Abstract
Oxidative DNA damage has been attributed to increased cancer incidence and premature aging phenotypes. Reactive oxygen species (ROS) are unavoidable byproducts of oxidative phosphorylation and are the major contributors of endogenous oxidative damage. To prevent the negative effects of ROS, cells have developed DNA repair mechanisms designed to specifically combat endogenous DNA modifications. The base excision repair (BER) pathway is primarily responsible for the repair of small non-helix distorting lesions and DNA single strand breaks. This repair pathway is found in all organisms, and in mammalian cells, consists of three related sub-pathways: short patch (SP-BER), long patch (LP-BER) and single strand break repair (SSBR). While much is known about nuclear BER, comparatively little is known about this pathway in the mitochondria, particularly the LP-BER and SSBR sub-pathways. There are a number of proteins that have recently been found to be involved in mitochondrial BER, including Cockayne syndrome proteins A and B (CSA and CSB), aprataxin (APTX), tryosyl-DNA phosphodiesterase 1 (TDP1), flap endonuclease 1 (FEN-1) and exonuclease G (EXOG). These significant advances in mitochondrial DNA repair may open new avenues in the management and treatment of a number of neurological disorders associated with mitochondrial dysfunction, and will be reviewed in further detail herein.
Keywords: DNA repair, Mitochondria, Aging, Oxidative DNA damage
1. Introduction
Constant genomic maintenance is essential for both the viability and longevity of an organism. Genomic instability can result in a variety of deleterious outcomes, including cancer and accelerated aging. At the cellular level, persistent DNA damage, caused by exposure to genotoxic agents or by inhibition of DNA repair, can lead to increased mutation and is a well-established trigger for apoptosis. Accumulated research over the last four decades has given us considerable insight into the numerous DNA repair mechanisms available to cells. However, the vast majority of previous DNA repair research has focused on maintenance of the nuclear genome, driven primarily by the field of cancer research. In mammalian cells, the nucleus is one of two organelles that have DNA. The other, the mitochondrion, has a 16.5 kb circular genome that encodes for many of the components required for oxidative phosphorylation. The two organelles have their genomes organized in very different ways. In nuclear DNA (nDNA), there is only a single copy of the entire 46 chromosome genome, of which approximately 93% of the sequence is non-coding. In comparison, the mitochondrion has multiple copies of mitochondrial DNA (mtDNA) per organelle, where virtually all the sequence is coding (Chen and Butow, 2005). Depending on energy requirements, each cell may contain up to 100 mitochondria and hence thousands of copies of mtDNA.
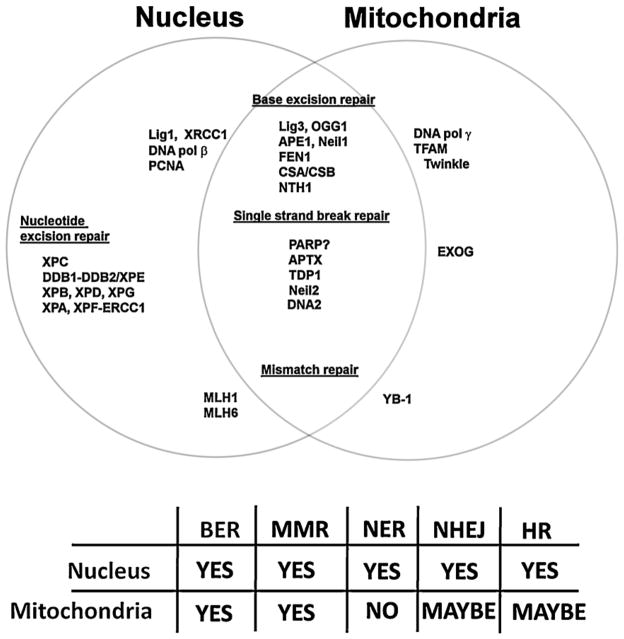
Despite each mitochondrion containing many copies of mtDNA, the mitochondrial genome does not encode any DNA repair proteins. Instead, the mitochondria rely solely on the nucleus for their entire DNA repair capability. As depicted in Fig. 1, many of the DNA repair proteins used by the mitochondria are also present in the nucleus, an aspect which makes delineating the effect of protein dysfunction considerably more difficult. The nucleus also encodes proteins involved strictly in mtDNA repair, with no direct nuclear homolog, such as DNA polymerase γ (Polγ) and the DNA helicase twinkle (Fig. 1); these proteins also play central roles in mtDNA replication, and as such, are not classified as primarily mtDNA repair enzymes and are outside the primary focus of this review. Mitochondria do not contain the full set of DNA repair strategies present in the nucleus, with nucleotide excision repair (NER) and its associated sub-pathways missing (Fig. 1) (Clayton et al., 1974; LeDoux et al., 1992). Also, either entirely absent or heavily attenuated are the double stranded break repair pathways, both homologous recombination and non-homologous end joining (Ashkenas, 1997; Cullinane and Bohr, 1998; Thyagarajan et al., 1996). Why mitochondria do not possess these more complex repair mechanisms is still open for speculation. The organelle may encounter less of the adducts processed by these mechanisms, or alternatively, the high energy demand and complicated mechanics of these processes may make DNA repair not essential for the mitochondria, since it has access to multiple copies of its genomes. Conversely, mitochondria do have an effective DNA base excision repair (BER) pathway, originally reported to be only a skeleton of the nuclear pathway, but now known to be comprised of the short-patch (SP-BER), long-patch (LP-BER) and single strand break repair (SSBR) sub-pathways that operate in the nucleus.
Fig. 1.
Nuclear and mitochondrial DNA repair shares common proteins. However not all nuclear DNA repair pathways are present in the mitochondria. However the BER pathway has a high degree of conservation between the two organelles, with most of the mitochondrial BER proteins, isoforms of nuclear relatives. Abbreviations are as follows: base excision repair (BER), miss match repair (MMR), nucleotide excision repair (NER), non-homologous end joining (NHEJ) and homologous recombination (HR).
A functional BER system is a requirement for viability. This pathway has a high degree of evolutionary conservation and appears to exist in all organisms regardless of complexity. Both the requirement for viability and the remarkable pathway conservation can no doubt be attributed to the spectrum of lesions that this pathway resolves. The base excision system is primarily responsible for the repair of non-helix distorting lesions, including oxidized, alkylated and deaminated bases, as well as base mismatches arising from mis-incorporation. The BER pathway is central in the repair of endogenous lesions, including those that occur as an inevitable byproduct of respiration [recently reviewed (Svilar et al., 2011)]. Oxidative phosphorylation occurs in the mitochondria, and as such, mtDNA is in close proximity to reactive oxygen species (ROS), and consequently is heavily bombarded with endogenous oxidative lesions (Hudson et al., 1998). While ROS have the ability to directly damage DNA bases, they can also cause and contribute to single strand DNA breaks (SSBs), believed to be the most common form of DNA damage. Apart from SSBs occurring due to oxidative attack, they are also an intermediate formed during BER. Unrepaired SSBs can cause replication and transcription stalling, increasing the potential of these lesions to be converted into the significantly more genotoxic double stranded DNA breaks (DSB) (McKinnon and Caldecott, 2007). In both the nucleus and the mitochondria, SSBs are repaired by the BER sub-pathway SSBR. The related BER and SSBR sub-pathways share common components, particularly in the later stages. As seen in Fig. 2, the combined SSBR/BER pathway generally involves four distinct events: lesion detection, lesion processing, DNA synthesis and finally ligation with the number of enzymes recruited being dependent on the type of lesion to be repaired. Advances have recently been made in our understanding of mtBER, particularly mtSSBR, and some of the significant updates are discussed below.
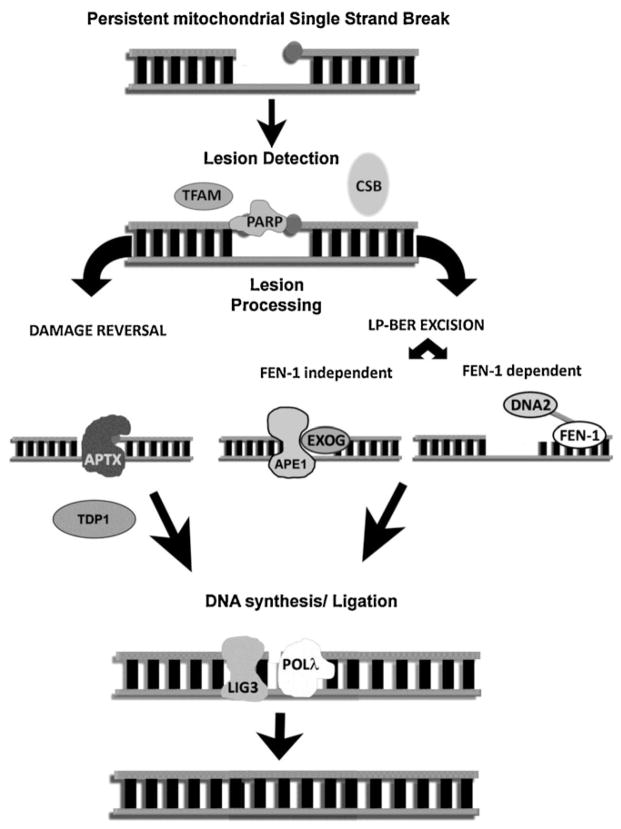
Fig. 2.
Resolution of persistent mitochondrial single strand breaks. A strand break with blocking groups at one or both exposed ends is unable to be repaired by simple SP-BER. The detection of these lesions may rely on more than one enzyme and potentially involves PARP, CSB or TFAM. The detection method may influence the type of lesion processing, either damage reversal, if a specific enzyme such as APTX or TDP1 are able to resolve the lesion, or damage excision, a more generic pathway that excises the DNA damage using the LP-BER pathway. In the mitochondria LP-BER dependant multi nucleotide excision appears to be driven by EXOG as part of an APE1 complex however the presence of FEN-1 and DNA2 in the mitochondria suggests a secondary LP-BER pathway, potentially to excise lesions resistant to EXOG activity. After the lesion has been resolved nucleotides are inserted by polymerase γ and the phosphodiester backbone re-ligated by ligase 3.
2. Lesion detection
In the nucleus, the resolution of SSBs begins with the detection of the lesion by the poly ADP ribose polymerase (PARP) family of proteins. After PARP encounters a strand break, it begins the synthesis of poly ADP ribose (PAR) chains, a signal for the recruitment of DNA repair proteins. In comparison, little is known about how mitochondria detect SSBs. There have been a number of reports demonstrating the occurrence of mitochondrial poly(ADP-ribosyl)ation of proteins (Du et al., 2003; Lai et al., 2008; Pankotai et al., 2009). In addition, in support of mitochondrial protein ribosylation, there are new reports of a poly ADP ribose glycohydrolase (PARG) isoform, an enzyme required to breakdown PAR polymers back to ADP-ribose units, that localizes exclusively to the mitochondria (Du et al., 2003; Mortusewicz et al., 2011; Niere et al., 2008; Whatcott et al., 2009). From these reports, it would seem reasonable to conclude that there is a mitochondrial isoform of PARP, which is able to detect SSBs and begin a repair cascade, mirroring the process in the nucleus. Consistently, numerous groups have published studies on PARP localization over the past twenty years, with evidence that mitochondria does indeed have an active PARP, including the latest from Rossi et al. (2009). This report showed that PARP-1 interacted with both the mitochondrial inner membrane protein mitofilin and DNA ligase 3, and was required to maintain mtDNA integrity. However, there have been an equal number of reports suggesting that PARP is not in the mitochondria, but is more likely a nuclear regulator of mtDNA repair, as suggested by Lapucci et al. (2011). Specifically, despite extensive efforts, this research group could not detect over-expressed PARP in the mitochondria of human cells, yet they found that PARP-1 regulated the expression of nuclear genes that encoded for mtDNA repair factors. Considering the large amount of published research in this area, it is unlikely that PARP is simply always in the mitochondria or not at all. More likely, there may be tissue specific, cell cycle or DNA damage specific mechanisms underlying the localization of PARP to the mitochondria that we are in the early stages of understanding. In the absence of PARP in the mitochondria, there are other mitochondrial proteins that could act as a lesion detector or a DNA repair recruiter to the sites of SSBs (Fig. 2). In particular, Canugovi et al. (2010) reported that TFAM showed a DNA binding preference to 8-oxoG containing substrate and TFAM binding inhibited the activity of a range of BER enzymes. This competitive inhibition supports the hypothesis that TFAM may modulate mtBER by competitively binding to oxidative lesions. TFAM binds tightly to mtDNA and may provide a chromatin like organization (Canugovi et al., 2010).
An emerging player in mtDNA repair is the Cockayne syndrome (CS) group of proteins. CSA and CSB have both been identified in the mitochondria (Kamenisch et al., 2010; Aamann et al., 2010). Mutations in either protein cause CS, a recessive segmental premature aging syndrome that shares many symptoms with mtDNA diseases. In the nucleus, these proteins participate in the NER sub-pathway, termed transcription coupled repair. CSB also plays a role in nuclear BER, particularly in the repair of oxidative DNA damage in the general genome (Stevnsner et al., 2008). Although its exact role in the mtBER pathway remains to be elucidated, CSB may be required to recruit and stabilize BER proteins to repair complexes associated with the inner mitochondrial membrane (Aamann et al., 2010). Importantly, both reports of CSB in mitochondria confirmed that the protein translocates to the mitochondria after treatment with an oxidative agent, suggesting that CSB is associated with mtDNA repair and not mtDNA replication.
3. Lesion processing
Lesion processing encompasses a variety of different activities to arrive at the same endpoint, that is, a substrate that is able to be repaired by the subsequent downstream steps. Lesion processing entails not only resolving damage to DNA bases, but also modification of strand break ends. Such “dirty breaks” can be particularly deleterious to a cell and often require specialized enzymes outside the range of the simple SP-BER pathway to be resolved. The field of mtSSBR is still young; however, developments have been made that extend our understanding of how mitochondria are able to correct persistent DNA strand breaks.
Aprataxin (APTX) removes 5′-AMP groups that arise from aborted DNA ligation reactions (Fig. 3) (Ahel et al., 2006; Rass et al., 2008; Seidle et al., 2005). Ataxia with ocular motor apraxia (AOA1) is a rare autosomal recessive disorder caused by mutations in the APTX gene that encodes aprataxin (Date et al., 2001; Moreira et al., 2001). The symptoms of AOA1 are similar to Ataxia-Telangiectasia (A-T), although AOA1 patients do not exhibit any of the peripheral symptoms of A-T. Moreover, unlike conditions that arise from DNA repair deficiencies, such as xeroderma pigmentosum, AOA1 patients show no increase in cancer incidence. Cells from AOA1 patients are also not particularly susceptible to genotoxic agents, including methyl methane sulphonate (MMS) and hydrogen peroxide (Clements et al., 2004; El-Khamisy et al., 2009). Indeed, the main function of aprataxin may not reside in the nucleus, as APTX knockout mice do not exhibit a clear defect in SSBR and aprataxin does not appear to re-localize to nuclear DNA lesions after exposure to a range of DNA damaging agents (Hirano et al., 2007). These observations led us to hypothesize that AOA1 may stem from mitochondrial dysfunction, rather than nuclear repair defect. In support, the symptoms of AOA1 are similar to other autosomal recessive cerebellar ataxias caused by defects in mtDNA maintenance, such as the mtDNA helicase twinkle and the DNA repair and replication protein Polγ (Finsterer, 2009; Hakonen et al., 2008; Nikali et al., 2005). Further evidence of a potential role of aprataxin in the mitochondria can be derived from patient muscle biopsies that showed reduced levels of the mitochondrial enzyme coenzyme Q10, despite displaying no functional defect in mitochondrial respiration (Le et al., 2003, 2007; Quinzii et al., 2005).
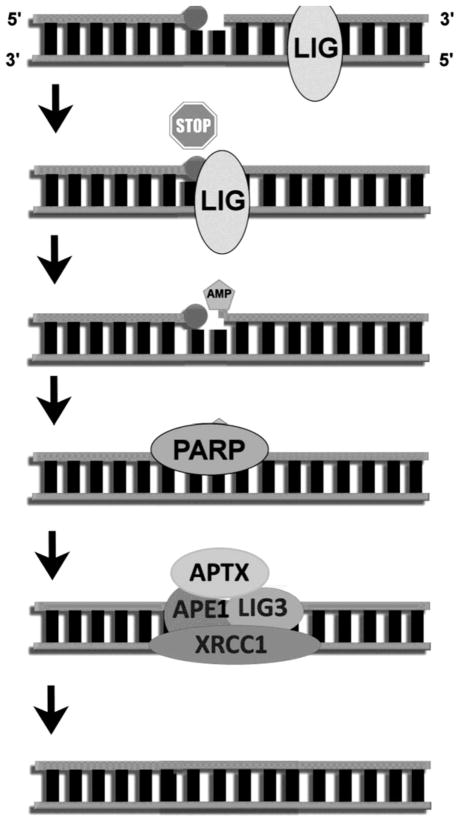
Fig. 3.
DNA–AMP complex formation and the resolution of the lesion in the nucleus. The AMP intermediate is formed when the DNA ligase encounters a strand break with a blocked 3′ end. The ligase protein does not recognize that this substrate is unligateable and begins ligation but is unable to complete the ligation because of the incompatible 3′ end. Ligation is prematurely terminated leaving a 5′-AMP residue. The break site is potentially recognized by PARP in the nucleus, signaling the recruitment of an APE-1 complex, consisting of ligase 3, XRCC-1 and the 5′-AMP resolving protein aprataxin. Aprataxin rolls back ligation, processing the 5′ AMP and leaving a SSB. This SSB is then able to be processed by BER.
Using Western blot analysis and immunochemistry, we showed that aprataxin is present in the mitochondria and that a putative mitochondrial localization signal on the N-terminus likely targets the protein to this organelle (Sykora et al., 2011). Interestingly, when transcript levels of APTX were assessed in various human brain regions, the cerebellum, the area of the brain most heavily affected in AOA1 patients, had the highest level of the regions tested. Mitochondrial dysfunction after aprataxin reduction was observed by a number of methods and in a range of relevant human cells, including primary muscle and terminally differentiated neurons. In particular, aprataxin-deficient cells had a higher level of endogenous ROS, a lower mtDNA copy number, and reduced numbers of mitochondria per cell. When the amount of endogenous DNA lesions was measured, it was found that mtDNA from aprataxin-deficient cells had a significantly higher lesion frequency, whereas nuclear DNA damage remained unchanged (Sykora et al., 2011). The result of these changes in mitochondrial function upon aprataxin knockdown was a breakdown and fragmentation of the mitochondrial network, leading us to deduce that aprataxin deficiency leads to mitochondrial dysfunction that may be central in the disease progression (Sykora et al., 2011).
Another end processing enzyme, tyrosyl-DNA phosphodiesterase (TDP1), has also recently been identified to have an important role in mtSSBR. TDP1 hydrolyzes the phosphodiester bond at a 3′ end of a SSB that is linked to a tyrosyl group of a protein moiety (Pouliot et al., 1999). This protein–DNA complex primarily occurs when topoisomerase I is trapped on DNA and represents a significant risk of inducing a DSB if left unresolved. Mutations in TDP1 cause the cerebellar ataxia, spinocerebellar ataxia with axonal neuropathy 1 (SCAN1), an autosomal recessive neurodegenerative disease with symptoms very similar to AOA1 (reviewed Caldecott, 2008). SCAN1 patients also have few, if any, extra neuronal symptoms and appear to have no increase in cancer incidence. However, unlike AOA1 patient cells, cells from SCAN1 patients are hypersensitive to the topoisomerase I inhibitor camptothecin and its analogs, and do accumulate unrepaired strand breaks in response to this genotoxin (El-Khamisy et al., 2005). Recently, Das et al. (2010) reported that TDP1 was present in mitochondria. Through Western analysis of cell fractionations and immunocytochemistry, TDP1 was identified in the mitochondria and confirmed to be the only enzyme in this organelle to possess the activity needed to process 3′-protein–DNA complexes. The potential that TDP1 has a greater role in mitochondria than in the nucleus may stem from functions other than DNA repair. In the mitochondria, TDP1 is involved with an isoform of DNA ligase 3, and was shown to be required for efficient repair of mtDNA damage (hydrogen peroxide induced). Our group has also observed TDP1 in mitochondrial fractions using Western analysis (unpublished observation). Of note, both research groups have seen that per microgram of protein, mitochondria actually have far more TDP1 than does the nucleus.
MtDNA repair was long believed to be limited to SP-BER. However, given the proximity of mtDNA to ROS, a subset of oxidative lesions, namely strand breaks with 5′ blocking ends, would require LP-BER processing. Three laboratories have recently characterized LP-BER in mitochondria and the role of a 5′ flap endonuclease (e.g. flap endonuclease 1 (FEN-1)) in this pathway. The original report of LP-BER activity in mitochondria came from Akbari et al. who reported that common lesions including uracil and AP sites could be repaired using SP-BER, as well as through a process that involved incorporation of several nucleotides (Akbari et al., 2008). Such a synthesis reaction creates a 5′ protruding flap via strand displacement that must be removed. The group confirmed the presence of a 5′ flap endonuclease activity, initially believed to be FEN-1, the major structure-specific endonuclease in the nucleus. However, in this study, FEN-1 depletion of mitochondrial extracts did not reduce the reported activity. In a subsequent report by Liu et al., robust LP-BER was confirmed in the mitochondria of mammalian cells, and FEN-1 was identified in mitochondrial extracts (Liu et al., 2008). We found that immunodepletion using FEN-1 specific antibody caused flap endonuclease activity to be diminished, but not abrogated, and that the activity could be restored by the addition of recombinant FEN-1 (Liu et al., 2008). Localization of FEN-1 within the mitochondrial nucleoid was confirmed by a protein/DNA cross-linking technique with purified mitochondria. Using Q-PCR, it was found that FEN-1 was required for efficient repair of hydrogen peroxide induced DNA damage in both the nucleus and the mitochondria. Recently, another group (Szczesny et al., 2008) also detected the presence of LP-BER, by the repair of tetrahydrofuran (THF) in mitochondrial extracts from mouse liver and kidney, as well as the human colorectal cancer cell line HCT116. In addition, they expressed FLAG-tagged APE1 in HCT116 cells and isolated an APE1-FLAG immunocomplex from mitochondrial extracts. This immunocomplex was found to successfully carry out repair of a THF containing LP-BER substrate. Importantly, the group could detect FEN-1 in mitochondrial extracts (Szczesny et al., 2008). However, after immunoprecipitating FEN-1 from mitochondrial extracts, they did not detect any of the SP-BER proteins found in the APE1 immunocomplex. Using a siRNA strategy to deplete FEN-1 levels, Szczesny et al. observed that mitochondrial LP-BER was only slightly reduced, suggesting that there is a FEN-1 independent pathway in mitochondria that resolves LP-BER substrates.
The existence of the FEN-1 dependent pathway is indirectly supported by the presence of another 5′ exonuclease, DNA2 (Duxin et al., 2009; Zheng et al., 2008). DNA2 also recognizes and cleaves 5′ flap single stranded DNA (ssDNA). Unlike FEN-1 it cannot cleave the overhang at the junction between the ssDNA and double stranded DNA, rather it leaves a residual 5′ ssDNA overhang that requires further processing by FEN-1. It was originally believed that DNA2 functioned in nuclear DNA maintenance similar to the yeast DNA2 homolog. However, Zheng et al. (2008) recently reported that human DNA2 is found primarily in the mitochondria where it interacts with Polγ and stimulates Polγ DNA synthesis. Using a LP-BER THF substrate, the group showed that it was possible to complete repair with only DNA2 or FEN-1, when added to a reaction mixture containing purified APE1, ligase 3 and Polγ. However, a tenfold increase in DNA repair activity was seen when both FEN1 and DNA2 were added to the same mixture. The result demonstrated that the activities of both enzymes are required for effective FEN1 dependant LP-BER in the mitochondria. Why two nucleases are required for the particular LP-BER pathway may stem from the strong strand displacement activity of Polγ when compared to the enzyme’s weak dRP lyase activity, as suggested in (Copeland and Longley, 2008). This strand displacement activity may frequently produce 5′ ssDNA flaps that cannot be processed by FEN-1 alone and require the additional activity of DNA2. More recently a second group has also confirmed the presence of DNA2 in mitochondria and showed that DNA2 depletion in human cells causes inhibition of mtDNA repair (Duxin et al., 2009). To summarize, mtDNA does have access to LP-BER, consisting of two separate sub-pathways (Fig. 2). The first sub-pathway is FEN-1 dependant and also involves DNA2. The second sub-pathway involves an APE1 LP-BER complex that functions independently of FEN-1. This APE1, Polγ and ligase 3 complex, can presumably resolve LP-BER substrates using a newly identified 5′ exo/endonuclease activity.
Cymerman et al. (2008) conducted the first in depth analysis of a human paralog of the yeast Nucp1 exo/endonuclease. The enzyme, which they named EXOG, had endonuclease activity that was greatest on single stranded DNA, as well as a strong 5′–3′ exonuclease activity. The relevance of this activity would not become apparent until years later when Tann et al. (2011) published their findings confirming EXOG as a major player in mitochondrial LP-BER. Specifically, they reported that depletion of EXOG triggers apoptosis due to mitochondrial dysfunction in a range of human cells. Importantly, the same effects were not seen when the cell was depleted of one of the other 5′ exo/endonucleases present in mitochondria, i.e. FEN-1 and DNA2. The mitochondrial dysfunction observed in EXOG depleted cells was also directly dependent on the presence of mtDNA, verified using Rh0 cells, suggesting that the enzyme maybe essential in the repair of mtDNA. The critical role for the protein in mtDNA maintenance was verified by Q-PCR, which showed that EXOG depleted cells had significantly less mtDNA amplification (Tann et al., 2011). Notably, only the mitochondrial genome was affected by EXOG depletion, as little effect was seen in the amplification of nuclear DNA, where FEN1 was found to play a major role. EXOG provided the vast majority of 5′ end processing activity in mitochondrial LP-BER/SSBR when tested using a THF substrate (Tann et al., 2011). Immunoprecipitation of FLAG-tagged EXOG revealed the enzyme to be in complex with polymerase γ, APE1 and ligase 3. The presence of EXOG in the mitochondria adds to our understanding of the mitochondrial LP-BER systems. However, it remains to be determined whether the enzyme is as active on other LP-BER substrates, including the 5′ AMP substrate persistent in APTX deficient cells. The findings that both FEN-1 and APTX are in mitochondria support the existence of SSBR or LP-BER intermediates that EXOG cannot resolve. Since the EndoG knockout mouse displays no apparent phenotype (David et al., 2006), it is of interest to see what will be the physical characteristics of an EXOG knockout mouse.
4. Lesion ligation
The final step in SSBR/BER, or any other DNA repair, recombination or replication event, is the re-ligation of the DNA phosphodiester backbone. In higher eukaryotes, there are three distinct DNA ligases that were originally attributed to different cellular events: Ligase 1 believed to function primarily in replication, ligase 3 in DNA repair, and ligase IV in non-homologous end joining. Knockout mice of ligases 1 and 3 are embronically lethal, a testament to the lack of redundancy between the ligase family members (Bentley et al., 1996; Puebla-Osorio et al., 2006). However, an isoform of ligase 3 is responsible for all ligation events in mitochondria (Lakshmipathy and Campbell, 2000, 2001), and until recently, it was unknown whether the embryonic lethality reported in ligase 3 knockout mice was a direct result of the enzyme’s role in nuclear and/or mtDNA maintenance. Gao et al. (2011) created conditional knockout mice that can be induced to express a DNA ligase 3 deficiency only in the nervous system. These mice were born alive, although quickly became growth retarded and ataxic, not surviving beyond three weeks of age. Since ligase 3 and XRCC1 are known to exist in a stable complex in the nucleus (Cappelli et al., 1997; Nash et al., 1997; Parsons et al., 2005; Taylor et al., 1998), it was striking that the ligase 3 defective mice had a phenotype that was dissimilar to the comparable XRCC1 conditional knockout mice (Lee et al., 2009). The neurodevelopmental defects of the ligase 3 deficient animals appeared to be associated with mitochondrial dysfunction, including decreased mtDNA, defects in mitochondrial respiration, and mitochondrial morphological abnormalities (Gao et al., 2011). Using the Comet assay, the group showed that ligase 3 was not required for efficient nuclear DNA repair after a range of DNA-damaging agent treatments. However, in the absence of ligase 1, ligase 3 participates in the repair of the nuclear DNA damage, indicating co-operation or substrate overlap between the two ligases. The role of ligase 3 in nuclear DNA repair was further investigated in an accompanying paper by Simsek et al. This group confirmed the previous results, finding that only mitochondrial ligase 3, and not nuclear ligase 3, is required for cellular viability (Simsek et al., 2011).
5. Conclusions
The ligase 3 conditional knock out mouse shows us what affect a breakdown in mtDNA maintenance can have in the brain. One particular feature of the ligase 3 mouse is the advanced atrophy of the cerebellum causing ataxia. This is a hallmark of many human neurological disorders classified as autosomal recessive cerebellar ataxias (ARCA). While these disorders have different underlying factors, dysfunctions in mitochondrial proteins, particularly mtDNA maintenance proteins, feature prominently. Autosomal recessive cerebellar disorders include diseases associated with mutations in twinkle and DNA polymerase γ. The lack of cerebellar ataxia in XRCC1 deficient mice highlights that it is not simply a general symptom of nuclear DNA repair breakdown. The recent finding that both APTX and TDP1 are found in the mitochondria further supports the notion that the cerebellum is heavily affected in mitochondrial disorders. Patients from both disease groups have severe atrophy of the cerebellum that results in progressive and debilitating ataxia. This indirectly provides evidence that AOA1 and SCAN1 are diseases caused by mitochondrial and not nuclear dysfunction.
As alluded to earlier, the presence of the same DNA repair protein in the nucleus and the mitochondria makes it difficult to separate the symptoms of a disease and identify which organelle may be particularly affected. An additional factor that also warrants mention is the re-localization of repair proteins between the two DNA containing organelles in response to increased levels of damage. Indeed, the localization of many of the above described mitochondrial repair proteins could be dependent on the balance of damage between the nucleus and the mitochondria, similar to the translocation of CSB to the mitochondria after stress (Aamann et al., 2010; Stevnsner et al., 2008). In support, the cellular localization of the glycoslyase protein Ntg1, could be modulated using damage inducing agents (Griffiths et al., 2009). Stress induced re-localization of proteins including PARP and potentially FEN-1 could offer an explanation for the disparate mitochondrial localization reported for these proteins.
The breakdown of mitochondrial DNA maintenance has been linked directly to an aging phenotype (Hiona et al., 2010; Kujoth et al., 2005). This supports the mitochondrial or “vicious” cycle of aging theory which predicts that dysfunction of mitochondrial DNA repair would lead to accelerated aging. An expanded understanding on the repair mechanisms available to mitochondria gives us insight into repair pathways that may compensate for one another, allowing a greater comprehension of overall mtDNA maintenance.
Acknowledgments
We thank Dr. Leslie Ho and Dr. Chandrika Canugovi for their critical reading of the manuscript. This research was supported entirely by the Intramural Research project Program at the NIH, National Institute of Aging.
Contributor Information
Peter Sykora, Email: sykorap@mail.nih.gov.
David M. Wilson, III, Email: wilsonda@mail.nih.gov.
References
- Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, III, Stevnsner T, Bohr VA. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Ashkenas J. Homologous recombination in human mitochondria? Am J Hum Genet. 1997;61:18. doi: 10.1086/513915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Canugovi C, Maynard S, Bayne AC, Sykora P, Tian J, de Souza-Pinto NC, Croteau DL, Bohr VA. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 2010;9:1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci U S A. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Copeland WC, Longley MJ. DNA2 resolves expanding flap in mitochondrial base excision repair. Mol Cell. 2008;32:457–458. doi: 10.1016/j.molcel.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane C, Bohr VA. DNA interstrand cross-links induced by psoralen are not repaired in mammalian mitochondria. Cancer Res. 1998;58:1400–1404. [PubMed] [Google Scholar]
- Cymerman IA, Chung I, Beckmann BM, Bujnicki JM, Meiss G. EXOG, a novel paralog of Endonuclease G in higher eukaryotes. Nucleic Acids Res. 2008;36:1369–1379. doi: 10.1093/nar/gkm1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BB, Dexheimer TS, Maddali K, Pommier Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci U S A. 2010;107:19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- David KK, Sasaki M, Yu SW, Dawson TM, Dawson VL. EndoG is dispensable in embryogenesis and apoptosis. Cell Death Differ. 2006;13:1147–1155. doi: 10.1038/sj.cdd.4401787. [DOI] [PubMed] [Google Scholar]
- Du L, Zhang X, Han YY, Burke NA, Kochanek PM, Watkins SC, Graham SH, Carcillo JA, Szabo C, Clark RS. Intra-mitochondrial poly(ADP-ribosylation) contributes to NAD+ depletion and cell death induced by oxidative stress. J Biol Chem. 2003;278:18426–18433. doi: 10.1074/jbc.M301295200. [DOI] [PubMed] [Google Scholar]
- Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human DNA2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Katyal S, Patel P, Ju L, McKinnon PJ, Caldecott KW. Synergistic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair (Amst) 2009;8:760–766. doi: 10.1016/j.dnarep.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- Finsterer J. Mitochondrial ataxias. Can J Neurol Sci. 2009;36:543–553. doi: 10.1017/s0317167100008027. [DOI] [PubMed] [Google Scholar]
- Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths LM, Swartzlander D, Meadows KL, Wilkinson KD, Corbett AH, Doetsch PW. Dynamic compartmentalization of base excision repair proteins in response to nuclear and mitochondrial oxidative stress. Mol Cell Biol. 2009;29:794–807. doi: 10.1128/MCB.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonen AH, Goffart S, Marjavaara S, Paetau A, Cooper H, Mattila K, Lampinen M, Sajantila A, Lonnqvist T, Spelbrink JN, Suomalainen A. Infantile-onset spinocerebellar ataxia and mitochondrial recessive ataxia syndrome are associated with neuronal complex I defect and mtDNA depletion. Hum Mol Genet. 2008;17:3822–3835. doi: 10.1093/hmg/ddn280. [DOI] [PubMed] [Google Scholar]
- Hiona A, Sanz A, Kujoth GC, Pamplona R, Seo AY, Hofer T, Someya S, Miyakawa T, Nakayama C, Samhan-Arias AK, Servais S, Barger JL, Portero-Otin M, Tanokura M, Prolla TA, Leeuwenburgh C. Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS One. 2010;5:e11468. doi: 10.1371/journal.pone.0011468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Yamamoto A, Mori T, Lan L, Iwamoto TA, Aoki M, Shimada K, Furiya Y, Kariya S, Asai H, Yasui A, Nishiwaki T, Imoto K, Kobayashi N, Kiriyama T, Nagata T, Konishi N, Itoyama Y, Ueno S. DNA single-strand break repair is impaired in aprataxin-related ataxia. Ann Neurol. 2007;61:162–174. doi: 10.1002/ana.21078. [DOI] [PubMed] [Google Scholar]
- Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dolle ME, Kuiper R, Majora M, Schaller M, van der Horst GT, van SH, Rocken M, Rapaport D, Krutmann J, Mullenders LH, Berneburg M. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med. 2010;207:379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van RH, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Lai Y, Chen Y, Watkins SC, Nathaniel PD, Guo F, Kochanek PM, Jenkins LW, Szabo C, Clark RS. Identification of poly-ADP-ribosylated mitochondrial proteins after traumatic brain injury. J Neurochem. 2008;104:1700–1711. doi: 10.1111/j.1471-4159.2007.05114.x. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 2000;28:3880–3886. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res. 2001;29:668–676. doi: 10.1093/nar/29.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapucci A, Pittelli M, Rapizzi E, Felici R, Moroni F, Chiarugi A. Poly(ADP-ribose) polymerase-1 is a nuclear epigenetic regulator of mitochondrial DNA repair and transcription. Mol Pharmacol. 2011;79:932–940. doi: 10.1124/mol.110.070110. [DOI] [PubMed] [Google Scholar]
- Le BI, Dubourg O, Benoist JF, Jardel C, Mochel F, Koenig M, Brice A, Lombes A, Durr A. Muscle coenzyme Q10 deficiencies in ataxia with oculomotor apraxia 1. Neurology. 2007;68:295–297. doi: 10.1212/01.wnl.0000252366.10731.43. [DOI] [PubMed] [Google Scholar]
- Le BI, Moreira MC, Rivaud-Pechoux S, Chamayou C, Ochsner F, Kuntzer T, Tardieu M, Said G, Habert MO, Demarquay G, Tannier C, Beis JM, Brice A, Koenig M, Durr A. Cerebellar ataxia with oculomotor apraxia type 1: clinical and genetic studies. Brain. 2003;126:2761–2772. doi: 10.1093/brain/awg283. [DOI] [PubMed] [Google Scholar]
- LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- Lee Y, Katyal S, Li Y, El-Khamisy SF, Russell HR, Caldecott KW, McKinnon PJ. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci. 2009;12:973–980. doi: 10.1038/nn.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, III, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O, Fouquerel E, Ame JC, Leonhardt H, Schreiber V. PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 2011;39:5045–5056. doi: 10.1093/nar/gkr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash RA, Caldecott KW, Barnes DE, Lindahl T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- Niere M, Kernstock S, Koch-Nolte F, Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol Cell Biol. 2008;28:814–824. doi: 10.1128/MCB.01766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikali K, Suomalainen A, Saharinen J, Kuokkanen M, Spelbrink JN, Lonnqvist T, Peltonen L. Infantile onset spinocerebellar ataxia is caused by recessive mutations in mitochondrial proteins Twinkle and Twinky. Hum Mol Genet. 2005;14:2981–2990. doi: 10.1093/hmg/ddi328. [DOI] [PubMed] [Google Scholar]
- Pankotai E, Lacza Z, Muranyi M, Szabo C. Intra-mitochondrial poly(ADP-ribosyl)ation: potential role for alpha-ketoglutarate dehydrogenase. Mitochondrion. 2009;9:159–164. doi: 10.1016/j.mito.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Parsons JL, Dianova II, Allinson SL, Dianov GL. DNA polymerase beta promotes recruitment of DNA ligase III alpha-XRCC1 to sites of base excision repair. Biochemistry. 2005;44:10613–10619. doi: 10.1021/bi050085m. [DOI] [PubMed] [Google Scholar]
- Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- Puebla-Osorio N, Lacey DB, Alt FW, Zhu C. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol Cell Biol. 2006;26:3935–3941. doi: 10.1128/MCB.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinzii CM, Kattah AG, Naini A, Akman HO, Mootha VK, DiMauro S, Hirano M. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology. 2005;64:539–541. doi: 10.1212/01.WNL.0000150588.75281.58. [DOI] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Molecular mechanism of DNA deadenylation by the neurological disease protein aprataxin. J Biol Chem. 2008;283:33994–34001. doi: 10.1074/jbc.M807124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi MN, Carbone M, Mostocotto C, Mancone C, Tripodi M, Maione R, Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem. 2009;284:31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidle HF, Bieganowski P, Brenner C. Disease-associated mutations inactivate AMP-lysine hydrolase activity of Aprataxin. J Biol Chem. 2005;280:20927–20931. doi: 10.1074/jbc.M502889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van HB, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevnsner T, Muftuoglu M, Aamann MD, Bohr VA. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech Ageing Dev. 2008;129:441–448. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svilar D, Goellner EM, Almeida KH, Sobol RW. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011;14:2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora P, Croteau DL, Bohr VA, Wilson DM., III Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc Natl Acad Sci U S A. 2011;108:7437–7442. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tann AW, Boldogh I, Meiss G, Qian W, Van HB, Mitra S, Szczesny B. Apoptosis induced by persistent single-strand breaks in the mitochondrial genome: critical role of EXOG (5′ exo/endonuclease) in their repair. J Biol Chem. 2011 doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RM, Wickstead B, Cronin S, Caldecott KW. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr Biol. 1998;8:877–880. doi: 10.1016/s0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Padua RA, Campbell C. Mammalian mitochondria possess homologous DNA recombination activity. J Biol Chem. 1996;271:27536–27543. doi: 10.1074/jbc.271.44.27536. [DOI] [PubMed] [Google Scholar]
- Whatcott CJ, Meyer-Ficca ML, Meyer RG, Jacobson MK. A specific isoform of poly(ADP-ribose) glycohydrolase is targeted to the mitochondrial matrix by a N-terminal mitochondrial targeting sequence. Exp Cell Res. 2009;315:3477–3485. doi: 10.1016/j.yexcr.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]