Abstract
Background
There is considerable heterogeneity in asthma treatment response.
Objective
To identify biomarkers of corticosteroid treatment response in children with asthma and evaluate the utility and mechanistic basis of these biomarkers.
Methods
Children (5–18 years) presenting to the Emergency Department (ED) with an acute asthma exacerbation were recruited and followed during hospitalization. Nasal epithelial cells were collected upon presentation to the ED (T0) and 18–24 hours later (T1) and T1/T0 gene expression ratios were analyzed to identify genes associated with good and poor corticosteroid treatment response phenotypes. The utility of these genes in discriminating between systemic corticosteroid treatment response groups was then tested prospectively in a new cohort of patients. A gene candidate (VNN1) that consistently distinguished good versus poor response phenotypes was further studied in an experimental asthma model and VNN1 promoter methylation was measured by bisulfite pyrosequencing in patients.
Results
VNN1 mRNA expression changes were associated with systemic corticosteroid treatment response in children with acute asthma and VNN1 was required for optimal response to corticosteroid treatment in an experimental asthma model. A CpG site within the VNN1 promoter was differentially methylated between good versus poor treatment response groups and methylation at this site correlated with VNN1 mRNA expression.
Conclusions
We have identified a biological basis for poor corticosteroid treatment response that can be used to distinguish a subgroup of asthmatic children who respond poorly to systemic corticosteroid treatment. VNN1 contributes to corticosteroid responsiveness and changes in VNN1 nasal epithelial mRNA expression and VNN1 promoter methylation may be clinically useful biomarkers of treatment response in asthmatic children.
Keywords: childhood asthma, biomarker, vanin-1, treatment response, phenotype
INTRODUCTION
Asthma affects 25.7 million people in the US including 7 million children1. Although patients suffering from asthma share similar clinical symptoms, the disease is heterogeneous2. This heterogeneity contributes to the difficulty in both studying and treating asthma. Nearly two-thirds of asthmatic children reported at least one attack in the past year3, highlighting the suboptimal management of childhood asthma4. The frequency of absent or incomplete efficacy in asthma treatment has been estimated to be 40–70%5.
Currently, systemic corticosteroid treatment is considered the most effective medication for control of chronic asthma and for rescue of acute exacerbation. To better understand this individual variation and identify biomarkers of systemic corticosteroid treatment response, transcriptional profiling of individual host responses is a necessary and fundamental next step. This approach has been used successfully to classify subphenotypes of asthma including treatment response phenotypes6–9. Previous studies have often utilized samples requiring bronchoscopy or induced sputum collection, which is not always feasible in clinical practice, especially in children with acute asthma exacerbation (AAE). In the present study, we utilized genome-wide expression profiling of nasal epithelial cells to identify genes whose temporal expression patterns (before and after treatment) consistently and reliably discriminated between systemic corticosteroid treatment response groups among children hospitalized for asthma exacerbation. Nasal epithelial cells can be readily sampled safely during an asthma attack10 and reflect changes observed in the bronchial airways of asthmatic children11. We identified and replicated a gene, VNN1, whose mRNA expression consistently discriminated between good and poor responders to systemic corticosteroid treatment. We pursued mechanistic studies in an experimental asthma model and in human samples.
METHODS
Subjects
After IRB approval, children diagnosed with asthma ages 5–18 years who presented to Cincinnati Children’s Hospital Medical Center (CCHMC) ED with AAE were recruited. Exclusion criteria are listed in the Online Repository. Of the 57 subjects consented, 21 were hospitalized for asthma exacerbation and 15 genome-wide mRNA expression data for both time points. These 15 patients were used as a discovery cohort to test the association between gene expression and systemic corticosteroid treatment response. To further validate the findings from the discovery cohort, a replication cohort of 25 children hospitalized for asthma were recruited. Eighteen children hospitalized for AAE were recruited for methylation studies, five of whom overlapped with the replication cohort.
Subjects provided demographic, environmental, asthma trigger information, and personal and family allergy and asthma history data. Parental report of current inhaled corticosteroid (ICS) controller medication (Asmanex®, Flovent®, Qvar®, Pulmicort®, Advair®, Dulera®, Symbicort®, etc.) was also collected. To assess baseline asthma symptom severity and control, a respiratory symptom score was calculated (based on frequency of wheeze, cough, shortness of breath, and chest tightness)12 and the age-specific Asthma Control Test™ score was collected13.
Treatment protocol and Treatment Response Definitions
Enrolled patients were treated according to the CCHMC evidence-based treatment protocol for inpatient asthma exacerbation14–16. The admitting physician determined the initial interval of albuterol treatments, which were subsequently spaced based on physician or respiratory therapist assessments. Patients received 2mg/kg/day of prednisone while hospitalized and inhaled corticosteroids were continued via mouthpiece. Length of stay (LOS) was calculated as the number of hours from the time the admission decision was made to the time the subject met clinical discharge criteria (see Online Repository). Good responders were defined as those with LOS≤24 hours and poor responders as those with LOS>24 hours.
Nasal epithelial cell sample collection and processing
Nasal epithelial samples were collected at two time points from each subject: (1) in the ED (T0) and (2) on the inpatient floor 18–24 hours after receiving corticosteroids in the ED (T1). The procedure, characterization of cell types, sample processing, and RNA isolation have been described previously10. Nasal samples collected contained >90% epithelial cells, similar to our previous findings10. Expression profiles were generated on the Affymetrix Human Gene 1.0 ST platform. Quantitative real time-PCR was used to validate and replicate candidate genes (see Online Repository). DNA isolation, bisulphite treatment, and pyrosequencing analysis of methylation levels of 5 CpG sites within the VNN1 promoter for the methylation cohort is detailed in the Online Repository.
Experimental Asthma Model
VNN1−/− mice and age and sex-matched wild-type BALB/c mice (see Online Repository) were exposed to intratracheal doses of HDM (20 ug in 50 ul saline) or saline 3 times a week for 3 weeks as previously described17. Mice were treated with intra-peritoneal dexamethasone (3mg/kg in dimethyl sulfoxide, (DMSO)) or DMSO (100 ul) for the last 5 days of the 3-week model. Twenty-four hours after the last HDM challenge, airway hyperresponsiveness (AHR) was assessed (see Online Repository), bronchoalveolar lavage fluid (BALF) was collected, processed, and inflammatory cells were quantified as previously described17.
Statistical Analysis
Detection of differentially expressed genes in the discovery set
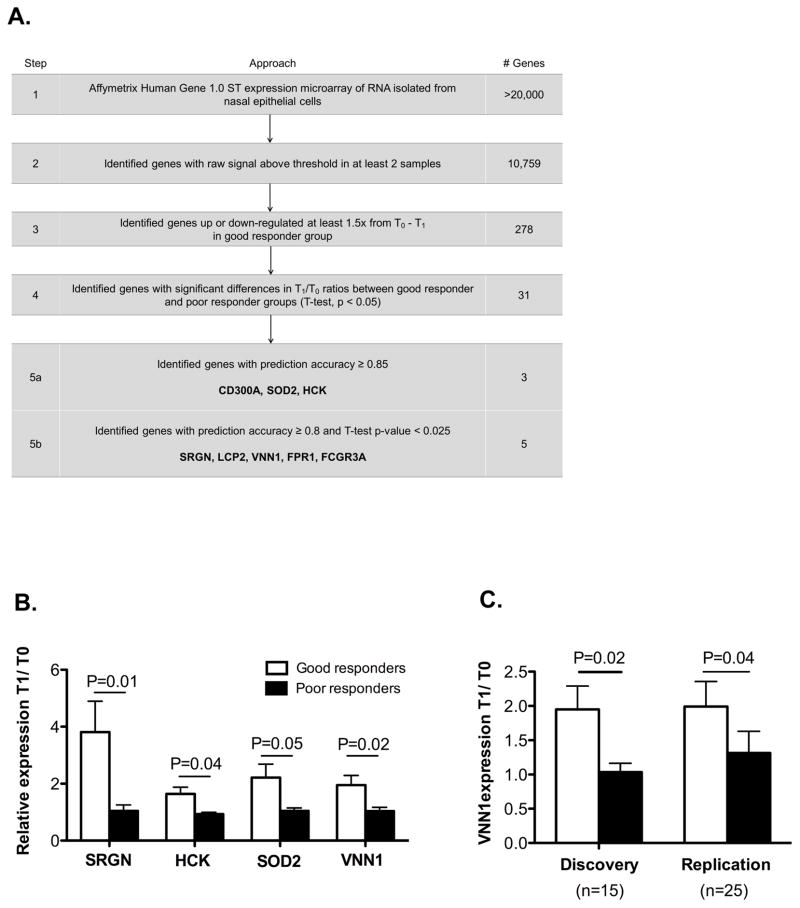
To identify candidate genes, we performed sequential filtering to balance concerns of type I and type II errors (Figure 1A). First, we sought to identify genes reliably expressed in nasal cells (raw signal above 100 in at least 2 samples). Next, we sought to identify genes responsive to treatment (T1/T0 ≥ 1.5 or T1/T0 ≤ 0.66; T1/T0: gene expression at T1 relative to that at T0). Then, we identified genes with significant differences in T1/T0 ratio between the good and poor responder groups. To minimize the risk of missing true associations, a p value threshold of 0.05 was employed as independent replication samples and complimentary biologic studies minimize the risk of false positive discovery. We then identified those genes with a high rate of prediction accuracy (≥0.80) through linear discriminant analysis. To validate these results, we performed qRT-PCR.
Figure 1.
(A) Overview of approach identifying genes whose expression in nasal epithelial cells discriminates pediatric asthma corticosteroid treatment response. (B) Expression of SOD2, HCK, SRGN, and VNN1 was quantified by qRT-PCR. (C) VNN1 expression was quantified in the discovery and replication cohorts. qRT-PCR results are expressed as average fold change at T1 relative to T0 with GAPDH as the internal control.
Microarray Data Analysis
Microarray cell image files were analyzed using GeneSpring GX software (Agilent Technologies, Santa Clara, CA). Probe level measurements were subject to initial background correction and normalization using GC-RMA. Transcript levels were normalized per chip to the 50th percentile, and per gene to median intensity.
Association Testing
In the discovery phase, we used t-tests (with log transformation) to identify genes between good and poor responders. Linear discriminant analysis18 was applied to find genes that best discriminated between good and poor responders.
For replication, we first examined whether there were differences between the discovery and replication cohorts that may introduce bias. Time of admission was significantly different between the discovery and replication cohorts. Thus, we matched our replication cohort to the discovery cohort based on month, T0 and T1 time using propensity scores19. Importantly, gene expression profiles were not considered in the matching process. After matching, we performed t-tests comparing the qPCR results from good and poor responders. A linear regression model was fitted to examine the association between the VNN1 mRNA expression change (T1/T0) and the continuous length of hospital stay (in hours) in the combined discovery and replication cohorts.
For the experimental asthma model, individual AHR, total BALF cell counts and eosinophil percentage in mice treated with HDM plus dexamethasone were compared to and normalized by the corresponding mean value in the HDM-treated group. Difference between the WT and VNN1−/− group was tested using non-parametric Mann Whitney test. A p-value < 0.05 was considered significant. Percentage reduction was used to present the corticosteroid response results.
For methylation analysis, Pearson correlation was used to measure the correlation between changes in mRNA expression (T1/T0) and DNA methylation (mT1-mT0) of VNN1. Fisher’s exact test was used to compare the difference in VNN1 DNA methylation between patients with good and poor treatment response. A p-value < 0.05 was considered significant.
RESULTS
Subjects
The discovery and replication cohorts were primarily male and non-white (Table 1). The discovery cohort was older than the replication cohort, but within each cohort there was no difference in age between the good and poor responders. There were no differences in individual parental reported asthma triggers (data not shown), mean baseline respiratory symptom frequency score, asthma control scores, or the proportion of patients presenting to the ED on a controller medicine between the discovery and replication cohorts (Table 1). By design, the discovery and replication cohorts were similar with respect to month admitted, T0 time, and T1 time (Table 1). The demographics and clinical features were also compared between the good and poor responders and no differences were detected (Table 2).
Table 1.
Description of discovery, replication and methylation cohorts.
| Discovery (n=15) | Replication (n=25) | p-valuea | Methylation (N=18) | |
|---|---|---|---|---|
|
| ||||
| Age mean (SD) | 13.4 (3.8) | 8.1 (2.8) | 0.0001b | 8.7 (4.3) |
| Age range | (7.4, 18.0) | (5.0,15.1) | (5.0,18.4) | |
| % Caucasian | 33.3% | 8.0% | 0.08d | 11.1% |
| % Male | 73.3% | 64.0% | 0.73d | 66.7% |
| Admission month range | (Apr, Dec) | (Mar Nov) | - | (Feb, Dec) |
| T0 sample time (24hr) | (9.3,20.8) | (10.0,21.4) | - | (10.0,18.3) |
| T1 sample time (24hr) | (8.1,16.7) | (8.5,17.3) | - | (7.7, 17.6) |
| ACT score (SD) | 16.0 (2.5) | 16.0 (4.4) | 0.98c | 16.3 (4.0) |
| Baseline average respiratory symptom score (median) (SD) | 1.5 (0.5) | 1.8 (0.9) | 0.18c | 1.8 (0.8) |
| % presenting to ED on ICS controller medicine | 26.7% | 32.0% | 1.00d | 55.6% |
ACT: asthma control test.
Comparison between discovery and replication populations.
Student’s t-test.
Mann-Whitney U test.
Fisher’s exact test.
Baseline average respiratory symptom score: representing average number of times per week the patient had coughing, wheezing, shortness of breath, or chest tightness and/or pain. A score was assigned for each of the four symptoms, and then an average was taken. A higher score represents higher symptom frequency. The values of the scores for each symptom are: 0 = Never; 1 = less than one time per week; 2 = 1–2 times per week; 3 = 3–5 times per week; 4 = 6–7 times per week.
Table 2.
Demographics and clinical features of good and poor responders.
| Good (n=21) | Poor (n=33) | p-valuea | |
|---|---|---|---|
|
| |||
| Age mean (SD) | 10.2 (4.2) | 9.9 (4.2) | 0.79c |
| % Caucasian | 20.0% | 15.2% | 0.64b |
| % Male | 70.0% | 69.7% | 0.98b |
| Admission month range | (Mar, Dec) | (Feb, Nov) | - |
| T0 sample time (24hr) | (10.0,20.5) | (9.3,21.4) | - |
| T1 sample time (24hr) | (9.8,14.8) | (7.7,17.6) | - |
| ACT score (SD) | 15.9 (4.0) | 16.2 (3.3) | 0.95c |
| Baseline average respiratory symptom score (median) (SD) | 1.7 (0.8) | 1.7 (0.7) | 0.55c |
| % presenting to ED on ICS controller medicine | 35.0% | 36.4% | 0.92b |
Comparison between good and poor responders.
Chi-squared test.
Mann-Whitney U test.
Baseline average respiratory symptom score: refer to Table 1.
Identification of genes differentially expressed between good and poor responder groups in discovery cohort
We used a multi-step filtering process to identify genes (Figure 1A). Starting with more than 20,000 genes, we identified 8 genes which were nominally significant (p ≤ 0.05) and had prediction accuracy of ≥0.80. Of these 8 genes, qRT-PCR expression of SOD2, HCK, SRGN, and VNN1 was significantly induced at T1 in the good compared to the poor responder group (Fig 1B). CD300A was not detectable in most samples and reliable results could not be achieved for LCP2, FPR1, and FCGR3A, due to low copy numbers.
VNN1 mRNA expression change predicts corticosteroid treatment response in the replication cohort
In order to substantiate our findings, we recruited an independent prospective cohort to serve as a replication. VNN1 mRNA expression was lower in the poor responder group compared to the good responder group (p=0.04, Figure 1C), replicating our findings from the discovery cohort (p=0.02, Figure 1C). Expression of SOD2, HCK, and SRGN was not significantly different between the treatment response groups (data not shown). We also evaluated length of stay as a continuous outcome in the combined discovery and replication cohorts. We observed an inverse association between the VNN1 mRNA expression change (T1/T0) and the length of hospital stay in hours. The results from the linear regression model show that for every unit increase in VNN1 expression, the stay length decreases by 7.7 hours (p=0.01), further supporting our findings.
In order to evaluate whether the observed VNN1 mRNA expression change was attributable to a baseline difference in VNN1 mRNA expression at T0, we compared VNN1 expression at T0 of all patients; no significant difference was detected (data not shown). To test whether the baseline ICS exposure was a confounding factor for the corticosteroid treatment response, we compared the proportion of subjects that presented to the ED on ICS between the good and poor responders; no significant difference was detected (Table 2).
Differential VNN1 methylation in response to corticosteroid treatment in good versus poor treatment response groups
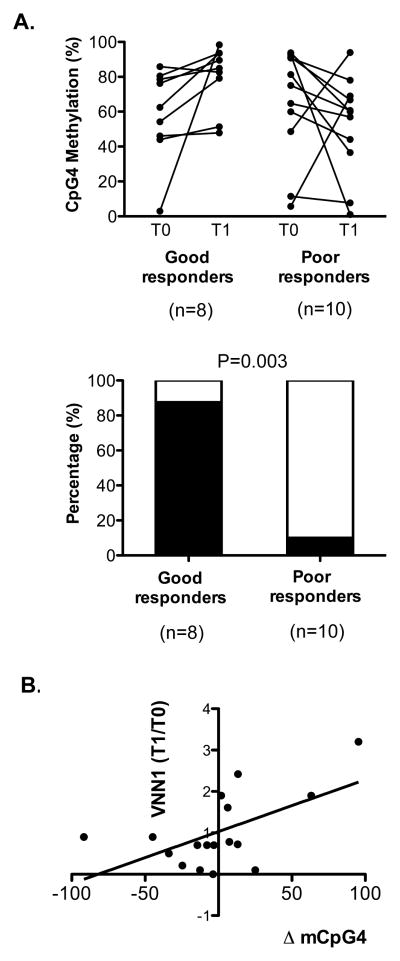
Since changes in VNN1 mRNA expression at T1/T0 were significantly associated with treatment response phenotypes in hospitalized asthmatic children and DNA methylation is an epigenetic mechanism regulating gene expression, we hypothesized that corticosteroid treatment may result in differential VNN1 methylation. To test this, we examined the methylation level of 5 CpG sites within the VNN1 promoter (defined as 2kb upstream from the transcription start site) in 20 patients (Table 1) with simultaneous nasal epithelial RNA and DNA samples collected at both time points (T0 and T1). The methylation level at the CpG4 site (1380bp upstream from TSS) trended to decrease in the poor responders, but increase in good responders following treatment. There was a significant difference in the percent of patients with increased versus decreased methylation at CpG4 at T1 compared to T0 in good versus poor responders (p=0.003, Fig 2A lower panel). Further, there was a positive correlation between the change in DNA methylation at CpG4 (mT1-mT0) and VNN1 mRNA expression (p=0.02, Pearson r=0.58, Fig 2B). These findings collectively suggest that methylation at the CpG4 site might be a crucial molecular event regulating VNN1 gene expression and modulating response to corticosteroid treatment.
Figure 2.
(A) Upper panel: CpG4 percent methylation at T0 vs. T1 in good (p=0.07) and poor responders (p=0.16). Lower panel: Percent of patients with increased (black) and decreased (white) mCpG4 at T1 vs. T0 in good and poor responders (**p=0.003). (B) Positive correlation between the percent change in VNN1 CpG4 methylation and fold change in VNN1 mRNA expression. *p=0.02, Pearson r=0.58.
VNN1−/− mice less responsive to dexamethasone treatment in an experimental asthma model
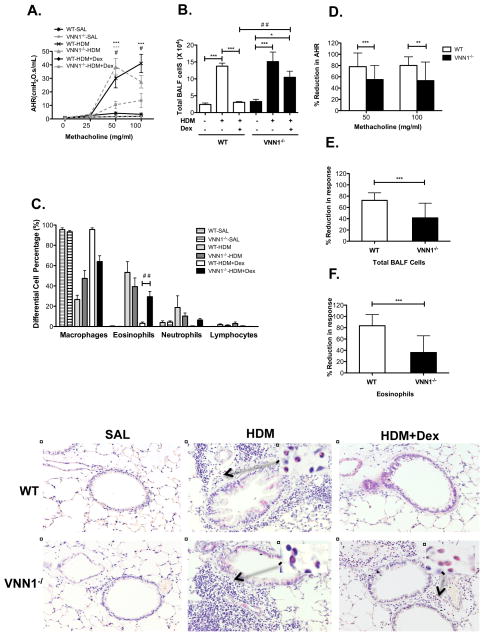
In order to more directly examine the role of VNN1 in the development of airway hyperresponsiveness and inflammation and corticosteroid treatment response, we studied VNN1−/− mice in an experimental asthma model. Repeated HDM exposure induced allergic airway inflammation and AHR in both WT and VNN1−/− mice, and the phenotype was comparable at higher doses of methacholine (50 and 100 mg/ml) (Fig 3A–C). At lower doses of methacholine (12.5 and 25 mg/ml), VNN1−/− mice exhibited significantly milder AHR compared to WT mice (data not shown), suggesting that VNN1 may contribute to the development of airway hyperresponsiveness. When we examined the response to dexamethasone treatment, VNN1 was required for optimal response to corticosteroid treatment. Dexamethasone significantly reduced AHR in WT mice, with an average percentage reduction of 78.1% (SD 24.2%) and 80.2% (SD 15.3%) at 50mg/ml and 100mg/ml methacholine challenge, respectively (Fig 3D). Dexamethasone also significantly alleviated airway inflammation in WT mice (Fig 3G, I, K), as reflected by a large reduction in total BALF cells (70.0%, SD 13.5 %) (Fig 3E) and eosinophils (83.6%, SD 19.6%) (Fig 3F). In contrast, VNN1−/− mice were significantly less responsive to dexamethasone. AHR was reduced by an average percentage of 55.2% (SD 24.9%) at 50mg/ml methacholine and 53.3% (SD 32.9%) at 100mg/ml methacholine (Fig 3D). Total BALF cells and eosinophils were only reduced by 40.9% (SD 26.0%) (Fig 3E) and 36.1% (SD 29.6%) (Fig 3F), respectively (p<0.001). Importantly, substantial numbers of residual eosinophils remained in the lung tissue after dexamethasone treatment (Fig 3H, J, L) in VNN1−/− mice.
Figure 3.
Representative (A) AHR, (B) Total BALF cell counts and (C) percent differential BALF cells in WT and VNN1−/− mice treated with saline, HDM or HDM plus dexamethasone (HDM+Dex) (n=6–8 per group). In (A): ***p<0.001 WT-HDM vs. WT-HDM+Dex; ***p<0.001 VNN1−/−-HDM vs. VNN1−/−-HDM+Dex; #p<0.05 WT-HDM+Dex vs. VNN1−/−-HDM+Dex. In (B–F): *p<0.05, ***p<0.001, ##p<0.01. Percentage reduction in (D) AHR, (E) total BALF cell counts and (F) percent BALF eosinophils in mice treated with HDM+Dex (n=20–28 per group). Data are from four independent experiments. H&E staining of lung tissues collected from WT (G,I,K) and VNN1−/− mice (H,J,L); mice treated with saline (G,H), HDM (I,J) and HDM plus dexamethasone (K,L) at a magnification of X200. Inserts: Infiltrated eosinophils at a magnification of x1000.
DISCUSSION
Children may have poorly controlled asthma for numerous reasons including lack of compliance with medications, socioeconomic barriers, suboptimal environments with numerous asthma triggers, and biologic causes. It is important to identify the underlying causes that contribute to poorly controlled asthma in each individual so that management strategies can be personalized to achieve the best outcomes. We have identified a biological basis for poor corticosteroid treatment response that can be used to distinguish a subgroup of children with asthma who respond poorly to treatment. VNN1 mRNA expression and promoter methylation were induced following corticosteroid treatment in nasal epithelial cells of asthmatic children who respond well to corticosteroid treatment, but not in those who were poorly responsive, i.e. difficult to treat during an AAE. Thus, these may be clinically useful biomarkers to identify children with a biologic etiology for poor corticosteroid response who would benefit from a different treatment plan. Further, based on animal studies, VNN1 contributes to corticosteroid responsiveness. Collectively, our studies suggest that targeting the VNN1 pathway may be a useful therapeutic strategy to enhance corticosteroid response among these difficult to treat patients.
Nasal epithelial expression of VNN1 discriminates between good and poor treatment responder phenotypes in children hospitalized for asthma exacerbation. Mechanistically, this is likely due to altered methylation at the CpG4 site of the VNN1 promoter in response to corticosteroids. We focused on children hospitalized for asthma because the inpatient setting provides a unique opportunity to characterize response to standardized treatment regimens for AAE. Since non-adherence to medications was not an issue during hospitalization, differences between subjects could be largely attributed to variation in individual host response to treatment. Although our findings stem from the hospital environment, they may have broad implications for difficult to treat patients who are not fully responsive to corticosteroid treatment and, as a consequence, do not easily achieve asthma control20. This group of patients is the most challenging and accounts for >50% of health care utilization related to this disease21, 22. Whether adherence to corticosteroid medication affects VNN1 mRNA expression and thus may impact asthma control in a chronic scenario is another interesting question worthy of further investigation.
A limitation of this study is that we did not have the data necessary to determine whether a given individual maintains his/her treatment response phenotype in subsequent exacerbations. Another limitation of our study is the lack of information regarding whether the protein level of VNN1 would be differentially expressed between the good and poor responders. The modest, yet reproducible alteration in mRNA expression might significantly affect the level or enzyme activity of VNN1 protein, which may subsequently cause variation in treatment response to corticosteroids.
Considerable advances have been made in recent years in identifying treatment response phenotypes of asthma that can help inform clinical decision-making. One phenotype, associated with high interleukin-13 expression7 and increased circulating periostin, is associated with improved response to an interleukin-13 inhibitor23 and to anti-IL-4 receptor alpha therapy24. Further, gene expression changes in response to inhaled corticosteroids in asthmatic adults identified that high baseline expression of three genes (CLCA1, periostin and serpinB2) and one gene (FKBP51) were associated with good or poor clinical response to corticosteroids, respectively6. In another study, expression of 6 genes in induced sputum discriminated between eosinophilic and neutrophilic asthma and predicted response to treatment with inhaled corticosteroids9 Our study adds to this small, but expanding list of biomarkers that can inform clinical decision making. A major advantage of our study is the use of nasal epithelial cells, which can be readily and easily sampled at the point of clinical care even in children experiencing an acute exacerbation.
Enhanced expression of VNN1 has been associated with multiple human diseases, including immune thrombocytopenia25, systemic lupus erythematosus26, and inflammatory bowel disease27, however, dysregulated VNN1 mRNA expression has not been reported in asthma. Consistent with this, the expression level of VNN1 in mouse lungs was not altered by repeated allergen or IL-13 in experimental models of asthma28, 29. Our animal studies reveal that VNN1 may play dual roles in asthma. On the one hand, absence of VNN1 prevents against development of asthma as evidenced by lower AHR at lower methacholine doses. This is not surprising based on the biological function of VNN1. Vanin-1 is an epithelial ectoenzyme with pantetheinase activity that provides cysteamine/cystamine to tissues and is implicated in redox homeostasis30, 31. VNN-1−/− mice have increased reduced glutathione stores and thus, may be more resistant to asthma development. Consistent with this, VNN-1−/− mice exhibit resistance to oxidative injury and reduced inflammatory responses to ROS inducers in thymus31. Nevertheless, even in the absence of VNN1, an asthma phenotype developed in the mice including severe AHR and airway inflammation indicating that VNN1 is not essential to asthma development. On the other hand, absence of the VNN1 gene resulted in resistance to corticosteroid treatment that was reflected by persistent AHR and inflammatory cells in the lungs despite treatment with dexamethasone in mice. Notably, eosinophils, a hallmark of pediatric severe therapy-resistant asthma32, persisted in the BALF and lungs of the VNN1−/− mice after dexamethasone treatment. Together, our findings suggest that VNN1 contributes to optimal host response to corticosteroid treatment.
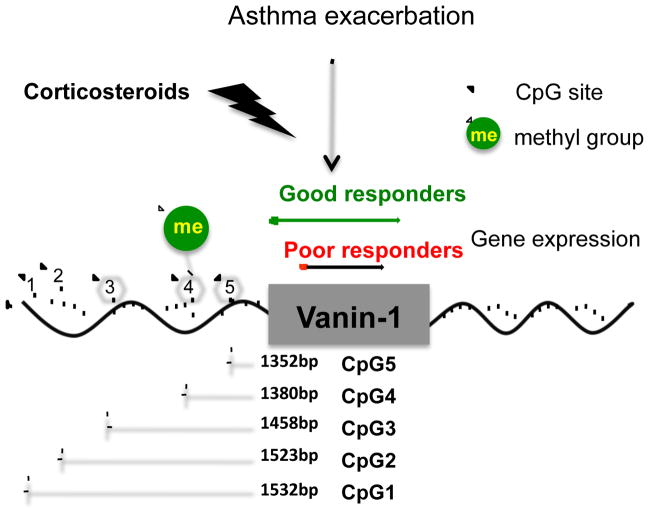
Differential methylation of the VNN1 promoter at CpG4 in response to corticosteroids is implicated as the mechanistic basis for the differential expression of VNN1 in good versus poor responders. Our study supports a model whereby corticosteroid treatment induces methylation of VNN1 at CpG4 and this leads to increased expression of VNN1 in good responders, but not poor responders (Figure 4). Methylation of CpG sites at promoter regions is generally believed to cause gene silencing33–35. However, positive correlations between promoter methylation and increased gene expression have been reported36,37. A recent study has verified numerous transcription factors with methylated CpG (mCpG)-dependent DNA binding activity38. Further, a zinc finger protein, CTCF, was found to bind to the promoter region of VNN1 gene in small airway epithelial cells39. CTCF can function as a chromatin insulator, repressing gene expression by blocking the interaction between gene promoters and enhancers40. For example, CTCF binds to the imprinting control region (ICR) of the Igf2/H19 locus and silences Igf2 expression via its enhancer-blocking activity. This activity is sensitive to DNA methylation as methylation of CpGs within the ICR abolishes the binding of CTCF and thus allows Igf2 expression41, 42. These findings provide possible explanations for our observation that the methylation level of the CpG4 motif was positively correlated with VNN1 expression. The mechanism by which increased VNN1 expression contributes to corticosteroid responsiveness remains to be elucidated. VNN1 has been shown to promote inflammation in murine intestinal epithelium by inhibiting expression and ligand-induced activation of PPARγ, an anti-inflammatory checkpoint, therefore, upregulating the expression of downstream proinflammatory target genes43. We did not observe any change in PPARγ expression in the lungs of VNN1−/− mice (data not shown). However, PPARγ may be dysregulated in specific cell types or an alternative downstream regulatory target of VNN1 other than PPARγ may be relevant in this phenotype. VNN1 deficiency did not impact the development of airway inflammation in an experimental asthma model, but it remains possible that the resolution of the asthma phenotype may be impacted.
Figure 4.
Proposed model. At baseline, the VNN1 gene is modestly expressed and this level of expression is not altered in stable asthma or acute asthma. During treatment for acute asthma exacerbation, corticosteroid treatment induces DNA methylation at the CpG4 site of the VNN1 gene promoter, enhancing the expression of VNN1 gene. Enhanced VNN1 expression contributes to optimal response to corticosteroid treatment.
In summary, nasal VNN1 expression may be a clinically useful biomarker to identify a subset of difficult to treat asthmatic children with a biologic etiology for poor corticosteroid response. Targeting the VNN1 pathway in this subset may be a useful therapeutic strategy to enhance corticosteroid response.
Supplementary Material
Clinical Implications.
VNN1 mRNA expression is induced following systemic corticosteroid treatment in asthmatic children who respond well to treatment, but not in those children who are poor treatment responders.
VNN1 is required to optimal response to corticosteroid treatment in experimental asthma.
VNN1 promoter methylation and expression are novel biomarkers of treatment response in asthmatic children.
Acknowledgments
We thank Cynthia Chappell for editorial assistance. We thank the physicians, nurses and staff of Cincinnati Children’s Hospital Medical Center Emergency Department and inpatient asthma team. We also thank all the patients and their families who participated in this study. This work was supported by NIH U19AI70235 (GKKH), NIH R21AI101375 (HJ) and a training grant fellowship from the National Institute of Environmental Health Sciences (T32 ES010957) to Chang Xiao.
Abbreviations
- AAE
Acute asthma exacerbation
- VNN1
Vanin-1
- IRB
Institutional review board
- CCHMC
Cincinnati Children’s Hospital Medical Center
- ED
Emergency department
- ICS
Inhaled corticosteroid
- LOS
Length of stay
- qRT-PCR
Quantitative real time polymerase chain reaction
- DNA
Deoxyribonucleic acid
- RNA
Ribonucleic acid
- HDM
House dust mite
- DMSO
dimethyl sulfoxide
- AHR
Airway hyperresponsiveness
- BALF
bronchoalveolar lavage fluid
- WT
Wild type
- VNN1−/−
Vanin-1 knockout
- SOD2
Superoxide dismutase 2
- HCK
Tyrosine-protein kinase
- SRGN
Serglycin
- CD300A
Cluster of Differentiation 300A
- LCP2
Lymphocyte cytosolic protein 2
- FPR1
Formyl peptide receptor 1
- FCGR3A
Low affinity immunoglobulin gamma Fc region receptor III-A
- IL-4
Interleukin 4
- IL-13
Interleukin 13
- CLCA1
Calcium-activated chloride channel regulator 1
- FKBP51
FK506 binding protein 5
- CTCF
CCCTC-Binding factor
- ICR
Imprinting control region
- PPARγ
peroxisome proliferator-activated receptor gamma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akinbami LJ, Moorman JE, Bailey C, Zahran HS, King M, Johnson CA, et al. Trends in asthma prevalence, health care use, and mortality in the United States, 2001–2010. NCHS Data Brief. 2012:1–8. [PubMed] [Google Scholar]
- 2.Bel EH. Mild asthma. N Engl J Med. 2013;369:2362. doi: 10.1056/NEJMc1313111. [DOI] [PubMed] [Google Scholar]
- 3.Fassl B, Nkoy F, Stone B, Srivastava R, Simon T, Uchida D, et al. The Joint Commission Children’s Asthma Care quality measures and asthma readmissions. Pediatrics. 2012;130:482–91. doi: 10.1542/peds.2011-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akinbami L Centers for Disease C, Prevention National Center for Health S. The state of childhood asthma, United States, 1980–2005. Adv Data. 2006:1–24. [PubMed] [Google Scholar]
- 5.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56:1054–70. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 6.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–63. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–95. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y, Zaas AK, Rao A, Dobigeon N, Woolf PJ, Veldman T, et al. Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza a infection. PLoS Genet. 2011;7:e1002234. doi: 10.1371/journal.pgen.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baines KJ, Simpson JL, Wood LG, Scott RJ, Fibbens NL, Powell H, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. J Allergy Clin Immunol. 2014;133:997–1007. doi: 10.1016/j.jaci.2013.12.1091. [DOI] [PubMed] [Google Scholar]
- 10.Guajardo JR, Schleifer KW, Daines MO, Ruddy RM, Aronow BJ, Wills-Karp M, et al. Altered gene expression profiles in nasal respiratory epithelium reflect stable versus acute childhood asthma. J Allergy Clin Immunol. 2005;115:243–51. doi: 10.1016/j.jaci.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 11.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014;133:670–8. e12. doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butsch Kovacic M, Biagini Myers JM, Lindsey M, Patterson T, Sauter S, Ericksen MB, et al. The Greater Cincinnati Pediatric Clinic Repository: A Novel Framework for Childhood Asthma and Allergy Research. Pediatr Allergy Immunol Pulmonol. 2012;25:104–13. doi: 10.1089/ped.2011.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu AH, Zeiger RS, Sorkness CA, Ostrom NK, Chipps BE, Rosa K, et al. The Childhood Asthma Control Test: retrospective determination and clinical validation of a cut point to identify children with very poorly controlled asthma. J Allergy Clin Immunol. 2010;126:267–73. 73 e1. doi: 10.1016/j.jaci.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 14.Evidence-based care guideline for management of acute exacerbation of asthma in children aged 0 to 18 years. Rockville MD: Agency for Healthcare Research and Quality (AHRQ).]; Available from http://www.guideline.gov/content.aspx?id=24528. [Google Scholar]
- 15.Muething SE. Improving patient outcomes by standardizing care. J Pediatr. 2005;147:568–70. doi: 10.1016/j.jpeds.2005.08.058. [DOI] [PubMed] [Google Scholar]
- 16.Gerhardt WE, Schoettker PJ, Donovan EF, Kotagal UR, Muething SE. Putting evidence-based clinical practice guidelines into practice: an academic pediatric center’s experience. Jt Comm J Qual Patient Saf. 2007;33:226–35. doi: 10.1016/s1553-7250(07)33027-4. [DOI] [PubMed] [Google Scholar]
- 17.Brandt EB, Kovacic MB, Lee GB, Gibson AM, Acciani TH, Le Cras TD, et al. Diesel exhaust particle induction of IL-17A contributes to severe asthma. J Allergy Clin Immunol. 2013;132:1194–204. e2. doi: 10.1016/j.jaci.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ripley BD. Pattern Recogition and Neural Networks. United Kingdom The Press Syndicate of the University of Cambridge; 1996. [Google Scholar]
- 19.Austin PC. An Introduction to Propensity Score Methods for Reducing the Effects of Confounding in Observational Studies. Multivariate Behav Res. 2011;46:399–424. doi: 10.1080/00273171.2011.568786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bousquet J, Mantzouranis E, Cruz AA, Ait-Khaled N, Baena-Cagnani CE, Bleecker ER, et al. Uniform definition of asthma severity, control, and exacerbations: document presented for the World Health Organization Consultation on Severe Asthma. J Allergy Clin Immunol. 2010;126:926–38. doi: 10.1016/j.jaci.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 21.Bell MC, Busse WW. Severe asthma: an expanding and mounting clinical challenge. J Allergy Clin Immunol Pract. 2013;1:110–21. doi: 10.1016/j.jaip.2013.01.005. quiz 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desai D, Brightling C. Cytokine and anti-cytokine therapy in asthma: ready for the clinic? Clin Exp Immunol. 2009;158:10–9. doi: 10.1111/j.1365-2249.2009.03998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–98. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 24.Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med. 2013;368:2455–66. doi: 10.1056/NEJMoa1304048. [DOI] [PubMed] [Google Scholar]
- 25.Zhang B, Lo C, Shen L, Sood R, Jones C, Cusmano-Ozog K, et al. The role of vanin-1 and oxidative stress-related pathways in distinguishing acute and chronic pediatric ITP. Blood. 2011;117:4569–79. doi: 10.1182/blood-2010-09-304931. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez-Munoz F, Amezcua-Guerra LM, Macias-Palacios M, Marquez-Velasco R, Bojalil R. Vanin-1 as a potential novel biomarker for active nephritis in systemic lupus erythematosus. Lupus. 2013;22:333–5. doi: 10.1177/0961203312474085. [DOI] [PubMed] [Google Scholar]
- 27.Gensollen T, Bourges C, Rihet P, Rostan A, Millet V, Noguchi T, et al. Functional polymorphisms in the regulatory regions of the VNN1 gene are associated with susceptibility to inflammatory bowel diseases. Inflamm Bowel Dis. 2013;19:2315–25. doi: 10.1097/MIB.0b013e3182a32b03. [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann N, Mishra A, King NE, Fulkerson PC, Doepker MP, Nikolaidis NM, et al. Transcript signatures in experimental asthma: identification of STAT6-dependent and -independent pathways. J Immunol. 2004;172:1815–24. doi: 10.4049/jimmunol.172.3.1815. [DOI] [PubMed] [Google Scholar]
- 29.Lewis CC, Aronow B, Hutton J, Santeliz J, Dienger K, Herman N, et al. Unique and overlapping gene expression patterns driven by IL-4 and IL-13 in the mouse lung. J Allergy Clin Immunol. 2009;123:795–804. e8. doi: 10.1016/j.jaci.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitari G, Malergue F, Martin F, Philippe JM, Massucci MT, Chabret C, et al. Pantetheinase activity of membrane-bound Vanin-1: lack of free cysteamine in tissues of Vanin-1 deficient mice. FEBS Lett. 2000;483:149–54. doi: 10.1016/s0014-5793(00)02110-4. [DOI] [PubMed] [Google Scholar]
- 31.Berruyer C, Martin FM, Castellano R, Macone A, Malergue F, Garrido-Urbani S, et al. Vanin-1−/− mice exhibit a glutathione-mediated tissue resistance to oxidative stress. Mol Cell Biol. 2004;24:7214–24. doi: 10.1128/MCB.24.16.7214-7224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bossley CJ, Fleming L, Gupta A, Regamey N, Frith J, Oates T, et al. Pediatric severe asthma is characterized by eosinophilia and remodeling without T(H)2 cytokines. J Allergy Clin Immunol. 2012;129:974–82. e13. doi: 10.1016/j.jaci.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–58. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 35.Nan X, Cross S, Bird A. Gene silencing by methyl-CpG-binding proteins. Novartis Found Symp. 1998;214:6–16. doi: 10.1002/9780470515501.ch2. discussion -21, 46–50. [DOI] [PubMed] [Google Scholar]
- 36.Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–42. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 37.Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15:R37. doi: 10.1186/gb-2014-15-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN, et al. DNA methylation presents distinct binding sites for human transcription factors. Elife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–8. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–96. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 41.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–9. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 42.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 43.Berruyer C, Pouyet L, Millet V, Martin FM, LeGoffic A, Canonici A, et al. Vanin-1 licenses inflammatory mediator production by gut epithelial cells and controls colitis by antagonizing peroxisome proliferator-activated receptor gamma activity. J Exp Med. 2006;203:2817–27. doi: 10.1084/jem.20061640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.