Interleukin-4 boosts the capacity of dendritic cells to present endogenous antigens on MHC II and to resist bacterial infection through a mechanism shown to be partially dependent on RUFY4 expression.
Abstract
Autophagy is a key degradative pathway coordinated by external cues, including starvation, oxidative stress, or pathogen detection. Rare are the molecules known to contribute mechanistically to the regulation of autophagy and expressed specifically in particular environmental contexts or in distinct cell types. Here, we unravel the role of RUN and FYVE domain–containing protein 4 (RUFY4) as a positive molecular regulator of macroautophagy in primary dendritic cells (DCs). We show that exposure to interleukin-4 (IL-4) during DC differentiation enhances autophagy flux through mTORC1 regulation and RUFY4 induction, which in turn actively promote LC3 degradation, Syntaxin 17–positive autophagosome formation, and lysosome tethering. Enhanced autophagy boosts endogenous antigen presentation by MHC II and allows host control of Brucella abortus replication in IL-4–treated DCs and in RUFY4-expressing cells. RUFY4 is therefore the first molecule characterized to date that promotes autophagy and influences endosome dynamics in a subset of immune cells.
Introduction
Cells rely on autophagy to survive diverse cellular insults such as nutrient depletion, accumulation of protein aggregates, damaged mitochondria, or intracellular bacteria (Deretic et al., 2013). Autophagy describes different eukaryotic mechanisms of protein degradation, which result in the transfer to and the degradation of organelles or cytosolic material in the lysosomes (Hamasaki et al., 2013b). Macroautophagy (here referred as autophagy) is controlled by specialized Atg factors that promote the genesis of autophagosomes, substrate recruitment, lysosome–autophagosome fusion, and final degradation of autolysosome contents. One of the earliest steps of autophagy involves the activation of the ULK1–ATG13–FIP200 protein complex at the surface of the ER for recruitment of the class III phosphatidylinositol-3-kinase vacuolar protein sorting 34 (VPS34), together with the Vps15–Beclin1–ATG14 complex (Mizushima, 2010). Phosphatidylinositol-3-phosphate (PtdIns(3)P) formation by VPS34 is absolutely required for autophagosome formation, and one of the major upstream regulators of this step is the mTORC1 complex, whose inactivation induces autophagic flux through ATG1/ULK1 activation in response to metabolic changes, starvation, or rapamycin treatment (Kim et al., 2011).
Like other membrane trafficking steps, autophagy is regulated by Rab GTPases, soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs), and tethering complexes. The role of Rab7 in autophagosome-lysosome fusion has been extensively studied (Tabata et al., 2010), whereas the Qa-SNARE Syntaxin 17 (STX17) has been proposed to mediate both autophagosome formation and fusion with lysosomes by binding to its partners SNAP29 and VAMP8 (Itakura et al., 2012; Hamasaki et al., 2013a). In their active form, GTPases and SNAREs are anchored to intracellular vesicles, subsequently regulating recruitment of various effector proteins in cooperation with the accumulation of specific lipids like PtdIns(3)P. RUN and FYVE domain–containing proteins (RUFYs), which contain an N-terminal RUN domain, shown to interact with small GTPases (Callebaut et al., 2001), and a PtdIns(3)P-interacting C-terminal FYVE domain, are good candidate effectors capable of controlling endocytosis or autophagy (Birkeland and Stenmark, 2004). A member of the RUFY family, the FYVE and coiled-coil domain containing 1 (FYCO1) has been demonstrated to bind LC3/ATG8, PtdIns(3)P, and Rab7 (Pankiv et al., 2010), linking autophagosomes to microtubule plus end–directed molecular motors.
Antigen presentation mostly consists of the direct presentation of proteasome-degraded cytosolic peptides by MHC class I and the presentation of internalized exogenous proteins by MHC class II. The closely related autophagy and endocytic degradative pathways are particularly important for the orchestration of immunity by dendritic cells (DCs) because MHC II–restricted antigen presentation is heavily dependent on endo/lysosomal proteolytic processing to achieve efficient peptide presentation to naive CD4+ T cells (Honey and Rudensky, 2003). Interestingly, cytosolic proteins have also been shown to use autophagy to transit from the cytosol to endosomes and enter the MHC II–restricted presentation pathway (Schmid et al., 2007; Münz, 2012), whereas endosomal function and exogenous antigen MHC II presentation are altered in Atg-deficient DCs (Lee et al., 2010). DCs are activated through microbial pattern recognition receptors such as Toll-like receptors (TLRs), which results in unmatched T cell priming capacities (Kawai and Akira, 2008). Autophagy functions in many aspects of innate immune signaling (Deretic et al., 2013) and can be enhanced by microbial stimulation, as shown for RAW264.7 macrophages by lipopolysaccharide (LPS; Delgado et al., 2008; Levine et al., 2011). Thus, DCs exposed to microbial stimuli and various polarizing cytokines offer a unique model to study autophagy regulation and identify novel factors controlling this function in mammalian cells.
We focus here on the regulation of autophagy in DCs responding to LPS and Interleukin-4 (IL-4) in vitro. We show that autophagy flux is reduced during the stimulation of primary DCs by LPS. This inhibition is a consequence of mTORC1 activation after TLR stimulation and results in a clear reduction of endogenous MHC II–restricted antigen presentation and accumulation of neosynthetized misfolded proteins in DC–aggresome-like induced structures (DALISs). We further demonstrate that IL-4–mediated differentiation of DCs is able to considerably augment basal autophagy flux, promoting endogenous antigen presentation and reducing the ability of the intracellular pathogen Brucella abortus to replicate. IL-4 exerts this function by interfering with mTOR signaling but also via the induction of RUFY4, a previously uncharacterized RUFY family member (Ivan et al., 2012). RUFY4 expression increases autophagy and leads to the reorganization of late endosomal compartments, thereby changing the overall endosome membrane dynamic during DC differentiation and revealing its functional role as a Rab7 effector and one of the few positive regulators of autophagy identified to date.
Results
TLR stimulation activates mTOR and suppresses autophagy in DCs
Engagement of the TLR4 pathway by LPS induces in bone marrow–derived DCs (bmDCs) the phosphorylation of mTORC1 targets, such as p70 ribosomal protein S6 kinase (p70S6K), ribosomal protein S6, translation factor inhibitor 4E-BP1, and, importantly, AMBRA1 at residue Ser52 (Fig. 1, A and B), as well as ULK1 kinase at residue Ser757 (see Fig. 3 A). mTORC1 inhibits autophagy through its recruitment into the Atg1/ULK1–mAtg13–FIP200 autophagy initiation complex and subsequent Ser757 phosphorylation of ULK1 (Mizushima, 2010; Kim et al., 2011) and Ser52 phosphorylation of AMBRA1, which interferes with ULK1 function (Nazio et al., 2013). LPS stimulation of mTORC1, in addition to enhancing global protein synthesis (Lelouard et al., 2007), likely reduces ULK1 activity and consequently basal levels of autophagy in activated DCs.
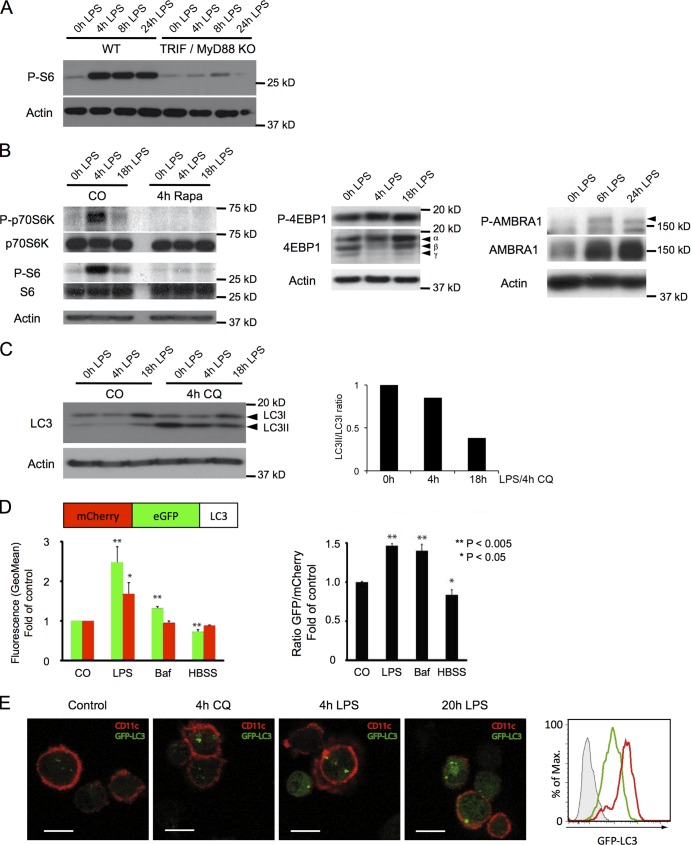
Figure 1.
The mTOR pathway is activated and autophagy is inhibited in DCs upon LPS exposure. (A) WT or MyD88- and TRIF-deficient (KO) DCs were stimulated with LPS as indicated, and S6 phosphorylation was analyzed by immunoblot. (B) DCs were stimulated with LPS and treated or not with rapamycin (4 h) as indicated. Cell lysates were analyzed by immunoblot for phosphorylation of p70-S6 kinase and S6 and total levels of p70, and S6 and actin served as controls (left). DCs were stimulated as indicated with LPS, and lysates were analyzed by immunoblot for phosphorylation of 4E-BP1 (Thr37/46; middle) and AMBRA1 (right; P-AMBRA1 is indicated by an arrowhead). Note the shift of 4-EBP1 from the nonphosphorylated γ form to the partially phosphorylated (hypophosphorylated) β and the hyperphosphorylated α forms (middle). (C) DCs were stimulated with LPS for the indicated time, and CQ was added for 4 h where specified. Cell lysates were analyzed by immunoblot for LC3 and actin (left). LC3I, nonlipidated form; LC3II, lipidated form. The relative LC3II accumulation was quantified using ImageJ (National Institutes of Health; right). The data shown are from a single representative experiment out of seven repeats. (D) DC maturation induces autophagy inhibition. DCs were transfected with mRNA encoding for mCherry-GFP-LC3 and incubated for 8 h in the presence of LPS or bafilomycin or starved in HBSS. GFP and mCherry fluorescence in CD11c+ cells was assessed by FACS (left). Plotted is the ratio of GFP fluorescence (fold over control) versus mCherry fluorescence (fold over control; right). Mean and SEM from three independent experiments and p-values (Student’s t test) are shown. (E) Autophagy inhibition is revealed by accumulation of GFP-LC3. DCs from GFP-LC3 transgenic mice were treated or not for 4 or 20 h with LPS or for 4 h with CQ, stained for CD11c, and analyzed for GFP fluorescence (left). B6 WT DCs (histogram filled with gray) or GFP-LC3 DCs were stimulated (red) or not (green) with LPS for 20 h, and GFP fluorescence was analyzed in CD11c+ cells by FACS (rightmost panel). The data shown are from a single representative experiment out of three repeats. Bars, 10 µm.
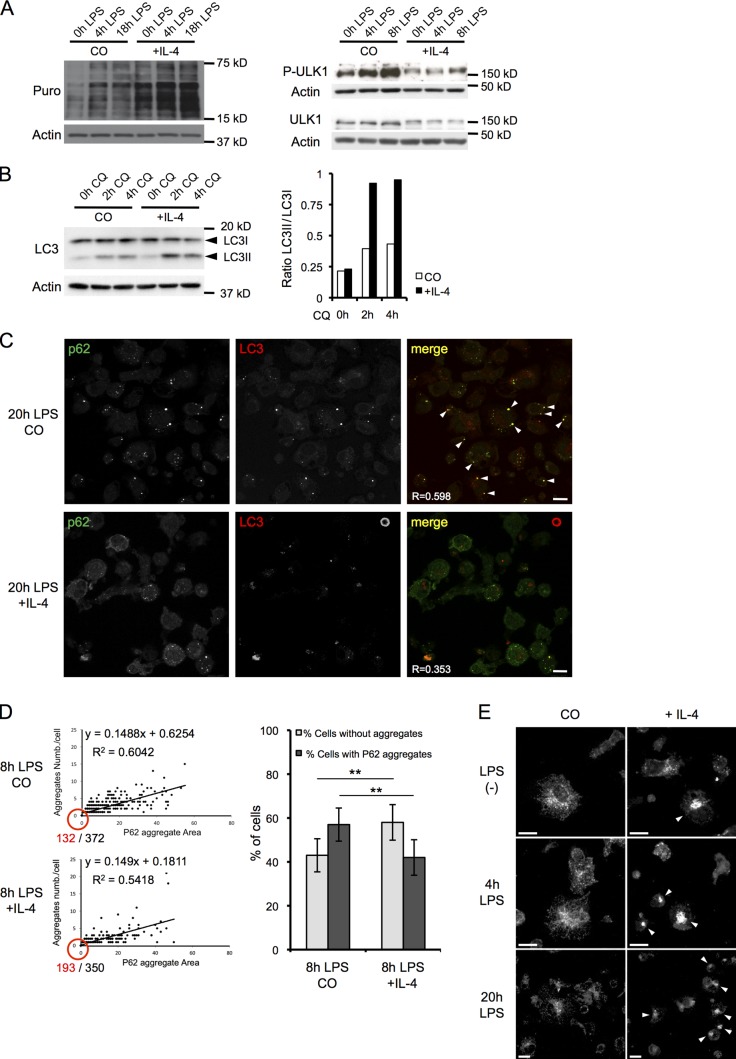
Figure 3.
IL-4 differentiation promotes autophagy and prevents DALIS formation in DCs. (A) IL-4 and control DCs were stimulated as indicated with LPS and were given 1 µg/ml puromycin for 10 min (left) before harvesting. Puromycin incorporation (left) or ULK-1 phosphorylation (right) was monitored by immunoblot analysis, representative of n = 3. (B) cDCs or IL-4–DCs were stimulated with LPS for 16 h, and CQ was added as indicated. Cell lysates were analyzed by immunoblot for LC3 and actin (left). The ratio of LC3II/LC3I was determined by densitometry analysis (right). The data shown are from a single representative experiment out of six repeats. (C) DCs generated in the presence or absence of IL-4 were activated with LPS for 20 h, and then p62 and LC3 localization was analyzed by ICM. p62 and LC3 overlaps in DALIS are indicated by arrowheads. R = Pearson’s correlation. (D) DCs generated in the presence or absence of IL-4 were activated with LPS for 8 h. The size of p62 aggregates (x axis) and the number of aggregates per cell (y axis) were quantified by automated video microscopy and plotted. The numbers of aggregate-negative cells (red) and total analyzed cell number (black) are shown at the origin of the plot (left). The proportion of cells with or without p62 aggregates is shown as bar graph (right). n = 350 and 372; mean and SEM from three independent experiments. **, P < 0.01. (E) DCs generated in the absence or presence of IL-4 were stimulated with LPS as indicated. LAMP2 localization, monitored by ICM analysis, revealed late endosome clustering in IL-4–DCs (arrowheads). (C and E) Bars, 10 µm.
TLR-dependent inhibition of autophagy was confirmed by comparing nonactivated immature DCs (iDCs), which accumulated the phosphatidylethanolamine-conjugated and autophagosome-associated LC3II isoform (Kabeya et al., 2004), with LPS-activated mature DCs (mDCs), in which the nonlipidated LC3I precursor was stabilized, denoting a reduced autophagy flux (Fig. 1 C). Inhibition of lysosomal proteolysis with chloroquine (CQ) promoted accumulation of LC3II in iDCs but not in mDCs, confirming the LPS-dependent inhibition of autophagic flux (Fig. 1 C). In transfected DCs expressing mCherry-eGFP tandem-tagged LC3, similarly to bafilomycin, LPS treatment increased the ratio of eGFP- versus mCherry-associated fluorescence, confirming a decrease in LC3 turnover (Fig. 1 D; Klionsky et al., 2012). In autophagy-active cells, GFP-LC3 fluorescence was rapidly quenched upon autophagosome-lysosome fusion (Klionsky et al., 2012), as observed by cytometry in iDCs expressing GFP-LC3 (Fig. 1 E), in which only few and weakly fluorescent GFP-positive structures were detected by microscopy in absence of CQ treatment. Conversely, upon LPS stimulation, a rapid accumulation of GFP-LC3 in organelles reminiscent of stalled autophagosomes was observed together with increased LC3-associated fluorescence levels (Fig. 1 E). Electron microscopy revealed the presence of partially fused autophagosomes with lysosomes containing undigested material in LPS-activated DCs, which is supportive of a globally reduced autophagic flux in mDCs (Fig. S1). Collectively, these results demonstrate that autophagy flux is reduced during DC activation and probably mediated by TLR-dependent mTORC1 activation.
Autophagy flux reduction leads to defective ribosomal protein (DRiP) accumulation in DALISs
Upon inhibition of autophagy in HeLa cells, neosynthetized DRiP processing is delayed, thereby promoting accumulation of these potential antigens in large ubiquitin-positive aggregates, termed ALISs (Wenger et al., 2012), together with the autophagy adaptors p62/SQTM1 and Nbr1 (Johansen and Lamark, 2011). We examined the distribution of p62 and NBR1 by confocal microscopy in mDCs and found that the two adaptors accumulated in DALISs, together with LC3, polyubiquitinated proteins (FK2), and puromycin-tagged DRiPs (puro-DRiPs; Fig. 2 A and Fig. S2 A; Lelouard et al., 2004). Autophagic flux inhibition in mDCs could therefore contribute to DRiP aggregation with p62 and LC3. Consistent with this hypothesis, the serine 403 phospho form of p62, which is increased upon autophagy inhibition and has enhanced affinity for polyubiquitinated proteins (Matsumoto et al., 2011), was found enriched in DALISs (Fig. S2 A). Pharmacological inhibition of autophagy using 3-methyl adenine (3-MA) strongly promoted DALIS formation in mDCs, whereas autophagy induction with the mTORC1 inhibitor rapamycin prevented p62 aggregation and DALIS assembly (Fig. 2 B). As DALIS formation requires active protein synthesis, we demonstrated that although the mRNA translation was reduced by 3-MA and rapamycin treatment, the drugs did not interfere with LPS-dependent induction of protein synthesis, which remained active throughout the time of drug exposure (Fig. 2 C), further suggesting that, in addition to increased protein synthesis, autophagy inhibition is absolutely key for DALIS formation. Polyubiquitinated protein aggregation and p62 expression have been linked to reactive oxygen species (ROS) expression and NRF-2 activation (Fujita et al., 2011); however, treatment of DCs with N-acetyl-l-cysteine (NAC) did not prevent DALIS formation in response to LPS (Fig. S2 B), indicating that ROS production is not required for TLR-driven aggregation of DRiPs with p62 in DCs. p62/Nbr1/ubiquitin-positive aggregates and DALISs are therefore identical structures, are also detectable in splenic CD11c+ DC subtypes purified from LPS-injected mice (Fig. S2 C), and are the likely manifestation of a reduced autophagic processing of neosynthetized proteins.
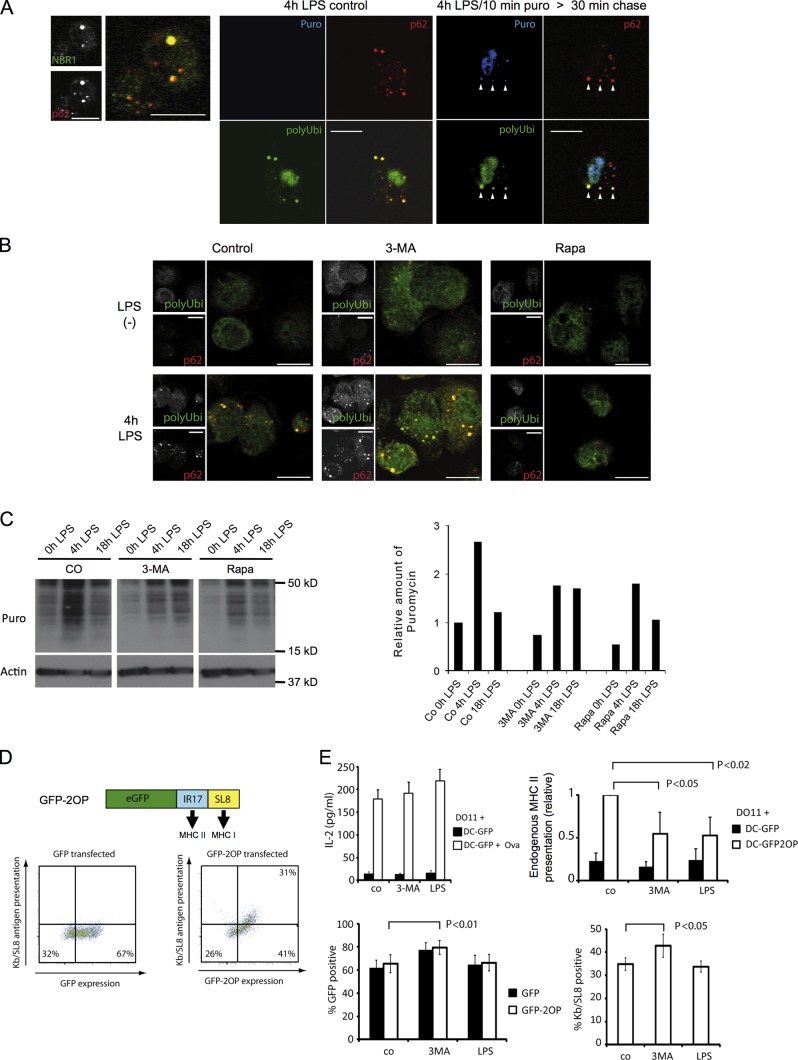
Figure 2.
Autophagy inhibition by DC activation leads to the aggregation of endogenous antigens in DALISs and down-regulation of MHC II presentation. (A) Immunofluorescence confocal microscopy (ICM) analysis for NBR1 and p62 was performed on LPS-stimulated DCs (left). LPS-stimulated DCs were given a 10-min pulse of 0.1 µg/µl puromycin (right) or left untreated (middle). 30 min later, cells were detergent extracted and fixed. Polyubiquitinylated proteins (polyUbi), p62, and puromycin-marked (Puro) proteins were analyzed by ICM. Arrowheads indicate the rapid localization of puromycin-labeled proteins with p62 in DALISs. (B) ICM analysis of polyubiquitinylated proteins and p62 was performed on LPS-stimulated (4 h) or control DCs, treated or not for 4 h with 3-MA or rapamycin. (A and B) Bars, 10 µm. (C) DCs were stimulated as indicated with LPS in the presence or absence of 3-MA or rapamycin and were given 1 µg/ml puromycin 10 min before harvesting and incorporation monitoring by immunoblot. Density quantification is shown on the right. The data shown are from a single representative experiment out of three repeats. (D) Schematic representation of the eGFP/double ovalbumin peptide (GFP-2OP) model antigen, containing the I-Ad–presented IR17 peptide (Ova 323–339) and the Kb-presented SL8 peptide (Ova 257–264). Representative FACS analysis (n = 4) of GFP fluorescence and Kb/SL8 antigen presentation (25D1 mAb) upon transfection of GFP or GFP-SL8 mRNA in CB6F1 DCs. (E, top left) GFP mRNA–transfected CB6F1 DCs were incubated with 0.3 µg/ml ovalbumin (delivered as immune complexes) alone or together with LPS or 3-MA for 8 h and used to stimulate DO.11 T cells. Mean and SEM from three independent experiments. (top right) CB6F1 DCs transfected for 8 h with eGFP or eGFP-SL8-OT2P mRNA were used to stimulate DO.11 T cells for 24 h. T cell activation was determined by IL-2 dosage. Mean and SEM from four experiments and p-values (Student’s t test) are shown. (bottom) CB6F1 DCs were transfected with GFP or with GFP-2OP mRNA and cultured for 8 h in the presence or absence of 3-MA or LPS, and antigen expression (% GFP positive) and Kb/SL8 surface appearance (% Kb/SL8 positive by 25D1 staining) were measured. Mean and SEM from three independent experiments.
Autophagy reduction in maturing DCs decreases endogenous MHC II presentation
Professional antigen-presenting cells use autophagy to process and load endogenous cytosolic antigens on MHC II (Schmid et al., 2007). A model antigen (GFP-2OP) was created by fusing eGFP together with two different ovalbumin-derived peptides, the I-Ad–restricted IR17 peptide (Ova 323–339) and the Kb-restricted SL8 peptide (Ova 257–264, SIINFEKL; Fig. 2 D). MHC I presentation of GFP-2OP is highly dependent on neosynthesis, misfolding, and rapid proteasome degradation (Wenger et al., 2012). Interestingly, inhibition of autophagy in HeLa cells promotes the accumulation of similar antigens in ALISs (Wenger et al., 2012) and delays their degradation, suggesting that in professional APCs, GFP-2OP will be processed and presented by MHC II molecules. GFP-2OP mRNA was electroporated in CB6F1 mouse DCs, resulting in strong antigen expression (GFP fluorescence) as well as MHC I Kb/SL8 presentation (Fig. 2 D). eGFP-expressing DCs exhibited a mature surface phenotype with high levels of CD86 and MHC II, which could be enhanced, along with IL-12 production, by LPS (Fig. S2, D and E). In transfected CD11C+ DCs, a portion of GFP was found aggregated together with p62 in DALISs, confirming the impact of DC activation on autophagic processing (Fig. S2 F). MHC II–restricted GFP-2OP presentation levels were quantified after IL-2 production by DO11.10 CD4+ T cells. Upon short-term autophagy inhibition by 3-MA or after LPS stimulation, a 50% reduction in IL-2 release by DO11.10 T cells was observed (Fig. 2 E), leaving both MHC I presentation of the same antigens and control MHC II presentation of exogenously added ovalbumin unaltered (Fig. 2 E).
IL-4 increases basal levels of autophagy in DCs
Circulating monocytes can give rise to DCs in steady state, but their contribution to the DC pool is by far greater under inflammatory conditions (Geissmann et al., 2010). The breadth and quality of the immune response is influenced by environmental cues, such as cytokine signals provided by neighboring tissues or cells. For example, in the context of allergic inflammation, exposure of macrophages or monocytes to the canonical Th2 cytokines IL-4 and IL-13 induces characteristic markers (M2 phenotype or alternative activation), along with changes in their metabolism, that enhance the synthesis of antiinflammatory factors (Van Dyken and Locksley, 2013). Human monocyte-derived DCs (MoDCs) have been shown to efficiently present LC3-associated antigens to CD4+ T cells with little influence of LPS-mediated activation (Schmid et al., 2007). Moreover, LPS-activated MoDCs display little co-aggregation of p62 together with polyubiquitination (FK1) in DALISs (Fig. S2 G), suggesting that MoDCs, conversely to mouse bmDCs, do not significantly reduce their autophagy flux in response to microbial activation. As the most relevant difference in the protocols used to differentiate MoDCs and bmDCs in vitro is the addition of IL-4 (Zhou and Tedder, 1996), we decided to compare autophagy and antigen presentation levels in mouse DCs differentiated in the presence of GM-CSF and IL-4, instead of GM-CSF alone (Wells et al., 2005).
As expected, CD11c+ DCs differentiated with GM-CSF and IL-4 (IL-4–DCs) exhibited slightly elevated CD86 and CD40 levels (Wells et al., 2005) but responded normally to LPS (Fig. S3 A). IL-4–DCs had a higher basal rate of protein synthesis, which was proportionally less augmented by LPS activation than in GM-CSF–DCs (Fig. 3 A). When phosphorylation of AMBRA1 and ULK1 was monitored, little Ser52 and Ser757 phosphorylation could be detected in LPS-activated IL-4–DCs (Fig. 3 A and Fig. S3 B). The same held true for rapamycin-sensitive rS6 phosphorylation (Fig. S3 B), indicating that, conversely to conventional GM-CSF–differentiated DCs, mTORC1 activation in response to TLR4 triggering is inhibited by IL-4 and constitutive autophagy levels are likely to be preserved in IL-4–differentiated DCs.
LC3 immunoblots indicated that autophagy flux was increased in IL-4–DCs (Fig. 3 B). When the fate LC3-binding proteins and DALISs were monitored by microscopy (Fig. 3 C) and quantified using automated imaging software (Fig. 3 D), diminished levels in numbers and size of p62 aggregates confirmed that autophagy was far more active in LPS-treated IL-4–DCs. By electron microscopy, IL-4–DCs displayed autophagosome populations with normal morphology, with a significant decrease in numbers of partially fused autophagosomes found in GM-CSF mDCs (Fig. S1). Activated IL-4–DCs also displayed an increase in the number of electron-dense late endosomal structures found in the cytosol. Immunofluorescence microscopy confirmed this observation and revealed that LAMP2-positive late endosome/lysosome pericentriolar clusters were increased in IL-4–DCs (Fig. 3 E), suggesting that both autophagosome and endosome dynamics are enhanced during differentiation with IL-4.
IL-4 facilitates the clearance of damaged mitochondria in mDCs
To avoid cellular damage, ROS-generating mitochondria are constantly removed by mitophagy, a specialized form of autophagy. We speculated that inhibition of mitophagy should lead to the accumulation of ROS-producing mitochondria upon LPS activation of DCs and a reduction of these same mitochondria in IL-4–DCs. We used mitochondria-specific fluorescent labels that distinguish respiring (MitoTracker deep red), total (MitoTracker green), and ROS-generating mitochondria (MitoSOX; Tal et al., 2009). Cytometry analysis (Fig. S3 C) showed that LPS exposure increased by 25% the number of cells containing damaged mitochondria and by 30% the number of cells producing mitochondrial ROS (Fig. S3 D). IL-4 exposure considerably decreased these proportions by augmenting damaged mitochondria clearance, as determined by the large cell fraction displaying reduced mitochondrial staining and the inhibition of this phenomenon in ATG5flx/flx-CD11C-cre DCs (Fig. S3 C; Lee et al., 2010). Interestingly, the difference between LPS-activated wild-type (WT) and ATG5−/− GM-CSF DCs was minimal, confirming the inhibitory effect of LPS on mitophagy in DCs. Therefore, LPS- and IL-4–mediated control of autophagy have important consequences on DC physiology, and IL-4 is also likely to exert its antiinflammatory role through enhanced mitophagy and an associated decrease in ROS production.
IL-4 differentiation facilitates T cell priming by endogenous antigen
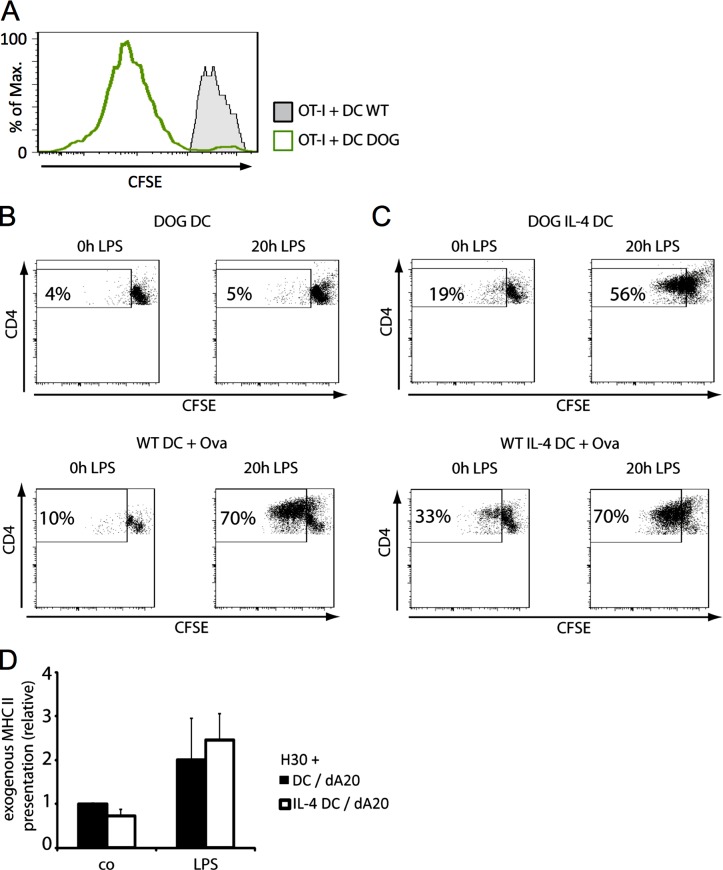
Upon IL-4–driven DC differentiation, a larger flow of endogenous antigens should be available for MHC II presentation. DCs, derived from DOG mice that express hDTR–Ova 140–386–eGFP, were used to present ovalbumin-derived peptides on H-2Kb to OT-I and on I-Ab to OT-II primary T cells (Hochweller et al., 2008). CFSE labeling dilution experiments indicated that OT-I CD8+ T cells were efficiently primed by DOG-DCs (Fig. 4 A), confirming the production of cytosolic ovalbumin peptides in these cells, which express this antigen in very low amounts difficult to visualize by other techniques (Hochweller et al., 2008). When MHC II presentation was investigated, GM-CSF DOG-DCs did not efficiently stimulate CFSE-labeled CD4+ OT-II cells (Fig. 4 B), independently of their activation state. Conversely, and despite low levels of antigen expression, immature IL-4–DOG-DCs weakly activated OT-II cells (Fig. 4 C), a phenomenon considerably enhanced by LPS stimulation and leading to CD4+ T cell divisions (Fig. 4 C). Exogenously added ovalbumin was only processed and presented efficiently upon LPS activation (Fig. 4 B), a situation also observed with Eα52-68–specific H30 T cell hybridomas, which failed to be activated by either type of iDCs fed with necrotic A20 B cells (Fig. 4 D).
Figure 4.
Endogenous MHC II presentation inefficiently primes primary T cells but is strongly enhanced in matured IL-4–DCs. (A) Immature WT or DOG DCs were used to stimulate OT-I T cells. CFSE dilution was assayed after 3 d of co-culture (n = 3). (B and C) Immature or mature DOG DCs treated with (C) or without IL-4 (B) were used to stimulate OT-II cells. CFSE dilution in CD4+ T cells was assayed after 3 d of co-culture (top). WT DCs incubated with 10 µg/ml ovalbumin treated with or without IL-4 were used to stimulate OT-II cells as a control of exogenous antigen presentation (bottom). The data shown are from a single representative experiment out of five repeats. (D) iDCs or mDCs generated in the presence or absence of IL-4 were harvested and incubated with freeze-killed A20 (dA20) cells at a 1:4 ratio. After 8 h, the DCs were used to stimulate H30 T cells, and TCR stimulation was evaluated by IL-2 production from H30 cells. Mean and SEM from four independent experiments are shown.
Short-term exposure to IL-4 does not impact autophagy in DCs
Short IL-4 exposure (2 h) inhibits starvation-induced autophagy in Raw and primary mouse macrophages (Harris et al., 2007). We therefore investigated whether the potentiation of autophagy in IL-4–DCs was the result of long-term differentiation and potentially cell specific. DCs and macrophages were generated in parallel cultures of mouse bone marrow in the presence of GM-CSF or M-CSF, to which IL-4 was added for 5 d or 2 h before LC3 processing analysis (Fig. S4 A). Interestingly, long-term IL-4 treatment enhanced autophagy in DCs much more than in macrophages, whereas short-term exposure had little effect on DCs and, as suggested previously, was moderately inhibitory for macrophages (Harris et al., 2007). Similar experiments performed on RAW macrophages indicated that IL-4 inhibits minimally autophagic flux after short-term exposure and had no effect after 24 h of treatment. Thus, increased autophagy in IL-4–DCs is dependent on both long-term differentiation with IL-4 and cell culture specificity because DCs and macrophages differentiated in vitro have remarkably distinct behaviors.
Transcriptomics analysis of DCs differentiated with IL-4
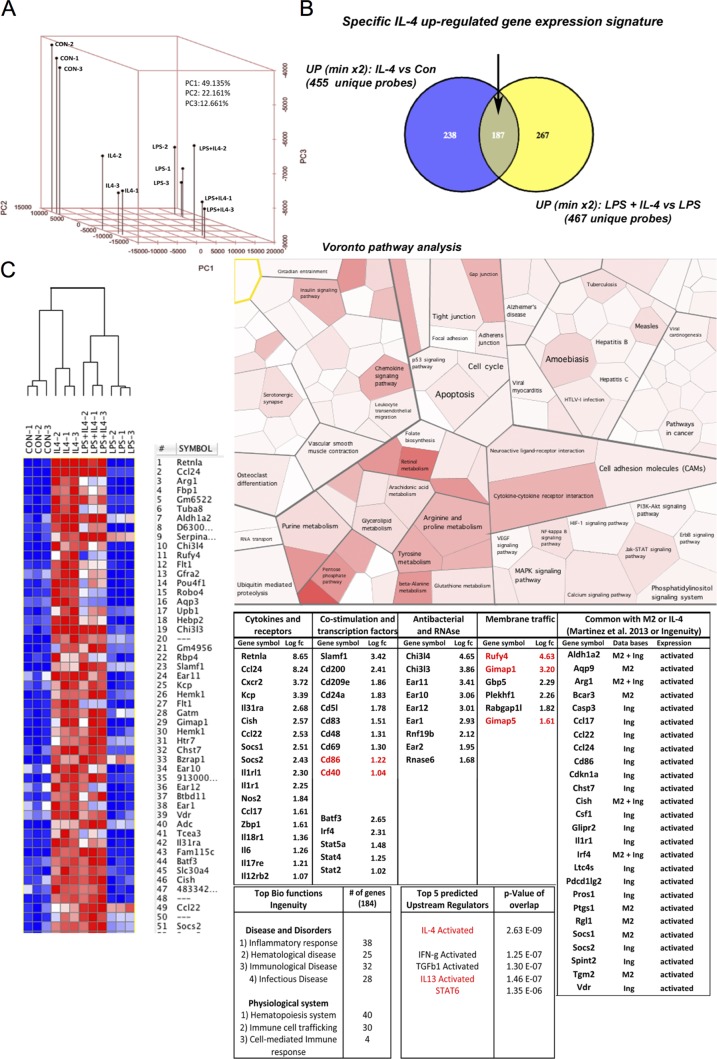
We initiated a comparative microarray-based genomic analysis of purified GM-CSF bm-CD11+ DCs differentiated or not in the presence of IL-4 to identify a transcriptional program capable of enhancing endosomes and/or autophagosomes dynamics. Principal component (PC) analysis (PCA) showed a close proximity of replicates (Fig. 5 A) with a clear partitioning of each experimental condition tested. PC1, corresponding to ‘‘LPS versus nonactivated (con),’’ represented 49% of the overall variability between microarrays. PC2, corresponding to ‘‘IL-4 treatment,’’ accounted for 22%, suggesting that LPS activation dominated the overall DC differential gene expression program. However, IL-4 treatment induced a specific response only moderately affected by LPS. Differentially expressed genes (DEGs) were selected on statistical significance (P < 0.1) and a fold change above 2 for up-regulated genes (Fig. 5 B) and below 0.33 for down-regulated genes (Fig. S5). After clustering and heat map representation of the top regulated genes, resistin-like-α, Ccl24, and arginase 1 were found to be the most up-regulated by IL-4, as previously described in macrophages (Van Dyken and Locksley, 2013). Venn diagrams showed that out of all probes tested, 187 DEGs defined a core IL-4 differential gene expression signature (Table S1). DEGs were submitted to two different pathway analyses, Voronto and Ingenuity Pathway Analysis (IPA) softwares. Voronto uses tessellation Voronoi diagrams to integrate expression data and hierarchical ontologies extracted from public databases (Santamaría and Pierre, 2012). The most significant pathways induced by IL-4 were linked to amino acid and lipid metabolisms as well as chemokine signaling. IPA identified the IL-4 signature list as constituted of immunologically relevant genes because top pathways were identified as inflammatory or immunological diseases. IL-4 was identified as the top predicted upstream regulator followed closely by IL-13 and STAT6, confirming the pertinence of our analysis. This prediction was mostly influenced by genes representing the hallmark of alternative versus classical macrophage activation, including Arginase 1 and anti–nematode and -fungal resistance genes such as chitinases (e.g., Chi3l4) and RNases (e.g., Ear11). When immunologically relevant genes were examined, mRNA levels of chemokines CCL24, CCL22, and IL-6 were all significantly augmented by IL-4, whereas levels of CD86 and CD40 were moderately up-regulated, as anticipated from cytometry (Fig. S3). As IL-4 shifts cellular resources away from the generation of molecules associated with proinflammatory cytokine and nitric oxide (NO) production, we extracted by Venn diagrams a list of 112 genes (common; Fig. S5 B) that were significantly down-regulated by IL-4 in all conditions tested. A majority of these DEGs belonged to immunological-relevant pathways (Fig. S5 C, yellow delineation), among which cytokines, cytokine receptors, and innate receptors were overrepresented. IL-1β and TNF, as well as TLR 1, 2, 6, and 7 and CD14 were down-modulated by IL-4 differentiation, confirming that IL-4 induces a bias in the capacity of DCs to sense microbes and promote inflammation. In agreement with these observations, upstream regulators of majorly IL-4–down-regulated genes included TNF, IL-6, and IL-1β.
Figure 5.
In silico analysis of IL-4–driven genes in mouse bmDCs. (A) PCA from multiple samples of DCs treated or untreated with IL-4 and/or LPS. (B) Venn diagram representation of genes up-regulated by IL-4 stimulation more than twofold. (C) Clustered heat map representation (left) and Voronto pathway analysis (right) of IL-4–driven genes are shown. (bottom right) IPA analysis of the top IL-4–induced genes organized by immune-relevant functions (fold change is indicated as Log Fc and genes mentioned in this paper are indicated in red), top biological functions, and upstream regulators are shown with the number of genes involved and associated p-value. A list of IL-4–induced genes found in common with previous studies from IPA (Ing) or Martinez et al. (2013) (M2) is presented on the right.
Up-regulated DEGs were compared with previously curated lists of IL-4–stimulated genes to identify novel IL-4–regulated genes. We restricted the comparison to the IPA and to a recently published analysis of the genes defining the core of alternatively activated mouse and human macrophages (Martinez et al., 2013). We could only find 26 genes from our list that were present in either of these two databases (Fig. 5 C), confirming that despite the ontogenic proximity of macrophages and GM-CSF–derived DCs, IL-4 induces a great heterogeneity in their respective gene expression program. When the IL-4–DC signature was compared with available monocyte/phagocyte gene signatures (Gautier et al., 2012), several genes defining the macrophage core signature were found to be down-regulated by IL-4, including CD64/FCGRI, emr1/F4/80, CD14, FCgRIII, Camk1, ARS, MR1, and the important macrophage differentiation transcription factor MAFB, suggesting that IL-4 augments GM-CSF–dependent differentiation of monocytes into DCs, as suggested by up-regulation of the Batf3 and IRF4 transcription factors, which are involved in DC development, as well as inflammation (Murphy et al., 2013). In contrast, 14 DEGs specific for lung macrophages (out of 108 genes Gautier et al., 2012) were found up-regulated in IL-4–DCs (429 genes), including Rufy4, Net1, Ear2, Ear1, Ear10, Cldn1, Kynu, Chi3l3, Flt1, Cd69, Olr1, Siglec5, Cdh1, and Mtmr7, suggesting the existence of a lung-specific IL-4–dependent signature in these macrophages.
Our analysis was primarily aimed at identifying molecules controlling endocytosis and autophagy in DCs. Among the small number of up-regulated genes (six) annotated to membrane traffic ontology, Rufy4, Gimap1, and Gimap5 stood out by their protein structural organization or anticipated function. The GTPases of the immunity-associated protein (GIMAP) family of putative GTPases function as nucleotide-regulated scaffolds on intracellular membranes (Saunders et al., 2010). RUFY4 is predicted to have a FYVE domain in its C-terminal portion (Fig. 6 A and Fig. S5 D) that could mediate interaction with PtdIns(3)P, separated by two coiled coil domains from its N-terminal RUN domain (Fig. 6 A). RUFY4 is thus a particularly interesting candidate for our study because RUFY proteins have been shown to influence endosomal spatial distribution (Ivan et al., 2012) and the closest RUFY4 paralogue, FYCO1 (Fig. S5 D), regulates autophagosome motility (Pankiv et al., 2010).
Figure 6.
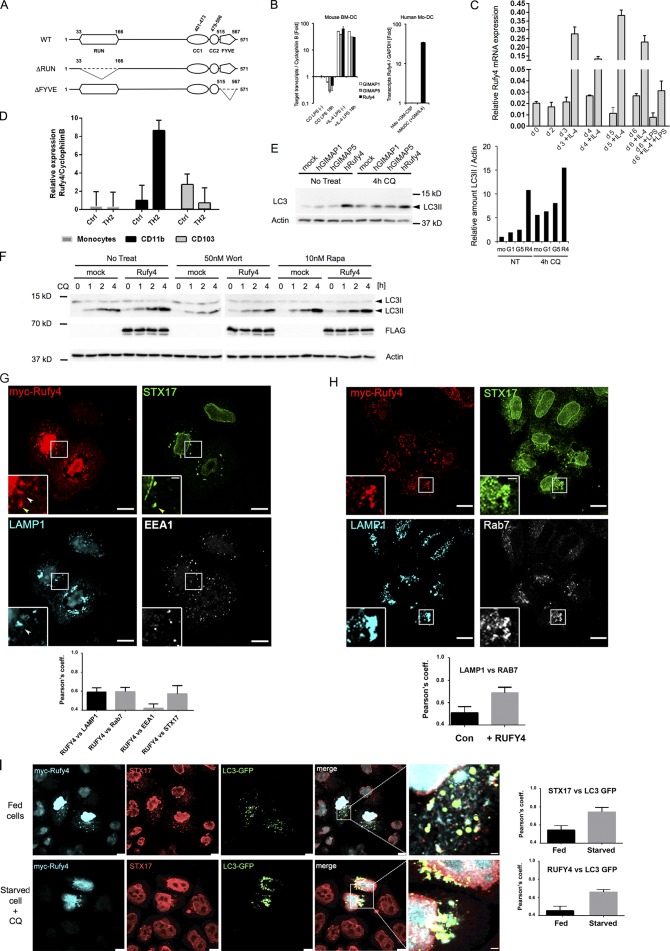
IL-4 drives the expression of Rufy4, a novel enhancer of autophagy. (A) Schematic representation of WT and RUN domain–less (ΔRUN) or FYVE domain–less (ΔFYVE) mutant of human Rufy4. Putative RUN/FYVE domains and coiled coil (CC) domains were calculated by using online prediction tools PROSITE and COILS server, respectively. (B) Induction of mouse GIMAP1, GIMAP5, and Rufy4 gene expression in IL-4–treated DCs was confirmed by qPCR (left). Rufy4 induction was also observed in human MoDCs, which was developed in the presence of GM-CSF and IL-4 (right). Mean and SEM from three independent experiments. (C) Time course of Rufy4 mRNA levels in mouse DCs generated in the presence or absence of IL-4. Rufy4 mRNA was monitored by qPCR at different days of culture (from day 0 to 6). The medium was replaced with fresh medium containing IL-4 on days 2 and 4. LPS was added on day 5 for 16 h. (D) Comparative qPCR for RUFY4 mRNA expression in different subsets of mouse lung DCs isolated from control and HDM-exposed animals. (C and D) The graphs show means ± SEM of triplicate determinations from one representative experiment. (E) HeLa cells were transfected with human GIMAP1, GIMAP5, or Rufy4 expression vector for 24 h in duplicate before treatment with 50 µM CQ for the last 4 h. Cell lysates were immunoblotted for LC3 and actin. One representative blot is shown (left; n = 3). Relative LC3 accumulation (LC3II/actin) was determined by densitometry analysis (right). (F) HEK293 cells were transfected with Flag-tagged human Rufy4 construct for 24 h in the absence or presence of 50 nM wortmannin or 10 nM rapamycin for the last 4 h. To monitor the time course of LC3II accumulation, 50 µM CQ was added for each group as indicated. (G and H) HeLa cells were transfected to express WT myc-hRufy4 (24 h). After immunostaining for myc tag and the indicated antigens, the cells were analyzed by ICM. Insets are magnifications of boxed representative colocalization areas. Arrowheads indicate accumulation of RUFY4-positive organelles either codistributing with Stx17 (yellow) or LAMP1 (white). Pearson’s coefficients (significant over 0.5) of colocalization among the indicated markers are plotted below the images. (I) HeLa cells were cotransfected with WT myc-hRufy4 and GFP-LC3 constructs (24 h), left untreated (top) or starved and treated with 50 µM CQ for the last 2 h (bottom). After pre-extraction in saponin, cells were fixed, permeabilized, and immunostained for myc tag and Stx17. Panels with enlarged views of the boxed regions in merged images are shown at the right. Pearson’s coefficients (significant over 0.5) of colocalization among indicated markers are plotted next to the images. Mean and SEM from three experiments, n = 25. Bars: (G and I) 10 µm; (H) 25 µm.
RUFY4 is induced in DCs differentiated in the presence of IL-4 both in vitro and in vivo
We next confirmed by quantitative PCR (qPCR) that Rufy4, Gimap1, and Gimap5 expression was induced by IL-4 in both mouse bmDCs and human MoDCs (Fig. 6 B). RUFY4 mRNA was induced 15- to 30-fold within the first days of IL-4 addition to the differentiation media (Fig. 6 C), whereas LPS treatment moderately decreased its expression. Given the difference in autophagy levels among DCs and macrophages differentiated in the presence of IL-4, we monitored RUFY4 expression in these two cell types as well as in RAW macrophages grown in the presence of IL-4 (Fig. S4 B). Strikingly, RUFY4 mRNA could only be detected in IL-4–DCs and not in macrophages, nor after short-term IL-4 exposure, suggesting that RUFY4 expression, which coincides with increased autophagy, is linked to both long-term IL-4 exposure and GM-CSF–induced differentiation.
Given this information, we explored in which physiological situations RUFY4 expression could be detected in DCs. In addition to bridging innate and adaptive immunity, lung DCs can also mount Th2 immune responses to harmless allergens, such as house dust mite (HDM) allergen and, in this way, contribute to immunopathology (Plantinga et al., 2013). Among DC subsets, CD11b+ conventional DCs (cDCs) and MoDCs were identified as important players in the allergic cascade, with CD11b+ cDCs being the principal subset inducing Th2 cell–mediated immunity characterized by the production of large quantities of IL-4 (Plantinga et al., 2013). Repeated HDM allergen inhalations by mice, followed by lung DC subset isolation, were performed to analyze RUFY4 expression. Comparative qPCR performed on the different DC subsets isolated from control or allergic mice demonstrated that RUFY4 mRNA expression was strongly augmented in CD11b+ cDCs of HDM-stimulated lungs (Fig. 6 D), whereas lower expression was detected in the CD103+ DC subset and allergic conditions did not augment RUFY4 mRNA expression in this cell type. Thus, in vivo, high RUFY4 expression correlates with Th2-related lung inflammation in the CD11b+ DC subset, confirming its potential relevance for DC-mediated physiopathology.
RUFY4, a novel positive regulator of autophagy
Ectopic expression of tagged RUFY4, GIMAP1, and GIMAP5 (Fig. 6 E) was performed before LC3 monitoring in cells treated with CQ for 4 h. RUFY4 expression promoted LC3II accumulation, contrasting with GIMAP1 and 5, which did not affect LC3 fate (Fig. 6 E and Fig. 7 A). We thus focused on RUFY4 expression and associated LC3 degradation by exposing transfected HEK293 cells to autophagy-interfering drugs (Fig. 6 F). Wortmannin treatment clearly inhibited LC3 processing and antagonized RUFY4 activity, suggesting that the putative PtdIns(3)P-binding FYVE domain of RUFY4 is functional. We generated a RUN-less and a FYVE-less form of RUFY4 (Fig. 6 A) to evaluate the importance of these domains (Fig. 7, A and B). Enhanced LC3II accumulation was observed upon RUFY4 expression but not with the RUN-less or the FYVE-less mutants, suggesting that both domains are necessary to enhance autophagy.
Figure 7.
RUFY4 and mutant expression in HeLa cells. (A) HEK293 cells were transfected with myc-tagged WT or ΔRUN and ΔFYVE mutant constructs for 24 h and treated with CQ as indicated. An empty plasmid (mock) was used as a negative control. Cell lysates were analyzed by immunoblot for LC3, myc tag, and actin. (B) Graph shows LC3II quantification over actin, mean ± SEM of three independent experiments. Student’s t test: *, P < 0.05; **, P < 0.01. (C–E) HeLa cells were transfected with myc-tagged WT, ΔRUN, or ΔFYVE mutant constructs for 24 h. Cells were subjected to pre-extraction with saponin, before fixation and staining for myc, LAMP1, and STX17 (D), EEA1 (C), or Rab7 (E). Representative confocal images are shown. Pearson’s coefficients (significant over 0.5) of colocalization among the indicated markers are plotted next to the images. Mean and SEM from three experiments, n = 25. Bars: (WT Rufy4) 10 µm; (ΔRUN and ΔFYVE) 25 µm. (D) Bar graphs indicate percentage of cells containing enlarged Stx17+ organelles among cells expressing (myc+) or not expressing (myc−) the exogenous Rufy4 proteins (n = 70). (F) Total lysate and immunoprecipitates of Myc-RUFY4 and mutants were submitted to immunoblot for myc, Rab7, and Rab9 detection. Rab7 was found to coimmunoprecipitate only with myc-Rufy4 but not ΔRUN and ΔFYVE mutants.
Ectopically expressed RUFY4 induced the aggregation of large membranous organelles, containing RUFY4 itself, and were either strongly positive for Stx17 (Fig. 6 G), the autophagosomal SNARE required for fusion with endosome/lysosome (Itakura et al., 2012; Takáts et al., 2013), or for LAMP1. EEA1-positive early endosomes remained unchanged by RUFY4 expression, which promoted the recruitment of Rab7 on LAMP1-positive endosomes and their clustering together with Stx17-containing organelles (Fig. 6 H). Cotransfection of RUFY4 and LC3 confirmed that RUFY4-induced Stx17-positive organelles are autophagosomes that actively recruit LC3, a phenomenon strongly amplified by starvation and CQ treatment (Fig. 6 I). The functionality of the RUN and FYVE domains present in RUFY4 was examined by microscopy. In the absence of the RUN domain, little difference was found with control cells, confirming our biochemical data and indicating that the RUN domain is absolutely critical for RUFY4 function (Fig. 7, C and D). The FYVE-less mutant did not localize to autophagosomes, nor promote Stx17 and Rab7 recruitment, but still induced perinuclear localization of LAMP1-positive endosomes (Fig. 7, C and D). In many occurrences, high levels of FYVE-less expression also promoted the collapse of large tubular and vesicular membrane structures (Fig. 7 E), with no apparent codistribution with LAMP1 and RAB7.
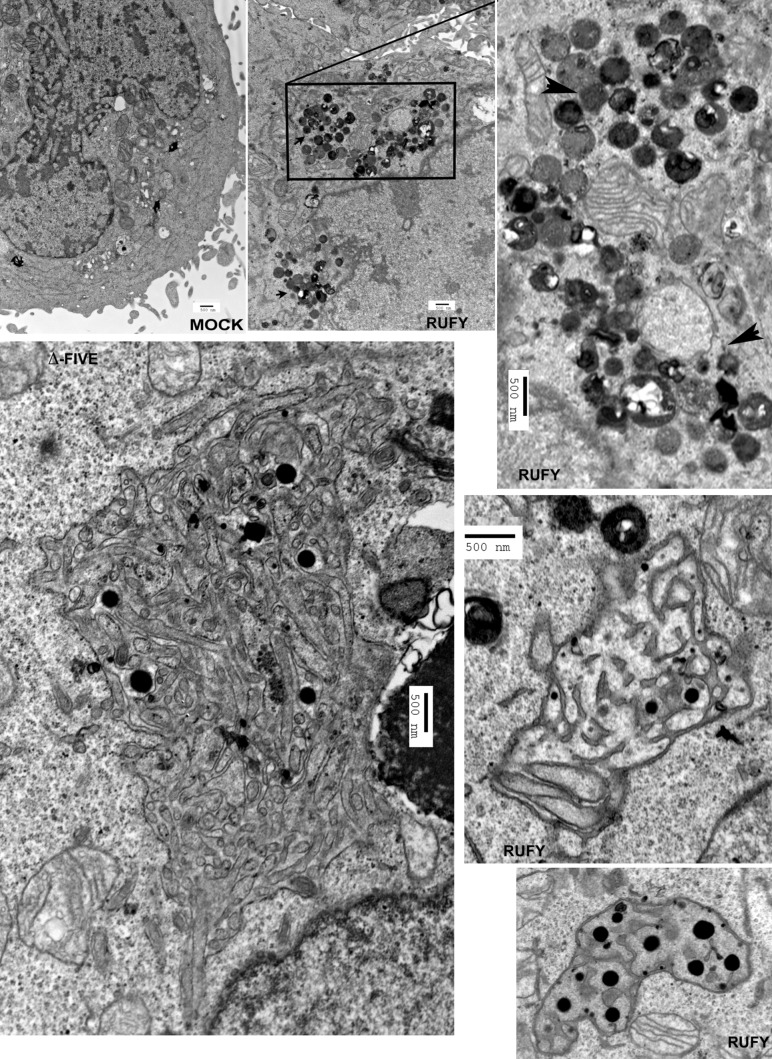
Given the active recruitment of RAB7 to RUFY4-positive organelles upon transfection, we investigated whether these two molecules could interact. Immunoprecipitation of transfected myc-tagged RUFY4 followed by immunoblot detection demonstrated that RUFY4 is able to interact biochemically with Rab7 but not Rab9 (Fig. 7 F). In agreement with confocal microscopy, interaction with Rab7 was not detected with the functional mutants, confirming that RUFY4 mediates part, but not all, of its effects through RAB7. By electron microscopy, lysosome and late endosome clustering in response to RUFY4 expression was detected in areas close to the nucleus (Fig. 8). These electron-dense organelles were found in juxtaposition to enlarged autophagosome-like organelles (Fig. 8), which were formed of an apparently continuous reticular network of double membranes carrying ribosomes on their cytosolic side and sometimes round electron-dense structures internally, possibly representing ferritin granules (Kishi-Itakura et al., 2014). The formation of these structures reached a paroxysm in FYVE-less mutant–transfected cells, in which giant reticulated “autophagosome-like” structures likely to be formed from ER membranes and containing different types of organelles and cytosolic material could be observed (Fig. 8). Thus, RUFY4 exerts its function by regulating the membrane dynamic of autophagosome growth, probably in liaison with Rab7 and other Rab activity. Moreover, RUFY4 also promotes the clustering and tethering of lysosomes, which could facilitate fusion with autophagosomes. Interestingly, this activity was not affected by nocodazole treatment (Fig. S4 D), suggesting that, differently from FYCO1, RUFY4 functions independently of the microtubule network and its associated molecular motors.
Figure 8.
Electron microscopy analysis of Rufy4-overexpressing HeLa cells. HeLa cells were transfected with human Rufy4 or ΔFYVE Rufy4 construct or with mock plasmid as control and were fixed with glutaraldehyde. Subcellular structures were analyzed by conventional electron microscopy. Boxed area is magnified on the right. Arrowheads indicate zones of lysosome clustering and tethering. Bars, 500 nm.
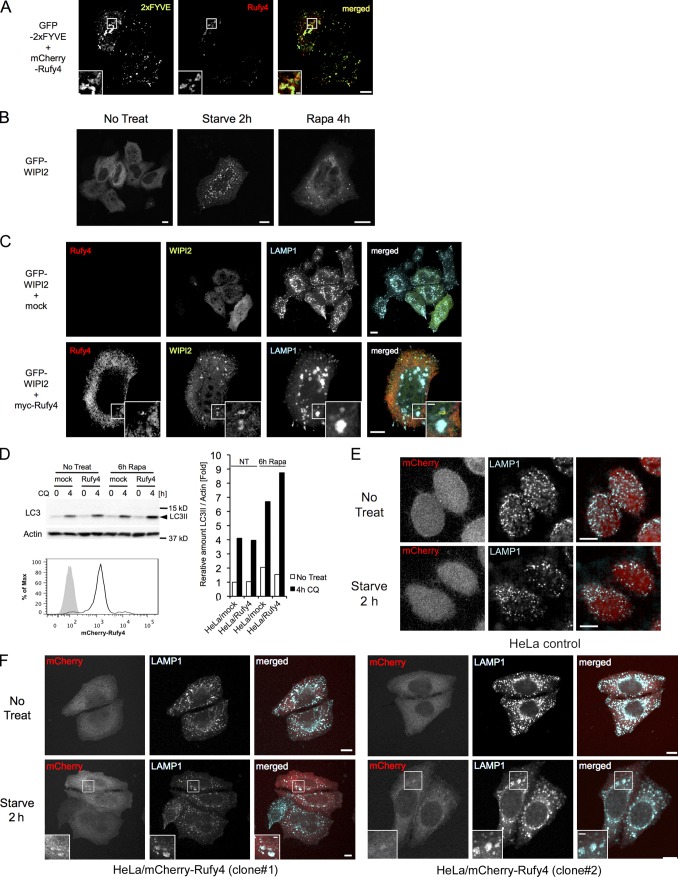
Production of PtdIns(3)P by the kinase Vps34 is key for autophagosome formation and recruitment of FYVE domain–bearing effectors, including Rubicon, DFcP1, and ALFY (Stein et al., 2003; Simonsen et al., 2004). The association of RUFY4 with PtdIns(3)P-containing organelles was monitored by confocal microscopy through its codistribution with overexpressed GFP-2×FYVE, which specifically associates with PtdIns(3)P-enriched domains (Simonsen et al., 2004). In cells coexpressing the two proteins, a strong degree of colocalization could be observed in most enlarged organelles (Fig. 9 A), confirming the enrichment in PtdIns(3)P of RUFY4-containing autophagosomes and late endosomes in vivo. WIPI2, an orthologue of ATG18, is an effector of PtdIns(3)P and is recruited to early autophagosomal structures to promote the formation of LC3-positive autophagosomes (Fig. 9 B; Polson et al., 2010). Interestingly, like starvation, RUFY4 expression induced WIPI2 punctae, which were found in direct vicinity with RUFY4-containing endo/lysosomes (Fig. 9 C), confirming that RUFY4 promotes autophagosome formation.
Figure 9.
Rufy4 colocalizes with PtdIns(3)P-enriched domains and promotes the formation of WIPI2 punctae. (A) HeLa cells were cotransfected with GFP-2×FYVE and mCherry-tagged human Rufy4 constructs for 24 h. The cells were fixed without permeabilization, and the cellular localization of these fluorescent proteins was monitored by confocal microscopy. (B) HeLa cells were transfected with GFP-tagged WIPI2 and monitored by confocal microscope. Punctate WIPI2 formation was observed upon autophagy induction by either starvation or 10 nM rapamycin treatment. (C) HeLa cells were cotransfected with GFP-tagged WIPI2 and control (top) or myc-tagged Rufy4 (bottom) constructs for 24 h. The cells were permeabilized with saponin-containing buffer and then stained for LAMP1. Representative ICM images are shown. (D) mCherry-tagged human Rufy4 constructs were stably transfected to HeLa cells. Bulk HeLa/mCherry-Rufy4 lines express moderate amounts of Rufy4, as shown by FACS analysis (bottom). mCherry-Rufy4–expressing cells were cultured in the absence or presence of 10 nM rapamycin for 6 h before monitoring LC3II accumulation; 50 µM CQ was added as indicated. The cell lysates were immunoblotted with LC3 and actin antibodies (top left). Relative accumulation of the LC3II form was determined over actin levels by densitometry analysis (right). A single representative experiment is presented out of three repeats. (E and F) Immunofluorescence microscopy shows that after a short 2-h starvation in HBSS, accumulation of enlarged LAMP1-positive structures is promoted in two independent stably transfected clones expressing low levels of mCherry-Rufy4 (F) but not in the mock-transfected control cells (E). (A, C, and F) Insets are magnifications of boxed representative colocalization areas. Bars, 10 µm.
Physiological RUFY4 expression together with appropriate intracellular signaling, such as mTOR inhibition, could thus considerably enhance the rate of autophagy flux, as observed in IL-4–DCs. To verify this hypothesis, we established stable HeLa cell lines expressing near physiological levels of a Cherry-tagged form of RUFY4. At steady state, the RUFY4-expressing line displayed no obvious difference in LC3 processing nor endo/lysosome dynamic compared with control cells (Fig. 9 D). However, upon rapamycin treatment or a short 2-h starvation period, enhanced LC3II processing and accumulation of enlarged LAMP1-positive structure were observed in the presence of moderate levels of mCherry-RUFY4 (Fig. 9, D–F). Thus, similarly to IL-4–treated DCs, alteration of mTOR signaling together with enhanced RUFY4 expression considerably increased autophagy and the endo/lysosomes dynamic.
RUFY4 silencing prevents Stx17 association with autophagosomes
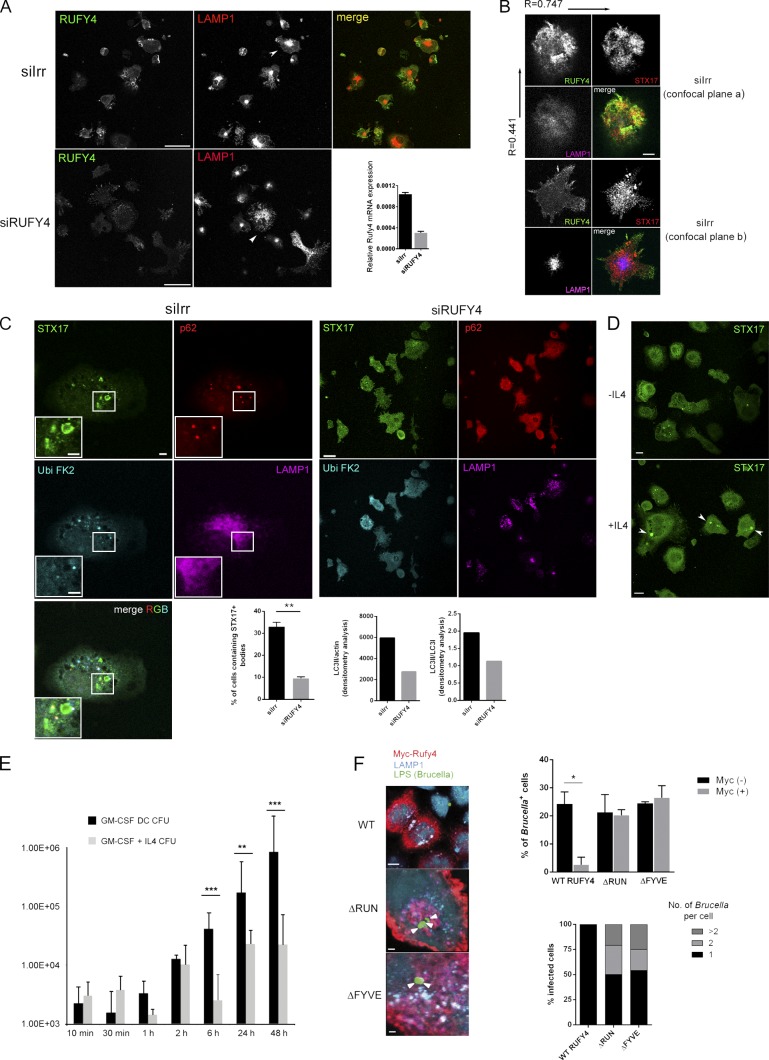
The impact of RUFY4 depletion in IL-4–DCs was monitored by examining the distribution of different autophagy markers and the processing of LC3 in the cells silenced for RUFY4 (Fig. 10). RUFY4 extinction was 70% complete 24 h after the introduction of relevant siRNA by nucleoporation, as established by qPCR and immunofluorescence (Fig. 10 A). RUFY4 is distributed in different subcellular locations, both in the cortical periphery of the cells and in more central locations, where it displayed a significant overlap with membrane-associated Stx17 (Pearson’s coefficient r = 0.75 in confocal plane a) but not LAMP1 (r = 0.42; Fig. 10 B). Upon Rufy4 silencing, late endosomes and lysosomes were more dispersed throughout the cell than their counterparts in IL-4–DCs (Fig. 10 A, arrowheads), whereas in the minority of activated control siRNA-treated IL-4–DCs displaying DALISs, Stx17 was found associated with P62 and polyubiquitin (FK2) in punctate structures reminiscent of autophagosomes but distinct from LAMP1-positive vesicles (Fig. 10 C). Stx17 association in clusters was only observed in IL-4–treated DCs, suggesting that enhanced autophagosome formation driven by IL-4 and RUFY4 promotes a strong membrane recruitment of Stx17 (Fig. 10 D, arrowheads). This hypothesis was confirmed upon Rufy4 silencing, during which Stx17 association with membranes and punctae was lost, and its distribution appeared to be cytosolic (Fig. 10 C). LC3II/actin and LC3II/LC3I ratios were both decreased by Rufy4 silencing (Fig. 10 C), confirming the importance of RUFY4 to maintain active autophagy in IL-4–DCs. These observations are completely in line with the results obtained in transfected HeLa cells and suggest further that RUFY4 acts together with STX17 as a positive regulator of autophagy and late endosome clustering.
Figure 10.
RUFY4 silencing influences STX17 distribution and inhibits autophagy and B. abortus replication. (A) IL-4–DCs treated with control (siIrr, top) or Rufy4-specific siRNA (siRUFY4, bottom) were grown in the presence of IL-4 and immunostained with anti-Rufy4 and anti-LAMP1 antibodies. Representative single confocal planes are shown. Late endosome clustering is indicated by arrowheads. The bar graph shows Rufy4 mRNA levels in control and Rufy4-silenced cells quantified by qPCR (means ± SEM of triplicate determinations from one representative experiment). (B) Control or Rufy4-silenced DCs were immunostained for Rufy4, LAMP1, and STX17. Two confocal imaging planes of the same cell are shown. (C) siIrr (left) and siRUFY4 (right) DCs were immunostained for STX17, p62, FK2, and LAMP1. Representative panels with enlarged views of the boxed region are shown. (bottom left) Graphical representation of the percentage of cells containing STX17-positive organelles in siRNA-treated DCs is shown. Bars show mean ± SEM of two independent experiments (n > 50); Student’s t test: **, P < 0.01. (bottom right) Quantitative representation of LC3II/actin and LC3II/LCI protein ratios detected by immunoblot in extracts from the same siRNA-treated cells processed for immunofluorescence (one representative experiment, n = 3). (D) Confocal immunofluorescence of bmDCs generated in the presence or absence of IL-4 and stained for STX17, which accumulates in large punctate structures (arrowheads). (E) bmDCs were grown in the presence or absence of IL-4 and infected for different time points with B. abortus. Bar graph indicates bacteria replication assessed by CFUs; Student’s t test: **, P < 0.01; ***, P < 0.001. Error bars indicate SEM (n > 3). (F) HeLa cells were transfected with WT myc-hRufy4 or the myc-tagged mutants ΔRUN and ΔFYVE (24 h) and infected with B. abortus for 3 h. Confocal microscopy for B. abortus LPS (arrowheads), myc, and LAMP1 was performed (left) and quantified for infection levels (right). Graphs show the percentage of cells containing B. abortus among cells expressing (myc+) or not (myc−) RUFY4 constructs (top) and mean number of bacteria per cell (bottom). B. abortus–containing cells were assigned to one of the following categories: cells containing one bacterium, cells containing two bacteria, or cells containing more than two bacteria. n = 50 from two independent experiments; Student’s t test: *, P < 0.05. Error bars indicate SEM. Bars: (A and D) 25 µm; (B) 5 µm; (C) 7.5 µm; (F, WT) 10 µm; (F, mutants) 2 µm.
RUFY4 expression prevents B. abortus replication
Brucellosis is a zoonotic disease that results from direct contact with infected animals or ingestion of contaminated food products. B. abortus virulence is dependent on its ability to survive and replicate within host cells. Once internalized, B. abortus is found within a compartment, the B. abortus–containing vacuole (BCV) that transiently interacts with early endosomes (Salcedo et al., 2008). However, BCVs do not normally fuse with lysosomes and instead fuse with membranes of the ER to establish a vacuole suited for its replication. At late stages of the B. abortus intracellular cycle, a post-ER replication compartment is initiated at the ER and converts BCVs into vacuoles with autophagic features. Importantly, BCV conversion into these vacuoles completes the intracellular cycle of B. abortus and promotes cell to cell spread. Macrophages and placental trophoblasts are two key targets of B. abortus infection, but we have shown that HeLa cells and more importantly mouse DCs also support the replication of this pathogen in vitro (Salcedo et al., 2008). B. abortus is therefore ideal to test whether increased classical autophagy flux and endosomal compartment reorganization upon DC differentiation in IL-4 or upon RUFY4 expression could interfere with its replication. We first analyzed the survival of the WT B. abortus 2308 strain in murine bmDCs by enumerating the colony-forming units (CFUs) at different times after infection of GM-CSF and IL-4–DCs (Fig. 10 E and Fig. S4 E). As expected, we observed in GM-CSF DCs an increase in the number of intracellular bacteria up to 48 h after inoculation, demonstrating that B. abortus is able to replicate within GM-CSF DCs. However, a completely different situation was observed with IL-4–DCs, which considerably reduced B. abortus replication at a late time of infection. As expected, this difference was not observed upon IL-4 addition at the time of infection (Fig. S4 E) and did not reflect the inability of IL-4–DCs to internalize bacteria because equivalent CFU formation was found immediately after inoculation, indicating an equal capacity to uptake B. abortus. To further evaluate the contribution of RUFY4 to prevent B. abortus replication, we transfected the different RUFY4 mutants in HeLa cells and monitored replication by microscopy at 6 h after infection. RUFY4 expression strongly prevented bacterial replication, whereas expression of the mutants did not (Fig. 10 F). In cells expressing the mutants, no difference in bacterial replication could be observed with control HeLa cells (Fig. 10 F), suggesting that the positive regulation of RUFY4 on autophagy is key in preventing infection. Therefore, IL-4 and RUFY4 have anti-intracellular bacteria activity, a situation that is likely to have a physiopathological relevance in different infection contexts.
Discussion
Few studies have addressed how IL-4 changes DC functional specificity (Wells et al., 2005). Our work reveals that DCs differentiated in the presence of IL-4 display considerable functional differences with “traditional” GM-CSF–differentiated cells, both at the transcriptional level and, perhaps more importantly, at the posttranslational level. We showed that autophagy and associated functions are positively influenced by IL-4 with the consequence of augmenting the processing of endogenous antigens and their MHC II–restricted presentation, which is normally decreased upon microbial sensing by DCs. Our data highlight that in addition to a reduction in mTOR activity, IL-4–dependent transcriptional activation of several genes, including rufy4, is likely required for maintaining a relatively high basal autophagic flux in cells exposed to IL-4. Independently of its immunological relevance, our work reveals for the first time the implication of RUFY4 in augmenting autophagy flux, promoting late endosomal and autophagosome membrane dynamic and preventing B. abortus replication in vitro.
The ability of DCs to present the same antigens on both MHC I and MHC II molecules in the absence of costimulatory molecules could confer them with the ability to simultaneously anergize autoreactive CD8+ and CD4+ T cells in the periphery. Conversely, the reduction of autophagy-dependent MHC II presentation upon DC activation could focus presentation on captured exogenous antigens, while preventing untimely activation of autoreactive CD4+ T cells. The efficient autophagy-dependent MHC II presentation previously reported in human MoDCs (Schmid et al., 2007) can be explained by the presence of IL-4 in the differentiation media and underlines the importance for monocytes of the nature of the cytokines and inflammatory mediators present at recruitment sites in vivo. Indeed, the presence or absence of Th2 cytokines will drive DC differentiation with relatively equivalent cell surface phenotypes but with very different cell biology and functional abilities, as observed in the lungs of asthmatic mice (Plantinga et al., 2013).
The signal cascade responsible for IL-4 induction of autophagy remains to be clarified, in particular in its capacity to prevent mTOR phosphorylation of ULK1 upon LPS detection. Upon IL-4 ligation, IL-4Rα heterodimerizes with the gamma common chain (γc) and signals via the insulin receptor substrate-1/2 (IRS-1/2) and STAT6 pathways (Heller et al., 2008). Although STAT6 is involved in IL-4–induced gene expression, IRS-1 signaling activates the type I PI3K pathway and subsequently the Akt/mTOR pathway, suggesting that IL-4, like LPS or other TLR agonists, could inhibit autophagy. However, Yamamoto et al. (2006) have reported that activation of IRS-2 leads to an autophagy-mediated clearance of accumulated poly-Q proteins, which occurs despite the activation of mTOR. IRS-2 recruitment by IL-4Rα could promote autophagy and change the balance among different transduction cascades. It has also been recently reported that upon viral infection of DCs, a decrease of the intracellular concentration of free arginine and a corresponding increase in citrulline occurs (Ravindran et al., 2014), whereas the deacetylated tRNA-sensing eIF2α kinase GCN2 was proposed to control autophagy and to regulate DC function. As these results were obtained with cells differentiated with IL-4, it is tempting to speculate that mTOR reprogramming and autophagy induction could be linked to arginase-1 expression, which by decreasing cytosolic amino acids concentration could influence mTOR activity.
The Rufy4 gene is located on chromosome 1 in mouse (Chr 2 in human), and its promotor region contains several potential STAT6 consensus binding sites (TRANSFAC database). Rufy4 is immediately adjacent to the CXCR2 gene, which is also strongly up-regulated by IL-4 treatment (Fig. 5 C), indicating that this area of the genome is transcriptionally active upon GM-CSF– and IL-4–induced differentiation. Rufy4 has appeared relatively late in evolution (reptile) and is clearly not part of the core autophagy machinery characterized in yeast (Deretic et al., 2013). Together with Rab7, RUFY4 is likely part of a regulatory module required for the fine tuning of autophagy in specific immune cells. RUFY4 overexpression increases autophagic flux, probably by stimulating both autophagosome generation and facilitating tethering with lysosomes. The association of RUFY4 with Rab7 and PtdIns(3)P-containing organelles is very similar to what is observed with Rubicon, whose overexpression leads to abnormal morphology of late endosomes. Rubicon mediates the simultaneous interaction between the Beclin 1–Vps34 and Rab7 (Tabata et al., 2010) and negatively regulates autophagosome and late endosome fusion (Matsunaga et al., 2009). Thus, Rufy4 and Rubicon could compete for the same effectors, such as Rab7, and have opposite effects for the control of autophagy.
Our in vitro experimental conditions mimic the differentiation in DCs of monocytes exposed or not to IL-4 for a relatively long period. Cells grown in these conditions initiate transcription programs linked to their long-term functional differentiation and not to a rapid response to a cytokine stimulus. This difference is also illustrated by the strong expression of RUFY4 mRNA in lung macrophages and not in other tissue-resident macrophages or those differentiated in vitro with M-CSF. (Gautier et al., 2012), suggesting that long-term exposure to a particular combination of cytokines in a given tissue environment contextualizes the basal cellular functions of resident immunocytes (Ginhoux and Jung, 2014). This situation could increase resistance to intracellular bacterial infection at potential sites of entry, as suggested by the relative protection conferred by RUFY4 against B. abortus infection. The reason of this inhibition remains to be established; however, the rapid kinetics of bacterial clearance suggests that exacerbated xenophagy could be key for this process. Our observations suggest the existence of a need for enhanced autophagy and endosome traffic in the frame of Th2 immune responses, including the MHC II presentation of cytosolic antigens and the regulation of metabolic homeostasis. Whether a deregulation of autophagy could contribute to the exacerbation of allergic diseases, such as asthma, that are critically dependent on IL-4 and DCs will have to be investigated in the future.
Materials and methods
Mice
Male C57BL/6 or CB6F1 mice 7–8 wk old were purchased from Charles River. C57BL/6J ATG5flox/flox/CD11c-Cre (Lee et al., 2010) mice were obtained from A. Iwasaki (Yale School of Medicine, New Haven, CT). C57BL/6J MyD88/TRIF−/− mice were obtained from L. Alexopoulou (CIML, Marseille, France). C57BL/6JJcl mice transgenic for GFP-LC3 under the MAP1 promotor were obtained from the RIKEN depository. C57BL/6J RAG2−/− TCR transgenics OT-I and OT-II mice were bred at the CIML animal facilities. C57BL/6J CD11c.DOG (DTR-OVA-eGFP) mice were from N. Garbi (DKFZ, Heidelberg, Germany). Experiments were conducted in accordance with institutional guidelines for animal care and use. Protocols have been approved by the French Provence ethical committee (number 04/2005).
Cell culture
bmDCs were cultured as described previously (Lelouard et al., 2002). IL-4 (PeproTech) at 20 ng/ml was used from day 2 of DC culture onward, where indicated. Bone marrow–derived macrophages and RAW cells were cultured as described previously (Harris et al., 2007), and 30 ng/ml IL-4 was added for the indicated times. HeLa and HEK293 cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal calf serum (Hyclone, PERBIO), at 37°C and 5% CO2. IL-12 production was determined by ELISA (eBioscience). Chemicals were used at the following concentrations: 10–50 nM rapamycin, 5 mM 3-MA, 100 ng/ml LPS, 50 µM CQ, and 0.1 µg/ml puromycin (all from Sigma-Aldrich). Lung DCs were purified from C57BL/6 mice exposed or not to HDM as described by Plantinga et al. (2013).
Antigen presentation experiments
Primary T cells were isolated from spleen and lymph nodes by magnetic depletion (Invitrogen), labeled with 2 µM CFSE where indicated, and co-cultured with 5,000 DCs at a 4:1 (T/DC) ratio. For T cell hybridoma activation by endogenous presentation, transfected DCs were fixed mildly (0.25% PFA, 2 min, RT) and cultured at 2:1 (T/DC) ratio with DO11 or H30 T cells for 24 h. For exogenous presentation experiments, DCs were harvested and plated (at 1:4 DC/A20 ratio) with freeze-killed A20 cells (I-Eα–positive B cell line, source of Ea52-68 peptide) or with ovalbumin immune complexes (0.5 µg/ml Ova, 2 µg/ml rabbit anti-Ova final) and cultured at 2:1 (T/DC) ratio with DO11 (Ova 329–337) on I-Ad–specific or H30 (Ea52-68) on I-Ab–specific T cells for 24 h. IL-2 production was determined by ELISA (eBioscience).
Immunodetection and immunoprecipitation
25–50 µg Triton X-100 soluble material or total protein from 5 × 104 cells (lysed directly in 2× reducing sample buffer, boiled, and homogenized using a 26-G needle) was separated by 3–15% gradient or 12% SDS-PAGE before immunoblotting and chemiluminescence detection (Thermo Fisher Scientific). Antibodies used in this study were rabbit polyclonal anti-p62, –phospho-S6, -S6, -p70S6k1, -4EBP1, and –phospho-Ulk1 (Ser 757) all from Santa Cruz Biotechnology, Inc.; and anti-Ulk1 (H240) and -ATG5 (N terminus) from Sigma-Aldrich. Mouse monoclonal anti–phospho-p70S6K1 (49D7), –phospho-4EBP1 (236B4), -myc (9B11 and 71D10), and -Stx17 (I-14) were all from Cell Signaling Technology; and anti-actin (AC-15) and -Rab7 were from Sigma-Aldrich. Other mouse monoclonal antibodies were anti-LC3 (2G6; nanoTools), -NBR1 (6B11; AbD Serotec), -polyubiquitin (FK2; Biomol), -SIINFEKL (IF10.2.2), -GFP (JL-8; Takara Bio Inc.), -CD11c (N418), and -puromycin (12D10; Schmidt et al., 2009); and brilliant violet 421 anti-LAMP1 (H4A3; BioLegend) and -EEA1 (BD). Rabbit polyclonal antibodies against AMBRA1 and phospho-AMBRA1 (ser52) were from EMD Millipore. Secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc., Molecular Probes, and Cell Signaling Technology. For immunofluorescence, cells on coverslips were fixed with 3.5% PFA and permeabilized with 0.1% Triton X-100. Where indicated, cells were permeabilized (0.2% Triton X-100, 5 min, on ice) before fixation (3.5% PFA), alternatively pre-extracted with 0.02% saponin in PBS (30 s, RT), fixed with 3.5% PFA (15 min, RT), and permeabilized with 0.1% Triton X-100 (15 min, RT) or cold methanol permeabilized for 2 min at −20°C after PFA fixation. Immunofluorescence and confocal microscopy were performed at 20°C with a LSM580 (Carl Zeiss) or a TCS SPX5 (Leica) using a 40× and 63× objective and accompanying imaging software. Fluorochrome or biotin directly coupled antibodies for FACS were CD8α (RM2201) from Invitrogen; I-A/I-E (M5/114.15.2), CD80 (16-10A1), CD4 (RM4-5), and DEC205 (205yekta) from eBioscience; and CD11c (N418), CD86 (GL-1), CD40 (HM40-3), and H-2Kb (HB-176) from BD. Anti-Kb/SL8 25D1.16 (Porgador et al., 1997) was purified and fluorochrome coupled using the Alexa Fluor 647 protein labeling kit (Invitrogen). Data were acquired on a FACS LSR II or Canto II (BD) and analyzed using FlowJo (Tree Star). For immunoprecipitation, 0.4 × 106 HeLa cells were transfected with mock or the Myc-tagged hRUFY4 (WT and mutants) plasmids for 24 h. The cells were then lysed (RIPA) and subjected to immunoprecipitation using the Cross-link Magnetic IP/Co-IP kit with anti-Myc (mouse IgG2a 9B11, 5 µg; Thermo Fisher Scientific). Immunoprecipitates and input control were blotted with rabbit anti-Rab7 (D95F2; Cell Signaling Technologies) and mouse IgG1 anti-Rab9 (Thermo Fisher Scientific).
DALIS scoring
Cells on coverslips were pre-extracted with cold 0.1% Triton-X for 2 min at 4°C before fixation with 3.5% PFA. Detection was performed with an anti-P62 (SQTM1; sc-25575; Santa Cruz Biotechnology, Inc.) and anti–rabbit Alexa Fluor A568 (A11011; Molecular Probes), mounted with Prolong Gold with DAPI (P-36931; Life Technologies). DALIS quantification was performed by mosaic image acquisition with an Axioplan video microscope (Carl Zeiss). Images were then analyzed using a multiparameter protocol based on the Physiology Analyst module of ASSAY Builder software (Carl Zeiss). This protocol allows the automatic recognition and characterization of the cell population and intensities, areas, and spot number in the cellular regions.
cDNA cloning, in vitro transcription, and gene transduction or silencing
All plasmids were generated using standard cloning, PCR, and fusion PCR techniques and based on pcDNA3.1 backbone (Invitrogen). The H-2Kb cDNA (gift from F. Momburg, DKFZ) was inserted into pcDNA3.1. The myc-tagged human GIMAP1 and GIMAP5 inserted in pCANmyc1 were gifts from G. Butcher (The Babraham Institute, Cambridge, England, UK). A plasmid for GFP-2OP in vitro transcription was generated using eGFP cloned in pcDNA/Zeo, as described previously (Wenger et al., 2012). 4 × 106 DCs were transfected with 20 µg mRNA using the mouse DC nucleofection kit (Amaxa). Total mRNA was purified using the RNeasy Mini kit (QIAGEN), and cDNAs were synthesized by SuperScript II system (Invitrogen) according to the manufacturers’ instruction. Mouse and human Rufy4 cDNA was amplified by nested-PCR using cDNA from MoDCs as template and then cloned to pcDNA3.1 vector with tagging with Flag (C terminus), myc (N terminus), or mCherry (N terminus). The ΔRUN and ΔFYVE domain mutants were obtained by PCR. Human WIPI2 cDNA was also cloned from human MoDC cDNA by conventional PCR method and then fused to the GFP gene at its N terminus in the pcDNA3.1 vector. The plasmid DNAs were introduced to the cell lines with use of JetPrime regent (Polyplus). ONE-TARGET plus siRNA targeting Rufy4 and ONE-TARGETING plus nontargeting siRNA (Thermo Fisher Scientific) were used as specific Rufy4 siRNA and as control siRNA, respectively. 4 × 106 IL-4–treated bmDCs were transfected at day 4 with 5 µM siRNA using the Nucleofector (Amaxa) according to the manufacturer’s instructions. Cells were collected 48 h after transfection and analyzed.
qPCR
Total RNAs were extracted and purified using the RNeasy Mini kit. 100 ng to 1 µg total RNA was subjected to reverse transcription using SuperScript II. Each gene transcript was quantified by SYBR Green method with 7500Fast (Applied Biosystems). The relative amount of each transcript was determined by normalizing to internal housekeeping gene expression. Primer sets were as follows: mouse Rufy4, 5′-TGGCCCCTGGACACTGCATA-3′ and 5′-AGAGCAGGCGTGGCAAACCA-3′; human Rufy4, 5′-TGCTCTCAATGGGGTGGCCT-3′ and 5′-GGGTTTGGGTGGAACTGGAGCA-3′; mouse GIMAP1, 5′-ATGCAGAGGCGCACCAGGTT-3′ and 5′-TGGCCACACCCAGGAAGACA-3′; mouse cyclophilin B, 5′-ATGTGGTTTTCGGCAAAGTT-3′ and 5′-TGACATCCTTCAGTGGCTTG-3′; mouse GAPDH, 5′-GGGTGTGAACCACGAGAAATA-3′ and 5′-CTTCCACAATGCCAAAGTTGT-3′; and human GAPDH, 5′-CAATGACCCCTTCATTGACC-3′ and 5′-GACAAGCTTCCCGTTCTCAG-3′.
Microarray and bioinformatics analyses
Equal amounts of RNA extract (500 ng) from each replicate were amplified and Cy3 labeled using the Low Input Quick Amp Labeling kit (Agilent Technologies). Hybridizations were performed according to the manufacturer’s instructions for One-Color Microarray-Based Gene Expression Analysis using Whole Mouse Genome GE 4 × 44K v2 microarrays (Agilent Technologies). Images were scanned on an Agilent G2565AA microarray scanner, and raw images were quantified using the Agilent Feature Extraction 10.5.1.1 software and the GE1_105_Dec08 protocol. The signal intensity was aligned and normalized between microarrays by centering the median of the signal distribution using BRB-ArrayTools v3.8.1. The microarray data have been submitted to the GEO database and can be accessed under accession no. GSE56929. Raw gene expression data obtained from the Agilent Feature Extraction software were background corrected using the “normexp” method and quantile normalized, using the limma package through Bioconductor in the R statistical environment (version 2.15.0). Quality control of the expression data was assessed by boxplots of raw expression data, density plots of the normalized data, and calculation of the Pearson’s correlation coefficients between arrays. The Cluster and Treeview softwares were used to classify samples according to the similarity of their gene expression profiles as assessed by hierarchical clustering with average linkage as a clustering method and Pearson correlation as a metric distance, considering the genes having a minimal fold change of two across all samples. PCA was computed with the MeV software across all probes, using the median-centered method and 10 neighbors for KNN imputation. The 3D plot was generated using the scatterplot3D package in the R statistical environment. Genes differentially expressed between each sample in a statistically significant manner were selected as having a fold change >2× and an FDR (Benjamini-Hochberg correction) <0.1 after empirical Bayes processing using the limma package. These gene lists were submitted to Venn software for comparisons or uploaded into IPA and Voronto (Santamaría and Pierre, 2012) to test for significant enrichment of functional annotations.
Statistical analysis
Data representing multiple experiments are displayed as mean ± SEM. Paired Student’s t test was used where indicated.
Bacterial infection and replication assays
Infections were performed with the smooth virulent B. abortus strain 2308 at a multiplicity of infection of 20:1 for all experiments. Bacteria were centrifuged onto bmDCs or HeLa at 400 g for 10 min at 4°C and then incubated for 30 min at 37°C with 5% CO2 atmosphere. Cells were gently washed twice with medium and then incubated for 1 h in medium supplemented with 100 µg/ml streptomycin to kill extracellular bacteria. Thereafter, the antibiotic concentration was decreased to 20 µg/ml. Control samples were always performed by incubating cells with media only and following the exact same procedure for infection. To monitor bacterial intracellular survival, infected cells were lysed with 0.1% Triton X-100 in H2O and serial dilutions plated onto TSB agar to enumerated CFUs.
Online supplemental material
Fig. S1 shows comparative electron micrographs of GM-CSF– and IL-4–differentiated and LPS-activated DCs. Fig. S2 shows DALIS formation in DCs exposed to different conditions. Fig. S3 shows that IL-4–induced signaling augments mitophagy and decreases mitochondrial ROS in DCs. Fig. S4 shows the consequences on macrophages of short-term IL-4 treatment and long-term differentiation. Fig. S5 shows in silico analysis of genes down-modulated by IL-4 differentiation. Table S1 is a list of up-regulated genes during IL-4–induced DC differentiation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501059/DC1.
Supplementary Material
Acknowledgments
The laboratory is supported by EU FP 7 Collaborative Project SYBARIS grant agreement number 242220; the Equipe labelisée Ligue Nationale Contre le Cancer; Agence Nationale de la Recherche grants ANR 10-BLAN-1236, ANR 10-BLAN-1308, ANR-12-BSV2-0025-01, and ANR-FCT 12-ISV3-0002-01; DCBiol Labex ANR-11-LABEX-0043 grant ANR-10-IDEX-0001-02 PSL*, and A*MIDEX project ANR-11-IDEX-0001-02 funded by the “Investissements d'Avenir” French government program. S. Terawaki is a Uehara Memorial Foundation fellow. S. Terawaki and F. Prete are Fondation de la Recherche Médicale (FRM) fellows. T. Wenger is a von Humboldt foundation and Institut National du Cancer fellow. T.-P. Vu Manh was supported by the PhylogenDC ANR grant. We acknowledge support from no. ANR-10-INBS-04-01 France Bio Imaging for advanced microscopy. Part of the research was also supported by FCT through the Institute for Biomedicine–iBiMED contract UID/BIM/04501/2013 and PTDC/IMI-IMU/3615/2014.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- 3-MA
- 3-methyl adenine
- BCV
- B. abortus–containing vacuole
- bmDC
- bone marrow–derived DC
- cDC
- conventional DC
- CFU
- colony-forming unit
- CQ
- chloroquine
- DALIS
- DC–aggresome-like induced structure
- DC
- dendritic cell
- DEG
- differentially expressed gene
- DRiP
- defective ribosomal protein
- HDM
- house dust mite
- ICM
- immunofluorescence confocal microscopy
- iDC
- immature DC
- LPS
- lipopolysaccharide
- mDC
- mature DC
- MoDC
- monocyte-derived DC
- PCA
- principal component analysis
- PtdIns(3)P
- phosphatidylinositol-3-phosphate
- qPCR
- quantitative PCR
- ROS
- reactive oxygen species
- RUFY
- RUN and FYVE domain–containing protein
- SNARE
- soluble N-ethylmaleimide–sensitive factor attachment protein receptor
- TLR
- Toll-like receptor
- WT
- wild type
References
- Birkeland H.C., and Stenmark H.. 2004. Protein targeting to endosomes and phagosomes via FYVE and PX domains. Curr. Top. Microbiol. Immunol. 282:89–115. [DOI] [PubMed] [Google Scholar]
- Callebaut I., de Gunzburg J., Goud B., and Mornon J.P.. 2001. RUN domains: a new family of domains involved in Ras-like GTPase signaling. Trends Biochem. Sci. 26:79–83. 10.1016/S0968-0004(00)01730-8 [DOI] [PubMed] [Google Scholar]
- Delgado M.A., Elmaoued R.A., Davis A.S., Kyei G., and Deretic V.. 2008. Toll-like receptors control autophagy. EMBO J. 27:1110–1121. 10.1038/emboj.2008.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Saitoh T., and Akira S.. 2013. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13:722–737. 10.1038/nri3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Maeda D., Xiao Q., and Srinivasula S.M.. 2011. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc. Natl. Acad. Sci. USA. 108:1427–1432. 10.1073/pnas.1014156108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. Immunological Genome Consortium . 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F., Manz M.G., Jung S., Sieweke M.H., Merad M., and Ley K.. 2010. Development of monocytes, macrophages, and dendritic cells. Science. 327:656–661. 10.1126/science.1178331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., and Jung S.. 2014. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14:392–404. 10.1038/nri3671 [DOI] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013a Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393. 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hamasaki M., Shibutani S.T., and Yoshimori T.. 2013b Up-to-date membrane biogenesis in the autophagosome formation. Curr. Opin. Cell Biol. 25:455–460. 10.1016/j.ceb.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Harris J., De Haro S.A., Master S.S., Keane J., Roberts E.A., Delgado M., and Deretic V.. 2007. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 27:505–517. 10.1016/j.immuni.2007.07.022 [DOI] [PubMed] [Google Scholar]
- Heller N.M., Qi X., Junttila I.S., Shirey K.A., Vogel S.N., Paul W.E., and Keegan A.D.. 2008. Type I IL-4Rs selectively activate IRS-2 to induce target gene expression in macrophages. Sci. Signal. 1:ra17 10.1126/scisignal.1164795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochweller K., Striegler J., Hämmerling G.J., and Garbi N.. 2008. A novel CD11c.DTR transgenic mouse for depletion of dendritic cells reveals their requirement for homeostatic proliferation of natural killer cells. Eur. J. Immunol. 38:2776–2783. 10.1002/eji.200838659 [DOI] [PubMed] [Google Scholar]
- Honey K., and Rudensky A.Y.. 2003. Lysosomal cysteine proteases regulate antigen presentation. Nat. Rev. Immunol. 3:472–482. 10.1038/nri1110 [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Ivan V., Martinez-Sanchez E., Sima L.E., Oorschot V., Klumperman J., Petrescu S.M., and van der Sluijs P.. 2012. AP-3 and Rabip4′ coordinately regulate spatial distribution of lysosomes. PLoS ONE. 7:e48142 10.1371/journal.pone.0048142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., and Lamark T.. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 7:279–296. 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., and Yoshimori T.. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117:2805–2812. 10.1242/jcs.01131 [DOI] [PubMed] [Google Scholar]
- Kawai T., and Akira S.. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1–20. 10.1196/annals.1443.020 [DOI] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., and Guan K.L.. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13:132–141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Itakura C., Koyama-Honda I., Itakura E., and Mizushima N.. 2014. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 127:4089–4102. 10.1242/jcs.156034 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 8:445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Mattei L.M., Steinberg B.E., Alberts P., Lee Y.H., Chervonsky A., Mizushima N., Grinstein S., and Iwasaki A.. 2010. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 32:227–239. 10.1016/j.immuni.2009.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelouard H., Gatti E., Cappello F., Gresser O., Camosseto V., and Pierre P.. 2002. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature. 417:177–182. 10.1038/417177a [DOI] [PubMed] [Google Scholar]
- Lelouard H., Ferrand V., Marguet D., Bania J., Camosseto V., David A., Gatti E., and Pierre P.. 2004. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J. Cell Biol. 164:667–675. 10.1083/jcb.200312073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelouard H., Schmidt E.K., Camosseto V., Clavarino G., Ceppi M., Hsu H.T., and Pierre P.. 2007. Regulation of translation is required for dendritic cell function and survival during activation. J. Cell Biol. 179:1427–1439. 10.1083/jcb.200707166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., and Virgin H.W.. 2011. Autophagy in immunity and inflammation. Nature. 469:323–335. 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F.O., Helming L., Milde R., Varin A., Melgert B.N., Draijer C., Thomas B., Fabbri M., Crawshaw A., Ho L.P., et al. 2013. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 121:e57–e69. 10.1182/blood-2012-06-436212 [DOI] [PubMed] [Google Scholar]
- Matsumoto G., Wada K., Okuno M., Kurosawa M., and Nukina N.. 2011. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell. 44:279–289. 10.1016/j.molcel.2011.07.039 [DOI] [PubMed] [Google Scholar]
- Matsunaga K., Noda T., and Yoshimori T.. 2009. Binding Rubicon to cross the Rubicon. Autophagy. 5:876–877. 10.4161/auto.9098 [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2010. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22:132–139. 10.1016/j.ceb.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Münz C. 2012. Antigen processing for MHC class II presentation via autophagy. Front. Immunol. 3:9 10.3389/fimmu.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.L., Tussiwand R., and Murphy K.M.. 2013. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 13:499–509. 10.1038/nri3470 [DOI] [PubMed] [Google Scholar]
- Nazio F., Strappazzon F., Antonioli M., Bielli P., Cianfanelli V., Bordi M., Gretzmeier C., Dengjel J., Piacentini M., Fimia G.M., and Cecconi F.. 2013. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 15:406–416. 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Alemu E.A., Brech A., Bruun J.A., Lamark T., Overvatn A., Bjørkøy G., and Johansen T.. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188:253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantinga M., Guilliams M., Vanheerswynghels M., Deswarte K., Branco-Madeira F., Toussaint W., Vanhoutte L., Neyt K., Killeen N., Malissen B., et al. 2013. Conventional and monocyte-derived CD11b+ dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 38:322–335. 10.1016/j.immuni.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Polson H.E., de Lartigue J., Rigden D.J., Reedijk M., Urbé S., Clague M.J., and Tooze S.A.. 2010. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 6:506–522. 10.4161/auto.6.4.11863 [DOI] [PubMed] [Google Scholar]
- Porgador A., Yewdell J.W., Deng Y., Bennink J.R., and Germain R.N.. 1997. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 6:715–726. 10.1016/S1074-7613(00)80447-1 [DOI] [PubMed] [Google Scholar]
- Ravindran R., Khan N., Nakaya H.I., Li S., Loebbermann J., Maddur M.S., Park Y., Jones D.P., Chappert P., Davoust J., et al. 2014. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 343:313–317. 10.1126/science.1246829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo S.P., Marchesini M.I., Lelouard H., Fugier E., Jolly G., Balor S., Muller A., Lapaque N., Demaria O., Alexopoulou L., et al. 2008. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 4:e21 10.1371/journal.ppat.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría R., and Pierre P.. 2012. Voronto: mapper for expression data to ontologies. Bioinformatics. 28:2281–2282. 10.1093/bioinformatics/bts428 [DOI] [PubMed] [Google Scholar]
- Saunders A., Webb L.M., Janas M.L., Hutchings A., Pascall J., Carter C., Pugh N., Morgan G., Turner M., and Butcher G.W.. 2010. Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood. 115:3249–3257. 10.1182/blood-2009-08-237586 [DOI] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., and Münz C.. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 26:79–92. 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E.K., Clavarino G., Ceppi M., and Pierre P.. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 6:275–277. 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- Simonsen A., Birkeland H.C., Gillooly D.J., Mizushima N., Kuma A., Yoshimori T., Slagsvold T., Brech A., and Stenmark H.. 2004. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 117:4239–4251. 10.1242/jcs.01287 [DOI] [PubMed] [Google Scholar]
- Stein M.P., Feng Y., Cooper K.L., Welford A.M., and Wandinger-Ness A.. 2003. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 4:754–771. 10.1034/j.1600-0854.2003.00133.x [DOI] [PubMed] [Google Scholar]
- Tabata K., Matsunaga K., Sakane A., Sasaki T., Noda T., and Yoshimori T.. 2010. Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol. Biol. Cell. 21:4162–4172. 10.1091/mbc.E10-06-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A.L., Hegedűs K., and Juhász G.. 2013. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201:531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal M.C., Sasai M., Lee H.K., Yordy B., Shadel G.S., and Iwasaki A.. 2009. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. USA. 106:2770–2775. 10.1073/pnas.0807694106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken S.J., and Locksley R.M.. 2013. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu. Rev. Immunol. 31:317–343. 10.1146/annurev-immunol-032712-095906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.W., Darling D., Farzaneh F., and Galea-Lauri J.. 2005. Influence of interleukin-4 on the phenotype and function of bone marrow-derived murine dendritic cells generated under serum-free conditions. Scand. J. Immunol. 61:251–259. 10.1111/j.1365-3083.2005.01556.x [DOI] [PubMed] [Google Scholar]
- Wenger T., Terawaki S., Camosseto V., Abdelrassoul R., Mies A., Catalan N., Claudio N., Clavarino G., de Gassart A., Rigotti F.A., et al. 2012. Autophagy inhibition promotes defective neosynthesized proteins storage in ALIS, and induces redirection toward proteasome processing and MHCI-restricted presentation. Autophagy. 8:350–363. 10.4161/auto.18806 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Cremona M.L., and Rothman J.E.. 2006. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J. Cell Biol. 172:719–731. 10.1083/jcb.200510065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.J., and Tedder T.F.. 1996. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proc. Natl. Acad. Sci. USA. 93:2588–2592. 10.1073/pnas.93.6.2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.