activities that are consistent with known WRN functions (Table 3). An example of this is the identification of the highly abundant lamina-associated polypeptide 2-alpha (LAP2A/TMPO) as a putative WRN-interacting protein. LAP2A is likely involved in the dynamics of higher order chromatin organization. Indeed, Werner syndrome cells display a distorted nuclear shape phenotype. 62 Another example is RPS3. RPS3 is a ribosomal protein with endonuclease activities involved in oxidative DNA damage recognition in cells.36–38 As a proof of principle, we confirmed that RPS3 is a new direct interactor of the endogenous WRN protein as it can be co-immunoprecipitated by an anti-WRN antibody. Similarly, WRN can be co-immunoprecipitated with endogenous RPS3. Future experiments will be aimed at determining the impact of the WRN/RPS3 functions in cells as RPS3 bound two catalytic regions of WRN.
Table 3.
Significant Statistical Biological Functions Associated with Proteins Identified by LC–MS/MS in eYFP-WRN Immunoprecipitates Treated with Nucleases As Revealed by PANTHERa
|
Nucleoside, nucleotide and nucleic acid metabolism (P < 0.001) RALY, HIST1H2AB, XRCC5, XRCC6, PRKDC, RPA1, RPA2, TOP1, HNRNPM, DDX3X, HIST1H4A, DDX21, DHX9, SSRP1, HIST1H1E, TRIM28, DDX1, LIG3, SFRS1, RBMX, HNRNPU, RFC4, SUPT16H, PARP-1, THOC1, BAT1, PELP1, TFAM. |
|
DNA metabolism (P < 0.001) RPA1, XRCC5, RPA2, TOP1, RFC4, XRCC6, LIG3, PRKDC, PARP-1. |
|
DNA repair (P < 0.001) XRCC5, RPA2, XRCC6, LIG3, PRKDC, PARP-1, RPS3. |
|
Pre-mRNA processing (P < 0.001) RALY, HNRNPM, DHX9, SFRS1, RBMX, HNRNPUL2. |
|
DNA replication (P < 0.005) RPA1, RPA2, TOP1, RFC4. |
|
Chromatin packaging and remodeling (P < 0.012) HIST1H2AB, HIST1H1E, HIST1H4A, TRIM28, HIST1H2BK. |
|
Protein biosynthesis and processing (P < 0.027) RPS3A, RPS9, RPL5, RPL11, RPS3, HERC2, HSPA5, CCT8. |
Not in this list: TMPO for nuclear structure; PELP1 and TFAM for transcription; VRK3 for signal transduction.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Canadian Institutes of Health Research to G.G.P.,J.-Y.M., and M.L.J.-Y.M. and M.L. are senior scholars from the Fonds de la Recherche en Santé du Québec.
Footnotes
ASSOCIATED CONTENT
Additional information as mentioned in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The authors declare that no competing interests exist.
REFERENCES
- 1.Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner’s syndrome: a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine. 1966;45(3):177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Salk D. Werner’s syndrome: a review of recent research with an analysis of connective tissue metabolism, growth control of cultured cells, and chromosomal aberrations. Hum. Genet. 1982;62(1):1–5. doi: 10.1007/BF00295598. [DOI] [PubMed] [Google Scholar]
- 3.Ozgenc A, Loeb LA. Current advances in unraveling the function of the Werner syndrome protein. Mutat. Res. 2005;577(1–2):237–251. doi: 10.1016/j.mrfmmm.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 4.Yu C, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner’s syndrome gene. Science. 1996;272(5259):258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 5.Schonberg S, Niermeijer MF, Bootsma D, Henderson E, German J. Werner’s syndrome: proliferation in vitro of clones of cells bearing chromosome translocations. Am. J. Hum. Genet. 1984;36(2):387–397. [PMC free article] [PubMed] [Google Scholar]
- 6.Scappaticci S, Forabosco A, Borroni G, Orecchia G, Fraccaro M. Clonal structural chromosomal rearrangements in lymphocytes of four patients with Werner’s syndrome. Ann. Genet. 1990;33(1):5–8. [PubMed] [Google Scholar]
- 7.Hanada K, Ukita T, Kohno Y, Saito K, Kato J-I, Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1997;94(8):3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1(1):80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prince PR, Emond MJ, Monnat RJ., Jr Loss of Werner syndrome protein function promotes aberrant mitotic recombination. Genes Dev. 2001;15(8):933–938. doi: 10.1101/gad.877001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in Werner syndrome. Mol. cell. Biol. 2002;22(20):6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orren DK, Brosh RM, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27(17):3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz VP, Zakian VA, Ogburn CE, McKay J, Jarzebowicz AA, Edland SD, Martin GM. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum. Genet. 1996;97(6):750–754. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- 13.Tahara H, Tokutake Y, Maeda S, Kataoka H, Watanabe T, Satoh M, Matsumoto T, Sugawara M, Ide T, Goto M, Furuichi Y, Sugimoto M. Abnormal telomere dynamics of B-lymphoblastoid cell strains from Werner’s syndrome patients transformed by Epstein-Barr virus. Oncogene. 1997;15(16):1911–1920. doi: 10.1038/sj.onc.1201377. [DOI] [PubMed] [Google Scholar]
- 14.Lebel M, Lavoie J, Gaudreault I, Bronsard M, Drouin R. Cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am. J. Pathol. 2003;162(5):1559–1569. doi: 10.1016/S0002-9440(10)64290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavoie J, Bronsard M, Lebel M, Drouin R. Mouse telomere analysis using an optimized primed in situ (PRINS) labeling technique. Chromosoma. 2003;111(7):438–444. doi: 10.1007/s00412-002-0225-1. [DOI] [PubMed] [Google Scholar]
- 16.Laud PR, Bailey SM, Multani AS, Kingsley C, Wu L, Pathak S, Lebel M, DePinho RA, Chang S. Elevated telomere-telomere recombination correlates with increased cellular immortalization and transformation in G5 Terc−/− Wrn−/− mouse cells. Genes Dev. 2005;19(21):2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greider CW. Telomere length regulation. Annu. Rev. Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 18.Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr V. A Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14(8):907–912. [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Comai L. Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J. Biol. Chem. 2000;275(37):28349–28352. doi: 10.1074/jbc.C000289200. [DOI] [PubMed] [Google Scholar]
- 20.Brosh RM, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274(26):18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 21.von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol. Cell. Biol. 2003;23(23):8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyng KJ, May A, Kolvraa S, Bohr V. A Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proc. Natl. Acad. Sci. U.S.A. 2003;100(21):12259–12264. doi: 10.1073/pnas.2130723100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turaga RVN, Paquet ER, Sild M, Vignard J, Garand C, Johnson FB, Masson J-Y, Lebel M. The Werner syndrome protein affects the expression of genes involved in adipogenesis and inflammation in addition to cell cycle and DNA damage responses. Cell Cycle. 2009;8(13):2080–2092. doi: 10.4161/cc.8.13.8925. [DOI] [PubMed] [Google Scholar]
- 24.Baynton K, Otterlei M, Bjørås M, von Kobbe C, Bohr VA, Seeberg E. WRN interacts physically and functionally with the recombination mediator protein RAD52. J. Biol. Chem. 2003;278(38):36476–36486. doi: 10.1074/jbc.M303885200. [DOI] [PubMed] [Google Scholar]
- 25.Buisson R, Dion-Côté A, Coulombe Y, Launay H, Cai H, Stasiak A, Stasiak A, Xia B, Masson J-Y. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. struct. Mol. Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 27.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 28.Haince JF, McDonald D, Rodrigue A, Déry U, Masson J-Y, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283(2):1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 29.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner Syndrome Helicase and Exonuclease Cooperate to Resolve Telomeric D Loops in a Manner Regulated by TRF1 and TRF2. Mol. Cell. 2004;14(6):763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Guay D, Gaudreault I, Massip L, Lebel M. Formation of a nuclear complex containing the p53 tumor suppressor, YB-1, and the Werner syndrome gene product in cells treated with UV light. Int. J. Biochem. Cell. Biol. 2006;38(8):1300–1313. doi: 10.1016/j.biocel.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J. Biol. Chem. 2001;276(41):38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 32.Karmakar P, Piotrowski J, Brosh RM, Jr, Sommers JA, Miller SP, Cheng WH, Snowden CM, Ramsden DA, Bohr VA. Werner protein is a target of DNA-dependent protein kinase In Vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J. Biol. Chem. 2002;277(21):18291–18302. doi: 10.1074/jbc.M111523200. [DOI] [PubMed] [Google Scholar]
- 33.Chakraborty P, Grosse F. WRN helicase unwinds Okazaki fragment-like hybrids in a reaction stimulated by the human DHX9 helicase. Nucleic Acids Res. 2010;38(14):4722–4730. doi: 10.1093/nar/gkq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen JC, Gray MD, Oshima J, Loeb LA. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998;26(12):2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood. 2008;112(4):1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SH, Lee JY, Kim J. Characterization of a wide range base-damage-endonuclease activity of mammalian rpS3. Biochem. Biophys. Res. Commun. 2005;328(4):962–967. doi: 10.1016/j.bbrc.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 37.Hegde V, Wang M, Mian IS, Spyres L, Deutsch WA. The high binding affinity of human ribosomal protein S3 to 7,8-dihydro-8-oxoguanine is abrogated by a single amino acid change. DNA Repair. 2006;5(7):810–815. doi: 10.1016/j.dnarep.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Yadavilli S, Hegde V, Deutsch W. A Translocation of human ribosomal protein S3 to sites of DNA damage is dependant on ERK-mediated phosphorylation following genotoxic stress. DNA Repair. 2007;6(10):1453–1462. doi: 10.1016/j.dnarep.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagano G, Zatterale A, Degan P, d’Ischia M, Kelly FJ, Pallardo FV, Calzone R, Castello G, Dunster C, Giudice A, Kilinc Y, Lloret A, Manini P, Masella R, Vuttariello E, Warnau M. In Vivo prooxidant state in Werner syndrome (WS): results from three WS patients and two WS heterozygotes. Free Radical Res. 2005;39(5):529–533. doi: 10.1080/10715760500092683. [DOI] [PubMed] [Google Scholar]
- 40.Massip L, Garand C, Paquet E, Cogger VC, Oreilly J, Tworek L, Hatherell A, Taylor CG, Thorin E, Zahradka P, Le Couteur DG, Lebel M. Vitamin C restores healthy aging in a mouse model for Werner Syndrome. FASEB J. 2010;24(1):158–172. doi: 10.1096/fj.09-137133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labbé A, Turaga RVN, Paquet ER, Garand C, Lebel M. Expression profiling of mouse embryonic fibroblasts with a deletion in the helicase domain of the Werner Syndrome gene homologue treated with peroxide. BMC Genomics. 2010;11:127. doi: 10.1186/1471-2164-11-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, Mitra S, Bohr V. A The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J. Biol. Chem. 2007;282(36):26591–602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 43.Pivonková H, Sebest P, Pecinka P, Tichá O, Nemcová K, Brázdová M, Jagelská EB, Brázda V, Fojta M. Selective binding of tumor suppressor p53 protein to topologically constrained DNA: Modulation by intercalative drugs. Biochem. Biophys. Res. Commun. 2010;393(4):894–899. doi: 10.1016/j.bbrc.2010.02.120. [DOI] [PubMed] [Google Scholar]
- 44.Kusumoto R, Dawut L, Marchetti C, Wan Lee J, Vindigni A, Ramsden D, Bohr VA. Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing. Biochemistry. 2008;47(28):7548–7556. doi: 10.1021/bi702325t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brosh RM, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20(20):5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma S, Sommers JA, Driscoll HC, Uzdilla L, Wilson TM, Brosh RM., Jr The exonucleolytic and endonucleolytic cleavage activities of human exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein. J. Biol. Chem. 2003;278(26):23487–23496. doi: 10.1074/jbc.M212798200. [DOI] [PubMed] [Google Scholar]
- 47.Ahn B, Harrigan JA, Indig FE, Wilson DM, III, Bohr VA. Regulation of WRN helicase activity in human base excision repair. J. Biol. Chem. 2004;279(51):53465–53474. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- 48.Cheng WH, Kusumoto R, Opresko PL, Sui X, Huang S, Nicolette ML, Paull TT, Campisi J, Seidman M, Bohr VA. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006;34(9):2751–2760. doi: 10.1093/nar/gkl362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr V. A Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J. Cell Sci. 2006;119(Pt 24):5137–5146. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- 50.Rajesh C, Gruver AM, Basrur V, Pittman DL. The interaction profile of homologous recombination repair proteins RAD51C, RAD51D and XRCC2 as determined by proteomic analysis. Proteomics. 2009;9(16):4071–4086. doi: 10.1002/pmic.200800977. [DOI] [PubMed] [Google Scholar]
- 51.Surtees JA, Alani E. Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J. Mol. Biol. 2006;360(3):523–536. doi: 10.1016/j.jmb.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 52.Saydam N, Kanagaraj R, Dietschy T, Garcia PL, Peña-Diaz J, Shevelev I, Stagljar I, Janscak P. Physical and functional interactions between Werner syndrome helicase and mismatch-repair initiation factors. Nucleic Acids Res. 2007;35(17):5706–5716. doi: 10.1093/nar/gkm500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16(5):571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 2004;36(6):636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 55.Strachan T. Cornelia de Lange Syndrome and the link between chromosomal function, DNA repair and developmental gene regulation. Curr. Opin. Genet. Dev. 2005;15(3):258–264. doi: 10.1016/j.gde.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 56.Revenkova E, Focarelli ML, Susani L, Paulis M, Bassi MT, Mannini L, Frattini A, Delia D, Krantz I, Vezzoni P, Jessberger R, Musio A. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum. Mol. Genet. 2009;18(3):418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamath-Loeb AS, Johansson E, Burgers PM, Loeb LA. Functional interaction between the Werner syndrome protein and DNA polymerase delta. Proc. Natl. Acad. Sci. U.S.A. 2000;97(9):4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 2007;104(25):10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balajee AS, Machwe A, May A, Gray MD, Oshima J, Martin GM, Nehlin JO, Brosh RM, Orren DK, Bohr VA. The Werner syndrome protein is involved in RNA polymerase II transcription. Mol. Biol. Cell. 1999;10(8):2655–2668. doi: 10.1091/mbc.10.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma A, Awasthi S, Harrod CK, Matlock EF, Khan S, Xu L, Chan S, Yang H, Thammavaram CK, Rasor RA, Burns DK, Skiest DJ, Van Lint C, Girard AM, McGee M, Monnat RJ, Jr, Harrod R. The Werner syndrome helicase is a cofactor for HIV-1 long terminal repeat transactivation and retroviral replication. J. Biol. Chem. 2007;282(16):12048–12057. doi: 10.1074/jbc.M608104200. [DOI] [PubMed] [Google Scholar]
- 61.Turaga RVN, Massip L, Chavez A, Johnson FB, Lebel M. Werner syndrome protein prevents DNA breaks upon chromatin structure alteration. Aging Cell. 2007;6(4):471–481. doi: 10.1111/j.1474-9726.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- 62.Adelfalk C, Scherthan H, Hirsch-Kauffmann M, Schweiger M. Nuclear deformation characterizes Werner syndrome cells. Cell Biol. Int. 2005;29(12):1032–1037. doi: 10.1016/j.cellbi.2005.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


