Abstract
Cockayne syndrome complementation group B (CSB) protein is engaged in transcription-coupled repair (TCR) of UV induced DNA damage and its deficiency leads to progressive multisystem degeneration and premature aging. Here, we show that human CSB-deficient cells are hypersensitive to physiological concentrations (1–10 µM) of a lipid peroxidation product, trans-4-hydroxy-2-nonenal (HNE), and in response to HNE they develop a higher level of sister chromatid exchanges (SCEs) in comparison to the wild-type cells. HNE-DNA adducts block in vitro transcription by T7 RNA polymerase, as well as by HeLa cell-free extracts. Treatment of wild-type cells with 1–20 µM HNE causes dephosphorylation of the CSB protein, which stimulates its ATPase activity necessary for TCR. However, high HNE concentrations (100–200 µM) inhibit in vitro CSB ATPase activity as well as the transcription machinery in HeLa cell-free extracts.
Cell lines expressing CSB protein mutated in different ATPase domains exhibit different sensitivities to HNE. The motif II mutant, which binds ATP, but is defective in ATP hydrolysis was as sensitive to HNE as CSB-null cells. In contrast, motif V mutant cells were as sensitive to HNE as were the cells bearing wild-type protein, while motif VI mutant cells showed intermediate sensitivity to HNE. These mutants exhibit decreased ATP binding, but retain residual ATPase activity. Homology modeling suggested that amino acids mutated in motifs II and VI are localized closer to the ATP binding site than amino acids mutated in ATPase motif V.
These results suggest that HNE-DNA adducts are extremely toxic endogenous DNA lesion, and that their processing involves CSB. When these lesions are not removed from the transcribed DNA strand due to CSB gene mutation or CSB protein inactivation by high, pathological HNE concentrations, they may contribute to accelerated aging.
Keywords: Lipid peroxidation, HNE adducts, Transcription inhibition, NER, CSB, CSB ATPase mutations
1. Introduction
One of the prominent aging theories proposes that a gradual accumulation of DNA damage during the cellular and organismal lifetime contributes to the aging process. This is supported by the observation that the degree of DNA oxidation correlates negatively with the maximum life span [1].
Cockayne syndrome (CS) is a degenerative human disease characterized by traits reminiscent of normal aging, such as neurological degeneration, cataracts and systemic growth failure. The majority of CS cases are caused by defects in the CS complementation group B (CSB) protein. The CSB gene encodes a 168 kDa protein belonging to the SWI2/SNF2 protein family. The CSB protein participates in transcription-coupled repair (TCR), global genome repair (GGR), base excision repair (BER) of certain types of oxidative DNA damage, e.g. 8-oxoG, as well as in general transcription [2]. CSB is also involved in the poly(ADP-ribose) polymerase-1 (PARP-1) mediated response to oxidative DNA damage [3], as well as in strand annealing and exchange, which might be engaged in mitotic recombination [4].
CSB protein contains an acidic region, a glycine-rich region, two putative nuclear localization signal sequences and an ATPase domain consisting of seven conserved motifs (I, Ia and II–VI) [5]. The SNF2-like ATPase domain is critical for CSB function in vivo [6,7]: DNA-dependent ATPase activity [8–10], ATP-dependent chromatin remodeling [11] and modulation of negative supercoiling of DNA [12].
The ATPase activity of the CSB protein is increased in the presence of DNA. The biggest effect is achieved with dsDNA containing a bubble or loop, corresponding to the open DNA structure during the nucleotide excision repair (NER) and transcription arrest [13].
The ATPase activity can also be modified by phosphorylation of the CSB protein. In vitro and in whole cell extracts, CSB was shown to be phosphorylated by casein kinase 2 (CK2) [13]. In response to hydrogen peroxide, CSB protein was also phosphorylated by c-Abl tyrosine kinase [14]. Phosphorylation of CSB, as well as poly(ADP-ribosyl) ation by PARP-1, lead to a decrease of the ATPase activity [3,13]. Correspondingly, dephosphorylation of CSB, which occurs after UV irradiation, results in an increase of the ATPase activity of CSB protein by approximately 40% [13].
During TCR, CSB protein is involved in the removal of stalled RNA polymerase from the damaged template and in recruitment of the NER machinery to repair the transcribed DNA strand. Recognized TCR substrates are UV photoproducts, and some bulky adducts, e.g. cisplatin intrastrand cross-links [13]. However, deficiency in repair of these types of DNA damage does not explain the premature aging, or the developmental and neurological CS symptoms. At least two explanations are possible: either other mechanisms than TCR may be involved in generating these symptoms in CS cells or not all endogenous TCR substrates are well characterized. CSA and CSB proteins are engaged in ubiquitination of RNA polymerase II, as well as several other proteins, including p21. CSB mutants accumulate p21 due to retarded protein turnover, which in consequence causes retardation of DNA replication and accumulation of DNA double strand breaks [15]. Among known endogenous DNA lesions, only few were unambiguously demonstrated to block transcription, and were suggested to be processed by TCR. These include, e.g. 3-methyladenine, which may arise from non-enzymatic DNA methylation by S-adenosylmethionine, thymine glycol, which is formed during DNA oxidation, and malondialdehyde-guanine adduct, which results from lipid peroxidation (LPO) [16].
Lipid peroxidation is implicated in aging as well as in the pathogenesis of numerous human diseases, including atherosclerosis, cancer, diabetes and arthritis [17]. Significant increase in the LPO level was found in skeletal muscle of old individuals [18], as well as in hepatocytes isolated from old ovariectomized rats [19].
One of the major LPO products is trans-4-hydroxy-2-nonenal (HNE) [20]. HNE is considered as a biomarker of oxidative stress in pathophysiological processes. For example, increased level of HNE accumulation was observed in brain tissue and in cerebrum of patients suffering from several neurodegenerative disorders like Alzheimer’s (AD), Parkinson’s, Pick’s, amyotrophic lateral sclerosis and Huntington diseases [21]. Interestingly, large amounts of HNE-modified proteins were found in the brains (globus pallidus) of CS patients [22].
Physiological concentrations of HNE vary from 0.1 to 3 µM, and can increase up to 50 µM or even milimolar values under oxidative stress [23,24]. Within cells, HNE binds primarily to thiols and proteins [25]. HNE also forms exocylic propano- and etheno-type adducts to DNA bases bearing six or seven carbon atom side chains [26,27]. Etheno-type adducts derive from HNE oxidation to epoxide by peroxides or atmospheric oxygen [27]. HNE also induces DNA intra- and interstrand cross-links [28], as well as DNA–protein cross-links [29] and protein adducts [29,30]. HNE-dG adducts were detected in DNA of non-exposed humans and rodents, which indicates their endogenous source [26,31,32].
We have previously shown that HNE-DNA adducts block replication, trigger recombination, base substitutions and frameshift mutations in ssM13 phage [27]. Other studies showed that in mammalian cells HNE increases the frequency of micronuclei, chromosomal aberrations, sister chromatid exchanges (SCEs) [33–35] and point mutations [36], already at low, physiological concentrations of 0.1–10 µM. HNE also exerts a clastogenic effect in human cells, possibly via inactivation of the functional SH groups in DNA polymerases [37].
HNE-DNA adducts were previously shown to be recognized by the bacterial and mammalian nucleotide excision repair (NER) proteins [38,39]. Here, we show that CSB protein is engaged in the processing of HNE-DNA adducts, and that low concentrations of HNE stimulate CSB dephosphorylation, and possibly transcription-coupled NER. Deficiency in this pathway may cause inhibition of transcription by unrepaired HNE-DNA adducts, as shown by in vitro experiments, thus leading to apoptosis. Accumulation of these lesions may be one of the possible mechanisms of the developmental and neurological features of CS patients, as well as premature aging.
2. Materials and methods
2.1. Cell lines and culture conditions
GM00038, a normal human diploid fibroblast cell line was grown in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 5% fetal bovine serum (FBS); GM1428, a Cockayne syndrome group B (CSB) human fibroblast line was grown in MEM with 10% FBS, vitamins, essential and non-essential amino acids (Invitrogen); CS1AN.S3.G2, an SV-40 transformed human CS-B fibroblast cell line [40] was grown in MEM with 15% FBS; CS1AN.S3.G2 cells transfected with the mammalian expression vector pcDNA3.1 alone (Invitrogen, San Diego, CA; abbreviated as pc3.1) and with pc3.1 containing the wild-type human CSB gene are designated as CS1AN/pc3.1 and CS1AN/pc3.1-CSBwt, respectively. CS1AN.S3.G2 cells transfected with pc3.1-CSB containing different mutations in the CSB gene are designed as CS1AN/pc3.1-CSBE646Q, CS1AN/pc3.1-CSBT912/913 V, CS1AN/pc3.1-CSBQ942E and CS1AN/pc3.1-CSB R946A, respectively. All CS1AN cell lines were grown in MEM supplemented with 15% FBS. CS1AN.S3.G2 cell lines transfected with pc3.1 were grown in media containing 400 µg/ml geneticin (Invitrogen) [7]. For all the cell lines, 100 U/ml penicillin and 0.1 mg/ml streptomycin was included in the growth medium and the cells were incubated at 37 °C and 5% CO2.
2.2. Mutagens
4-hydroxy-2-nonenal (HNE) was synthesized in the form of dimethylacetal derivative as described before [41]. Prior to use, HNE dimethylacetal was hydrolyzed to aldehyde as described [27], dissolved in serum-free media and used immediately.
2.3. Colony formation assay
Exponentially growing cells were trypsinized, seeded on 10 cm2 dishes (300–1000 cells per plate) and allowed to attach. The cells were treated with varying doses of HNE (1–40 µM) in serum-free medium for 2 h, 37 °C, 5% CO2. After treatment, cells were rinsed with serum-free medium and cell colonies were cultured in a fresh, serum-supplemented medium for 10 days. The colonies were washed with phosphate-buffered saline (PBS) and fixed with 3.7% formaldehyde for 10 min at RT. The cells were then washed with PBS and stained with crystal violet (0.1% in dH2O) for 2 h at RT. Colony-formation ability was calculated based on the plating efficiency of treated cells versus the plating efficiency of untreated control cells.
2.4. Sister chromatid exchanges (SCEs)
For the determination of SCEs, exponentially growing cells were cultured in standard growth medium containing 20 µg/ml 5-bromo-2-deoxyuridine (BrdU) for one cell cycle. Cells were then exposed to HNE (0–3 µM) in serum-free medium for 2 h. The cells were then rinsed with PBS and allowed to grow in a fresh medium until the end of the second cell cycle. Further procedure was performed as described [42]. For SCEs analysis 90 metaphases were scored for each cell line and HNE dose. Three independent experiments were performed. SCE frequencies were compared using the two-tailed Student’s t-test.
2.5. In vitro transcription assays
2.5.1. In vitro assay using T7 RNA polymerase
Plasmid DNA containing T7 RNA promoter (pTZ19R, Fermentas) was linearized by SmaI restriction endonuclease, purified using clean-up column and modified with HNE (0.1–30 mM), pH 5.5, for 24 h at 37 °C. DNA was ethanol precipitated and resuspended in diethyl pyrocarbonate (DEPC) treated water. Transcription assay was performed using Fermentas transcription kit (K0412) and reaction product was visualized by 1.2% agarose gel electrophoresis.
2.5.2. In vitro assay using HeLa cell-free extract
HeLa cell-free extract was prepared according to the method of Tanaka et al. [43], modified by Vodenicharov et al. [44] Plasmid DNA containing SV40 RNA promoter and enhancer driving a luciferase reporter gene (pGL3-Control Vector, Promega) was linearized by XbaI restriction endonuclease, purified using clean-up column (Promega) and modified with HNE (0.1–30 mM), pH 5.5, for 24 h at 37 °C. DNA was purified using clean-up column. Transcription assay was performed using 100 µg HeLa cell-free extract, NTP set, RiboLock RNase Inhibitor and transcription buffer from Fermentas transcription kit (K0412). Reaction product was visualized by 1.2% agarose gel electrophoresis.
2.5.3. In vitro assay using HeLa cell-free extract modified with HNE
HeLa cell-free extract was incubated with HNE (0.05–5 mM) for 2 h on ice. After incubation the extract was immediately used for in vitro transcription assay using XbaI linearized pGL3-Control Vector as described above.
2.6. CSB protein phosphorylation status
2.6.1. CSB dephosphorylation in cells
GM00038 cells were plated and allowed to grow for 48 h. Subsequently, cells (2 × 106–5 × 106) were incubated with phosphate-free DMEM (Invitrogen) supplemented with 5% dialyzed FBS for 1 h to exhaust the pool of endogenous phosphate, and then treated with 1–20 µM HNE for 30 min. The cells were supplemented with fresh phosphate-free medium containing 100 µCi 32P inorganic phosphate (Pi) (Hartmann Analytic) for 4 h. After termination of radiolabeling by washing twice with ice-cold PBS, cells were lysed in RIPA (radioimmunoprecipitation assay) buffer (150 mM NaCl, 1% Triton, 0.1% SDS, 10 mM Tris pH 8, 0.5% sodium deoxycholate, 0.2 mM PMSF, 20 µg/ml aprotinin, 10 µg/ml leupeptine, 1 mM Na-orthovanadate and 10 U/ml DNase) and the lysate was centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatant was pre-cleared with protein A magnetic beads (New England Biolabs), and incubated with 5 µg of polyclonal anti-ERCC6 (A. F. Schützdeller, Germany) for 1 h at 4 °C. Protein A magnetic beads were incubated with immune complexes for 1 h at 4 °C, and beads were collected using the accompanying magnet. The beads were washed three times and proteins were analyzed by SDS-PAGE and visualized by a Molecular Imager and silver stain.
2.6.2. Casein kinase 2 activity in solution
Human recombinant casein kinase2, CK2α protein, was purified as previously described [45]. CK2α protein (100 ng) was modified with varying HNE concentrations (10, 100 and 200 µM) for 10 min at RT. The CK2 kinase in-solution assay was performed in a final volume of 50 µl. The incubation mixture contained 100 ng of purified human recombinant CK2α, 75 µg of casein, 20 mM Tris–HCl, pH 7.5, 20 mM MgCl2, 50 µM γ [32P]ATP (100–200 cpm pmol−1). After 20 min of incubation at 30 °C, 40 µl of the assay mixture with casein was spotted onto 3 mm filter strip, and then the filters were washed five times for 10 min in 5% TCA. Next, the filters were washed in 96% ethanol, dried, and the radioactivity was measured in a scintillation counter.
2.7. ATPase activity of the CSB protein
Recombinant CSB wt protein used for the assay was purified using a modified method of Citterio et al. [10]. CSB protein (50 ng) was modified with varying HNE concentrations (10, 100 and 200 µM) for 10 min at 30 °C. Reactions were performed in standard CSB ATPase buffer, without dithiothreitol (DTT). After incubation, excess of HNE was deactivated by addition of 1 µl of 1 mM DTT to the final concentration of 0.1 mM DTT, and modified protein was immediately used for ATPase assay. ATPase activity was measured as described [13]. Reaction mixture (total volume 10 µl) contained 150 ng of dsDNA cofactor bearing a bubble structure (15 nt), 50 ng CSB protein, and 1 µCi [γ-32P] ATP (3000 Ci/mmol; Amersham Biosciences) in 20 mM Tris–HCl pH 7.5, 4 mM MgCl2, 1 µM ATP, 40 µg/ml BSA, 1 mM DTT buffer. Reactions were incubated for 1 h at 30°C and stopped by the addition of 5 µl 0.5 M EDTA. One microliter of each sample was analyzed on a polyethylenimine-cellulose thin layer chromatography plate (PEI Cellulose F, Merck) developed in 2 M LiCl and 0.5 M formic acid. Plates were analyzed by phosphorImaging and the ATPase activity of CSB was analyzed using Image Quant software (Molecular Dynamics). Results from at least three experiments are reported.
2.8. Homology modeling of CSB protein ATPase domain
The structure of the CSB helicase domain was modeled using zebrafish Rad54 as a template (PDB code 1z3i) [46]. Both template structure and sequence alignment have been found using ModBase service [47]. The CSB 483–1079 region was aligned with Rad54 123–733 fragment (e-value 4e−95). The model of a CSB SWI2/SNF2 chromatin-remodeling domain (helicase ATP-binding and helicase C-terminal domains) was built using Modeler software. A model of ATPgammaS molecule was docked manually into the putative binding site of the CSB model.
3. Results
3.1. HNE toxicity for CSB mutants
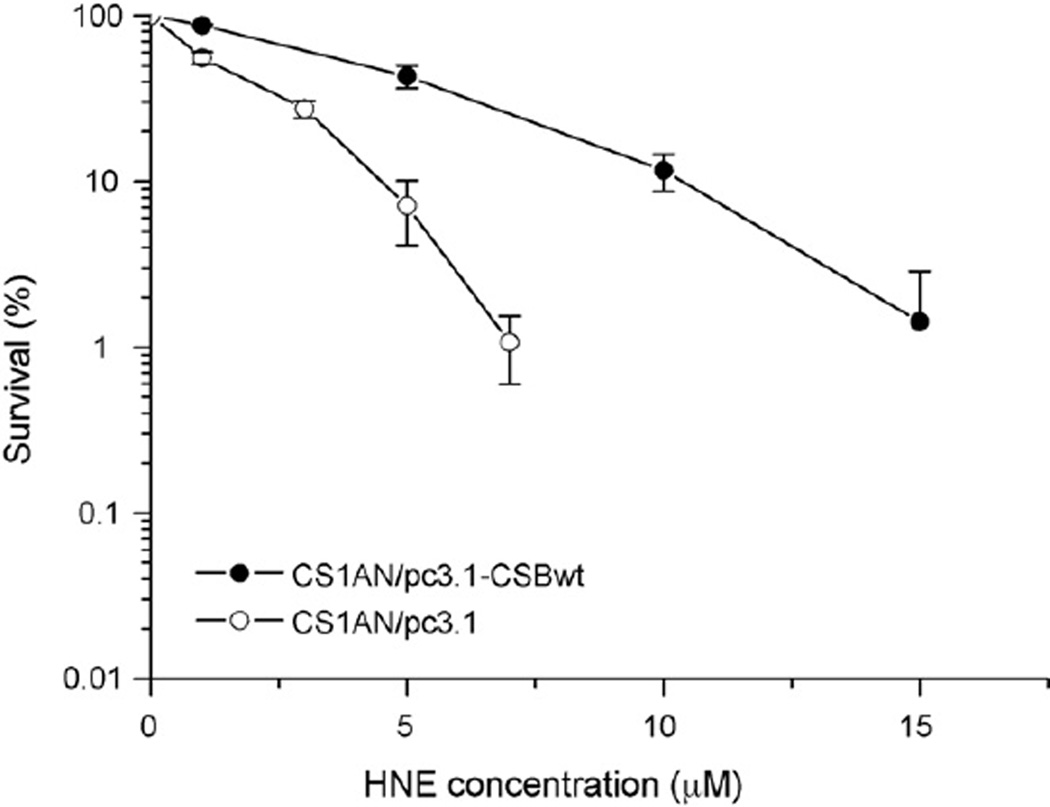
To investigate the role of CSB in the processing of HNE-DNA adducts, we studied its cyto- and genotoxic properties in CSB-proficient and -deficient cell lines. HNE appeared to be highly toxic for both lines, however, CSB-deficient cells were more sensitive to HNE than wild-type cells (Fig. 1). IC50 for CSB-deficient cells was 1.3 µM, while for isogenic CSB-proficient cells it was 4.4 µM HNE.
Fig. 1.
Clonogenic survival of transformed CSB-deficient, CS1AN/pc3.1, (○) and CSB-proficient, CS1AN/pc3.1-CSBwt (●) cell lines after 2 h of treatment with HNE. HNE survival curve represents the mean of three independent experiments. Error bars represent the standard errors of the mean.
3.2. Sister chromatid exchanges (SCEs)
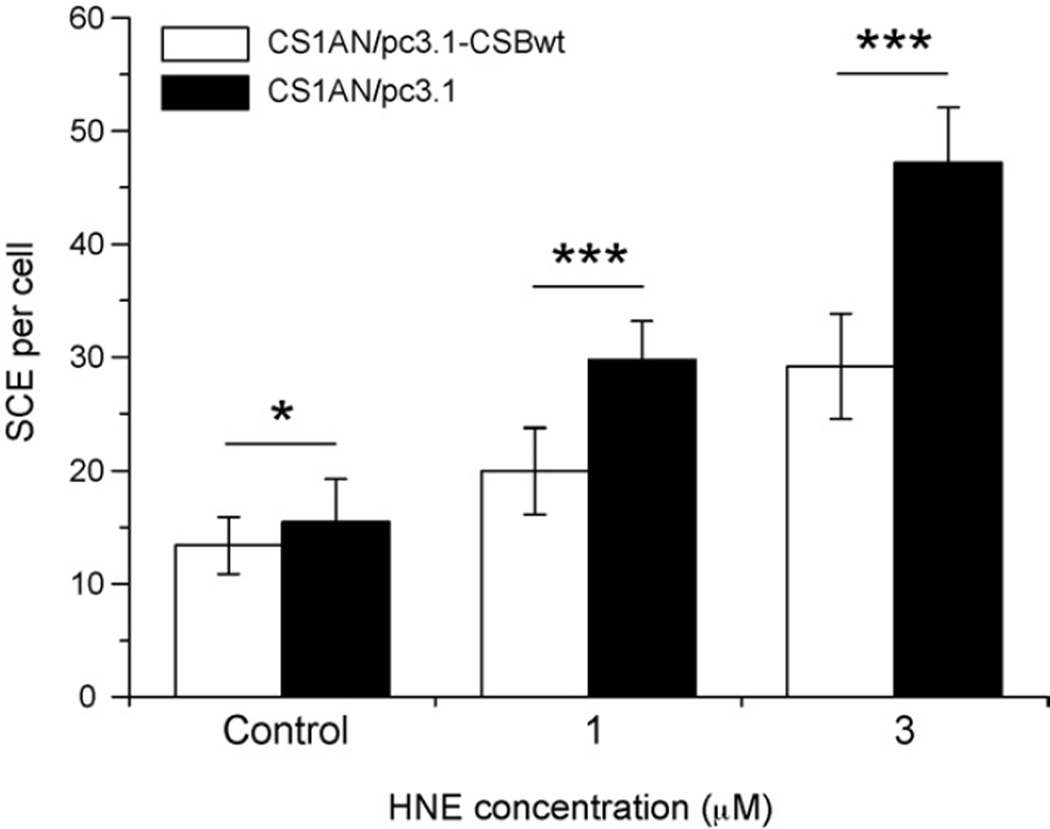
HNE induced SCEs in a dose-dependent manner in both CSB-proficient and -deficient cell lines. However, significantly higher number of SCEs was induced in CSB-deficient cells (30 SCEs/cell at 1 µM HNE, and 45 SCEs/cell at 3 µM HNE) than in their wild-type counterparts (20 SCEs/cell at 1 µM HNE, and 28 SCEs/cell at 3 µM HNE) (Fig. 2). These results suggest that lack of functional CSB protein increases homologous recombination after HNE treatment.
Fig. 2.
Sister chromatid exchanges (SCEs) induced by HNE in CSB-deficient, CS1AN/pc3.1 (black bars) and CSB-proficient, CS1AN/pc3.1-CSBwt (white bars) cell lines. Error bars represent standard errors of the mean. Statistically significant differences are indicated as follows: *p < 0.05; ***p < 0.0005 (Student’s t-test).
3.3. Transcription inhibition
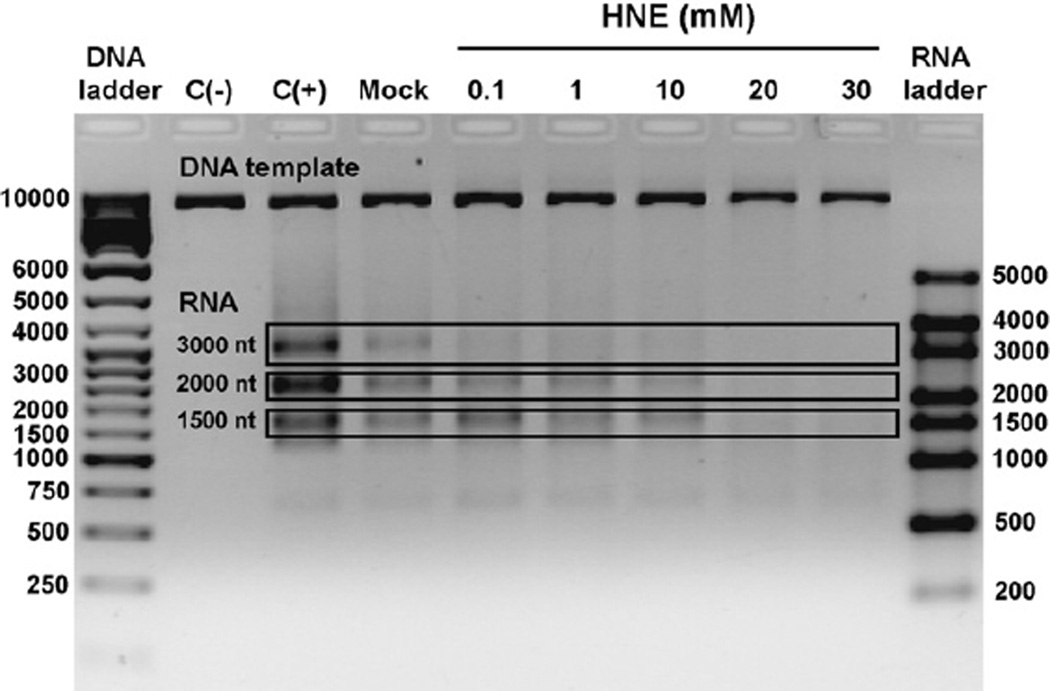
In order to further clarify whether HNE-DNA adducts may be substrates for TCR, we investigated whether their presence in DNA could inhibit transcription. T7 RNA polymerase generated RNA products of 1500, 2000 and 3000 ribonucleotides on undamaged linearized pTZ19R plasmid (Fig. 3). Modification of the DNA template with HNE caused inhibition of transcription in vitro by T7 RNA polymerase. Already at 0.1 mM HNE, formation of 3000 nt transcript was inhibited, whereas transcription of two other products, 1500 and 2000, was gradually decreased upon template modification with 0.1–10 mM HNE. Modification of the template with 20 mM HNE completely abolished RNA synthesis (Fig. 3).
Fig. 3.
In vitro transcription by T7 RNA polymerase on undamaged templates and the template modified with HNE. Lanes:C(−), negative control, transcription reaction performed in the absence of T7 RNA polymerase; C(+), positive control, transcription performed on undamaged DNA; Mock, control transcription performed on undamaged DNA that has been treated as HNE modified DNA but without HNE; HNE, transcription performed on DNA modified with different HNE concentrations (0.1–30 mM for 24 h, pH 5.5 at 37 °C).
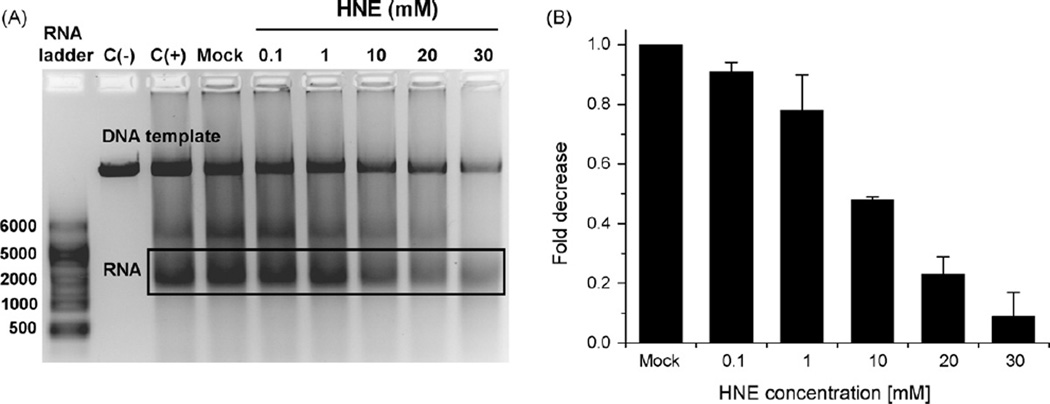
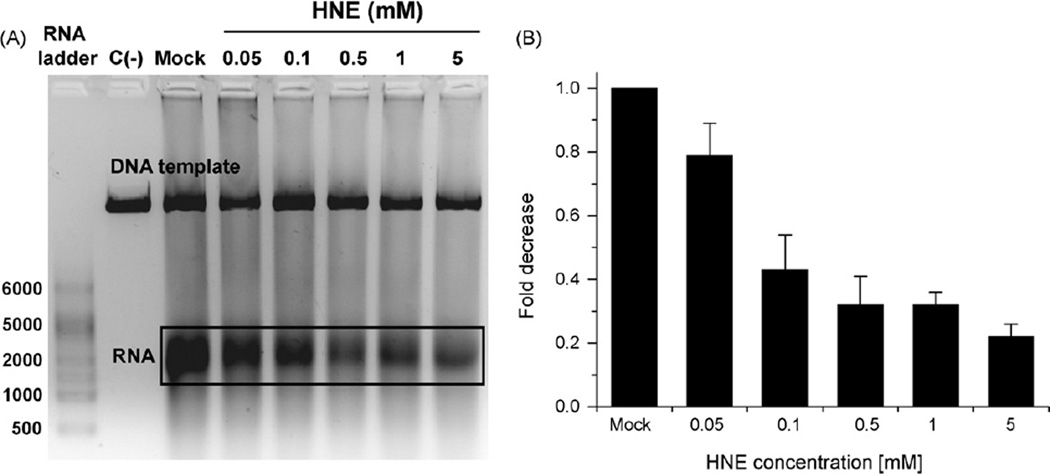
In order to investigate whether HNE affects transcription also in mammalian cells, linearized pGL3 plasmid carrying promoter for luciferase gene was modified in vitro with 0.1–30 mM HNE (24 h at 37 °C, pH 5.5) and used as a template for transcription by HeLa cell-free extracts. The amount of transcript decreased gradually with increasing HNE concentration (Fig. 4A). It is, however, indispensable to note that HNE-modified plasmid was partially degraded by the HeLa cell-free extract, most probably due to numerous incisions caused by DNA repair enzymes recognizing damaged DNA. For the quantification of transcription inhibition we thus measured the ratio of the transcript quantity to the quantity of the template, and subsequently compared these values at different HNE concentrations (Fig. 4B). The identity of transcription product was confirmed by digestion with RNase A (not shown).
Fig. 4.
In vitro transcription by HeLa cell-free extract on undamaged templates and on the templates reacted with HNE. Lanes: C(−), negative control, transcription reaction performed in the absence of HeLa cell-free extract; C(+), positive control, transcription performed on undamaged DNA; Mock, control transcription performed on DNA that has been treated as HNE modified DNA but without HNE; HNE, transcription performed on DNA modified with different HNE concentrations (0.1–30 mM for 24 h, pH 5.5 at 37 °C).
We also wanted to know whether transcription machinery itself may be affected by HNE. When HeLa cell-free extract was pretreated with HNE (2 h on ice), transcript quantity also gradually decreased; at 0.1 mM HNE reaching ~40% of the activity of HNE non-treated extract, and at 5 mM, only ~20% (Fig. 5). Under these conditions only marginal template degradation was observed; however, for comparison, the quantification was done as in Fig. 4B.
Fig. 5.
In vitro transcription activity by unmodified and HNE modified HeLa cell-free extract on undamaged templates. Lanes: C(−), negative control, transcription reaction performed in the absence of HeLa cell-free extract; Mock, control transcription performed by the extract that has been treated as HNE modified extract but without HNE; HNE, transcription performed by extracts modified with different HNE concentrations (0.05–5 mM for 2 h, on ice).
These results suggest that HNE may inhibit transcription in human cells by inhibition of RNA polymerization when HNE-DNA adducts are present in the template, but also by direct inhibition of transcription enzymes. Enzymes inhibition occurs, however, at very high HNE concentrations.
3.4. CSB protein dephosphorylation in cells treated with HNE
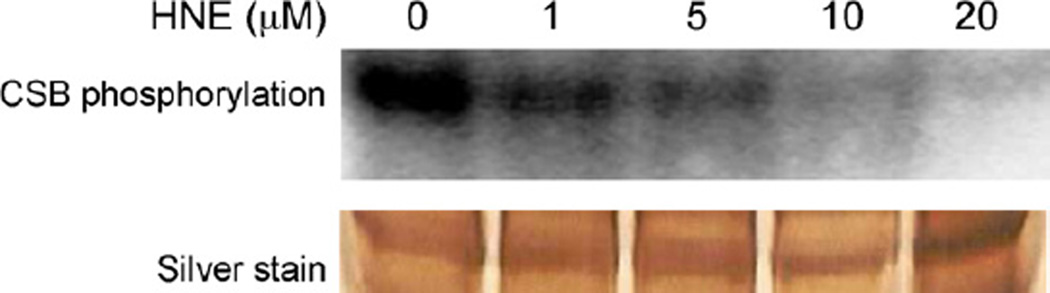
After UV irradiation of mammalian cells, the CSB protein undergoes dephosphorylation. This stimulates its ATPase activity and possibly favors switching to the TCR pathway [13].We observed that treatment of cells with HNE triggered the CSB dephosphorylation in a dose-dependent manner. A significant decrease in CSB phosphorylation was observed already at the lowest HNE concentration (1 µM), and at 20 µM HNE almost all protein was dephosphorylated (Fig. 6).
Fig. 6.
Dephosphorylation of CSB protein upon treatment of cells with HNE, autoradiography (upper panel) and silver staining of the CSB protein. GM00038 (wt) cells were exposed to the indicated doses of HNE for 30 min and incubated for 4 h with 100 µCi of 32P phosphate. The cells were lysed and immunoprecipitated with antibody against CSB. Immunoprecipitated proteins were analyzed by PAGE. The pictures are representatives of three independent experiments.
The observed effect was probably not due to the decreased phosphorylation of the CSB protein, since the activity of the main CSB kinase, casein kinase 2 was not affected in vitro by high 100–200 µM HNE concentrations (Table 1).
Table 1.
Effect of CK2 kinase pre-incubation with HNE on its enzymatic activity.
| HNE concentration (µM) | % Activity (AV ± SD) |
|---|---|
| 0 | 100 ± 0 |
| 10 | 96 ± 20 |
| 100 | 95 ± 9 |
| 200 | 100 ± 3 |
3.5. Inhibition of CSB ATPase activity by HNE
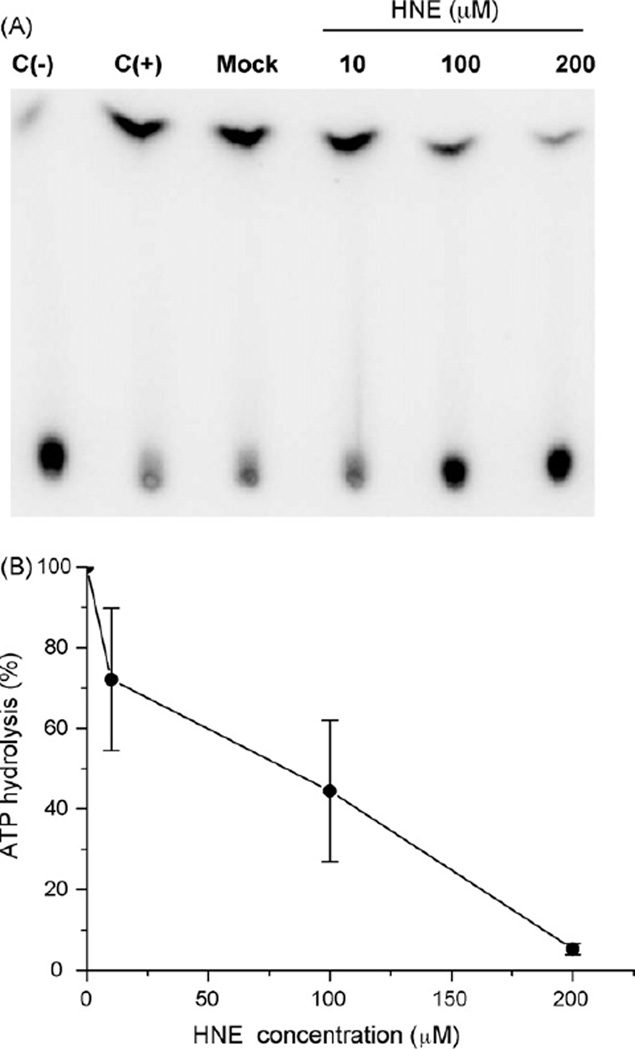
It was previously shown, that HNE may directly affect the activity of some DNA repair proteins. Treatment of cells with HNE inhibited excision of benzo[a]pyrene adducts and UV dimers from DNA, probably by HNE binding to one or several NER proteins [48] and inhibition of the NER machinery. Here, we investigated whether HNE is able to inactivate the ATPase activity of CSB protein in vitro. We observed significant inhibition of ATP hydrolysis using CSB protein pre-treated with HNE, however, only when higher HNE doses were used (100–200 µM) (Fig. 7A). At 10 µM HNE, a slight reduction of CSB ATPase activity was observed (72% of the ATP underwent hydrolysis). At 100 µM HNE, 44% ATP was hydrolyzed to inorganic phosphate, and at 200 µM HNE, 4% (Fig. 7B).
Fig. 7.
Inhibition of CSB ATPase activity after HNE treatment. (A) Thin layer chromatography with CSB ATPase reactions. Lanes: C(−), negative control, ATPase reaction performed in the absence of recombinant CSB wt protein; C(+), positive control, ATPase reaction performed in the presence of recombinant CSB wt protein; Mock, control reaction performed with unmodified recombinant CSB wt protein that has been treated as HNE modified protein, but without HNE; [10, 100, 200], ATPase reaction performed with recombinant CSB wt protein modified by different HNE concentrations: 10, 100 or 200 µM. (B) Percent of ATP hydrolysis incision from the data shown in (A). Data points are the means of at least three independent experiments with SDs indicated by error bars.
3.6. Cell lines bearing mutations in different motifs of the CSB ATPase domain respond differentially to HNE treatment
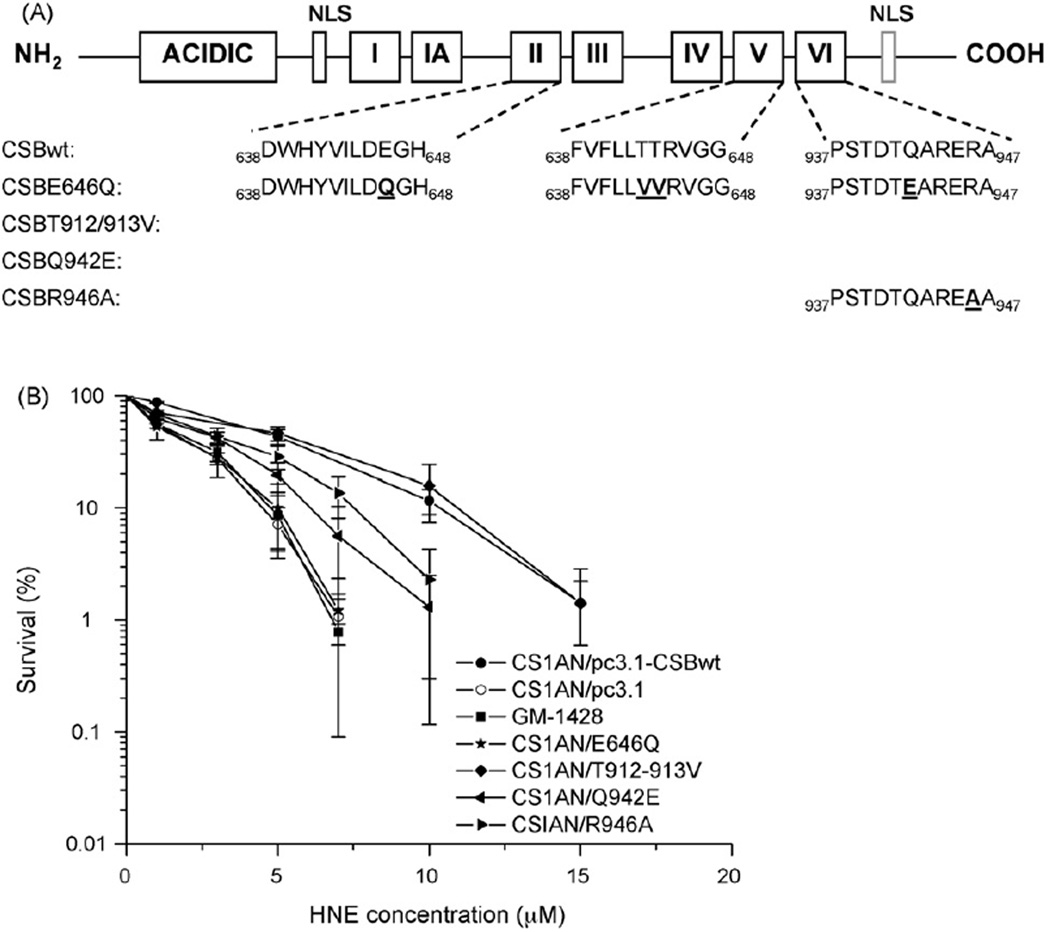
To analyze the role of the ATPase activity of CSB on the repair of HNE-induced DNA lesions, we used cell lines expressing CSB proteins mutated in different conserved motifs of the ATPase domain (Fig. 8A). CSB protein mutated in the ATPase motif II, V or VI is affected in the ATPase activity, and cells bearing these mutations are sensitive to UV and oxidative stress [7,13]. The motif II mutant (E646Q) appeared to be defective in TCR and GGR CSB function [49]. Motif V (T912–913V) and motif VI (Q942E and R946A) mutants are probably defective in base excision repair (BER) [49,50]. In our study, the motif II mutant was as sensitive to HNE as CSB null cells transformed with the empty vector. Both cell lines expressing CSB protein mutated in motif VI of the ATPase showed intermediate sensitivity to HNE, while the sensitivity of motif V mutant to HNE resembled that of the wild-type line (Fig. 8B). Thus, the CSB ATPase domain involved in TCR seems to be important for processing of HNE-DNA adducts, while domains which are involved in BER are less or not engaged. It should be mentioned that although unsubstituted ethenoadducts are repaired by BER [51], our previous studies failed to detect these lesions in nucleosides reacted with HNE in non-oxidizing and oxidizing conditions [27].
Fig. 8.
(A) Schematic representation of the CSB protein and CSB amino acid sequence of conserved regions in ATPase motifs II, V, and VI. Amino acids mutated in CSB in the different cell lines used are indicated in bold underlined letters. (B) Comparison of the sensitivity of cell lines bearing different mutations in CSB protein towards HNE. The data are the means of three independent experiments. Error bars represent SD of the mean.
3.7. Homology modeling of the CSB protein
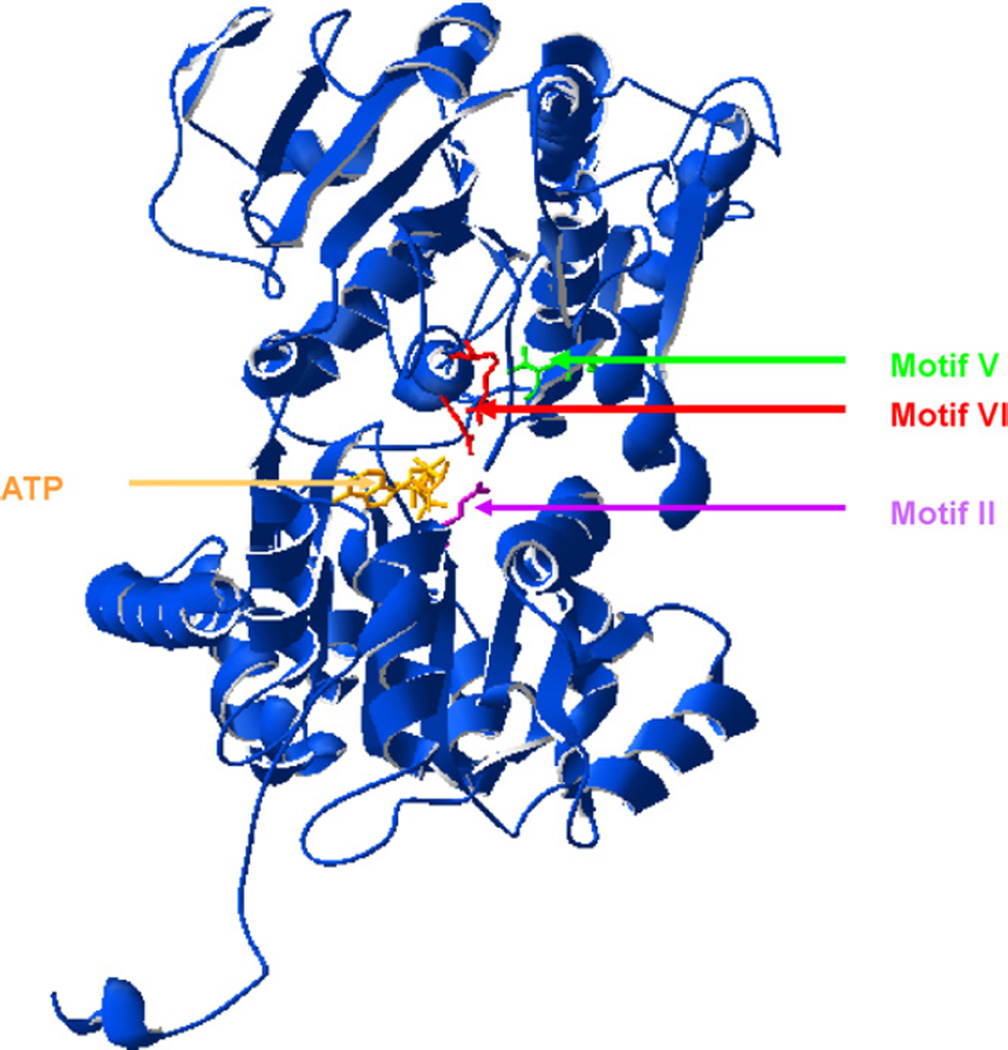
So far, no proteins of the SNF2 family, which CSB belongs to, were crystallized. Based on the model of zebra fish Rad 54 protein [46] we constructed a model of the CSB protein ATPase domain (Fig. 9). The mutations in the CSB ATPase domain carried by the investigated cell lines were mapped into the tri-dimensional structure of CSB protein. Structural analysis suggests that the amino acids changed in ATPase motifs II and VI are located closer (2.79–10.5 Å) to the ATP binding site than the amino acids mutated in motif V (10.98–16.11 Å) (Fig. 9).
Fig. 9.
Homology model of the ATPase motif of the CSB protein with ATP molecule in ATP binding site. The amino acid at the site of 646 E→Q mutation (domain II) is marked in violet, at 912 and 913 TT→VV site (domain V) in green, at 942 Q→E and 946 R→A (domain VI) in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
4. Discussion
Cockayne syndrome (CS) is an inherited neurodevelopmental disorder with progeroid features. The genes responsible for CS have been implicated in a variety of DNA repair- and transcription-related pathways [52]. Among them, it was found that the level of 8-oxoG in DNA is increased in CSB-deficient cells and that its repair is delayed [53]. However, the nature of the molecular defect in CS remains elusive. Particularly, modulation of chromatin metabolism and apoptosis by cellular metabolism still deserve attention. Massive protein carbonylation caused by oxidative stress and LPO in tissues of aging people and patients with neurodegenerative disorders [54], prompted us to investigate the effect of one of the major LPO products, HNE, on CSB protein activities in vitro and in human cells.
Here, we show that CSB−/− cells are hypersensitive to low physiological concentrations of HNE, and in response to HNE, they develop higher number of SCEs than their wild-type counterparts (Figs. 1 and 2). This suggests that HNE-DNA adducts may block replication and trigger recombination at stalled replication forks, and that lack of functional CSB protein increases recombination after HNE treatment. CSB protein plays a major role in initiation of the repair of the transcribed DNA strand, by recruiting histone acetyltransferase p300 and NER proteins to RNA polymerase II stalled at a DNA lesion. This initiates repair of the transcribed DNA strand and decreases the number of potential sites of recombination caused by DNA damage [55].
Our in vitro data demonstrate, that HNE-DNA adducts, when present in the template, inhibit RNA synthesis by T7 RNA polymerase (Fig. 3), already at 0.1 mM HNE, for the longest transcription product. A brief estimation of adduct number can be proposed, based on literature data. On average, 1.2 HNE-dG adducts per 1000 bp were quantified in plasmid DNA modified in vitro with 192 mM HNE for 20 h [56]. Thus, in our study at 20 mM HNE, where no transcription product was observed, approximately 0.12 HNE-dG adduct/1000 bp, which equals 0.69 per 5.8 kb plasmid, is expected. This rough estimation suggests that HNE-DNA adducts may constitute a very strong hindrance to RNA synthesis by T7 RNA polymerase.
We also observed inhibition of transcription when the reporter luciferase gene was transcribed by HeLa cell-free extracts from HNE-modified template (Fig. 4) as well as when HNE pre-treated extract proteins transcribed unmodified plasmid DNA (Fig. 5). When extract was incubated with HNE, 50 µM HNE was sufficient to retard transcription by ~20%, whereas, when DNA was modified, only at 1 mM HNE a comparable inhibition of transcription was observed. HNE concentration in humans may reach quite high values. For example, in human monocytes the basic HNE level equals 1 µM, but when monocytes are fed with malaria pigment, hemozoin, HNE may increase up to 46 µM, and in kidney tubular cells may be even higher [24]. Thus, in stress conditions HNE effect on transcription machinery may be non-negligible. HNE reacts poorly with DNA and very well with proteins, which could explain differences between HNE concentration necessary to inhibit transcription when either DNA or cellular extract was reacted with HNE. However, in cells, HNE oxidation will result in formation of epoxide, which readily reacts with DNA [26,34]. Thus, our results suggest that transcription inhibition by HNE in human cells may have dual impacts, by creating a steric hindrance for RNA polymerase by bulky HNE-DNA adducts, and by transcription machinery inactivation due to modification by HNE. The latter mechanism may be of limited importance, since occurs only at very high HNE concentrations.
We also observed dose-dependent dephosphorylation of CSB protein in cells incubated with HNE (Fig. 6), similar to the effect of UV. UV light stimulates dephosphorylation of the CSB protein by protein phosphatase I, which increases the CSB ATPase activity, and in consequence activates TCR [13]. In our study, decreased CSB phosphorylation was not likely to be caused by inhibition of CSB phosphorylation, since the activity of the main CSB kinase, CK2 was not affected in vitro by high HNE concentrations up to 200 µM (Table 1). The molecular mechanism of CSB dephosphorylation is not explained. Since HNE may bind both to DNA and to proteins, it is not clear whether the signal comes from damaged DNA and the change of chromatin structure, or from direct interaction of HNE with specific proteins. If the signal comes from damaged DNA, HNE is quite efficient in inducing CSB dephosphorylation. At 10 µM HNE, the CSB dephosphorylation was almost complete (Fig. 6). Based on literature data [56], 0.06 HNE-dG adducts/Mb are expected in this DNA (and 0.06 HNE-dC adducts/Mb, since cytosine is as efficiently modified by HNE as guanine, while adenine and thymine much less efficiently [26]). 6 J/m2 of UVC, inducing approximately 45 CPD/Mb [57], triggered CSB dephosphorylation comparable to 10 µM HNE in wild-type cells [13]. One should, however, bear in mind that HNE easily undergoes further oxidation by cellular metabolism, which significantly increases its affinity to DNA, so our calculations may be underestimated. The endogenous level of HNE-dG adducts in humans is quite high, e.g. in human colon epithelium was estimated for 0.27–0.35 HNE-dG/Mb [58]. These brief estimations suggest that HNE may be at least as potent as UV to trigger CSB dephosphorylation.
Previous studies showed that NER activity is inhibited by HNE [48]. Affected proteins were, however, not identified. One cannot exclude that HNE treatment of cells deficient in TCR, could inhibit proteins participating in GGR, increasing the sensitivity of the cells to HNE. In experiments conducted by Feng et al. [48] only high HNE concentrations (50–100 µM) exerted this inhibitory effect. We observed that the ATPase activity of the CSB protein is inactivated in vitro by HNE (Fig. 7). Significant inhibition occurred only at very high HNE concentrations (100 and 200 µM) that are rare in living organisms [23].However, CSB protein was incubated only for 15 min with HNE. Since LPO products are quite stable in cells, inhibition of CSB protein and TCR in aged individuals, in which LPO is enhanced, might reach a significant extent.
The strong toxic and recombinogenic properties of HNE in CSB cells may also be caused by its ability to trigger formation of inter-strand DNA–DNA cross-links [59]. Richards et al. [60] showed that CSB participates in cross-link repair in Chinese hamster ovary cells. DNA inter-strand cross-links can be repaired by both NER pathways, GGR and TCR, as well as by homologous recombination. It is likely that CSB is involved in the repair of cross-links at multiple cell cycle phases, including G1 for incision and S phase for recombination. It is also possible that in response to DNA damage, CSB links both TCR and recombination.
Recently, it was found that the CSB protein has strand annealing and exchange activities, which might be engaged in mitotic recombination and/or restoration of stalled replication fork due to encountered damage [4]. This activity is stimulated by dephosphorylation of CSB, however, ATP binding inhibits its ssDNA annealing activity [4], and ATP hydrolysis is not required for strand annealing and exchange.
In order to get a better insight into the mechanism of HNE-DNA adduct processing by the CSB protein, we have investigated the sensitivity to HNE of CSB-null cell lines bearing plasmids with the CSB gene mutated in different ATPase motifs. These cell lines were previously shown to efficiently express all mutated proteins [13]. The ATPase motif II-mutant of CSB can bind ATP and DNA [13], but is completely deprived of the ATPase activity. The motif II-mutant is very sensitive to UV and 4-NQO and is unable to recover RNA synthesis after UV and 4-NQO, thus appears defective in TCR-NER. The motif II-mutated cell line was as sensitive to HNE as CSB-null cell line (Fig. 8). The homology model of the CSB protein showed that the mutated amino acid in CSB motif II is located in close vicinity to the ATP binding site and may be engaged in ATP binding and/or hydrolysis, necessary for TCR, but not for BER (Fig. 9). Studies of the mitochondrial repair of oxidative damage in CSB motif II-mutant human cells by Stevnsner et al. [61], failed to find an influence of the CSB ATPase activity on 8-oxoG repair in vitro or in vivo. Mutants in ATPase motifs V and VI of CSB protein exhibit an almost complete inhibition of the ATPase activity, reduced ATP binding in vitro, UV-induced apoptosis, and do not recover RNA synthesis after UV [62]. Both these mutants are defective in BER. We observed that motif V-mutant was similarly sensitive to HNE as the wild-type cell line, while motif VI-mutant showed an intermediate sensitivity between that of the CSB null cell line and the wild-type cells (Fig. 8). Homology modeling suggests that the amino acids changed in ATPase motifs II and VI are located closer to the ATP binding site than the amino acids mutated in motif V (Fig. 9). Mutations in motifs II and VI reverse or neutralize the charge at the ATP binding site (Glu→Gln; Glu→Gln or Arg→Ala). Mutations in motif V create a more hydrophobic environment of the ATP binding site (Thr→Val). Whether and how such change in hydrophobicity may change CSB ATPase activity in contact with DNA containing hydrophobic HNE adducts, remains to be established. However, no difference in sensitivity to HNE was observed between the wild-type cells and CSB motif V mutants. An alternative explanation may be that mutations in motif V, in contrast to those in motifs II and VI, do not interfere with an alternative repair mechanism, e.g. recombination, which may supplement the deficient TCR and/or GGR. This, however, needs further studies.
The obtained data suggest that HNE-induced DNA adducts that are endogenous lesions trigger replication and transcription arrest. These lesions may be processed by homologous recombination and possibly by TCR, since they require an active CSB protein. In normal cells, low HNE concentrations activate TCR via CSB protein dephosphorylation. In contrast, at very high concentrations HNE may inhibit TCR, probably by a direct adduction to CSB protein leading to inactivation of the enzyme, that may accelerate cell death. Hence, HNE-induced DNA damage may play a role in neurological and developmental abnormalities and premature aging observed in CS patients.
Acknowledgments
This work was supported by the grants of the Polish Ministry of Science and Higher Education N N303 328834 (awarded for years 2001–2008) and N N303 391436 (awarded for years 2009–2012), Danish Cancer Society (grant no: DP03131) and the Lundbeck Foundation (grant no: 329/06). M.C. was supported by The Carlsberg Foundation (04/0169/20).
Abbreviations
- HNE
trans-4-hydroxy-2-nonenal
- LPO
lipid peroxidation
- MMS
methyl methanesulfonate
- CSB
Cockayne syndrome group B
- NER
nucleotide excision repair
- GGR
global genome repair
- TCR
transcription-coupled repair
- HR
homologous recombination
Footnotes
Conflict of interest
None.
References
- 1.Foksinski M, Rozalski R, Guz J, Ruszkowska B, Sztukowska P, Piwowarski M, Klungland A, Olinski R. Urinary excretion of DNA repair products correlates with metabolic rates as well as with maximum life spans of different mammalian species. Free Radical Biology & Medicine. 2004;37:1449–1454. doi: 10.1016/j.freeradbiomed.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Stevnsner T, Muftuoglu M, Aamann MD, Bohr VA. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mechanisms of Ageing and Development. 2008;129:441–448. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorslund T, von Kobbe C, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Molecular & Cellular Biology. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muftuoglu M, Sharma S, Thorslund T, Stevnsner T, Soerensen MM, Brosh RM, Jr, Bohr VA. Cockayne syndrome group B protein has novel strand annealing and exchange activities. Nucleic Acids Research. 2006;34:295–304. doi: 10.1093/nar/gkj410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 6.Brosh RM, Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De Santis L, Bohr VA. The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Molecular Biology of the Cell. 1999;10:3583–3594. doi: 10.1091/mbc.10.11.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muftuoglu M, Selzer R, Tuo J, Brosh RM, Jr, Bohr VA. Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene. 2002;283:27–40. doi: 10.1016/s0378-1119(01)00870-8. [DOI] [PubMed] [Google Scholar]
- 8.Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. Journal of Biological Chemistry. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 9.Tantin D, Kansal A, Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Molecular & Cellular Biology. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. Journal of Biological Chemistry. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 11.Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Molecular & Cellular Biology. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beerens N, Hoeijmakers JH, Kanaar R, Vermeulen W, Wyman C. The CSB protein actively wraps DNA. Journal of Biological Chemistry. 2005;280:4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- 13.Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM, Jr, Bohr VA. Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Research. 2003;31:963–973. doi: 10.1093/nar/gkg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imam SZ, Indig FE, Cheng WH, Saxena SP, Stevnsner T, Kufe D, Bohr VA. Cockayne syndrome protein B interacts with and is phosphorylated by c-Abl tyrosine kinase. Nucleic Acids Research. 2007;35:4941–4951. doi: 10.1093/nar/gkm386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleaver JE, Hefner E, Laposa RR, Karentz D, Marti T. Cockayne syndrome exhibits dysregulation of p21 and other gene products that may be independent of transcription-coupled repair. Neuroscience. 2007;145:1300–1308. doi: 10.1016/j.neuroscience.2006.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tornaletti S. Transcription arrest at DNA damage sites. Mutation Research. 2005;577:131–145. doi: 10.1016/j.mrfmmm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Halliwell B. Free radicals, reactive oxygen species and human disease: a critical evaluation with special reference to atherosclerosis. British Journal of Experimental Pathology. 1989;70:737–757. [PMC free article] [PubMed] [Google Scholar]
- 18.Pansarasa O, Bertorelli L, Vecchiet J, Felzani G, Marzatico F. Age-dependent changes of antioxidant activities and markers of free radical damage in human skeletal muscle. Free Radical Biology & Medicine. 1999;27:617–622. doi: 10.1016/s0891-5849(99)00108-2. [DOI] [PubMed] [Google Scholar]
- 19.Castillo C, Salazar V, Ariznavarreta C, Vara E, Tresguerres JA. Effect of isoflavone administration on age-related hepatocyte changes in old ovariectomized femal Wistar rats. Phytomedicine. 2006;13:468–476. doi: 10.1016/j.phymed.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Bartsch H, Nair J. Oxidative stress and lipid peroxidation-derived DNA-lesions in inflammation driven carcinogenesis. Cancer Detection & Prevention. 2004;28:385–391. doi: 10.1016/j.cdp.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Zarkovic K. 4-hydroxynonenal and neurodegenerative diseases. Molecular Aspects of Medicine. 2003;24:293–303. doi: 10.1016/s0098-2997(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi M, Itoh M, Araki S, Kumada S, Shioda K, Tamagawa K, Mizutani T, Morimatsu Y, Minagawa M, Oda M. Oxidative stress and disturbed glutamate transport in hereditary nucleotide repair disorders. Journal of Neuropathology & Experimental Neurology. 2001;60:350–356. doi: 10.1093/jnen/60.4.350. [DOI] [PubMed] [Google Scholar]
- 23.Dianzani MU, Barrera G, Parola M. 4-Hydroxy-2,3-nonenal as a signal for cell function and differentiation. Acta Biochimica Polonica. 1999;46:61–75. [PubMed] [Google Scholar]
- 24.Poli G, Schaur RJ, Siems WG, Leonarduzzi G. 4-hydroxynonenal: a membrane lipid oxidation product of medicinal interest. Medicinal Research Reviews. 2008;28:569–631. doi: 10.1002/med.20117. [DOI] [PubMed] [Google Scholar]
- 25.Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Molecular Aspects of Medicine. 2003;24:149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 26.Chung FL, Zhang L, Ocando JE, Nath RG. Role of 1, N2-propanodeoxyguanosine adducts as endogenous DNA lesions in rodents and humans. IARC Scientific Publications. 1999:45–54. [PubMed] [Google Scholar]
- 27.Kowalczyk P, Ciesla JM, Komisarski M, Kusmierek JT, Tudek B. Long-chain adducts oftrans-4-hydroxy-2-nonenal to DNA bases cause recombination, base substitutions and frameshift mutations in M13 phage. Mutation Research. 2004;550:33–48. doi: 10.1016/j.mrfmmm.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA interchain cross-links formed by acrolein and crotonaldehyde. Journal of the American Chemical Society. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 29.Kurtz AJ, Lloyd RS. 1,N2-deoxyguanosine adducts of acrolein, crotonaldehyde, andtrans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. Journal of Biological Chemistry. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 30.Uchida K, Kanematsu M, Sakai K, Matsuda T, Hattori N, Mizuno Y, Suzuki D, Miyata T, Noguchi N, Niki E, Osawa T. Protein-bound acrolein: potential markers for oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:4882–4887. doi: 10.1073/pnas.95.9.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wacker M, Schuler D, Wanek P, Eder E. Development of a (32)P-postlabeling method for the detection of 1,N(2)-propanodeoxyguanosine adducts oftrans-4-hydroxy-2-nonenalin vivo. Chemical Research in Toxicology. 2000;13:1165–1173. doi: 10.1021/tx000058r. [DOI] [PubMed] [Google Scholar]
- 32.Chung FL, Nath RG, Ocando J, Nishikawa A, Zhang L. Deoxyguanosine adducts of t-4-hydroxy-2-nonenal are endogenous DNA lesions in rodents and humans: detection and potential sources. Cancer Research. 2000;60:1507–1511. [PubMed] [Google Scholar]
- 33.Esterbauer H, Eckl P, Ortner A. Possible mutagens derived from lipids and lipid precursors. Mutation Research. 1990;238:223–233. doi: 10.1016/0165-1110(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 34.Eckl PM, Ortner A, Esterbauer H. Genotoxic properties of 4-hydroxyalkenals and analogous aldehydes. Mutation Research. 1993;290:183–192. doi: 10.1016/0027-5107(93)90158-c. [DOI] [PubMed] [Google Scholar]
- 35.Karlhuber GM, Bauer HC, Eckl PM. Cytotoxic and genotoxic effects of 4-hydroxynonenal in cerebral endothelial cells. Mutation Research. 1997;381:209–216. doi: 10.1016/s0027-5107(97)00170-x. [DOI] [PubMed] [Google Scholar]
- 36.Cajelli E, Ferraris A, Brambilla G. Mutagenicity of 4-hydroxynonenal in V79 Chinese hamster cells. Mutation Research. 1987;190:169–171. doi: 10.1016/0165-7992(87)90050-9. [DOI] [PubMed] [Google Scholar]
- 37.Emerit I, Khan SH, Esterbauer H. Hydroxynonenal, a component of clastogenic factors? Free Radical Biology & Medicine. 1991;10:371–377. doi: 10.1016/0891-5849(91)90045-5. [DOI] [PubMed] [Google Scholar]
- 38.Hu W, Feng Z, Eveleigh J, Iyer G, Pan J, Amin S, Chung FL, Tang MS. The major lipid peroxidation product,trans-4-hydroxy-2-nonenal, preferentially forms DNA adducts at codon 249 of human p53 gene, a unique mutational hotspot in hepatocellular carcinoma. Carcinogenesis. 2002;23:1781–1789. doi: 10.1093/carcin/23.11.1781. [DOI] [PubMed] [Google Scholar]
- 39.Chung FL, Pan J, Choudhury S, Roy R, Hu W, Tang MS. Formation oftrans-4-hydroxy-2-nonenal- and other enal-derived cyclic DNA adducts fromomega-3 and omega-6 polyunsaturated fatty acids and their roles in DNA repair and human p53 gene mutation. Mutation Research. 2003;531:25–36. doi: 10.1016/j.mrfmmm.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Mayne LV, Priestley A, James MR, Burke JF. Efficient immortalization and morphological transformation of human fibroblasts by transfection with SV40 DNA linked to a dominant marker. Experimental Cell Research. 1986;162:530–538. doi: 10.1016/0014-4827(86)90356-3. [DOI] [PubMed] [Google Scholar]
- 41.Chandra A, Srivastava SK. A synthesis of 4-hydroxy-2-trans-nonenal and 4-(3H) 4-hydroxy-2-trans-nonenal. Lipids. 1997;32:779–782. doi: 10.1007/s11745-997-0100-6. [DOI] [PubMed] [Google Scholar]
- 42.Perry P, Wolff S. New Giemsa method for the differential staining of sister chromatids. Nature. 1974;251:156–158. doi: 10.1038/251156a0. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka M, Lai JS, Herr W. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell. 1992;68:755–767. doi: 10.1016/0092-8674(92)90150-b. [DOI] [PubMed] [Google Scholar]
- 44.Vodenicharov MD, Sallmann FR, Satoh MS, Poirier GG. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Research. 2000;28:3887–3896. doi: 10.1093/nar/28.20.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grankowski N, Boldyreff B, Issinger OG. Isolation and characterization of recombinant human casein kinase II subunits alpha and beta from bacteria. European Journal of Biochemistry. 1991;198:25–30. doi: 10.1111/j.1432-1033.1991.tb15982.x. [DOI] [PubMed] [Google Scholar]
- 46.Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nature Structural & Molecular Biology. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 47.Pieper U, Eswar N, Braberg H, Madhusudhan MS, Davis FP, Stuart AC, Mirkovic N, Rossi A, Marti-Renom MA, Fiser A, Webb B, Greenblatt D, Huang CC, Ferrin TE, Sali A. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Research. 2004;32:D217–D222. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng Z, Hu W, Tang MS. Trans-4-hydroxy-2-nonenal inhibits nucleotide excision repair in human cells: a possible mechanism for lipid peroxidation-induced carcinogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8598–8602. doi: 10.1073/pnas.0402794101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Licht CL, Stevnsner T, Bohr VA. Cockayne syndrome group B cellular and biochemical functions. American Journal of Human Genetics. 2003;73:1217–1239. doi: 10.1086/380399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuo J, Muftuoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM, Jr, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. Journal of Biological Chemistry. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 51.Gros L, Ishchenko AA, Saparbaev M. Enzymology of repair of etheno-adducts. Mutation Research. 2003;531:219–229. doi: 10.1016/j.mrfmmm.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 52.Kleppa L, Kanavin OJ, Klungland A, Stromme P. A novel splice site mutation in the Cockayne syndrome group A gene in two siblings with Cockayne syndrome. Neuroscience. 2007;145:1397–1406. doi: 10.1016/j.neuroscience.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 53.D’Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM, Orioli D, Frosina G, Zambruno G, Dizdaroglu M, Stefanini M, Dogliotti E. The role of CSA in the response to oxidative DNA damage in human cells. Oncogene. 2007;26:4336–4343. doi: 10.1038/sj.onc.1210232. [DOI] [PubMed] [Google Scholar]
- 54.Kim CH, Zou Y, Kim DH, Kim ND, Yu BP, Chung HY. Proteomic analysis of nitrated and 4-hydroxy-2-nonenal-modified serum proteins during aging. Journals of Gerontology Series A–Biological Sciences & Medical Sciences. 2006;61:332–338. doi: 10.1093/gerona/61.4.332. [DOI] [PubMed] [Google Scholar]
- 55.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase IIin vivo. Molecular Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 56.Choudhury S, Pan J, Amin S, Chung FL, Roy R. Repair kinetics oftrans-4-hydroxynonenal-induced cyclic 1,N2-propanodeoxyguanine DNA adducts by human cell nuclear extracts. Biochemistry. 2004;43:7514–7521. doi: 10.1021/bi049877r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wacker M, Wanek P, Eder E, Hylla S, Gostner A, Scheppach W. Effect of enzyme-resistant starch on formation of 1,N(2)-propanodeoxyguanosine adducts oftrans-4-hydroxy-2-nonenal and cell proliferation in the colonic mucosa of healthy volunteers. Cancer Epidemiology, Biomarkers & Prevention. 2002;11:915–920. [PubMed] [Google Scholar]
- 59.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radical Biology & Medicine. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 60.Richards S, Liu ST, Majumdar A, Liu JL, Nairn RS, Bernier M, Maher V, Seidman MM. Triplex targeted genomic crosslinks enter separable deletion and base substitution pathways. Nucleic Acids Research. 2005;33:5382–5393. doi: 10.1093/nar/gki851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stevnsner T, Nyaga S, de Souza-Pinto NC, van der Horst GT, Gorgels TG, Hogue BA, Thorslund T, Bohr VA. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene. 2002;21:8675–8682. doi: 10.1038/sj.onc.1205994. [DOI] [PubMed] [Google Scholar]
- 62.Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM, Jr, Bohr VA. Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Research. 2002;30:782–793. doi: 10.1093/nar/30.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]