A chimeric ligand–gated channel is activated by the ligands that activate both parent channels, which gate it through two distinct mechanisms.
Abstract
Recent high resolution structures of several pentameric ligand–gated ion channels have provided unprecedented details of their molecular architecture. However, the conformational dynamics and structural rearrangements that underlie gating and allosteric modulation remain poorly understood. We used a combination of electrophysiology, double electron–electron resonance (DEER) spectroscopy, and x-ray crystallography to investigate activation mechanisms in a novel functional chimera with the extracellular domain (ECD) of amine-gated Erwinia chrysanthemi ligand–gated ion channel, which is activated by primary amines, and the transmembrane domain of Gloeobacter violaceus ligand–gated ion channel, which is activated by protons. We found that the chimera was independently gated by primary amines and by protons. The crystal structure of the chimera in its resting state, at pH 7.0 and in the absence of primary amines, revealed a closed-pore conformation and an ECD that is twisted with respect to the transmembrane region. Amine- and pH-induced conformational changes measured by DEER spectroscopy showed that the chimera exhibits a dual mode of gating that preserves the distinct conformational changes of the parent channels. Collectively, our findings shed light on both conserved and divergent features of gating mechanisms in this class of channels, and will facilitate the design of better allosteric modulators.
INTRODUCTION
Pentameric ligand–gated ion channels (pLGICs) mediate fast synaptic transmissions in the central and peripheral nervous systems, and the vertebrate members of this family include the nicotinic acetylcholine (ACh) receptors (nAChRs), the γ–aminobutyric acid receptors (GABAARs), glycine receptors (GlyRs), and serotonin receptors (5HT3Rs) (Sine and Engel, 2006; Thompson et al., 2010; Corringer et al., 2012; daCosta and Baenziger, 2013). pLGICs play a key role in neuronal function associated with learning and memory, and dysfunctions in these channels underlie several neurodegenerative diseases including Alzheimer’s disease, Parkinson’s disease, schizophrenia, epilepsies, myasthenia gravis, and congenital myasthenic syndromes. pLGICs are targets for a wide range of therapeutics including general and local anesthetics, alcohols, benzodiazepines, neurosteroids, and barbiturates. Upon neurotransmitter binding, pLGICs rapidly switch between the closed, open, and desensitized states. These conformational transitions are governed by a timely orchestration of protein motions over a 60-Å distance along the length of the channel. Drugs and modulators enhance or inhibit channel activity by allosterically shifting the equilibrium toward the open or closed/desensitized conformation, respectively. However, the molecular details underlying gating and drug modulation are still unclear and remain areas of intense research.
Our understanding of pLGICs is entering a new phase, as there are now high resolution structures of several members of the family: the invertebrate glutamate-gated chloride channel (Hibbs and Gouaux, 2011), the human β3-GABAAR (Miller and Aricescu, 2014), the mouse 5-HT3 (Hassaine et al., 2014), and the prokaryotic homologues Gloeobacter violaceus ligand–gated ion channel (GLIC; pH gated) (Bocquet et al., 2009; Hilf and Dutzler, 2009) and Erwinia chrysanthemi ligand–gated ion channel (ELIC; primary amine gated) (Hilf and Dutzler, 2008). These findings lay the foundation for an in-depth mechanistic understanding of ligand recognition, channel gating, and drug modulation. The prokaryotic homologues, GLIC and ELIC, have served as excellent structural surrogates in describing protein motions underlying channel gating and in identifying conserved drug-binding regions. In GLIC, details of the conformational changes associated with channel activation come from crystal structures of the channel in the putative open and closed conformations (Bocquet et al., 2009; Hilf and Dutzler, 2009; Sauguet et al., 2013, 2014) and from electron paramagnetic resonance (EPR) spectroscopy studies of the channel embedded in membranes (Velisetty et al., 2012, 2014; Dellisanti et al., 2013; Chakrapani, 2015). Collectively, these studies reveal that during transitions from the closed to the open/desensitized states, there are extensive changes in quaternary structure, domain interfaces, solvent and lipid accessibility, and overall channel dynamics. On the other hand, although ELIC structures have provided a comprehensive view of agonist and modulator-binding sites (Zimmermann and Dutzler, 2011; Pan et al., 2012; Spurny et al., 2012; Ulens et al., 2014), the conformational changes that would be associated with gating or modulation are not evident in these structures. In fact, ELIC mutants with dramatically altered functionality revealed little changes in the crystal structures (Gonzalez-Gutierrez et al., 2012). A recent 19F nuclear magnetic resonance study on the pore-lining region of ELIC suggests that the channel opens via expansion of the extracellular half of M2 and impedes conduction in the desensitized state through a compression of the intracellular half (Kinde et al., 2015). Although this mechanism of activation and desensitization are in remarkable agreement with the previously proposed model for GLIC (Velisetty et al., 2012), and supported by the GABA crystal structure (Miller and Aricescu, 2014), it is noteworthy that the range of protein motions observed in ELIC are distinctly smaller and only occur locally. The difference in global dynamics despite the conservation of key mechanistic events is intriguing and may represent fundamental variations among different members of the family. Investigating the molecular basis for such disparity in conformational dynamics will further strengthen our understanding of functioning and modulation within members of pLGICs.
Here, we have taken an integrated approach toward understanding the structure, dynamics, and function of pLGICs using x-ray crystallography, EPR spectroscopy, and electrophysiological measurements. We first investigated ligand-induced conformational changes in ELICs by EPR spectroscopy. A key finding of our study is that the loop C (capping the neurotransmitter-binding site) movements in response to binding agonists and antagonists are distinct and in contrast to the canonical motions observed in other members of the family. To further understand how diverse conformational changes at the extracellular domain (ECD) led to opening of the channel pore, and to reconcile the observed differences in structural dynamics between ELIC and GLIC, we engineered a functional chimera by fusing the ELIC-ECD with the GLIC–transmembrane domain (TMD). Interestingly, the chimeric channel responds to both primary amines and protons. This finding suggests that despite limited conservation of the domain interface and differences in ligand-induced protein motions, the allosteric communication between the two domains is preserved. We determined a 4.6-Å structure of the ELIC-GLIC chimera in the resting state, which shows that the overall architecture of the parent channels is unperturbed. Because several key interdomain interactions, previously implicated in gating, were absent in this construct, global protein motions rather than pairwise interactions appear to govern gating. EPR spectroscopic measurements in the ELIC-GLIC chimera revealed that conformational changes in response to binding primary amines or protons emulate those of the parent channel throughout the chimera, in particular, large global conformational changes elicited by pH similar to GLIC and smaller and more local changes mediated by primary amines similar to ELIC.
MATERIALS AND METHODS
Cloning and functional measurements in oocytes
The ELIC-GLIC chimera was engineered by fusing the ECD of ELIC with the TMD of GLIC. The positions selected for fusion were Asn (200 in ELIC) at the end of the ECD and Tyr (194 in GLIC) in the pre-M1 region. The ELIC and GLIC clones were provided by R. Dutzler (University of Zürich, Zürich, Switzerland; Hilf and Dutzler, 2008, 2009). DNA encoding the chimera was inserted into the pTLN vector for oocyte expression and confirmed by DNA sequencing. The DNA was then linearized with the Mlu1 restriction enzyme overnight at 37°C. The RNA was synthesized using the mMessage mMachine kit (Ambion), purified with RNAeasy (QIAGEN), and injected (5–15 ng) into Xenopus laevis oocytes (stages V–VI). Control ooctyes were injected with the same volume of water to verify that endogenous currents were not present. Oocytes were maintained at 18°C in OR3 media (Leibovitz’s media containing glutamate, 500 U each of penicillin and streptomycin, with pH adjusted to 7.5 and osmolarity adjusted to 200–205 mOsm; Gibco). Two-electrode voltage-clamp experiments were then performed at room temperature 2–5 d after injection. An oocyte clamp (OC-725; Warner Instruments) was used for the measurements, and the current was sampled and digitized at 500 Hz with a Digidata 1332A (Molecular Devices). The traces were analyzed by Clampfit 10.2 (Molecular Devices). Oocytes were clamped at a holding potential of −60 mV, and current traces were recorded in response to ligand application. Solutions were changed using a syringe pump perfusion system flowing at a rate of 4–10 ml/min. The electrophysiological solutions contain 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES (neutral pH buffer) or 5 mM sodium citrate (acidic pH buffer). For patch-clamp recordings, currents were elicited in response ligands delivered using a fast solution exchanger (switch time 2 ms; RCS-200) fed by gravity (Bio-Logic). Currents were measured using Axopatch 200B, digitized at a sampling frequency of 10 kHz, and analyzed using Clampfit 10.2 (Molecular Devices).
Cloning and protein expression
The chimeric gene was cloned into a modified pET26b vector that carried an N-terminal maltose-binding protein. The chimeric protein was expressed and purified as described previously (Hilf and Dutzler, 2008; Bocquet et al., 2009; Velisetty and Chakrapani, 2012). In brief, BL21 (DE3) Escherichia coli cells transformed with the construct were grown in terrific broth media containing 50 µg/ml kanamycin at 37°C to an OD600 of 1.0. Cells were induced with 0.2 mM isopropyl 1-thio-β-d-galactopyranoside (Gold Biotechnology) overnight at 18°C. Membranes were prepared by homogenizing the cells in buffer A (150 mM NaCl and 20 mM Tris base, pH 7.4) with protease inhibitors and centrifuging at 100,000 g for 45 min. Membranes were solubilized in buffer A using 40 mM n-dodecyl-β-d-maltopyranoside (DDM; Anatrace) at 4°C. The chimera was purified by binding to amylose resin and eluting with 20 mM maltose. The maltose-binding protein tag was cleaved with human rhinovirus 3C protease (GE Healthcare), and the chimeric protein was separated using size-exclusion chromatography on a Superdex 20/200 column (GE Healthcare) with buffer A and 0.5 mM DDM. For ELIC, the purification process was identical, except that the chromatography was done with 1.2 mM n-undecyl-β-d-maltopyranoside (UDM; Anatrace) as the solubilizing detergent.
Membrane reconstitution and electrophysiology
Electrophysiological measurements were made by patch-clamp recordings in channel-reconstituted liposomes prepared as described previously (Delcour et al., 1989; Cortes et al., 2001; Chakrapani et al., 2007; Velisetty and Chakrapani, 2012). Purified protein was reconstituted into preformed asolectin vesicles by diluting in 150 mM NaCl and 10 mM HEPES, pH 7.0 (reconstitution buffer). Detergent was removed by incubating the proteoliposome suspension with Bio-Beads (Bio-Rad Laboratories). The suspension was centrifuged at 100,000 g for 1 h, and the pellet was resuspended in reconstitution buffer. A drop of the proteoliposome was placed on a glass slide and dried overnight in a desiccator at 4°C. The sample was then rehydrated with 20 µl buffer, which yielded giant liposomes. Channels were reconstituted in 1:10,000 protein/lipid (molar ratio) for macroscopic currents. Currents were measured using inside-out patch clamp of proteoliposomes in symmetrical NaCl, unless stated otherwise. All experiments were performed at room temperature. Recording pipettes were pulled from thin-walled borosilicate glass and heat polished to a resistance of 1.5–2 MΩ and filled with 150 mM NaCl and 10 mM HEPES, pH 8.0. Low pH was obtained using 10 mM sodium citrate buffer. Currents were elicited in response to concentration jumps using a fast solution exchanger (switch time, 2 ms) fed by gravity (RCS-200; Bio-Logic). Currents were measured using Axopatch 200B, digitized at a sampling frequency of 10 kHz, and analyzed using Clampfit 10.2 (Molecular Devices).
Site-directed spin labeling
The WT chimera is intrinsically Cys-less and therefore was directly used as a template for generation of single-Cys mutants. The two native cysteines in ELIC (Cys300 and Cys313 in the TMD) were mutated to serine residue. Mutant proteins were expressed and purified similar to the WT channels. Purified mutants were labeled with a methanethiosulfonate spin probe, (1-oxyl-2,2,5,5-tetramethylpyrrolidin-3-yl) methyl methanethiosulfonate (MTSSL; Toronto Research), at a 10:1 label/protein molar ratio and incubated on ice for 30 min, after which a fivefold molar excess of the MTSSL was added and further incubated for 2 h for better labeling efficiency (Velisetty et al., 2012). The labeled protein was then purified by size-exclusion chromatography on a Superdex 20/200 column (GE Healthcare) in buffer A supplemented with 0.5 mM DDM. Spin-labeled samples were reconstituted at a 1:3,000 protein/lipid molar ratio in asolectin, incubated with Bio-Beads (Bio-Rad Laboratories) to remove solubilizing detergent, and centrifuged to obtain a pellet of the proteoliposomes.
EPR spectroscopy and analysis
Continuous wave (CW)-EPR spectroscopy measurements were performed at room temperature on a spectrometer (EMX X-band; Bruker) equipped with a dielectric resonator and a gas-permeable TPX plastic capillary. First-derivative absorption spectra were recorded with an incident microwave (power of 2.0 mW, modulation frequency of 100 kHz, and modulation amplitude of 1.0 G). The EPR signal is normalized to the total number of spins in the sample by dividing the spectra by the peak-to-peak value of the double integral (which is proportional to the total number of spins). Our analyses were centered on two types of dynamic EPR structural information (Farahbakhsh et al., 1992; Altenbach et al., 2005). The first is the mobility of the spin probe, calculated as the inverse of the central line width of the first-derivative absorption spectra (ΔHo−1). Spin-probe mobility is governed both by the local steric contacts in the immediate vicinity of the probe and by the flexibility of the backbone to which it is attached (Mchaourab et al., 1996). The second is spin-probe solvent accessibility evaluated by collisional relaxation methods. Here, polar Ni(II) ethylenediamine diacetic acid (NiEDDA) serves to evaluate the extent of water exposure (Farahbakhsh et al., 1992; Gross and Hubbell, 2002). The accessibility parameter (Π) is estimated from power saturation experiments in which the vertical peak-to-peak amplitude of the central line of the first-derivative EPR spectra is measured as a function of increasing incident microwave power (Farahbakhsh et al., 1992). Conformational changes were measured by equilibrating the sample with appropriate buffers (pH and ligand) in a 42°C water bath. The samples were centrifuged, and the process was repeated three times to ensure complete buffer exchange. Reversibility of structural changes was ensured by switching back to pH 7.0.
Double electron–electron resonance (DEER) measurements
Intersubunit distances (<50 Å) were measured using DEER methods (Jeschke et al., 2002; Zou and Mchaourab, 2010) for spin-labeled samples (∼100 µM and 10 µl) in buffer A with 0.5 mM DDM and supplemented with 30% (wt/vol) glycerol (for cryoprotection). Dipolar time-evolution data were obtained at 83 K using a standard DEER four-pulse protocol, (π/2)mw1 − τ1 − (π)mw1 − τ1 − (π)mw2 − τ2 − (π)mw1 − τ2 − echo (Pannier et al., 2000), on a 580 pulsed EPR spectrometer (Bruker) operating at Q-band frequency (33.9 GHz). The pulse lengths for (π/2)mw1 and (π)mw1 were 10 and 20 ns, respectively, and 40 ns for (π)mw2. DEER signals were background-corrected assuming a 3-D homogeneous background and analyzed by the Tikhonov regularization in the DeerAnalysis software (Chiang et al., 2005; Jeschke et al., 2006) to determine average distances and distributions in distance. DEER data were collected in detergent samples because of the limitations associated with these measurements in liposomes arising from numerous factors, including lower sensitivity, higher background contribution through intermolecular interactions, and significant reduction in the accessible distance range (<50 Å). In general, for longer distances (>50 Å), dipolar evolution times can be insufficient, causing uncertainty in the widths of the distance distributions. The expected Cβ-Cβ distances for the studied positions are in the 30–70-Å range, which is beyond the detection limit for the liposome samples. Although lipids could potentially modify channel properties, for the measurements we have made, there is essentially no difference in spectral changes in detergents and liposomes (Fig. S7).
Simulations
For mutants studied by DEER experiments, inter-label distance distributions were simulated with ELIC structures (Protein Data Bank [PDB] accession no. 3RQU, apo ELIC, and PDB accession no. 4A98, benzodiazepine-bound ELIC) based on analysis of spin-label rotamers using the multiscale modeling of macromolecular systems (MMM) software package (Polyhach et al., 2007, 2011). Rotamer library calculations were conducted at 83 K.
Isothermal titration calorimetry
Binding affinity of ELIC and the WT ELIC-GLIC chimera for the agonist (3-amino propanol [3-AP]) was measured by isothermal titration experiments with MicroCal ITC200 (GE Healthcare). Measurements were made at constant temperature (25°C). The sample cell was filled with purified protein at a concentration of 25–50 µM in the gel-filtration buffer (100 mM NaCl, 20 mM Tris buffer, and 1.2 mM UDM [ELIC] or 0.5 mM DDM [ELIC-GLIC]). The injection syringe was filled with 8–10 mM amino propanol solution in the gel-filtration buffer. 20 successive injections of 2 µl each were titrated into the cell with constant stirring at 500 rpm. An equilibration time of 180 s was set between consecutive injections. The data were fit to a single site–binding isotherm using Origin software (OriginLab Corporation).
Crystallization
The WT ELIC-GLIC chimera in 100 mM NaCl, 10 mM Tris, pH 7.4, and 0.5 mM DDM was concentrated to between 9 and 10 mg/ml with a cutoff concentrator (50 kD; Amicon Ultra; EMD Millipore). Before crystallization setup, the protein was supplemented with 0.5 mg/ml E. coli polar extract (Avanti Polar Lipids, Inc.) and incubated on ice for 1 h. The protein was crystallized at 4°C by sitting drop vapor diffusion in Cryschem plates (Hampton Research) with a 1:1 mixture (1 µl each) of protein and reservoir solution (200 mM ammonium sulfate, 50 mM sodium ADA, pH 6.7–7.6, and 7.5–10% PEG4000). Crystals initially formed after 1 wk and typically took 2–3 wk to reach full size. The crystals were cryoprotected by adding 6 µl reservoir solution supplemented with 30% ethylene glycol to the drop, and directly frozen in liquid nitrogen using appropriately sized microloops (Mitegen) or cryoloops (Hampton Research).
Structure determination
X-ray diffraction data were acquired on NE/CAT beamlines 24ID-C and 24ID-E at the Advanced Photon Source at Argonne National Laboratory. The data were indexed using XDS (Kabsch, 2010) and further processed using programs within the CCP4 suite (Collaborative Computational Project, Number 4, 1994). Because of anisotropy and poor data quality, it was necessary to merge six datasets to obtain an optimally complete dataset. The crystals belong to space group C2221 with one pentamer in the asymmetric unit. Initial phases were obtained by molecular replacement using PHASER (McCoy, 2007) with the ELIC-ECD (PDB accession no. 2VL0; Hilf and Dutzler, 2008) and the GLIC-TMD (PDB accession no. 4HFI; Sauguet et al., 2013) crystal structures as search models. During later rounds of refinement, we substituted the ECD of PDB accession no. 2YN6 (Zimmermann et al., 2012) because of better geometry. The GLIC structure at pH 4.0 was chosen as the search model in comparison to the structure at pH 7.0 because of its higher resolution (2.4 vs. 4.35 Å).To minimize model bias within the pore region of the channel, the first round of refinement omitted the M2 helix, which was then rebuilt manually into the m|Fo|-D|Fc| omit density. The initial model was refined with REFMAC5 (Murshudov et al., 1997) and PHENIX (Adams et al., 2010), and BUSTER (Blanc et al., 2004) was used for the final rounds with fivefold NCS restraints. Refinement was followed by model building/fitting in COOT (Emsley et al., 2010). On each successive cycle of model building/refinement, we analyzed the structure with MolProbity software (Chen et al., 2010). The average B factor for the ECD and TMD in previous ELIC structures exhibits significantly higher B values for the ECD than the TMD; because of the modest data resolution, the B factors were modeled with a combination of fixed B factors (100 Å2 for the TMD and 150 Å2 for the ECD) and a single overall TLS term for the TMD and ECD of each model (10 total TLS groups). MOLE software (Petřek et al., 2007) was used to compute pore radius profiles, and PISA software (Krissinel and Henrick, 2007) was used to determine solvent-occluded surface area at the ECD–TMD interface.
Accession numbers
The coordinates and structure factors of the ELIC-GLIC chimera have been deposited in the PDB under accession number 4YEU.
Online supplemental material
Table S1 shows data refinement and statistics for the ELIC-GLIC chimera crystal structure. Fig. S1 shows changes in loop C dynamics and accessibility upon binding agonist and competitive antagonist. Fig. S2 shows current traces from patch-clamp measurements (in oocytes and in liposomes) of ELIC, GLIC, and the ELIC-GLIC chimera. Fig. S3 shows gel-filtration profiles of the ELIC-GLIC chimera. Fig. S4 displays the electron density map in key regions of the ELIC-GLIC chimera. Fig. S5 highlights the observed quaternary twist in the ELIC-GLIC chimera ECD. Fig. S6 demonstrates that the agonist sensitivity of the ELIC-GLIC chimera is similar to that of ELIC. Fig. S7 shows that the ligand-induced spectral changes at Ser189 measured in detergent and in liposomes are identical. Fig. S8 shows spectral changes in the ELIC-GLIC chimera as a function of ligand concentration. Fig. S9 shows rotamer simulation for Ser189 using MMM. Fig. S10 shows rotamer simulation for loop F mutations. Fig. S11 is a plot of ΔHo−1 at various positions in the apo, 3-AP, and pH 3.0 conditions. Fig. S12 shows changes in the ECD–TMD interface dynamics in the ELIC-GLIC chimera in comparison to ELIC and GLIC. Fig. S13 demonstrates that the I9′A substitution rescues the function of the H11′F mutation both in GLIC and in the ELIC-GLIC chimera. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201511478/DC1.
RESULTS
Agonist-evoked conformational changes in ELIC measured by EPR spectroscopy
Considerable insights into the conformational changes during activation in GLIC come from crystal structures at neutral and acidic pH as well as from EPR spectroscopic measurements (Bocquet et al., 2009; Velisetty et al., 2012, 2014; Dellisanti et al., 2013; Sauguet et al., 2013, 2014). In contrast, we have little understanding of gating conformational changes in ELIC because agonist-bound ELIC structures are identical to those of the apo structures. However, ACh (a competitive antagonist) has previously been shown to inhibit ELIC currents, and the ACh-bound ELIC structure reveals loop C in a “closed” conformation with a 6-Å inward movement that caps the ACh-binding site (Pan et al., 2012; Fig. 1 A). Such inward loop C motions have been associated with agonist-mediated changes in pLGICs and the ACh-binding protein, although in comparison to other pLGICs, ELIC loop C is longer and less structured. In ELICs, ACh could prevent channel opening either by eliciting changes in the binding site opposite to those mediated by the agonist or by promoting agonist-like ECD motions that fail to successfully open the channel. To distinguish between the two possibilities and measure ligand-induced structural changes, single-Cys mutations in the loop C region (Fig. 1 A) were labeled with MTSSL and probe dynamics were monitored by CW-EPR spectroscopy at room temperature.
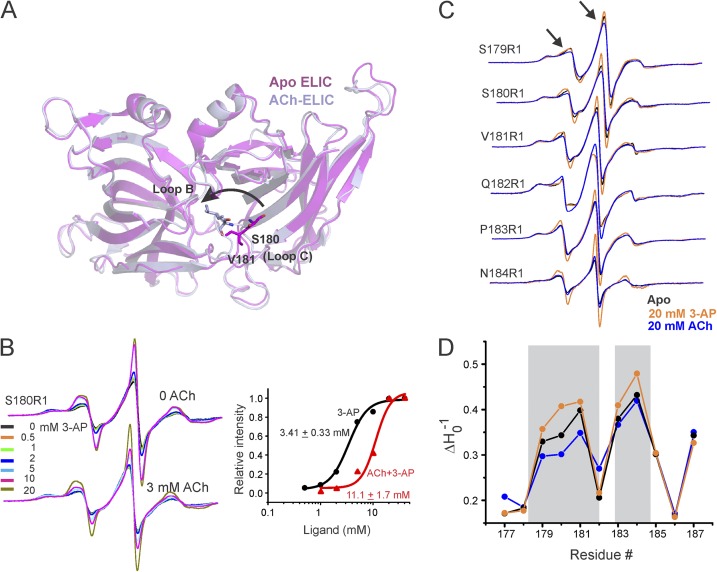
Figure 1.
Agonist- and antagonist-evoked conformational changes in the ELIC-ECD. (A) Superimposition of apo-ELIC (PDB accession no. 3RQU) and ACh-ELIC (PDB accession no. 3RQW) structures shows an inward (counterclockwise) motion of loop C. (B) Concentration-dependent changes in the EPR line shape and amplitude measured at position Ser180 in the presence and absence of 3 mM ACh. In each case, the spectra are normalized to the total number of spin (see Materials and methods). A plot of peak amplitude (of the central line) as a functional 3-AP concentration (right). (C) Spin-normalized CW-EPR spectra of representative loop C residues in apo state and in the presence of ACh or 3-AP. (D) Changes in probe mobility of loop C positions in the three conditions. The gray boxes highlight regions displaying prominent changes.
We began by measuring ligand-induced spectral changes at the tip of loop C (position Ser180), where the most prominent change in conformation was observed between the apo-ELIC and ACh-ELIC structures (Fig. 1 A, arrow). As a function of increasing agonist concentration (3-AP), the spectra for S180R1 shows an increase in amplitude accompanied by a corresponding decrease in spectral broadening (Fig. 1 B), which reflects increased dynamics at this position upon 3-AP binding. However, in the presence of 3 mM ACh, spectral changes induced by 3-AP are inhibited, and the dose–response curve shows a rightward shift in comparison to that measured in the absence of ACh (Fig. 1 B). These results demonstrate that agonist and competitive antagonist evoke distinct conformational changes at Ser180. Curiously, agonist binding mobilizes this position and thereby alludes to a potentially distinct agonist-induced motion.
To further understand the agonist- and antagonist-induced changes in loop C dynamics, we investigated structural changes along the length of loop C in ELIC. Measurements were made in the unliganded (apo) state and under steady-state liganded (20 mM 3-AP) conditions, where the channel would be expected to be in the resting and desensitized conformations, respectively. Because of the transient nature of the open conformation, it cannot be probed under steady-state EPR conditions. In addition, we also measured the effect of 20 mM ACh at each of these positions. In the presence of 3-AP, several positions within loop C (179, 180, 181, 183, and 184) showed an increase in probe mobility accompanied by an increase in signal amplitude (Figs. 1, C and D, and S1 A). In addition, the accessibility of these positions to water-soluble NiEDDA also increases upon binding 3-AP (Fig. S1 B), indicating an overall increase in local dynamics and solvent exposure upon activation. This movement is in contrast to the restricted dynamics and immobilization of loop C in GLIC when activated by protons (Velisetty and Chakrapani, 2012). Interestingly, most of the 3-AP–mediated changes are reversed when the binding site is occupied instead by ACh, as noted by a decrease in mobility at positions 179, 180, and 181, and minimal changes at 183, 184, and 187 (Figs. 1 B and S1 A). These findings demonstrate that the conformational flexibility of loop C in ELIC is increased when bound to agonist and decreased when bound to antagonist. A more restricted loop C in the ACh-bound state is consistent with the closed conformation of this region in the ELIC-ACh crystal (Pan et al., 2012). A decrease in the average B factor of loop C was also noted for this structure revealing reduced flexibility in the ACh-bound form. Furthermore, NiEDDA measurements in the ACh-bound state are in agreement with the solvent-accessible surface area calculated for loop C residues in the ACh-bound ELIC (PDB accession no. 3RQW) structure (Fig. S1 B, bottom).
Collectively, these findings strongly suggest that unlike in other members of the pLGIC family, increases in loop C dynamics in ELIC are associated with channel activation. Although ELIC structures crystallized with agonists do not reveal major conformational changes, an outward extension of loop C (∼7 Å away from the tip of loop B) was reported for ELIC bound to bromo-flurazepam, a positive modulator of GABAAR (Spurny et al., 2012).
Engineering a functional ELIC-GLIC chimera
Besides contrasting loop C motions reported here, there are several aspects of ELIC gating that defy the general trend observed in pLGICs (Gonzalez-Gutierrez et al., 2012; Pan et al., 2012; Spurny et al., 2012; Gonzalez-Gutierrez and Grosman, 2015; Kinde et al., 2015). Particularly, agonists and antagonists induce relatively smaller conformational changes in the ECD–TMD interface and the TMD as reported by x-ray crystallography condition and nuclear magnetic resonance/EPR spectroscopy. To better understand the mechanism of conformational coupling between the ECD and TMD in ligand recognition and channel activation, as well as to examine the compatibility of such different channels, we engineered a chimeric channel between ELIC-ECD and GLIC-TMD (Fig. 2 A). ELIC and GLIC sequences show an overall 22% sequence identity with considerable variability at the domain interfaces. Despite limited overall sequence identity across the family, chimeras constructed by swapping the ECD and the TMD between different pLGIC family members have been shown to be functional (Eiselé et al., 1993; Cooper et al., 1999; Grutter et al., 2005; Duret et al., 2011; Tillman et al., 2014; Moraga-Cid et al., 2015).
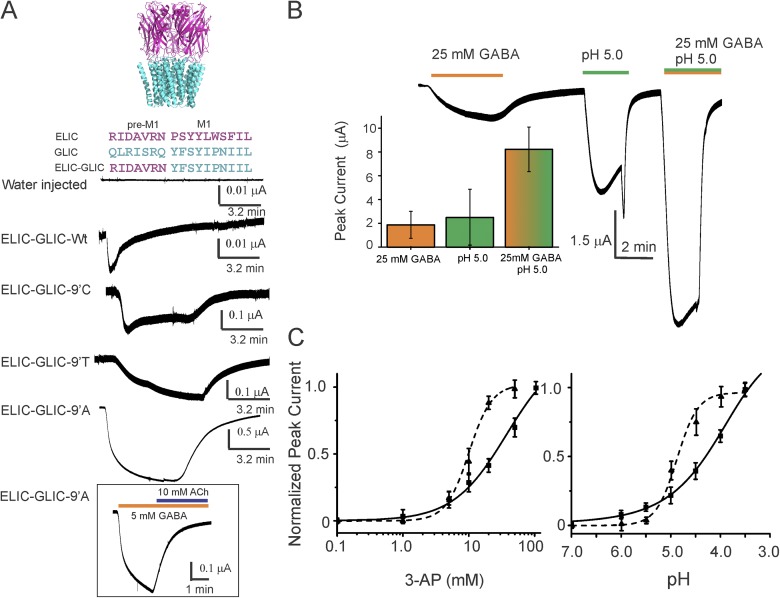
Figure 2.
Functional characterization of the ELIC-GLIC chimera. (A) A schematic representation of the chimera based on GLIC (PDB accession no. 4HFI) and ELIC (PDB accession no. 3RQU) structures, and the sequence alignment highlighting the region where the ELIC-ECD was fused to the GLIC-TMD (top). Agonist-induced (25 mM 3-AP) 13-min current response in oocytes measured by two-electrode voltage clamp (bottom). Inset shows (B) current traces for the chimera 9′Ala mutant in response to activation by acid (pH 5.0), agonist (25 mM GABA), and a combination of both. The plot shows peak response for the three conditions. The error bars denote standard deviation (n = 3). (C) Dose–response curves for the 9′Ala mutant (solid line) in the presence of 3-AP (dashed line, compared with ELIC) or acid (dashed line, comparison with GLIC). Currents are expressed as a fraction of the maximal response. The error bars denote standard deviation (n > 5). The EC50 (3-AP) and pH50 values are 32.8 ± 9.1 mM and 4.22 ± 0.11, and the corresponding Hill coefficients are 1.1 ± 0.2 and 9.1 ± 2.1, respectively. The EC50 (3-AP) and Hill coefficient for WT ELIC are 10.02 ± 0.05 mM and 2.5 ± 0.3. The EC50 (pH) and Hill coefficient for WT GLIC are 4.89 ± 0.06 mM and 2.0 ± 0.1. The error bars for WT denote standard deviation (n > 3).
To determine the functionality of the ELIC-GLIC chimera, the mRNA encoding the chimera was injected into oocytes, and the functional properties were assessed by two-electrode voltage-clamp measurements. We observed small, rapidly desensitizing currents in response to activation by 25 mM 3-AP (Fig. 2 A, bottom). A faster macroscopic current decay was also seen in excised outside-out oocyte patches (Fig. S2). A small current response in the ELIC-GLIC chimera may also arise from poor membrane expression and/or weaker ligand sensitivity. Similar to the observed effects in GLIC (Bocquet et al., 2007; Gonzalez-Gutierrez and Grosman, 2010), polar and small-chain amino acids (Cys, Thr, and Ala) at the 9′ position in M2 (Ile240) dramatically increased agonist-evoked responses with slowed macroscopic desensitization, with the Ala mutation causing the most prominent effect (Fig. 2 A, bottom). Furthermore, as in ELIC, agonist-induced currents were inhibited in the presence of ACh (Fig. 2 A, inset). However, to our surprise, the ELIC-GLIC chimera was activated not only by primary amines but also by extracellular protons (Fig. 2 B). Although the precise location of the pH sensor in GLIC is unknown, this result was unexpected, as the GLIC-ECD conferred pH sensitivity to the GLIC-GlyR chimera (Duret et al., 2011). The ELIC-GLIC chimera at I9′A activates with an estimated EC50 of 32.8 mM for 3-AP and pH50 of 4.22 (Fig. 2 C). In the oocyte measurements, 3-AP concentrations >100 mM or pH conditions <3.5 led to unreliable currents because of issues with osmolality differences. The individual agonist sensitivity of the chimera to acid or amine is lower than our measured responses for GLIC (pH50 of 4.9 ± 0.1) and ELIC (EC50 of 10.0 ± 0.9 mM), respectively.
Even though the sequence similarity of the ECD–TMD interface between ELIC and GLIC is low, the chimera was functional with no perturbation to the interface and preserves activation by both protons and primary amines. To better understand the molecular basis for multi-ligand gating, structural and dynamic studies were undertaken on the WT ELIC-GLIC chimera.
Crystal structure of the ELIC-GLIC chimera in the resting conformation
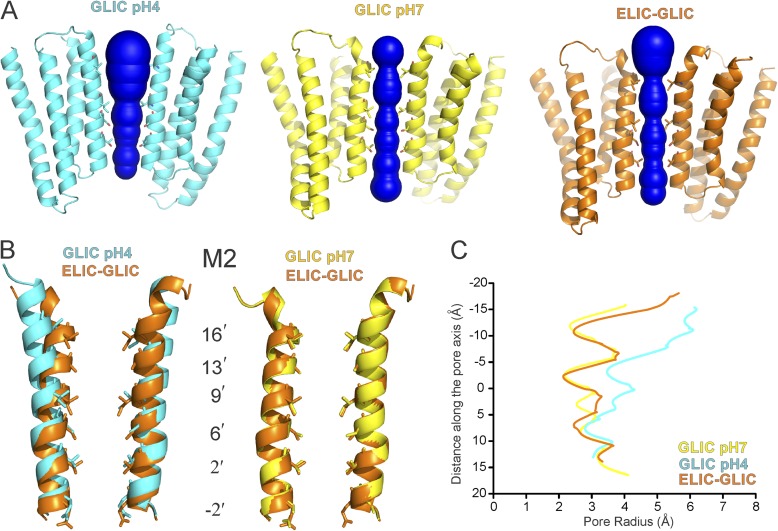
The gene encoding the WT ELIC-GLIC chimera was expressed in E. coli and purified as a stable monodisperse pentameric population (Fig. S3). In agreement with oocyte measurements, the purified chimera when reconstituted into liposomes showed current response to both primary amines and protons (Fig. S2). We then crystallized the chimera at neutral pH and in the absence of 3-AP, conditions that are expected to stabilize the resting state of the channel. The crystals belong to space group C2221, and the structure (Table S1) was determined at 4.6-Å resolution by molecular replacement using the ELIC-ECD (apo state) and the GLIC-TMD (at pH 4.0, open conformation) crystal structures as search models (Hilf and Dutzler, 2008; Sauguet et al., 2013). Although this resolution does not allow us to unambiguously assign individual side-chain orientations or rearrangements of the flexible loop regions, large positional changes at the backbone level in the TMD or global reorientation of the ECD and TMD with respect to the TMD are readily observable (Fig. S4). The electron density quality was better for the TMD in comparison to the ECD, and the positions of the transmembrane helices were clearly observable. Extensive effort was made to ensure that model bias did not affect the structure of the pore (Fig. S4; see Materials and methods). Compared with the GLIC-TMD at pH 4.0 (Figs. 3 A, left, and 4, A and B), the M2 helices in the extracellular end were detached from M3 and bent toward the pore axis forming a tight occlusion, a conformation reminiscent of the closed GLIC structure at pH 7.0 (Sauguet et al., 2014). The closed and open GLIC structures show that the TMD interacts with the ECD primarily through the M2–M3 linker and β6–β7 and β1–β2 loops. Interestingly, the overall positions of the transmembrane helices and the M2–M3 loop were identical to that of the GLIC-TMD at pH 7.0 (a Cα root mean square deviation [RMSD] of 0.35 Å, in comparison to GLIC-TMD at pH 4.0 with a RMSD of 0.69 Å; RMSD calculated for residues 200–315 in chains A–E; Fig. 3, A, right, and B). An analysis of the pore along M2 shows a near-identical diameter along the entire length as that of the closed GLIC (Fig. 4 C).
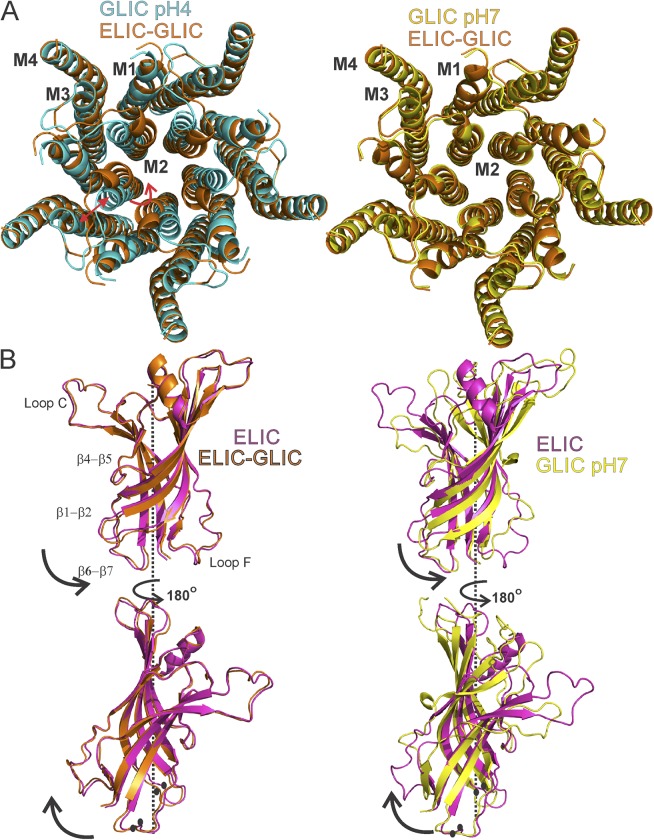
Figure 3.
Structural differences in the ELIC-GLIC chimera compared with ELIC and GLIC. (A) Top view of TMDs comparing the ELIC-GLIC chimera with GLIC structures in the closed (pH 7.0; PDB accession no. 4NPQ) and open (pH 4.0; PDB accession no. 4HFI) forms. Red arrows highlight positional differences. (B) Superimposition of the ELIC (PDB accession no. 2YN6), GLIC (pH 7.0; PDB accession no. 4NPQ), and ELIC-GLIC structures at the level of the ECD. The ECD pentamer comprising of residues 11–200 was superimposed at the Cα level. Black arrows show the direction of twist from ELIC to ELIC-GLIC (left) and from ELIC to GLIC (right). Key regions showing difference in position are marked by black circles.
Figure 4.
Pore profile of the ELIC-GLIC chimera and GLIC structures. (A) Water-accessible region of the pore as determined by the MOLE PyMOL plugin (Petřek et al., 2007). Two subunits are shown, with residues lining the ion permeation pathway represented as sticks. (B) A close-up view of diagonal M2 helices. (C) Pore radius along the channel axis in the ELIC-GLIC chimera and in the GLIC-closed (PDB accession no. 4NPQ) and -open (PDB accession no. 4HFI) forms calculated using MOLE software (Petřek et al., 2007).
Superposition with apo-ELIC-ECD (RMSD, residues 11–199, 0.73 Å) reveals that the chimera-ECD has undergone a rotation about the central symmetry axis (Fig. 3 B, left). The extent of twisting is more pronounced at the ECD–TMD interface, where the inner β strands are rotated counterclockwise in a concerted manner (Fig. S5). An alignment of apo-ELIC and GLIC–pH 7.0 structures shows that the GLIC-ECD is twisted in a similar manner and along the same direction as the ELIC-GLIC chimera (Fig. 3 B, right). Between apo-ELIC and the ELIC-GLIC chimera, the twist leads to a Cα-Cα displacement of 1.5 Å at loop 2 (Asn27) and of 2.1 Å at loop 7 (Phe121); the corresponding difference between the apo-ELIC and GLIC–pH 7.0 are 3.3 and 2.6 Å, respectively (Fig. 3 B, black circles). There is a varying extent of relative β-strand twists among pLGIC structures either related to their phylogenetic origin or to the crystallographically captured conformational states (Hassaine et al., 2014; Miller and Aricescu, 2014; Sauguet et al., 2014). Furthermore, we observe during refinement consistently higher B factors in the ECD than in the TMD, suggesting an increase in the structural flexibility of this domain in the ELIC-GLIC chimera (see Materials and methods). Although the reported ELIC structures have fairly low B factors and therefore a rigid ECD, the GLIC ECD at pH 7.0 has a considerably higher B factor. It is likely that the closed GLIC pore may contribute both to the twisting of ECD and the increase in its flexibility. Curiously enough, despite limited conservation of the ECD–TMD interface between ELIC and GLIC, the chimeric channel appears to show tight coupling of the two domains, with a solvent-inaccessible area of 746 Å2, which is in fact larger than ELIC (501 Å2) but comparable to GLIC (756 Å2) structure.
Collectively, our structure reveals a channel with a twist in the ELIC-ECD and a TMD that has a remarkably well-conserved structure compared with the GLIC-TMD at pH 7.0. A complete understanding of gating in the ELIC-GLIC chimera will require structures in the presence of 3-AP and under acidic conditions. However, obtaining well-diffracting crystals under these conditions has been challenging, and we therefore used site-directed spin labeling and EPR spectroscopy to determine the extent of conformational changes upon ligand activation.
Agonist-evoked conformational changes measured by EPR spectroscopy
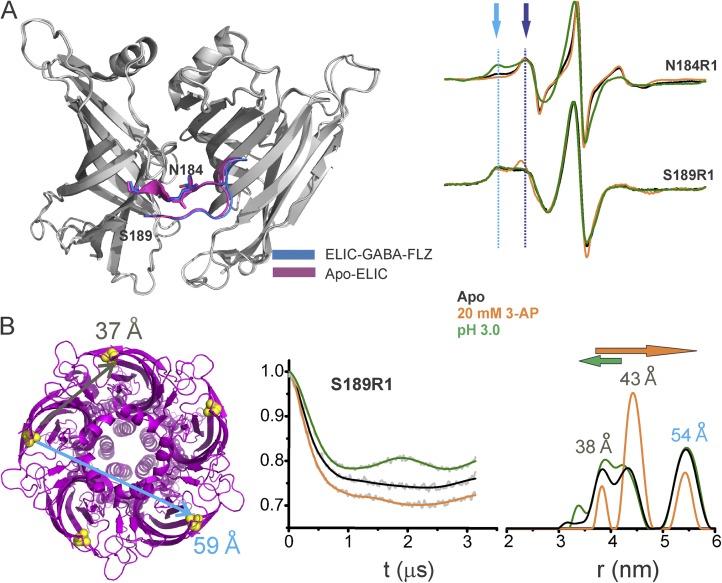
We first examined the ECD in the WT ELIC-GLIC chimera to see if it behaves similarly to WT ELIC. The equilibrium agonist binding measured by isothermal titration calorimetry showed that there are no significant changes in the binding constant for the two channels (Fig. S6). Furthermore, at position Asn184 in loop C, 3-AP evoked a remarkably similar spectral change by increasing both probe mobility and signal amplitude (Figs. S6 and 5 A). In fact, at 20 mM 3-AP, the N184R1 spectra for ELIC and the ELIC-GLIC chimera were identical (Fig. S6). Interestingly, acidic pH has the opposite effect, leading to extensive broadening of the spectra, suggesting an immobilization of loop C (Fig. 5 A). A closer inspection of the line shape shows two spectral components: an immobile population at low field and a mobile population at high field strength (Fig. 5 A, indicated by arrows). Although the amplitude of the immobile component decreases in the presence of 3-AP, its amplitude is increased under acidic conditions.
Figure 5.
Loop C movements in the ELIC-GLIC chimera. (A) An overlay of ELIC-ECD in the apo (PDB accession no. 3RQU)- and GABA-flurazepam (PDB accession no. 4A96)–bound forms, with residues Asn184 and Ser189 highlighted (left). CW-EPR line shapes for the loop C positions in the apo conformation, and in the presence of 20 mM 3-AP, or at pH 3.0. The light and dark blue arrows highlight the immobile and mobile components of the spectra, respectively. (B) ELIC structure showing the location of Ser189 and corresponding cβ-cβ distances for the adjacent and nonadjacent subunits (left). Background-subtracted DEER echo intensity is plotted against evolution time and fit using model-free Tikhonov regularization. The corresponding interspin distance distribution (right). The arrows highlight the direction of change (orange, 3-AP; green, pH 3.0).
To further quantify these differential motions, we measured distance changes with DEER experiments. Because there are five spin labels within the pentamer, at least two distances are expected: one corresponding to the adjacent subunits and the other from nonadjacent subunits. Because the Cβ-Cβ diagonal distances of loop C residues are beyond the DEER detection range (<60 Å), the closest neighboring position in the β10 strand (S189; Fig. 5 A) was chosen for investigation. Although movements at S189 are smaller relative to loop C, the overall direction of line-shape changes for the 3-AP and pH 3.0 conditions is similar to that of loop C residues (Fig. 5 A). The DEER signal decays and the corresponding probability distributions for S189R1 are shown in Fig. 5 B. In the apo state, there are three distinct peaks of which the first two short-distance peaks (between 35–45 Å) represent distances between adjacent subunits, and the third peak corresponds to the nonadjacent distance. The two components arise from either alternate rotameric spin orientations or from two distinct loop C conformations in equilibrium. Although one would expect two populations for the nonadjacent distance as well, reliable measurements beyond 60 Å are infeasible under our experimental conditions as a result of technical difficulties with background correction. In remarkable agreement with the CW line-shape changes, 3-AP increases the relative population of the longer distance peak (second peak), whereas protons shift the equilibrium toward short distance (first peak).
To further gain insights into the origin of multiple components in the DEER distance distribution, we estimated distance distributions from crystal structures (using spin-label rotamer library within the MMM software package; Polyhach et al., 2007, 2011) and compared them with experimental distance distributions (see Materials and methods). For the apo-ELIC structure (PDB accession no. 3RQU), the predicted distances fell within the DEER distribution and show the presence of two rotameric populations for the spin label. Because the agonist brought about rather small loop C changes in crystal structures, we used the benzodiazepine-bound ELIC structure (PDB accession no. 4A98) for simulations. Loop C in this structure adopts an extended outward conformation, and consistently the distance distribution for S189R1 is predicted to switch toward a longer distance (Fig. S9). Although changes in the β10 strand appear to be subtle, they seem to faithfully follow large movements at the tip of loop C. In addition, ligand-induced changes in the local environment around the probe may also contribute to the rotameric changes at S189R1. Collectively, findings from CW and DEER measurements demonstrate that 3-AP binding favors an extended loop C conformation in the ELIC-GLIC chimera, whereas protons mediate a more contracted orientation of this region, although both ligands activate the channel.
The dual activation by different ligands makes the chimera an ideal framework to study how diverse conformational changes in loop C could couple to channel activation. Studies in the AChBP and several members of the family including GLIC have shown that loop F, similar to loop C, displays significant conformational flexibility and large ligand–induced conformational changes (Celie et al., 2004; Bourne et al., 2005; Hansen et al., 2005; Dellisanti et al., 2013; Sauguet et al., 2014; Velisetty et al., 2014). However, the role of these motions in channel gating has been questioned by several groups (Khatri et al., 2009). In GLIC, loop F was shown to be both less dynamic and solvent inaccessible under acidic pH, and DEER measurements showed a large inward motion (Dellisanti et al., 2013; Velisetty et al., 2014). Consistently, the GLIC structure at pH 7.0 shows a more flexible and disordered loop F oriented outward compared with that of the pH 4.0 structure (Sauguet et al., 2014). Interestingly, the AChR-AChBP chimera shows loop F interacting with loop C and hence suggesting that their motions could be coupled (Li et al., 2011), and the differing motions in loop C in our chimera allow us to test whether this is the case.
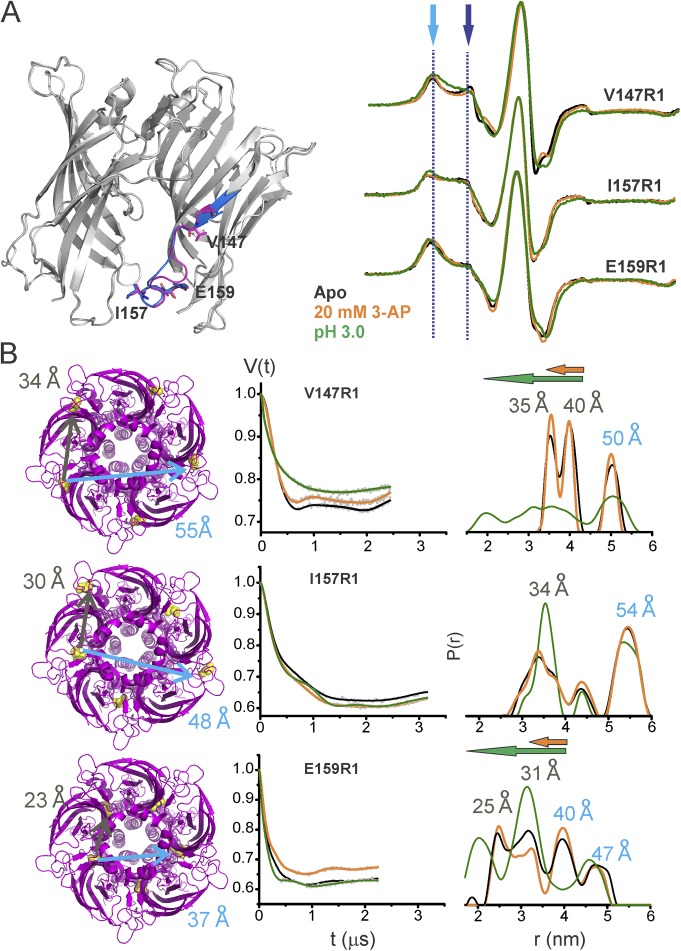
Conformational differences in loop F in the apo state and in the presence of 3-AP or protons are shown in Fig. 6 A. As with loop C, in the apo state, two spectral components were seen for positions Val147 and Glu159 in the CW spectra and were more evident in distance distributions from DEER (Fig. 6 B). Notice that for Glu159, which had the shortest Cβ-Cβ distance among the measured mutants, both components of the adjacent and nonadjacent distances can be clearly seen. Rotamer simulations for spin label at these positions in the apo-ELIC structure (PDB accession no. 3RQU) were performed to gain additional insight into loop F dynamics. They show multicomponent distributions and are largely consistent with the dynamic nature of loop F (Fig. S10). There are, however, differences between the predicted width of the distributions and the DEER measurements. At the two ends of loop F (Val147 and Glu159), the measured distances are longer than the calculated distribution, whereas at Ile157, the DEER distances are relatively shorter. Note that the additional peaks in DEER distribution may arise from multiple orientations sampled by loop F.
Figure 6.
3-AP– and pH-induced conformational changes in loop F of the ELIC-GLIC chimera. (A) Locations of examined residues in ELIC-GLIC (left) and spin-normalized CW-EPR spectra of the residues in the apo state, with 20 mM 3-AP, and at pH 3.0 (right). (B) The corresponding distances mapped on the ELIC structure (PDB accession no. 3RQU; left). The DEER echo and the distance distribution under the three conditions (right). The peak values for distance distribution are marked for the apo state. The arrows highlight the direction of change (orange, 3-AP; green, pH 3.0).
CW-EPR under acidic conditions at three loop F positions showed spectral broadening, whereas the effect of 3-AP was modest (Figs. 6 and S11). Further increases in 3-AP concentration did not lead to additional changes (Fig. S8). DEER data for V147R1 and E159R1 showed that both 3-AP and protons led to a shift toward shorter distances, although the proton-induced effect at these positions was much more pronounced than that of 3-AP. The effect of either agonist for Ile157 was marginal both in CW as well as in DEER, suggesting that this position undergoes minimal change from the resting position during activation. In contrast to the effect on loop C, both 3-AP and acid had the same direction of change, revealing an inward displacement of loop F toward the central pore axis during activation, with protons exerting a larger degree of displacement. An immobilization and inward motion of loop F is similar to the proposed motion during GLIC activation (Dellisanti et al., 2013; Sauguet et al., 2014; Velisetty et al., 2014).
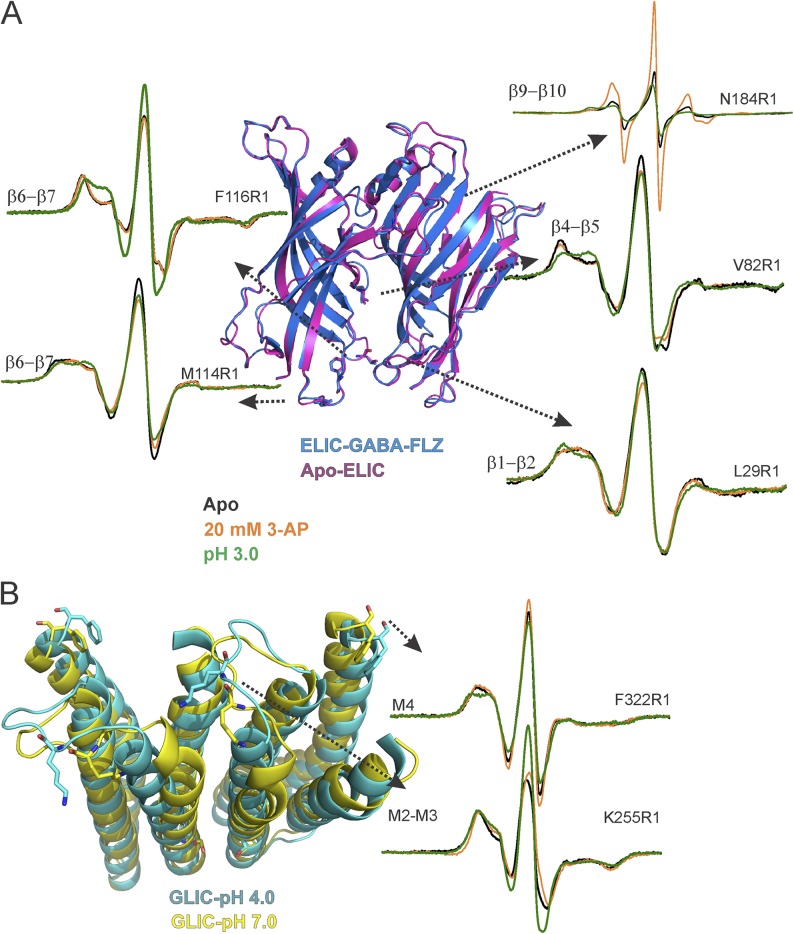
To further understand the similarities and differences in gating by the two different ligands in the chimera, we then measured line-shape changes at representative positions in loops β1–β2, β4–β5, and β6–β7 in the ECD (Fig. 7 A) and in the M2–M3 linker and the C-terminal end of M4 in the TMD (Fig. 7 B). These positions were chosen based on extensive conformational changes observed in the corresponding positions in GLIC (Velisetty et al., 2014). The spectra at Val82 in the β4–β5 linker shows a small increase in mobility with 3-AP and a much larger increase in mobility at pH 3.0. For L29R1 in the β1–β2 loop, 3-AP causes a marginal decrease in mobility, whereas at low pH, this effect is much more pronounced. Similarly, for the two positions in the β6–β7 loop, Met114 and Phe116, protons elicited stronger changes in line shape, a decrease in mobility at M114, and an increase at Phe116. On the other hand, 3-AP caused a small decrease at Met114, and at Phe116 there was essentially no change.
Figure 7.
Structural rearrangements at the domain interface in the ELIC-GLIC chimera. (A) Overlays of Apo-ELIC (PDB accession no. 3RQU) and ELIC-GABA-flurazepam (PDB accession no. 4A96) (left) and spin-normalized CW spectra of interface domain residues L29R1 (β1–β2 loop), V82R1 (β4–β5 loop), M114R1, and F116R1 (β6–β7 loop) in the apo state, in the presence of 20 mM 3-AP, or at pH 3.0 (right). (B) Overlay of GLIC structures in the closed (PDB accession no. 4NPQ) and open conformations (PDB accession no. 4HFI; left). CW-EPR spectra of K255R1 (M2–M3 linker) and F322R1 (M4) in the apo state, in the presence of 20 mM 3-AP, and at pH 3.0 (right).
Similar to the agonist-induced effects on the ECD, residues in the TMD underwent larger conformational changes in the presence of protons compared with 3-AP. At the M2–M3 loop, K255R1 (corresponding to K248 in GLIC) showed strong spectral broadening, likely from dipolar coupling caused by the close proximity of spin probes (Cβ-Cβ distance of ∼17 Å; Velisetty et al., 2014). Low pH caused an increase in amplitude and a decrease in spectral broadening in the chimera, consistent with an outward motion of this region as reported in GLIC (Sauguet et al., 2014; Velisetty et al., 2014). The effect of 3-AP was weaker in comparison. At the C-terminal end of M4, F322R1 (corresponding to F315 in GLIC) shows a partially mobile spectra. At pH 3.0, the F322R1 spectra is more immobile, and in the presence of 3-AP, it shows minimal change.
We further evaluated spectral differences at the ECD–TMD interface in the chimera and those at the equivalent positions in ELIC and GLIC. In contrast to loop C, where the apo and liganded spectra of the chimera are identical to ELIC, positions at the ECD–TMD interface show small differences between the chimera and the parent channels (Fig. S12). Although differences in the local environment of the probe in the two channels may contribute to variations in the spectral lines shape, these results are consistent with a twist in the chimera ECD with respect to the TMD. Particularly, apo spectra of both L29R1 (β1-β2) and F116R1 (β6-β7) are more immobile in the ELIC-GLIC chimera than in ELIC (Fig. S12 A). If the chimera ECD were to twist in the direction of the pH 7.0–GLIC structure (Fig. 3 B), one would expect these residues to be in a similar environment as that in closed GLIC. Indeed, the spectra at corresponding positions in GLIC are more immobile and overlay more closely with those from the chimera (Fig. S12 A, inset). In both ELIC and the chimera, 3-AP–induced conformational changes in the β1-β2 and β6-β7 loops are small, indicating that these regions undergo minimal change between the closed and desensitized states.
From the TMD side of the interface, the strong spectral broadening at the N-terminal end of the M2–M3 linker is similar to GLIC, and also consistent with the closed GLIC structure (Fig. S12 B). However, for F322R1 in the unliganded state, this position in GLIC is much more constrained than the chimera (Velisetty et al., 2014). Interestingly, the C-terminal end of M4 in GLIC is closely associated with the ECD–TMD interface, whereas this region in ELIC is separated from the interface (Hilf and Dutzler, 2008). Consistently, in the chimera, a small outward displacement of M4 can be observed (Fig. S12). Although the functional significance of this outward M4 orientation has yet to be characterized, in ELIC this state is implicated to be an “uncoupled” or inactive conformation (daCosta and Baenziger, 2013). In response to acid, the M2–M3 linker moves outward in both the chimera and GLIC with minimal differences, and the tip of M4 also adopts an identical extended conformation in both the two channels (Fig. S12 B).
We therefore conclude that, despite the fact that both 3-AP and protons gate the ELIC-GLIC chimera, the extent of mobility changes upon gating depend on the type of ligand, and tend to mirror the parent channel; in response to acid, the domain interfaces undergo large conformational rearrangement, similar to what has been reported for GLIC, whereas in response to 3-AP, modest changes are observed, analogous to effects observed in ELIC.
DISCUSSION
The conservation of the molecular architecture of the pLGIC is now well established. Chimeric channels offer a great deal of insight into the modular features of the individual component domains and long-range allosteric communication across these domains. Several functional chimeric constructs within the pLGIC have been reported: GLIC-α1GlyR (Duret et al., 2011; Moraga-Cid et al., 2015), ELIC-α7-nAChR (Tillman et al., 2014), α7nAChR-α1GlyR (Grutter et al., 2005), α4β2-nAChR-5HT3R (Cooper et al., 1999), and α7-nAChR-5HT3R (Eiselé et al., 1993), where the ECD of the former channel was fused to the TMD of the latter. In addition, fusion of the 5HT3R-TMD with the AChBP yields a functional channel (Bouzat et al., 2004). In each case, the chimeric channels exhibited the ligand dependence of the ECD and the selectivity, conductance, and pharmacological sensitivities characteristic of the TMD. It is noteworthy that the efficiency of ECD–TMD coupling in many cases was optimized by extensively modifying the interface to mimic either of the parent channels. Here, our engineered chimeric channel was functional with an unmodified interface, although the macroscopic currents desensitized more rapidly than either of the parent channels. It is interesting that the ECD–TMD interface region, in addition to playing a key role in the binding–gating coupling, has also been shown to drastically affect desensitization kinetics (Bouzat et al., 2008). Mutations substituting polar side chains at the M2 9′ position slowed macroscopic desensitization (Bocquet et al., 2007; Gonzalez-Gutierrez et al., 2012) and enabled measurement of robust ligand-dependent currents.
Location of the pH sensor in GLIC
Gating of the ELIC-GLIC chimera by protons in addition to amines clearly demonstrates that the GLIC-TMD can serve as a pH sensor on its own. The GLIC-ECD was previously implicated to be the pH sensor based on findings from GLIC-GlyR chimera, where the channel was shown to gate in response to external pH changes (Duret et al., 2011). However, protonation of His11′ in the M2 helix was also proposed to be essential for stabilizing the open channel through an intrasubunit hydrogen bond with Ile35′ in M3 (Prevost et al., 2012), as mutating this residue to His11′F eliminated GLIC’s pH response (Wang et al., 2012; Rienzo et al., 2014). Although the lack of function can arise because protonation of His11′ is essential to channel opening, it is also possible that the His11′ protonation simply enhances intrinsic pH sensitivity and lowers the barrier to reach the open state. If the latter were the case, the H11′F mutant would simply have a shifted pH dependence. Indeed, the functionality of GLIC His11′ can be rescued in the presence of I9′A background mutation (Fig. S13). Furthermore, the I9′A mutation also rescues the pH dependence of the H11′F in the ELIC-GLIC chimera (Fig. S13). The residual pH dependence in the absence of GLIC-ECD and the His11′ side chain demonstrates that although these two entities contribute to proton sensitivity, there are clearly other ionizable residues that are responsible for pH-dependent gating. Consistent with this idea, the crystal structure of GLIC H11′F at acidic pH reveals a locally closed conformation, where besides a change in the upper part of M2, the ECD and most of the TMD adopts a conformation akin to the open GLIC structure (Prevost et al., 2012).
Loop C motion: Is directionality coupled to channel opening?
At the structural level, we show here that loop C in ELIC and in the ELIC-GLIC chimera is mobile and moves outward upon binding agonist. These motions are inhibited and further reversed in the presence of a competitive antagonist, ACh. These findings are consistent with the orientation of loop C in the ELIC structures bound to ACh or GABA/flurazepam (Spurny et al., 2012). However, this direction of loop C motion clearly contradicts the general trend observed in AChBP and other members of the pLGIC family (Celie et al., 2004; Hansen et al., 2005; Mukhtasimova et al., 2009; Li et al., 2011; Unwin and Fujiyoshi, 2012), where an inward or outward movement of loop C has been attributed to agonism or antagonism, respectively (Celie et al., 2004, 2005; Bourne et al., 2005; Hansen et al., 2005; Brams et al., 2011; Li et al., 2011). The inward or capping motion has been proposed to be an essential feature of the transduction mechanism that links agonist binding to channel gating at the pore (Lee and Sine, 2005; Mukhtasimova et al., 2005; Lape et al., 2008). The prior conservation of inward loop C motion across different channel types of diverse origin led to the implication that this movement is somehow coupled to channel opening. Even in GLIC, which lacks a canonical ligand-binding pocket, loop C undergoes a similar constriction under acidic pH (Velisetty and Chakrapani, 2012; Sauguet et al., 2014). ELIC therefore appears to be the first reported outlier of this trend. Interestingly, in the ELIC-GLIC chimera, protons and 3-AP lead to contrasting loop C dynamics and motional directions, yet both result in channel opening. The opposite motions in the chimera suggest that although loop C moves in a ligand-specific manner, motions in this region may be associated with only ligand binding and not directly coupled to channel opening. Not surprisingly, replacement of the entire loop C sequence by Gly residues in nAChR (Purohit and Auerbach, 2013) or truncation of this region in GLIC (Gonzalez-Gutierrez and Grosman, 2010) had little effect on intrinsic channel opening.
The structural changes at loop C are different from those in the rest of the ELIC and chimera channels in two regards. First, the ligand-induced changes in loop C are much larger than ligand-induced changes elsewhere in the channel, either in ELIC or in the chimera. Second, in the chimera, this is the only region that exhibits opposing EPR mobility changes in response to acid and ligand (broader peaks or lower mobility with acid, sharper peaks or higher mobility with ligand). Everywhere else in the chimera, acid and the ligand move in similar directions, although with different magnitudes of change.
In loop F, for both our chimera, GLIC, and other members of the family, activation universally induces an inward constriction, suggesting that the motions of loop F and loop C are not coupled as a part of channel opening. These differences introduce the possibility that the changes brought about in loop F are simply a result of the quaternary motions in the ECD, including the twist of the β strands that we observe in the resting state of our chimera. Furthermore, DEER measurements show that loop F can coexist in two distinct orientations in the resting state (inward and outward), and this equilibrium is shifted in a ligand-dependent manner. This is in line with the allosteric models of pLGIC gating, where the ligand merely shifts the preexisting equilibrium between the resting and activated states. However, in the ELIC-GLIC chimera, different ligands induce different magnitudes of the structural change. The pH-elicited changes in loop F and the rest of the ECD–TMD interface are much more pronounced than those by 3-AP, with the pH inducing larger mobility changes than the ligand.
Therefore, when considered with the additional fact that changes throughout the channel are broad in the presence of acid and limited in the presence of 3-AP (Fig. 7), we conclude that the agonist-induced changes in the ECD and TMD of the ELIC-GLIC chimera seem to reflect the behavior of their parent channels, with large global conformational changes in GLIC and more local and subtle changes in ELIC.
Structural conservation in pLGIC
In general agreement with the EPR studies and functional data, the structure of the ELIC-GLIC chimera in its apo conformation revealed no major changes in the overall architecture compared with the parent channels. The overall arrangement of the TMD and the orientation of M2 in the ELIC-GLIC chimera are similar to GLIC at pH 7.0. Given the absence of either activating ligand in the crystallization conditions, it is no surprise that the channel is closed; however, the quaternary rearrangements including a twist in the ECD are intriguing. In all ELIC structures determined to date, the ELIC-ECD has been remarkably resilient to conformational changes (with the exception of loop C; Zimmermann and Dutzler, 2011; Gonzalez-Gutierrez et al., 2012; Pan et al., 2012; Spurny et al., 2012; Ulens et al., 2014), and the twist of the ECD relative to the TMD therefore represents the largest motion ever observed for the protein backbone of the ELIC channel, under any condition with any ligand. This confirms that the ELIC-ECD is capable of undergoing a twisting motion similar to what has been observed in GLIC, at least in this chimeric context. This suggests that the observed conformational changes in the ECD are driven, at least in part, by the TMD and underscore the two-way communication between the domains in these channels. It is remarkable that crystal structures of pLGIC differ most prominently in the extent of twisting of the ECD β sheets with respect to the pore domain (Hilf and Dutzler, 2008, 2009; Bocquet et al., 2009; Hibbs and Gouaux, 2011; Hassaine et al., 2014; Miller and Aricescu, 2014). These differences in the observed twist have been implicated to be either associated with the different crystallographically trapped conformations (closed, open, or desensitized) or caused by their different phylogenetic origins (from prokaryotes through humans).
Functional chimeric channels, particularly those without any modification to the interface, including the ELIC-GLIC chimera, emphasize the fact that domain movements critical to gating are essentially conserved across the family, even though pairwise interactions across domains may not be preserved (Xiu et al., 2005). Yet, exactly what these conserved domain motions are remains unclear. In our chimera, we found that both the amine-induced activation pathway in ELIC and the proton-mediated pathway in GLIC are preserved, giving us the advantage of direct comparison of conserved and divergent features in the two mechanisms. Conformational changes that are divergent are clearly not critical for pore opening, whereas conserved features suggest a core gating mechanism.
To summarize, the chimeric channel, rather than being gated by a single mechanism, appears to be a channel that is gated by not only two different ligands, but two different mechanisms (as revealed by the different magnitudes of mobility changes in response to the different ligands). As a result, the chimera allowed us to specifically identify gating conformational changes that are either conserved or divergent between ELIC and GLIC (which share only 20% sequence identity). Contrary to the general belief in the field (held for over 15 years), we find that agonist-driven inward motion of loop C is not necessary for channel opening. Although the direction of loop C motions is sensitive to the nature of the ligand bound, its motion is not coupled to the channel gate. However, our results suggest that the twisting motion of the ECD β strands is potentially the conserved mechanism of communication between the ECD and TMD, and thereby our chimera is functional even in the absence of conserved ECD–TMD interactions. The high level of broad structural conservation in the cys-loop family suggests that this general topology can support many different pathways of gating, although some key elements must be preserved.
GLIC and ELIC show distinct pharmacological profiles, and therefore this chimera sets the stage for a thorough understanding of how the two gating pathways are differentially altered by allosteric modulators. Of course, the disadvantage of using these chimeras is that fusing different regions of a channel together invariably leads to differences in both the sensitivity and nature of pharmacological interactions and may complicate structural interpretation. Future and past studies on chimeric channels, including this one, should give greater thought to the possibility of the cys-loop topology supporting a range of gating and modulation mechanisms.
Supplementary Material
Acknowledgments
We are very grateful to the members of the Chakrapani laboratory for critical reading and comments on the manuscript. We thank Dr. Walter F. Boron for unrestricted access of the oocyte rig. ELIC and GLIC clones were a generous gift from Dr. Raimund Dutzler. We are also profoundly appreciative of the help and assistance provided by the staff at the NE/CAT beamlines 24ID-C and 24ID-E at the Argonne National Laboratory.
This work was supported by National Institutes of Health (NIH) grant 1R01GM108921 and the American Heart Association NCRP Scientist Development grant 12SDG12070069 to S. Chakrapani; NIH grants U54-GM087519 and S10RR027091 to H.S. Mchaourab; NIH grant R00EY019718 to D.T. Lodowski; and NIH grant R01DK81567 to W.F.B. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by a grant from the National Institute of General Medical Sciences (P41 GM103403) from the NIH. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract number DE-AC02-06CH11357.
The authors declare no competing financial interests.
Sharona E. Gordon served as editor.
Footnotes
Abbreviations used in this paper:
- 3-AP
- 3-amino propanol
- ACh
- acetylcholine
- CW
- continuous wave
- DDM
- n-dodecyl-β-d-maltopyranoside
- DEER
- double electron–electron resonance
- ECD
- extracellular domain
- ELIC
- Erwinia chrysanthemi ligand–gated ion channel
- EPR
- electron paramagnetic resonance
- GLIC
- Gloeobacter violaceus ligand–gated ion channel
- GlyR
- glycine receptor
- MMM
- multiscale modeling of macromolecular systems
- MTSSL
- (1-oxyl-2,2,5,5-tetramethylpyrrolidin-3-yl) methyl methanethiosulfonate
- nAChR
- nicotinic ACh receptor
- NiEDDA
- Ni(II) ethylenediamine diacetic acid
- pLGIC
- pentameric ligand–gated ion channel
- RMSD
- root mean square deviation
- TMD
- transmembrane domain
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach C., Froncisz W., Hemker R., Mchaourab H., and Hubbell W.L.. 2005. Accessibility of nitroxide side chains: Absolute Heisenberg exchange rates from power saturation EPR. Biophys. J. 89:2103–2112. 10.1529/biophysj.105.059063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S.M., and Bricogne G.. 2004. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60:2210–2221. 10.1107/S0907444904016427 [DOI] [PubMed] [Google Scholar]
- Bocquet N., Prado de Carvalho L., Cartaud J., Neyton J., Le Poupon C., Taly A., Grutter T., Changeux J.P., and Corringer P.J.. 2007. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature. 445:116–119. 10.1038/nature05371 [DOI] [PubMed] [Google Scholar]
- Bocquet N., Nury H., Baaden M., Le Poupon C., Changeux J.P., Delarue M., and Corringer P.J.. 2009. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 457:111–114. 10.1038/nature07462 [DOI] [PubMed] [Google Scholar]
- Bourne Y., Talley T.T., Hansen S.B., Taylor P., and Marchot P.. 2005. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. EMBO J. 24:1512–1522. 10.1038/sj.emboj.7600620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzat C., Gumilar F., Spitzmaul G., Wang H.L., Rayes D., Hansen S.B., Taylor P., and Sine S.M.. 2004. Coupling of agonist binding to channel gating in an ACh-binding protein linked to an ion channel. Nature. 430:896–900. 10.1038/nature02753 [DOI] [PubMed] [Google Scholar]
- Bouzat C., Bartos M., Corradi J., and Sine S.M.. 2008. The interface between extracellular and transmembrane domains of homomeric Cys-loop receptors governs open-channel lifetime and rate of desensitization. J. Neurosci. 28:7808–7819. 10.1523/JNEUROSCI.0448-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brams M., Pandya A., Kuzmin D., van Elk R., Krijnen L., Yakel J.L., Tsetlin V., Smit A.B., and Ulens C.. 2011. A structural and mutagenic blueprint for molecular recognition of strychnine and d-tubocurarine by different cys-loop receptors. PLoS Biol. 9:e1001034 10.1371/journal.pbio.1001034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celie P.H., van Rossum-Fikkert S.E., van Dijk W.J., Brejc K., Smit A.B., and Sixma T.K.. 2004. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 41:907–914. 10.1016/S0896-6273(04)00115-1 [DOI] [PubMed] [Google Scholar]
- Celie P.H., Kasheverov I.E., Mordvintsev D.Y., Hogg R.C., van Nierop P., van Elk R., van Rossum-Fikkert S.E., Zhmak M.N., Bertrand D., Tsetlin V., et al. 2005. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha-conotoxin PnIA variant. Nat. Struct. Mol. Biol. 12:582–588. 10.1038/nsmb951 [DOI] [PubMed] [Google Scholar]
- Chakrapani S. 2015. EPR studies of gating mechanisms in ion channels. Methods Enzymol. 557:279–306. 10.1016/bs.mie.2014.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrapani S., Cordero-Morales J.F., and Perozo E.. 2007. A quantitative description of KcsA gating I: Macroscopic currents. J. Gen. Physiol. 130:465–478. 10.1085/jgp.200709843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B. III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., and Richardson D.C.. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y.W., Borbat P.P., and Freed J.H.. 2005. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. 172:279–295. 10.1016/j.jmr.2004.10.012 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- Cooper S.T., Harkness P.C., Baker E.R., and Millar N.S.. 1999. Up-regulation of cell-surface alpha4beta2 neuronal nicotinic receptors by lower temperature and expression of chimeric subunits. J. Biol. Chem. 274:27145–27152. 10.1074/jbc.274.38.27145 [DOI] [PubMed] [Google Scholar]
- Corringer P.J., Poitevin F., Prevost M.S., Sauguet L., Delarue M., and Changeux J.P.. 2012. Structure and pharmacology of pentameric receptor channels: From bacteria to brain. Structure. 20:941–956. 10.1016/j.str.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Cortes D.M., Cuello L.G., and Perozo E.. 2001. Molecular architecture of full-length KcsA: Role of cytoplasmic domains in ion permeation and activation gating. J. Gen. Physiol. 117:165–180. 10.1085/jgp.117.2.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- daCosta C.J., and Baenziger J.E.. 2013. Gating of pentameric ligand-gated ion channels: Structural insights and ambiguities. Structure. 21:1271–1283. 10.1016/j.str.2013.06.019 [DOI] [PubMed] [Google Scholar]
- Delcour A.H., Martinac B., Adler J., and Kung C.. 1989. Modified reconstitution method used in patch-clamp studies of Escherichia coli ion channels. Biophys. J. 56:631–636. 10.1016/S0006-3495(89)82710-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellisanti C.D., Ghosh B., Hanson S.M., Raspanti J.M., Grant V.A., Diarra G.M., Schuh A.M., Satyshur K., Klug C.S., and Czajkowski C.. 2013. Site-directed spin labeling reveals pentameric ligand-gated ion channel gating motions. PLoS Biol. 11:e1001714 10.1371/journal.pbio.1001714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret G., Van Renterghem C., Weng Y., Prevost M., Moraga-Cid G., Huon C., Sonner J.M., and Corringer P.J.. 2011. Functional prokaryotic-eukaryotic chimera from the pentameric ligand-gated ion channel family. Proc. Natl. Acad. Sci. USA. 108:12143–12148. 10.1073/pnas.1104494108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiselé J.L., Bertrand S., Galzi J.L., Devillers-Thiéry A., Changeux J.P., and Bertrand D.. 1993. Chimaeric nicotinic-serotonergic receptor combines distinct ligand binding and channel specificities. Nature. 366:479–483. 10.1038/366479a0 [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farahbakhsh Z.T., Altenbach C., and Hubbell W.L.. 1992. Spin labeled cysteines as sensors for protein-lipid interaction and conformation in rhodopsin. Photochem. Photobiol. 56:1019–1033. 10.1111/j.1751-1097.1992.tb09725.x [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gutierrez G., and Grosman C.. 2010. Bridging the gap between structural models of nicotinic receptor superfamily ion channels and their corresponding functional states. J. Mol. Biol. 403:693–705. 10.1016/j.jmb.2010.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gutierrez G., and Grosman C.. 2015. The atypical cation-conduction and gating properties of ELIC underscore the marked functional versatility of the pentameric ligand-gated ion-channel fold. J. Gen. Physiol. 146:15–36. 10.1085/jgp.201411333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gutierrez G., Lukk T., Agarwal V., Papke D., Nair S.K., and Grosman C.. 2012. Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals. Proc. Natl. Acad. Sci. USA. 109:6331–6336. 10.1073/pnas.1119268109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A., and Hubbell W.L.. 2002. Identification of protein side chains near the membrane-aqueous interface: A site-directed spin labeling study of KcsA. Biochemistry. 41:1123–1128. 10.1021/bi015828s [DOI] [PubMed] [Google Scholar]
- Grutter T., de Carvalho L.P., Dufresne V., Taly A., Edelstein S.J., and Changeux J.P.. 2005. Molecular tuning of fast gating in pentameric ligand-gated ion channels. Proc. Natl. Acad. Sci. USA. 102:18207–18212. 10.1073/pnas.0509024102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.B., Sulzenbacher G., Huxford T., Marchot P., Taylor P., and Bourne Y.. 2005. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 24:3635–3646. 10.1038/sj.emboj.7600828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassaine G., Deluz C., Grasso L., Wyss R., Tol M.B., Hovius R., Graff A., Stahlberg H., Tomizaki T., Desmyter A., et al. 2014. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 512:276–281. 10.1038/nature13552 [DOI] [PubMed] [Google Scholar]
- Hibbs R.E., and Gouaux E.. 2011. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 474:54–60. 10.1038/nature10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilf R.J., and Dutzler R.. 2008. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 452:375–379. 10.1038/nature06717 [DOI] [PubMed] [Google Scholar]
- Hilf R.J., and Dutzler R.. 2009. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 457:115–118. 10.1038/nature07461 [DOI] [PubMed] [Google Scholar]
- Jeschke G., Koch A., Jonas U., and Godt A.. 2002. Direct conversion of EPR dipolar time evolution data to distance distributions. J. Magn. Reson. 155:72–82. 10.1006/jmre.2001.2498 [DOI] [PubMed] [Google Scholar]
- Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C.R., Hilger D., and Jung H.. 2006. DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 30:473–498. 10.1007/BF03166213 [DOI] [Google Scholar]
- Kabsch W. 2010. Xds. Acta Crystallogr. D Biol. Crystallogr. 66:125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri A., Sedelnikova A., and Weiss D.S.. 2009. Structural rearrangements in loop F of the GABA receptor signal ligand binding, not channel activation. Biophys. J. 96:45–55. 10.1016/j.bpj.2008.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde M.N., Chen Q., Lawless M.J., Mowrey D.D., Xu J., Saxena S., Xu Y., and Tang P.. 2015. Conformational changes underlying desensitization of the pentameric ligand-gated ion channel ELIC. Structure. 23:995–1004. 10.1016/j.str.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., and Henrick K.. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Lape R., Colquhoun D., and Sivilotti L.G.. 2008. On the nature of partial agonism in the nicotinic receptor superfamily. Nature. 454:722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.Y., and Sine S.M.. 2005. Principal pathway coupling agonist binding to channel gating in nicotinic receptors. Nature. 438:243–247. 10.1038/nature04156 [DOI] [PubMed] [Google Scholar]
- Li S.X., Huang S., Bren N., Noridomi K., Dellisanti C.D., Sine S.M., and Chen L.. 2011. Ligand-binding domain of an α7-nicotinic receptor chimera and its complex with agonist. Nat. Neurosci. 14:1253–1259. 10.1038/nn.2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63:32–41. 10.1107/S0907444906045975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mchaourab H.S., Lietzow M.A., Hideg K., and Hubbell W.L.. 1996. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 35:7692–7704. 10.1021/bi960482k [DOI] [PubMed] [Google Scholar]
- Miller P.S., and Aricescu A.R.. 2014. Crystal structure of a human GABAA receptor. Nature. 512:270–275. 10.1038/nature13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga-Cid G., Sauguet L., Huon C., Malherbe L., Girard-Blanc C., Petres S., Murail S., Taly A., Baaden M., Delarue M., and Corringer P.J.. 2015. Allosteric and hyperekplexic mutant phenotypes investigated on an α1 glycine receptor transmembrane structure. Proc. Natl. Acad. Sci. USA. 112:2865–2870. 10.1073/pnas.1417864112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtasimova N., Free C., and Sine S.M.. 2005. Initial coupling of binding to gating mediated by conserved residues in the muscle nicotinic receptor. J. Gen. Physiol. 126:23–39. 10.1085/jgp.200509283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtasimova N., Lee W.Y., Wang H.L., and Sine S.M.. 2009. Detection and trapping of intermediate states priming nicotinic receptor channel opening. Nature. 459:451–454. 10.1038/nature07923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., and Dodson E.J.. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Pan J., Chen Q., Willenbring D., Yoshida K., Tillman T., Kashlan O.B., Cohen A., Kong X.P., Xu Y., and Tang P.. 2012. Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine. Nat. Commun. 3:714 10.1038/ncomms1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannier M., Veit S., Godt A., Jeschke G., and Spiess H.W.. 2000. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 142:331–340. 10.1006/jmre.1999.1944 [DOI] [PubMed] [Google Scholar]
- Petřek M., Kosinová P., Koca J., and Otyepka M.. 2007. MOLE: A Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure. 15:1357–1363. 10.1016/j.str.2007.10.007 [DOI] [PubMed] [Google Scholar]
- Polyhach Y., Godt A., Bauer C., and Jeschke G.. 2007. Spin pair geometry revealed by high-field DEER in the presence of conformational distributions. J. Magn. Reson. 185:118–129. 10.1016/j.jmr.2006.11.012 [DOI] [PubMed] [Google Scholar]
- Polyhach Y., Bordignon E., and Jeschke G.. 2011. Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 13:2356–2366. 10.1039/C0CP01865A [DOI] [PubMed] [Google Scholar]
- Prevost M.S., Sauguet L., Nury H., Van Renterghem C., Huon C., Poitevin F., Baaden M., Delarue M., and Corringer P.J.. 2012. A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat. Struct. Mol. Biol. 19:642–649. 10.1038/nsmb.2307 [DOI] [PubMed] [Google Scholar]
- Purohit P., and Auerbach A.. 2013. Loop C and the mechanism of acetylcholine receptor–channel gating. J. Gen. Physiol. 141:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rienzo M., Lummis S.C., and Dougherty D.A.. 2014. Structural requirements in the transmembrane domain of GLIC revealed by incorporation of noncanonical histidine analogs. Chem. Biol. 21:1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauguet L., Poitevin F., Murail S., Van Renterghem C., Moraga-Cid G., Malherbe L., Thompson A.W., Koehl P., Corringer P.J., Baaden M., and Delarue M.. 2013. Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J. 32:728–741. 10.1038/emboj.2013.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauguet L., Shahsavar A., Poitevin F., Huon C., Menny A., Nemecz À., Haouz A., Changeux J.P., Corringer P.J., and Delarue M.. 2014. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl. Acad. Sci. USA. 111:966–971. 10.1073/pnas.1314997111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sine S.M., and Engel A.G.. 2006. Recent advances in Cys-loop receptor structure and function. Nature. 440:448–455. 10.1038/nature04708 [DOI] [PubMed] [Google Scholar]
- Spurny R., Ramerstorfer J., Price K., Brams M., Ernst M., Nury H., Verheij M., Legrand P., Bertrand D., Bertrand S., et al. 2012. Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc. Natl. Acad. Sci. USA. 109:E3028–E3034. 10.1073/pnas.1208208109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.J., Lester H.A., and Lummis S.C.. 2010. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 43:449–499. 10.1017/S0033583510000168 [DOI] [PubMed] [Google Scholar]
- Tillman T.S., Seyoum E., Mowrey D.D., Xu Y., and Tang P.. 2014. ELIC-α7 nicotinic acetylcholine receptor (α7nAChR) chimeras reveal a prominent role of the extracellular-transmembrane domain interface in allosteric modulation. J. Biol. Chem. 289:13851–13857. 10.1074/jbc.M113.524611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulens C., Spurny R., Thompson A.J., Alqazzaz M., Debaveye S., Han L., Price K., Villalgordo J.M., Tresadern G., Lynch J.W., and Lummis S.C.. 2014. The prokaryote ligand-gated ion channel ELIC captured in a pore blocker-bound conformation by the Alzheimer’s disease drug memantine. Structure. 22:1399–1407. 10.1016/j.str.2014.07.013 [DOI] [PubMed] [Google Scholar]
- Unwin N., and Fujiyoshi Y.. 2012. Gating movement of acetylcholine receptor caught by plunge-freezing. J. Mol. Biol. 422:617–634. 10.1016/j.jmb.2012.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velisetty P., and Chakrapani S.. 2012. Desensitization mechanism in prokaryotic ligand-gated ion channel. J. Biol. Chem. 287:18467–18477. 10.1074/jbc.M112.348045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velisetty P., Chalamalasetti S.V., and Chakrapani S.. 2012. Conformational transitions underlying pore opening and desensitization in membrane-embedded Gloeobacter violaceus ligand-gated ion channel (GLIC). J. Biol. Chem. 287:36864–36872. 10.1074/jbc.M112.401067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velisetty P., Chalamalasetti S.V., and Chakrapani S.. 2014. Structural basis for allosteric coupling at the membrane-protein interface in Gloeobacter violaceus ligand-gated ion channel (GLIC). J. Biol. Chem. 289:3013–3025. 10.1074/jbc.M113.523050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.L., Cheng X., and Sine S.M.. 2012. Intramembrane proton binding site linked to activation of bacterial pentameric ion channel. J. Biol. Chem. 287:6482–6489. 10.1074/jbc.M111.305839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu X., Hanek A.P., Wang J., Lester H.A., and Dougherty D.A.. 2005. A unified view of the role of electrostatic interactions in modulating the gating of Cys loop receptors. J. Biol. Chem. 280:41655–41666. 10.1074/jbc.M508635200 [DOI] [PubMed] [Google Scholar]
- Zimmermann I., and Dutzler R.. 2011. Ligand activation of the prokaryotic pentameric ligand-gated ion channel ELIC. PLoS Biol. 9:e1001101 10.1371/journal.pbio.1001101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann I., Marabelli A., Bertozzi C., Sivilotti L.G., and Dutzler R.. 2012. Inhibition of the prokaryotic pentameric ligand-gated ion channel ELIC by divalent cations. PLoS Biol. 10:e1001429 10.1371/journal.pbio.1001429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P., and Mchaourab H.S.. 2010. Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophys. J. 98:L18–L20. 10.1016/j.bpj.2009.12.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.