Abstract
Droplet digital PCR (ddPCR) can be used to detect low frequency mutations in oncogene-driven lung cancer. The range of KRAS point mutations observed in NSCLC necessitates a multiplex approach to efficient mutation detection in circulating DNA. Here we report the design and optimisation of three discriminatory ddPCR multiplex assays investigating nine different KRAS mutations using PrimePCR™ ddPCR™ Mutation Assays and the Bio-Rad QX100 system. Together these mutations account for 95% of the nucleotide changes found in KRAS in human cancer. Multiplex reactions were optimised on genomic DNA extracted from KRAS mutant cell lines and tested on DNA extracted from fixed tumour tissue from a cohort of lung cancer patients without prior knowledge of the specific KRAS genotype. The multiplex ddPCR assays had a limit of detection of better than 1 mutant KRAS molecule in 2,000 wild-type KRAS molecules, which compared favourably with a limit of detection of 1 in 50 for next generation sequencing and 1 in 10 for Sanger sequencing. Multiplex ddPCR assays thus provide a highly efficient methodology to identify KRAS mutations in lung adenocarcinoma.
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide [1] and over 20 000 cases of non-small cell lung cancer (NSCLC) were diagnosed in the UK in 2012 [2]. The most frequently mutated oncogenes in lung adenocarcinoma are the RAS family GTPases and EGFR (25% and 15% respectively [3]). Knowledge of the molecular profile of advanced lung adenocarcinoma is critical for therapeutic decision-making [4], particularly in the use of EGFR and ALK tyrosine kinase inhibitors [5,6]. The presence of a KRAS mutation may also be of therapeutic relevance if MEK inhibitor and taxane combinations prove efficacious in this patient cohort [7]. Obtaining adequate tumour tissue for conclusive genotyping in lung cancer can be problematic, however [8]. Droplet digital PCR (ddPCR) is a sensitive method of quantitative mutation detection [9,10] that has the potential to accurately genotype patient-derived material from a small amount of starting material.
Detection of KRAS hotspot mutations by ddPCR has been limited by the variety of potential alleles within adjacent loci, although some KRAS mutations occur more commonly in cancer than others (Tables 1 and 2). The four commonest mutations account for 80% of all KRAS nucleotide changes found in human cancers (85% of all changes in NSCLC), while the nine commonest mutations account for 95% of all changes and 97.5% of changes in NSCLC. Fluorophore—labelled digital PCR probes complement a particular mutant DNA sequence and so only detect one specific KRAS mutation. Using these assays in duplex with a probe to detect the mutant allele and a probe to detect the wild-type allele allow mutant allele fraction calculation for a given mutation, but this approach requires potentially multiple assays and the use of more material before correct identification of the genotype. Development of a multiplex assay combining several different mutant probes in the same reaction is therefore an attractive alternative. Multiplex KRAS digital PCR assays have been described using the RainDrop™ Digital PCR system (RainDance Technologies, Billerica, Massachusetts, USA) in advanced colorectal cancer [11] but not as yet with the Bio-Rad QX100 system, an affordable digital PCR system with commercially-available probes for several KRAS mutations, nor has a multiplex tool been used in lung cancer.
Table 1. KRAS mutations observed in human cancer in order of decreasing frequency (http://www.sanger.co.uk/cosmic; accessed 14th July 2015).
| KRAS genotype | Mutation | Cases | Frequency |
|---|---|---|---|
| G12D | c.35G>A | 10719 | 33.8% |
| G12V | c.35G>T | 7138 | 22.5% |
| G13D | c.38G>A | 3959 | 12.5% |
| G12C | c.34G>T | 3713 | 11.7% |
| G12A | c.35G>C | 1694 | 5.3% |
| G12S | c.34G>A | 1507 | 4.7% |
| G12R | c.34G>C | 1024 | 3.2% |
| G13C | c.37G>T | 276 | 0.9% |
| Q61H | c.183A>C | 144 | 0.5% |
Table 2. KRAS mutations observed in human NSCLC in order of decreasing frequency (http://www.sanger.co.uk/cosmic; accessed 14th July 2015).
| KRAS genotype | Mutation | Cases | Frequency |
|---|---|---|---|
| G12C | c.34G>T | 604 | 36.7% |
| G12V | c.35G>T | 350 | 21.3% |
| G12D | c.35G>A | 336 | 20.4% |
| G12A | c.35G>C | 107 | 6.5% |
| G13C | c.37G>T | 54 | 3.3% |
| G12S | c.34G>A | 53 | 3.2% |
| G13D | c.38G>A | 43 | 2.6% |
| G12R | c.34G>C | 36 | 2.2% |
| Q61H | c.183A>C | 20 | 1.2% |
We set out to design multiplex digital PCR assays which would accurately identify nine different KRAS mutations and to demonstrate the application of these assays to patient-derived material using the Bio-Rad QX100 system.
Materials and Methods
Digital PCR probe assays
Each digital PCR probe is an oligonucleotide specific to the region of interest with a 5’ fluorophore and a 3’ quencher. The fluorophore is either HEX for probes specific to wild-type sequences or FAM for mutant probes. KRAS c.35G>T (G12V; cat no. dHsaCP2500592), c.35G>A (G12D; cat no. dHsaCP2500596), c.35G>C (G12A; cat no. dHsaCP2500586), c.34G>A (G12S; cat no. dHsaCP2500588), c.34G>C (G12R; cat no. dHsaCP2500590), c.34G>T (G12C; dHsaCP2500584), c.37G>T (G13C; cat no. dHsaCP2500595), c.38G>A (G13D; cat no. dHsaCP2500598), c.183A>C (Q61H; cat no. dHsaCP2000133), WT for c.35G>T (WT for G12V; cat no. dHsaCP2500593), WT for c.35G>A (WT for G12D; cat no. dHsaCP2000002), WT for c.35G>C (WT for G12A; cat no. dHsaCP2000004), WT for c.34G>A (WT for G12S; cat no. dHsaCP2000012), WT for c.34G>C (WT for G12R; cat no. dHsaCP2000010), WT for c.34G>T (WT for G12C; cat no. dHsaCP2000008), WT for c.37G>T (WT for G13C; cat no. dHsaCP2500595), WT for c.38G>A (WT for G13D; cat no. dHsaCP2000014) and WT for c.183A>C (WT for Q61H; cat no. dHsaCP2000132) 20x PrimePCR™ ddPCR™ Mutation Assays (containing both primers and ddPCR probes) were purchased from Bio-Rad (Bio-Rad Laboratories, Hercules, California, USA) and used in this study.
Assay optimisation and DNA quantification
Digital PCR assays were optimised using cell line genomic DNA (gDNA) or synthetic oligonucleotides as appropriate. Cell lines used for this study were authenticated using STR profiling by the Cell Services facility at the Cancer Research UK London Research Institute and the Institute of Cancer Research. These included NCI-H727 (KRAS G12V, lung), NCI-H358 (KRAS G12C, lung), A549 (KRAS G12S, lung), SK-LU-1 (KRAS G12D, lung), A427 (KRAS G12D, lung), HCT-116 (KRAS G13D, colon) and NCI-H1975 (KRAS WT, lung). All cell lines were provided directly by Cancer Research UK Cell Services with the exception of HCT-116 (American Type Culture Collection, Rockville, Maryland, USA; cat. no ATCC®CCL-247TM). Cell gDNA was extracted using the DNeasy™ Blood and Tissue Kit (Qiagen, Venlo, Limburg, Netherlands) as per manufacturer’s instructions. All DNA used in subsequent digital PCR reactions, except synthetic oligonucleotides, was quantified on a QX100 ddPCR system using human RNase P TaqMan® Copy Number Reference Assay (Life Technologies Corporation, Carlsbad, California, USA). 1 μL of eluate was added to a digital PCR reaction containing 10 μL ddPCR Supermix for probes (Bio-Rad) and 1 μL of TaqMan Copy Number Reference Assay, human, RNase P (Life Technologies) on a total volume of 20 μL. The reaction was partitioned into ~14,000 droplets per sample in a QX-100 droplet generator according to manufacturer’s instructions. Emulsified PCR reactions were run on a 96 well plate (Eppendorf, Stevenage, UK) on a G-Storm GS4 thermal cycler (G-Storm, Somerton, Somerset, UK) incubating the plates at 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec, followed by 10 min incubation at 98°C. The temperature ramp increment was 2.5°C/sec for all steps. Plates were read on a Bio-Rad QX-100 droplet reader using QuantaSoft v1.4.0.99 software (Bio-Rad). At least one negative control wells with no DNA were included in every run. The amount of amplifiable RNase P DNA was quantified from the concentration provided by the software. After quantification, cell line DNA was digested using Hind 3-HF™ restriction enzyme (New England BioLabs, Ipswich, Massachusetts, USA) at 37°C for 1 hour.
Droplet digital PCR for KRAS mutation detection
PCR reaction mixtures were prepared in a total volume of 20 μL containing: 10 μL 2x Supermix for Probes without dUTP (Bio-Rad), relevant primer probe assays, DNA (variable volume) and nuclease-free water (Life Technologies Corporation, Carlsbad, California, USA). Multiple wells were used if required to analyse the necessary DNA eluate. At least 500 pg equivalent of DNA eluate was analysed in each multiplex assay for each patient. 20 μL of PCR reaction mixture was transferred to a sample well in a disposable droplet generator cassette (Bio-Rad). 70 μL of droplet generation oil (Bio-Rad) was then loaded into the oil well for each channel and the cassette loaded into a QX100 Droplet Generator (Bio-Rad). The droplets were then transferred to a 96-well PCR plate. Emulsified PCR reactions were run on a G-Storm GS4 thermal cycler incubating the plates at 95°C for 10 min followed by 40 cycles of 94°C for 30 sec and 54°C for 60 sec, followed by 10 min incubation at 98°C. The temperature ramp increment was 2.5°C/sec for all steps. Plates were read on a Bio-Rad QX-100 droplet reader using QuantaSoft v1.4.0.99 software from Bio-Rad. At least one negative control well with no DNA was included in every run. Each well was then read using the QX100 Droplet Reader (Bio-Rad) and droplets analysed for emission in the HEX or FAM wavelengths. Two-dimensional amplitude plots displaying measured HEX and FAM amplitude for each droplet show four main droplet populations; droplets containing no amplified DNA, droplets with only mutant DNA and high FAM amplitude, droplets with only wild-type DNA and high HEX amplitude and droplets with both wild-type and mutant amplified DNA and high FAM and HEX amplitude. If multiple copies of wild-type and mutant DNA are contained in the same droplet, droplets with higher FAM and HEX amplitude will be read. These droplets have been excluded from any analyses throughout this study.
Digital PCR analysis
To assess the mutant allele fraction, the concentration of mutant DNA (copies of mutant DNA per droplet) was estimated from the Poisson distribution. Number of mutant copies per droplet Mmu = -ln (1-(nmu/n)), where nmu = number of droplets positive for mutant FAM probe and n = total number of droplets. The DNA concentration in the reaction was estimated as follows: MDNAconc = -ln (1-(nDNAcon/n)), where nDNAconc = number of droplets positive for mutant FAM probe and/or wild-type HEX probe and n = total number of droplets. The mutant allele fraction = Mmu/ MDNAconc. The measured mutant allele frequency describes the mutant allele fraction expressed as a percentage.
FFPE tissue DNA access and extraction
Formalin fixed paraffin embedded (FFPE) tumour samples from NSCLC patients surplus to clinical care were collected with written patient consent at The Royal Marsden NHS Foundation Trust. FFPE tissue DNA was extracted (after macrodissection if required to ensure >10% tumour content) using the Qiagen DNA FFPE Tissue Kit (Qiagen) as per manufacturer’s instructions. The eluted DNA was stored at -20°C.
Ethics statement
All patients provided written consent to this study (ethics approval 13/LO/1389, NRES Committee London-Central), which incorporated surplus somatic DNA from the Cancer Research UK Stratified Medicine Programme (ethics approval 11/EE/0202, NRES Committee East of England).
Sanger sequencing
The region of interest was amplified using 10 μM forward (5’-TATTATAAGGCCTGCTGAAAATG-3’) and reverse (5’-TTGGATCATATTCGTCCACAA-3’) primers, 10 μL Amplitaq Gold® PCR Master Mix (Life Technologies) and 3 ng of template FFPE—derived DNA or 5ng gDNA. PCR conditions were as per manufacturer’s recommendations. Purified PCR product then underwent a further PCR step using chain-terminating dideoxynucleotides (BigDye® 1.0, Life Technologies) and subsequent DNA sequence was read on an automated sequencer.
Ion torrent proton sequencing
Sequencing libraries were prepared with the Ion AmpliSeq™ Cancer Hotspot Panel v2 (Life Technologies) using the Ion AmpliSeq Library Preparation protocol with 3-5ng of DNA, according to manufacturer’s instructions. Following barcoding, libraries were quantified using qPCR and diluted to 100 pM. Libraries were templated with the Ion OneTouch2 system (Life Technologies) and sequenced on a PI chip using the Ion PI OT2 200 Kit (Life Technologies), 520 flows and an average amplicon length of 112 bases to a mean depth of x2721307. The sequencing resulted in 45272–11767856 reads per sample.
Ion torrent Variant caller v4.0-r73742 with no Hotspot region and configuration “Germ Line Low Stringency” was used for calling variants. Read counts for all positions were computed using pileup (SAMtools v1.1 [12]) and this data was analysed for possible variants using custom Perl and R scripts. Variants at > 3% reported by both analysis methods and not reported in 1000 Genomes Project database (www.1000genomes.org) were identified as possible somatic mutations. The data was cross referenced against the Cosmic database v70 (cancer.sanger.ac.uk) to identify possible hotspot mutations.
Statistical analysis
Linear regression was performed using the formula r2 = 1-(SSreg/SStot) where SSreg refers to the sum of the squares of distances to best-fit linear regression line and SStot refers to the sum of the squares of vertical distances from the null hypothesis (y = mean of all y values) using GraphPad Prism version 6.0a (GraphPad Software, La Jolla, California, USA).
KRAS multiplex assay specificity
92 wells of KRAS wild-type gDNA were analysed with each multiplex. The limit of detection was set at 0.05% to reflect our measured results with spiked KRAS mutant gDNA species (S8 Fig). Individual droplets were clustered according to their HEX amplitude into two groups using the kMeans algorithm. A threshold for mutants was estimated based on 1.5 times the median of the FAM values of those drops that belong to the cluster with higher HEX categories. False positive ‘mutant’ droplets were classified as droplets that were in the lower HEX cluster, but whose FAM value exceeds the threshold to reflect the FAM and HEX amplitudes measured for any of the KRAS mutant species tested for in that multiplex assay. A false positive result was defined as three false positive ‘mutant’ droplets as per the Rare Mutation Detection Best Practices Guidelines [13]. To assess the specificity, we calculated the binomial probability of achieving fewer than three ‘mutant’ droplets per 6000 wild-type droplets for each multiplex assay.
Results
KRAS mutation duplex ddPCR
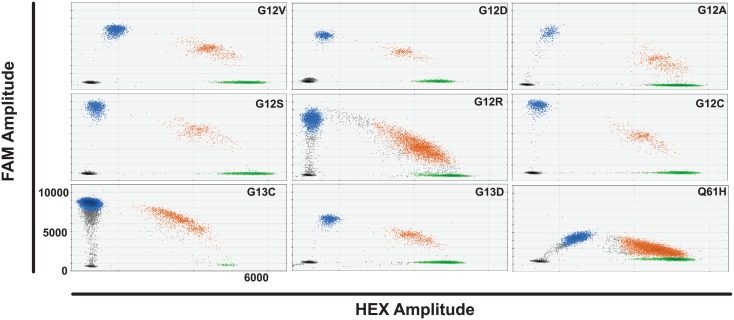
We tested all KRAS ddPCR assays separately across an annealing temperature gradient to optimise thermocycling conditions. Each assay was tested with the appropriate gDNA or oligonucleotide alone and then in duplex with wild-type and KRAS mutant DNA and both relevant FAM and HEX probes present. Decreasing annealing temperature increased FAM amplitude of the mutant probe to a plateau at 54°C for KRAS G12V, D, A, S, R and C and G13C and 13D probes (Fig 1). The KRAS Q61H probe had a further minimal increase in FAM amplitude at 53.4°C as compared to 54°C. All probes tested showed good separation of the four different droplet groups at 54°C, allowing clear identification and quantification of different DNA populations.
Fig 1. KRAS duplex assays at optimal annealing temperature.
Droplet populations observed for each duplex assay tested with wild-type and relevant mutant cell line gDNA or oligonucleotide at the optimal annealing temperature e.g. G12V panel top left shows droplet populations seen with WT for G12V assay, G12V assay, NCI-H727 gDNA and NCI-H1975 gDNA present. HEX amplitude is up to 6000 on the x axis and FAM amplitude up to 11000 on the y-axis of each panel. Key: Black drops- empty droplets, blue- mutant DNA FAM positive droplets, green- wild-type DNA HEX positive droplets, brown—wild-type and mutant DNA double positive droplets.
KRAS multiplex ddPCR assay design
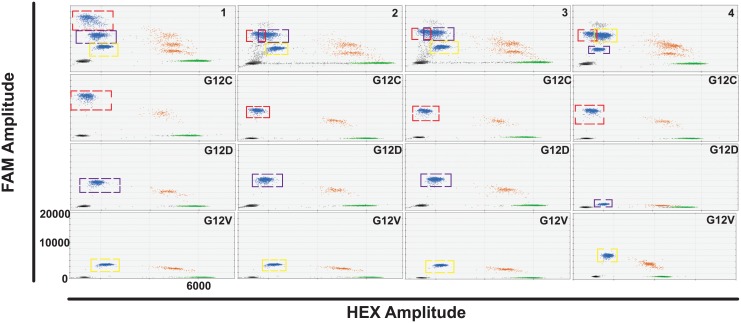
We designed a digital PCR-based multiplex tool to screen for the most common KRAS mutations in lung adenocarcinoma; G12C, G12D and G12V (http://www.sanger.co.uk/cosmic). Wild-type probe assays for each mutation were tested in combination with varying concentrations of mutant probe assays, giving rise to mutant droplet populations of varying FAM amplitude (Fig 2). The HEX amplitude of all wild-type probe assays was found to be very similar and so the WT for G12C, WT for G12V and WT for G12D assays were selected as references for all subsequent KRAS G12 and G13 mutant assays. Of the different combinations of mutant assays trialled, the multiplex assay which gave best separation of the droplet populations used 900 nM primers and 500 nM G12C probe, 562.5 nM primers and 312.5 nM G12D probe and 225 nM primers and 125 nM G12V probe (Fig 2, top left panel).
Fig 2. Four different KRAS multiplex digital PCR assays combining G12C, G12V and G12D mutant assays and corresponding duplex assays.
KRAS G12C mutant droplet populations are indicated by a red dashed square, KRAS G12D mutant populations by a blue dashed square and KRAS G12V mutant populations by a yellow dashed square. Each multiplex assay, combining all relevant FAM and HEX assays, KRAS WT gDNA and G12V, D and C mutant gDNA, is shown in the top panel. The corresponding duplex assay for each mutation, using the same FAM and HEX assay concentration with KRAS WT DNA and the appropriate KRAS mutant DNA present, is shown in the panels below each multiplex assay. Multiplex 1 (top left panel) is an assay combination of 900 nM primers and 500 nM G12C probe, 562.5 nM primers and 312.5 nM G12D probe and 225 nM primers and 125 nM G12V probe with 450 nM primers and 250 nM WT for G12C probe. Multiplex 2 uses the same concentration of G12V and WT for G12C assay as Multiplex 1 but also contains 450 nM primers and 250 nM G12C probe and 675 nM primers and 375 nM G12D probe. Multiplex 3 uses the same concentration of KRAS mutant assays as in Multiplex 2 but with the WT for G12V probe assay present, used at the same concentration as the WT for G12C assay in Multiplex 1. Multiplex 4 is an assay combination of the G12C assay as in Multiplex 2 with 225 nM primers and 125 nM G12D probe and 675 nM primers and 375 nM G12V probe with the WT for G12D assay at the same concentration as the WT for G12C assay in Multiplex 1. Key: black- empty droplets, blue- mutant DNA FAM positive droplets, green- wild-type DNA HEX positive droplets, brown—wild-type and mutant DNA double positive droplets. The same scale is used for all panels (HEX amplitude up to 7000 and FAM amplitude up to 22000).
The ability to test for multiple KRAS mutations would help to detect sub-clonal populations by applying the multiplex approach to clinical samples on which starting material is scarce. To model this, we employed cell-line derived gDNA for the three mutations and tested it with each probe in duplex to ensure specificity (S1 Fig). In the presence of the G12D mutant probe and KRAS G12D, V and C mutant DNA, a second mutant population of droplets was identified at lower FAM amplitude to the G12D mutant DNA population (red hatched box, left uppermost panel). This population was not observed when each mutant DNA species was tested in duplex with the G12D probe (left second panel). A similar second mutant population was observed with the G12V mutant probe and all three mutant DNA species as compared with each mutant DNA in duplex (left lower panels). This second population, particularly as seen with the G12D mutant probe, may fall at the expected FAM amplitude of a different KRAS mutation in the multiplex assay and lead to false positive mutation detection. To further explore this, FFPE tissue DNA from an individual known to have a KRAS 12/13 mutation (F124), as verified by Cobas® testing (Cobas® KRAS Mutation Test, Roche Molecular Systems, Inc., Branchburg, NJ, USA) was analysed (S1 Fig, right panels). The multiplex assay identified a mutant droplet population with FAM amplitude that was interpreted as a G12D or G12V mutation. When the sample was tested with each duplex assay individually, no mutant population in either G12V or G12C was observed, whilst a population with a lower FAM amplitude than expected with the G12D mutant assay was observed. On subsequent sequencing and further development of the digital PCR analysis of the tissue DNA, this sample was later identified as KRAS G12A mutant (Table 3).
Table 3. Detection of KRAS mutant clones in FFPE tissue DNA using next generation sequencing (NGS) and KRAS multiplex digital PCR assays.
| ID | KRAS genotype | NGS mutant allele frequency (%) | Digital PCR multiplex mutant allele frequency (%) |
|---|---|---|---|
| S007 | G12C | 30.4 | 29.5 |
| S027 | G12C | 48 | 53.8 |
| F007 | G12D | 4 | 4.6 |
| S002 | G12A | 32 | 30 |
| S010 | G12C | ND | 0.3 |
| G12A | 8.8 | 8.7 | |
| S011 | G12V | 33.1 | 32.8 |
| G12D | ND | 0.05 | |
| S018 | G12F | 29 | 29.5 |
| S030 | G13C | 17 | 18.6 |
| S035 | G12C | 30 | 19.8 |
| F124 | G12A | 23 | 23.7 |
| F130 | G12V | 28 | 34.2 |
Key: ND- not detected.
Cross-reactivity of KRAS probe assays on combination
Due to the close similarities between the DNA sequences of the various KRAS mutations, significant cross-reactivity was noted between probes designed for mutations within the same region. This was particularly evident with probes designed for substitutions at the same nucleotide i.e. KRAS G12V, D and A and KRAS G12S, R and C. The cross-reactivity of the probe assay with ‘mismatched’ DNA carrying a different KRAS mutation (S2 and S3 Figs) results in a droplet population of varying FAM and VIC amplitudes relatively close to the empty droplet population and another population close to the true wild-type droplet population. The position of these additional droplet populations dictates the design of multiplex assays to reduce potential misinterpretation of KRAS genotypes. Therefore, three multiplex assays were devised to each combine a probe for a mutation at nucleotide position 35, a probe for a mutation at position 34 and a probe for a mutation at either position 37, 38 or 183.
KRAS multiplex ddPCR assay optimisation
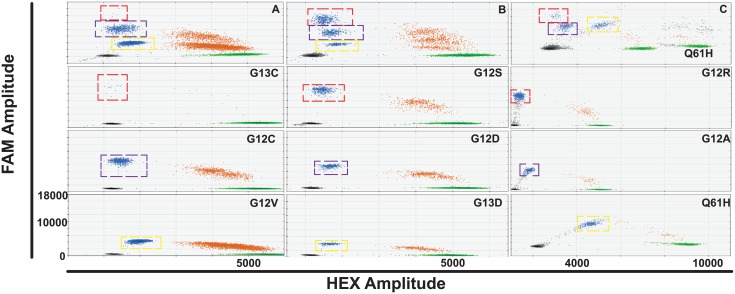
Multiplex A is a combination of FAM assays for KRAS G13C, G12C and G12V. Several different combinations of mutant probe concentrations were tested and the use of a wild-type probe for G12C and G13C at varying concentrations (S4 Fig). There was significant overlap between the HEX amplitude of the wild-type droplet populations and due to the proximity of the relevant nucleotide bases and the estimated length of the probe, it was felt that 450 nM primers and 250 nM G12C, V or D wild-type probe was adequate for quantification of both G12 and G13 wild-type populations. The optimal multiplex assay (S4 Fig, top left panel) comprised 900 nM primers and 500 nM G13C mutant probe, 450 nM primers and 250 nM G12C probe and 225 nM primers and 125 nM G12V probe. Multiplex B is a combination of FAM assays for KRAS G12S, G12D and G13D. Of all the concentrations tested, the optimal combination of mutant probe assays was 675 nM primers and 375 nM G12S probe, 450 nM primers and 250 nM G12D probe and 225 nM primers and 125 nM G13D probe (S5 Fig, top left panel). Multiplex C analyses KRAS mutations at G12R, G12A and Q61H. It requires a wild-type probe for both the G12/13 position and the Q61 position for accurate quantification of mutant allele frequency. The assay was optimised with 450 nM primers and 250 nM G12C probe and 900 nM primers and 500 nM Q61H probe as the wild-type assays. The best separation of the mutant droplet populations was achieved using 675 nM primers and 375 nM G12R probe, 450 nM primers and 250 nM G12A probe and 900 nM primers and 500 nM Q61H mutant probe (S6 Fig, top left panel).
All optimised multiplex assays (Fig 3) were tested for cross-reactivity with various species of KRAS mutant DNA (S7 Fig). There is no overlap between the desired droplet populations and the populations for the other KRAS mutant DNA species in all three multiplexes. In addition, the position of droplet populations due to cross-reactivity is highly reproducible and can be used to start to identify which KRAS genotype is present in a multiplex assay containing other mutant probes.
Fig 3. KRAS multiplex digital PCR assays A-C and corresponding duplex assays.
Multiplex A (top left panel) is an assay combination of 900 nM primers and 500 nM G13C probe (red dashed square), 450 nM primers and 250 nM G12C probe (blue dashed square) and 225 nM primers and 125 nM G12V probe (yellow dashed square). Multiplex B (top middle panel) is an assay combination of 675 nM primers and 375 nM G12S probe (red dashed square), 450 nM primers and 250 nM G12D probe (blue dashed square) and 225 nM primers and 125 nM G13D probe (yellow dashed square). Multiplex C (top right panel) is an assay combination of 675 nM primers and 375 nM G12R probe (red dashed square), 450 nM primers and 250 nM G12A probe (blue dashed square) and 900 nM primers and 500 nM Q61H probe (yellow dashed square). Multiplex C has 900 nM primers and 500 nM Q61H wild-type probe in addition to a G12C wild-type assay. All other wild-type droplet populations shown, except in the Q61H duplex assay, are 450 nM primers and 250 nM G12C wild-type probe. All panels in the left and centre columns show a FAM amplitude up to 18000 and an HEX amplitude up to 6000. Panels in the right column have a FAM amplitude up to 18000 and a HEX amplitude up to 11000.
KRAS multiplex ddPCR assay characterisation
Decreasing amounts of NCI-H358 (KRAS G12C) and A549 (KRAS G12S) gDNA were spiked into KRAS wild-type gDNA (NCI-H1975) and tested in the appropriate KRAS multiplex assay. The limit of detection for KRAS G12C mutant DNA in multiplex A was 0.03% and for KRAS G12S DNA in multiplex B was 0.045% (S8 Fig, top panels) The measured mutant allele frequency correlated well with decreasing amounts of spiked KRAS mutant gDNA in multiplexes A and B (r2 = 0.9992 and 0.0998 respectively), demonstrating the linearity of mutation detection in the multiplex assays. Mutant allele frequency showed little intra-well variability for either multiplex assay and high reproducibility between two different operators on three alternate days for a range of allele frequencies (S8 Fig, middle and bottom panels).
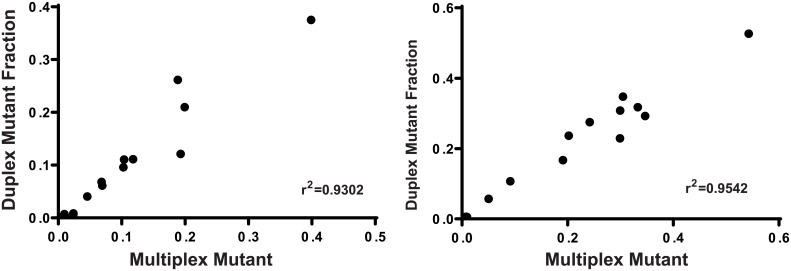
Similar KRAS allele frequency was observed using multiplex or duplex assays in both cell line DNA (single species KRAS mutant gDNA and combinations of KRAS mutant gDNA at varying allele frequencies) or oligonucleotides and FFPE tissue DNA (Fig 4; r2 = 0.9302 and 0.9542 respectively).
Fig 4. Correlation of mutant DNA fraction detected in multiplex and duplex assays using cell line gDNA or oligonucleotides (left) and in FFPE tissue DNA (right).
We analysed multiple wells of NCI-H1975 KRAS wild-type gDNA with each of the three KRAS multiplex assays and calculated the probability of false positive mutation detection, setting the limit of detection at 0.05% to reflect our measured results with spiked KRAS mutant gDNA species (S8 Fig). The specificity of each multiplex is 99.99995%, 99.99857% and 99.85578% for multiplexes A, B and C respectively (S1 Table).
Comparison of KRAS multiplex assay mutant detection with Sanger sequencing
A range of samples containing decreasing amounts of NCI-H358 (KRAS G12C) or A549 (KRAS G12S) gDNA spiked into KRAS wild-type DNA (NCI-H1975) were simultaneously analysed using digital PCR and Sanger sequencing. KRAS G12C mutant DNA was detectable using multiplex A down to a mutant allele frequency of 0.2%, whereas the mutant peak on the chromatogram is only visible at a mutant allele frequency of 31.5% and not below 10% (S9 Fig, top panels). KRAS G12S mutant DNA remains detectable by digital PCR using multiplex B at a mutant allele frequency of 0.1%, but is only visible using Sanger sequencing at 17%. (S9 Fig, lower panels).
Detection of KRAS mutations in FFPE tissue DNA
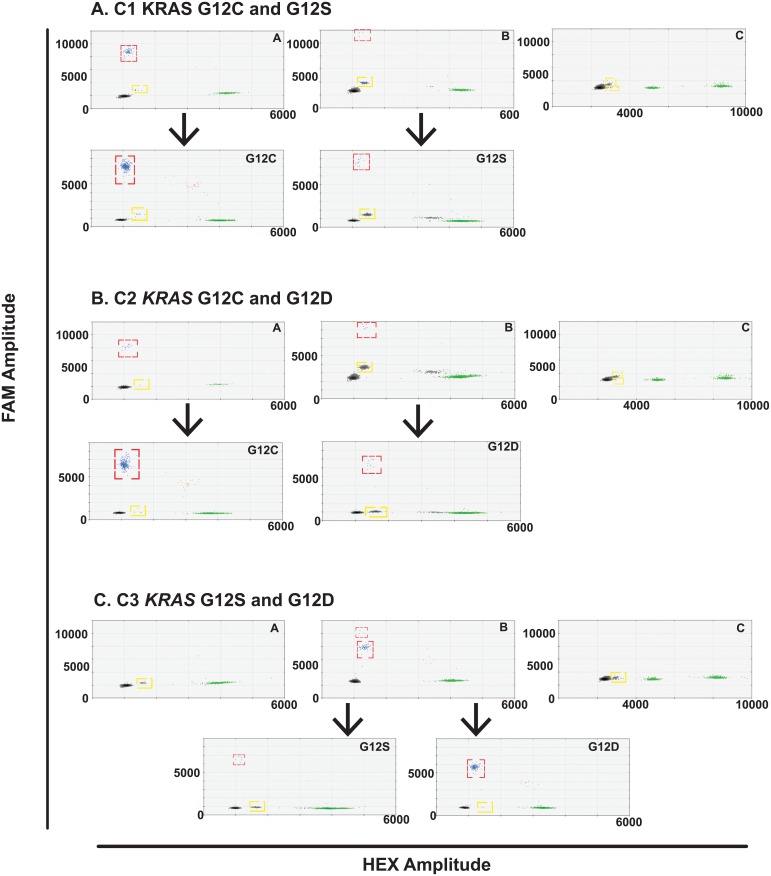
FFPE tissue DNA extracted from 11 cases of advanced KRAS mutant NSCLC (S2 Table) was analysed using each multiplex assay. The presence of a KRAS 12/13 mutation was known from prior Cobas® testing (Cobas® KRAS Mutation Test, Roche) but investigators were blinded to the specific KRAS genotype. At least one KRAS mutation was detected in each case and two cases had two detectable KRAS clones (Fig 5). Mutations were confirmed with the appropriate duplex assay. The low frequency G12D mutation in case S011 may represent FFPE artefact due to the frequent deamination of guanine nucleotides during the tissue preservation process [14]. The G12F mutation in S018 was identified by subsequent sequencing of the tissue sample after observation of the cross-reactivity droplet population in all multiplex assays. In addition, two species of KRAS mutant gDNA were spiked into a background of KRAS wild-type DNA (NCI-H1975) gDNA to create three samples containing a major and minor KRAS clone; C1 is a combination of NCI-H358 (KRAS G12C) and A549 (KRAS G12S) gDNA, C2 is a combination of NCI-H358 (KRAS G12C) and A427 (KRAS G12D) gDNA and C3 is a combination of A427 (KRAS G12D) and A549 (KRAS G12S) gDNA. All KRAS species were detectable using the KRAS multiplex assays, including the minor clones down to a mutant allele frequency of 0.3% measured for the KRAS G12D mutant DNA in sample C3 (Fig 6).
Fig 5. KRAS mutant FFPE tissue DNA analysis using multiplex and duplex assays to detect KRAS mutant clones.
All samples, except for S011, were analysed with multiplexes A, B and C (upper panels) and the KRAS mutation detected was subsequently confirmed with the appropriate duplex assay (lower panels). Mutant DNA droplet populations are highlighted with a red dashed square. Droplet populations caused by cross-reactivity with a KRAS mutant DNA species not present in the multiplex are indicated by a yellow dashed square.
Fig 6. KRAS mutant biclonal gDNA analysis using multiplex and duplex assays to detect KRAS mutant clones.
All samples were analysed with multiplexes A, B and C (upper panels) and the KRAS mutation detected was subsequently confirmed with the appropriate duplex assay (lower panels). Mutant DNA droplet populations are highlighted with a red dashed square. Droplet populations caused by cross-reactivity with a KRAS mutant DNA species not present in the multiplex are indicated by a yellow dashed square.
Comparison of KRAS multiplex assay mutant detection with next generation sequencing
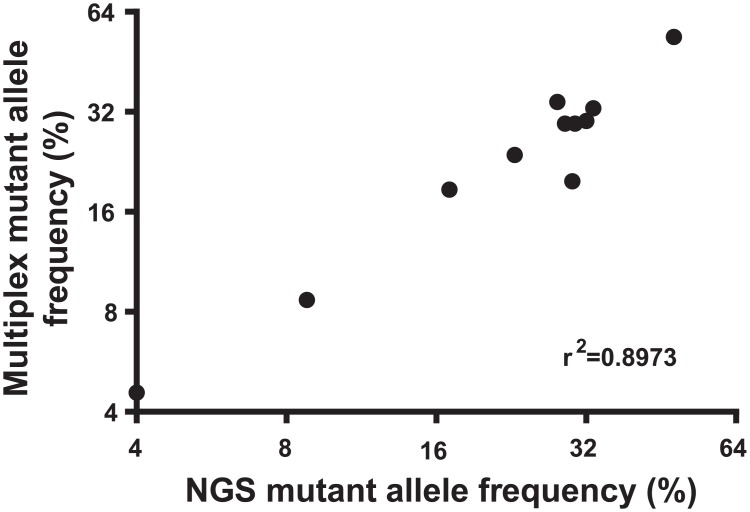
FFPE tissue DNA from all 11 cases and gDNA from the 3 biclonal cell line DNA samples were sequenced using the Ion Ampliseq Cancer Hotspot Panel v2 on the Ion ProtonTM platform (Life Technologies) to analyse the KRAS 12 and 13 loci. Next generation sequencing could not detect KRAS mutant allele frequencies below 4% in tissue DNA and none of the minor clones below 2% were detectable in the biclonal gDNA samples in either of the duplicate library preparations (Tables 3 and 4). The frequency of KRAS mutant alleles that were detectable on next generation sequencing correlate well with those measured in the KRAS multiplex digital PCR assays (Fig 7, r2 = 0.8973).
Table 4. Detection of KRAS mutant clones in biclonal gDNA samples using next generation sequencing (NGS) and KRAS multiplex digital PCR assays.
Two individual library preparations were analysed for each sample.
| ID | KRAS genotype | Library 1 NGS mutant allele frequency (%) | Library 2 NGS mutant allele frequency (%) | Digital PCR mutant allele frequency (%) |
|---|---|---|---|---|
| C1 | G12C | 25 | 23 | 19.4 |
| G12S | ND | ND | 1.9 | |
| C2 | G12C | 26 | 26 | 18.4 |
| G12D | ND | ND | 0.49 | |
| C3 | G12D | 9 | 10 | 9.8 |
| G12D | ND | ND | 0.58 |
Key: ND- not detected.
Fig 7. Correlation of NGS KRAS mutant allele frequency with digital PCR KRAS mutant allele frequency detected in the appropriate multiplex assay for FFPE tissue DNA and cell line gDNA samples.
Discussion
We have used commercially available primer probe assays for nine common KRAS mutations to design and optimize a multiplex assay approach to genotype KRAS mutant cancers using limited material using digital droplet PCR. The Bio-Rad QX200 system has been used to analyse copy number variation in a multiplexed fashion with EvaGreen DNA binding dye [15] but this is the first report of KRAS mutation detection multiplex assays on the QX100 system that are based on varying concentrations of the primer probe assays.
These multiplex assays permit the identification of the specific KRAS genotype and we have observed that this can be deduced from the reproducible droplet populations caused by cross-reactivity in any of the multiplex assays as well as the positive mutant droplet populations at the predicted FAM amplitude. These multiplexes can therefore be used to identify the presence of rarer KRAS mutations at the G12/13 and Q61 positions other than those already tested for in the multiplex assays, as demonstrated with case S018.
Sub-clonal mutations can also be detected using this ddPCR approach to KRAS mutation detection. In two cases, S010 and S011, two different KRAS mutations were detected both in multiplex and duplex. We have noted, however, that all the mutations tested in multiplex B, are G>A nucleotide changes. Low frequency single nucleotide changes as an artifact of the tissue preservation process have been observed [16]. The KRAS G12D mutation observed in S011 (mutant allele frequency 0.05%) may therefore be a false positive result. The low frequency KRAS G12C mutation in S010, however, is a G>T change and therefore much more likely to be a true result.
We have demonstrated that the multiplex assays are more sensitive than Sanger and next generation sequencing and highly specific. The assays have linearity down to a limit of detection of 0.03–0.045%. Due to the false positive signal seen from FFPE tissue on NGS below mutant allele frequencies of 1.5%[17], the limit of detection is at best 1 mutant KRAS molecule in 50 wild-type KRAS molecules. The KRAS G12S minor clone in the C1 gDNA sample that was measured at 1.9% on multiplex digital PCR, for example, was not detectable on NGS in either of two independent library preparations. Sanger sequencing, in comparison, has a limit of detection of only 1 in 10 and over a range of mutant allele frequencies, a sensitivity of 20%. Digital PCR maintains a very high specificity despite this increased sensitivity and is approaching the 100% specificity seen with NGS [17]. The accuracy of KRAS multiplex ddPCR is reflected by the correlation observed with the mutant allele frequency detected by Ion Torrent ProtonTM sequencing, an established method of determining this variable. In addition, we have demonstrated the robustness of the multiplex assays across a range of mutant allele frequencies both within the same experiment and between operators on three non-consecutive days.
We have described the combination and optimization of commercially available digital PCR assays to develop novel multiplex assays that allow the accurate detection and discrimination of KRAS genotypes in patient-derived material. Multiplexing of assays reduces the amount of clinical material required for testing, which will be particularly important in settings where the proportion of mutant relative to wild-type DNA is likely to be low, such as with circulating free tumour DNA from plasma samples. The three multiplexed assays described here between them will identify the nucleotide changes in 95% of the cancer cases where the KRAS gene is mutated.
Supporting Information
Left panels: KRAS G12V, G12C and G12D mutant DNA tested with 562.5 nM primers and 312.5 nM G12D FAM probe in one well (top left panel) or in three separate wells (i.e. one well G12V DNA with G12D probe assay etc.). Wells with only one type of KRAS mutant DNA are merged (left second panel) to compare with the single well containing all DNA species. KRAS G12V, G12C and G12D mutant DNA tested with 225 nM primers and 125 nM G12V FAM probe in one well (left third panel) or in three separate wells (wells merged, left bottom panel). A second droplet population with lower FAM amplitude is detected when more than one species of KRAS mutant DNA is present (red dashed box). Right panels: KRAS 12/13 FFPE tissue DNA sample (F124) tested with multiplex assay (top right) and then 3 separate duplex assays as indicated. The same scale is used for all panels (HEX amplitude up to 7000 and FAM amplitude up to 11000).
(EPS)
Duplex assays for different KRAS mutations (horizontal) with different KRAS mutation variants (vertical). More cross-reactivity, indicated by a droplet population near the empty droplets with relatively low FAM and HEX amplitude and a population near the true wild-type population (examples in red dashed square in top right panel), was seen between probes and DNA for the same nucleotide position in exon 2 of KRAS. Each panel shows FAM amplitude of up to 10000 and HEX amplitude between 1000 and 6000.
(EPS)
Duplex assays for different KRAS mutations (horizontal) with different KRAS mutation variants (vertical). More cross-reactivity, indicated by a droplet population near the empty droplets with relatively low FAM and HEX amplitude and a population near the true wild-type population (examples in red dashed square in top right panel), was seen between probes and DNA for the same nucleotide position in exon 2 of KRAS. Key: X—omitted panel of duplex assay with correct probe and KRAS mutation variant as higher magnification. Each panel shows FAM amplitude of up to 3000 and HEX amplitude between 1000 and 6000.
(EPS)
G13C populations are indicated by a red dashed square, G12C populations by a blue dashed square and G12V droplet populations are indicated by a yellow dashed square. Multiplex 1 (top left panel) is an assay combination of 900 nM primers and 500 nM G13C probe, 450 nM primers and 250 nM G12C probe and 225 nM primers and 125 nM G12V probe, with 900 nM primers and 500 nM WT for G12C probe and 450 nM primers and 250 nM WT for G13C probe. Multiplex 2 (centre left) uses the same FAM assay concentrations as in Multiplex 1 with 450 nM primers and 250 nM WT for G12C probe and 900 nM primers and 500 nM WT for G13C probe. Multiplex 3 (centre right) is a combination of 450 nM primers and 250 nM G13C probe and 900 nM primers and 500 nM G12C probe with the same G12V and WT G12C and G13C assays as in Multiplex 1. Multiple 4 uses the same FAM assays concentrations as Multiplex 3 with the WT for G12C and G13C assay concentrations as in Multiplex 2. Each panel shows a FAM amplitude up to 18000 and a HEX amplitude up to 12000.
(EPS)
G12S droplet populations are indicated by a red dashed square, G12D populations by a blue dashed square, and G13D populations by a yellow dashed square. Multiplex 1 (top left panel) is an assay combination of 675 nM primers and 375 nM G12S probe, 450 nM primers and 250 nM G12D probe and 225 nM primers and 125 nM G13D probe with 225 nM primers and 125 nM WT for G12C probe and 450 nM primers and 250 nM WT for G13D probe. Multiplex 2 (top middle panel) is a FAM assay combination of 225 nM primers and 125 nM G12S probe, 675 nM primers and 375 nM G12D probe and 450 nM primers and 250 nM G13D probe with the same WT for G12C and G13D assay concentrations as in Multiplex 1. Multiplex 3 (top right panel) is a combination of, 450 nM primers and 250 nM G12S probe, 675 nM primers and 375 nM G12D probe and the same FAM 13D assay concentration as in Multiplex 1. WT assay concentrations are 450 nM primers and 250 nM WT for G12C probe and 225 nM primers and 125 nM WT for G13D probe. Each panel shows a FAM amplitude up to 18000 and a HEX amplitude up to 9000.
(EPS)
G12R droplet populations are indicated by a red dashed square, G12A populations by a blue dashed square and Q61H populations by a yellow dashed square. Multiplex 1 (top left panel) is an assay combination of 675 nM primers and 375 nM G12R probe, 450 nM primers and 250 nM G12A probe and 900 nM primers and 500 nM Q61H probe with 450 nM primers and 250 nM WT for G12C probe and 900 nM primers and 500 nM WT for Q61H probe. Multiplexes 2–4 use the same WT assay concentrations. Multiplex 2 (centre left panel) is a FAM assay combination of 450 nM primers and 250 nM G12R probe and 675 nM primers and 375 nM G12A probe with the same FAM Q61H assay concentration as in Multiplex 1. Multiplex 3 (centre right panel) is an assay combination of 900 nM primers and 500 nM G12R probe, 225 nM primers and 125 nM G12A probe and 450 nM primers and 250 nM Q61H probe. Multiplex 4 (top right panel) is an assay combination of 450 nM primers and 250 nM G12A probe and 225 nM primers and 125 nM Q61H probe with the same FAM G12R assay as in Multiplex 3. Each panel shows a FAM amplitude up to 16000 and a HEX amplitude up to 11000.
(EPS)
Droplet populations seen with the addition of cell line DNA or oligonucleotides containing different KRAS mutation variants and analysed with multiplexes A, B and C. Multiplex A is an assay combination of G13C, G12C and G12V. Multiplex B is an assay combination of G12S, G12D and G13D. Multiplex C is an assay combination of G12R, G12A and Q61H. Panel A1 –multiplex A with KRAS G12V, C, D, S and R DNA; panel A2 –multiplex A with KRAS G12A DNA; panel A3- multiplex A with KRAS G13C and G13D DNA. Panel B1 –multiplex B with KRAS G12V, C, D, S and R DNA; panel B2 –multiplex B with KRAS G12A DNA; panel B3- multiplex B with KRAS G13C and G13D DNA. Panel C—multiplex C with KRAS G12V, C, D, S, R, A, 13C, 13D and Q61H DNA. Panels A1-3 and B1-3 show a FAM amplitude up to 18000 and an HEX amplitude up to 7000. Panel C has a FAM amplitude up to 18000 and a HEX amplitude up to 10000. Key: * = KRAS G12V mutant DNA, ¶ = G12D, ⌃ = G12C, ★ = G12S, ♦ = G12R, • = G12A, § = G13C, ■ = G13D; black- empty droplets, blue- mutant DNA FAM positive droplets, green- wild-type DNA HEX positive droplets, brown—wild-type and mutant DNA double positive droplets.
(EPS)
Left panels: Multiplex A tested with decreasing amounts of KRAS mutant G12C DNA spiked in against a KRAS wild-type DNA background; right panels: Multiplex B tested with decreasing amounts of KRAS mutant G12S DNA against a KRAS wild-type DNA background. Top panels: Correlation of amount of input KRAS mutant DNA with measured mutant allele frequency. Middle panels: Mutant allele frequency per well as compared with mean mutant allele frequency across all wells for a range of allele frequencies (0.05–4% multiplex A and KRAS G12C, 0.08–3% multiplex B and KRAS G12S). Bottom panels: Mutant allele frequency measured on three non-adjacent days by two different operators across a range of amounts of spiked KRAS mutant DNA. Key: Triangle = Day 1, square = Day 2, circle = Day 3.
(EPS)
KRAS G12C mutant DNA samples at allele frequencies ranging from 0.2–31.5% analysed by digital PCR in KRAS multiplex assay A (upper panels) or Sanger sequencing (upper chromatograms). KRAS G12S mutant DNA samples at allele frequencies ranging from 0.1–17% analysed by digital PCR in KRAS multiplex assay B (lower panels) or Sanger sequencing (lower chromatograms). All panels in the upper row show a FAM amplitude up to 10000 and a HEX amplitude up to 6000. Panels in the lower row have a FAM amplitude up to 14000 and a HEX amplitude up to 11000. Key: black- empty droplets, blue- mutant DNA FAM positive droplets, green- wild-type DNA HEX positive droplets, brown—wild-type and mutant DNA double positive droplets; black arrow—mutant nucleotide not discernible from background signal on Sanger sequencing, red arrow—mutant thymine nucleotide detectable using Sanger sequencing, green arrow—mutant adenine nucleotide detectable using Sanger sequencing.
(EPS)
(DOC)
Key: F- former smoker, S- smoker, N- never smoker, Unk- unknown.
(DOC)
Acknowledgments
We acknowledge NHS funding to the NIHR Biomedical Research Centre at The Royal Marsden and the ICR. We gratefully acknowledge the contribution to this publication made by Cancer Research UK, Stratified Medicine Programme. We also acknowledge material support from Dr Madeleine Hewish. This research was supported in part by the Revere Charitable Trust, an unrestricted educational grant from Pierre-Fabre Ltd, funding from the European Research Council Advanced Grant RASTARGET and funding from the Wellcome Trust Senior Investigator Award 103799/Z/14/Z.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors gratefully acknowledge the support of this work by the National Health Service (England), through funding to the National Institute for Health Research Biomedical Research Centre at The Royal Marsden Hospital. They also gratefully acknowledge funding from the Institute of Cancer Research and Cancer Research UK, Stratified Medicine Programme. This research was also supported in part by the Revere Charitable Trust, an unrestricted educational grant from Pierre-Fabre Ltd., the European Research Council Advanced Grant RASTARGET and the Wellcome Trust Senior Investigator Award, 103799/Z/14/Z. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin D M (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917. 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 2.Health and Social Care Information Centre (2013) National Lung Cancer Audit Report 2013. [Internet] 26 Nov 2013. Available: http://www.hscic.gov.uk/hospital-care. Accessed 14 Jul 2015.
- 3. Pao W, Girard N (2011) New driver mutations in non-small-cell lung cancer. Lancet Oncology 12: 175–180. 10.1016/S1470-2045(10)70087-5 [DOI] [PubMed] [Google Scholar]
- 4. Leighl NB, Rekhtman N, Biermann WA, Huang J, Mino-Kenudson M, Ramalingam SS, et al. (2014) Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: american society of clinical oncology endorsement of the college of American pathologists/international association for the study of lung cancer/association for molecular pathology guideline. Journal of Clinical Oncology 32: 3673–3679. 10.1200/JCO.2014.57.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sequist LV, Yang JC-H, Yamamoto N, O'Byrne K, Hirsh V, Mok T, et al. (2013) Phase III Study of Afatinib or Cisplatin Plus Pemetrexed in Patients With Metastatic Lung Adenocarcinoma With EGFR Mutations. Journal of Clinical Oncology 31: 3327–3334. 10.1200/jco.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 6. Popat S, Mok T, Yang JC-H, Wu Y-L, Lungershausen J, Stammberger U, et al. (2014) Afatinib in the treatment of EGFR mutation-positive NSCLC—a network meta-analysis. Lung Cancer 85: 230–238. 10.1016/j.lungcan.2014.05.007 [DOI] [PubMed] [Google Scholar]
- 7. Jänne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, et al. (2012) Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncology 14: 38–47. 10.1016/S1470-2045(12)70489-8 [DOI] [PubMed] [Google Scholar]
- 8. Leary AF, de Castro DG, Nicholson AG, Ashley S, Wotherspoon A, O’Brien MER, et al. (2012) Establishing an EGFR mutation screening service for non-small cell lung cancer—sample quality criteria and candidate histological predictors. Eur J Cancer 48: 61–67. [DOI] [PubMed] [Google Scholar]
- 9. Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, et al. (2013) Absolute quantification by droplet digital PCR versus analog real-time PCR. Nature Methods 10: 10003–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA, et al. (2011) Evaluation of Digital PCR for Absolute DNA Quantification. Anal Chem 83: 6474–6484. 10.1021/ac103230c [DOI] [PubMed] [Google Scholar]
- 11. Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, et al. (2013) Multiplex Picodroplet Digital PCR to Detect KRAS Mutations in Circulating DNA from the Plasma of Colorectal Cancer Patients. Clinical Chemistry. 10.1373/clinchem.2013.206359 [DOI] [PubMed] [Google Scholar]
- 12. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. Oxford University Press; 2009;25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rare Mutation Detection Best Practices Guidelines [Internet]. 1 Feb 2015, Available: http://www.biorad.com/webroot/web/pdf/lsr/literature/Bulletin_6628.pdf. Accessed 26 Feb 2015.
- 14. Gallegos Ruiz MI, Floor K, Rijmen F, Grunberg K, Rodriguez JA, Giaccone G. EGFR and KRAS mutation analysis in non-small cell lung cancer: comparison of paraffin embedded versus frozen specimens. Cell Oncol. 2007;29: 257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McDermott GP, Do D, Litterst CM, Maar D, Hindson CM, Steenblock ER, et al. Multiplexed target detection using DNA-binding dye chemistry in droplet digital PCR. Anal Chem. 2013;85: 11619–11627. 10.1021/ac403061n [DOI] [PubMed] [Google Scholar]
- 16. Do H, Wong SQ, Li J, Dobrovic A. Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates. Clinical Chemistry. 2013;59: 1376–1383. 10.1373/clinchem.2012.202390 [DOI] [PubMed] [Google Scholar]
- 17. Lin M-T, Mosier SL, Thiess M, Beierl KF, Debeljak M, Tseng L-H, et al. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol. American Society for Clinical Pathology; 2014;141: 856–866. 10.1309/AJCPMWGWGO34EGOD [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Left panels: KRAS G12V, G12C and G12D mutant DNA tested with 562.5 nM primers and 312.5 nM G12D FAM probe in one well (top left panel) or in three separate wells (i.e. one well G12V DNA with G12D probe assay etc.). Wells with only one type of KRAS mutant DNA are merged (left second panel) to compare with the single well containing all DNA species. KRAS G12V, G12C and G12D mutant DNA tested with 225 nM primers and 125 nM G12V FAM probe in one well (left third panel) or in three separate wells (wells merged, left bottom panel). A second droplet population with lower FAM amplitude is detected when more than one species of KRAS mutant DNA is present (red dashed box). Right panels: KRAS 12/13 FFPE tissue DNA sample (F124) tested with multiplex assay (top right) and then 3 separate duplex assays as indicated. The same scale is used for all panels (HEX amplitude up to 7000 and FAM amplitude up to 11000).
(EPS)
Duplex assays for different KRAS mutations (horizontal) with different KRAS mutation variants (vertical). More cross-reactivity, indicated by a droplet population near the empty droplets with relatively low FAM and HEX amplitude and a population near the true wild-type population (examples in red dashed square in top right panel), was seen between probes and DNA for the same nucleotide position in exon 2 of KRAS. Each panel shows FAM amplitude of up to 10000 and HEX amplitude between 1000 and 6000.
(EPS)
Duplex assays for different KRAS mutations (horizontal) with different KRAS mutation variants (vertical). More cross-reactivity, indicated by a droplet population near the empty droplets with relatively low FAM and HEX amplitude and a population near the true wild-type population (examples in red dashed square in top right panel), was seen between probes and DNA for the same nucleotide position in exon 2 of KRAS. Key: X—omitted panel of duplex assay with correct probe and KRAS mutation variant as higher magnification. Each panel shows FAM amplitude of up to 3000 and HEX amplitude between 1000 and 6000.
(EPS)
G13C populations are indicated by a red dashed square, G12C populations by a blue dashed square and G12V droplet populations are indicated by a yellow dashed square. Multiplex 1 (top left panel) is an assay combination of 900 nM primers and 500 nM G13C probe, 450 nM primers and 250 nM G12C probe and 225 nM primers and 125 nM G12V probe, with 900 nM primers and 500 nM WT for G12C probe and 450 nM primers and 250 nM WT for G13C probe. Multiplex 2 (centre left) uses the same FAM assay concentrations as in Multiplex 1 with 450 nM primers and 250 nM WT for G12C probe and 900 nM primers and 500 nM WT for G13C probe. Multiplex 3 (centre right) is a combination of 450 nM primers and 250 nM G13C probe and 900 nM primers and 500 nM G12C probe with the same G12V and WT G12C and G13C assays as in Multiplex 1. Multiple 4 uses the same FAM assays concentrations as Multiplex 3 with the WT for G12C and G13C assay concentrations as in Multiplex 2. Each panel shows a FAM amplitude up to 18000 and a HEX amplitude up to 12000.
(EPS)
G12S droplet populations are indicated by a red dashed square, G12D populations by a blue dashed square, and G13D populations by a yellow dashed square. Multiplex 1 (top left panel) is an assay combination of 675 nM primers and 375 nM G12S probe, 450 nM primers and 250 nM G12D probe and 225 nM primers and 125 nM G13D probe with 225 nM primers and 125 nM WT for G12C probe and 450 nM primers and 250 nM WT for G13D probe. Multiplex 2 (top middle panel) is a FAM assay combination of 225 nM primers and 125 nM G12S probe, 675 nM primers and 375 nM G12D probe and 450 nM primers and 250 nM G13D probe with the same WT for G12C and G13D assay concentrations as in Multiplex 1. Multiplex 3 (top right panel) is a combination of, 450 nM primers and 250 nM G12S probe, 675 nM primers and 375 nM G12D probe and the same FAM 13D assay concentration as in Multiplex 1. WT assay concentrations are 450 nM primers and 250 nM WT for G12C probe and 225 nM primers and 125 nM WT for G13D probe. Each panel shows a FAM amplitude up to 18000 and a HEX amplitude up to 9000.
(EPS)
G12R droplet populations are indicated by a red dashed square, G12A populations by a blue dashed square and Q61H populations by a yellow dashed square. Multiplex 1 (top left panel) is an assay combination of 675 nM primers and 375 nM G12R probe, 450 nM primers and 250 nM G12A probe and 900 nM primers and 500 nM Q61H probe with 450 nM primers and 250 nM WT for G12C probe and 900 nM primers and 500 nM WT for Q61H probe. Multiplexes 2–4 use the same WT assay concentrations. Multiplex 2 (centre left panel) is a FAM assay combination of 450 nM primers and 250 nM G12R probe and 675 nM primers and 375 nM G12A probe with the same FAM Q61H assay concentration as in Multiplex 1. Multiplex 3 (centre right panel) is an assay combination of 900 nM primers and 500 nM G12R probe, 225 nM primers and 125 nM G12A probe and 450 nM primers and 250 nM Q61H probe. Multiplex 4 (top right panel) is an assay combination of 450 nM primers and 250 nM G12A probe and 225 nM primers and 125 nM Q61H probe with the same FAM G12R assay as in Multiplex 3. Each panel shows a FAM amplitude up to 16000 and a HEX amplitude up to 11000.
(EPS)
Droplet populations seen with the addition of cell line DNA or oligonucleotides containing different KRAS mutation variants and analysed with multiplexes A, B and C. Multiplex A is an assay combination of G13C, G12C and G12V. Multiplex B is an assay combination of G12S, G12D and G13D. Multiplex C is an assay combination of G12R, G12A and Q61H. Panel A1 –multiplex A with KRAS G12V, C, D, S and R DNA; panel A2 –multiplex A with KRAS G12A DNA; panel A3- multiplex A with KRAS G13C and G13D DNA. Panel B1 –multiplex B with KRAS G12V, C, D, S and R DNA; panel B2 –multiplex B with KRAS G12A DNA; panel B3- multiplex B with KRAS G13C and G13D DNA. Panel C—multiplex C with KRAS G12V, C, D, S, R, A, 13C, 13D and Q61H DNA. Panels A1-3 and B1-3 show a FAM amplitude up to 18000 and an HEX amplitude up to 7000. Panel C has a FAM amplitude up to 18000 and a HEX amplitude up to 10000. Key: * = KRAS G12V mutant DNA, ¶ = G12D, ⌃ = G12C, ★ = G12S, ♦ = G12R, • = G12A, § = G13C, ■ = G13D; black- empty droplets, blue- mutant DNA FAM positive droplets, green- wild-type DNA HEX positive droplets, brown—wild-type and mutant DNA double positive droplets.
(EPS)
Left panels: Multiplex A tested with decreasing amounts of KRAS mutant G12C DNA spiked in against a KRAS wild-type DNA background; right panels: Multiplex B tested with decreasing amounts of KRAS mutant G12S DNA against a KRAS wild-type DNA background. Top panels: Correlation of amount of input KRAS mutant DNA with measured mutant allele frequency. Middle panels: Mutant allele frequency per well as compared with mean mutant allele frequency across all wells for a range of allele frequencies (0.05–4% multiplex A and KRAS G12C, 0.08–3% multiplex B and KRAS G12S). Bottom panels: Mutant allele frequency measured on three non-adjacent days by two different operators across a range of amounts of spiked KRAS mutant DNA. Key: Triangle = Day 1, square = Day 2, circle = Day 3.
(EPS)
KRAS G12C mutant DNA samples at allele frequencies ranging from 0.2–31.5% analysed by digital PCR in KRAS multiplex assay A (upper panels) or Sanger sequencing (upper chromatograms). KRAS G12S mutant DNA samples at allele frequencies ranging from 0.1–17% analysed by digital PCR in KRAS multiplex assay B (lower panels) or Sanger sequencing (lower chromatograms). All panels in the upper row show a FAM amplitude up to 10000 and a HEX amplitude up to 6000. Panels in the lower row have a FAM amplitude up to 14000 and a HEX amplitude up to 11000. Key: black- empty droplets, blue- mutant DNA FAM positive droplets, green- wild-type DNA HEX positive droplets, brown—wild-type and mutant DNA double positive droplets; black arrow—mutant nucleotide not discernible from background signal on Sanger sequencing, red arrow—mutant thymine nucleotide detectable using Sanger sequencing, green arrow—mutant adenine nucleotide detectable using Sanger sequencing.
(EPS)
(DOC)
Key: F- former smoker, S- smoker, N- never smoker, Unk- unknown.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.