K63-linked ubiquitination of GPCRs mediated by the NEDD4-2 E3 ubiquitin ligase regulates recruitment of a TAB1–TAB2 complex on endosomes and stimulates p38 MAPK through a noncanonical pathway, which is critical for endothelial barrier disruption.
Abstract
Protease-activated receptor 1 (PAR1) is a G protein–coupled receptor (GPCR) for thrombin and promotes inflammatory responses through multiple pathways including p38 mitogen-activated protein kinase signaling. The mechanisms that govern PAR1-induced p38 activation remain unclear. Here, we define an atypical ubiquitin-dependent pathway for p38 activation used by PAR1 that regulates endothelial barrier permeability. Activated PAR1 K63-linked ubiquitination is mediated by the NEDD4-2 E3 ubiquitin ligase and initiated recruitment of transforming growth factor-β–activated protein kinase-1 binding protein-2 (TAB2). The ubiquitin-binding domain of TAB2 was essential for recruitment to PAR1-containing endosomes. TAB2 associated with TAB1, which induced p38 activation independent of MKK3 and MKK6. The P2Y1 purinergic GPCR also stimulated p38 activation via NEDD4-2–mediated ubiquitination and TAB1–TAB2. TAB1–TAB2-dependent p38 activation was critical for PAR1-promoted endothelial barrier permeability in vitro, and p38 signaling was required for PAR1-induced vascular leakage in vivo. These studies define an atypical ubiquitin-mediated signaling pathway used by a subset of GPCRs that regulates endosomal p38 signaling and endothelial barrier disruption.
Introduction
G protein–coupled receptors (GPCRs) control a vast number of physiological responses including inflammation. The coagulant protease thrombin is generated during vascular inflammation, and promotes endothelial barrier disruption through activation of protease-activated receptor 1 (PAR1; Coughlin, 1994; Soh et al., 2010). Thrombin-activated PAR1 couples to G12/13 and to Gq signaling effectors RhoA, Ca2+, and PKC to induce endothelial barrier permeability in vitro (McLaughlin et al., 2005; Komarova et al., 2007; Soh and Trejo, 2011). Studies using endothelium-specific knockout mice have shown that Gq/11 but not G12/13 is critical for regulating endothelial barrier permeability in response to activation by many GPCRs including PAR1 (Korhonen et al., 2009). Signaling by p38 MAPK has also been implicated in thrombin-induced endothelial barrier permeability (Borbiev et al., 2004), but precisely how PAR1 mediates p38 activation is not known.
All four p38 isoforms (α, β, γ, and δ) are activated through the canonical three-tiered kinase cascade mediated by upstream MAP3Ks. The canonical cascade converges on two MAP2Ks—MKK3 and MKK6—that phosphorylate and activate p38 (Raingeaud et al., 1996). However, p38-α isoform can also be activated through a noncanonical pathway mediated by direct binding of transforming growth factor-β–activated protein kinase-1 binding protein 1 (TAB1) to p38-α, bypassing the requirement for MAP2Ks. TAB1 promotes a conformational change in p38, enabling autophosphorylation and activation (Ge et al., 2002; De Nicola et al., 2013). However, whether GPCRs activate p38 through autophosphorylation and the functional implications of this type of noncanonical p38 signaling is not known.
Posttranslational modifications are essential for regulating GPCR function. Phosphorylation of GPCRs mediates the recruitment of adaptor and regulatory proteins that initiate desensitization and internalization. In addition to phosphorylation, many GPCRs are modified with ubiquitin. Ubiquitination of GPCRs is best known to function as a sorting signal for lysosomal degradation (Marchese and Trejo, 2013). However, not all GPCRs require ubiquitination for sorting to lysosomes (Dores and Trejo, 2014). We previously showed that activated PAR1 is trafficked to lysosomes through an ubiquitin-independent pathway, mediated by the adaptor protein ALIX (Wolfe et al., 2007; Dores et al., 2012). ALIX binds directly to PAR1 via a conserved YPXnL motif and facilitates sorting into multivesicular bodies/lysosomes (Dores et al., 2012). We also identified a subset of GPCRs with a conserved YPXnL motif including the P2Y1 purinergic receptor that may use a similar lysosomal sorting pathway (Dores et al., 2012). These studies suggest that ubiquitination of certain GPCRs may serve a function distinct from lysosomal sorting.
Given that ubiquitination is not required for PAR1 lysosomal sorting, we sought to determine if receptor ubiquitination modulated signaling responses such as p38 MAPK activation. Here, we report that thrombin activation of PAR1 induces noncanonical p38 activation through autophosphorylation via a ubiquitin and TAB1–TAB2–dependent pathway on endosomes. We further establish that noncanonical p38 signaling induced by PAR1 is critical for the regulation of endothelial barrier permeability in vitro and vascular leakage in vivo. The ubiquitin- and TAB1–TAB2–dependent p38 activation pathway is conserved for the G protein–coupled purinergic P2Y1 receptor. These findings reveal an atypical function for ubiquitination of GPCRs in the initiation of noncanonical p38 signaling that is critical for the regulation of endothelial barrier disruption.
Results
Ubiquitination of PAR1 is critical for p38 MAPK activation
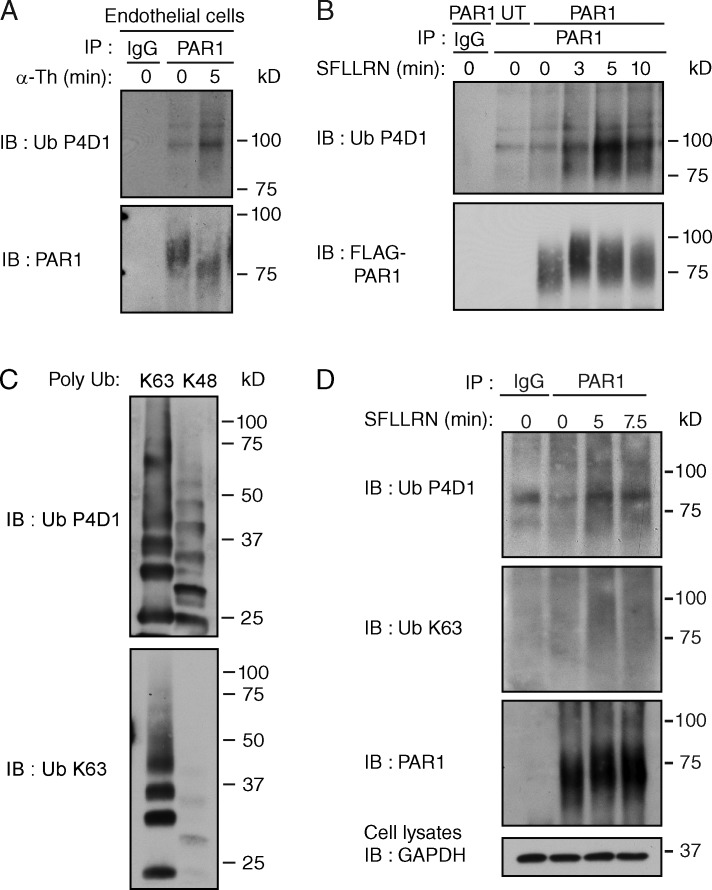
We previously showed that agonist stimulation of a ubiquitination-deficient PAR1 “0K” mutant in which cytoplasmic lysines (K) were converted to arginine (R) displayed comparable changes in Gq-mediated phosphoinositide hydrolysis, internalization, and lysosomal degradation (Wolfe et al., 2007; Dores et al., 2012). These studies indicate that ubiquitination of PAR1 is not essential for lysosomal sorting. Despite these observations, we found that PAR1 wild type (WT) expressed natively in endothelial cells or exogenously in HeLa cells is ubiquitinated within minutes of agonist stimulation detected as a high molecular weight species using an anti-pan ubiquitin antibody (Fig. 1, A and B). To determine the nature of PAR1 ubiquitination, we used an anti-K63 ubiquitin antibody that specifically detects K63-linked ubiquitin chains (Fig. 1 C). Similar to detection by the anti-pan ubiquitin antibody, the anti-K63 ubiquitin specific antibody detected an increase in PAR1 ubiquitination after minutes of agonist stimulation (Fig. 1 D). To ensure that the ubiquitin signal detected is not due to coassociated ubiquitinated proteins, cells were stimulated with agonist and processed using a hot lysis method (Mukai et al., 2010). Agonist induced a similar increase in PAR1 ubiquitination under hot lysis conditions (Fig. S1 A). These findings suggest that activated PAR1 is modified by K63-linked ubiquitin. However, ubiquitination of PAR1 is not required for lysosomal sorting (Dores et al., 2012), suggesting that it may serve a distinct function.
Figure 1.
Activated PAR1 is modified with K63-linked ubiquitin. (A) Ubiquitination of endogenous PAR1 detected in endothelial cells after stimulation with 10 nM α-Th. (B) Ubiquitination of FLAG-tagged PAR1 WT expressed in HeLa cells after stimulation with 100 µM SFLLRN. (C) Poly K63- or K48-linked ubiquitin chains (300 ng each) detected with anti-pan ubiquitin (Ub) P4D1 or anti-K63 Ub specific antibodies. (D) Ubiquitination of FLAG-tagged PAR1 in HeLa cells after stimulation with 100 µM SFLLRN. See Fig. S1 A.
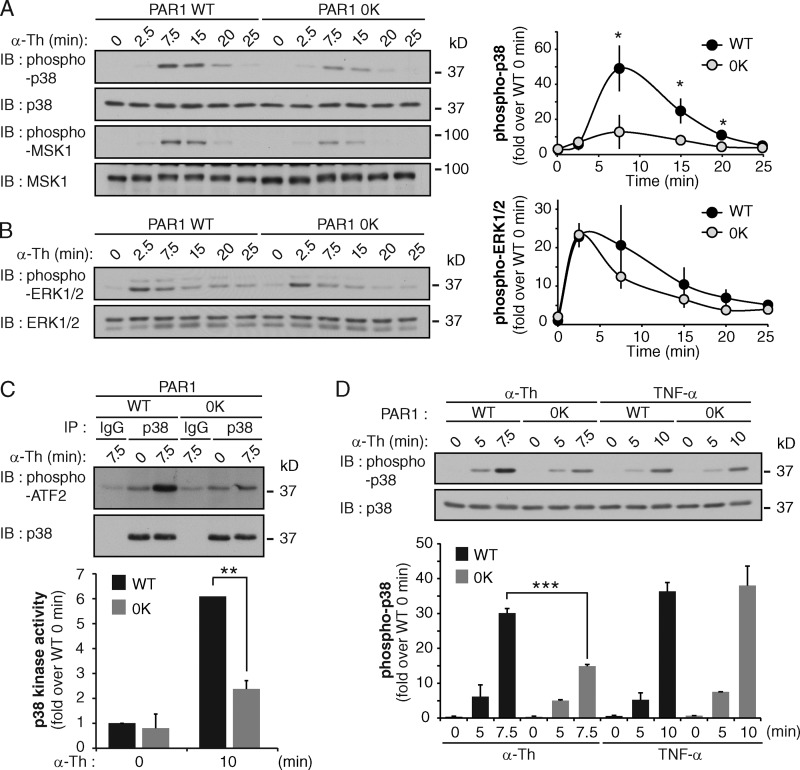
To examine the function of PAR1 ubiquitination, we first used PAR1 WT and 0K ubiquitin-deficient mutant stably expressed in HeLa cells (Wolfe et al., 2007; Dores et al., 2012) and examined various MAPK signaling cascades after agonist stimulation. PAR1 WT and 0K mutant are expressed similarly in these cell lines (Fig. S1, B and C). Activation of PAR1 WT with thrombin caused a significant increase in p38 phosphorylation at 7.5 min (Fig. 2 A); however, the response was markedly reduced in PAR1 0K–expressing cells (Fig. 2 A). Phosphorylation of mitogen- and stress-activated protein kinase 1 (MSK1), a downstream p38 substrate, was also reduced in thrombin-treated PAR1 0K cells compared with WT cells (Fig. 2 A). However, both thrombin-stimulated PAR1 WT and 0K cells displayed rapid and comparable increases in extracellular signal regulated kinase 1 and 2 (ERK1/2) phosphorylation (Fig. 2 B) and Akt phosphorylation (Fig. S1 D). In vitro p38 kinase activity assays also showed a significant loss in thrombin-induced p38 activation in PAR1 0K versus WT cells (Fig. 2 C). In contrast to thrombin, the cytokine tumor necrosis factor-α (TNF) triggered a comparable increase in p38 phosphorylation in both PAR1 WT and 0K HeLa cells (Fig. 2 D), indicating that p38 activation is not globally impaired in the PAR1 0K cell line.
Figure 2.
Ubiquitination of PAR1 is required for p38 activation. (A and B) HeLa cells expressing FLAG-PAR1 WT or ubiquitin-deficient (0K) mutant were stimulated with 10 nM α-Th, and p38, MSK-1, and ERK1/2 phosphorylation was determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05). See Fig. S1 (B–D). (C) PAR1 WT and 0K HeLa cells were incubated with 10 nM α-Th, and p38 kinase activity was determined in vitro. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (**, P < 0.01). (D) HeLa cells expressing PAR1 WT or 0K were stimulated with 10 nM α-Th or 16 nM TNF, and p38 phosphorylation was determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (***, P < 0.001).
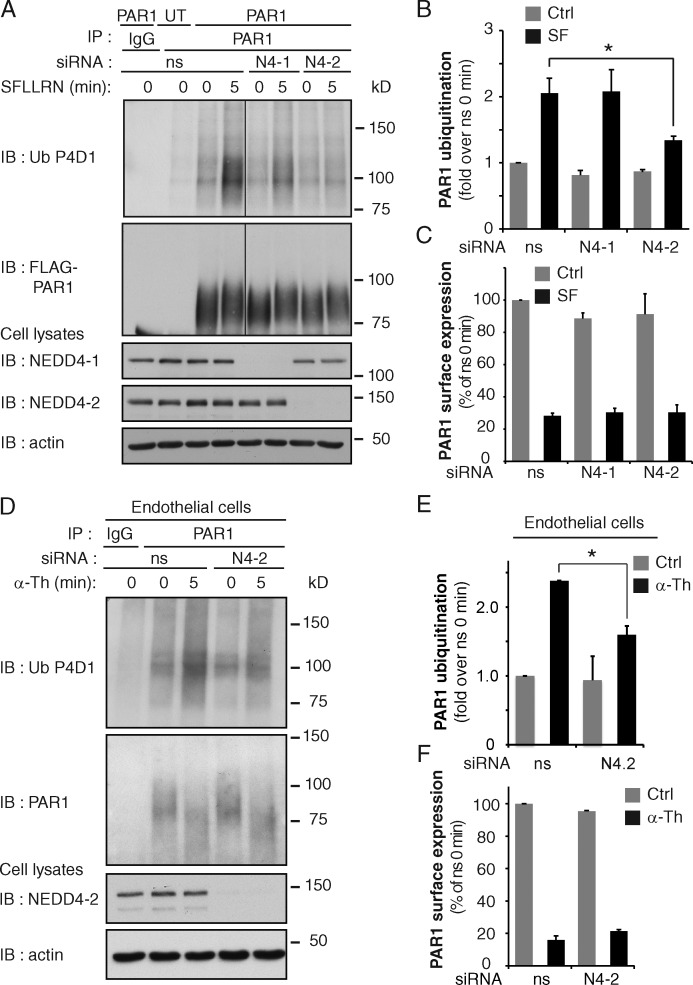
We next sought to identify the E3 ubiquitin ligase responsible for PAR1 ubiquitination using siRNA-mediated depletion of neural precursor cell expressed developmentally down-regulated protein 4 (NEDD4) family members. The NEDD4 family of E3 ubiquitin ligases is known to mediate ubiquitination of GPCRs (Marchese and Trejo, 2013). HeLa cells expressing PAR1 WT were transiently transfected with siRNA targeting NEDD4 E3 ubiquitin ligase family members including AIP4, NEDD4-1, NEDD4-2, WWP1, and WWP2 or nonspecific (ns) siRNA. After 72 h, cells were stimulated with agonist, PAR1 was immunoprecipitated, and endogenous ubiquitin was detected. Of the NEDD4 family members tested, only siRNA-mediated depletion of NEDD4-2 significantly reduced agonist-induced PAR1 ubiquitination (Fig. 3, A and B; and Fig. S2 A). Similar effects were observed with different NEDD4-2 specific siRNAs (Fig. S2, B and C). However, the loss of NEDD4-2 expression did not affect basal PAR1 surface expression or agonist-promoted internalization (Figs. 3 C and S2 D). Depletion of NEDD4-2 also disrupted thrombin-induced ubiquitination of endogenous PAR1 in endothelial cells (Fig. 3, D and E), whereas basal PAR1 surface expression and agonist-stimulated internalization remained unchanged (Fig. 3 F). These findings suggest that NEDD4-2 is a critical mediator of agonist-induced PAR1 ubiquitination.
Figure 3.
Activated PAR1 ubiquitination is mediated by NEDD4-2. (A) PAR1 WT or untransfected (UT) HeLa cells were transfected with ns, NEDD4-1 (N4-1), or NEDD4-2 (N4-2) siRNA and stimulated with 100 µM SFLLRN. Then PAR1 ubiquitination was detected. See Fig. S2 (A–C). (B) Quantitation of PAR1 ubiquitination. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05). (C) PAR1 surface expression and internalization detected in HeLa cells transfected with ns, N4-1, and N4-2 siRNA after 10 min of stimulation with 100 µM SFLLRN. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test. (D) PAR1 ubiquitination detected in endothelial cells transfected with ns or N4-2 siRNAs after stimulation with 10 nM α-Th. (E) PAR1 ubiquitination was quantitated. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05). (F) PAR1 surface expression and internalization determined in endothelial cells transfected with ns or N4-2 siRNA after 5 min of 10 nM α-Th. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test.
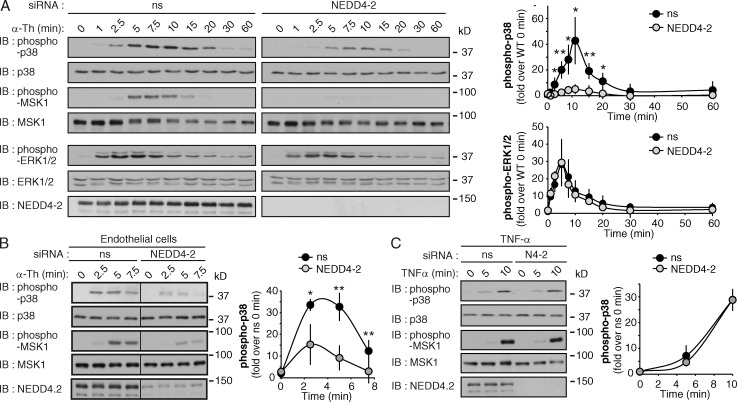
To further explore the role of PAR1 ubiquitination in p38 activation, NEDD4-2 was depleted from PAR1 WT HeLa cells. NEDD4-2 knockdown significantly inhibited thrombin-dependent p38 activation at various time points compared with control siRNA transfection (Fig. 4 A). Similar effects were observed in cells transfected with different NEDD4-2–specific siRNAs and stimulated with thrombin or peptide agonist (Fig. S2, E and F). Thrombin-stimulated MSK1 phosphorylation was also reduced in NEDD4-2–deficient cells (Fig. 4 A), whereas thrombin-dependent ERK1/2 activation was comparable in both control and NEDD4-2–depleted cells (Fig. 4 A). Depletion of endothelial cell NEDD4-2 expression also inhibited thrombin-induced p38 and MSK1 phosphorylation (Fig. 4 B). In contrast to thrombin, TNF-induced p38 and MSK1 phosphorylation were similar in control and NEDD4-2–depleted HeLa cells (Fig. 4 C), indicating that p38 activation is not globally disrupted in NEDD4-2–deficient cells. Together, these data suggest that NEDD4-2–mediated ubiquitination of PAR1 is important for thrombin-induced p38 activation.
Figure 4.
Thrombin-induced p38 activation requires NEDD4-2 expression. (A) PAR1-expressing HeLa cells transfected with ns or NEDD4-2 siRNA were stimulated with 10 nM α-Th, and phosphorylation of p38, MSK1, and ERK1/2 was determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05; **, P < 0.01). See Fig. S2 (D–F). (B) Endothelial cells were transfected with ns or NEDD4-2 siRNAs and incubated with 10 nM α-Th. Phosphorylation of p38 and MSK1 was then determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05; **, P < 0.01). (C) PAR1 HeLa cells transfected with ns or NEDD4-2 (N4-2) siRNAs were treated with 16 nM TNF, and phosphorylation of p38 and MSK1 was determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test.
Thrombin induces p38 autophosphorylation
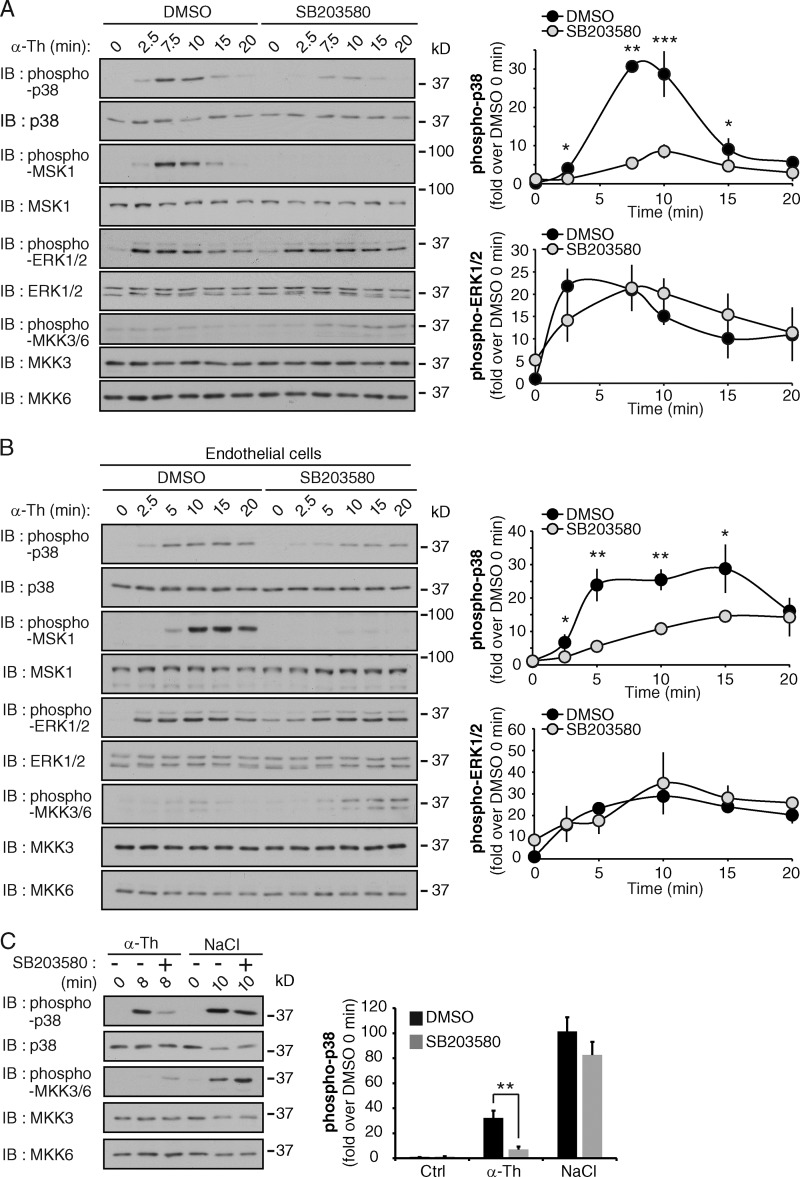
To determine if thrombin activates p38 through the canonical three-tiered kinase cascade, we used the p38 inhibitor SB203580, which specifically inhibits the catalytic activity of p38-α and -β isoforms without blocking p38 phosphorylation mediated by the upstream kinases (Ge et al., 2002). PAR1 WT HeLa cells or endothelial cells were pretreated with SB203580 before addition of an agonist. Thrombin-stimulated p38 and MSK1 phosphorylation was virtually abolished in HeLa and endothelial cells pretreated with SB203580 (Fig. 5, A and B; and Fig. S3 A), whereas ERK1/2 phosphorylation was not affected (Fig. 5, A and B). Similar results were observed with the structurally related p38 inhibitor SB202190 (Fig. S3 B). Moreover, thrombin failed to increase phosphorylation of MKK3 and MKK6 in both cell types (Fig. 5, A and B). However, MKK3/MKK6 phosphorylation was detectable after incubation with hyperosmotic NaCl, which triggered robust MKK3/MKK6 phosphorylation in SB203580-treated and nontreated HeLa cells (Fig. 5 C). In addition, unlike thrombin-dependent p38 activation, NaCl-induced p38 phosphorylation was not inhibited by SB203580 (Fig. 5 C). These findings suggest that thrombin-induced p38 activation may occur through autophosphorylation and be independent of MKK3 and MKK6.
Figure 5.
Thrombin-activated PAR1 induces p38 autophosphorylation. (A and B) PAR1-expressing HeLa cells (A) and endothelial cells (B) were pretreated with 5 µM SB203580 for 20 min and then stimulated with 10 nM α-Th, and phosphorylation of p38, MSK1, ERK1/2, MKK3, and MKK6 was determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). See Fig. S3 (A and B). (C) PAR1 HeLa cells were pretreated with SB203580 as described above and then incubated with 10 nM α-Th and 400 µM NaCl, and phosphorylation of p38, MKK3, and MKK6 was determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (**, P < 0.01).
Ubiquitination of activated PAR1 initiates formation of a TAB2–TAB1–p38 signaling complex on endosomes
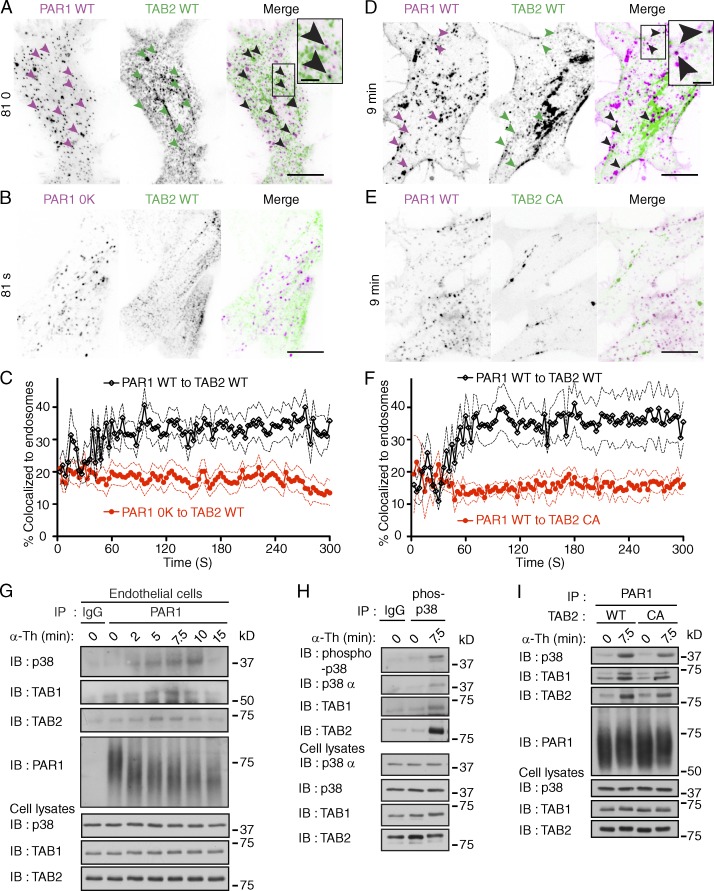
Previous studies showed that the direct binding of TAB1 to p38-α MAPK induces autophosphorylation and activation (Ge et al., 2002; De Nicola et al., 2013), but whether GPCRs induce p38 activation through this pathway is not known. TAB2 is an adaptor protein that associates with TAB1 (Bouwmeester et al., 2004) and contains an Npl4 zinc finger (NZF) domain that binds K63-linked ubiquitin (Kulathu et al., 2009). The ubiquitin-binding properties of TAB2 suggested that it might link TAB1 to ubiquitinated PAR1 to promote p38 activation. To investigate a role for TAB1 and TAB2 in PAR1 signaling, we first examined if TAB2 was recruited to PAR1 on endosomes through a ubiquitin-dependent pathway using TIRF and confocal microscopy in live cells. Stimulation of PAR1 WT HeLa cells cotransfected with TAB2 WT fused to tdTomato resulted in the rapid formation of PAR1 endocytic puncta that displayed substantial colocalization with TAB2 WT (Fig. 6, A and C) and early endosome antigen 1 (EEA1), a marker of early endosomes (Fig. S4, A–D). However, TAB2 WT failed to associate with ubiquitination-deficient PAR1 0K mutant after agonist addition (Fig. 6, B and C). To test whether the ubiquitin-binding capacity of TAB2 was also necessary for recruitment to activated PAR1 WT, the TAB2 CA mutant defective in ubiquitin binding was used (Kanayama et al., 2004). In contrast to TAB2 WT, the TAB2 CA mutant fused to tdTomato failed to colocalize with activated PAR1 on endocytic punctae (Fig. 6, D–F). These findings suggest that the capacity of TAB2 to bind to ubiquitin is critical for its association with activated PAR1 on endosomes.
Figure 6.
Activation of PAR1 initiates assembly of an ubiquitin-dependent TAB2–TAB1–p38 signaling complex on endosomes. (A and B) PAR1 WT or 0K mutant coexpressed with TAB2 WT tdTomato in HEK293 cells were stimulated with 100 µM SFLLRN. Images of live cells at 81 s are shown. Arrowheads show PAR1 WT and TAB2 WT containing punctae. PAR1 WT and TAB2 WT colocalization is indicated by black punctae in the merged image. Insets are magnifications of the boxed areas. Bars: (main panels) 10 µm; (insets) 2.5 µm. See Fig. S4 (A–D). (C) Quantification of TAB2 WT tdTomato colocalization with PAR1 WT (black lines) or 0K mutant (red lines) on endosomes induced by 100 µM SFLLRN. Solid lines represent the mean and the dashed lines represent the standard deviation. n > 5 cells. (D and E) PAR1 WT coexpressed with either TAB2 WT or CA mutant tdTomato in HEK293 cells were stimulated with 100 µM SFLLRN. Arrowheads show PAR1 WT and TAB2 WT positive punctae at 9 min. PAR1 WT and TAB WT containing punctae are indicated by black punctae in the merged image. Insets are magnifications of boxed areas. Bars: (main panels) 10 µm; (insets) 2.5 µm. (F) Quantification of PAR1 WT colocalization with either TAB2 WT (black lines) or CA mutant (red lines) tdTomato on endosomes induced by 100 µM SFLLRN. Solid lines represent the mean and the broken lines represent the standard deviation. n > 5 cells. (G) Endogenous PAR1 was immunoprecipitated from endothelial cells stimulated with 10 nM α-Th, and coassociated p38, TAB1, and TAB2 were determined. (H) Phosphorylated p38 was immunoprecipitated from PAR1 HeLa cells stimulated with 10 nM α-Th, and coprecipitated p38-α, TAB1, and TAB2 was detected. (I) PAR1-expressing HeLa cells coexpressing TAB2 WT or CA mutant were stimulated with 10 nM α-Th, and coassociated p38, TAB1, and TAB2 were determined.
To test the hypothesis that activation of PAR1 promotes assembly of a TAB–p38 signaling complex, endogenous PAR1 was immunoprecipitated from thrombin-stimulated endothelial cells, and coassociated proteins were examined. TAB1 and TAB2 coassociated with activated PAR1 within minutes of thrombin stimulation (Fig. 6 G), which also induced the recruitment of p38 MAPK (Fig. 6 G). To confirm p38 complex assembly, PAR1 WT HeLa cells were incubated with thrombin, phospho-p38 was immunoprecipitated, and coassociated proteins were determined. Phospho-p38-α isoform, TAB1, and TAB2 were detected in immunoprecipitates from cells stimulated with thrombin but not in unstimulated cells or IgG control (Fig. 6 H), which is consistent with thrombin-induced formation of a TAB2–TAB1–p38-α signaling complex. To test whether the ubiquitin-binding capacity of TAB2 was necessary for activated PAR1-induced TAB2–TAB1–p38 signaling complex formation, the TAB2 CA mutant defective in ubiquitin binding was used (Kanayama et al., 2004). The formation of agonist-promoted PAR1-induced TAB1–TAB2 p38 signaling complex was reduced in cells exogenously expressing TAB2 CA mutant compared with WT TAB2 (Fig. 6 I). These findings suggest that PAR1 ubiquitination and the ubiquitin binding capacity of TAB2 are required for formation of a p38 MAPK endosomal signaling complex.
TAB1 and TAB2 are critical mediators of thrombin-induced p38 activation
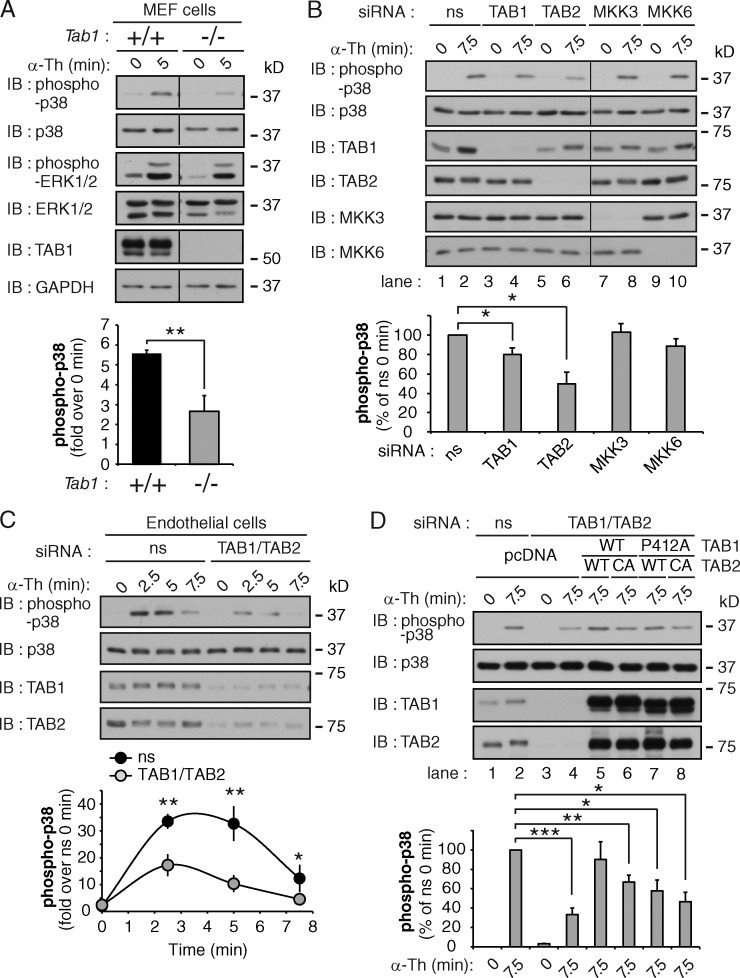
To define the function of TAB1 in thrombin-stimulated p38 signaling, we used mouse embryonic fibroblasts (MEFs) isolated from WT and Tab1−/− homozygous mutant embryos (Inagaki et al., 2008). WT MEFs exhibited a marked increase in p38 activation after thrombin stimulation, whereas p38 activation was significantly reduced in Tab1−/−-deficient MEFs (Fig. 7 A). In contrast, thrombin-induced ERK1/2 phosphorylation was comparable in WT and Tab1−/−-deficient MEFs (Fig. 7 A), indicating that the loss of TAB1 expression specifically disrupts thrombin-dependent p38 MAPK activation.
Figure 7.
TAB1 and TAB2 are required for activated PAR1-induced p38 activation. (A) Tab1 WT (+/+) and null (−/−) MEFs were stimulated with 10 nM α-Th, and phosphorylation of p38 and ERK1/2 was determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (**, P < 0.01). (B) PAR1 HeLa cells were transiently transfected with ns, TAB1, TAB2, MKK3, or MKK6 siRNAs, and stimulated with 10 nM α-Th. p38 phosphorylation was then determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05). See Fig. S4 E. (C) Endothelial cells transfected with ns, TAB1, and TAB2 siRNA were stimulated with 10 nM α-Th, and p38 phosphorylation was examined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05; **, P < 0.01). (D) PAR1 HeLa cells were cotransfected with ns or TAB1 and TAB2 siRNA together with pcDNA vector (lanes 1–4); siRNA-resistant TAB1 WT, TAB2 WT, or CA mutant (lanes 5–6); or siRNA-resistant TAB1 P412A mutant, TAB2 WT, or CA mutant (lanes 7–8); and stimulated with 10 nM α-Th. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Next, we examined the role of TAB1 and TAB2 versus MKK3 and MKK6 in thrombin-induced p38 activation. TAB1, TAB2, MKK3, or MKK6 were depleted in PAR1 WT HeLa cells using siRNA. The loss of either MKK3 or MKK6 had no significant effect on thrombin-stimulated p38 activation compared with ns siRNA-transfected cells (Fig. 7 B, lanes 7–10), which suggests that neither MKK3 nor MKK6 expression are critical mediators of the pathway. Because codepletion of MKK3 and MKK6 by siRNA resulted in a marked loss in TAB1 expression (Fig. S4 E), we were unable to accurately assess effects on p38 activation. However, in contrast to the loss of either MKK3 or MKK6 expression, depletion of TAB1 alone caused a modest but significant decrease in thrombin-stimulated p38 activation, whereas loss of TAB2 expression resulted in a marked reduction in p38 activation without affecting MKK3 or MKK6 expression (Fig. 7 B, lanes 1–6). Thrombin-promoted p38 activation was also significantly reduced in endothelial cells depleted of TAB1 and TAB2 expression (Fig. 7 C), indicating that TAB1 and TAB2 are critical mediators of thrombin-induced p38 activation.
To confirm that the observed siRNA effects are specific, we performed rescue experiments using siRNA-resistant TAB1 and TAB2. Co-depletion of TAB1 and TAB2 caused a marked inhibition of thrombin-dependent p38 activation (Fig. 7 D, lanes 1–4). However, coexpression of siRNA-resistant TAB1 WT and TAB2 WT resulted in significant recovery of thrombin-stimulated p38 phosphorylation that was comparable to ns siRNA-transfected cells (Fig. 7 D, lanes 2, 4, and 5). These findings indicate that WT TAB1 and TAB2 expression are sufficient to restore thrombin-dependent p38 activation. In contrast, coexpression of a TAB1 proline 412 to alanine (P412A) mutant that blocks interaction with p38-α (Zhou et al., 2006) together with TAB2 WT or TAB1 WT together with TAB2 CA mutant failed to rescue thrombin-induced p38 activation (Fig. 7 D, lanes 5–8). These results suggest that in addition to PAR1 ubiquitination, the TAB1 p38 binding motif and ubiquitin-binding properties of TAB2 are required for thrombin-induced p38 activation.
Thrombin-induced p38 MAPK signaling regulates TAB1 stability
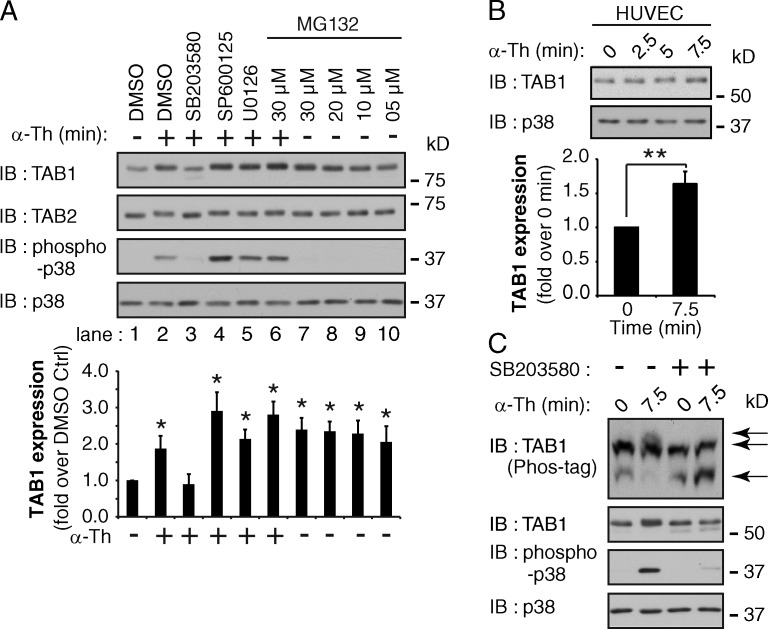
TAB1 activity is modulated by phosphorylation mediated by various MAPKs including p38, JNK, and ERK1/2 after cytokine stimulation (Cheung et al., 2003; Mendoza et al., 2008). However, the mechanisms that control TAB1 activity in response to GPCR activation are not known. We found that thrombin activation of PAR1 caused a significant approximately twofold increase in TAB1 expression (Fig. 8 A, lanes 1 and 2), whereas TAB2 expression remained unchanged, suggesting that TAB1 protein stability is dynamically regulated. A significant increase in TAB1 expression was also induced after thrombin stimulation of endogenous PAR1 in endothelial cells (Fig. 8 B). To determine whether MAPKs regulate TAB1 expression in response to thrombin stimulation, HeLa cells were pretreated with p38 (SB203580), JNK (SP600125), or MEK1/2 (U0126) inhibitors and TAB1 expression was assessed. SB203580 blocked both thrombin-mediated p38 activation and increased TAB1 expression (Fig. 8 A, lanes 1–3), whereas the JNK and MEK1/2 inhibitors did not (Fig. 8 A, lanes 1–5). Phos-tag gels were then used to determine whether thrombin affected the phosphorylation status of TAB1 via p38 activation. Thrombin induced a marked shift in TAB1 mobility that was virtually abolished by SB203580 pretreatment (Fig. 8 C), indicating that p38 is required for TAB1 phosphorylation. To examine whether regulation of TAB1 expression is mediated by rapid degradation in the proteasome, HeLa cells were pretreated with the proteasomal inhibitor MG132. Incubation with various concentrations of MG132 resulted in a 2.5-fold increase in basal TAB1 protein expression compared with DMSO control (Fig. 8 A, lanes 1 and 7–10) that was not further increased by thrombin stimulation (Fig. 8 A, lanes 2, 6, and 7). These results suggest that TAB1 expression is dynamically regulated through p38-mediated phosphorylation and proteasomal degradation. We speculate that TAB1 is protected from proteasomal degradation after thrombin activation of PAR1 possibly through recruitment into the TAB1–TAB2–p38 endosomal signaling complex.
Figure 8.
Activation of PAR1 increases TAB1 expression through a p38-dependent pathway. (A) PAR1 HeLa cells were pretreated for 30 min with DMSO, 5 µM SB203580, 20 µM SP600125, 10 µM U0126, or varying MG123 concentrations and then stimulated with 10 nM α-Th. TAB1 and TAB2 expression and p38 phosphorylation were then detected. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05). (B) HUVECs were treated with 10 nM α-Th, and TAB1 expression was determined. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (**, P < 0.01). (C) PAR1 HeLa cells were pretreated with DMSO or 5 µM SB203580 for 30 min and stimulated with 10 nM α-Th. Cell lysates were resolved on Phos-tag gels to detect TAB1 mobility (indicated by arrows) or SDS-PAGE to detect TAB1 and p38.
Ubiquitin-dependent regulation of p38 activation is conserved for GPCRs
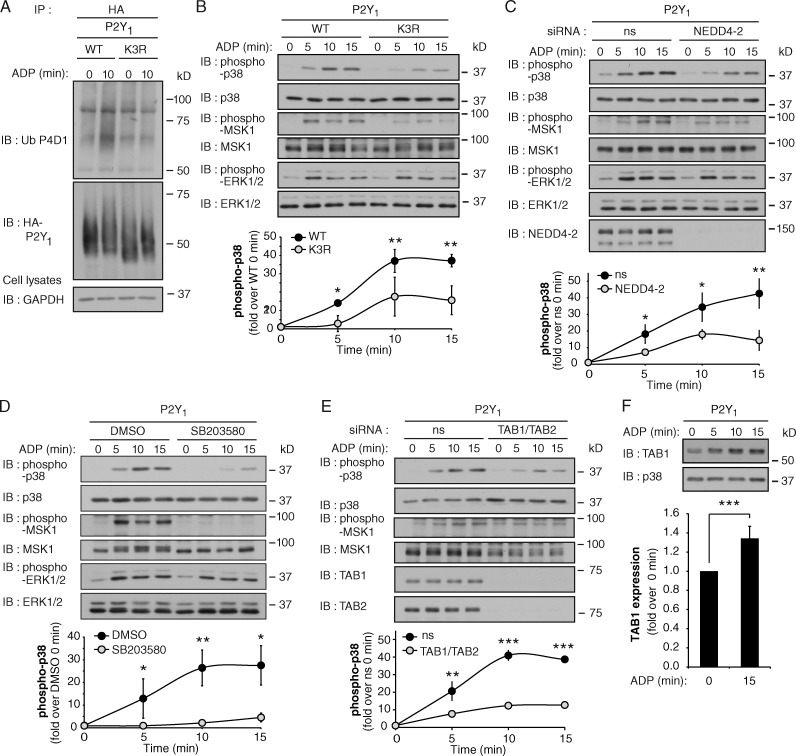
We hypothesize that similar to PAR1, GPCRs containing ALIX-binding YPXnL motifs may be modified with ubiquitin to facilitate assembly of a p38 signaling complex rather than lysosomal degradation. The P2Y1 receptor signals in response to the adenine nucleotide ADP and contains a highly conserved YPXnL motif (Dores et al., 2012). P2Y1 is also expressed in endothelial cells and promotes inflammatory signaling (Zemskov et al., 2011). We therefore examined whether ubiquitination of the purinergic P2Y1 receptor is required for p38 MAP kinase activation. Agonist activation of P2Y1 expressed in HeLa resulted in a marked increase in receptor ubiquitination (Figs. 9 A and S5 A). To determine the sites of P2Y1 ubiquitination, lysine K341, K353, and K366 residues in the cytoplasmic tail were mutated to arginine (R) and designated “K3R.” HeLa cells expressing similar amounts of P2Y1 WT and K3R at the cell surface (Fig. S5 B) were then stimulated with ADP and receptor ubiquitination was assessed. In contrast to WT P2Y1, ADP failed to increase ubiquitination of the P2Y1 K3R mutant receptor (Fig. 9 A), indicating that the cytoplasmic tail lysine residues are the major sites of agonist-induced P2Y1 receptor ubiquitination.
Figure 9.
P2Y1 receptor-induced p38 activation is mediated by a ubiquitin- and TAB1–TAB2-dependent pathway. (A) HA-tagged P2Y1 WT or K3R mutant expressed in HeLa cells were stimulated with 10 µM ADP and ubiquitination was detected. See Fig. S5 (A–C). (B) HA-P2Y1 WT or K3R HeLa cells were stimulated with 10 µM ADP, and p38, MSK1, and ERK1/2 phosphorylation was detected. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05; **, P < 0.01). (C) HA-P2Y1 HeLa cells transfected with ns or NEDD4-2 siRNA were treated with 10 µM ADP, and p38, MSK1, and ERK1/2 phosphorylation was detected. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05; **, P < 0.01). See Fig. S5 (D and E). (D) HA-P2Y1 HeLa cells pretreated with DMSO or 5 µM SB203580 for 20 min were stimulated with 10 µM ADP, and p38, MSK1, and ERK1/2 phosphorylation was detected. Data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (*, P < 0.05; **, P < 0.01). (E) HA-P2Y1 HeLa cells transfected with ns or TAB1 and TAB2 siRNA were stimulated with 10 µM ADP, and p38 and MSK1 phosphorylation was detected. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (**, P < 0.01; ***, P < 0.001). See Fig. S5 F. (F) HA-P2Y1 HeLa cells were stimulated with 10 µM ADP, and TAB1 expression was detected. The data (mean ± SD [error bars], n = 3) were analyzed using a Student’s t test (***, P < 0.001).
We next examined the capacity of the ubiquitin-deficient P2Y1 K3R mutant to promote p38 signaling. ADP stimulation of P2Y1 WT HeLa cells resulted in a significant increase in phosphorylation of p38 and MSK1 compared with untreated and untransfected HeLa cells (Figs. 9 B and S5 C). In contrast to P2Y1 WT, activation of p38 and MSK1 by the P2Y1 K3R mutant was markedly reduced (Fig. 9 B), whereas ERK1/2 signaling remained intact. We next examined whether P2Y1 ubiquitination is mediated by NEDD4-2 and regulates p38 activation using siRNA-targeted depletion of NEDD4-2. In NEDD4-2–deficient cells, ADP-stimulated ubiquitination of P2Y1 was significantly reduced compared with control siRNA-transfected cells (Fig. S5 D), whereas the expression of P2Y1 was not affected (Fig. S5, D and E). These results indicate that ADP-induced P2Y1 ubiquitination is mediated by NEDD4-2. A significant reduction in ADP-dependent phosphorylation of p38 and MSK1, but not ERK1/2, was also observed in NEDD4-2 deficient cells (Fig. 9 C). These findings suggest that ubiquitination of P2Y1 is required for p38 activation, similar to PAR1.
To investigate whether P2Y1 activation of p38 MAPK occurs through autophosphorylation, we used the p38 inhibitor SB203580. ADP stimulation of p38 and MSK1 phosphorylation were significantly inhibited in SB203580-treated P2Y1 WT HeLa cells compared with control cells (Fig. 9 D), whereas ERK1/2 activation was not affected (Fig. 9 D), indicating that P2Y1-mediated p38 activation occurs through autophosphorylation. Next, the function of TAB1 and TAB2 in P2Y1-induced p38 activation was examined using siRNA-targeted depletion. A significant decrease in ADP-stimulated p38 and MSK1 phosphorylation was detected in cells deficient in TAB1 and TAB2 expression (Fig. 9 E), which expressed comparable amounts of cell surface P2Y1 to the control cells (Fig. S5 F). In addition, a significant increase in TAB1 protein expression was detected after ADP activation of P2Y1 (Fig. 9 F). These findings provide evidence that agonist-induced ubiquitination of a subset of GPCRs is essential for the initiation of p38 autophosphorylation and activation mediated through a noncanonical TAB1- and TAB2-dependent pathway.
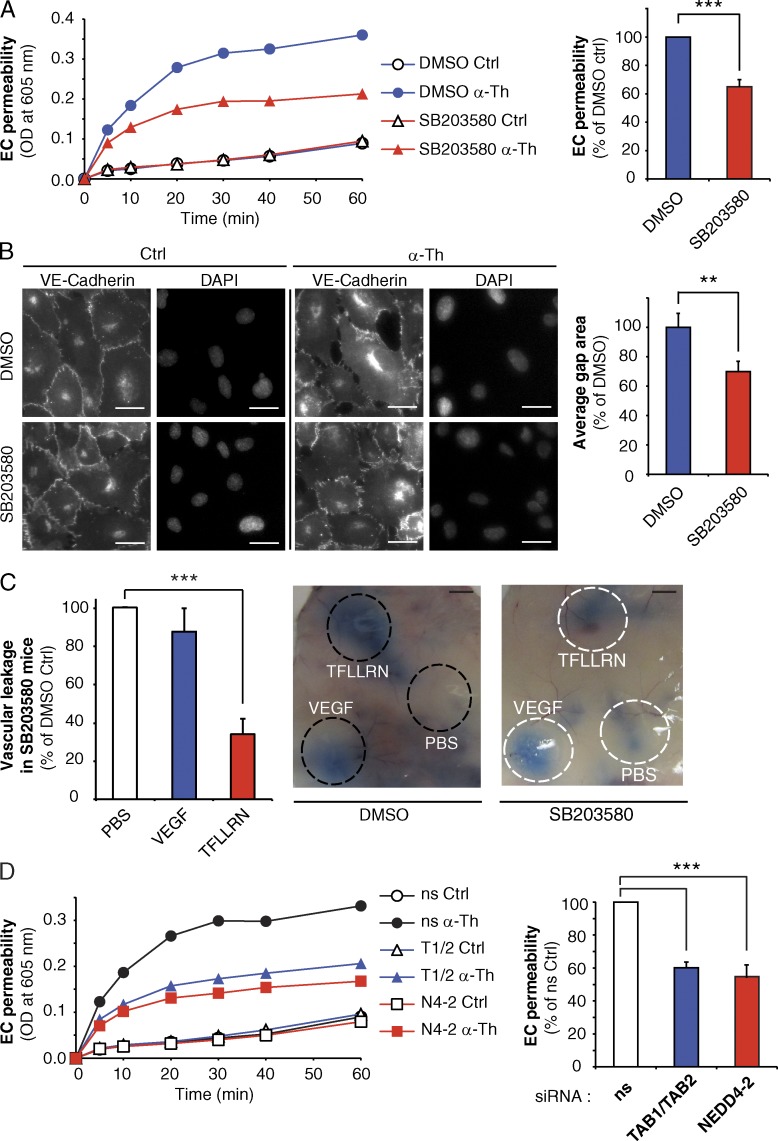
PAR1 regulates endothelial barrier permeability through the atypical TAB2–TAB1–p38 signaling pathway
To assess the function of PAR1-induced ubiquitin and TAB1–TAB2 p38 activation pathway, we examined whether inhibition of p38 autophosphorylation affected thrombin-induced endothelial permeability in vitro. Confluent monolayers of human endothelial cells were pretreated with SB203580 or DMSO and then incubated with thrombin. Thrombin induced a rapid increase in endothelial barrier permeability in control cells (Fig. 10 A), whereas pretreatment with the SB203580 resulted in a significant inhibition of thrombin-induced permeability (Fig. 10 A). To confirm these results, we examined thrombin-stimulated HUVEC monolayer gap formation by immunofluorescence microscopy using VE-cadherin as a marker of adherens junctions. Pretreatment of human umbilical vein endothelial cells (HUVECs) with SB203580 caused significant inhibition of thrombin-induced gap formation (Fig. 10 B). These findings suggest that p38 mediates thrombin-induced endothelial barrier disruption. Next, we examined whether p38 signaling is required for activated PAR1-induced vascular permeability in vivo using mice. The extent of vascular leakage measured by the “Miles assay” flux of Evans blue–conjugated albumin was minimal in mice pretreated with SB203580 or DMSO injected with PBS (Fig. 10 C). However, intradermal injection of the PAR1-specific agonist peptide TFLLRN induced a marked increase in leakage of Evans blue compared with PBS injected control (Fig. 10 C), which was virtually ablated in mice pretreated with SB203580. In contrast, SB203580 failed to affect VEGF-induced vascular leakage (Fig. 10 C), suggesting that p38 inhibition is specific to PAR1. Together these findings indicate that p38 is a critical mediator of activated PAR1-induced endothelial barrier disruption in vitro and in vivo.
Figure 10.
PAR1-mediated endothelial barrier disruption requires p38 signaling, NEDD4-2, TAB1 and TAB2. (A) Endothelial cells pretreated with DMSO or 5 µM SB203580 for 20 min were stimulated with 10 nM α-Th, and permeability was determined. The data in bar graphs (mean ± SD [error bars], n = 3) from 30 min is representative of three independent experiments and were analyzed using a Student’s t test (***, P < 0.001). (B) HUVECs pretreated with DMSO or 5 µM SB203580 for 20 min were stimulated with 10 nM α-Th for 10 min. Cells were immunostained for VE-cadherin and gap formation was determined. The data (mean ± SD [error bars], n = 4) were analyzed using a Student’s t test (**, P < 0.01). Bars, 10 µm. (C) Mice received intraperitoneal injection of PBS or 1 µM SB203580 before injection of PBS, 4 ng/µl VEGF, or 1 µg/µl TFLLRN. The data (mean ± SD [error bars], n = 10 mice) from three independent experiments were analyzed using a Student’s t test (***, P < 0.001). Bars, 5 mm. (D) Endothelial cells transfected with ns, NEDD4-2 (N4-2), or TAB1 and TAB2 (T1/T2) siRNA were stimulated with 10 nM α-Th, and permeability was determined. Data (mean ± SD [error bars], n = 3) in bar graphs are representative of three independent experiments at 30 min, and were analyzed using a Student’s t test (***, P < 0.001).
We next examined whether TAB1, TAB2, and NEDD4-2 mediators of noncanonical p38 activation induced by activated PAR1 regulate endothelial barrier permeability in vitro. The extent of basal permeability detected in TAB1–TAB2 or NEDD4-2–deficient endothelial cells was comparable to ns siRNA-treated control cells (Fig. 10 D). However, thrombin-induced endothelial barrier permeability was significantly inhibited in endothelial cells lacking TAB1 and TAB2 or NEDD4-2 expression (Fig. 10 D). Collectively these studies strongly suggest that activated PAR1-stimulated p38 activation occurs via an atypical TAB1–TAB2–NEDD4-2–mediated pathway that is important for the regulation of endothelial barrier disruption.
Discussion
In this study we define a novel signaling pathway by which ubiquitination of GPCRs is linked to noncanonical activation of p38 MAPK. We show that ubiquitination of PAR1 is mediated by the NEDD4-2 E3 ligase and initiates formation of a TAB2–TAB1–p38 signaling complex that occurs on endosomes. In addition, ubiquitin and TAB-dependent activation of p38 is required for thrombin-induced endothelial barrier permeability in vitro. We further demonstrate that signaling by p38 MAPK is critical for PAR1-stimulated vascular leakage in vivo. Induction of noncanonical p38 signaling is not limited to PAR1 since we found that ubiquitination of the purinergic GPCR P2Y1 is also required for TAB-dependent activation of p38 MAPK. Together these findings establish for the first time a novel function for ubiquitination of GPCRs in the regulation of p38 MAPK signaling.
Although the canonical three-tiered kinase cascade is implicated in p38 MAPK activation induced by many GPCRs, the precise mechanism by which inflammatory GPCRs regulate p38 activation has not been clearly defined. Noncanonical p38 activation is induced by direct binding of TAB1 to the C-terminal domain of p38 and requires a critical proline residue P412 of TAB1 (Ge et al., 2002; Zhou et al., 2006). TAB1 induces a conformational change in p38 that triggers autophosphorylation of threonine-180 and tyrosine-182 residues present in the activation loop, which are the same sites phosphorylated by MKK3 and MKK6 in the canonical pathway (De Nicola et al., 2013). However, unlike MKK3 and MKK6, which activate all four p38 isoforms, TAB1 specifically activates the p38-α isoform (Ge et al., 2002; Remy et al., 2010). We discovered that both PAR1 and P2Y1 promote p38 activation through autophosphorylation, since the process was inhibited by SB203580, which blocks p38-α and p38-β activity but does not perturb phosphorylation of p38 mediated by upstream MAPKs (Davies et al., 2000; Ge et al., 2002). The role of TAB1 in p38 activation induced by PAR1 was demonstrated using TAB1-deficient MEFs and was supported further for both PAR1 and P2Y1 using siRNA-mediated ablation of TAB1 expression. We also failed to detect phosphorylation of MKK3 or MKK6 after PAR1 activation, and depletion of MKK3 or MKK6 by siRNA had no effect on thrombin-induced p38 activation. In addition, siRNA knockdown/rescue studies indicated that expression of siRNA-resistant TAB1 WT was sufficient to restore p38 activation in response to thrombin, whereas the TAB1 P412A mutant was not. These findings are the first to show that a subset of GPCRs activates p38 MAPK through a noncanonical pathway mediated by TAB1-dependent p38 autophosphorylation.
Ubiquitination of GPCRs is known to function in lysosomal sorting (Dores and Trejo, 2014). However, not all GPCRs require ubiquitination for lysosomal trafficking, including PAR1 (Dores et al., 2012). Here we show an atypical function for ubiquitination of a subset of GPCRs that is critical for initiating TAB-dependent p38 activation. We found that PAR1 rendered ubiquitin-deficient by either mutation of critical cytoplasmic lysine residues (Wolfe et al., 2007) or through siRNA-mediated depletion of NEDD4-2 resulted in significant inhibition of agonist-promoted p38 activation. Similarly, activation of p38 MAPK by the purinergic P2Y1 receptor requires ubiquitination mediated by E3 ligase NEDD4-2. Under these conditions, ERK1/2 signaling remained intact, indicating that GPCR ubiquitination is selective for activation of specific signaling pathways. We further demonstrate that K63-ubiquitin chains are conjugated to PAR1, consistent with NEDD4-2’s capacity to modify substrate proteins with K63-linked ubiquitin (Vina-Vilaseca and Sorkin, 2010). However, it is not clear why ubiquitin conjugation to PAR1 does not function in sorting the receptor to lysosomes. Our data indicate that the association of TAB2, an adaptor protein that binds to K63-linked ubiquitin chains via its NZF domain (Kulathu et al., 2009), with PAR1 early in the endocytic pathway requires its ubiquitin-binding domain and may prevent PAR1 interaction with ESCRT components on early endosomes. We also found that WT TAB2 is rapidly recruited to activated PAR1 WT but not to the 0K ubiquitin-deficient mutant and remains associated with the receptor during peak p38 activation on endosomes based on TIRF microscopy and coimmunoprecipitation experiments. In contrast, the capacity of a TAB2 NZF CA mutant defective in ubiquitin binding to associate with PAR1 and restore p38 signaling in response to thrombin stimulation was markedly reduced. These studies suggest that the ubiquitin-binding capacity of TAB2 is essential for association with ubiquitinated PAR1 and is critical for TAB1-dependent p38 activation. However, these studies cannot exclude the possibility that TAB2 associates indirectly with PAR1 via an intermediary ubiquitin-binding protein. While TAB2 and TAB1 have been shown to coassociate (Bouwmeester et al., 2004), the mechanisms that govern their coassociation and function in propagation of p38 signaling remain to be determined.
The mechanisms that regulate TAB1 function during GPCR-stimulated noncanonical p38 activation are not known. We discovered that TAB1 expression was significantly increased after activation of PAR1. Several studies indicate that TAB1 is phosphorylated on C-terminal serine and threonine residues by JNK, ERK1/2, and p38 MAPK (Cheung et al., 2003; Mendoza et al., 2008). We show that PAR1-induced increase in TAB1 expression is mediated by p38 and not by JNK or ERK1/2. Moreover, p38 phosphorylates TAB1 in response to PAR1 activation, which appears to inhibit its rapid degradation by the proteasome. In previous studies, TAB1 phosphorylation by p38 was shown to inhibit its ability to bind to TAK1, an upstream MAP3K (Cheung et al., 2003). Inhibition of the TAB1 phosphatase DUSP14 also blocked TAB1 dephosphorylation and reduced TAK1 activation (Yang et al., 2014). These studies, and our results, suggest that phosphorylation of TAB1 by p38 may specify its function in the noncanonical p38 pathway and likely limit its role in TAK1-dependent activation of MKK3 and MKK6. Intriguingly, MG132 incubation alone caused substantial accumulation of TAB1 protein in the absence of stimulation, indicating that the TAB1 protein displays high basal turnover. An increase in TAB1 expression alone however was not sufficient to initiate p38 activation, but rather required activation of PAR1. These studies suggest that TAB1 is dynamically regulated through phosphorylation and degradation. The recruitment of TAB1 into a signaling complex with activated and ubiquitinated PAR1 on endosomes appears to stabilize TAB1 expression and is critical for p38 activation.
The physiological importance of TAB1-dependent autophosphorylation and activation of p38 has been demonstrated in disease contexts such as interleukin-12 production in macrophages during infection (Kim et al., 2005); myeloid light-chain induced cardiotoxicity (amyloidosis) in rat, mice, and zebrafish models (Shi et al., 2010; Mishra et al., 2013); and skin inflammation (Theivanthiran et al., 2015). In skin, the E3 ligase Itch was shown to regulate p38 signaling by controlling the ubiquitination and degradation of TAB1 (Theivanthiran et al., 2015). Murine models of myocardial ischemia have also shown that p38-α activation is regulated through TAB1-dependent autophosphorylation and contributes to disease progression (Tanno et al., 2003; De Nicola et al., 2013; Wang et al., 2013). Importantly, thrombin generation and PAR1 have also been implicated in myocardial ischemia, but whether TAB1-dependent p38 activation functions in this pathway is not known. Our findings indicate that TAB2-, TAB1-, and NEDD4-2–dependent p38 signaling is critical for activated PAR1-stimulated endothelial barrier permeability in vitro and that PAR1-induced p38 signaling is essential for vascular leakage in vivo. However, the mechanisms by which TAB1-dependent p38 signaling specifically regulates endothelial barrier disruption require further exploration. Both Gq and G12/13 have been linked to thrombin-induced endothelial barrier permeability in vitro (Komarova et al., 2007). In a previous study, Gq was shown to be critical for vascular leakage induced by PAR1 and other GPCRs in vivo, rather than G12/13 (Korhonen et al., 2009). PAR1 coupling to Gq leads to intracellular Ca2+ mobilization and PKC activation. PKC regulates the actin cytoskeleton and disassembly of adherens junctions, which promotes endothelial barrier permeability (Komarova et al., 2007). It is not known if components of the TAB–p38 signaling pathway are modulated by thrombin-induced mobilization of intracellular Ca2+ or PKC activation, and this will be investigated. In addition, the mechanism by which PAR1-induced TAB1-stimulated p38 MAPK activation promotes endothelial barrier permeability is not clear but may involve direct effects on adherens junction components that lead to disassembly and is an important area for future study.
Materials and methods
Antibodies and reagents
PAR1 agonist peptides (SFLLRN and TFLLRN) were synthesized and purified by reverse-phase high-pressure liquid chromatography at Tufts University Core Facility. α-Thrombin (α-Th) was purchased from Enzyme Research Laboratories. ADP was from Acros Organics. TNF-α was obtained from EMD Millipore. VEGF was purchased from PeproTech. Polyclonal rabbit anti-FLAG and -HA antibodies and mouse IgG antibody were from Rockland Immunochemicals. Mouse monoclonal P4D1 anti-ubiquitin and polyclonal rabbit anti-p38 antibodies were from Santa Cruz Biotechnology, Inc. Monoclonal anti-PAR1 WEDE antibody was from Beckman Coulter. The monoclonal mouse anti-HA antibody was from Covance. The mouse M2 anti-FLAG and -actin antibodies were from Sigma-Aldrich. Polyclonal and monoclonal rabbit anti–phospho-ATF2, Akt, phospho-Akt, K63 polyubiquitin, MSK1, phospho-MSK1, MKK3, phospho-MKK3, MKK6 and phospho-MKK6, NEDD4-1, NEDD4L (also known as NEDD4-2), p38, phospho-p38, p38α, p42/44 MAPK, phospho-p42/44 MAPK, VE-cadherin antibodies, and ATF2 fusion protein were from Cell Signaling Technology. Monoclonal mouse anti-EEA1 was from BD. Monoclonal mouse anti-GAPDH antibody was from GeneTex. HRP-conjugated goat–anti-rabbit and goat–anti-mouse antibodies were from Bio-Rad Laboratories. Alexa Fluor 488–, 594–, and 647–conjugated secondary antibodies were from Life Technologies. SB203580 and SB202190 (p38 inhibitors) and U0126 (MEK inhibitor) were from LC laboratories. SP600125 (JNK inhibitor) and MG132 (proteasomal inhibitor) were from Sigma-Aldrich. Phos-Tag was purchased from Wako Pure Chemical Industries. Poly K48 and K63 ubiquitin chains were from Boston Biochem.
Plasmids and cells
N-terminal FLAG-tagged human PAR1 WT and FLAG-PAR1 0K mutant cDNAs were cloned into the mammalian expression vector pBJ, and N-terminal HA-tagged PAR1 WT was cloned into pcDNA3.1 as described previously (Trejo et al., 2000; Wolfe et al., 2007; Lin and Trejo, 2013). Human TAB1 pCMV, and TAB2 pCMV were provided by J. Ninomiya-Tsuji (North Carolina State University, Raleigh, NC). N-terminal HA-tagged human P2Y1 pcDNA was obtained from R. Nicholas (University of North Carolina). TAB2 Cysteine (C) 670 to alanine (A) and C673A pCMV (termed TAB2-CA), TAB1 proline 412 to A termed “TAB1 P412” pCMV, HA P2Y1 lysines (K) 341, K353, and K366 mutated to arginine (R), and pcDNA termed “P2Y1 K3R” mutants were generated by QuikChange site-directed mutagenesis (Agilent Technologies) and confirmed by dideoxy sequencing. The siRNA-resistant TAB1 WT and P412A mutant pCMV and TAB2 WT and CA mutant pCMV mutants were also generated by site-directed mutagenesis of nucleotides t373c, g375c, t378g, and g381a for TAB1 and c1044a, t1045a, c1046g, and c1050t for TAB2, and confirmed by sequencing. TAB2 WT and TAB2 CA mutant and N-terminal tdTomato were subcloned into pcDNA3.1 using standard techniques. HUVECs were from Lonza Ltd. and maintained according to the manufacturer’s instructions. HUVEC-derived EA.hy926 cells and HeLa cells were cultured as described previously (Wolfe et al., 2007; Russo et al., 2009). HeLa cells stably expressing FLAG-PAR1 WT or FLAG-PAR1 0K mutant pBJ were generated by cotransfection with hygromycin resistance vector, and screened by cell surface ELISA. Cells were grown and maintained as described previously (Wolfe et al., 2007). Immortalized MEFs from WT (Tab1+/+) and mutant (Tab1−/−) embryos isolated from heterozygous Tab1 floxed mice, in which the loxP sites flanked exons 9 and 10 to remove the C-terminal region of TAB1 protein, were maintained as previously described (Inagaki et al., 2008). HEK293 cells were cultured as described previously (Soohoo and Puthenveedu, 2013).
Transfections and siRNA
Cells were transfected with plasmids and siRNAs using Lipofectamine 2000 and Oligofectamine, respectively, according to the manufacturer’s instructions (Life Technologies). NEDD4.1, NEDD4L (NEDD4.2) SMARTpool, WWP1 SMARTpool, and AIP4 (5′-GGUGACAAAGAGCCAACAGAG-3′) were used at 50 nM and purchased from GE Healthcare. WWP2 Stealth siRNA was used at 50 nM and obtained from Life Technologies. NEDD4-2 #7 siRNA (5′-AAGAAUAUCGCUGGAGACUCU-3′) and #9 (5′-AAGAUCAUAACACAAAGACUA-3’) siRNA were used at 25 nM. TAB1 (5′-CGGCUAUGAUGGCAACCGA-3′) used at 12.5 nM, MKK3 (5′-GGAUAUCCUGCAUGUCCAA-3′) used at 12.5 nM, MKK6 (5′-GGGCCACCGUGAACUCACA-3′) used at 50 nM, TAB2 siRNA (5′-CCUCCAGCACUUCCUCUUC-3′) used at 50 nM, and ns siRNA (5′-GGCUACGUCCAGGAGCGCACC-3′) were all obtained from QIAGEN. The TAB knockdown rescue experiments were performed by transfecting HeLa cells with ns or TAB1–TAB2-specific siRNAs for 48 h. Cells were then washed with starvation buffer DMEM containing 1 mg/ml BSA, 10 mM Hepes, and 1 mM CaCl2 and transfected with either 150 ng of pcDNA3.1 or 150 ng of TAB1 WT or P412A pCMV combined with either TAB2 WT or TAB2 CA mutant using Lipofectamine 2000 and incubated for an additional 24 h.
MAPK signaling assays
Serum-starved cells were treated under various conditions. Equivalent amounts of cell lysates were resolved by SDS-PAGE, transferred to membranes, and probed with specific antibodies. Membranes were developed by chemiluminescence and quantified by densitometry. In vitro p38 kinase assays were performed according to the manufacturer’s instructions (Cell Signaling Technology).
Phos-tag gels
Phosphorylation of p38 MAPK was detected using Phos-tag gels containing 100 µM Phos-Tag acrylamide and 100 µM MgCl2 according to the manufacturer’s instructions (Wako Pure Chemical Industries).
Immunoprecipitations and ubiquitination
PAR1 and P2Y1 receptor immunoprecipitation and ubiquitination experiments were performed essentially as described previously (Chen et al., 2011; Dores et al., 2012). HeLa cells or endothelial EA.hy926 cells were grown in 6 cm dishes and starved overnight. Serum-starved cells were stimulated with agonists for the indicated times at 37°C. Cells were lysed in RIPA buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS with 50 mM β-glycerophosphate, 10 µg/ml leupeptin, aprotinin, trypsin protease inhibitor, pepstatin, 100 µg/ml benzamide, and 20 mM N-ethylmaleimide. Samples were passed through a 21 GA needle and cleared by centrifugation. Protein concentrations were determined with a bicinchoninic acid assay and equivalent amounts of lysates were used for immunoprecipitations using anti-PAR1 WEDE or anti-HA antibodies. Immunoprecipitates were eluted with 2× Laemmli sample buffer containing 200 mM dithiothreitol. Coimmunoprecipitations were performed as described above except that cells were lysed in Triton lysis buffer containing 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% Triton X-100, 5 mM EDTA, 50 mM NaF, and 50 mM β-glycerophosphate, supplemented with protease inhibitors and 20 mM N-ethylmaleimide and processed as described for ubiquitination immunoprecipitations.
Confocal and TIRF microscopy
HUVECs grown on coverslips were stimulated with agonists, fixed with 4% paraformaldehyde, permeabilized with methanol, and immunostained with a primary rabbit anti–VE-cadherin antibody, secondary anti–rabbit Alexa Fluor 488 antibody, and costained with 4’,6-diamidino-2-phenylindole. HeLa cells expressing FLAG-PAR1 grown on coverslips were stimulated with agonists, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and immunostained with a primary rabbit anti-TAB2 and mouse anti-EEA1 antibodies, and secondary anti–rabbit Alexa Fluor 488 and anti–mouse Alexa Fluor 647 antibodies. Images for both the HUVEC and HeLa cell experiments were acquired using a two-color microscope (IX81 ZDC2; Olympus) equipped with an UPlanFL N 20×/0.50 NA objective lens (1.6 NA; Olympus) and a CoolSNAP H2Q CCD camera (Photometrics). Images were collected using MetaMorph 7.7 software (Molecular Devices). To quantify gap junctions, binary masks were generated for all images using a fluorescence intensity threshold that allowed for discrimination between cells and gaps. MetaMorph’s Integrated Morphometry Analysis tool was used to calculate the mean gap area for each image’s binary mask. Data from four independent experiments were analyzed, with each experiment including 16 images per treatment. Average gap areas were normalized to the thrombin/DMSO condition. Pearson’s correlation coefficients (r) for quantifying colocalization of PAR1 with either EEA1 or TAB2 tdTomato were calculated for 12 different cells from multiple independent experiments using SlideBook 4.2 software. TIRF live-cell imaging was performed in HEK293 cells expressing PAR1 and TAB2 tdTomato. PAR1 was labeled with Alexa Fluor 488–conjugated M1-FLAG antibody. Imaging was performed at 37°C using an inverted microscope (Eclipse Ti; Nikon) equipped with a 100× 1.49 NA TIRF objective lens in a temperature and CO2 controlled chamber. The confocal experiments were performed using the 100× objective lens and images were acquired every 30 s. The images were acquired using cell imaging software (Andor iQ) on an electron-multiplying charge-coupled device camera (iXon +897; Andor) with 488 and 561 solid-state lasers as light sources as described previously (Soohoo and Puthenveedu, 2013). Data were analyzed using ImageJ, Imaris, and MATLAB software (MathWorks). Objective-based detection of PAR1 and TAB2 objects was identified using the Spots tracking algorithm in Imaris (Bitplane). Spots were identified using local background subtraction and filtered using the quality filter. Each object for the confocal images was manually verified while blinded from the other channel. TIRF images were not all manually verified, but were optimally filtered. Colocalization was determined by using the Matlab Imaris plug-in Colocalize Spots with a threshold of 0.5 voxels. This is an object-based colocalization plug-in that considers two spots to be colocalized when their centers are 0.5 voxels apart or closer. The number of colocalized spots over the total number of spots per time frame was plotted to give percent colocalization.
Cell surface ELISA
Cell surface expression of PAR1 and P2Y1 receptor was measured by ELISA (Soto and Trejo, 2010). Cells were fixed with 4% paraformaldehyde before incubation with primary antibody followed by secondary HRP-conjugated antibody. The amount of antibody bound to the cell surface was determined by incubation with one-step 2,2’-azinobis-3-ethylbenzthiazoline-6-sulfonic acid (Thermo Fisher Scientific) substrate for 10–20 min at room temperature. An aliquot was removed and the absorbance at 405 nm was determined using a microplate reader (SpectraMax Plus; Molecular Devices).
Endothelial barrier permeability
Endothelial barrier permeability was quantified by measuring the flux of Evans blue–bound BSA as described previously (Russo et al., 2009). Endothelial EA.hy926 cells were seeded into 3.0 µM transwell permeability support chambers (Corning) and grown for 3–5 d until confluent. The cells were starved overnight and treated as indicated. Evans blue conjugated to BSA was added to the upper chamber after 10 min of agonist stimulation. Samples were removed from the lower chamber at the indicated time points and the amount of Evans blue diffusion was quantified by measuring the absorbance at 605 nm using a microplate reader (SpectraMax Plus; Molecular Devices).
Vascular permeability assay
Vascular permeability was measured in vivo as described previously with minor modifications (Korhonen et al., 2009). In brief, 8-wk-old CD1/CD1 female mice were anesthetized and injected intraperitoneally with 100 µl of PBS or 1 µM SB203580. After 1 h, mice were anesthetized again and injected in the tail vein with 200 µl of 1.0% Evan’s blue–0.1% BSA diluted in PBS. After 1 min, 50 µl of 0.1% BSA in PBS, 4 ng/ µl VEGF, or 1 µg/µl TFLLRN (PAR1-specific agonist peptide) were injected intradermally into separate areas of the shaved back skin of the mouse. The mice were sacrificed 10 min after injection and 8 mm of skin containing the site of injection was removed. The skin biopsies were incubated in 500 µl of formamide at 65°C for 24 h, and the amount of extracted Evan’s blue dye was measured using a spectrophotometer at OD 595 nm. Animal studies were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Diego.
Statistical analysis
Data were analyzed using Prism software (version 4.4; GraphPad Software). Statistical significance was determined by performing a Student’s t test.
Online supplemental material
Fig. S1 shows PAR1 ubiquitination with the hot lysis method, PAR1 WT and 0K mutant expression, and α-Th–induced Akt phosphorylation in PAR1 WT and 0K HeLa cells. Fig. S2 shows E3 ubiquitin ligase mini-screen and loss of α-Th and SFLLRN-induced PAR1 ubiquitination in HeLa cells transfected with NEDD4-2 SMARTpool and individual siRNAs. Fig. S3 shows inhibition of α-Th–stimulated p38 phosphorylation in HUVECs pretreated with SB203580 and PAR1 HeLa cells pretreated with SB202190. Fig. S4 shows PAR1, TAB2, and EEA1 colocalization and TAB1 expression in HeLa cells depleted of MKK3 and MKK6 expression. Fig. S5 shows ADP-induced HA-P2Y1 WT ubiquitination but not 3KR mutant ubiquitination and surface expression in HeLa cells. ADP-induced p38 activation in P2Y1 WT HeLa cells but not in untransfected cells is also shown. P2Y1 receptor surface expression is not changed in NEDD4-2 and TAB1–TAB2 siRNA depleted HeLa cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201504007/DC1.
Supplementary Material
Acknowledgments
We thank members of the Trejo laboratory for comments and advice and Joshua Olson for assisting with the vascular permeability assays.
This work was supported by National Institutes of Health R01 GM090689 (to J. Trejo) and American Heart Association (AHA) Grant-In-Aid 18630018 (to J. Trejo). N.J. Grimsey was supported by an AHA Postdoctoral Fellowship, T.H. Smith is supported by a F31 NHLBI Predoctoral Fellowship, and B. Aguilar is a San Diego IRACDA Fellow supported by National Institutes of Health K12 GM06852.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- EEA1
- early endosome antigen 1
- ERK1/2
- extracellular signal regulated kinase 1/2
- HUVEC
- human umbilical vein endothelial cell
- GPCR
- G protein–coupled receptor
- MEF
- mouse embryonic fibroblast
- MSK1
- mitogen- and stress-activated protein kinase 1
- NEDD4
- neural precursor cell expressed developmentally down-regulated protein 4
- NZF
- Npl4 zinc finger
- ns
- nonspecific
- PAR1
- protease-activated receptor 1
- α-Th
- α-thrombin
- TAB
- transforming growth factor-β–activated protein kinase-1 binding protein
- WT
- wild type
References
- Borbiev T., Birukova A., Liu F., Nurmukhambetova S., Gerthoffer W.T., Garcia J.G., and Verin A.D.. 2004. p38 MAP kinase-dependent regulation of endothelial cell permeability. Am. J. Physiol. Lung Cell. Mol. Physiol. 287:L911–L918. 10.1152/ajplung.00372.2003 [DOI] [PubMed] [Google Scholar]
- Bouwmeester T., Bauch A., Ruffner H., Angrand P.O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., et al. 2004. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat. Cell Biol. 6:97–105. 10.1038/ncb1086 [DOI] [PubMed] [Google Scholar]
- Chen B., Dores M.R., Grimsey N., Canto I., Barker B.L., and Trejo J.. 2011. Adaptor protein complex-2 (AP-2) and epsin-1 mediate protease-activated receptor-1 internalization via phosphorylation- and ubiquitination-dependent sorting signals. J. Biol. Chem. 286:40760–40770. 10.1074/jbc.M111.299776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P.C., Campbell D.G., Nebreda A.R., and Cohen P.. 2003. Feedback control of the protein kinase TAK1 by SAPK2a/p38α. EMBO J. 22:5793–5805. 10.1093/emboj/cdg552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin S.R. 1994. Molecular mechanisms of thrombin signaling. Semin. Hematol. 31:270–277. [PubMed] [Google Scholar]
- Davies S.P., Reddy H., Caivano M., and Cohen P.. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95–105. 10.1042/bj3510095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nicola G.F., Martin E.D., Chaikuad A., Bassi R., Clark J., Martino L., Verma S., Sicard P., Tata R., Atkinson R.A., et al. 2013. Mechanism and consequence of the autoactivation of p38α mitogen-activated protein kinase promoted by TAB1. Nat. Struct. Mol. Biol. 20:1182–1190. 10.1038/nsmb.2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores M.R., and Trejo J.. 2014. Atypical regulation of G protein-coupled receptor intracellular trafficking by ubiquitination. Curr. Opin. Cell Biol. 27:44–50. 10.1016/j.ceb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores M.R., Chen B., Lin H., Soh U.J., Paing M.M., Montagne W.A., Meerloo T., and Trejo J.. 2012. ALIX binds a YPX3L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J. Cell Biol. 197:407–419. 10.1083/jcb.201110031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge B., Gram H., Di Padova F., Huang B., New L., Ulevitch R.J., Luo Y., and Han J.. 2002. MAPKK-independent activation of p38α mediated by TAB1-dependent autophosphorylation of p38α. Science. 295:1291–1294. 10.1126/science.1067289 [DOI] [PubMed] [Google Scholar]
- Inagaki M., Omori E., Kim J.Y., Komatsu Y., Scott G., Ray M.K., Yamada G., Matsumoto K., Mishina Y., and Ninomiya-Tsuji J.. 2008. TAK1-binding protein 1, TAB1, mediates osmotic stress-induced TAK1 activation but is dispensable for TAK1-mediated cytokine signaling. J. Biol. Chem. 283:33080–33086. 10.1074/jbc.M807574200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A., Seth R.B., Sun L., Ea C.K., Hong M., Shaito A., Chiu Y.H., Deng L., and Chen Z.J.. 2004. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell. 15:535–548. 10.1016/j.molcel.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Kim L., Del Rio L., Butcher B.A., Mogensen T.H., Paludan S.R., Flavell R.A., and Denkers E.Y.. 2005. p38 MAPK autophosphorylation drives macrophage IL-12 production during intracellular infection. J. Immunol. 174:4178–4184. 10.4049/jimmunol.174.7.4178 [DOI] [PubMed] [Google Scholar]
- Komarova Y.A., Mehta D., and Malik A.B.. 2007. Dual regulation of endothelial junctional permeability. Sci. STKE. 2007:re8 10.1126/stke.4122007re8 [DOI] [PubMed] [Google Scholar]
- Korhonen H., Fisslthaler B., Moers A., Wirth A., Habermehl D., Wieland T., Schütz G., Wettschureck N., Fleming I., and Offermanns S.. 2009. Anaphylactic shock depends on endothelial Gq/G11. J. Exp. Med. 206:411–420. 10.1084/jem.20082150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y., Akutsu M., Bremm A., Hofmann K., and Komander D.. 2009. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 16:1328–1330. 10.1038/nsmb.1731 [DOI] [PubMed] [Google Scholar]
- Lin H., and Trejo J.. 2013. Transactivation of the PAR1-PAR2 heterodimer by thrombin elicits β-arrestin-mediated endosomal signaling. J. Biol. Chem. 288:11203–11215. 10.1074/jbc.M112.439950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A., and Trejo J.. 2013. Ubiquitin-dependent regulation of G protein-coupled receptor trafficking and signaling. Cell. Signal. 25:707–716. 10.1016/j.cellsig.2012.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J.N., Shen L., Holinstat M., Brooks J.D., Dibenedetto E., and Hamm H.E.. 2005. Functional selectivity of G protein signaling by agonist peptides and thrombin for the protease-activated receptor-1. J. Biol. Chem. 280:25048–25059. 10.1074/jbc.M414090200 [DOI] [PubMed] [Google Scholar]
- Mendoza H., Campbell D.G., Burness K., Hastie J., Ronkina N., Shim J.H., Arthur J.S., Davis R.J., Gaestel M., Johnson G.L., et al. 2008. Roles for TAB1 in regulating the IL-1-dependent phosphorylation of the TAB3 regulatory subunit and activity of the TAK1 complex. Biochem. J. 409:711–722. 10.1042/BJ20071149 [DOI] [PubMed] [Google Scholar]
- Mishra S., Guan J., Plovie E., Seldin D.C., Connors L.H., Merlini G., Falk R.H., MacRae C.A., and Liao R.. 2013. Human amyloidogenic light chain proteins result in cardiac dysfunction, cell death, and early mortality in zebrafish. Am. J. Physiol. Heart Circ. Physiol. 305:H95–H103. 10.1152/ajpheart.00186.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai A., Yamamoto-Hino M., Awano W., Watanabe W., Komada M., and Goto S.. 2010. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 29:2114–2125. 10.1038/emboj.2010.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raingeaud J., Whitmarsh A.J., Barrett T., Dérijard B., and Davis R.J.. 1996. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16:1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy G., Risco A.M., Iñesta-Vaquera F.A., González-Terán B., Sabio G., Davis R.J., and Cuenda A.. 2010. Differential activation of p38MAPK isoforms by MKK6 and MKK3. Cell. Signal. 22:660–667. 10.1016/j.cellsig.2009.11.020 [DOI] [PubMed] [Google Scholar]
- Russo A., Soh U.J., Paing M.M., Arora P., and Trejo J.. 2009. Caveolae are required for protease-selective signaling by protease-activated receptor-1. Proc. Natl. Acad. Sci. USA. 106:6393–6397. 10.1073/pnas.0810687106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Guan J., Jiang B., Brenner D.A., Del Monte F., Ward J.E., Connors L.H., Sawyer D.B., Semigran M.J., Macgillivray T.E., et al. 2010. Amyloidogenic light chains induce cardiomyocyte contractile dysfunction and apoptosis via a non-canonical p38α MAPK pathway. Proc. Natl. Acad. Sci. USA. 107:4188–4193. 10.1073/pnas.0912263107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh U.J., and Trejo J.. 2011. Activated protein C promotes protease-activated receptor-1 cytoprotective signaling through β-arrestin and dishevelled-2 scaffolds. Proc. Natl. Acad. Sci. USA. 108:E1372–E1380. 10.1073/pnas.1112482108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh U.J., Dores M.R., Chen B., and Trejo J.. 2010. Signal transduction by protease-activated receptors. Br. J. Pharmacol. 160:191–203. 10.1111/j.1476-5381.2010.00705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soohoo A.L., and Puthenveedu M.A.. 2013. Divergent modes for cargo-mediated control of clathrin-coated pit dynamics. Mol. Biol. Cell. 24:1725–1734: S1–S12. 10.1091/mbc.E12-07-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto A.G., and Trejo J.. 2010. N-linked glycosylation of protease-activated receptor-1 second extracellular loop: a critical determinant for ligand-induced receptor activation and internalization. J. Biol. Chem. 285:18781–18793. 10.1074/jbc.M110.111088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno M., Bassi R., Gorog D.A., Saurin A.T., Jiang J., Heads R.J., Martin J.L., Davis R.J., Flavell R.A., and Marber M.S.. 2003. Diverse mechanisms of myocardial p38 mitogen-activated protein kinase activation: evidence for MKK-independent activation by a TAB1-associated mechanism contributing to injury during myocardial ischemia. Circ. Res. 93:254–261. 10.1161/01.RES.0000083490.43943.85 [DOI] [PubMed] [Google Scholar]
- Theivanthiran B., Kathania M., Zeng M., Anguiano E., Basrur V., Vandergriff T., Pascual V., Wei W.Z., Massoumi R., and Venuprasad K.. 2015. The E3 ubiquitin ligase Itch inhibits p38α signaling and skin inflammation through the ubiquitylation of Tab1. Sci. Signal. 8:ra22 10.1126/scisignal.2005903 [DOI] [PubMed] [Google Scholar]
- Trejo J., Altschuler Y., Fu H.W., Mostov K.E., and Coughlin S.R.. 2000. Protease-activated receptor-1 down-regulation: a mutant HeLa cell line suggests novel requirements for PAR1 phosphorylation and recruitment to clathrin-coated pits. J. Biol. Chem. 275:31255–31265. 10.1074/jbc.M003770200 [DOI] [PubMed] [Google Scholar]
- Vina-Vilaseca A., and Sorkin A.. 2010. Lysine 63-linked polyubiquitination of the dopamine transporter requires WW3 and WW4 domains of Nedd4-2 and UBE2D ubiquitin-conjugating enzymes. J. Biol. Chem. 285:7645–7656. 10.1074/jbc.M109.058990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Feng J., Wang J., Zhang X., Zhang D., Zhu T., Wang W., Wang X., Jin J., Cao J., et al. 2013. Disruption of TAB1/p38α interaction using a cell-permeable peptide limits myocardial ischemia/reperfusion injury. Mol. Ther. 21:1668–1677. 10.1038/mt.2013.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B.L., Marchese A., and Trejo J.. 2007. Ubiquitination differentially regulates clathrin-dependent internalization of protease-activated receptor-1. J. Cell Biol. 177:905–916. 10.1083/jcb.200610154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.Y., Li J.P., Chiu L.L., Lan J.L., Chen D.Y., Chuang H.C., Huang C.Y., and Tan T.H.. 2014. Dual-specificity phosphatase 14 (DUSP14/MKP6) negatively regulates TCR signaling by inhibiting TAB1 activation. J. Immunol. 192:1547–1557. 10.4049/jimmunol.1300989 [DOI] [PubMed] [Google Scholar]
- Zemskov E., Lucas R., Verin A.D., and Umapathy N.S.. 2011. P2Y receptors as regulators of lung endothelial barrier integrity. J. Cardiovasc. Dis. Res. 2:14–22. 10.4103/0975-3583.78582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Zheng M., Chen J., Xie C., Kolatkar A.R., Zarubin T., Ye Z., Akella R., Lin S., Goldsmith E.J., and Han J.. 2006. Determinants that control the specific interactions between TAB1 and p38alpha. Mol. Cell. Biol. 26:3824–3834. 10.1128/MCB.26.10.3824-3834.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.