Abstract
Genetic variation in DNA repair genes can modulate DNA repair capacity and may be related to cancer risk. However, study findings have been inconsistent. Inheritance of variant DNA repair genes is believed to influence individual susceptibility to the development of environmental cancer. Reliable knowledge on which the base excision repair (BER) sequence variants are associated with cancer risk would help elucidate the mechanism of cancer. Given that most of the previous studies had inadequate statistical power, we have conducted a systematic review on sequence variants in three important BER proteins. Here, we review published studies on the association between polymorphism in candidate BER genes and cancer risk. We focused on three key BER genes: 8-oxoguanine DNA glycosylase (OGG1), apurinic/apyrimidinic endonuclease (APE1/APEX1) and x-ray repair cross-complementing group 1 (XRCC1). These specific DNA repair genes were selected because of their critical role in maintaining genome integrity and, based on previous studies, suggesting that single-nucleotide polymorphisms (SNPs) in these genes have protective or deleterious effects on cancer risk. A total of 136 articles in the December 13, 2010 MEDLINE database (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/pubmed/) reporting polymorphism in OGG1, XRCC1 or APE1 genes were analyzed. Many of the reported SNPs had diverse association with specific human cancers. For example, there was a positive association between the OGG1 Ser326Cys variant and gastric and lung cancer, while the XRCC1 Arg399Gln variant was associated with reduced cancer risk. Gene–environment interactions have been noted and may be important for colorectal and lung cancer risk and possibly other human cancers.
Keywords: OGG1, XRCC1, APE1, cancer, gene polymorphism, DNA repair, DNA damage
Introduction
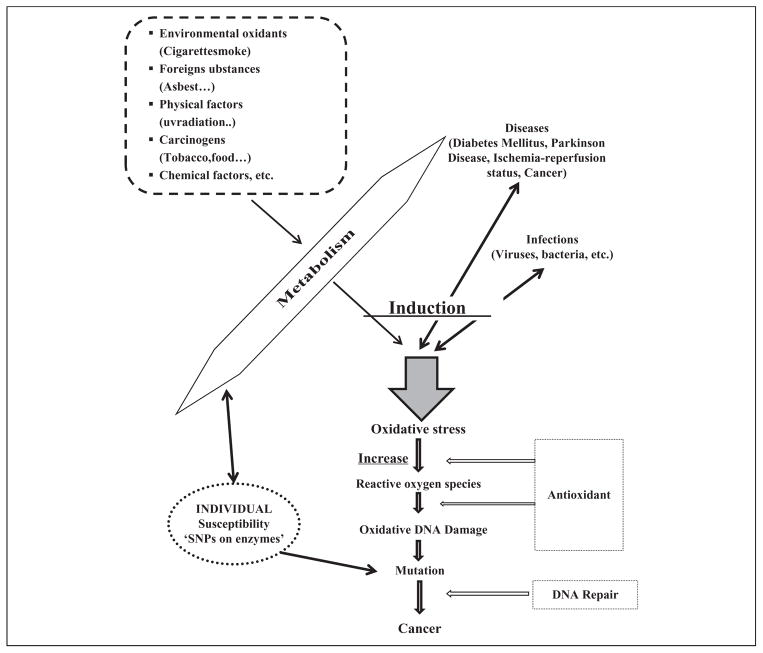
There are two main types of DNA damages, endogenous and exogenous. Endogenous DNA damage is primarily caused by reactive oxygen species (ROS), which are the by-products of normal cellular metabolism. Exogenous DNA damage is caused by the environmental agents such as ultraviolet light, ionizing radiation and chemicals.1 Some oxidative DNA lesions form at a high rate. For example, it is estimated that 100–500 8-hydroxyguanine (8-oxoG) lesions are formed per day in a human cell,2 and formamidopyrimidine lesions (2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine) form at a similar or higher rate in cells exposed to oxidative stress.3 Oxidative damage to DNA, which also includes sugar modifications and strand breaks, can lead to transcription or replication defects, mutations, genomic instability and ultimately cellular dysfunction. In turn, cell stress pathways can induce apoptosis or support unregulated cell growth4,5 (Figure 1) through the activation of oncogenes or inactivation of tumor suppressor genes.6
Figure 1.
The relationship between oxidative DNA damage, carcinogenesis and environmental factors and their link with individual susceptibility.
Environmental chemicals and cancer; tobacco smoke as a model compound
Most cancers are the result of the interactions of multiple genes and environmental factors. Tobacco smoke is a good example of an environmental risk factor for lung and other cancers.7–9 Tobacco use is by far the most recognized link between exposure to known carcinogens and death from cancer, and like the aflatoxin–hepatocellular carcinoma relationship, can be considered a model for understanding mechanisms of cancer induction by exogenous chemical carcinogens.10,11 Tobacco smoke consists of at least 3800 chemical constituents,12 including benzo[a]pyrene (BaP) and small chemical carcinogens, such as formaldehyde, ethylene oxide, benzene, 1,3-butadiene and many others. After biotransformation, the metabolites can induce oxidative stress or can covalently bind to DNA, leading to DNA modifications that can promote mutagenic bypass if unrepaired.13 Tobacco smoke-induced DNA damage is repaired predominantly by base excision repair (BER, see more in The BER Pathway section) and nucleotide excision repair (NER).
In addition to DNA repair, xenobiotic metabolizing enzymes, that is, cytochromes, glutathione-S-transferase (GST) and N-acetyl-transferase, can modify the risk of exposure. However, in the absence of exposure, the presence or absence of these genetic risk factors is irrelevant. For example, both GSTM1 and GSTT1 genes can be deleted and their corresponding conjugating activity is absent without increased cancer risk, whereas functional polymorphisms in these genes may play a significant role in tobacco-related cancer susceptibility.14 There are also associations between DNA repair gene polymorphism and lung cancer, presumably because the genetic defects decrease capacity to remove potentially mutagenic BaP DNA adducts or other DNA lesions introduced by tobacco smoke.15–21 Antioxidant defense systems and multiple DNA repair pathways have evolved to protect cells from oxidative stress and oxidative DNA damaging agents, acting as first and second lines of defense, respectively.22,23
The BER pathway
BER is critical for genome maintenance, and defects in BER are related to cancer risk and premature aging.24 BER is the main pathway for the repair of apurinic and apyrimidinic (AP) sites.25 DNA single-strand breaks as well as 8-oxoG, Fapy lesions or other base or sugar modifications arise via oxidation, alkylation or spontaneous decay.26 BER is classically initiated by a DNA glycosylase, which cleaves the N-glycosyl bond, releasing the substrate base and generating an abasic site. The abasic site is then cleaved by an AP lyase or AP endonuclease and, at instances when repair is successfully executed, the one base gap in the cleaved DNA strand is filled-in by a DNA polymerase and ligated by a DNA ligase.1,27
Genetic susceptibility to cancer and DNA repair
Single-nucleotide polymorphisms (SNPs) are the DNA base variants present in the human population at a frequency >1%.28,29 The nonsynonymous coding SNPs (nsSNPs) and SNPs in regulatory regions are believed to have the highest impact on phenotype. Both nsSNPs and regulatory SNPs in DNA repair genes can result in reduced DNA repair capacity, which can underlie a higher mutation rate and increased cancer risk.30 If a polymorphism is in a repair gene that plays a pivotal role in the removal of oxidative DNA damage, for instance, the resulting reduced repair activity will increase the possibility of disease, particularly in the face of oxidative stress. Studies on polymorphisms, therefore, provide an opportunity to discover candidate susceptibility alleles. However, the functional outcome and phenotypic effect of some polymorphisms are unknown. This review focuses on the phenotypic effects of variants in three genes, which have important roles in BER, in the development of carcinogenesis.
This review examines possible effects on cancer risk from polymorphisms in three BER proteins: 8-oxoguanine DNA glycosylase 1 (OGG1), AP endonuclease 1 (APE1), and x-ray repair cross complementing group 1 (XRCC1). The OGG1 gene is located at 3p25 and encodes the major repair enzyme for 8-oxoG. 1245C>G is a well-characterized OGG1 polymorphism that results in substitution of serine with cysteine at codon 326.31 Molecular epidemiologic case-control studies suggest that the hOGG1Cys326 allele is associated with a higher risk of developing several types of cancers.32–36 In addition to catalyzing excision of 8-oxoG from DNA, OGG1 possesses the ability to incise at abasic sites via an AP lyase activity. It has been demonstrated that the OGG1 Cys326 protein repairs 8-oxoG inefficiently, leading to the hypothesis that this variant may contribute to a higher cellular mutation rate.37
APE1 (also known as APE, APEX, HAP1 and REF-1) plays an essential role in BER. APE1 initiates repair of abasic sites in DNA by hydrolyzing the phosphodiester backbone 5′ to an AP site.38 Known sequence variants in APE1 include an amino acid change from aspartic acid to glutamic acid (Asp148Glu) in exon 5 that may be associated with hypersensitivity to ionizing radiation and cancer risk.38–40 However, the Asp148Glu substitution did not alter APE1 AP site incision activity in vitro.41 APE1 also plays a role in regulating the DNA binding capacity of several transcription factors and thus gene expression efficiency. As will be discussed further below, some studies show an influence of APE1 polymorphism on the prognosis of ovarian, gastrooesophageal and pancreatico-biliary cancer.42
XRCC1 is an important scaffold protein that interacts with the enzymatic factors such as polyadenosine diphosphate (ADP)-ribose polymerase, DNA ligase III and DNA polymerase β to facilitate efficient repair of DNA single-strand breaks.43 XRCC1 is located at 19q13.2 and consists of 17 exons encoding a 633 amino acid protein. A common genetic variant in XRCC1 (C>T) in exon 6 results in an Arg to Trp (rs25487) substitution at codon 194, while another polymorphism (G>A) in exon 10 changes amino acid Arg to Gln in codon 39944 and yet another polymorphism (G>A) in exon 9 changes amino acid Arg to His (rs25489) in codon 280.45
A variant XRCC1 gene may alter protein function and repair capacity, thus leading to genetic instability and carcinogenesis. However, the current data on the consequences of the 194 and 399 amino acid variants on XRCC1 function are complex.37 Nevertheless, since any of the nsSNPs described above may represent functional polymorphisms, the possible influence of the polymorphic variants in OGG1, XRCC1 and APE1 on cancer risk is reviewed in further detail herein (Figure 2).
Figure 2.
The roles of OGG1, XRCC1 and APE1 protein in base excision repair.
Materials and methods
As will be discussed in much detail later, SNPs in some BER genes appear to confer greater susceptibility to environmentally induced cancers. While recent studies have begun to evaluate the effect of SNPs on the DNA repair and metabolizing genes on the treatment outcome, such analyses have not been included in this review. In this review, we conducted a PubMed search of published articles using the keywords ‘OGG1, Ser326Cys or APE1, Asp148Glu or XRCC1, Arg399Gln, Arg194Trp, Arg280His and cancer’. A total of 136 articles were identified in the December 13, 2010 MEDLINE database (National Center for Biotechnology Information, PubMed database: http://www.ncbi.nlm.nih.gov/pubmed/). These specific gene polymorphisms were selected because of their high frequency and because of the large number of epidemiological studies examining their association with multiple cancers. Only quantitative studies were considered. Abstracts from scientific meetings were not reviewed. Studies with a sample size of <100 were excluded. There was no selection for specific cancer types. Odds ratios (ORs) for selected polymorphisms were tabulated and compared, primarily from case-control studies. Figure 3 presents inclusion/exclusion criteria. Forty-one articles did not meet the inclusion criteria (such as, studies on disease polymorphism, response to drug therapy polymorphism, occupational exposure to chemical polymorphisms, genotoxicity assays–polymorphism associations, etc.). Of the 136 articles, 76 met inclusion criteria. The articles included studies on 15 types of cancer (Figure 4) and their association with OGG1, APE1 or XRCC1 gene polymorphisms. Of all the studies, 55%were conducted in Asian populations and 45% were conducted in Caucasian populations. Most of the studies were hospital-based. Some cancer types were pooled and defined as one cancer type: oral/pharyngeal cancers and laryngeal cancers were pooled and defined as upper aerodigestive cancer (UADC); colon and colorectal cancer were pooled and defined as colon cancer; biliary tract and gallbladder cancer were pooled and defined as biliary tract cancer; mesothelioma, lung and non-small-cell lung carcinoma (NSCLC) were pooled and defined as lung cancer; precancerous gastric lesions, gastric cancer and gastric cardiac adenocarcinoma (ADC) were pooled and defined as gastric cancer; cervix, endometriosis and uterine leiomyoma were pooled and defined as gynecological cancer. In some articles, many different gene polymorphisms (more than 15 gene polymorphisms) and one cancer type association were studied. For these articles, only data relevant to OGG1, APE1 and XRCC1 SNPs are discussed in detail herein. Results of each study are given in Tables 1 to 8 and are discussed below.
Figure 3.
Flow chart of selection of studies.
Figure 4.
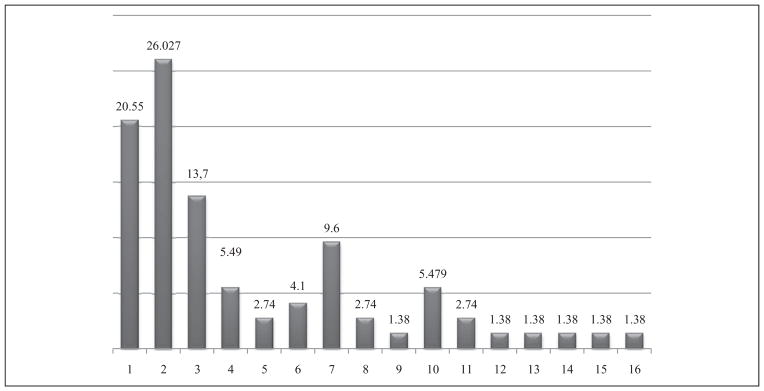
Distribution in cancer types (%) among individuals in the population discussed in this review. 1. Colon cancer; 2. Lung cancer; 3. Gastric cancer; 4. Bladder cancer; 5. Biliary tract cancer; 6. Upper aerodigestive cancer; 7. Breast cancer; 8. ALL; 9. Head and neck cancer; 10. Gynecological cancers; 11. Skin cancer; 12. Hepatocarcinoma; 13. Tyroid cancer; 14. Brain cancer; 15. Prostate cancer; 16. Non-Hodgkin lymphoma. Oral/pharyngeal cancers and laryngeal cancers were pooled and defined as upper aerodigestive cancer. Colon and colorectal cancer were pooled and defined as colon cancer. Biliary tract and gallbladder cancer were pooled and defined as biliary tract cancer. Mesothelioma, lung and non-small-cell lung carcinoma were pooled and defined as lung cancer. Precancerous gastric lesions, gastric cancer and gastric cardiac adenocarcinoma were pooled and defined as gastric cancer. Cervix, endometriosis and uterine leiomyoma were pooled and defined as gynecological cancer.
Table 1.
Summary of findings from case–control studies of genetic polymorphisms in DNA repair genes (OGG1, APE1 and XRCC1) and their interactions with environmental factors and colorectal cancers including colon, rectum and colorectal
| Study population | No. of cases/controls | Main effects of polymorphisms
|
Gene–environment interaction | References | ||
|---|---|---|---|---|---|---|
| OGG1 Ser326Cys | XRCC1 (Arg399Gln, Arg194Trp, Arg280His) | APE1Asp148Glu | ||||
| American (H) | 1582/1951 | 1.3-fold IR (Cys/Cys and Ser/Cys + Cys/Cys) | NA (Arg/Gln) 1.2-fold IR (Arg/Trp + Trp/Trp) -(Arg/His) | – | – | Curtin et al.46 |
| Korean (H) | 125/247 | NA | – | – | S; 2.5- to 3-fold IR (Cys/Cys) RM; 4.5-fold IR (2 times/week) |
Kim et al.47 |
| Norwegian (H) | 166/974 | RR (Cys/Cys) (adenoma, ADC) | – | – | – | Hansen et al.48 |
| Norwegian (H) | 1100/399 | – | NA (C, ADC) | RR (Arg/Gln) IR (Arg/His) |
– | Skjelbred et al.49 |
| Spanish (H) | 377/329 | 2-fold IR (Cys/Cys) | NA (Arg/Trp) | 3-fold IR (Asp/Glu) | – | Moreno et al.50 |
| Czech Republic (H) | 532/532 | 6- to 7-fold IR (OGG1 Cys/Cys + APE1 Glu + Glu) | NA (Arg/Gln, Arg/Trp) - (Arg/His) | IR (Colon, colorectal) (Asp/Glu) | S; 4- and 5-fold IR (Cys/Cys) | Pardini et al.51 |
| Japanese (H) | 68/121 | NA | NA | 2.3-fold IR (Asp/Glu + Glu/Glu) | S; 5-fold IR (Asp/Glu + Glu/Glu) | Kasahara et al.52 |
| Chinese (P) | 303/1159 294/1120 | NA | NA (Arg/Gln, Arg/Trp) -(Arg/His) | – | S;1.5- to 2-fold IR (Arg/Trp, Trp/Trp) | Stern et al.53 |
| Turkish (H) | 110/116 | NA | – | – | NA | Engin et al.54 |
| Indian (H) | 302/291 | – | 1.6-fold IR (Rectal Cancer in males, Gln399 allele) -(Arg/His, Arg/Trp) | – | S;1- to 1.5-fold IR (Arg/Arg) 2-fold IR rectal cancer for vegetarians |
Wang et al.55 |
–: Unreported; NA; no association; IR: increased risk; RR: reduced risk; S: smoking; RM: red meat; H: hospital based study; P: population based study.
Table 8.
Summary of findings from case–control studies of genetic polymorphisms in DNA repair genes (OGG1, APE1 and XRCC1) and their interactions with environmental factors and different cancer types (skin, ALL, prostate, NHL, brain, lung, UADC, ALL)
| cancer Type | Study population | No. of cases/controls | Main effects of polymorphisms
|
Gene–environment interaction | References | ||
|---|---|---|---|---|---|---|---|
| OGG1 Ser/Cys | XRCC1 (Arg/Gln, Arg/Trp, Arg/His) | APE1Asp/Glu | |||||
| Skin | Korean (H) | 212/207 | – | RR (Trp/Trp) SCC IR(Arg/Gln, Arg/Gln + Gln/Gln) BCC |
– | – | Kang et al.4 |
| ALL | Mexican (H) | 120/120 | – | NA | – | – | Meza-Espinoza et al.97 |
| Brain, lung, UADC, ALL | European (H) | 567/1094 | – | 2.5-fold IR (Gln/Gln) (ALL), NA; Others | – | – | Matullo et al.99 |
| Prostate | Korean (H) | 207/235 | – | 4-fold IR (Arg/Gln, Arg/Gln + Gln/Gln) | – | – | Xu et al.101 |
| NHL | Chinese (H) | 221/254 | – | 3-fold IR (Arg/Gln + Gln/Gln, Arg/Trp + Trp/Trp, Arg/His + His/His) | – | – | Liu et al.105 |
| HCC | Indian (H) | – | – | 2.27- and 4.95-fold IR (Arg/Trp and Arg/His) | – | – | Kiran et al.106 |
| Brain | Caucasian (H) | 1225/1560 | – | NA (Arg/Gln, Arg/Trp, Arg/His) | – | – | Kiuru et al.108 |
| DTC | Caucasian (H) | – | – | IR (Trp/Trp) DTC 4.5-fold IR LN |
NA | – | Chiang et al.109 |
ALL: acute lymphoblastic leukemia; NHL: non-Hodgkin lymphoma; HCC: hepatocellular carcinoma; GBC: gall bladder cancer; DTC: differentiated thyroid carcinoma; LN6: regional lymph node; SCC: squamous cell carcinoma; BCC: basal cell carcinoma; –: unreported; NA; no association; IR: increased risk; RR: reduced risk; S: smoking; RM: red meat; H: hospital based study; P: population based study.
Results
Colorectal cancers
Table 1 summarizes the associations of gene polymorphisms (OGG1 Ser326Cys, APE1 Asp148Glu, XRCC1 Arg399Gln, Arg280His and Arg194Trp) and colorectal cancer. Gu et al. conducted a meta-analysis of 27 published studies that included 12,432 cancer cases and 17,349 controls. Their overall results suggested that the variant genotypes were associated with a moderately increased risk of all cancer types (OR = 1.09, 95% CI = 1.01–1.18 for TG vs. TT; OR = 1.08, 95% CI = 1.00–1.18 for GG/TG vs. TT). The meta-analysis also supported that the APE1 Asp148Glu polymorphism is a low-penetrance risk factor for cancer development.43 Curtin et al. genotyped colon cancer cases and matched population-based controls for BER variants in OGG1 (Ser326Cys) and XRCC1 (Arg399Gln, Arg280His and Arg194Trp). No association was found between the selected polymorphisms and overall colon cancer susceptibility. However, a twofold increased risk of carrying the proto-oncogene serine/threonine protein kinase BRAF V600E allele was observed in current and former cigarette smokers for the OGG1 Cys326Cys polymorphism (OR = 2.2; 95% CI = 1.02–4.9). XRCC1 Arg194Trp gene polymorphism was associated with a modestly increased risk of TP53 mutations in smokers (OR = 1.4; 95% CI = 1.02–1.9). Collectively, these findings suggest that OGG1 Ser326Cys and Arg194Trp cosegregate with other colon cancer-associated mutations.46
Kim et al. investigated the effect of OGG1 on the risk of colon cancer in the Korean population and found that the OGG1 Ser326Cys polymorphism did not alter the overall OR for colon cancer. However, subgroup analysis revealed a trend for increased OR with smoking or frequent meat intake in the Cys/Cys carriers.47
The study of Hansen et al. did not support Kim et al.’s results. Hansen et al. found no association between OGG1 Ser326Cys polymorphism and the risk of colorectal adenomas, regardless of the degree of dysplasia in a Norwegian population. Furthermore, OGG1 Ser326Cys gene polymorphism had a protective effect (OR = 0.56; 95% CI = 0.32–0.97 Ser/Ser vs. Ser/Cys) against the risk of colorectal cancer. Since meat consumption and smoking are frequent in the Norwegian population, different gene–environment interactions are likely in both the types of population.48 In another cohort study, in a Norwegian population, the association between XRCC1 gene polymorphisms (Arg194Trp, Arg280His and Arg399Gln) and the risk of colorectal adenomas and carcinomas was examined. This study demonstrated increased risk of advanced colorectal neoplasia in individuals with the XRCC1 Arg280His gene polymorphism and a reduced risk associated with the Arg399Gln gene polymorphism.49
Moreno et al. investigated whether common polymorphisms in DNA repair genes correlate with colorectal cancer risk or patient prognosis and response to chemotherapy. In fact, adjuvant chemotherapy causes an acute increase in DNA damage, suggesting that the DNA repair capacity in specific cancer cells, tumors or individuals may influence the therapeutic efficacy of the selected DNA-interactive drugs. In the study, the incidence of 28 common SNPs in 15 DNA repair genes was determined in DNA from colorectal cancer patients. SNPs were examined in the following genes: BER (OGG1, LIG3, APE, POLB, XRCC1, PCNA and MUTYH), NER (ERCC1, ERCC4 and ERCC5), double-strand break repair (XRCC2, XRCC3 and XRCC9) and direct reversal repair (MGMT). The findings suggest that, among the BER genes, in contrast to the results of Hansen et al., homozygous carriers of OGG1 Ser/Cys had a statistically significant twofold higher risk of colorectal cancer than the carriers of the wild type allele (OR = 2.31; 95%CI = 1.05–5.09). The APE1 Asp148Glu variant was also associated with an increased risk of colorectal cancer (OR = 3.04; 95% CI = 1.38–6.71 for APE1 Asp148Glu).50
In a subsequent study, no association was found with APE1 in colorectal cancer, but the risk was increased in colon cancer for the Glu/Glu genotype. For OGG1 Ser326Cys, a significantly increased risk was observed in smokers. Interestingly, the joint effect of variant alleles of APE1 Asp/Glu and hOGG1 Ser/Cys showed increased colorectal cancer risk (OR = 6.37; 95% CI = 1.40–29.02).51 However, different results were obtained in Korean (OR = 1.72; 95% CI = 1.12–2.76 Ser/Cys, only meat intake),47 Japanese (OR = 1.43; 95% CI = 0.69–2.95 for Ser/Cys + Cys/Cys) and Chinese populations (OR = 1.4; 95% CI = 0.9–2.2 for Ser/Cys; OR = 1.4; 95% CI = 0.9–2.1 for Cys/Cys).52,53 Another study in a Caucasian population showed no association of the OGG1 Cys/Cys genotype (OR = 0.963; 95% CI = 0.446–2.079) with the risk of cancer progression, despite the elevated oxidative stress in these cancer patients.54
Wang et al. analyzed the distributions of three SNPs (XRCC1 Arg399Gln, XRCC3 Thr241Met and XPD Lys751Gln) in DNA repair genes, assessed the association of these genetic polymorphisms with colon and rectal cancer susceptibility, as well as evaluated the interactions of gene–gene and gene–environment in a case-control study of an Indian population. No association was observed between the XRCC1 Arg399Gln gene polymorphism and cancer risk (OR = 1.44; 95% CI = 0.76–2.75 for colon; OR = 1.40; 95% CI = 0.96–2.06 for rectal; OR = 1.41; 95% CI = 0.99–2.03 colorectal cancer). However, the XRCC1 Gln399 allele was associated with a significantly increased rectal cancer risk among men (OR = 1.65; 95% CI = 1.04–2.64), indicating a gender effect of this allele on colorectal cancer. Furthermore, individuals carrying XRCC1 Gln399, XRCC3 Thr/Thr (double-strand break repair) and XPD Gln751 demonstrated the highest colorectal cancer risk (OR = 3.52; 95% CI = 1.43–9.44).55
Environmental factors, such as diet, lifestyle and environmental pollution, have important impact on the development of colorectal cancer. Thus, Wang et al. also evaluated the interaction of gene–smoking or gene–alcohol on rectal or colorectal cancer risk. The XRCC1 Arg399Gln gene polymorphism was not found to be interacting significantly with tobacco consumption on cancer susceptibility (interaction p > 0.05, respectively). Conversely, there was a positive association of the XRCC1 Gln399 allele with rectal (OR = 1.56; 95% CI = 1.05–2.33) and colorectal (OR = 1.61; 95% CI = 1.11–2.34) cancer among non-drinkers, and there was a weak evidence that XRCC1 Arg/Arg genotype increased the risk of rectal (OR = 1.93; 95% CI = 0.94–4.04) or colorectal (OR = 1.91; 95% CI = 0.96–3.86) cancer among drinkers (p = 0.05 for rectal cancer and 0.03 for colorectal cancer). Alcohol intake did not affect the results of the other genetic polymorphisms, that is, XRCC3 Thr241Met and XPD Lys715Gln (interaction p > 0.05, respectively). Finally, based on a small proportion of vegetarians in their study group, Wang et al. argued that the insufficient nutrition may explain some of the increased risk of rectal cancer (OR = 1.83; 95% CI = 1.04–3.26). Meta-analysis with a large population confirmed the previous studies, showing no association between XRCC1 gene polymorphisms and colorectal cancer.56 Additional studies similarly found no association between XRCC1 Arg399Gln gene polymorphism and colorectal cancer risk.46,50,52,53
Lung cancer
Table 2 summarizes the associations of gene polymorphisms and lung cancer. Lung cancer is one of the most common cancers and is the leading cause of cancer death worldwide. In 2006, there were an estimated 1,352,000 new lung cancer cases and 1,179,000 lung cancer deaths. The main types of lung cancer are small-cell lung carcinoma and NSCLC. There are three main subtypes: squamous cell lung carcinoma (SCLC), ADC and large-cell lung carcinoma. This distinction is important for therapy. SCLC is most strongly associated with smoking, while ADC is least strongly associated with smoking and the most common lung cancer in nonsmokers. Thus, genetic as well as environmental factors should be considered in developing and applying interventions.33
Table 2.
Summary of findings from case–control studies of genetic polymorphisms in DNA repair genes (OGG1, APE1 and XRCC1) and their interactions with environmental factors and lung cancer
| Study population | No. of Cases/Controls | Main effects of polymorphisms
|
Gene–environment interaction | References | ||
|---|---|---|---|---|---|---|
| OGG1 Ser326Cys | XRCC1 (Arg399Gln, Arg194Trp, Arg280His) | APE1Asp148Glu | ||||
| German (H) | 463/460 | NA | 2-fold IR (Arg/Gln + Gln/Gln) -(Arg/His, Arg/Trp) | RR (Glu/Glu) | – | Popanda et al.59 |
| Chinese (H) | 350/350 | – | 2-fold IR (Arg/Gln, Gln/Gln) NA (Arg/His, Arg/Trp) |
– | – | Li et al.8 |
| Danish(H) | 256/269 | NA | NA (Arg/Gln, Arg/His);-(Arg/Trp) | – | – | Vogel et al.60 |
| Japanese (H) | 108/121 | NA (ADC, SCC) | – | – | – | Miyaishi et al.19 |
| Turkish (H) | 165/250 | NA | – | – | IE (Cys/Cys) (heavy smokers) | Karahalil et al.61 |
| Japanese (H) | 1097/394 | IR (Cys/Cys) | – | – | S; 1.5-fold IR (Ser/Cys) | Kohno et al.33 |
| Caucasian Asian (H) | 2569/4178 | 1.5-fold IR (Cys/Cys) | NA | NA | – | Hung et al.62 |
| Norwegian (H) | 326/386 | 1.6-fold IR (Cys/Cys) | NA (Arg/Gln, Arg/His, Arg/Trp) | NA | – | Zieolddiny et al.63 |
| Chinese (P) | –/1281 | NA | 1.5-fold IR (Arg/Arp + Trp/Trp) NA (Arg/Gln);-(Arg/His) |
NA | – | Li et al. 73 |
| Belgian (H) | 220/350 | NA | NA | IR (Asp/Glu Glu/Glu) | – | DeRuyck et al.66 |
| Chinese (H) | 247/253 | – | RR (Arg/His + His/His) | – | NA | Yin et al.67 |
SCC: squamous cell carcinoma; BCC: basal cell carcinoma; ADC: adenocarcinoma; C: carcinoma; –: unreported; NA: no association; IR: increased risk; RR: reduced risk; S: smoking; D: drinking; RM: red meat; H: hospital based study; P: population based study.
The International Lung Cancer Consortium analyzed the correlation between DNA repair gene variants and lung cancer susceptibility using the data from 14 studies for 18 sequence variants in 12 DNA repair genes (APE1, OGG1, XRCC1, XRCC2, XRCC3, ERCC1, XPD, XPF, XPA, XPG, MGMT and TP53). Six studies were conducted in European countries, four in the United States and four in Asia or the Pacific Islands. Seven studies used hospital-based controls, seven studies used population-based controls and one study used a nested cohort study design. The number of subjects in these studies ranged from 2073 to 13,955. Relevant to the review here, we focused on the BER variants in OGG1, APE1 and XRCC1. OGG1 Ser326Cys homozygous variant individuals were associated with lung cancer risk in Caucasians (OR = 1.34; 95% CI = 1.01–1.79) based on 2569 cases and 4178 controls in four studies; however, this association was lacking in Asian populations.18,32,57,58 OGG1 Ser326Cys also showed a differential effect in non-Hispanic Whites in one study.32 Popanda et al. found reduced risk of lung cancer associated with APE1 Glu148Glu in Caucasians. When APE1 Glu148Glu and XRCC1 Gln399Gln were combined, ORs were reduced (OR = 0.69; 95%CI = 0.28–1.73), especially in the SCLC group (OR = 0.39; 95%CI = 0.10–1.53). For homozygous individuals carrying the Glu variant of APE1, a protective effect was found (OR = 0.77; 95% CI = 0.51–1.16). Popanda et al. found that XRCC1 Arg/Gln and Gln/Gln genotypes, when pooled and considered together, had higher lung cancer risk (adjusted OR = 2.15; CI 95% = 1.08–4.28) than XRCC1 Arg/Arg homozygotes in an Italian population.59 Similar observations were made in a Chinese hospital based case-control study.8 In this study, individuals with a homozygous XRCC1 Gln/Gln genotype (OR = 1.75; 95% CI = 1.02–3.01) exhibited a slightly higher risk of lung cancer overall and the adjusted OR was increased in the cases with ADC (OR = 2.62; 95% CI = 1.44–4.79). No joint effect of cooking oil fume and Gln399 was observed on lung cancer risk in women nonsmokers.8 No significant association was reported between OGG1 Ser326Cys and lung cancer risk.
In the first prospective population-based study of Danes, there were no associations between XRCC1 Arg280His and Arg399Gln or OGG1 Ser326Cys gene polymorphisms and risk of lung cancer.60 Miyaishi et al. also did not find a significantly increased risk of lung ADC or SCLC with respect to OGG1 genotype (OR = 1.39; 95% CI = 0.61–3.19; Ser/Ser vs. Cys/Cys for ADC and OR = 1.22; 95% CI = 0.40–3.75; Ser/Ser vs. Cys/Cys for SCLC).19 A larger hospital-based study performed in Turkey generally supported these results, although the OGG1 Ser326Cys polymorphism had a reduced lung cancer risk in smokers (Cys/Cys vs. Ser/Ser; adjusted OR = 0.62; 95% CI = 0.26–1.43).61 Kohno et al. found that the effect of the OGG1 Ser326Cys gene polymorphism on lung ADC risk was not modified by smoking in this study population.33 However, some lung cancer studies have suggested higher lung cancer risk associated with OGG1 Cys/Cys vs. Ser/Ser,33,62,63 yet did not find an association between the XRCC1 Arg399Gln gene polymorphism and lung cancer risk. These inconsistencies highlight the difficulties associated with comparing different genotypes and different cancer subtypes.64
Nonsmokers who develop lung cancer provide a good model for investigating the effects of genetic polymorphisms. A hospital-based case-control (n = 55 lung cancers and n = 74 controls) study was conducted to evaluate the impact of polymorphisms on lung cancer risk in nonsmoking Chinese women. Although the sample size was small, this study was valuable because it only involved nonsmokers. A significant protective effect of the variant T allele of XRCC1 (Trp194) was observed (T vs. C, OR = 0.51; 95% CI = 0.29–0.90, p = 0.02). Women carrying at least one variant T allele of XRCC1 had a decreased risk of lung cancer compared with homozygous Arg194 wild type carriers (OR = 0.46; 95% CI = 0.22–0.96, p = 0.04).65 Protective effects of the variant T allele of XRCC1 Trp194 have been observed by De Ruyck et al. among Caucasian Belgians, and they reported that XRCC1 Arg/Trp heterozygotes have a significantly reduced risk of lung cancer (OR = 0.32, p = 0.024).66
Yin et al. found a significantly decreased risk of lung cancer associated with the Arg/His or His/His genotype of XRCC1 at position 280 (OR = 0.38; 95% CI = 0.19–0.75, Arg/His + His/His vs. Arg/Arg, p = 0.005) in a larger population of ‘never-smokers’, while there was no interaction between Arg/His genotype and smoking.67 In a population-based cohort of 57,053 Danes, messenger RNA (mRNA) levels of OGG1, NEIL1 (nei endonuclease VIII-like 1 (Escherichia coli)), MUTYH (mutY Homolog (E. coli)) and NUDT and subsequent lung cancer risk was evaluated. Overall, mRNA levels of BER genes were not predictors of lung cancer risk. However, subjects with the OGG1 Cys/Cys genotype had a reduced expression level of OGG1 mRNA, and there was a positive association between the level and risk of lung cancer. Assuming that the amino acid substitution reduces repair capacity, as has been demonstrated in several independent studies,68,69 this observation might indicate that the expression of OGG1 is regulated by the level of oxidative DNA damage. Finally, the intracellular level of OGG1 Cys326 protein was directly correlated with urinary excretion of 8-oxoG, which could be interpreted as a biomarker of exposure to oxidative stress.9
Gastrointestinal cancer
Table 3 summarizes the associations of gene polymorphisms and gastrointestinal cancer. Gastric cancer is the second leading cause of cancer death and the fourth most common epithelial neoplasia worldwide, with the highest incidence in China, Japan and Eastern European countries.70 According to the GLOBOCAN 2008 data, summary statistics indicate that the gastric cancer is fifth in males among the five most frequent cancers (prostate, lung, colorectal, bladder and stomach), whereas no gastric cancer is observed in females.71 A variety of factors including sodium intake, Helicobacter pylori infection and smoking induce inflammation in the stomach. An inflammatory cell infiltrate in the gastric mucosa increases the production of ROS, which causes potentially mutagenic 8-oxoG lesions in the DNA. In mammalian cells, 8-oxoG is repaired by BER enzymes such as OGG1, MYH and NEIL1. In addition, MTH1, a part of the defense system against oxidative damage, removes 8-oxo-dGTP from the nucleotide pool, thus preventing incorporation of the base lesion into DNA during replication. OGG1-c.977C>G (Ser326Cys) was associated with a difference in 8-oxoG repair capacity, and MTH1-c.247G>A (Val83Met) has been reported to be associated with increased risk of gastric cancer. These results suggest that SNPs OGG1-c.977C>G (Ser326Cys) and MTH1-c.247G>A (Val83Met) may lead to the accumulation of 8-oxoG in the gastric mucosa.72
Table 3.
Summary of findings from case–control studies of genetic polymorphisms in DNA repair genes (OGG1, APE1 and XRCC1) and their interactions with environmental factors and gastrointestinal cancer
| Study population | No. of cases/controls | Main effects of polymorphisms
|
Gene–environment interaction | References | ||
|---|---|---|---|---|---|---|
| OGG1 Ser326Cys | XRCC1 (Arg399Gln, Arg194Trp, Arg280His) | APE1Asp148Glu | ||||
| Chinese (H) | 1281 | IR; (Ser/Cys + Cys/Cys) | PE; (Arg/Trp + Trp/Trp) NA (Arg/Gln), -(Arg/His) |
NA | S; 1.5-fold IR (Ser/Cys + Cys/cys) | Li et al.73 |
| Korean (H) | 190/172 | RR; (Arg/Trp + Trp/Trp) | Lee et al. 74 | |||
| Indian (H) | 108/195 | NA | – | – | S; 8-fold IR(Ser/Cys); NA, salted tea | Malik et al.75 |
| Chinese (H) | 455/650 | – | NA (Arg/Trp), NA (Arg/Gln) -(Arg/His) | – | S; 1.5-fold IR (Arg/His + His/His) | Yan et al.76 |
| Chinese (H) | 190/180 | – | IR (Arg/Trp) NA (Arg/His) -(Arg/Gln) | – | S; IR (Arg/His + His/His) | Yuan et al.78 |
–: Unreported; NA; no association; IR: increased risk; RR: reduced risk; PE: protective effect; S: smoking; H: hospital based study.
Gastric Cancer includes gastric cardiac cardioma.
Li et al. conducted a population-based cohort study in China to evaluate the association between H. pylori-associated precancerous gastric lesions and the following polymorphisms: XRCC1, Arg399Gln and Arg194Trp; poly-ADP-ribose polymerase, Val762Ala; OGG1 Ser326Cys and APE1 Asp148Glu. An inverse association was found between XRCC1 Arg/Trp + Trp/Trp and progression of gastric lesions, suggesting that Trp194 allele may play a protective role in this population. The OGG1 Ser/Cys + Cys/Cys genotype was associated with an increased risk of progression in subjects with intestinal metaplasia or dysplasia at baseline, indicating that the Cys326 allele was a deleterious allele in this high-risk population.73 In the Korean population, a similar protective effect of Trp194 was observed.74 In particular, the frequencies of Trp194 or Gln399 homozygotes were similar in the cases and controls and were not associated with a risk of gastric cancer upon adjustment for age and sex. However, with the Arg194Arg genotype as the reference group, the combined genotype (Arg/Trp and Trp/Trp) specific OR was 0.66 (95%CI = 0.44–1.00, p = 0:052), indicative of the 194Trp allele operating as a protective allele for gastric cancer.76
Malik et al. assessed the role of OGG1 Ser326Cys in susceptibility to gastric cancer in Kashmir valley, where the incidence is three to six times higher than various metropolis cancer registries in India. No association was observed. However, smoking habit showed significantly higher risk in gastric cancer patients harboring the OGG1 Ser/Cys genotype (OR = 8.975; 95% CI = 5.156–15.622; p = 0.0001), suggesting that the gene–environment interactions are more important in the development of gastrointestinal cancer compared with colorectal cancer. Higher consumption of salted tea was also found to be associated with increased risk of gastric cancer. Individuals that consumed salted tea in a range of two to eight cups per day and individuals that consumed salted tea of more than four cups per day were regarded as high salted tea consumers. However, when taking potential gene–environment interactions into account, there was no association of OGG1 Ser326Cys gene polymorphism with smoking and salted tea consumption. Therefore, the authors concluded that although the OGG1 Ser326Cys gene polymorphism is reported to be a risk factor in some cancers, it does not appear to play a major role in gastric cancer development in Kashmir valley.75
Yan et al. did not find any significant difference in allele and genotype distributions of XRCC1 Arg/Trp or Arg/Gln genotypes between gastric cardia adenocarcinoma (GCA) and controls. However, smokers with the His allele at codon 280 had a significantly increased risk of GCA.76 Since the Arg280His variant occurs in the proliferating cell nuclear antigen binding region,77 the transition from a positively charged Arg to His within the conserved region could alter specific XRCC1 functions. For the XRCC1 Arg194Trp gene polymorphism, no association was found for the risk of GCA,76 consistent with previous results conducted in China and Korean.73,74 In contrast, Yuan et al. found that the XRCC1 Trp194 allele significantly increased the risk of gastric cancer and also associated with the risk of gastric cardia carcinoma and promoted distant metastasis of gastric cancer.78 Thus, there are conflicting results on the association between gastric cancer and XRCC1 or OGG1 gene polymorphisms. These discrepancies may reflect differences in the study population or cancer location. Indeed, noncardia gastric cancer risk is positively associated with H. pylori infection, but cardia cancer is not. Ethnic differences may also influence gastric cancer risk. Palli et al. carried out a multigenic study on gastric cancer with polymorphic DNA repair and metabolic genes. A significant interaction of the APE1 wild type genotype was observed with either the single GSTT1 or double GSTM1–GSTT1 null genotype or the XPA mutant allele, with APE1 being protective for cancer susceptibility.79
Bladder cancer
Table 4 summarizes the associations of gene polymorphisms and bladder cancer. In our previous work, we found the OGG1 Cys/Cys genotype to be more frequent among bladder cancer patients (OR = 2.41; 95% CI = 1.36–4.25). However, no association was observed in the Arg399Gln gene polymorphism and the risk of bladder cancer (OR = 0.72; 95% CI = 0.41–1.26).80 Huang et al. observed similar results regarding the XRCC1 Arg399Gln gene polymorphism, but did not find any association with the OGG1 Ser326Cys gene polymorphism and bladder cancer risk.64
Table 4.
Summary of findings from case–control studies of genetic polymorphisms in DNA repair genes (OGG1, APE1 and XRCC1) and their interactions with environmental factors and bladder cancer
| Study population | No. of cases/controls | Main effects of polymorphisms
|
Gene–environment interaction | References | ||
|---|---|---|---|---|---|---|
| OGG1 Ser326Cys | XRCC1 (Arg399Gln, Arg194Trp, Arg280His) | APE1Asp148Glu | ||||
| Turkish (H) | 100/100 | 2.5-fold IR (Cys/Cys) | NA | – | Not interaction with smoking | Karahalil et al.80 |
| Caucasian (H) | 609/613 | NA | PE (Arg/Trp + Trp/Trp) | NA | IR (Cys/Cys) (ever smokers) No interactions (Others) | Huang et al.64 |
| American (H) | 539/524 | NA | NA (Arg/Gln, Arg/Trp) -(Arg/His) | – | RR (Ser/Cys) (ever smokers) | Huang et al.64 |
–: Unreported; NA; no association; IR: increased risk; RR: reduced risk; PE: protective effect; S: smoking; RM: red meat; H: hospital based study.
Breast cancer
Table 5 summarizes the associations of gene polymorphisms and breast cancer. Studies suggest that breast cancer is initiated by somatic mutations that may have been introduced during error-prone repair of estrogen-induced abasic sites.81 However, SNPs may modulate breast cancer risk, including XRCC1 Arg194Trp and Arg399Gln, APE1 Asp148Glu and OGG1 Ser326Cys. Sigurdson et al. evaluated 19 SNPs in eight genes involved in BER (e.g. XRCC1 and APE1), interactions with BRCA1 (BRIPI, ZNF350 and BRCA2) or growth regulation (TGFβ1 and IGFBP3). XRCC1 Arg194Trp and Arg399Gln polymorphisms were associated with increased breast cancer risk (XRCC1 Arg/Arg vs. Arg/Trp + Trp/Trp, cumulative risk up to age 70 years, risk ratio (RR) = 2.3, 95% CI = 1.3–3.8; Arg/Arg vs. Gln/Gln, RR = 1.9, 95% CI = 1.1–3.9, respectively). However, APE1 Asp148Glu was associated with a significantly lower risk than the homozygous genotype (RR = 0.2, 95% CI = 0.1–0.5, Asp/Asp vs. Glu/Glu, cumulative risk up to age 70 years, p < 0.001). No associations were observed for the polymorphisms in the other genes noted above. Moreover, the study indicated that some XRCC1 variants in combination with BRCA1 are low penetrance breast cancer risk alleles.82 Recently, a study was conducted by Ali et al. on US women who carried XRCC1 Arg399Gln variant, but not the commonly reported APE1 Glu148 variant. This study reported that XRCC1 Gln399 may enhance the risk of breast cancer.81
Table 5.
Summary of findings from case–control studies of genetic polymorphisms in DNA repair genes (OGG1, APE1 and XRCC1) and their interactions with environmental factors and breast cancer
| Study population | No. of cases/controls | Main effects of polymorphisms
|
Gene–environment interaction | References | ||
|---|---|---|---|---|---|---|
| OGG1 Ser326Cys | XRCC1 (Arg399Gln, Arg194Trp, Arg280His) | APE1Asp148Glu | ||||
| American (H) | 748/2240 | – | 2.3-fold IR; (Arg/Trp + Trp/Trp) 1.3-fold IR (Gln/Gln) |
IE;(Glu/Glu) | Sigurdson et al.82 | |
| Caucasian (H) African-American (AA)(H) | 336/416 63/78 | – | NA | 1.5-fold IR (Asp/Glu + Glu/Glu) | – | Smith et al.83 |
| USA (P) Poland | 1571/1784 1470/1366 | NA | NA | NA | No change (Pre and post-menapausal status) | Zhang et al. 84 |
| Danish (H) | 425/434 | NA | – | – | – | Vogel et al.85 |
| Taiwanese (H) | 506/424 | NA | NA (Arg/Trp, Arg/His) -(Arg/Gln) | NA | – | Sangrajrang et al.86 |
–: Unreported; NA; no association; IR: increased risk; RR: reduced risk; S: smoking; D: drinking; RM: red meat; H: hospital based study; P: population based study
Although individual DNA repair genotypes might have a small effect, combined nsSNPs in multiple DNA repair pathways may have a larger effect on cancer risk. Smith et al. analyzed 18 nsSNPs in four DNA repair pathways in Caucasians and African Americans to evaluate potential associations with breast cancer risk. Their data suggested that in Caucasians, breast cancer risk was significantly associated with APE1 Asp/Glu genotype (OR = 1.44; 95% CI = 1.03–2.00, Asp/Glu + Glu/Glu); this association was not observed in African Americans due to limited sample size.83
Two large population-based studies (one in the United States and one in Poland), a hospital-based cohort study in Denmark and one hospital-based study in Taiwan found no significant association between OGG1 Cys/Cys (Cys/Cys vs. Ser/Ser; adjusted OR = 1.00; 95% CI = 0.7–1.4 and OR = 0.98; 95% CI = 0.52–1.86; Ser/Ser vs. Cys/Cys for cohort study) or XRCC1 Gln/Gln (Gln/Gln vs. Arg/Arg, adjusted OR = 0.8; 95% CI = 0.7–1.109) and breast cancer risk.84 A smaller Taiwan hospital-based breast cancer study concurred with this result83 data were adjusted for age, body mass index (BMI), family history of breast cancer in first-degree relatives, age at menarche, age at first full-term pregnancy, parity, age at menopause, menopausal status, hormone replacement therapy and highest level of education. In the Taiwan population, a significantly increased breast cancer risk was found for the Cys/Cys genotype (Cys/Cys vs. Ser/Ser; OR = 2.05; 95% CI = 1.14–3.69) in postmenopausal women.84–86 Yuan et al. investigated the effect of OGG1 Ser326Cys gene polymorphism on the susceptibility to breast cancer with data collected in a meta-study, since the published studies on OGG1 Ser326Cys polymorphism and breast cancer risk had inconsistent conclusions. The meta-analysis suggested that the hOGG1 Cys326 allele is associated with significant protection against breast cancer in European women.87
Gynecologic cancers and disorders
Table 6 summarizes the associations of gene polymorphisms and gynecologic cancers and/or disorders. Cervical cancer, including squamous cell carcinoma (SCC), ADC and adenosquamous cell carcinoma (ADSC) subtypes, is the third most common cancer worldwide. Cervical cancer is associated with persistent human papillomavirus infection in the minority of cases. Polymorphisms in the OGG1 and XRCC1 genes were not associated with cervical cancer risk.88 In a hospital-based epidemiologic study conducted in Japan, there was no significant association between the XRCC1 Arg399Gln gene polymorphism and risk of cervical cancer, when all histologic subtypes were included.88 However, ADC/ADSC was associated with Arg/Gln (OR = 2.98; 95% CI = 1.11–8.01 for Gln/Gln and OR = 2.18; 95% CI = 1.11–4.26 for Arg/Gln + Gln/Gln in ADC and ADSC), whereas SCC was not. Similar data for Gln/Gln were observed in never smokers (OR = 3.85; 95% CI = 1.28–11.59, p = 0.017), but not among smokers.88
Table 6.
Summary of findings from case–control studies of genetic polymorphisms in DNA repair genes (OGG1, APE1 and XRCC1) and their interactions with environmental factors and gynecological (GyC) cancer
| Study population | No. of cases/controls | Main effects of polymorphisms
|
Gene–environment interaction | References | ||
|---|---|---|---|---|---|---|
| OGG1 Ser326Cys | XRCC1 (Arg399Gln, Arg194Trp, Arg280His) | APE1Asp148Glu | ||||
| Japanese (H)a | 131/320 | NA | 2- to 3-fold IR (Arg/Gln and Arg/Gln + Gln/Gln; ADC/ADSC)- | – | NS; 3.85-fold IR | Niwa et al. 88 |
| Chineseb (H) | 136/140 | – | 2-fold IR (Arg/His) NA; (Arg/Gln, Arg/Trp) |
– | – | Yang et al.89 |
ADC: adenocarcinoma; ADSC: adenosquamous cell carcinoma; –: Unreported; NA; no association; IR: increased risk; RR: reduced risk; S: smoking; NS: never smoker; H: hospital based study.
Cervical cancer.
Uterine leiomyoma.
Endometriosis is a common, benign, estrogen-dependent, chronic gynecological disorder. Although multiple theories exist regarding the etiology of endometriosis, the exact pathogenesis remains uncertain. A recent study on a Turkish population showed no association between XRCC1 Arg399Gln, XRCC3 Thr241Met, XPD Lys751Gln, XPD Asp104His, APE1 Asp148Glu and hOGG1 Ser326Cys and endometriosis risk.42 However, the study sample size was very small (52 patients and 110 controls).
Uterine leiomyoma is the most common gynecological benign tumor of the myometrium. It is a significant cause of pelvic pain, abnormal uterine bleeding, infertility and pregnancy complications. Many factors including age, family genetics, emotional status, hormone levels and environmental factors may contribute to the development of this disease. One study investigated whether the different XRCC1 codon polymorphisms were related to the occurrence of uterine leiomyoma in a Chinese population.89 These investigators found a significant risk associated with codon 280 (OR = 3.633; 95% CI = 2.147–6.148; Arg/Arg vs. Arg/His), but no association with codons 399 and 194. However, the sample size again was small (n = 136), and further studies are therefore needed.89
Upper aerodigestive cancer
Table 7 summarizes the associations of gene polymorphisms and UADC. Elahi et al. measured OGG1 expression and the association between OGG1 polymorphism and aerodigestive tract cancer. Significantly increased risk of orolaryngeal cancer was observed with decreased hOGG1 expression in all the tissues (tongue, tonsil, mouth, larynx and esophagus).36 Furthermore, although no association between OGG1 genotype and orolaryngeal cancer risk was found in never smokers, significantly increased risk was observed in Cys/Cys smokers.36 Hashimoto et al. confirmed this latter observation regarding the relationship of Cys326 and smoking status with cancer susceptibility (OR = 8.10; 95% CI = 1.06–61.73 for OGG1 Cys/Cys).90 For XRCC1, the polymorphic variants at codons 399, 194 and 280 were associated with increased risk of oral cancer in an Indian population. Moreover, smokers and betel quid chewers with the variant allele of XRCC1 at position 399 also exhibited increased risk of oral cancer.91 The XRCC1 Arg194Trp gene polymorphism was associated with increased risk of head and neck cancer in Korean and Hungarian populations,92–94 whereas no associations were reported for the other XRCC1 polymorphisms. In a separate study, Kowalski et al. observed an increase risk with only Arg/Trp plus Trp/Trp genotypes at codon 194.93 In the US population, no association was found with APE1 Asp148Glu and XRCC1 Arg399Gln, Arg280Trp and Arg194Trp gene polymorphisms and head and neck cancer risk.94
Table 7.
Summary of findings from case–control studies of genetic polymorphisms in DNA repair genes (OGG1, APE1 and XRCC1) and their interactions with environmental factors and upper aero digestive cancer (UADCa)
| Study population | No. of cases/controls | Main effects of polymorphisms
|
Gene–environment interaction | References | ||
|---|---|---|---|---|---|---|
| OGG1 Ser326Cys | XRCC1 (Arg399Gln, Arg194Trp, Arg280His) | APE1Asp148Glu | ||||
| Japanese (H) | 192a | NA | – | – | S; 8-fold IR (Cys/Cys); D; NA | Hashimoto et al.90 |
| Indian (H) | 200/110 | – | 2- to 13-fold IR (Gln/Gln, Arg/Gin + Gln/Gln, Arg/Trp, Arg/Trp + Trp/Trp) | – | S; IR (Arg/Gln + Gln/Gln) | Ramachandran et al.91 |
| Korean (H) | 147/168 | – | 2.6-fold IR (Arg/Trp, Trp/Trp) | – | – | Tae et al.92 |
| Polish (H) | 92/124 | – | IR (Arg194Trp + Arg399Arg) NA others |
NA | Kowalski et al.93 | |
| American (H) | 830/854 | – | NA (Arg/Gln) | NA (Asp/Glu) | NA | Li et al.94 |
UADC: upper aero digestive cancer including oral, pharynx, laryngeal, esophageal, head and neck; –: unreported; NA; no association; IR: increased risk; S: smoking; D: drinking; H: hospital based study.
Case only design.
The others
Table 8 summarizes the associations of gene polymorphisms and different cancer types (skin, acute lymphoblastic leukemia (ALL), head and neck, prostate, non-Hodgkin Lymphoma (NHL), brain, lung and UADC), with further discussion in each of the sections to follow.
Skin cancer
Sun exposure, but not history of sunburn, is recognized as an important risk factor for skin cancer development. Kang et al. found lower risk of SCC associated with XRCC1 Arg194Trp and higher risk of basal cell carcinoma (BCC) associated with the Arg399Gln variant.4 Another study suggests that XRCC1 Arg280His and Arg399Gln genotypes are risk factors for skin cancer.95 A well-designed nested case-control study undertaken by the Danish prospective cohort ‘Diet, Cancer and Health’ involved 319 cases carefully matched with 319 controls recruited from a cohort of 57,053 Danes. However, no association was found between the OGG1 Ser326Cys and risk of BCC (OR = 0.98, 95% CI = 0.70–1.37; for Ser326Cys, OR = 1.09; 95% CI = 0.59–2.03 for Cys/Cys). The lack of association may reflect low environmental exposure to sunlight in Denmark or selection of the OGG1 SNP and this skin cancer subtype.60
Acute lymphoblastic leukemia
ALL is characterized by improper development and differentiation of lymphoid progenitor cells. It is the most common childhood cancer with a peak at 2 ± 5 years of age.96 The influence of the Arg194Trp, Arg280His and Arg399Gln polymorphisms of XRCC1 on the development of childhood ALL was investigated in 120 patients and 120 controls in Mexico. No significant difference was observed between patients and controls (OR = 1.71, 95% CI = 0.65–4.48 for Arg/Arg vs. Gln/Gln, OR = 2.15; 95% CI = 0.52–8.89 for Arg/Arg vs. Trp/Trp and OR = 2.02; 95% CI = 0.18–22.72 for Arg/Arg vs. His/His).97 The lack of association for the XRCC1 Trp194 and Gln399 variants with ALL was similarly observed in Turkish children.98 However, Matullo et al. found a 2.5-fold increased risk of ALL in patients carrying the Gln/Gln genotype.99
Prostate cancer
Prostate cancer (PCa) is one of the most common malignancies in men in the Western countries and accounts for 31% of new annual cancer cases in men. The incidence of PCa varies greatly with race and geography.100 Xu et al. investigated individual and joint effects of the three polymorphisms in XRCC1 on PCa risk. The194 Arg/Trp genotype was associated with lower risk of PCa than wild type, while Arg280His had no impact on PCa risk. Compared with wild type, Arg399Gln variant alone was associated with a statistically significant increased risk of PCa (adjusted OR: 3.19; 95% CI = 1.48–6.88). When the three polymorphisms (Arg194Trp, Arg280His and Arg399Gln) were evaluated in combination, subjects with a Arg/Arg, Arg/His and Arg/Gln genotype had a significantly higher risk of PCa (adjusted OR = 4.31; 95% CI = 1.24–14.99).101
Non-Hodgkin lymphoma
NHL represents a heterogeneous group of lymphocytic disorders ranging in aggressiveness from indolent to highly aggressive and rapidly proliferative. Although it is the fifth cause of cancer mortality in the United States, NHL remains poorly understood and is largely incurable.102 Well-known risk factors include family history, immune dysfunction (e.g. autoimmune diseases, immune deficiency syndromes and iatrogenic immunosuppression after organ transplant), immune stimulation and infection (e.g. human T-lymphotrophic virus type I and human immunodeficiency virus). Occupational and environmental exposures have also been proposed as risk factors for NHL.103 SNP variants of XRCC1 Arg399Gln, Arg194Trp and Arg280His were not significantly related to the risk of NHL subtypes, including diffuse large B cell lymphoma or follicular lymphoma. However, two combinations of SNP variants (Arg/Gln + Gln/Gln or Arg/Trp + Trp/Trp or Arg/His + His/His vs. wild genotype) showed a trend of increased risk, suggesting an ‘additive’ effect. Regarding smoking status, individuals with at least one Arg399Gln variant allele have a threefold increase in follicular lymphoma risk (adjusted OR = 3.01; 95% CI = 1.16–7.82) compared with heavy smokers with the wild type genotype.104 However, as these past studies involved about 200 cases and controls, larger scale studies are needed.
Hepatocellular carcinoma
Hepatitis B virus (HBV) and hepatitis C virus (HCV) increase chromosomal instability and insertional mutagenesis and are primary risk factors for hepatocellular carcinoma (HCC). Approximately 30–50% of HBV-related deaths are attributable to HCC; however, only 2.5% of HCV-infected individuals develop HCC. The association between XRCC1 (codons 194, 280 and 399) and hepatitis virus-related HCC was investigated in an Indian population. The homozygous variant genotypes at codons 194 and 399 were protective for HCC risk (OR = 0.78 and 0.33, respectively). Arg/Trp and Arg/His heterozygosity at positions 194 and 399 increased risk to approximately twofold (OR = 2.27; 95% CI = 1.01–5.08; p < 0.001) or fivefold (OR = 4.95; 95% CI = 2.48–9.89; p < 0.001), respectively, for HCC, and twofold (OR = 2.25; 95% CI = 1.02–5.00; p < 0.05) and sixfold (OR = 6.27; 95% CI = 3.13–12.58), respectively, for viral hepatitis in healthy subjects. The Arg280His variant genotype had a twofold increased risk of HCC. Arg/Trp + Arg/His and Arg/Gln + Arg/His combinations were 35.96- and 5.28-fold more frequently associated with HCC, respectively. Gln/Gln at codon 399 was negatively associated with HCC. No association was observed for the risk of HCC and chronic viral hepatitis B and C.105
Gall bladder tract cancers
The gallbladder is susceptible to ROS-induced DNA damage, lipid peroxidation and biliary stasis. Since XRCC1 and OGG1 play important roles in the repair of oxidative DNA damage. Srivastava et al. investigated the role of OGG1 Ser326Cys and XRCC1 Arg399Gln gene polymorphisms in gallbladder cancer (GBC) susceptibility. The frequency of the variant OGG1 Cys/Cys genotype in female GBC patients was significantly higher than in controls (OR = 5.92; 95% CI = 1.20–29.13). However, this significance vanished when Ser/Cys + Cys/Cys genotypes were combined and compared with wild type. Disease risk due to the OGG1 Ser326Cys polymorphism in female GBC patients may be due to gallstone complications, which are more predominant in females, or may reflect the larger sample size of this group. In GBC, the Cys allele may be recessive, so that homozygote carriers could potentially have increased risk of the disease.44 The frequency of XRCC1 Arg/Gln and Gln/Gln genotypes for codon 399 was significantly different from controls (p = 0.039 and 0.003, respectively) and conferred low risk of GBC; a trend test was also significant (p = 0.001 for trend). Another study on biliary system cancer was performed by Huang et al. They found that XRCC1 Arg/Gln and OGG1 Cys/Cys genotypes were unrelated to GBC risk and suggested that the replication in future epidemiological studies is required to confirm the association of biliary tract cancer with the polymorphisms in OGG1 and XRCC1 genes.106
Brain tumors
XRCC1 Arg399Gln, Arg194Trp and Arg280His gene polymorphisms were not significantly associated with the risk of brain tumors; however, homozygous Gln/Gln showed a nearly statistically significant association with risk of all brain tumor types (i.e. glioma, glioblastoma and meningioma; OR = 1.32; 95% CI = 0.97–1.81; OR = 1.48; 95% CI = 0.98–2.24 and OR = 1.34; 95% CI, 0.96–1.86, respectively). There was no significant difference in the risk of brain tumors between carriers and noncarriers of the variant XRCC1 alleles Trp194 and His280.107
Thyroid carcinoma
Thyroid cancer is the most prevalent endocrine malignancy, and the incidence rate has increased in recent decades. The relationship between Arg399Gln, Arg280His, Arg194Trp in XRCC1 or Asp148Glu in APE1 and the risk of differentiated thyroid carcinoma and regional lymph node (LN) metastasis was examined. Only XRCC1 Arg194Trp was identified as having a possible association with the risk of thyroid cancer and LN, which may be increased by SNPs in other DNA repair genes.108
Discussion
DNA damage is generated by a variety of endogenous and exogenous chemical and physical agents. To prevent the potentially mutagenic or lethal consequences of DNA modifications, cells have evolved a series of complementary yet distinct DNA repair processes. The BER pathway copes with most spontaneous forms of DNA damage, including oxidative lesions such as base modifications, abasic sites and single-strand breaks. Not surprisingly, there is evidence that deficiencies in BER are associated with elevated risk of cancer, neurodegenerative disease and premature aging phenotypes. Thus, for the past decade or more, epidemiologists have determined the association of specific SNPs in BER genes with susceptibility to a range of cancer types, with or without a relevant exposure. We present in this review a summary of several key molecular epidemiology studies focused on the OGG1 Ser326Cys, APE1 Asp148Glu and XRCC1 Arg399Gln, Arg280His and Arg194Trp polymorphic variants. Based on the articles reviewed herein, we conclude the following.
No association was observed between XRCC1 Arg399Gln, Arg280His and Arg194Trp gene polymorphisms and colorectal cancer risk; however, OGG1 Ser326Cys and APE1 Asp148Glu increased colorectal cancer risk in some instances. Smoking and red meat consumption modifies the increased risk of colorectal cancer associated with the OGG1 Cys/Cys genotype. The gene–environment interaction between current smoking and APE1 (Glu/Glu and Asp/Glu + Glu/Glu) and XRCC1 (Arg/Trp) gene polymorphisms was statistically significant for colorectal cancer risk. Smoking is associated with significantly increased cancer risk in subjects with XRCC1 Arg399Gln genotype when combined with XRCC3 Thr241Thr and XPD 751Gln allele. The intake of meat and animal fat was consistently associated with the increased risk of large-bowel cancer. It was suggested that the abundant iron after red meat intake and inadequate activity of bile pigments to act as iron chelators would stimulate the Fenton reaction, which produces superoxide radicals. In addition, excessive fat intake increases the concentration of bile acids, which act as cytotoxic surfactants and can damage the colon.
Gene–environment interactions are much more important in the development of gastrointestinal cancer than colorectal cancer. While no association was reported between BER gene polymorphisms (i.e. OGG1 Ser326Cys, APE1 Asp148Glu and XRCC1 Arg399Gln, Arg194Trp and Arg280His) and gastrointestinal cancer, when adjusted for smoking status, alcohol and red meat, the interaction of genotypes with lifestyle factors (such as environmental risks, dietary habits and cigarette smoking) significantly increased the risk of development of gastrointestinal cancer. However, there are conflicting results on the association between gastrointestinal cancer and these gene polymorphisms due to the differences in the study population or cancer location.
The effect of OGG1 polymorphism on lung cancer risk differs in Caucasian and Asian individuals. OGG1 polymorphism has a significant impact on lung cancer risk in Caucasians, yet not in Asians. OGG1 Ser/Cys polymorphism is protective in smokers in a few studies, but not in all studies performed to date. One possible explanation for the protective effect is that the cells carrying the protective allele incur more DNA damage and undergo apoptosis more frequently than cells with the nonprotective allele. Some XRCC1 gene polymorphisms have a protective effect in nonsmokers in comparison with smokers.
Bladder and lung cancers are smoking-related cancers. Strong associations were reported between OGG1 Cys/Cys genotype and bladder cancer in smoker subjects. Conversely, no association was reported with nonsmokers and the APE1 Asp148Glu gene polymorphism. These conclusions are, however, based on a limited number of studies.
OGG1 Ser326Cys polymorphism and breast cancer risk had an inconsistent relationship. The risk of breast cancer was increased when the XRCC1 Arg/Gln and Arg/Trp genotypes were combined with BRCA1. Similarly, the risk of gynecological cancer increased with the XRCC1 Arg399Gln gene polymorphism. Notably, the codon 399 variant of XRCC1 lies near the BRCT-1 domain, a region that facilitates a biologically relevant interaction with PARP-1 with extensive homology to BRCA1.109 The conclusions regarding breast cancer risk and BER polymorphisms are also based on a limited number of studies.
UADC is another smoking related cancer type. Thus, smoking interacts strongly the risk of UADC in individuals with OGG1 Ser326Cys and XRCC1 Arg399Gln or Arg194Trp polymorphism.
For skin cancer and other cancers (Table 8), the most studied polymorphisms are XRCC1 Arg399Gln, Arg194Trp and Arg280His. A two-to fourfold increased cancer risk has been observed with these polymorphisms and each of these cancer types. Cancer subtypes are important and modulate the risk of cancer. For example, while Arg/Trp genotype reduces the risk of SCC, Arg/Gln genotype increases the risk of BCC. For GBC, a strong association with OGG1 Cys/Cys in females was reported. However, this association disappeared when genotypes were combined (Ser/Cys + Cys/Cys).
It still remains somewhat unclear whether the polymorphisms in OGG1 Ser326Cys, APE1 Asp148Glu or XRCC1 Arg399Gln, Arg280His and Arg194Trp directly or indirectly affect either protein levels or activities, which would be instrumental in modifying risk of different cancer types37 (see further discussion in the study by Wilson et al.37). However, while we advocate prospective examination of the associations between polymorphisms and cancer risk, it is always difficult to unambiguously incorporate environmental risk factor data (smoking, drinking, diet, etc.) into gene polymorphism–cancer association analysis due to uncertainties about the past. Thus, in some studies, alternative definitions for the environmental risk factors were used to try to resolve this problem. Nevertheless, simple and common study designs to evaluate gene polymorphism–cancer association have been the case-control approach. This method is easier and cheaper than other alternative protocols, although it may cause substantial bias and spurious associations as most required expectations cannot be met. Also, case-control studies on genetically homogenous or heterogynous populations may modify the results of the analysis.
Epidemiological investigations of DNA repair polymorphisms must be adequately designed with respect to sample size and the selection of polymorphisms and due to the small sample size of most of the studies to date, inconsistent results can be expected. Only six studies considered in this review had a sample size more than 1000,12,33,46,62,84 and these studies would be expected to give more accurate results with greater statistical power. In general, a large sample size reduces false-positive and false-negative results and provides more reliable data. It can, however, be very difficult to enroll enough patients for comprehensive association studies. Moreover, it is difficult to carry out multicentric hospital- and population-based studies in some countries. Furthermore, in most developing countries, follow-up studies and use of survey instruments are difficult. This leads to a small sample size in studies that are performed in developing countries. For example, in Turkey, it is very hard to get enough samples to carry out Genome Wide Association Studies (GWAS) or to obtain good statistics showing cancer frequencies.
In addition to the rationale, targeted SNP association studies described herein, GWAS are an important approach for discovering genetic variants influencing cancer risk. GWAS permits interrogation of the entire human genome at levels of resolution previously unattainable, in thousands of unrelated individuals, while many other approaches are biased by specific hypotheses regarding disease mechanism. On the other hand, to rule out false positive GWAS results, statistical analyses must be held to very high stringency and study replication is essential to account for biased representation in the study population and/or genotyping errors. For this reason, meta-analyses may be more valuable. However, meta-analyses also has limitations, such as (a) eligibility criteria for inclusion of subjects and sources of controls may differ among studies; (b) it is difficult to rule out incipient cancer in the controls and (c) gene–gene and gene–environment interactions may not be addressed because of the lack of sufficient data. Another weak aspect of GWAS is that many polymorphisms have different distributions and frequencies in different populations or geographic regions and consequently, association studies in mixed populations may produce false positive results. Confounding factors such as diet, smoking status, hormonal status and BMI should be considered in all GWAS studies. The selection of gene variants is also important and should consider functional outcome and phenotypic effect. Gene–gene interactions and their synergistic or additive effects should be considered as well as cancer type, stage or grade, patient ethnicity and population structure. DNA repair activity, when considered, should be quantified using the most accurate available methods, and/or multiple methods, where possible. mRNA or protein levels can also be used for additional evidence of DNA repair capacity.
Conclusion
In conclusion, to fully understand the role of DNA damage and DNA repair in the carcinogenesis, prospective studies are needed and additional genes should be analyzed. Such studies should investigate mRNA expression, DNA damage levels and DNA repair capacity and should consider gene–environment and gene–gene interactions.
Acknowledgments
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Dizdaroglu M, Olinski R, Doroshow JR, Akman SA. Modification of DNA bases in chromatin of intact target human cells by activated human polymorphonuclear leukocytes. Cancer Res. 1993;53:1269–1272. [PubMed] [Google Scholar]
- 4.Kang SY, Lee KG, Lee W, Shim JY, Ji SI, Chung KW, et al. Polymorphisms in the DNA repair gene XRCC1 associated with basal cell carcinoma and squamous cell carcinoma of the skin in a Korean population. Cancer Sci. 2007;98:716–712. doi: 10.1111/j.1349-7006.2007.00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halliwell B, Gutteridge JM. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 6.Hong WG, Bast RC, Hait WN, Kufe DW, Pollock RE, Weichselbaum RR, et al. Inflamation and Cancer. 8. Chapter 19. Connecticut, USA: People’s Medical Publishing House (PMPH); 2010. [Google Scholar]
- 7.Sugimura H, Kohno T, Wakai K, Nagura K, Genka K, Igarashi H, et al. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1999;8:669–674. [PubMed] [Google Scholar]
- 8.Li M, Yin Z, Guan P, Li X, Cui Z, Zhang J, et al. XRCC1 polymorphisms, cooking oil fume and lung cancer in Chinese women nonsmokers. Lung Cancer. 2008;62:145–151. doi: 10.1016/j.lungcan.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Hatt L, Loft S, Risom L, Møller P, Sørensen M, Raaschou-Nielsen O, et al. OGG1 expression and OGG1 Ser326Cys polymorphism and risk of lung cancer in a prospective study. Mutat Res. 2008;639:45–54. doi: 10.1016/j.mrfmmm.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Wogan GN, Hecht SS, Felton JS, Conney AH, Loebe LA. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14:473–486. doi: 10.1016/j.semcancer.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Thomas D. Gene–environment-wide association studies: emerging approaches, nature reviews. Genetics. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker RR, Massey ED, Smith G. An overview of the effects of tobacco ingredients on smoke chemistry and toxicity. Food Chem Toxicol. 2004;42:S53–S83. doi: 10.1016/j.fct.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Liang J, Jiang T, Yao RY, Liu ZM, Lv HY, Qi WW. The combination of ERCC1 and XRCC1 gene polymorphisms better predicts clinical outcome to oxaliplatin-based chemotherapy in metastatic colorectal cancer. Cancer Chemother Pharmacol. 2010;66:493–500. doi: 10.1007/s00280-009-1186-3. [DOI] [PubMed] [Google Scholar]
- 14.Taioli E. Gene-environment interaction in tobacco-related cancers. Carcinogenesis. 2008;29:1467–1474. doi: 10.1093/carcin/bgn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu NY, Wang HC, Wang CH, Chang CL, Chiu CF, Lee HZ, et al. Lung cancer susceptibility and genetic polymorphism of DNA repair gene XRCC4 in Taiwan. Cancer Biomar. 2009;5:159–165. doi: 10.3233/CBM-2009-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agaçhan B, Küçükhüseyin O, Aksoy P, Turna A, Yaylim I, Görmüs U, et al. Apurinic/apyrimidinic endonuclease (APE1) gene polymorphisms and lung cancer risk in relation to tobacco smoking. Anticancer Res. 2009;29:2417–2420. [PubMed] [Google Scholar]
- 17.Hu Z, Ma H, Lu D, Zhou J, Chen Y, Xu L, et al. A promoter polymorphism (−77T>C) of DNA repair gene XRCC1 is associated with risk of lung cancer in relation to tobacco smoking. Pharmacogenet Genomics. 2005;15:457–463. doi: 10.1097/01.fpc.0000167329.85163.0d. [DOI] [PubMed] [Google Scholar]
- 18.Le Marchand L, Donlon T, Lum-Jones A, Seifried A, Wilkens LR. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:409–412. [PubMed] [Google Scholar]
- 19.Miyaishi A, Osawa K, Osawa Y, Inoue N, Yoshida K, Kasahara M, et al. MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J Exp Clin Cancer Res. 2009;28:10–15. doi: 10.1186/1756-9966-28-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arizono K, Osada Y, Kuroda Y. DNA repair gene hOGG1 codon 326 and XRCC1 codon 399 polymorphisms and bladder cancer risk in a Japanese population. Jpn J Clin Oncol. 2008;38:186–191. doi: 10.1093/jjco/hym176. [DOI] [PubMed] [Google Scholar]
- 21.Pachouri SS, Sobti RC, Kaur P, Singh J. Contrasting impact of DNA repair gene XRCC1 polymorphisms Arg399Gln and Arg194Trp on the risk of lung cancer in the north-Indian population. DNA Cell Biol. 2007;26:186–191. doi: 10.1089/dna.2006.9999. [DOI] [PubMed] [Google Scholar]
- 22.Karihtala P, Soini Y. Reactive oxygen species and antioxidant mechanisms in human tissues and their relation to malignancies. APMIS. 2007;115:81–103. doi: 10.1111/j.1600-0463.2007.apm_514.x. [DOI] [PubMed] [Google Scholar]
- 23.Halliwell B. Biochemistry of oxidative stress. Biochem Soc Transact. 2007;35:1147–1150. doi: 10.1042/BST0351147. [DOI] [PubMed] [Google Scholar]
- 24.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basso D, Navaglia F, Fogar P, Zambon CF, Greco E, Schiavon S, et al. DNA repair pathways and mitochondrial DNA mutations in gastrointestinal carcinogenesis. Clin Chim Acta. 2007;381:50–55. doi: 10.1016/j.cca.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 26.Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 27.Loft S, Poulsen HE. Cancer risk and oxidative DNA damage in man. J Mol Med. 1996;74:297–312. doi: 10.1007/BF00207507. [DOI] [PubMed] [Google Scholar]
- 28.Kwok PY, Gu Z. Single nucleotide polymorphism libraries:why and how are we building them? Mol Med Today. 1999;5:538–543. doi: 10.1016/s1357-4310(99)01601-9. [DOI] [PubMed] [Google Scholar]
- 29.Collins FS, Guyer MS, Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 30.Butkiewicz D, Rusin M, Enewold L, Shields PG, Chorazy M, Harris CC. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis. 2001;22:593–597. doi: 10.1093/carcin/22.4.593. [DOI] [PubMed] [Google Scholar]
- 31.Park HW, Kim IJ, Kang HC, Jang SG, Ahn SA, Lee JS. The hOGG1 Ser326Cys polymorphism is not associated with colorectal cancer. Risk J Epidemiol. 2007;17:156–160. doi: 10.2188/jea.17.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung RJ, Brennan P, Canzian F, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, et al. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. J Natl Cancer Inst. 2005;97:567–576. doi: 10.1093/jnci/dji101. [DOI] [PubMed] [Google Scholar]
- 33.Kohno T, Kunitoh H, Toyama K, Yamamoto S, Kuchiba A, Saito D, et al. Association of the OGG1–326Cys polymorphism with lung adenocarcinoma risk. Cancer Sci. 2006;97:724–728. doi: 10.1111/j.1349-7006.2006.00240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farinati F, Cardin R, Bortolami M, Nitti D, Basso D, de Bernard M, et al. Oxidative DNA damage in gastric cancer: CagA status and OGG1 gene polymorphism. Int J Cancer. 2008;123:51–55. doi: 10.1002/ijc.23473. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Elahi A, Pow-Sang J, Lazarus P, Park J. Association between polymorphism of human oxoguanine glycosylase 1 and risk of prostate cancer. J Urol. 2003;170:2471–2474. doi: 10.1097/01.ju.0000087498.23008.bb. [DOI] [PubMed] [Google Scholar]
- 36.Elahi A, Zheng Z, Park J, Eyring K, McCaffrey T, Lazarus P. The human OGG1 DNA repair enzyme and its association with orolaryngeal cancer risk. Carcinogenesis. 2002;23:1229–1234. doi: 10.1093/carcin/23.7.1229. [DOI] [PubMed] [Google Scholar]
- 37.Wilson DM, 3rd, Kim D, Berquist BR, Sigurdson AJ. Variation in base excision repair capacity. Mutat Res. 2011;711:100–112. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo YL, Jou YS, Hsiao CF, Chang GC, Tsai Y, Su WC, et al. A polymorphism in the APE1 gene promoter is associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:223–229. doi: 10.1158/1055-9965.EPI-08-0749. [DOI] [PubMed] [Google Scholar]
- 39.Au WW. Heritable susceptibility factors for the development of cancer. J Radiat Res (Tokyo) 2006;47B:13–17. doi: 10.1269/jrr.47.b13. [DOI] [PubMed] [Google Scholar]
- 40.Chen PL, Yeh KT, Tsai YY, Koeh H, Liu YL, Lee H, et al. XRCC1, but not APE1 and hOGG1 gene polymorphisms is a risk factor for pterygium. Mol Vis. 2010;16:991–996. [PMC free article] [PubMed] [Google Scholar]
- 41.Hadi MZ, Coleman MA, Fidelis K, Mohrenweiser HW, Wilson DM., 3rd Functional characterization of APE1 variants identified in the human population. Nucleic Acids Res. 2000;28:3871–3879. doi: 10.1093/nar/28.20.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Attar R, Cacina C, Sozen S, Attar E, Agachan B. DNA repair genes in endometriosis. Genet Mol Res. 2010;9:629–636. doi: 10.4238/vol9-2gmr779. [DOI] [PubMed] [Google Scholar]
- 43.Shim HJ, Yun JY, Hwang JE, Bae WK, Cho SH, Lee JH, et al. BRCA1 and XRCC1 polymorphisms associated with survival in advanced gastric cancer treated with taxane and cisplatin. Cancer Sci. 2010;101:1247–1254. doi: 10.1111/j.1349-7006.2010.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srivastava A, Srivastava K, Pandey SN, Choudhuri G, Mittal B. Single-nucleotide polymorphisms of DNA repair genes OGG1 and XRCC1: association with gallbladder cancer in North Indian. Ann Surg Oncol. 2009;16:1695–1703. doi: 10.1245/s10434-009-0354-3. [DOI] [PubMed] [Google Scholar]
- 45.Gu D, Wang M, Wang M, Zhang Z, Chen J. The DNA repair gene APE1 T1349G polymorphism and cancer risk: a meta-analysis of 27 case-control studies. Mutagenesis. 2009;24:507–512. doi: 10.1093/mutage/gep036. [DOI] [PubMed] [Google Scholar]
- 46.Curtin K, Samowitz WS, Wolff RK, Ulrich CM, Caan BJ, Potter JD, et al. Assessing tumor mutations to gain insight into base excision repair sequence polymorphisms and smoking in colon cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3384–3388. doi: 10.1158/1055-9965.EPI-09-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JI, Park YJ, Kim KH, Kim JI, Song BJ, Lee MS, et al. hOGG1 Ser326Cys polymorphism modifies the significance of the environmental risk factor for colon cancer. World J Gastroenterol. 2003;9:956–960. doi: 10.3748/wjg.v9.i5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen R, Saebø M, Skjelbred CF, Nexø BA, Hagen PC, Bock G, et al. IGPX Pro198Leu and OGG1 Ser326Cys polymorphisms and risk of development of colorectal adenomas and colorectal cancer. Cancer Lett. 2005;229:85–91. doi: 10.1016/j.canlet.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 49.Skjelbred CF, Saebø M, Wallin H, Nexø BA, Hagen PC, Lothe IM. Polymorphisms of the XRCC1, XRCC3 and XPD genes and risk of colorectal adenoma and carcinoma, in a Norwegian cohort: a case control study. BMC Cancer. 2006;6:67. doi: 10.1186/1471-2407-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–2108. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- 51.Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, et al. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008;638:146–153. doi: 10.1016/j.mrfmmm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Kasahara M, Osawa K, Yoshida K, Miyaishi A, Osawa Y, Inoue N, et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J Exp Clin Cancer Res. 2008;27:49–58. doi: 10.1186/1756-9966-27-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stern MC, Conti DV, Siegmund KD, Corral R, Yuan JM, Koh WP, et al. DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese Health Study. Cancer Epidemiol Biomarkers Prev. 2007;16:2363–2372. doi: 10.1158/1055-9965.EPI-07-0268. [DOI] [PubMed] [Google Scholar]
- 54.Engin AB, Karahalil B, Engin A, Karakaya AE. Oxidative stress, Helicobacter pylori, and OGG1 Ser326Cys, XPC Lys939Gln, and XPD Lys751Gln polymorphisms in a Turkish population with colorectal carcinoma. Genet Test Mol Biomarkers. 2010;14:559–564. doi: 10.1089/gtmb.2009.0195. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Zhao Y, Jiang J, Gajalakshmi V, Kuriki K, Nakamura S, et al. Polymorphisms in DNA repair genes XRCC1, XRCC3 and XPD, and colorectal cancer risk: a case-control study in an Indian population. J Cancer Res Clin Oncol. 2010;136:1517–1525. doi: 10.1007/s00432-010-0809-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang B, Wang D, Huang G, Zhang C, Xu DH, Zhou W. XRCC1 polymorphisms and risk of colorectal cancer: a meta-analysis. Int J Colorectal Dis. 2010;25:313–321. doi: 10.1007/s00384-009-0866-0. [DOI] [PubMed] [Google Scholar]
- 57.Peluso M, Hainaut P, Airoldi L, Autrup H, Dunning A, Garte S, et al. Methodology of laboratory measurements in prospective studies on gene-environment interactions: the experience of GenAir. Mutat Res. 2005;574:92–104. doi: 10.1016/j.mrfmmm.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 58.Feyler A, Voho A, Bouchardy C, Kuokkanen K, Dayer P, Hirvonen A, et al. Point: myeloperoxidase -463G–>A polymorphism and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1550–1554. [PubMed] [Google Scholar]
- 59.Popanda O, Schattenberg T, Phong CT, Butkiewicz D, Risch A, Edler L, et al. Specific combinations of DNA repair gene variants and increased risk for non-small cell lung cancer. Carcinogenesis. 2004;25:2433–2441. doi: 10.1093/carcin/bgh264. [DOI] [PubMed] [Google Scholar]
- 60.Vogel U, Olsen A, Wallin H, Overvad K, Tjønneland A, et al. No association between OGG1 Ser326Cys and risk of basal cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2004;13:1680–1681. [PubMed] [Google Scholar]
- 61.Karahalil B, Emerce E, Koçer B, Han S, Alkiş N, Karakaya AE. The association of OGG1 Ser326Cys polymorphism and urinary 8-OHdG levels with lung cancer susceptibility: a hospital-based case-control study in Turkey. Arh Hig Rada Toksikol. 2008;59:241–250. doi: 10.2478/10004-1254-59-2008-1924. [DOI] [PubMed] [Google Scholar]
- 62.Hung RJ, Christiani DC, Risch A, Popanda O, Haugen A, Zienolddiny S, et al. International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol Biomarkers Prev. 2008;17:3081–3089. doi: 10.1158/1055-9965.EPI-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;527:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 64.Huang M, Dinney CP, Lin X, Lin J, Grossman HB, Wu X. High-order interactions among genetic variants in DNA base excision repair pathway genes and smoking in bladder cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2007;16:84–91. doi: 10.1158/1055-9965.EPI-06-0712. [DOI] [PubMed] [Google Scholar]
- 65.Yin J, Vogel U, Ma Y, Qi R, Wang H. Association of DNA repair gene XRCC1 and lung cancer susceptibility among nonsmoking Chinese women. Cancer Genet Cytogenet. 2009;188:26–31. doi: 10.1016/j.cancergencyto.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 66.De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K, et al. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631:101–110. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 67.Yin J, Vogel U, Ma Y, Qi R, Sun Z, Wang H. The DNA repair gene XRCC1 and genetic susceptibility of lung cancer in a northeastern Chinese population. Lung Cancer. 2007;56:153–160. doi: 10.1016/j.lungcan.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Bravard A, Vacher M, Moritz E, Vaslin L, Hall J, Epe B, et al. Oxidation status of human OGG1-S326C polymorphic variant determines cellular DNA repair capacity. Cancer Res. 2009;69:3642–3649. doi: 10.1158/0008-5472.CAN-08-3943. [DOI] [PubMed] [Google Scholar]
- 69.Aka P, Mateuca R, Buchet JP, Thierens H, Kirsch-Volders M. Are genetic polymorphisms in OGG1, XRCC1 and XRCC3 genes predictive for the DNA strand break repair phenotype and genotoxicity in workers exposed to low dose ionising radiations? Mutat Res. 2004;556:169–181. doi: 10.1016/j.mrfmmm.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 70.Parkin DM, Bray F, Ferlay J, Pisani P. Global Cancer Statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 71. [accessed 12 June 2010]; http://globocan.iarc.fr/factsheets/populations/factsheet.asp?uno=968#KEY.
- 72.Goto M, Shinmura K, Yamada H, Tsuneyoshi T, Sugimura H. OGG1, MYH and MTH1 gene variants identified in gastric cancer patients exhibiting both 8-hydroxy-2′-deoxyguanosine accumulation and low inflammatory cell infiltration in their gastric mucosa. J Genet. 2008;7:181–186. doi: 10.1007/s12041-008-0028-0. [DOI] [PubMed] [Google Scholar]
- 73.Li WQ, Zhang L, Ma JL, Zhang Y, Li JY, Pan KF, et al. Association between genetic polymorphisms of DNA base excision repair genes and evolution of precancerous gastric lesions in a Chinese population. Carcinogenesis. 2009;30:500–505. doi: 10.1093/carcin/bgp018. [DOI] [PubMed] [Google Scholar]
- 74.Lee SG, Kim B, Choi J, Kim C, Lee I, Song K. Genetic polymorphisms of XRCC1 and risk of gastric cancer. Cancer Lett. 2002;187:53–60. doi: 10.1016/s0304-3835(02)00381-6. [DOI] [PubMed] [Google Scholar]
- 75.Malik MA, Zargar SA, Mittal B. Lack of influence of DNA repair gene OGG1 codon 326 polymorphisms of gastric cancer risk in the Kashmir valley. Res Comm Asian Pacific J Cancer Prev. 2010;11:165–168. [PubMed] [Google Scholar]
- 76.Yan L, Yanan D, Donglan S, Na W, Rongmiao Z, Zhifeng C. Polymorphisms of XRCC1 gene and risk of gastric cardiac adenocarcinomades. Dis Esophagus. 2009;22:396–401. doi: 10.1111/j.1442-2050.2008.00912.x. [DOI] [PubMed] [Google Scholar]
- 77.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM., III XRCC1 colocalizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan T, Deng S, Chen M, Chen W, Lu W, Huang H, et al. Association of DNA repairgene XRCC1 and XPD polymorphisms with genetic susceptibility to gastric cancer in a Chinese population. Cancer Epidemiol. 2011;35:170–174. doi: 10.1016/j.canep.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 79.Palli D, Polidoro S, D’Errico M, Saieva C, Guarrera S, Calcagnile AS, et al. Polymorphic DNA repair and metabolic genes: a multigenic study on gastric cancer. Mutagenesis. 2010;25:569–575. doi: 10.1093/mutage/geq042. [DOI] [PubMed] [Google Scholar]
- 80.Karahalil B, Kocabas NA, Ozçelik T. DNA repair gene polymorphisms and bladder cancer susceptibility in a Turkish population. Anticancer Res. 2006;26:4955–498. [PubMed] [Google Scholar]
- 81.Ali MF, Meza JL, Rogan EG, Chakravarti D. Prevalence of BER gene polymorphisms in sporadic breast cancer. Oncol Rep. 2008;19:1033–1038. [PubMed] [Google Scholar]
- 82.Sigurdson AJ, Hauptmann M, Chatterjee N, Alexander BH, Doody MM, Rutter JL, et al. Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes. BMC Cancer. 2004;4:9–20. doi: 10.1186/1471-2407-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132–2138. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Newcomb PA, Egan KM, Titus-Ernstoff L, Chanock S, Welch R, et al. Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:353–358. doi: 10.1158/1055-9965.EPI-05-0653. [DOI] [PubMed] [Google Scholar]
- 85.Vogel U, Nexø BA, Olsen A, Thomsen B, Jacobsen NR, Wallin H, et al. No association between OGG1 Ser326Cys polymorphism and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:170–171. [PubMed] [Google Scholar]
- 86.Sangrajrang S, Schmezer P, Burkholder I, Waas P, Boffetta P, Brennan P, et al. Polymorphisms in three base excision repair genes and breast cancer risk in Thai women. Breast Cancer Res Treat. 2008;111:279–288. doi: 10.1007/s10549-007-9773-7. [DOI] [PubMed] [Google Scholar]
- 87.Yuan W, Xu L, Feng Y, Yang Y, Chen W, Wang J, et al. The hOGG1 Ser326Cys polymorphism and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;122:835–842. doi: 10.1007/s10549-009-0722-5. [DOI] [PubMed] [Google Scholar]
- 88.Niwa Y, Matsuo K, Ito H, Hirose K, Tajima K, Nakanishi T, et al. Association of XRCC1 Arg399Gln and OGG1 Ser326Cys polymorphisms with the risk of cervical cancer in Japanese subjects. Gynecol Oncol. 2005;99:43–49. doi: 10.1016/j.ygyno.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 89.Yang Y, Zhai XD, Gao LB, Li SL, Wang Z, Chen GD. Genetic polymorphisms of DNA repair gene XRCC1 and risk of uterine leiomyoma. Mol Cell Biochem. 2010;338:143–147. doi: 10.1007/s11010-009-0347-3. [DOI] [PubMed] [Google Scholar]
- 90.Hashimoto T, Uchida K, Okayama N, Imate Y, Suehiro Y, Hamanaka Y, et al. Interaction of OGG1 Ser326Cys polymorphism with cigarette smoking in head and neck squamous cell carcinoma. Mol Carcinogen. 2006;45:344–348. doi: 10.1002/mc.20140. [DOI] [PubMed] [Google Scholar]
- 91.Ramachandran S, Ramadas K, Hariharan R, Rejnish R, Kumar M, Pillai R. Single nucleotide polymorphisms of DNA repair genes XRCC1 and XPD and its molecular mapping in Indian oral cancer. Oral Oncol. 2006;2:350–362. doi: 10.1016/j.oraloncology.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 92.Tae K, Lee HS, Park BJ, Park CW, Kim KR, Cho HY, et al. Association of DNA repair gene XRCC1 polymorphisms with head and neck cancer in Korean population. Int J Cancer. 2004;111:805–808. doi: 10.1002/ijc.20338. [DOI] [PubMed] [Google Scholar]
- 93.Kowalski M, Przybylowska K, Rusin P, Olszewski J, Morawiec-Sztandera A, Bielecka-Kowalska A, et al. Genetic polymorphisms in DNA base excision repair gene XRCC1 and the risk of squamous cell carcinoma of the head and neck. J Exp Clin Cancer Res. 2009;28:37. doi: 10.1186/1756-9966-28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li C, Hu Z, Lu J, Liu Z, Wang L, El-Naggar AK, et al. Genetic polymorphisms in DNA base-excision repair genes ADPRT, XRCC1, and APE1 and the risk of squamous cell carcinoma of the head and neck. Cancer. 2007;110:867–875. doi: 10.1002/cncr.22861. [DOI] [PubMed] [Google Scholar]
- 95.Lee KG, Lee W, Shim JY, Ji SI, Chung KW, Chung YK, et al. Polymorphisms in the DNA repair gene XRCC1 associated with basal cell carcinoma and squamous cell carcinoma of the skin in a Korean population. Cancer Sci. 2007;98:716–720. doi: 10.1111/j.1349-7006.2007.00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stanczyk M, Sliwinski T, Cuchra M, Zubowska M, Bielecka-Kowalska A, Kowalski M, et al. The association of polymorphisms in DNA base excision repair genes XRCC1, OGG1 and MUTYH with the risk of childhood acute lymphoblastic leukemia. Mol Biol Rep. 2001;38:445–451. doi: 10.1007/s11033-010-0127-x. [DOI] [PubMed] [Google Scholar]
- 97.Meza-Espinoza JP, Peralta-Leal V, Gutierrez-Angulo M, Macias-Gomez N, Ayala-Madrigal ML, Barros-Nuñez P, et al. XRCC1 polymorphisms and haplotypes in Mexican patients with acute lymphoblastic leukemia. Genet Mol Res. 2009;8:1451–1458. doi: 10.4238/vol8-4gmr687. [DOI] [PubMed] [Google Scholar]
- 98.Batar B, Guven M, Baris S, Celkan T. DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res. 2009;33:759–763. doi: 10.1016/j.leukres.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 99.Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, Garte S, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort stud. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 100.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 101.Xu Z, Hua LX, Qian LX, Yang J, Wang XR, Zhang W, et al. Relationship between XRCC1 polymorphisms and susceptibility to prostate cancer in men from Han, Southern China. Asian J Androl. 2007;9:331–338. doi: 10.1111/j.1745-7262.2007.00263.x. [DOI] [PubMed] [Google Scholar]
- 102.Han X, Li Y, Huang J, Zhang Y, Holford T, Lan Q, et al. Identification of predictive pathways for non-hodgkin lymphoma prognosis. Cancer Inform. 2010;9:281–292. doi: 10.4137/CIN.S6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shen M, Purdue MP, Kricker A, Lan Q, Grulich AE, Vajdic CM, et al. Polymorphisms in DNA repair genes and risk of non-Hodgkin’s lymphoma in New South Wales, Australia. Haematologica. 2007;92:1180–1185. doi: 10.3324/haematol.11324. [DOI] [PubMed] [Google Scholar]
- 104.Liu J, Song B, Wang Z, Song X, Shi Y, Zheng J, et al. DNA repair gene XRCC1 polymorphisms and non-Hodgkin lymphoma risk in a Chinese population. Cancer Genet Cytogenet. 2009;191:67–72. doi: 10.1016/j.cancergencyto.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 105.Kiran M, Saxena R, Chawla YK, Kaur J. Polymorphism of DNA repair gene XRCC1 and hepatitis-related hepatocellular carcinoma risk in Indian population. Mol Cell Biochem. 2009;327:7–13. doi: 10.1007/s11010-009-0035-3. [DOI] [PubMed] [Google Scholar]
- 106.Huang WY, Gao YT, Rashid A, Sakoda LC, Deng J, Shen MC, et al. Selected base excision repair gene polymorphisms and susceptibility to biliary tract cancer and biliary stones: a population-based case–control study in China. Carcinogenesis. 2008;29:100–105. doi: 10.1093/carcin/bgm247. [DOI] [PubMed] [Google Scholar]
- 107.Kiuru A, Lindholm C, Heinävaara S, Ilus T, Jokinen P, Haapasalo H, et al. XRCC1 and XRCC3 variants and risk of glioma and meningioma. J Neurooncol. 2008;88:135–142. doi: 10.1007/s11060-008-9556-y. [DOI] [PubMed] [Google Scholar]
- 108.Chiang FY, Wu CW, Hsiao PJ, Kuo WR, Lee KW, Lin JC, et al. Association between polymorphisms in DNA base excision repair genes XRCC1, APE1, and ADPRT and differentiated thyroid carcinoma. Clin Cancer Res. 2008;14:5919–5924. doi: 10.1158/1078-0432.CCR-08-0906. [DOI] [PubMed] [Google Scholar]
- 109.Chiang CC, Tsai YY, Bau DT, Cheng YW, Tseng SH, Wang RF, et al. Pterygium and genetic polymorphisms of the DNA repair enzymes XRCC1, XPA, and XPD. Mol Vis. 2010;16:698–704. [PMC free article] [PubMed] [Google Scholar]