Abstract
Imaging based clinical trials can benefit from a solution to efficiently collect, analyze, and distribute multimedia data at various stages within the workflow. Currently, the data management needs of these trials are typically addressed with custom-built systems. However, software development of the custom- built systems for versatile workflows can be resource-consuming. To address these challenges, we present a system with a workflow engine for imaging based clinical trials. The system enables a project coordinator to build a data collection and management system specifically related to study protocol workflow without programming. Web Access to DICOM Objects (WADO) module with novel features is integrated to further facilitate imaging related study. The system was initially evaluated by an imaging based rehabilitation clinical trial. The evaluation shows that the cost of the development of system can be much reduced compared to the custom-built system. By providing a solution to customize a system and automate the workflow, the system will save on development time and reduce errors especially for imaging clinical trials.
Keywords: Imaging-based clinical trials, Workflow Engine, System Framework, Imaging Informatics System
1. BACKGROUND
Clinical trials have rapidly evolved during the past decade. According to U.S. National Institutes of Health, the total registered clinical studies rose from 10,241 in 2003 to 158,829 in 2013 [1]. Specifically, imaging techniques have been used widely in clinical trials to track and evaluate biomarkers [2]. Massive amounts of imaging data are produced during the clinical trial and the workflow has become more and more complex. Moreover, since these data are collected from different departments, collaboration between healthcare staff plays a more essential role and substantially impacts the overall efficiency of healthcare [3]. Unfortunately, the cost of collecting and managing the large volumes of data and communication processes has negatively impacted the productivity.
Electronic data collection (EDC) has been proven to reduce the monitoring and data management cost in comparison to paper data collection (PDC) [4]. For years, hospital information systems (HIS) and picture archiving and communication systems (PACS) are prevalent in hospital environment for data management. In contrast to the hospital environment, academic research data are usually stored in disparate environments and processed with distinct and highly customized, single silo workflows. Conventional PACS designed for the hospital environment is limited in the flexibility for various research- related workflows [5]. Furthermore, clinical trial workflows are usually agile and need to be adapted to disparate research methods that require the data management to be customized. Typically, the need for clinical trial workflow is addressed by custom-developed data management systems. However, custom- built software development can be time-consuming and resource-intensive, and most of small-scale clinical trials are limited for the cost of a custom-built informatics system. Data capture database development platforms, such as REDCap [6], are widely used for small-scale trials as a framework. Such software is mainly textual form based and not integrated with imaging data and multimedia data. In such systems, the workflow view is not presented, activities are not automated and relations between forms cannot be defined. As a result, a solution to provide efficient workflow development and refinement along with data management and decision support to serve the small-scale imaging-based clinical trials is urgently needed.
Workflow automation technology has been demonstrated to improve the efficiency of business workflow for over ten years [7]. In the business and industry field, workflow automation products enable the company to create a workflow with a variety of components, mostly electronic forms, and utilize the workflow to manage the data flow in their production line. The advantage of the workflow automation software is that it allows user to create a data management system based on specific workflow without software development. Additionally, the workflow engine is able to process a sequence of steps automatically based on the predefined rules. For example, a workflow requires the data management system to parse the data, store the data to the database and email the reports to related staff once the data is received. With the workflow engine, this set of actions can be easily defined. The avoidance of the software development reduces the cost and effort in the business model and minimizes the potential errors in human action [8].
The workflow engine concept has already been introduced in healthcare, e.g. Huser et al presented a system for clinical decision support in 2011[9]. However, there is no similar system designed for imaging based clinical trials. Compared with business model, imaging based clinical trials require more compatibility with multimedia data [10], more intuitive workflows, and particular analysis of imaging biomarkers. To address the challenges in the imaging-based clinical trial, we present a novel prototype imaging-informatics based prototype system named IWEIS (imaging workflow engine informatics system) to support imaging-based clinical trials.
As a case study of the imaging informatics system, the IWEIS prototype was implemented and applied to an imaging-based stroke rehabilitation phase I clinical trial named Dose Optimization for Stroke Evaluation (DOSE). This paper will discuss IWEIS based on this DOSE clinical trial. The DOSE clinical trial aims to understand the optimal dose of a principle-based rehabilitation intervention for stroke [11]. The trial aims to recruit 40 subjects in 4 years. During the trial, complex multi-media data including laboratory-based measurements such as Wolf Motor Function test (WMFT) and behavioral questionnaires, such as Motor Activity Log (MAL) will be collected. Imaging studies, such as structural MRI, Transcranial Magnetic Stimulation (TMS), and Diffusion Tensor Imaging (DTI) will also be conducted and collected. As an important component, the imaging data will be used to track the changes in stroke lesion size and location through the physical therapy treatment. Potential correlation of lesion size and rehabilitation progress will also be investigated.
By utilizing IWEIS, the deployment of the DOSE informatics system will be different from traditional methods. Figure 1 demonstrates the traditional scenario and the proposed new scenario. In the traditional scenario, after the design of the study, clinical designers need to set up multiple meetings with software developers to explain the requirements. This step is critical and could be time-consuming as the software developers may not have adequate clinical trial knowledge. The software development step, which is carried out by software developers, normally lasts several weeks to several months. The final step, quality assurance of the software, requires the collaboration between clinical researchers and software developers. Sometimes the system can have a lot of deficiencies due to developer’s lack of the knowledge of the clinical trial. In the IWEIS scenario, clinical researchers are able to deploy a system without software engineers. That means clinical researchers controls all the development processes and the system is deployed by people with thorough understanding of the clinical trial. Therefore, the IWEIS scenario improves the efficiency compared to traditional scenario. In figure 1, red stages show negative impact on the cost of time and resources with traditional scenario, while green stages shows where IWEIS can reduce of the time and cost of the system deployment.
Figure 1.
Traditional scenario and IWEIS scenario in the development of clinical trial informatics system. Stages with negative impacts on the cost of system development are marked as red, and stages with positive impacts are marked as green. This figure shows where the challenges are in the traditional scenario and where the IWEIS scenario will improve the efficiency of the system development.
2. Methods
IWEIS was developed in Hypertext Preprocessor (PHP) 5.3.10 along with MySQL 5.5.32 and Apache server 2.2 under the HTML5 standard and deployed within a Linux Environment. The IWEIS software also adopts a variety of open-source software such as JQuery [12], a prevalent JavaScript library, Diagramo [13], a web-based flowchart software with pure HTML 5, and Twitter bootstrap [14], an intuitive and powerful front-end style framework for web development etc. To meet privacy requirements, the system requires a SSL certificate and authentication with username and password.
The system is developed based on a two-tier architecture (See Figure 2). The base layer is comprised of a MySQL database, a file storage system, a DICOM (Digital Imaging and Communications in Medicine) viewer and the Apache web based server. The base layer provides necessary services for system operations and stores all the data to be collected. The second tier consists of two main components: the designer module and the application module. Each module incorporates a web-based graphical user interface (GUI) with a unique Uniform Resource Locator (URL). The designer module is the back-end view of the system, which allows a project designer to deploy and customize the informatics system. Once an instance of the system is deployed, the application module shows the front-end view of the created imaging informatics system, allowing client users to collect and manage the data by the workflow of the clinical trial.
Figure 2.
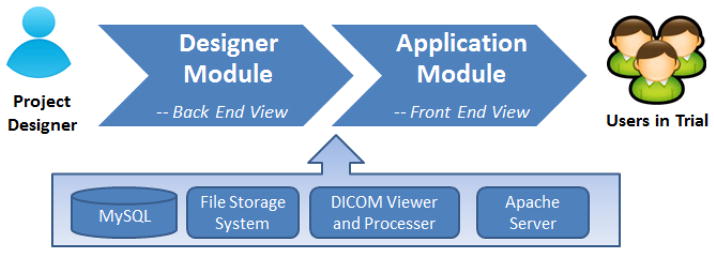
Schematic presentation of the IWEIS system architecture. The system architecture is a two-tier layer comprised of a base layer with four main components. In the second tier, the designer module is utilized for deployment and the application module is utilized as the imaging informatics based system for front end users in clinical trial applications.
2.1. System Components
2.1.1. The Designer Module
The designer module is the back end of the system that can create and deploy the information system instance. Typically, the deployment of the proposed system for an imaging based clinical trial is comprised of 5 steps.
A principle investigator (PI) designs the trial with specific workflows and requirements. In the DOSE trial, the PI defines the data to be collected in each stage and each staff’s tasks in each stage.
Develop the workflow with the IWEIS workflow designer. In DOSE trial, the manager is able to build the workflow with the workflow painter in IWEIS.
Define tasks in each stage of the workflow. In the DOSE trial, the manager is able to create a variety of tasks that need to be done in each stage. The tasks include collecting form data, upload imaging data etc.
Assign the rules in the workflow engine. The rules are used to automate some processes and assist in the decision making. In the DOSE trial, rules are used to assist screening patients. The criteria for screening are entered as rules, and the system shows if a patient is satisfied with these rules. The rules for restrict tasks access can also be defined. Firstly, the arrows in the workflow define the basic rules. Each stage can only be accessed when all the previous stages are completed. Secondly, rules can be set to each task. User can define a set of prerequisite tasks that must be completed or approved before access of the target task.
The front-end system is generated automatically and ready for use for the clinical trial.
Once a clinical trial workflow has been designed (step 1), the project manager is able to develop the workflow-based imaging informatics system using the designer module. The designer module is composed of three main components, which are related to three main steps (step 2–4).
Component 1: Workflow Designer
The designer module has an integrated workflow designer utilizing a thin-client HTML5 based workflow diagram painter (see figure 3) for users to design the workflow (step 2). The painter has a menu bar and three panels. Users are able to pick a shape from the left panel to draw on the canvas. Functions including showing grids, group/ungroup, text and customizing styles are also available within the tool. The canvas in figure 3 illustrates the current workflow for the DOSE trial as the first initial application example. In the workflow, each shape stands for a stage of the workflow trial. By choosing a specific shape and color, each shape can be used to indicate similar types of stages within the workflow. In each stage, the user is capable of combining one or several workflow tasks within the stage by selecting the shape and clicking the button “Edit contents” (Arrow B). By clicking “Engine Rules” (Arrow A), the user is also able to configure the rules for each shape in the workflow.
Figure 3.

Workflow Designer Module in the back end. The user is able to pick a shape in the left panel, drag and drop it into the canvas. The canvas illustrates a sample workflow design example of the DOSE rehabilitation clinical trial. Arrow A links to the engine rules page. Arrow B allows the user to edit contents in each stage of the workflow.
Component 2: Workflow Stage Editor and Tasks Builder
The next step is to define tasks in each workflow stage (step 3). The stage editor enables user to build one or more tasks in each stage. As long as a task is created in a stage, the task will be shown in the same stage of the created instance system for data collection and data presentation. A toolbox with five task builders is provided in the stage editor.
General Tasks Builder
-
Digital Forms Builder
The textual forms are the most widely used method for data collection within the clinical trial. IWEIS provides a digital form builder tool. To further facilitate the production of forms, a template library is also incorporated. The user is able to create a template, load a template from the library, and customize the form from the loaded template.
-
File Uploader Builder
Typically, a number of files relevant to the clinical trial need to be stored for review and other uses. For example, within the DOSE clinical trial, the patients’ signed HIPAA (The Health Insurance Portability and Accountability Act) authorization and patient’s motion sensor data during physical therapy must be stored and associated with each patient and specific workflow stages. The builder allows user to create a file uploading and downloading tool within any of the workflow stages.
Multimedia Tasks Builder
-
Medical Images task builder
This task builder enables the user to create an uploader and viewer to upload DICOM format files and view the DICOM studies through a zero-footprint Web Access to DICOM Objects (WADO) viewer. To respect patient privacy and abide by HIPAA’s Privacy Rule, an anonymizer is integrated with the uploader and DICOM headers of all uploaded images will be de-identified automatically. User can also link the images task with a textual form for collecting results of the images analysis. Additionally, user could upload template image as a mask to assist image interpretation. More details are further discussed in the imaging section within the Application module.
-
Video and Audio plugin builder
Video and audios are commonly used file format for clinical trials. This builder allows user to create a video uploader and viewer, which enables the user to upload a video and then review it with a video player.
In summary, in step 3, the user defines the data that needs to be collected in each stage and establishes the data collection tools.
Component 3: Rules Manager
Rules are essential for workflow engine systems. Within the clinical trial environment, the rules are less complicated than in the business environment. However, well-defined rules are needed to automate certain processes, provide decision-support, reduce human error, and minimize resource overhead. Based on the common requirements within clinical trial environments, the IWEIS prototype supports two categories of rules. The first category is workflow stage rules, which is built-in within the workflow engine. In the workflow diagram painter, the user is able to link stages within the clinical trial workflow by an arrow. These arrows define the sequence of the study, and a stage can only be accessed if the previous stages are completed. In other words, the user is not allowed to skip prerequisite stages during the clinical trial. The second category is task rules, which are applied to specific tasks in a workflow stage. There are three types of task rules: 1) Completion restriction rules; 2) Approval restriction rules; and 3) Complex requirements which will be described further below.
Completion restrictions: The task access is restricted unless specific prerequisite tasks are completed. IWEIS allows user to select the completion-required specific tasks for each task.
Approval restrictions: The task access is restricted unless specific tasks are approved by quality assurance (QA) users.
Complex restrictions: The workflow stage access is restricted by a set of rules, including mathematical comparisons of a value within tasks or complex states. For example, a task can be restricted by two rules. Suppose the first rule is limited by the “Age” value in the “Profile” form. The participant’s age must be less than a value defined by the user. The second rule is limited by the “stroke length” value in the “Phone Screen” form. The subject’s stroke length must be greater than a certain time defined by the user. Only if the subject’s data meet both rules, this task can be accessed.
In addition to the application of the rules discussed before, the rules can also be used as a decision support tool. For example, within the DOSE trial, a patient can only be enrolled if he/she satisfied all requirements within the screening workflow stage, e.g. age must be greater than 21, stroke severity is within a range etc. The complex restrictions are used as restrictions for the access of the enrollment form of the subject which would be the next step in the clinical workflow. Instead of manually looking through all the conditions and verifying the enrollment by the study coordinator, the system automatically shows unsatisfied requirements and prevents the enrollment of the subject resulting in fewer potential human errors.
2.1.2. The Application Module
After the deployment of the system, the application module creates an actual instance of the system that was designed in the designer module. The newly created system will be used to create new patients and enter data in the real clinical trial. The homepage of the system is a dashboard for users. The dashboard is composed of two sections, my tasks and patient worklist. The user with permissions is also able to create a new patient for the study from dashboard. The “my tasks” section shows all tasks that are ready for the user to process. The workflow engine checks all the rules of the tasks and pushes the available task to the user in charge. Pick a task will lead to the data entry page of that stage of the workflow for the patient. The patient worklist section shows all enrolled patient and link to patient profile page. The patient profile page is designed to show the study workflow with status tracker since a workflow showing the overview of the study process for each patient will benefit data collection and monitoring in clinical trial.
To utilize the system, the first step is to create a new subject with a subject ID. Creating a new subject or choosing one of the subjects from the worklist leads to the subject profile page with all the data collection links activated. The left part of figure 4 illustrates the subject profile page with the clinical trial workflow example. In each stage of the workflow, a progress indicator shows the number of tasks in the workflow stage that have been completed. Clicking the workflow stage results in a popup widow with all the tasks related to that particular stage. In the right part of figure 4, the first task shows a text-based digital form with a rule restriction. The rule is not met since a prerequisite task has not been completed. The second task shows a link for uploading videos and a list of the videos already uploaded. The uploaded video can be downloaded or streamed for viewing. The third task shows the link to the DICOM WADO viewer. An upload button allows the user to upload DICOM images, anonymize the DICOM header, and extract the header information into the database for efficient queries. The worklist within the viewer shows all the images uploaded. These images can be viewed through a zero-footprint DICOM WADO web viewer or downloaded to the local client for further image processing analysis. Task 4 illustrates the file uploader task.
Figure 4.
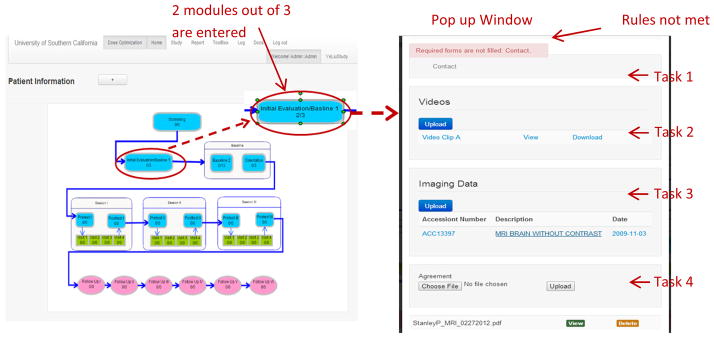
Screenshot of Patient profile page with integrated workflow in the front end. The left panel illustrates the patient’s page. In each stage of the workflow, the number shows the completion status of the modules. Clicking the stage leads to a popup window with all the modules in that stage (right panel). If the rules for a stage are not satisfied, the workflow stage will be inaccessible.
2.2. System Features
2.2.1. Images Analysis Module
Imaging data have been used widely as the biomarkers in current medicine. More and more clinical trials use imaging biomarkers as evidences to detect subtle changes. For example, the lesion size, rate of growth and location detected in brain images can be used to predict the severity of the stroke. As a result, a platform that can combine the traditional textual data and the imaging data will be beneficial to researchers in clinical trials. The IWEIS system has integrated a vendor neutral imaging data analysis tool, the Images Analysis Module, which could upload, view, and facilitate data analysis online.
The Images Analysis Module includes both a DICOM uploader and a WADO (Web Access to DICOM Objects) viewer. The DICOM uploader allows loading of image studies either from a CD or a local client machine. In order to comply with HIPAA, all images are de-identified and transmitted under Secure Sockets Layer (SSL) to the system. Once uploaded, all data will be parsed and the header information will be extracted to a database for efficient query.
To facilitate the analysis of the imaging data, IWEIS utilizes a vendor neutral WADO viewer developed by Imaging Processing and Informatics Laboratory (IPILab) at the University of Southern California for online image viewing [15]. The viewer is a stand-alone PHP-MySQL-Apache based software package. A PHP DICOM parser library NanoDICOM [16] is utilized to process the DICOM files. Images are stored in the file system and accessible by WADO identifiers through Apache. An interface was developed and the viewer is integrated with the IWEIS system. In the image viewer (See Figure 5), the user can navigate through all the image series within each study and change the layout of the viewer. The user is also able to scroll, pan, zoom, change window/level and show textual information extracted from DICOM header. Different from traditional image viewers, this viewer is a zero-footprint solution and no client side plugin is needed. This is extremely advantageous due to the common restrictions on computers used within the clinical environment and where most of the clinical trials personnel can benefit from the portability of the image viewer.
Figure 5.

Web-based zero footprint DICOM WADO viewer. The user is able to combine a self-designed data collection form with the images. Therefore user is able to read the image, fill out the reading result with a customized form and send it to database. The viewer also provides annotation, measurement, and ROI marker tool to assist the identification of lesions or other pathologies related to the clinical trial.
Distinguished from other image viewing software, IWEIS facilitates the analysis of the images by novel features. Similar to other software, measuring, ROI (Region of Interest) marking and annotation tools are available with the image viewer. Figure 5 illustrates an example screenshot where the diameter of the lesion is being measured and a ROI markup annotation of the lesion has been added. The measurement and markup results are stored within the database for future retrieval. Despite these traditional tools, Images Analysis Module is able to link a data collection form with the image viewer. In the practical application, radiologists usually need to record the image reading results into a textual form and send it to the database. IWEIS allows the user to create a textual data entry form in the back end and link it with the images task. Once an image task is linked with the form, the user is able to show the image and textual form side by side (see figure 5). Therefore, the user is able to collect the image reading results conveniently with a customized form. Based on the data collected by the forms, user is able to compare longitudinal reading results of the same patients and investigate the rate of growth of the lesion.
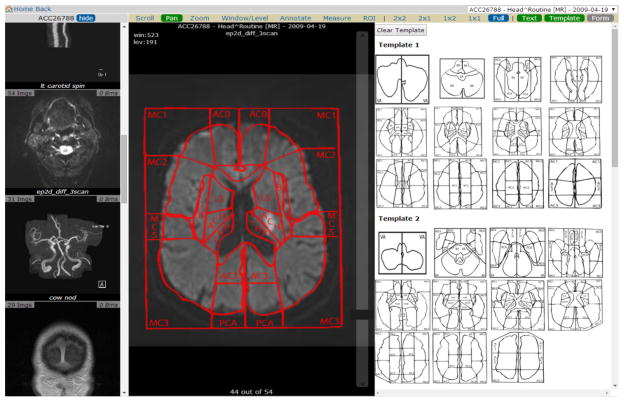
In order to assist the identification of the location of lesion, the Images Analysis Module is equipped with a template masking tool. To use this tool, the user needs to upload a PNG format semi-transparent template image, and the tool allows user to overlay the template on top of the image. Figure 6 illustrate an example of the tool. The user adapts published vascular territory templates [17, 18] and uploads these template images. At the right part of the window, all templates uploaded are shown. The user can pick any of the templates and overlay it on top of the image. User can also zoom in, zoom out and rotate the template mask to match the image better. By matching the template with the images, the identification of the lesion location within the organ will be easier and more accurate.
Figure 6.
Template masking tool. On the right part of the window, a library of sample templates (Templates adapted from Damasio et al.[17], courtesy of Dr. Matthew Edwardson) uploaded by user can be chosen. The uploaded templates can be shown on top of the image and can be rotated, zoomed in and zoomed out. Overlaying the template on top of images facilitate the identification of lesion location.
2.2.2. System Modification and Expansion
As discussed before, one of the challenges is that sometimes the researchers need to redesign or improve the informatics system even after some patients have been enrolled. For instance, after the treatment of several subjects, clinical researchers may find out that some forms need to be expanded and some stages of the workflow need to be modified. In the traditional scenario, clinical researchers need to contact the software developer and explain the requirements. The software developer needs to back up the data and reprogram the system. This process may take several days to several weeks, and the cost will be high. Sometimes, the modification will be expensive and buggy if the system is not designed flexibly at the beginning. To address this challenge, IWEIS system has several mechanisms in various situations.
Tasks Level
All the task tables have a “stage ID” data field and stores which stage it belongs to. When a task needs to be moved to another stage, the “stage ID” is updated and hereby the task is moved at the application end. New tasks can also be created in any stage. Deletion of a task will result in hiding the tasks in the workflow and the application site. However, the data that have already been collected will not be lost and can be retrieved at any time.
Workflow Level
The workflow level modification means that new stages will be inserted into the workflow or some stages will be moved to other positions in the workflow. To implement this feature, a “stage sequence” table is utilized to store the previous and next stage for each stage. When the workflow is modified, the database will be backed up before the modification, and the “stage sequence” table is updated. The tasks will be associated with the original stages no matter where they are moved. Therefore the tasks in each stage will not be affected and the collected data will be retained. To remove a stage, the user needs to move or delete all the tasks in the stage, and delete the stage, which will removed it from the “stage sequence” table.
Rules Level
Users can set new rules or update existing rules. Once the rules have been updated, the system will check all the rules with the collected data and warn all the violations under the new rule system. The new rule system will also be used in future data collection to prevent errors.
Data Fields Level
The data fields in each task can also be updated. User is able to insert new data field, update data field name and delete a data field. Deletion the data field result in hiding the data field from the tasks, while the data is still preserved and can be retrieved. After updating the data fields, the system will also automatically run the rule system to search for any violation of the predefined rules. Warnings will be shown for violations found.
2.2.3. Data Query and Presentation
IWEIS is more than a data collection tool or digital form builder. To further simplify the process for data analysis, IWEIS provides multiple solutions for data mining and presentation. To avoid potential data exposure, only approved users with data retrieval rights are able to access the data mining tools. The data can be queried in three ways. A query by patient ID leads to all the relevant data categorized by workflow stages in a patient record manner similar to an Electronic Medical Record (EMR). A query by the task and workflow stage leads to a cross-project sheet with all patients’ data in the task group. A comprehensive query allows user to specify multiple search criteria. In the comprehensive query mode, the user can pick values from any of the tasks or data collected from the digital forms and set search conditions for them. For example, in DOSE clinical trial, the users may need to search the patients with stroke history for more than 3 years that have an MRI scanned at the initial evaluation stage. The query would be the “stroke length” value in the “history and demographics” form to be greater than 3 years, and the images in the initial evaluation stage that have been uploaded. The results return a list of patients that satisfy the conditions.
The data retrieved can be displayed online or downloaded to the local client machine. Text-based digital form data will be converted to an excel sheet for download and further analysis. Video and audio files are able to be streamed online or downloaded for post-processing. The imaging data can be viewed through the DICOM WADO viewer discussed above and can be compressed as a zip file for download.
2.2.4. Quality Assurance
Clinical trials are usually under the monitoring of sponsors for quality control. A clinical trial has to be performed complying with standards and the data has to be collected with highest quality. To facilitate the quality assurance, the IWEIS adopts several mechanisms. Firstly, to be compliant with Title 21 CFR Part 11, all the activities are logged automatically. The log can be queried by username or downloaded directly from the graphical user interface. Secondly, data collected has to be approved at least one time (Please see Figure 7). During the data collection process, the data need to be reviewed by the data collector when they entered the data. After that, project managers are able to define additional reviews from other users for each task group. This step is optional and can be defined based on the importance of data and the scale of trials. The system also provides an option of randomly selecting part of data for review, which is designed for large scale clinical trials, since it is unfeasible to review all the data in some cases. Thirdly, the system allows creating a user group (QA group) account to review all the data. This mechanism aims to provide an account for third party reviewers.
Figure 7.
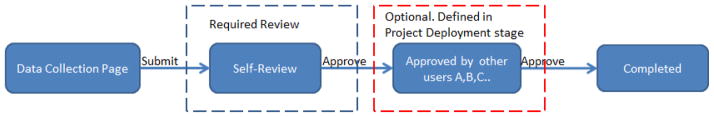
Data collection review flowchart. For each data collection, the data entry person is required to review and approve the data to complete the data entry. Moreover, the study manager is also able to define if the data needs to be approved by other users, such as supervisors or a QA group to complete the data entry.
3. Evaluation
The IWEIS was evaluated with the DOSE clinical trial. The DOSE trial aims to collect 40 subjects in a four year period and currently 30 subjects have been enrolled. The subjects are randomized into four dosage groups to receive treatment with physical therapy. A baseline evaluation, three sessions of treatments, and six follow-ups are conducted for each patient. The data involved includes digital questionnaires, MRI and DTI images, videos, and motion sensor data. These data need to be collected and associated with the various stages in the workflow. Currently, a custom-built system is developed and being used for DOSE trial [19]. To evaluate the IWEIS system, we created an instance in IWEIS and compared it with no-system situation and custom-built system situation.
As shown in table 1, the development time and the financial cost with IWEIS are much less than a custom-built system. To develop a custom-built system, the clinical trial leader needs to look for a developer, define the needs, communicate with the developer and review the delivered systems. Typically, the communication can be challenging for both clinical researchers and software developers. Miscommunication will result in a delay in delivery and more expenditure of man hours in re-development. In contrast, IWEIS allows researchers to deploy the system directly and eliminate the communication with developers. Thus, the development/refinement cycles timelines are reduced significantly. In DOSE trial, the custom-built system takes about 5 meetings and 6 months with a part-time developer for the development. During the development of the custom-built system, regular meetings were also held for progress report. In contrast, the deployment of IWEIS system takes a few days without any meeting.
Table 1.
Comparison of IWEIS system with no-system situation and custom-built system situation in DOSE clinical trial
| Tasks | No Electronic System | IWEIS System | Custom-built System |
|---|---|---|---|
| Pre-Development Meeting | NA | 0 times | ~ 5 times |
| System Development | NA | 3 days | 6 months with a part time developer |
| System Modification | NA | 1–8 hours | 1 day – 2 weeks with a part time developer |
| Advanced Features | NA | No | Yes |
| Financial Cost | NA | ~ $0 | ~ $5000 |
| Data Collection | Paper based, 1–2 minutes per question | 1–2 minutes per question. | 1–2 minutes per question |
| Imaging Data | Stored in CD | Uploaded to system in minutes | Uploaded to system in minutes |
| Textual data query | Manually look through paper forms, 1–20 minutes | < 1 second by database query | < 1 second by database query |
| Imaging data query | Look through CDs manually. 1–20 minutes | <1 second by database query | <1 second by database query |
| Collaborations between staff | Need to deliver paperwork physically. Up to several days | Instantly. Users can login to see the tasks need to process | Instantly. Users can login to see tasks need to process |
| Accidental Mistakes | Non-avoidable due to human errors | Accidental mistakes will be avoided if rules are set | Avoidable |
Another common situation is that clinical trials are easy to change in workflow and forms. Therefore, the flexibility of a system is another essential feature. In the experience of the DOSE clinical trial, there were quite a number of modifications of forms and requirements after the development started. For example, in the system development with DOSE trial, some forms were revised after the custom-system was developed. This modification demands communication with the developer, a re-programming of the software, a revising of the database, and a final review from the clinical trial researchers. Thus a custom- built system is able to be modified, but requires nontrivial efforts. Hence, the cost of a modifying a custom-built system is high and may not be appropriate choice for small-scale clinical trials. In contrast, the IWEIS system allows user to modify the form without re-programming. The data is backed up automatically, and the modification cost only several hours. IWEIS rules system’s warning of violations also facilitate the validation of the system modification. In DOSE trial, the modification of the custom-built system takes a day to 2 weeks depending on the complexity, while the IWEIS system takes hours for basic modification. However, the custom-built system is able to realize advanced features but IWEIS system is not. For example, in the DOSE trial, the custom-system is able to provide an interactive data collection tool for treatment and is able to clips videos automatically, while IWEIS system is limited in these functions.
From the functional perspective, both IWEIS and the custom-built system significantly reduce the time in data retrieval and collaboration compared with no-system situation. Without an electronic system, the data retrieval cost up to several hours for manually looking through paperwork or imaging CDs. With an electronic system, a simple data query is able to retrieve textual data and multimedia data instantly. The collaboration cost is also significantly reduced. Without an electronic system, physically delivery of paperwork and CDs may delay the progress for days. With an electronic system, the physical delivery is not necessary and system pushes tasks to user in charge promptly.
Based on the evaluation, the prototype IWEIS may have less functionality than a custom-built system today, but it is capable to meet regular needs with much less resources, which is beneficial for small scale clinical trials. In addition, once these generic tools are developed, they can be easily integrated into IWEIS and thus provide the same additional functionality as the custom-built system.
4. Discussion
Most of the imaging informatics systems are designed for hospital environment and not suitable for clinical trials. In the field of clinical trials, data capture software such as XNAT [20] and REDCap are the main software used to address the data management needs. However, current solutions are inadequate for imaging based clinical trials. Therefore IWEIS system aims to provide a solution specifically for imaging based clinical trials.
In imaging based trials, imaging biomarkers are used as evidences and researchers normally need to acquire information like lesion size, location, rate of growth. The reading results need to be collected in forms for longitudinal studies. Due to such characteristics of imaging based clinical trials, IWEIS provides a web viewer with measuring tool, side-by-side forms and template masking tool. Once the images reading results are collected, the user could query all the data for each patient and investigate the changes through different stages of the trial.
A comparison between IWEIS prototype and existing similar software REDCap and XNAT is illustrated in table 2. All three software are free of programming for the deployment. However, firstly, REDCap only supports textual data, and XNAT only supports imaging data. IWEIS supports textual data, imaging data and multimedia data. Secondly, In REDCap and XNAT, the workflow concept is not illustrated. The data in REDCap and XNAT managed on a “visit” manner. The data is organized by visit number and there are no intuitive views of the how the clinical trial look like. IWEIS presents the workflow and provides a workflow based manner for data collection, presentation, and data query. In IWEIS, the data are linked by the workflow. Instead of providing a set of non-related forms, IWEIS presents the workflow and status indicator in each subject profile page. Visit based data management is also supported in IWEIS. Thirdly, in IWEIS, rules can be setup to reduce errors and provide decision support. REDCap integrates a basic method for data validation while IWEIS integrates a rule system which allows setting up more complex rules. Fourthly, although all three systems allow flexibly modification of the system, IWEIS’ rule system helps validation and reducing errors during the system modification and expansion. Therefore, IWEIS is able to further facilitate the data capture and management for small scale imaging based clinical trials.
Table 2.
Comparison of IWEIS with existing similar software
| IWEIS | REDCap | XNAT | |
|---|---|---|---|
| Non-programming Deployment | Yes | Yes | Yes |
| Textual Data | Yes | Yes | No |
| Imaging Data upload and Display | Yes | No | Yes |
| Multimedia Data | Yes | No | No |
| Data security and access | Yes | Yes | Yes |
| Management by workflow | Yes | No | No |
| Management by visits | Yes | Yes | Yes |
| Rules Engine | Yes | Yes | No |
| Automate data collection by workflow | Yes | No | No |
| System Modification/Expansion | Yes | Yes | Yes |
In practice, to support a collaborative clinical trial in a large hospital, the administrator needs to plan thoroughly before the actual deployment of the system. The protocol, workflow, related personnel and each person’s responsibility and access to tasks need to be defined carefully. The deployed system can be utilized even in the pilot study for testing. In some cases the patients are screened from the HIS/EHR database, hence the prior background data needs to be retrieved. The system also needs to be hosted on a secure server behind firewall. The training for users is necessary as the users are also involved in other studies or normal clinical environment, which may be confusing and thus, reduce efficiency of the overall clinical workflow.
In addition to improving the efficiency, physicians can also benefit from the system. First of all, the physicians will benefit from the system by saving time on administrative work and focus on the diagnosis and treatment. Moreover, they also gain a platform for future data mining. The advantage of the IWEIS system is that it collects all the data related to the clinical trial from a variety of sources and store the information in a centralized database. This is a significant step in implementing a generalized platform for data mining and decision support. In the future, new patients can be matched to the system through data mining to produce decision support for the new patients’ treatment plan. Data analysis toolbox modules are also planned to be integrated with the system which allows researcher to perform statistical analysis online.
5. Limitations and Future Work
Clinical trials usually enroll patients by advertisement or by screening from HIS (Hospital Information System) or EHR (electronic Health Records system) database. In the second case, some data like prior background data, patient registry data can be reused. Hence an interface between the IWEIS system and HIS/EHR can benefit clinical trials by auto-population of the background data. In the subsequent version, the system will be integrated with HIS/EHR system and be able to receive HL7 messages for patient- specific health data. Once a patient is enrolled, the system will auto-populate the database based on HIS/EHR and reduce the unnecessary efforts for collection of redundant data. The system will also be equipped with a DICOM receiver and be able to communicate with PACS directly. In addition, the current system is developed as a web-based zero footprint application. Therefore, we can integrate with current web-based HIS/EHR through a patient portal.
Compared with custom systems, IWEIS system are limited in functionality and advanced features. Custom systems usually are better designed in software architecture and can meet the specific needs from an individual clinical trial. Moreover, in the generalized system, when the workflow and rules become complex, the possibility of the occurrence of software bugs increases during the integration of various stages and workflow modification. Therefore, development of a generalized system is more complex than a custom system and needs to be tested with more clinical trials with a variety of situations. However, as discussed in the introduction, custom system are more time consuming to develop while the deployment of IWEIS system is able to reduce the cost for development and keep general functionalities. In the long term, with more advanced features developed in subsequent versions of the software, the IWEIS will finally be capable to support complex clinical trials like custom systems.
The workflow expansion and modification feature provides more flexibility than custom systems. However, it may also bring data inconsistency issue and possible conflicts in rules. For example, the participants’ datasets collected before the modification of the workflow may be different from new participants which may come after the modification of the system. Another possible example is that new rules require the participant to collect new data while the existing participant has already passed the collection time for new data. In these cases, the researcher needs to mark it as “not applicable” or “missing data” to continue.
This is an inherent issue when the workflow is modified in middle of the clinical trial. To address this data inconsistency issue, the future work of the system will be the development of a new feature that can back up various versions of workflow and link the workflow versions with data versions. This feature will facilitate researchers to analyze the data with an intuitive view of the corresponding workflow and help resolve the potential data inconsistency issue.
6. Conclusion
Workflow engines have been applied widely across the business field for years. Complex clinical trials can also benefit from workflow engines to address the demands for data collection, analysis, and distribution. To address these special requirements of imaging-based clinical trials, this paper discussed a novel prototype system with a workflow engine. The system was developed and evaluated for an imaging-based clinical rehabilitation trial named as Dose Optimization of Stroke Evaluation (DOSE) trial and the features of the system have been discussed. By providing flexibility in building and tailoring the workflow in various stages of clinical trials, the system will ultimately save time and reduce errors for clinical trials.
We present a system with an integrated workflow engine to support imaging-based clinical trials
The system allows deployment and modification of a data management system without extensive programming
Medical Imaging features are integrated together with informatics data management
The system is evaluated by a phase I rehabilitation clinical trial with imaging studies
Acknowledgments
This work was supported by NIH/NICHD R01HD065438
Footnotes
Conflict of Interest Statement
Ximing Wang
None Declared
Brent J Liu, PhD
None Declared
Clarisa Martinez, DPT
None Declared
Xuejun Zhang, PhD
None Declared
Carolee J Winstein, PhD
None Declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ximing Wang, Image Processing and Informatics Lab, Department of Biomedical Engineering, Viterbi School of Engineering, University of Southern California, Los Angeles, CA 90089.
Brent J Liu, Image Processing and Informatics Lab, Department of Biomedical Engineering, Viterbi School of Engineering, University of Southern California, Los Angeles, CA 90089.
Clarisa Martinez, Division of Biokinesiology & Physical Therapy, University of Southern California, Los Angeles, CA 90089.
Xuejun Zhang, School of Computer, Electronics and Information, Guangxi University, Nanning, Guangxi 530004, P. R. China.
Carolee J Winstein, Division of Biokinesiology & Physical Therapy, University of Southern California, Los Angeles, CA 90089.
References
- 1.ClinicalTrials.gov. http://clinicaltrials.gov/ct2/resources/trends.
- 2.Erickson BJ, Buckner JC. Imaging in Clinical Trials. Cancer Informatics. 2007;4:13–18. [PMC free article] [PubMed] [Google Scholar]
- 3.Parker J, Coiera E. Improving Clinical Communication: A View from Psychology. Journal of the American Medical Informatics Associationı: JAMIA. 2000;7(5):453–461. doi: 10.1136/jamia.2000.0070453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavlović Ivan, Kern Tomaž, Miklavčič Damijan. Comparison of paper-based and electronic data collection process in clinical trials: Costs simulation study. Contemporary Clinical Trials. 2009 Jul;30(4):300–16. doi: 10.1016/j.cct.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Huang HK. PACS and Imaging Informatics: Principles and Applications. John Wiley & Sons; Hoboken, New Jersey: 2010. [Google Scholar]
- 6.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research Electronic Data Capture (REDCap) - A metadata-driven methodology and workflow process for providing translational research informatics support. Journal of biomedical informatics. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko Ryan KL. A computer scientist’s introductory guide to business process management (BPM) Crossroads. 2009 Jun;15(4):11–18. doi: 10.1145/1558897.1558901. [DOI] [Google Scholar]
- 8.Wang Ximing, Martinez Clarisa, Wang Jing, Liu Ye, Liu Brent. Development of a user customizable imaging informatics-based intelligent workflow engine system to enhance rehabilitation clinical trials. Proc. SPIE 9039, Medical Imaging 2014: PACS and Imaging Informatics: Next Generation and Innovations; March 19, 2014; p. 90390G. [DOI] [Google Scholar]
- 9.Huser Vojtech, Rasmussen Luke V, Oberg Ryan, Starren Justin B. Implementation of workflow engine technology to deliver basic clinical decision support functionality. BMC Medical Research Methodology. 2011;11:43. doi: 10.1186/1471-2288-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Ximing, Verma Sneha, Qin Yi, Sterling Josh, Zhou Alyssa, Zhang Jeffrey, Martinez Clarisa, Casebeer Narissa, Koh Hyunwook, Winstein Carolee, Liu Brent. Imaging informatics-based multimedia ePR system for data management and decision support in rehabilitation research. Proc. SPIE 8674, Medical Imaging 2013: Advanced PACS-based Imaging Informatics and Therapeutic Applications; March 29, 2013; p. 86740P. [DOI] [Google Scholar]
- 11.DOSE clinical trial website. http://pt.usc.edu/ongoing_studies/stroke/
- 12.JQuery. http://jquery.com/
- 13.Diagramo. http://diagramo.com/
- 14.Twitter Bootstrap. http://getbootstrap.com/
- 15.Wang Ximing, Documet Jorge, Garrison Kathleen A, Winstein Carolee J, Liu Brent. A multimedia comprehensive informatics system with decision support tools for a multi-site collaboration research of stroke rehabilitation. Proc. SPIE 8319, Medical Imaging 2012: Advanced PACS-based Imaging Informatics and Therapeutic Applications; February 23, 2012; p. 83190U. [DOI] [Google Scholar]
- 16.NanoDICOM. PHP DICOM Toolkit. http://www.nanodicom.org/
- 17.Damasio Hanna, MD, Damasio Antonio., MD, PhD . Lesion Analysis in Neuropsychology. Oxford University Press; 1989. [Google Scholar]
- 18.Tatu Laurent, MD, Moulin Thierry, MD, Bogousslavsky Julien, MD, Duvernoy Henri., MD Arterial Territories of the Human Brain cerebral hemispheres. Neurology. 1998 Jun;50(6):1699–1708. doi: 10.1212/wnl.50.6.1699. [DOI] [PubMed] [Google Scholar]
- 19.Liu Brent J, Winstein Carolee, Wang Ximing, Konersman Matt, Martinez Clarisa, Schweighofer Nicolas. An imaging informatics-based ePR (electronic patient record) system for providing decision support in evaluating dose optimization in stroke rehabilitation. Proc. SPIE 8319, Medical Imaging 2012: Advanced PACS-based Imaging Informatics and Therapeutic Applications; February 23, 2012; p. 83190V. [DOI] [Google Scholar]
- 20.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 5(1):11–33. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]