Abstract
Preserving the integrity of the DNA double helix is crucial for the maintenance of genomic stability. Therefore, DNA double-strand breaks represent a serious threat to cells. In this review, we describe the two major strategies used to repair double strand breaks: non-homologous end joining and homologous recombination, emphasizing the mutagenic aspects of each. We focus on emerging evidence that homologous recombination, long thought to be an error-free repair process, can in fact be highly mutagenic, particularly in contexts requiring large amounts of DNA synthesis. Recent investigations have begun to illuminate the molecular mechanisms by which error-prone double-strand break repair can create major genomic changes, such as translocations and complex chromosome rearrangements. We highlight these studies and discuss proposed models that may explain some of the more extreme genetic changes observed in human cancers and congenital disorders.
Keywords: End joining, Homologous recombination, Mutagenesis, Break induced replication, Translesion polymerases
DNA double-strand breaks create challenges to genome integrity and opportunities for genetic diversity
DNA double-strand breaks (DSBs) are chromosome lesions with high mutagenic potential. They can be caused by a number of exogenous factors and endogenous processes, including exposure to high-energy radiation, movement of transposable elements, and the collapse of DNA replication forks (reviewed in Mehta and Haber, 2014). In contrast to other types of DNA damage, which typically alter just one strand of the double helix and can be accurately repaired using the other strand as a template, DSBs result in the loss of integrity of both complementary strands. This poses a unique challenge to the cell and results in more opportunities for inaccurate repair. Importantly, error-prone repair of DSBs can alter the DNA sequence and result in loss of genetic information, occasionally leading to extreme genomic instability that characterizes cancer and certain other human diseases.
DSBs are also important intermediates in many adaptive processes, including meiotic recombination in most sexually-reproducing organisms and mating type switching in budding and fission yeasts. In many vertebrates, programmed DSBs are also created during the assembly of immunoglobulin antigen receptor genes by V(D)J recombination and during immunoglobulin heavy chain class switching (reviewed in Boboila et al., 2012). Furthermore, many genome editing techniques, including those that utilize zinc finger nucleases, TALENs, and CRISPR-Cas, involve double-strand break intermediates that must be repaired by the cell (reviewed in Cox et al., 2015). Depending on the type of repair employed, repair of these engineered breaks can proceed faithfully or result in mutations and/or genome modifications. Thus, a thorough understanding of the factors that may promote error-prone DSB repair is important to be able to drive these processes towards a desired outcome.
End joining and homologous recombination can both be mutagenic
Cells use two general types of mechanisms to repair DSBs (Figure 1). The first set of mechanisms, collectively referred to as non-homologous end-joining (NHEJ) repair, is characterized by rejoining of broken ends without the use of extensive homology. NHEJ is frequently associated with the presence of small insertions and deletions (indels) at the break site. Although NHEJ is usually regarded as error-prone, its actual propensity towards inaccurate repair may be overestimated, since accurate NHEJ repair is genetically silent in many of the standard assays that have been used to study it (Betermier et al., 2014). It is now clear that at least two subtypes of NHEJ operate in many cells. Generally, these are referred to as classical non-homologous end joining (C-NHEJ) and alternative non-homologous end joining (A-NHEJ). As we describe in the next section, these two types of repair have very different consequences for genome integrity.
Figure 1. Mutagenic potential of different double-strand break repair mechanisms.
Double-strand breaks can be repaired via either non-homologous end joining (NHEJ) or homologous recombination (HR) mechanisms. While classical NHEJ (C-NHEJ) can result in perfect repair, small insertions/deletions are also possible. Microhomology-mediated end joining (MMEJ) is a deletional repair mechanism, while other forms of alternative end joining (A-NHEJ) always result in changes to the DNA sequence. Homologous recombination that proceeds via double Holliday junction intermediates (DSBR) or synthesis-dependent strand annealing (SDSA) are potentially mutagenic, especially if template switching occurs, while break-induced replication (BIR) is highly mutagenic and can lead to complex chromosomal aberrations.
In contrast to NHEJ, repair of DSBs by homologous recombination (HR) involves the copying of DNA from a homologous template. HR is typically described as an error-free repair mechanism. However, studies from budding yeast have shown that HR can be up to 1000-fold more mutagenic than normal DNA replication (Deem et al., 2011; Hicks et al., 2010). The types of mutations observed in the yeast systems following HR repair include base pair substitutions, indels, and complex chromosome rearrangements. In addition, analysis of duplication and triplication events from human populations suggests that various forms of HR repair may be responsible for genome-destabilizing events that lead to cancers and inherited disorders (Chen et al., 2010). Thus, it is becoming clear that HR repair has the capacity to be highly mutagenic, at least in certain contexts.
In this review, we highlight recent studies that demonstrate the mutagenic capacity of various types of DSB repair, with a focus on mechanistic insights and models that can explain the observed mutagenicity of A-NHEJ and homologous recombination. We analyze the evidence for three potential sources of HR-induced mutagenesis: (1) the vulnerability of single-stranded DNA to damaging agents, (2) the use of error-prone translesion DNA polymerases during various stages of repair synthesis, and (3) the non-processive nature of HR repair synthesis, which provides frequent opportunities for DNA polymerase slippage and template switching. These template switches, when they involve synthesis from homeologous or microhomologous templates, can lead to the introduction of non-synonymous sequences and/or coupled deletions and insertions. Finally, we highlight how both aberrant end joining and homologous recombination may contribute to the chromosome rearrangements and genomic instability observed in many human diseases, particularly cancer.
End-joining repair encompasses several mechanisms with varying levels of mutagenic potential
Classical non-homologous end joining, or C-NHEJ, has evolved as a rapid and efficient way to repair DSBs. Typically, it is viewed as an error-prone mechanism that generates small sequence changes near the DSB site (Figure 2), perhaps because of its documented mutagenic role in V(D)J recombination during the diversification of antibodies in the mammalian immune system (Boboila et al., 2012). It has been argued that the perceived mutagenic nature of C-NHEJ is actually due to its flexibility and proficiency in dealing with a wide range of DSB structures (Betermier et al., 2014). Nonetheless, C-NHEJ is able to accurately restore the DNA duplex to its original form, especially if the DNA ends to be ligated are complementary. Such genetically silent outcomes are difficult to measure in a chromosomal context but can be quantified in assays involving recircularization of restriction-enzyme cut plasmid DNA (Boulton and Jackson, 1996).
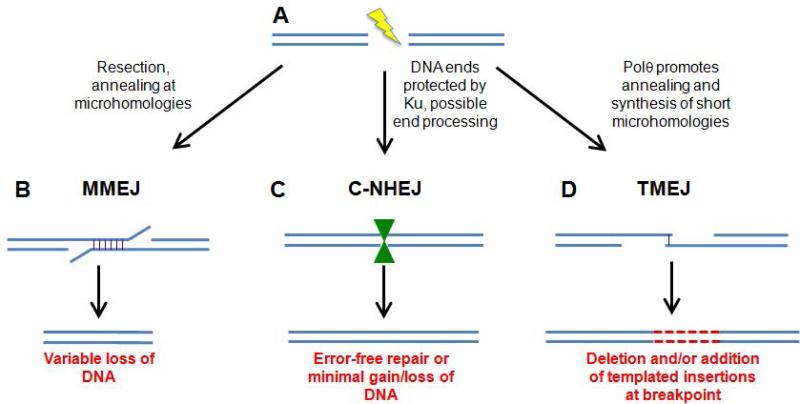
Figure 2. Mechanisms and outcomes of Non-Homologous End Joining (NHEJ).
A. Creation of a DNA double-stranded break. B. Repair via microhomology-mediated end joining (MMEJ). The break is resected (or ends are unwound) and exposed microhomologous sequences anneal (vertical lines). Repair is completed by flap removal, fill-in synthesis, and DNA ligation. C. Repair by classical non-homologous end joining (C-NHEJ). Binding of the Ku heterodimer to DNA ends protects the DNA from extensive resection or unwinding. If necessary, processing enzymes such as nucleases and polymerases are recruited. Completion of repair depends on DNA Ligase 4. Green hourglass represents the protein complex (Ku plus other proteins) that synapse and process the DNA ends. D. Repair by polymerase theta-mediated end joining (TMEJ). Short regions of homology are extended by Polθ. If no further processing occurs, the result is a deletion (not shown). Following synthesis, multiple rounds of unwinding, reannealing, and Polθ-dependent synthesis can lead to the addition of templated insertions resulting in simple deletions. Repair outcomes of additional small templated insertions (represented as red, dashed lines).
C-NHEJ requires, at a minimum, the Ku70/80 proteins that bind to the broken ends and prevent extensive DNA resection and the DNA ligase 4/XRCC4/XLF complex that is needed for the final ligation step (reviewed in Waters et al., 2014; Williams et al., 2014). C-NHEJ in vertebrates also utilizes DNA-PKcs, which employs its kinase signaling activity to autophosphorylate and to signal for the recruitment of other C-NHEJ proteins (Jette and Lees-Miller, 2015). A number of C-NHEJ proteins are also utilized for the processing and repair of non-cohesive or complex DNA ends, including the Artemis endonuclease (Riballo et al., 2004), the X-family polymerases mu and lambda (Nick McElhinny et al., 2005), and the newly-identified XRCC4 paralog PAXX (Xing et al., 2015).
Cells in which C-NHEJ is blocked due to mutation or chemical inhibition of one of the core C-NHEJ components are still able to repair DSBs through one or more alternative end-joining mechanisms (A-NHEJ) (reviewed in Deriano and Roth, 2013). In one type of A-NHEJ, termed microhomology-mediated end joining (MMEJ), repair initiates by resection or unwinding of double-stranded DNA to expose short, single-strand microhomologies on the order of 6-20 nucleotides (Figure 2B) (Ma et al., 2003; McVey and Lee, 2008). These sequences subsequently anneal and repair is completed by cleavage of overhanging 3’ flaps, synthesis to fill single-stranded gaps, and ligation. In contrast to C-NHEJ, MMEJ does not require DNA ligase 4, but instead utilizes DNA ligase 1 in yeast (Ma et al., 2003) and DNA ligases 1 and 3 in metazoans in the rejoining step (Paul et al., 2013). Poly (ADP-ribose) polymerase (PARP) has also been shown to be important for MMEJ repair (Wang et al., 2006), although its exact role is unclear. By its very nature, MMEJ is always mutagenic, resulting in deletions of varying lengths and loss of genetic information.
A more extreme version of MMEJ exists in the single-strand annealing (SSA) pathway of DSB repair. SSA is sometimes classified as a homologous recombination repair mechanism because SSA and HR share some common genetic requirements and initial mechanistic steps (Fishman-Lobell et al., 1992). During SSA, extensive resection occurs, revealing single-stranded, complementary DNA ends of ~25 base pairs to multiple kilobases. As with MMEJ, these ends anneal and repair is completed by flap cleavage and ligation. The genetic requirements for MMEJ and SSA partially, but not fully, overlap. Resection is needed for both mechanisms, but the annealing, synthesis, and processing stages may involve specific proteins and repair complexes (Decottignies, 2013).
While MMEJ and SSA always create repair products containing simple deletions, A-NHEJ mechanisms can also result in repair junctions with complex insertions/deletions (indels), where the insertions appear to be perfect or imperfect repeats of nearby flanking sequences. Multiple studies have now identified translesion DNA polymerase theta (Polθ) as a crucial player in this form of A-NHEJ, sometimes referred to as synthesis-dependent microhomology-mediated end joining (SD-MMEJ) (Yu and McVey, 2010) or theta-mediated end joining (TMEJ) (Koole et al., 2014). Polθ appears to play multiple roles in TMEJ (Figure 2D), including bridging of the broken ends, possibly at microhomologous sequences, and both template-dependent and independent synthesis of new DNA that may be used as nascent microhomologies during the annealing step (Kent et al., 2015; Yousefzadeh et al., 2014; Zahn et al., 2015). Intriguingly, Polθ expression is upregulated in a variety of human cancers (Higgins et al., 2010; Kawamura et al., 2004; Lemee et al., 2010). Recent studies suggest that Polθ-dependent TMEJ becomes essential when other DSB repair pathways, such as homologous recombination, are compromised (Ceccaldi et al., 2015; Kawamura et al., 2004; Mateos-Gomez et al., 2015). It will be important to determine the true extent to which Polθ-mediated processes are responsible for the mutagenesis observed during end-joining repair in humans and other organisms.
Homologous recombination repair—consult and copy
In contrast to NHEJ, the hallmark of homologous recombination repair is its reliance on a homologous template in order to recover genetic information that may be lost during the initial breakage step or subsequent DNA processing. Many HR models have been proposed, based on a combination of genetic and biophysical analyses of recombination intermediates and repair products (Figure 3). All of these models begin with 5’✧3’ resection of the broken ends to produce 3’ single-stranded DNA (ssDNA). Resection is initiated by the action of the Sae2/CtIP nuclease in combination with a complex of the Mre11, Rad50, and Xrs2/Nbs1 proteins (reviewed in Symington and Gautier, 2011). The short stretches of single-stranded DNA can then be acted upon by additional proteins that carry out more extensive resection, creating multiple kilobases of ssDNA (Figure 3B). Following resection, the single-stranded DNA is bound by replication protein A (RPA), which protects it from further degradation. Before HR can proceed, RPA must be displaced and the Rad51 recombinase protein must be loaded onto the single-stranded DNA ends, creating a nucleoprotein filament. Rad51 loading and stabilization is assisted by the Rad52 protein and other factors in budding yeast and by the BRCA1/PALB2/BRCA2 complex in metazoans (reviewed in Heyer, 2007; Prakash et al., 2015).
Figure 3. Opportunities for mutagenesis during homologous recombination repair of a DNA double-strand break.
A. Creation of a DNA double-strand break. B. Resection of broken ends creates 3’ single-stranded DNA that can be easily damaged. C. One-ended strand invasion into a homologous template creates a displacement (D)-loop. Synthesis during D-loop extension is potentially mutagenic. Initial strand invasion can also occur at homeologous or microhomologous sequences, resulting in insertion/deletion repair products. D. During synthesis-dependent strand annealing (SDSA), D-loop dissociation and annealing of the nascent strand with single-stranded DNA from the broken chromosome is followed by potentially mutagenic single-stranded gap filling and ligation. E. Alternatively, two-ended invasion and synthesis leads to double Holliday junction (dHJ) formation. Resolution of the dJHs can create crossover (shown) or non-crossover (not shown) products. Asterisks indicate new potential sites of mutagenesis at each step.
Once formed, the nucleoprotein filament engages in a search for a homologous repair template. The mechanism and exact nature of this homology search remains under intensive investigation. In most contexts, a sister chromatid is the preferred HR repair template, due to the presence of cohesin proteins that hold sister chromatids in close physical proximity during S phase and G2, when HR is most active (Mehta et al., 2013). However, the homology search can take place across the entire genome. As described below, this increases the chances of successful repair but also increases the opportunities for inappropriate pairing and mutagenesis.
Following successful strand invasion, a displacement loop (D-loop) is formed, in which the 3’ end of the broken chromosome pairs with one strand of the homologous template and concomitantly displaces the other strand (Figure 3C). The 3’ end can then be extended by one or more DNA polymerases. As repair synthesis proceeds and the D-loop is extended, the HR repair pathways can diverge. According to the double-strand break repair (DSBR) model, if the displaced strand of the expanded D-loop begins to pair with the other 3’ single-stranded tail, second end capture ensures (Figure 3E). Subsequent ligation of the nascent strands to the other broken ends results in the formation of double Holliday junctions. These can be cleaved by endonucleases to form either crossover or non-crossover products (Wyatt and West, 2014), or they can be dissolved through combined helicase/topoisomerase action (Bizard and Hickson, 2014).
An alternative to the DSBR model involves the unwinding of the nascent strand from the D-loop, followed by its annealing to the other 3’ end of the broken chromosome and filling of single-stranded gaps (Figure 3D). This type of repair, termed synthesis-dependent strand annealing (SDSA), results in non-crossover outcomes (Morrical, 2015). In contrast to bulk DNA replication, SDSA involves conservative DNA synthesis, as the presence of newly synthesized DNA is confined to the original, broken chromosome while the template DNA is unchanged (Ira et al., 2006). Notably, following D-loop unwinding, the nascent strand has an opportunity to anneal with sequences other than those found at the other end of the broken chromosome. This promiscuity can contribute greatly to the mutagenesis that is associated with SDSA.
In cases of one-ended chromosomal breaks caused by replication fork collapse or shortening of telomeres, another HR mechanism called break-induced replication (BIR) can be used for repair (Figure 4) (reviewed in Malkova and Ira, 2013). DNA synthesis during BIR is conservative, similar to SDSA (Donnianni and Symington, 2013). However, the amount of synthesis during BIR is much more extensive, consisting of up to hundreds of kilobases. Experiments conducted largely in S. cerevisiae suggest a model in which DNA copying proceeds via a migrating D-loop (Saini et al., 2013) with second-strand synthesis significantly delayed relative to the initial strand, although the length of the delay prior to second strand synthesis is not known.
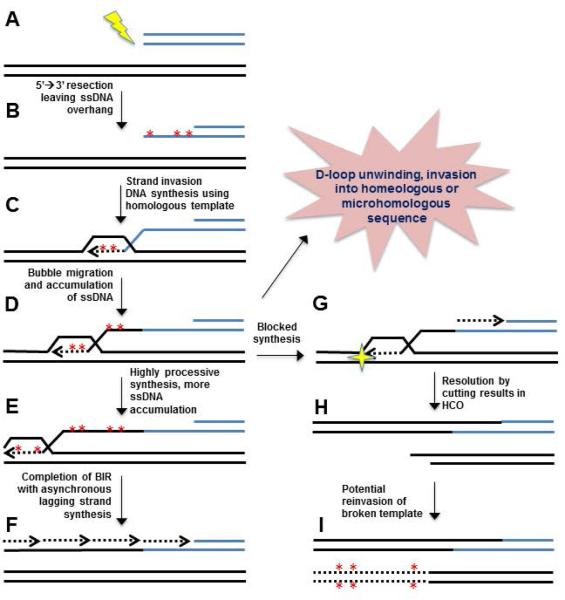
Figure 4. Opportunities for mutagenesis during break-induced replication (BIR).
A. Creation of a one-ended double-strand break. B. Resection of broken ends creates 3’ single-stranded DNA (ssDNA) that can be easily damaged. C. One-ended strand invasion into a homologous template creates a displacement (D)-loop. Synthesis during D-loop extension in BIR is highly mutagenic. D. Migration of the D-loop results in extensive accumulation of ssDNA. Unwinding of the D-loop prior to completion of synthesis and re-invasion into homeologous or microhomologous sequences can result in insertion/deletion mutations. E. Processive synthesis up to hundreds of kilobases to the end of a chromosome and accumulation of more ssDNA, which is easily damaged. F. Completion of repair via lagging strand synthesis. G. DNA damage or genetic impairment of BIR can stall repair synthesis. H. Subsequent resolution of the blocked replication fork results in a half crossover (HCO) event. I. Re-invasion of the broken template can result in subsequent rounds of BIR. Asterisks indicate new potential sites of mutagenesis at each successive step.
The first BIR assays in budding yeast involved transformation of a linearized vector, followed by BIR using a homologous sequence on chromosome III as a template to generate a stable chromosome fragment (Davis and Symington, 2004). These studies established that like SDSA, BIR is Rad52-dependent. BIR efficiency is greatly affected by loss of Rad51, although Rad51-independent BIR can occur at a low frequency (Davis and Symington, 2004; Ira and Haber, 2002).
Studies of BIR conducted in diploid strains revealed differences in the kinetics of BIR and SDSA. Using Southern blotting and PCR analysis of synchronously-induced breaks, Malkova et al. showed that SDSA repair of a two-ended DSB is typically completed in less than two hours and does not activate the G2/M DNA damage checkpoint (Malkova et al., 2005). However, the initiation of BIR takes several additional hours and occurs in the context of an active G2/M checkpoint, suggesting fundamental mechanistic differences between SDSA and BIR.
Thorough genetic analysis of SDSA and BIR reveals both similarities and differences in their genetic requirements (Table 1). While both share a need for certain proteins involved in the initiation of replication, including Dpb11 and Sld2/3, BIR has an additional requirement for the replicative helicase proteins, including the GINS complex, Cdc45, and the MCM complex. However, BIR does not need the Cdc6 or ORC proteins that allow DNA synthesis to initiate at replication origins (Lydeard et al., 2010). In addition, BIR, but not SDSA, utilizes DNA polymerase alpha (Polα). Intriguingly, BIR also requires Pol32, a non-essential subunit shared by Polδ and translesion polymerase zeta (Polζ), and the 3’✧5’ DNA helicase Pif1 (Lydeard et al., 2007; Saini et al., 2013; Wilson et al., 2013). Notably, Pol32 and Pif1 are not required for normal DNA replication. Although the exact functions of Pol32 and Pif1 in BIR are not well understood, one possibility is that Pol32 is acting to promote the processivity of Polδ, while Pif1 may be important for the progression of the mobile D-loop.
Table 1.
Genetic requirements of SDSA vs. BIR in S. cerevisiae
| Protein(s) | Required for SDSA? |
Required for BIR? | References |
|---|---|---|---|
| Rad52 | Yes | Yes | (Davis and Symington, 2004; Ira and Haber, 2002) |
| Rad51 | Usually, but not always |
Usually, but not always |
(Davis and Symington, 2004; Ira and Haber, 2002) |
| PCNA | Yes | Yes | (Holmes and Haber, 1999; Lydeard et al., 2010) |
| Dpb11, Sld2/3 | Yes | Yes | (Hicks et al., 2010; Lydeard et al., 2010) |
| Replicative helicase (Cdc45, GINS, MCMs) |
No | Yes | (Hicks et al., 2010; Lydeard et al., 2010; Wang et al., 2004) |
| Cdt1 | No | Yes | (Lydeard et al., 2010) |
| Cdc6/0RC | No | No | (Lydeard et al., 2010) |
| Pol δ/ε | Yes | Yes | (Hicks et al.,2010; Lydeard et al., 2010) |
| Pol α | No | Yes | (Lydeard et al., 2010) |
| Pol 32 (nonessential subunit of Pol δ and Pol ζ) |
No | Yes | (Deem et al., 2008; Lydeard et al., 2010) |
| Pif1 helicase | No | Yes | (Saini et al., 2013; Wilson et al., 2013) |
While most BIR studies have been conducted in yeast, there is evidence that BIR mechanisms exist and may be conserved in higher eukaryotes. One early study, which showed a complex rearrangement in a hemophilia patient that affected the copy number of several genes, including the Factor VIII gene responsible for hemophilia, attributed the copy number variations to a BIR-induced gross chromosomal rearrangement (Sheen et al., 2007). In a more recent study, BIR-like recombination events were observed in an osteosarcoma cell line in which replication stress was induced by cyclin E overexpression (Costantino et al., 2014). The majority of these events were dependent on POLD3, which encodes the human homolog of yeast Pol32. Thus, it is possible that some copy number alterations of up to 200 kb in human cells may result from a Pol32-dependent BIR process.
What processes are responsible for mutagenesis during SDSA?
Some of the first evidence that homologous recombination may be error-prone came from investigations that took advantage of the S. cerevisiae mating type switching system. Mating type in budding yeast is determined by the presence of one of two distinct alleles at the MAT locus. Haploid yeast cells can switch their mating type by using the site-specific HO endonuclease to induce a DSB at the MAT locus. Following induction of a DSB, SDSA is used to copy the opposite mating type information from one of two intrachromosomal donors (Haber, 2012). Many labs have adapted this system, placing the HO recognition sites at various places in the yeast genome and expressing HO under the control of inducible promoters.
In one of the earlier studies that used this system, a DSB was created at an HO site placed 0.3 kb away from various revertible trp1 mutant alleles. The reversion of these trp1 alleles was 100-fold higher in cells that suffered an induced DSB (Strathern et al., 1995). Interestingly, while Rev3, the catalytic subunit of error-prone Polζ, was not required for efficient homologous repair, it was responsible for about 95% of base-substitution mutations (Holbeck and Strathern, 1997). However, Rev3 was not responsible for frameshifts in this assay, suggesting that one or more DNA polymerases, in addition to Polζ, could be contributing to mutagenesis during HR. The most parsimonious interpretation of these results is that Polζ, supported by other translesion polymerases, is responsible for much of the single-strand gap filling that occurs at the end of SDSA.
Unexpectedly, it now appears that DNA synthesis that occurs during the extension of the D-loop intermediate during SDSA can also be error-prone. Support for this model came from experiments utilizing a mechanism similar to mating type switching, but with a modification in which the donor locus was altered to include the homeologous Kluveromyces lactis URA3 gene, which shares 73% identity with S. cerevisiae URA3 (Hicks et al., 2010). Following HO induction at the MAT locus, SDSA occurred efficiently to yield a wild-type URA3 gene at MAT, but rare Ura-deficient mutants could be selected. Astoundingly, the rate of DSB-induced mutagenesis was increased up to 1400-fold over spontaneous mutation rates (Hicks et al., 2010).
Most of the observed mutations in this study were base pair substitutions and frameshift mutations. In addition, more complex mutations were observed. These included multiple examples of template switching in the context of imperfect palindromes, along with apparent interchromosomal template switches that involved the use of 2-17 base pair microhomologies during the template switching process. These types of mutations provide the strongest support for the potential unstable nature of the D-loop intermediate in SDSA.
Using proofreading-deficient alleles of replicative polymerases Polδ and Polε, the researchers determined that most of the HR-induced mutations were due to the actions of these polymerases. Notably, a mutation in the catalytic subunit of Polδ eliminated all (−1) frameshift mutations and complex events involving template switching, suggesting that Polδ plays a significant role in specific mutational outcomes during SDSA.
A subsequent study from the same lab showed that interchromosomal template switching during gene conversion can occur at a rate of 0.3% when switching occurs between regions of limited homology, and that the switching can occur at a variety of microhomologous junctions (Tsaponina and Haber, 2014). Template switching relies heavily on the Rdh54 protein, a chromatin remodeling factor which plays a role in meiotic recombination in yeast but is not involved in normal SDSA. Although the exact role of Rdh54 in template switching is not well understood, its identification shows that the process of template switching during SDSA is regulated by proteins other than just the DNA polymerases themselves.
The question of whether or not SDSA is mutagenic in other organisms remains an important one to address. Germane to this issue is the degree of functional conservation of SDSA repair factors between yeast and other eukaryotes. Most of the proteins involved in the initial resection, strand invasion, and D-loop formation steps of SDSA do appear to be functionally conserved in C. elegans, Drosophila, and mammals. However, while the involvement of error-prone translesion synthesis (TLS) polymerases in the bulk of initial repair synthesis appears to be restricted in yeast, several studies from metazoan systems suggest that translesion polymerases may participate in synthesis during D-loop extension. Human TLS polymerase eta (Polη) can extend D-loop intermediates in vitro (McIlwraith et al., 2005) and chicken DT40 lymphocytes lacking Polη are defective in gene conversion (Kawamoto et al., 2005). In Drosophila, mutation of Polη or Polζ hampers the efficiency of SDSA, and mutation of both Polη and Polζ or Rev1 alone results in increased repair synthesis, suggesting that replicative and translesion polymerases may compete for access to D-loop intermediates (Kane et al., 2012). Finally, knockdown of Polζ or REV1 in HeLa cells results in reduced efficiencies of HR repair, as measured by sensitivity to DSB-induced agents and an HR reporter construct (Sharma et al., 2012). Together, these studies suggest that translesion polymerases do play important roles in SDSA in some organisms. What remains to be determined is to what extent and in what contexts their use may promote SDSA-induced mutagenesis
What processes are responsible for mutagenesis during BIR?
As with SDSA, evidence that BIR can be error-prone has largely been obtained from investigations in budding yeast. One widely used system employs disomic budding yeast strains with a galactose-inducible HO cut site at the MAT locus on one copy of chromosome III and a second uncut copy of chromosome III that serves as a donor during BIR (Deem et al., 2008; Deem et al., 2011). Following DSB induction, repair synthesis initiates and copies more than 100 kb to the end of the donor chromosome. Mutagenesis can be quantified using telomere-proximal reporters on the donor chromosome placed at MAT, 16 kb, and 36 kb away from the break site. Remarkably, in this system the mutation rate for BIR is increased 25-2800 fold relative to spontaneous rates (Deem et al., 2011). Although the reporters used in this system could only detect frameshift mutations, other types of mutations, including template switches similar to those observed in SDSA, have also been observed during BIR repair (Anand et al., 2014).
SDSA and BIR are similar in that Polδ appears to be the main repair polymerase and driver of mutagenesis during normal BIR (Wilson et al., 2013). However, in cases where BIR is compromised by loss of Pol32 or Pif1, mutagenic repair can still occur (Deem et al., 2008; Vasan et al., 2014; Wilson et al., 2013), and in pif1 mutants this backup repair appears to rely heavily on Polζ (A. Malkova, personal communication).
In yeast strains lacking Pol32, which is required for efficient BIR, the frequency of repair outcomes called ‘half crossovers’ increases dramatically (Figure 4) (Deem et al., 2008). Half crossovers are thought to arise when repair synthesis stalls during BIR and the recombination intermediates are cleaved by structure-specific endonucleases, resulting in a crossover where one of the products is eventually lost (hence the term ‘half crossover’). Occasionally, one of the cleaved products can reinvade into its intact crossover partner, initiating another round of BIR and generating additional mutations.
One likely cause of mutagenesis during BIR could be the presence of intermediates containing extended tracts of ssDNA, which may be prone to DNA damage. In support of this, ssDNA has been shown to be hypermutable when exposed to base alkylating agents such as methyl methanesulfonate (MMS), which has a mutational signature specific to ssDNA (Yang et al., 2010; Yang et al., 2008). This hypothesis was recently tested by Sakofsky et al., who induced BIR in yeast strains that were exposed to different concentrations of MMS (Sakofsky et al., 2014). Whole genome sequencing showed that more than 50% of the BIR products exhibited mutation clusters from 4 kb to 115 kb in length in the area BIR was expected to occur, but not in other regions of the genome. Higher concentrations of MMS resulted in “complex clusters” that may have been created during multiple rounds of half crossovers and secondary BIR events (Vasan et al., 2014).
Similar to SDSA, BIR is also prone to template switching. Interestingly, this template switching is dependent upon the FancM helicase ortholog Mph1 (Smith et al., 2007; Stafa et al., 2014). If the template switches occur between dispersed repeated sequences, chromosome rearrangements can result. In an elegant series of experiments, Anand et al. showed that multiple template switches can occur during BIR, giving rise to complex rearrangements in a process termed microhomology-mediated break induced replication (MM-BIR). While the initial strand invasion that establishes BIR is Rad51-dependent, subsequent template switches involving homeologous or microhomologous sequences do not require Rad51, but are instead reliant on Rdh54 and may involve a mechanism similar to the template switches that occur during SDSA (Anand et al., 2014). Interestingly, MM-BIR has been postulated to drive copy number variations (CNVs) and chromosomal rearrangements seen in human cancers and inherited diseases, due to the presence of small microhomologies at the junctions of the chromosomal breaks (Hastings et al., 2009).
A summary of mutagenic mechanisms that operate during HR repair
Based on the survey of SDSA and BIR studies described above, there are three likely reasons for the increased rates of mutagenesis that accompany SDSA and BIR. The first is the persistence of long stretches of ssDNA, which are created either during resection or, particularly in the case of BIR, through repair synthesis. This ssDNA is highly vulnerable to attack by various DNA damaging agents and subsequent restoration of the ssDNA to double-stranded DNA can result in the fixation of mutations. The second mutagenic aspect of HR is the repair synthesis that takes place in the context of the D-loop. Here, Polδ has been primarily implicated as the mutagenic culprit in yeast, especially for BIR. Why Polδ should be so much more error-prone in the context of HR synthesis compared to normal DNA replication is an intriguing question. The answer to this question may partially reside in the third mutagenic aspect of HR repair, which is an increased likelihood of template switching. The remarkable aspect of HR template switching is that the microhomology required for switching can be quite small (on the order of just a few bases).
Overall, the picture that emerges is one in which DNA synthesis during SDSA and BIR is fundamentally different from normal DNA replication. In both processes, DNA synthesis is not highly processive and the polymerase is prone to slippage and stalling, which may act to promote dissociation of the D-loop intermediate and subsequent invasion into homeologous templates. Frequent template switching then results in increased frequencies of mutagenesis and repair products with unique mutational spectra. As described in the next section, these mutational spectra are now being observed in human genomes, suggesting that at least some of the mechanisms behind DSB-induced mutagenesis observed in model systems might operate in humans.
Error-prone double-strand break repair contributes to genome instability observed in cancers and other human diseases
Deep sequencing of cancer genomes has revealed the extreme amount of genetic diversity that exists in different cancers, including point mutations, translocations, copy number variations, and complex genomic rearrangements. Many of these genomic changes can be attributed to inaccurate or inappropriate repair of DSBs. For example, in cases where two or more DSBs are formed simultaneously, end joining between heterologous chromosomes can result in chromosome fusions and translocations (reviewed in Iarovaia et al., 2014). In one study conducted using mouse embryonic stem cells, translocations were induced following cutting by multiple zinc finger nucleases (ZFNs). These translocations were largely dependent upon an A-NHEJ pathway, involving CtIP and DNA ligase 3, which utilizes microhomologous sequences during the rejoining process (Simsek et al., 2011; Zhang and Jasin, 2011). The presence of C-NHEJ factors, such as DNA ligase 4, suppressed these translocations. Notably, in the absence of Ligase 3, residual translocations were observed that depended on DNA ligase 1 but did not utilize long microhomologies, suggesting that there may be more than one A-NHEJ mechanism for translocation formation in mice (Simsek et al., 2011).
Strikingly, the ligase requirement for translocation formation in humans appears to be reversed in mice. In a recent study using human cells, chromosome translocations were created using a combination of designer nucleases, including ZFNs, TALENs, and CRISPR/Cas9 (Ghezraoui et al., 2014). The sequence signatures at the translocation breakpoints were changed in cells depleted for C-NHEJ factors such as DNA ligase 4 and Xrcc4, with larger deletions and greater use of microhomology, consistent with an alternative NHEJ signature. Thus, it appears that in human cells the primary driver of DSB-induced translocations may be C-NHEJ.
A more extreme case of error-prone DSB repair operating in cancer cells can be observed in chromothripsis, a recently described phenomenon characterized by massive chromosome translocations, rearrangements, and inaccurate “stitching” together of chromosome pieces. The first instance of chromothripsis was observed in a patient with chronic lymphocytic leukemia who possessed 42 somatic genomic rearrangements within the long arm of chromosome 4 (Stephens et al., 2011). Since then, hundreds of examples of potential chromothripsis events have been reported (Cai et al., 2014). Although controversy remains about exactly what types of events constitute chromothripsis, it has been proposed that the signature of a single catastrophic event leading to chromothripsis is distinct from other types of sequential rearrangements and can be summarized by specific criteria: the identification of many rearrangements in a single area such as a chromosome or chromosome arm, alternating copy number states within the rearrangements, and clustered breakpoints within one or several chromosomes that appear to originate from double-strand breaks (Maher and Wilson, 2012; Zhang et al., 2013). It bears noting that chromothripsis is not isolated to cancer genomes, as chromothripsis-like events have also been observed in normal human development and may contribute to genetic abnormalities resulting in birth defects (Kloosterman et al., 2011).
While the underlying mechanisms of chromothripsis are still not fully understood, the general consensus is that the shattering process is initiated by the creation of multiple DSBs, which are then repaired by end joining. One idea is that exogenous damage, such as ionizing radiation, could occur within a localized area while chromosomes are tightly compacted during mitosis, resulting in concentrated areas of DSBs in one or more chromosomes/chromosome arms. The complexity of the rearrangements and the fact that the repair junctions possess little to no homology suggest that the main mechanism used to repair the many breaks is either C-NHEJ, A-NHEJ, or some combination of the two. As one example, genomic analysis of ten individuals with congenital diseases who exhibited multiple complex translocations and rearrangements characteristic of chromothripsis revealed that the breakpoints possessed signatures of both NHEJ and MMEJ (Kloosterman et al., 2012).
In addition to chromothripsis, other types of complex duplication, triplication, and insertion/deletions events have been described in both cancers and in inherited genomic disorders. These types of rearrangements, sometimes described as chromoanasynthesis, involves a gain of chromosomal copy number, usually involving small fragments of one or more chromosomes resulting in clustered rearrangements (Hastings et al., 2009; Liu et al., 2011). The genesis of the types of complex rearrangements seen in chromoanasynthesis is difficult to explain by simple end-joining repair. Such events may instead occur by mechanisms involving replication fork stalling and fork restoration via template switching (FoSTeS), or by MM-BIR. FoSTeS was first proposed as an explanation for the breakpoint sequences observed in cells of patients with Pelizaeus-Merzbacher Disease (Lee et al., 2007).The FoSTeS model postulates fork stalling and disengagement of the lagging strand, invasion into a nearby replication fork, and annealing at microhomologies with low processivity polymerization and many template switches, resulting in complex rearrangements. MM-BIR, as discussed previously, is thought to occur after a replication fork collapses at a single stranded DNA break or nick and proceeds via invasion at microhomologies, followed by highly processive and mutagenic replication, potentially with multiple template switches. Both mechanisms are thought to be the cause of CNVs and chromosomal rearrangements observed in many human cancers and certain diseases (Forment et al., 2012; Zhang et al., 2009).
In addition to end-joining and FoSTeS/MM-BIR, cells may use other non-conservative repair processes in situations where chromosome shattering results in a large number of DSBs (Table 2) (reviewed in Colnaghi et al., 2011). Furthermore, the initial repair process may lead to more deleterious mutations and additional DSBs that can initiate more rounds of chromosome rearrangements, resulting in highly complex mutational patterns. Many types of DSB repair could be involved, including processes such as single-strand annealing (SSA) and mutagenic gene conversion or BIR (reviewed in Chen et al., 2010). As one example, an increase or decrease in rates of homologous recombination in multiple myeloma cell lines led to increases in loss of heterozygosity and ongoing rearrangements as seen in chromothripsis (Shammas et al., 2009). Non-allelic homologous recombination (NAHR) may also be a prevalent repair pathway for instances where multiple chromosome breaks occur, as this type of repair occurs via recombination between paralagous low copy repeats leading to deletions, reciprocal duplications, and inversions. NAHR can also lead to isochromosome formation, recurrent CNVs, and chromosomal rearrangements, which are all hallmarks of chromothripsis (Chen et al., 2010).
Table 2.
Models used to explain the formation of chromosome translocations, gross chromosomal rearrangements and copy number variations observed in cancers and congenital diseases
| Proposed mechanism of repair |
Explanation of mechanism contributing to mutational signatures |
References |
|---|---|---|
| Classical non- homologous end joining (C-NHEJ) |
Joining of DSBs via no or little microhomology, results in reciprocal translocations observed in human cells. |
(Ghezraoui et al., 2014) |
| Alternative non- homologous end joining (A-NHEJ) |
Joining of DSBs using microhomologies, results in reciprocal translocations observed in mice. |
(Simsek et al., 2011; Zhang and Jasin, 2011) |
| Homologous recombination (HR) |
Elevated rates of homologous recombination in multiple myeloma cell lines lead to increased incidences of loss of heterozygosity and ongoing rearrangements (as seen in chromothripsis). |
(Shammas et al., 2009) |
| SDSA or BIR with template switching |
Initiation of DSB repair by SDSA or BIR is followed by frequent intra- or interchromosomal template switching to complete repair of the break. |
(Anand et al., 2014; Hicks et al., 2010; Tsaponina and Haber, 2014) |
| Single strand annealing (SSA) |
Following DSB formation, resection occurs, followed by annealing at direct repeats flanking the DSB and flap removal. Single- strand gap filling and ligation results in large deletions. |
(Chen et al., 2010) |
| Nonallelic homologous recombination (NAHR) |
Recombination occurs between paralagous low copy repeats (LCRs)/ segmental duplications; in trans leads to deletion and reciprocal duplications, in cis results in an inversion. Leads to recurrent copy number variants and chromosomal rearrangements. |
(Liu et al., 2012) |
| Fork stalling template switching (FoSTeS) |
Stalling of replication fork at a DNA lesion leads to polymerase dissocation and invasion of nascent strand into a nearby replication fork at regions of microhomology, followed by low processivity polymerization. Complex rearrangements result from serial template switches. |
(Lange et al., 2011; Lee et al., 2007; Liu et al., 2011) |
| Microhomology- mediated break- induced replication (MMBIR) |
Collapsed replication fork at single stranded DNA break or nick, invasion at microhomologies, followed by highly processive and mutagenic replication. Leads to complex rearrangements. |
(Hastings et al., 2009; Liu et al., 2011) |
What are the most important questions to answer about error-prone DSB repair?
It is now clear that the inaccurate repair of double-strand breaks can serve as a driving force behind genomic change and genetic instability. While the molecular mechanisms underlying DSB-induced mutagenesis are starting to come into focus, many important issues remain to be resolved. First, from a mechanistic standpoint, we would like to know and understand the rules that govern the use of homologous and microhomologous sequences during template switching in SDSA and BIR. Second, besides the requirement for Pol32 and Pif1, what other genetic factors distinguish BIR from both SDSA and normal DNA replication in yeast? Along these lines, are the mechanisms of SDSA and BIR fully conserved in other eukaryotes, or are there crucial differences that have evolved due to genome size or structure? Finally, what are the relative contributions of C-NHEJ, A-NHEJ, and HR repair processes to phenomena such as chromothripsis, chromoanasynthesis, and complex genome rearrangements that are observed in the context of both normal development and disease? Future studies that combine experiments in model organisms with deep sequencing and genomic studies in humans should allow us to make rapid progress towards addressing these questions.
Acknowledgments
The authors would like to thank Sarah Dykstra for comments that greatly improved this review and Jim Haber, Kirill Lobachev, and members of the McVey lab for helpful discussions. Research in the McVey lab is supported by grants R01GM092866 and P01GM105473 from the NIH.
REFERENCES
- Anand RP, Tsaponina O, Greenwell PW, Lee CS, Du W, Petes TD, Haber JE. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 2014;28(21):2394–2406. doi: 10.1101/gad.250258.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10(1):e1004086.. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizard AH, Hickson ID. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol. 2014;6(7):a016477.. doi: 10.1101/cshperspect.a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24(23):4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Kumar N, Bagheri HC, von Mering C, Robinson MD, Baudis M. Chromothripsis-like patterns are recurring but heterogeneously distributed features in a survey of 22,347 cancer genome screens. BMC Genomics. 2014;15:82. doi: 10.1186/1471-2164-15-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O'Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D'Andrea AD. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518(7538):258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Ferec C, Kehrer-Sawatzki H, Patrinos GP. Genomic rearrangements in inherited disease and cancer. Semin Cancer Biol. 2010 doi: 10.1016/j.semcancer.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Colnaghi R, Carpenter G, Volker M, O'Driscoll M. The consequences of structural genomic alterations in humans: genomic disorders, genomic instability and cancer. Semin Cell Dev Biol. 2011;22(8):875–885. doi: 10.1016/j.semcdb.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD. Break-Induced Replication Repair of Damaged Forks Induces Genomic Duplications in Human Cells. Science. 2014;343(6166):88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21(2):121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol Cell Biol. 2004;24(6):2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A. Alternative end-joining mechanisms: a historical perspective. Frontiers in genetics. 2013;4:48. doi: 10.3389/fgene.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Barker K, Vanhulle K, Downing B, Vayl A, Malkova A. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics. 2008;179(4):1845–1860. doi: 10.1534/genetics.108.087940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9(2):e1000594.. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- Donnianni RA, Symington LS. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci U S A. 2013;110(33):13475–13480. doi: 10.1073/pnas.1309800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J, Rudin N, Haber JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12(3):1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12(10):663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, Jasin M, Brunet E. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55(6):829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191(1):33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5(1):e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD. Biochemistry of eukaryotic homologous recombination. Top Curr Genet. 2007;17:95–133. doi: 10.1007/978-3-540-71021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329(5987):82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins GS, Harris AL, Prevo R, Helleday T, McKenna WG, Buffa FM. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget. 2010;1(3):175–184. doi: 10.18632/oncotarget.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147(3):1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96(3):415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Iarovaia OV, Rubtsov M, Ioudinkova E, Tsfasman T, Razin SV, Vassetzky YS. Dynamics of double strand breaks and chromosomal translocations. Mol Cancer. 2014;13:249. doi: 10.1186/1476-4598-13-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol Cell Biol. 2002;22(18):6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Satory D, Haber JE. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol Cell Biol. 2006;26(24):9424–9429. doi: 10.1128/MCB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette N, Lees-Miller SP. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol. 2015;117(2-3):194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DP, Shusterman M, Rong Y, McVey M. Competition between replicative and translesion polymerases during homologous recombination repair in Drosophila. PLoS Genet. 2012;8(4):e1002659. doi: 10.1371/journal.pgen.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20(5):793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, Honda I, Sakiyama S, Tagawa M, J OW. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. International journal of cancer. 2004;109(1):9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol. 2015;22(3):230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, Cuppen E. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20(10):1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, Ballarati L, Vergult S, Giardino D, Hansson K, Ruivenkamp CA, Jager M, van Haeringen A, Ippel EF, Haaf T, Passarge E, Hochstenbach R, Menten B, Larizza L, Guryev V, Poot M, Cuppen E. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 2012;1(6):648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lemee F, Bergoglio V, Fernandez-Vidal A, Machado-Silva A, Pillaire MJ, Bieth A, Gentil C, Baker L, Martin AL, Leduc C, Lam E, Magdeleine E, Filleron T, Oumouhou N, Kaina B, Seki M, Grimal F, Lacroix-Triki M, Thompson A, Roche H, Bourdon JC, Wood RD, Hoffmann JS, Cazaux C. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci U S A. 2010;107(30):13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22(3):211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, Bacino CA, Campos-Acevedo LD, Delgado MR, Freedenberg D, Garnica A, Grebe TA, Hernandez-Almaguer D, Immken L, Lalani SR, McLean SD, Northrup H, Scaglia F, Strathearn L, Trapane P, Kang SH, Patel A, Cheung SW, Hastings PJ, Stankiewicz P, Lupski JR, Bi W. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146(6):889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448(7155):820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM, Haber JE. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24(11):1133–1144. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23(23):8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Wilson RK. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148(1-2):29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev. 2013;23(3):271–279. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol. 2005;25(3):933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518(7538):254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20(5):783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24(11):529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6(9):a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta GD, Kumar R, Srivastava S, Ghosh SK. Cohesin: functions beyond sister chromatid cohesion. FEBS letters. 2013;587(15):2299–2312. doi: 10.1016/j.febslet.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Morrical SW. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol. 2015;7(2):a016444. doi: 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19(3):357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Paul K, Wang M, Mladenov E, Bencsik-Theilen A, Bednar T, Wu W, Arakawa H, Iliakis G. DNA ligases I and III cooperate in alternative non-homologous end-joining in vertebrates. PLoS ONE. 2013;8(3):e59505. doi: 10.1371/journal.pone.0059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng W, Jasin M. Homologous Recombination and Human Health: The Roles of BRCA1, BRCA2, and Associated Proteins. Cold Spring Harb Perspect Biol. 2015;7(4) doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16(5):715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502(7471):389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Malkova A. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7(5):1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas MA, Shmookler Reis RJ, Koley H, Batchu RB, Li C, Munshi NC. Dysfunctional homologous recombination mediates genomic instability and progression in myeloma. Blood. 2009;113(10):2290–2297. doi: 10.1182/blood-2007-05-089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn JY, Glover TW, Canman CE. REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic Acids Res. 2012;40(2):682–691. doi: 10.1093/nar/gkr769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen CR, Jewell UR, Morris CM, Brennan SO, Ferec C, George PM, Smith MP, Chen JM. Double complex mutations involving F8 and FUNDC2 caused by distinct break-induced replication. Hum Mutat. 2007;28(12):1198–1206. doi: 10.1002/humu.20591. [DOI] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M. DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation. PLoS Genet. 2011;7(6):e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447(7140):102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Stafa A, Donnianni RA, Timashev LA, Lam AF, Symington LS. Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics. 2014;196(4):1017–1028. doi: 10.1534/genetics.114.162297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, Campbell PJ. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140(3):965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tsaponina O, Haber JE. Frequent Interchromosomal Template Switches during Gene Conversion in S. cerevisiae. Mol Cell. 2014;55(4):615–625. doi: 10.1016/j.molcel.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan S, Deem A, Ramakrishnan S, Argueso JL, Malkova A. Cascades of genetic instability resulting from compromised break-induced replication. PLoS Genet. 2014;10(2):e1004119. doi: 10.1371/journal.pgen.1004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34(21):6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ira G, Tercero JA, Holmes AM, Diffley JF, Haber JE. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24(16):6891–6899. doi: 10.1128/MCB.24.16.6891-6899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CA, Strande NT, Wyatt DW, Pryor JM, Ramsden DA. Nonhomologous end joining: a good solution for bad ends. DNA Repair (Amst) 2014;17:39–51. doi: 10.1016/j.dnarep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst) 2014;17:110–120. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, Ira G. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502(7471):393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, West SC. Holliday junction resolvases. Cold Spring Harb Perspect Biol. 2014;6(9):a023192. doi: 10.1101/cshperspect.a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Yang M, Huo W, Feng F, Wei L, Jiang W, Ning S, Yan Z, Li W, Wang Q, Hou M, Dong C, Guo R, Gao G, Ji J, Zha S, Lan L, Liang H, Xu D. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat Commun. 2015;6:6233. doi: 10.1038/ncomms7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair (Amst) 2010;9(8):914–921. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4(11):e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefzadeh MJ, Wyatt DW, Takata K, Mu Y, Hensley SC, Tomida J, Bylund GO, Doublie S, Johansson E, Ramsden DA, McBride KM, Wood RD. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10(10):e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38(17):5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn KE, Averill AM, Aller P, Wood RD, Doublie S. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol. 2015;22(4):304–311. doi: 10.1038/nsmb.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27(23):2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25(7):298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18(1):80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]