Abstract
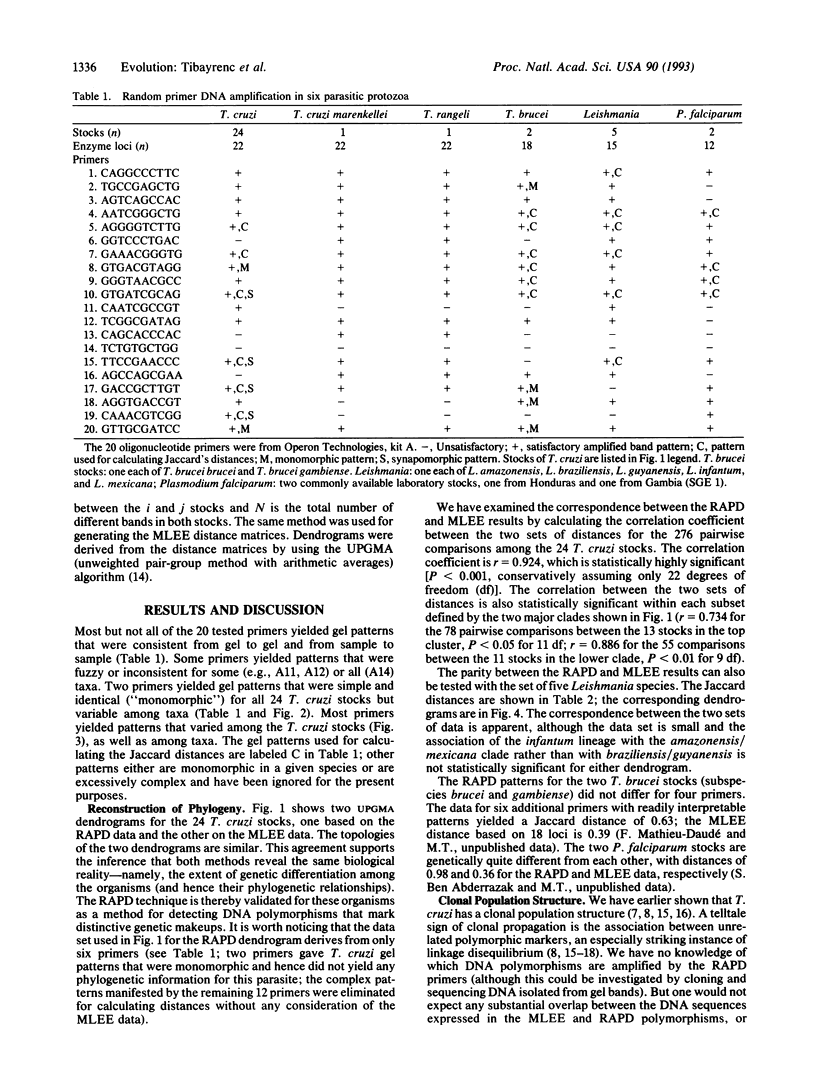
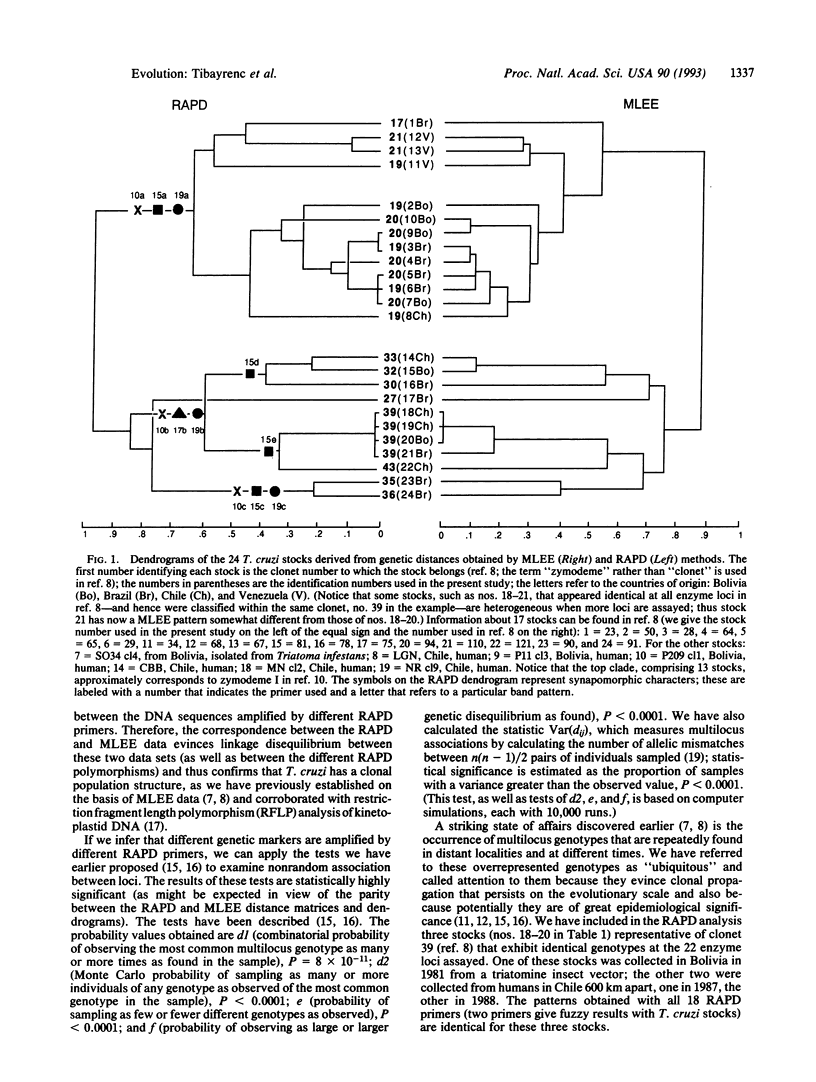
We have assayed genetic polymorphisms in several species of parasitic protozoa by means of random amplified polymorphic DNA (RAPD). One goal was to ascertain the suitability of RAPD markers for investigating genetic and evolutionary problems, particularly in organisms, such as the parasitic protozoa, unsuitable for traditional methods of genetic analysis. Another goal was to test certain hypotheses concerning Trypanosoma cruzi, and other protozoa, that have been established by multilocus enzyme electrophoresis. The RAPD results corroborate the hypothesis that the population structure of T. cruzi is clonal and yield a phylogeny of the clonal lineages in agreement with the one obtained by enzyme electrophoresis. This parity between the two sets of results confirms that RAPD markers are reliable genetic markers. The RAPD markers are also suitable for reconstructing species phylogenies and as diagnostic characters of species and subspecific lineages. The number of DNA polymorphisms that can be detected by the RAPD method seems virtually unlimited, since the number of primers can be increased effectively at will. The RAPD method is well suited for investigating genetic and evolutionary questions in certain organisms, because it is cost effective and demands no previous genetic knowledge about the organism.
Full text
PDF
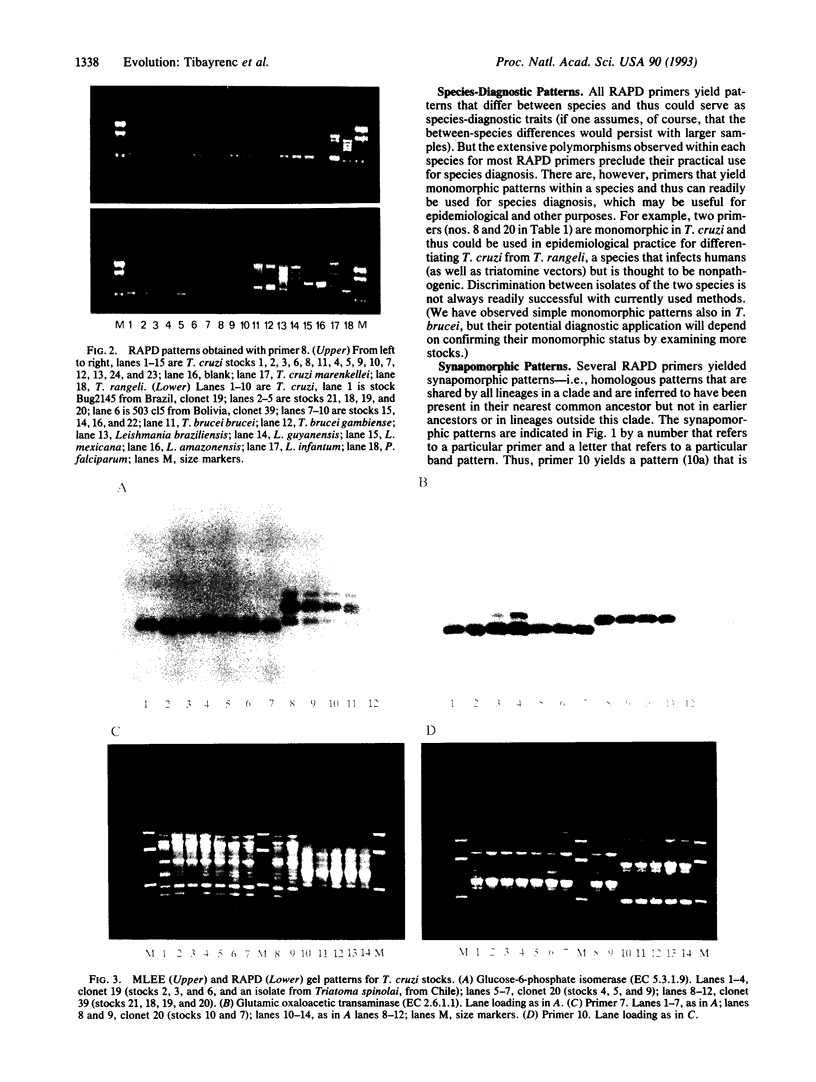
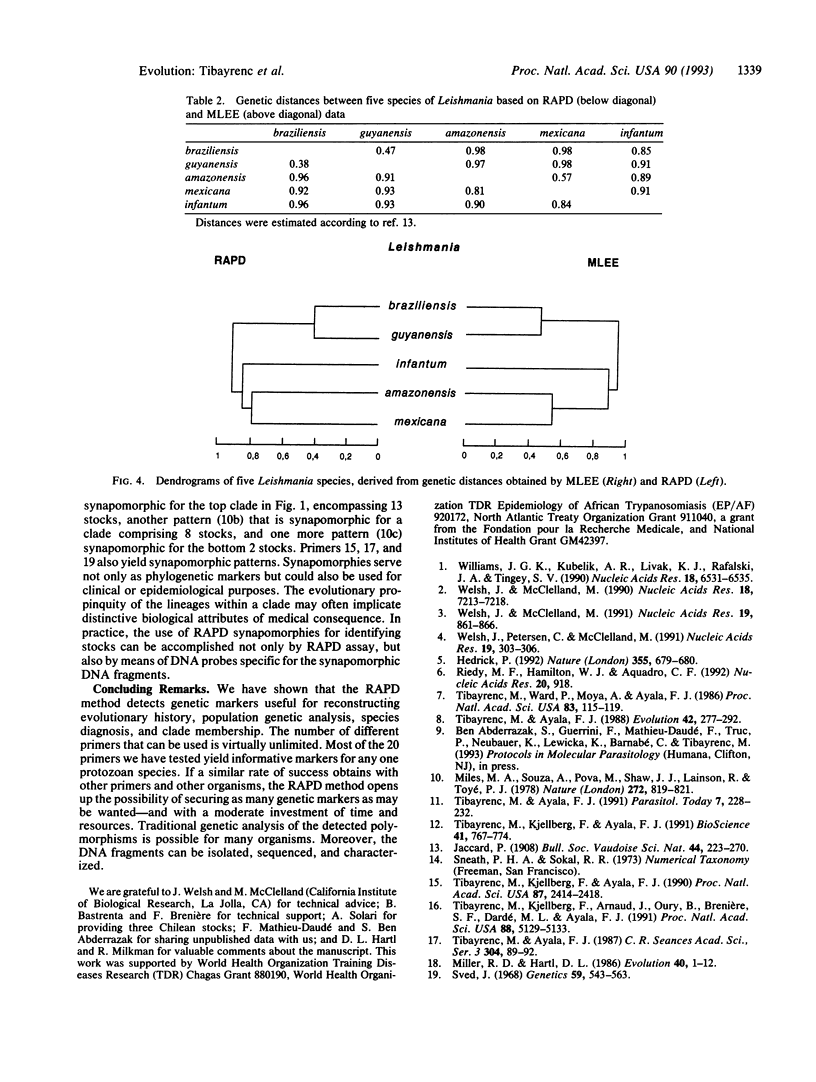
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Miles M. A., Souza A., Povoa M., Shaw J. J., Lainson R., Toye P. J. Isozymic heterogeneity of Trypanosoma cruzi in the first autochthonous patients with Chagas' disease in Amazonian Brazil. Nature. 1978 Apr 27;272(5656):819–821. doi: 10.1038/272819a0. [DOI] [PubMed] [Google Scholar]
- Riedy M. F., Hamilton W. J., 3rd, Aquadro C. F. Excess of non-parental bands in offspring from known primate pedigrees assayed using RAPD PCR. Nucleic Acids Res. 1992 Feb 25;20(4):918–918. doi: 10.1093/nar/20.4.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J. A. The stability of linked systems of loci with a small population size. Genetics. 1968 Aug;59(4):543–563. doi: 10.1093/genetics/59.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F. J. Forte corrélation entre classification isoenzymatique et variabilité de l'ADN kinétoplastique chez Trypanosoma cruzi. C R Acad Sci III. 1987;304(4):89–92. [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F. J. Towards a population genetics of microorganisms: The clonal theory of parasitic protozoa. Parasitol Today. 1991 Sep;7(9):228–232. doi: 10.1016/0169-4758(91)90234-f. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M., Kjellberg F., Arnaud J., Oury B., Brenière S. F., Dardé M. L., Ayala F. J. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Kjellberg F., Ayala F. J. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Ward P., Moya A., Ayala F. J. Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proc Natl Acad Sci U S A. 1986 Jan;83(1):115–119. doi: 10.1073/pnas.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 1991 Feb 25;19(4):861–866. doi: 10.1093/nar/19.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., Petersen C., McClelland M. Polymorphisms generated by arbitrarily primed PCR in the mouse: application to strain identification and genetic mapping. Nucleic Acids Res. 1991 Jan 25;19(2):303–306. doi: 10.1093/nar/19.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]