Our results demonstrate that E-cadherin/αE-catenin chimeras homodimerize and do not mimic αE-catenin in the native CCC, and imply that both CCC-bound monomer and cytosolic homodimer αE-catenin are required for strong cell adhesion.
Abstract
As part of the E-cadherin–β-catenin–αE-catenin complex (CCC), mammalian αE-catenin binds F-actin weakly in the absence of force, whereas cytosolic αE-catenin forms a homodimer that interacts more strongly with F-actin. It has been concluded that cytosolic αE-catenin homodimer is not important for intercellular adhesion because E-cadherin/αE-catenin chimeras thought to mimic the CCC are sufficient to induce cell–cell adhesion. We show that, unlike αE-catenin in the CCC, these chimeras homodimerize, bind F-actin strongly, and inhibit the Arp2/3 complex, all of which are properties of the αE-catenin homodimer. To more accurately mimic the junctional CCC, we designed a constitutively monomeric chimera, and show that E-cadherin–dependent cell adhesion is weaker in cells expressing this chimera compared with cells in which αE-catenin homodimers are present. Our results demonstrate that E-cadherin/αE-catenin chimeras used previously do not mimic αE-catenin in the native CCC, and imply that both CCC-bound monomer and cytosolic homodimer αE-catenin are required for strong cell–cell adhesion.
Introduction
The adherens junction (AJ) is essential for the development and maintenance of tissue integrity (Gumbiner, 1996; Collinet and Lecuit, 2013). Formation of the AJ is directed by the cadherin–catenin complex (CCC), which in epithelial cells comprises E-cadherin that binds β-catenin, which in turn recruits the F-actin binding and bundling protein αE-catenin. At cell–cell junctions, αE-catenin is thought to link the CCC to F-actin (Watabe-Uchida et al., 1998; Vasioukhin et al., 2001; Pokutta and Weis, 2007). Although early in vitro studies failed to reconstitute binding of the CCC to F-actin (Drees et al., 2005a; Yamada et al., 2005), recent studies showed that force is required to strengthen this interaction (Buckley et al., 2014). Mammalian αE-catenin also forms a homodimer that binds and bundles F-actin, and inhibits Arp2/3 and cofilin activities (Drees et al., 2005a; Benjamin et al., 2010; Hansen et al., 2013). αE-Catenin homodimerization and β-catenin binding are mutually exclusive and are mediated by a common domain in the N terminus of αE-catenin (Koslov et al., 1997; Pokutta and Weis, 2000). Thus, mammalian αE-catenin exists in distinct cellular pools: (a) membrane-tethered, monomeric αE-catenin bound directly to E-cadherin/β-catenin, and (b) cytoplasmic monomer and homodimer.
The function of junctional αE-catenin in the CCC has been studied with an E-cadherin/αE-catenin chimera, designated E-cadΔ70/α (Nagafuchi et al., 1994; Fig. 1 A). In fibroblast L cells, which lack endogenous E-cadherin, expression of E-cadΔ70/α induced cell–cell adhesion, which required the C-terminal actin-binding domain (ABD) of αE-catenin (Nagafuchi et al., 1994). Subsequent studies used E-cadΔ70/α and a full-length chimera containing the entire cytoplasmic tail of E-cadherin (E-cad/α) to induce cell–cell adhesion in a variety of cell types and tissues in vitro and in vivo (Ozawa, 1998; Ozawa and Kemler, 1998; Imamura et al., 1999; Gottardi et al., 2001; Winter et al., 2003; Pacquelet and Rørth, 2005; Qin et al., 2005; Abe and Takeichi, 2008; Noda et al., 2010; Ozono et al., 2011; Schulte et al., 2011; Maiden and Hardin, 2011; Sarpal et al., 2012; Shih and Yamada, 2012; Twiss et al., 2012; Thomas et al., 2013; Desai et al., 2013; Küppers et al., 2013; Dartsch et al., 2014). The consensus conclusions from these experiments are that membrane-tethered, monomeric αE-catenin is sufficient for intercellular adhesion by linking the CCC to the actin cytoskeleton, and that neither a cytoplasmic pool of αE-catenin nor αE-catenin homodimers are required (Ozono et al., 2011; Sarpal et al., 2012; Desai et al., 2013; Thomas et al., 2013). However, these interpretations overlook the fact that (a) αE-catenin bound to β-catenin in the CCC has a distinct conformation and different properties compared with free monomeric αE-catenin, and (b) the possibility that the E-cadΔ70/α chimera homodimerizes, a property of mammalian αE-catenin that normally occurs in the cytosol. Therefore, we tested whether E-cadΔ70/α is functionally equivalent to αE-catenin bound to β-catenin in the CCC, and whether homodimerization of αE-catenin is dispensable for E-cadherin–mediated cell–cell adhesion.
Figure 1.
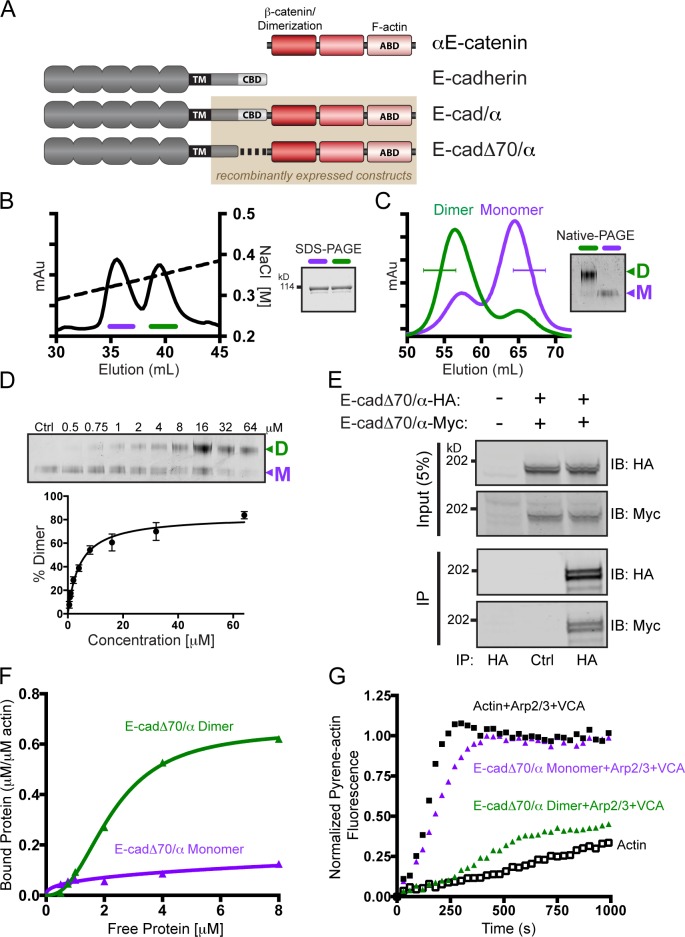
E-cadΔ70/α homodimerization is required for robust interaction with F-actin. (A) Schematic representation of the E-cadherin/αE-catenin chimeras. CBD, β-catenin-binding domain. (B) Ion exchange chromatography (IEC) of recombinant E-cadΔ70/α, and SDS-PAGE of protein from the resulting two peaks (fractions indicated in purple and green) stained with Coomassie Brilliant Blue (CBB). (C) Superdex 200 size exclusion chromatography of the two peaks from the IEC shown in B. Fractions indicated with a bracket were pooled and analyzed by Native-PAGE, and stained with CBB. (D) CBB stained Native-PAGE of increasing concentrations of monomeric E-cadΔ70/α chimera incubated for 16 h at 37°C. Ctrl, purified monomeric chimera. Quantification of the percentage of dimerization with standard deviation from three independent experiments. (E) Coimmunoprecipitation of Myc-tagged E-cadΔ70/α with HA-tagged E-cadΔ70/α from transfected L cells. Immunoprecipitated proteins were separated by SDS-PAGE and immunoblotted for HA and Myc. A representative image of three independent experiments is shown. (F) High-speed co-sedimentation of F-actin with E-cadΔ70/α monomer (purple) or homodimer (green). The data shown are from a single representative experiment out of three independent experiments. (G) Pyrene–actin polymerization assay with 10% pyrene–actin (white), with Arp2/3 complex and WASp-VCA (black), and either 8 µM E-cadΔ70/α homodimer (green) or monomer (purple). The data shown are from a single representative experiment out of three independent experiments.
Results and discussion
E-cadΔ70/α forms a homodimer in vitro and in mammalian cells
For in vitro analysis, we purified and analyzed the oligomeric state and functions of recombinant E-cadΔ70/α lacking the extracellular and transmembrane (TM) domains of E-cadherin (Fig. 1 A, brown box). Limited trypsin digestion of purified E-cadΔ70/α generated proteolytic products similar to those of αE-catenin (Fig. S1 A; Pokutta and Weis, 2000; Drees et al., 2005a; Kwiatkowski et al., 2010), indicating that fusion of the unstructured E-cadherin cytoplasmic tail to αE-catenin did not alter the conformation of αE-catenin in the chimera.
Ion exchange chromatography (IEC) of E-cadΔ70/α resulted in two peaks (Fig. 1 B). Subsequent size exclusion chromatography (SEC) of proteins from the first and second IEC peaks resulted in smaller (purple) and larger (green) apparent molecular mass proteins, respectively, that also separated into two protein bands upon Native-PAGE (Fig. 1 C). These separation profiles are similar to those of αE-catenin monomer and homodimer. The two oligomeric states of E-cadΔ70/α were confirmed by measuring the hydrodynamic radii (Rh) by dynamic light scattering, which were Rh = 7.1 for the first SEC peak and Rh = 5.95 for the second SEC peak. To determine whether monomeric E-cadΔ70/α converts into a homodimer, increasing concentrations of purified E-cadΔ70/α monomer were incubated for 16 hours at 37°C to reach equilibrium, and then analyzed by Native-PAGE. Monomeric E-cadΔ70/α started to homodimerize at single μM concentrations (Fig. 1 D). IEC, SEC, and Native-PAGE also showed that a full-length chimera (E-cad/α), which contains the entire cytoplasmic tail of E-cadherin fused to αE-catenin (Fig. 1 A), formed a monomer and homodimer (Fig. S1, B–D).
To determine whether E-cadΔ70/α formed homodimers in mammalian cells, we co-expressed E-cadΔ70/α containing the extracellular and TM domains of E-cadherin (Fig. 1 A) tagged at the C terminus with either a HA or Myc epitope for differential identification with specific antibodies. HA immunoprecipitation revealed that E-cadΔ70/α-HA and E-cadΔ70/α-Myc coimmunoprecipitated from L cells (Fig. 1 E) and HEK293 cells (Fig. S1 E). This coimmunoprecipitation was maintained in lysates from cells grown in a low Ca2+ conditions (Fig. S1 F), indicating that coimmunoprecipitation was not a result of homotypic binding by the E-cadherin extracellular domain. Altogether, these results show that E-cadΔ70/α formed a homodimer in vitro and in mammalian cells.
E-cadΔ70/α homodimer binds F-actin and inhibits Arp2/3
It has been assumed that E-cadΔ70/α contains monomeric αE-catenin and mimics the binding of the native CCC to F-actin because E-cadΔ70/α without the αE-catenin ABD does not induce cell adhesion (Nagafuchi et al., 1994; Imamura et al., 1999). Having now shown that E-cadΔ70/α forms a monomer and homodimer, we tested which oligomeric state bound F-actin in an F-actin co-sedimentation assay (Fig. 1 F). E-cadΔ70/α homodimer bound F-actin with a Kd of 1.6 µM, comparable to the αE-catenin homodimer/F-actin Kd of 1.0 µM (Hansen et al., 2013). In contrast, little of the E-cadΔ70/α monomer bound to F-actin in this assay. In addition, the E-cadΔ70/α homodimer, but not the monomer, inhibited Arp2/3-mediated actin polymerization at a concentration of 8 µM (Fig. 1 G), similar to αE-catenin homodimer (Drees et al., 2005a). These observations further demonstrate that the properties of the E-cadΔ70/α chimera do not mimic monomeric αE-catenin in the native CCC.
Complex formation between E-cadherin/αE-catenin chimeras and β-catenin
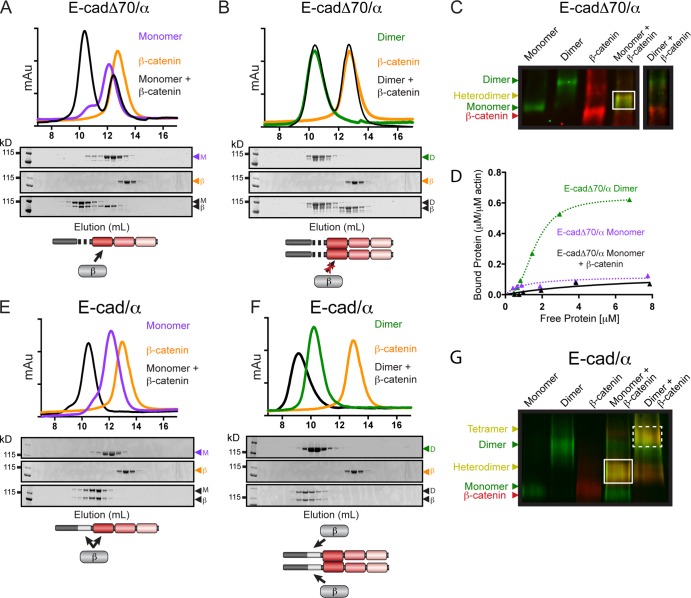
αE-catenin is recruited to the AJ by a preassembled E-cadherin–β-catenin complex (Hinck et al., 1994; Chen et al., 1999), which prevents αE-catenin homodimerization and decreases its apparent affinity for F-actin (Pokutta and Weis, 2000; Drees et al., 2005a; Miller et al., 2013). Therefore, we explored the interaction between β-catenin and the different oligomeric states of the chimeras.
IEC/SEC purified E-cadΔ70/α monomer (Fig. 2 A) or homodimer (Fig. 2 B) and β-catenin were incubated in a 1:1 molar ratio and subjected to SEC, and then eluted proteins were analyzed by SDS-PAGE (Fig. 2, A and B) and Native-PAGE (Fig. 2 C). E-cadΔ70/α monomer formed a heterodimer with β-catenin (Fig. 2 A, black peak, and Fig. 2 C), whereas E-cadΔ70/α homodimer and β-catenin did not bind (Fig. 2 B, black peak; and Fig. 2 C). Isothermal titration calorimetry (ITC) measurements demonstrated that monomeric E-cadΔ70/α and β-catenin formed a 1:1 stoichiometric complex (Kd = 37 nM). In contrast, a 10-fold molar excess of β-catenin did not interact with the E-cadΔ70/α homodimer, indicating that the homodimer is kinetically trapped (Table 1 and Fig. S1 G) like αE-catenin homodimer (Pokutta et al., 2014). Finally, binding of β-catenin reduced the binding affinity of E-cadΔ70/α for F-actin to a level even lower than that of monomeric E-cadΔ70/α (Fig. 2 D). We conclude that E-cadΔ70/α monomer, but not E-cadΔ70/α homodimer, forms a complex with β-catenin, and that this complex has a low apparent affinity for F-actin.
Figure 2.
Complex formation between E-cadherin/αE-catenin chimeras and β-catenin. Superdex 200 gel filtration chromatography of complex formation between β-catenin and E-cadΔ70/α monomer (A), E-cadΔ70/α homodimer (B), E-cad/α monomer (E), or E-cad/α homodimer (F). Proteins were incubated at 25°C either by themselves (colored lines) or together in a 1:1 molar ratio (black lines), and fractions from the individual S200 runs were analyzed by SDS-PAGE and CBB staining. A schematic representation of the complex formed between each chimera and β-catenin is shown. Native-PAGE of monomers and homodimers of E-cadΔ70/α (C) or E-cad/α (G) incubated with β-catenin and immunoblotted for αE-catenin (green) and β-catenin (red). Complex formation is indicated by a shift in band migration and co-fluorescence with both αE-catenin and β-catenin antibodies (heterodimer, solid box; tetramer, dashed box). Note that all proteins in C were run on the same Native-PAGE gel, but the brightness for the last lane was adjusted independently, as indicated. The gel images shown (C and G) are representative of four independent experiments. (D) High-speed co-sedimentation assay of E-cadΔ70/α-β-catenin heterodimer (black triangle) with F-actin. The data shown are from a single representative experiment out of three independent experiments. E-cadΔ70/α dimer (green triangle) and E-cadΔ70/α monomer (purple triangle) from F are shown for comparison.
Table 1. . ITC measurements of E-cadΔ70/α monomer, homodimer, and E-cad/α monomer interaction with β-catenin.
| Proteins | Kd | ΔH | TΔS |
ΔG |
|---|---|---|---|---|
| nM | kcal mol−1 | kcal mol−1 | kcal mol−1 | |
| E-cadΔ70/α (monomer) | 36.6 ± 4.1 | −16.9 ± 0.2 | −6.8 | −10.1 |
| E-cadΔ70/α (dimer) | NB | – | – | – |
| E-cadα (monomer) | 74.0 ± 2.9 | −58.0 ± 1.0 | −48.3 | −9.7 |
| αE-catenin (monomer)a | 23.4 ± 3.7 | −16.1 ± 0.7 | −5.7 | −10.4 |
| E-cadherin cyto tailb | 46.0 ± 4.4 | −39.2 ±0.2 | −29.0 | −10.2 |
The error reported on the measurement is the SD of the nonlinear least square fit. NB, no detectable binding.
aMeasurement previously published (Pokutta et al., 2014).
bMeasurement previously published (Choi et al., 2006).
The full-length E-cad/α chimera might interact differently with β-catenin because it contains two β-catenin–binding sites, one in the E-cadherin catenin-binding domain (CBD) and one in the N terminus of αE-catenin (Fig. 1 A). Both IEC/SEC purified E-cad/α monomer and homodimer bound β-catenin as shown by the shift in their SEC elution profiles (Fig. 2, E and F, black lines), and their decreased electrophoretic mobilities in Native-PAGE (Fig. 2 G, white boxes). A 1:1 binding stoichiometry between E-cad/α and β-catenin was determined by ITC (Table 1 and Fig. S1 G). The enthalpy change (ΔH) for this reaction (−58 kcal mol−1) was approximately equal to the sum of the enthalpy changes for β-catenin binding to the E-cadherin cytoplasmic tail (−39 kcal mol−1) and monomeric αE-catenin (−16 kcal mol−1; Table 1; Choi et al., 2006; Pokutta et al., 2014). These results indicate that a single molecule of β-catenin bound to monomeric E-cad/α, and support the prediction that in the native CCC the C-terminus of E-cadherin is positioned in close proximity to the N terminus of αE-catenin (Pokutta et al., 2014).
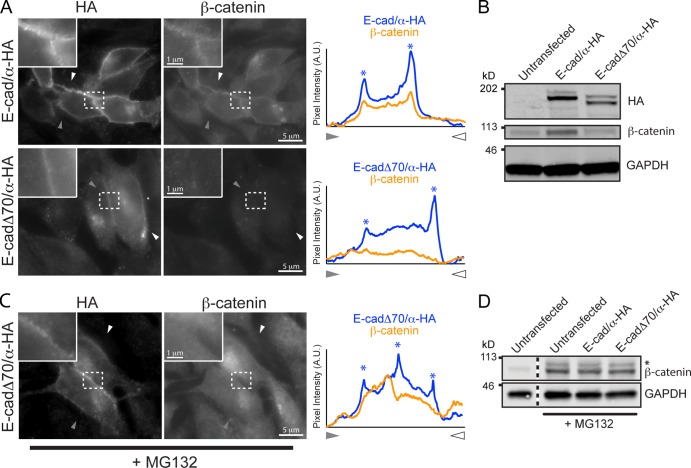
E-cadΔ70/α is a homodimer at the plasma membrane of mammalian cells
To visualize the oligomeric state of E-cadΔ70/α in L cells, we took advantage of the differential binding of β-catenin to E-cadΔ70/α and E-cad/α homodimers. β-Catenin bound to E-cadΔ70/α monomer but not to the kinetically trapped E-cadΔ70/α homodimer (Fig. 2, A–C), whereas it bound to either E-cad/α monomer or homodimer (Fig. 2, E–G) which therefore served as a positive control for β-catenin recruitment. L cells were transfected with HA-tagged E-cad/α or E-cadΔ70/α and co-stained with HA and β-catenin antibodies. β-Catenin colocalized with E-cad/α, but not E-cadΔ70/α, at the plasma membrane of cell–cell contacts (Fig. 3 A). To exclude the possibility that the lack of β-catenin colocalization with E-cadΔ70/α was caused by the lower expression level of β-catenin in these cells (Fig. 3 B), we inhibited proteasomal degradation with MG132 (Fig. 3 D), and still did not detect colocalization at cell–cell contacts (Fig. 3 C). We conclude that the majority of E-cadΔ70/α localized at the plasma membrane of L cell contacts is a homodimer.
Figure 3.
E-cadΔ70/α localizes to the plasma membrane as a homodimer. (A) Immunofluorescence of endogenous β-catenin in L cells expressing HA-tagged E-cad/α or E-cadΔ70/α. Line scans performed between the indicated arrowheads; the pixel intensity for HA and β-catenin immunofluorescence are shown. Cell–cell junctions indicated with asterisks. (B) Western blot of total cell lysates of L cells expressing E-cad/α-HA or E-cadΔ70/α-HA. β-Catenin level increases threefold when E-cad/α is expressed. GAPDH, loading control. (C) Immunofluorescence staining of endogenous β-catenin in L cells expressing HA-tagged E-cadΔ70/α treated with MG132 to stabilize β-catenin to levels similar to E-cad/α expressing cells. (D) Western blot of total cell lysates of L cells expressing E-cad/α-HA or E-cadΔ70/α-HA treated with MG132 (25 µM, 6 h), showing similar levels of β-catenin. Phosphorylated β-catenin becomes visible upon MG132 treatment (indicated by an asterisk).
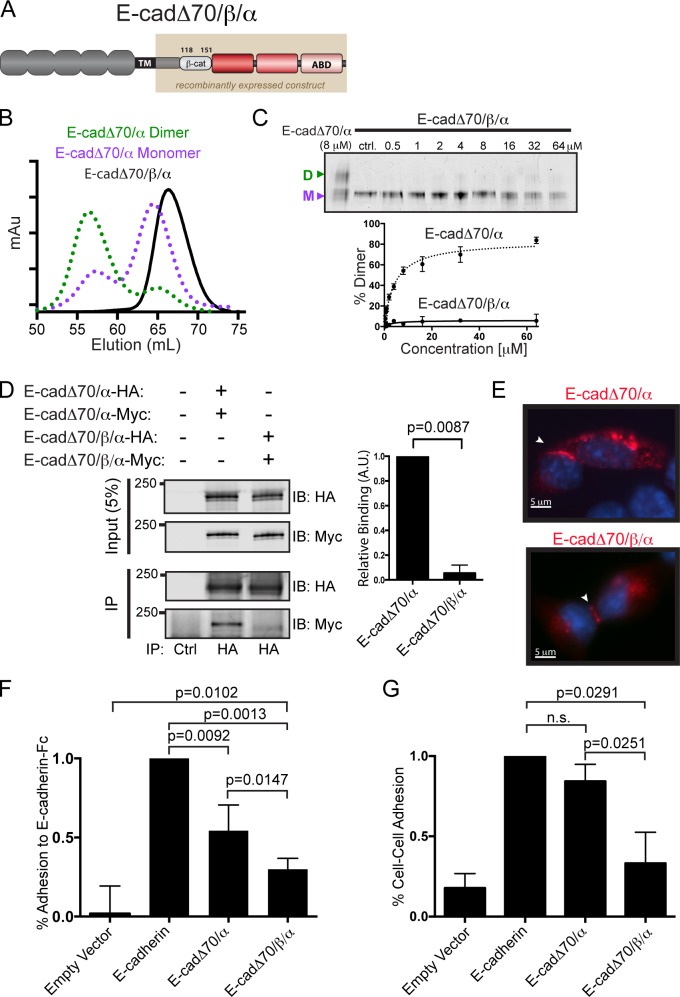
A constitutive monomeric E-cadherin/αE-catenin chimera induces weak E-cadherin–mediated cell adhesion
To test whether homodimerization of αE-catenin is dispensable for E-cadherin–mediated cell adhesion, we designed a constitutively monomeric chimera by inserting the minimal αE-catenin binding sequence of β-catenin in between E-cadherin and αE-catenin in E-cadΔ70/α to generate E-cadΔ70/β/α (Fig. 4 A). The N terminus of αE-catenin within the chimera presumably binds to this part of β-catenin, as shown with the previously characterized α/β-catenin fusion protein (Pokutta and Weis, 2000; Yamada et al., 2005), thereby blocking homodimerization.
Figure 4.
Reduced E-cadherin–mediated cell adhesion by a constitutively monomeric E-cadherin/αE-catenin chimera. (A) Schematic representation of E-cadΔ70/β/α, in which residues 118–151 of β-catenin are inserted in between E-cadherin and αE-catenin. (B) SEC of E-cadΔ70/β/α (black line), E-cadΔ70/α dimer (green dotted line), and monomer (purple dotted line). (C) CBB-stained Native-PAGE of increasing concentrations of E-cadΔ70/β/α that were incubated for 16 h at 37°C. Quantification of the percent dimerization of E-cadΔ70/β/α compared with E-cadΔ70/α. n = 3. (D) Coimmunoprecipitation of Myc tagged with HA-tagged chimeras, using either E-cadΔ70/α or E-cadΔ70/β/α. Upon immunoprecipitation with HA antibodies, proteins were separated by SDS-PAGE and immunoblotted for HA and Myc. Quantification of the relative binding between differentially tagged E-cadΔ70/α and E-cadΔ70/β/α is shown. n = 4. (E) Immunofluorescence of E-cadΔ70/α and E-cadΔ70/β/α in L cells with an antibody to the extracellular domain of E-cadherin. (F) Adhesion to E-cadherin-Fc of L cells transfected with E-cadherin-GFP, E-cadΔ70/α, E-cadΔ70/β/α, or empty vector control, together with luciferase. Background adhesion to a no calcium control was subtracted, and the mean percentage of adherent cells relative to the total input luciferase signal and normalized to E-cadherin-GFP values is shown. n = 5 with standard deviation. An unpaired Student’s t test was performed for statistical analysis. (G) Hanging drop assay of L cells transfected with E-cadherin-GFP, E-cadΔ70/α, E-cadΔ70/β/α, or empty vector control showing the percentage of clusters of four or more cells, normalized to levels observed with E-cadherin. n = 3, with standard deviation. An unpaired Student’s t test was performed for statistical analysis.
As predicted, E-cadΔ70/β/α eluted as one peak on SEC similar to monomeric E-cadΔ70/α (Fig. 4 B). Monomeric E-cadΔ70/β/α was more sensitive to tryptic digestion compared with E-cadΔ70/α, E-cad/α, or αE-catenin, all of which could homodimerize to produce different proteolytic protection patterns (Fig. S1 A). E-cadΔ70/β/α migrated as a monomer in Native-PAGE, even at 64 µM, a concentration sufficient to convert nearly all monomeric E-cadΔ70/α to homodimer (Fig. 4 C). Finally, HA-tagged and Myc-tagged E-cadΔ70/β/α did not coimmunoprecipitate from an L cell lysate (Fig. 4 D). Thus, E-cadΔ70/β/α is a monomer and does not homodimerize.
Because αE-catenin in the chimera is fused to E-cadherin, its cellular distribution is uncoupled from its oligomeric state, and we expected that insertion of β-catenin residues would only affect homodimerization, but not its cellular localization. Immunofluorescence staining of E-cadΔ70/β/α expressed in L cells showed that E-cadΔ70/β/α localized at cell–cell contacts similar to E-cadΔ70/α (Fig. 4 E). FACS analysis of L cells expressing E-cadΔ70/β/α, E-cadΔ70/α, or E-cadherin with an E-cadherin extracellular domain antibody verified that surface expression levels of all constructs were similar (geo mean of 0.97 ± 0.18 [E-cadΔ70/α] and 1.06 ± 0.09 [E-cadΔ70/β/α] relative to E-cadherin).
Having shown that E-cadΔ70/β/α is a monomer, we tested whether it could induce E-cadherin–dependent cell adhesion compared with E-cadΔ70/α or E-cadherin. L cells were cotransfected with luciferase (to measure adhesion selectively in transfected cells) and E-cadΔ70/β/α, E-cadΔ70/α, or E-cadherin. Importantly, ectopic expression of E-cadherin, but not expression of either chimera lacking the E-cadherin CBD, resulted in the stabilization of both β- and αE-catenin (Fig. S2 A), and the accumulation of a cytoplasmic pool of αE-catenin containing monomers, homodimers, and α/β-catenin heterodimers (Fig. S2 B; Nagafuchi et al., 1994). Adhesion was measured by attachment of cells to an E-cadherin-Fc–coated surface, and cells expressing monomeric E-cadΔ70/β/α had significantly less cadherin-mediated adhesion than cells expressing either E-cadΔ70/α or E-cadherin (Fig. 4 F). These results were confirmed independently with a hanging drop cell–cell adhesion assay, in which expression of E-cadΔ70/α and E-cadherin formed trituration-resistant cell clusters, whereas E-cadΔ70/β/α induced significantly less cell–cell adhesion (Fig. 4 G).
Altogether, we conclude that cell–cell adhesion was strongest when αE-catenin formed homodimers, either at the plasma membrane through E-cadΔ70/α homodimerization, or in the cytosol through αE-catenin accumulation by E-cadherin expression. Nonetheless, monomeric E-cadΔ70/β/α chimera induced some E-cadherin–mediated adhesion, implying that even in the absence of αE-catenin homodimer, junctional αE-catenin is able to induce adhesion, presumably through force-dependent binding to F-actin (Buckley et al., 2014) or indirect linkage to F-actin by binding other actin-binding proteins such as EPLIN or vinculin (Weiss et al., 1998; Abe and Takeichi, 2008).
Reinterpreting E-cadherin/αE-catenin chimeras and the role of αE-catenin homodimers in cell–cell adhesion
We conclude E-cadherin/αE-catenin chimeras used previously do not mimic αE-catenin in the native CCC, and our results imply that both junctional monomer and perijunctional homodimer pools of αE-catenin act together to induce strong intercellular adhesion. Previous work showed that E-cadherin/αE-catenin chimeras lacking the N-terminal β-catenin/homodimerization domain of αE-catenin induced cell–cell adhesion (Gottardi et al., 2001; Desai et al., 2013; Thomas et al., 2013). Analogously, intercellular adhesion could be induced by E-cadherin chimeras comprising only the αE-catenin ABD, or the ABD of other actin binding proteins such as EPLIN or utrophin (Nagafuchi et al., 1994; Winter et al., 2003; Kobielak et al., 2004;Abe and Takeichi, 2008; Hansen et al., 2013; Hong et al., 2013). These results have been used as the strongest line of evidence that αE-catenin homodimerization is not required for cell–cell adhesion. However, these chimeras lack allosteric regulation of the αE-catenin–F-actin interaction, and therefore exhibit increased association with the actin cytoskeleton compared with the native CCC (Nagafuchi et al., 1994; Rimm et al., 1995; Schulte et al., 2011; Dartsch et al., 2014). Thus, chimeras lacking the N-terminal domain αE-catenin or containing only the ABD of αE-catenin (or any other actin binding protein) have different F-actin–binding properties and are not functionally equivalent to the native CCC.
Our data indicate that αE-catenin homodimers contribute to cell–cell adhesion. Perijunctional αE-catenin homodimers may function to locally control actin dynamics necessary for cell–cell adhesion (Drees et al., 2005a; Benjamin et al., 2010; Hansen et al., 2013). αE-catenin bound to β-catenin in the CCC may also locally exchange with peri-junctional αE-catenin to form homodimers. This hypothesis is supported by the findings that simply concentrating α-catenin at the junctional cortex by fusion to PAR-3 or Echinoid does not rescue adhesion defects in Drosophila α-catenin–null cells (Desai et al., 2013). β-Catenin, in addition to its role in directly linking αE-catenin to E-cadherin in the CCC, is the likely candidate for regulating the interplay between junctional and perijunctional αE-catenin, and thereby the F-actin–binding activity of αE-catenin, which is lost in E-cadherin/αE-catenin chimeras.
Materials and methods
Constructs
Full-length mouse E-cadherin cytoplasmic tail (residues 736–884; NM_009864.2) and the cytoplasmic tail lacking the C-terminal 70 aa (residues 736–815) were fused N-terminally to mouse αE-catenin (NM_009818) and cloned into the pGEX-TEV bacterial expression vector (Epoch Life Science). The resulting constructs were termed E-cad/α and E-cadΔ70/α, respectively. For mammalian expression, full-length canine E-cadherin (NM_001287125.1) was fused N-terminally to mouse αE-catenin, and cloned into pcDNA3.1(+)−3xHA or pcDNA3.1(+)−MycHis vectors (Epoch Life Science). Deletions and insertions were generated using either site directed mutagenesis (QuikChange II XL Site-Directed Mutagenesis kit; Agilent Technologies) or DNA ligation with phosphorylated primers (Roche). To generate E-cadΔ70/β/α for mammalian expression, residues 118–151 of mouse β-catenin (NM_007614) containing both an N- and C-terminal 5-Glycine spacer was inserted between E-cadherin and αE-catenin in pcDNA3.1(+)−E-cadΔ70/α-3xHA, using phosphorylated primers. Full-length mouse β-catenin and αE-catenin were cloned into the pGEX-TEV bacterial expression vector, and full-length canine E-cadherin was cloned into pEGFP-N1 as previously described (Yamada et al., 2005). pCMV-luciferase was described previously (Ponsioen et al., 2009).
Antibodies and other reagents
The following commercially available antibodies and dyes were used: β-catenin (mouse, 610154; BD); HA (mouse, HA.11, MMS-101P; Covance; rabbit, H6908; Sigma-Aldrich); Myc (mouse, 9E10, MMS-150P; Covance); GAPDH (mouse, AB8245; Abcam); DECMA (rat, U3254; Sigma-Aldrich); anti–human IgG Fc specific (goat, I2136; Sigma-Aldrich); RR1, mouse, which was generated in-house and recognizes the canine E-cadherin extracellular domain as has been described previously (Marrs et al., 1993); DαM568 (A10037, Thermo Fisher Scientific); Hoechst 33342 (H-3570; Molecular Probes); and Coomassie Brilliant Blue G-250 (20279; Thermo Fisher Scientific). The rabbit αE-catenin antibody which recognizes full-length αE-catenin has been described previously (Näthke et al., 1994), and Alexa Fluor–conjugated secondary antibodies were used for immunofluorescence staining and immunoblotting.
Cell lines
L cells and HEK293 cells were maintained in DMEM with 1 g/l sodium bicarbonate, 4.5 g/l glucose, 10% FBS (Atlas Biologicals), 5% penicillin, streptomycin, and kanamycin. L cells were transfected at 70% confluency with Lipofectamine LTX (Thermo Fisher Scientific), Lipofectamine 3000 (Thermo Fisher Scientific), and HEK293 cells with Xtremegene HP (Roche) according to the manufacturer’s protocol, and experiments were performed 48 h after transfection.
Recombinant protein expression and purification
Constructs were expressed in BL21 (DE3) Codon Plus Escherichia coli cells and purified as described previously (Pokutta and Weis, 2000). In short, 1-l cultures were induced with 1 mM IPTG for 6 h at 30°C, proteins were isolated on glutathione agarose beads, and the protein was subsequently cleaved off the GST tag with tobacco etch virus (TEV) protease. Proteins were applied to a MonoQ anion exchange column (GE) and eluted in 20 mM Tris, pH 8.0, 1 mM DTT, and a NaCl gradient from 0 to 1 M. Proteins were further purified by Superdex 200 gel filtration chromatography (GE Healthcare) in Hepes buffer (20 mM Hepes, pH 8.0, 150 mM NaCl, and 1 mM DTT). Monomeric and homodimeric protein fractions were pooled separately, verified for purity by SDS-PAGE and Coomassie Blue (CBB) staining and oligomeric state purity by Native-PAGE and CBB staining, and stored at 4°C for up to 2 wk. For analytical size exclusion chromatography, proteins were incubated at 10 µM at 25°C for 1 h, either individually or in a 1:1 molar ratio. 200 µl of sample was analyzed by analytical SEC performed at 4°C on a Superdex 200 column (GE Healthcare) in 20 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, and 1 mM DTT. 0.5-ml fractions were collected and separated by SDS-PAGE followed by CBB staining, and gels were analyzed using a LICOR Odyssey imaging system (LI-COR Biosciences).
Limited trypsin proteolysis
15 µM M. musculus αE-catenin and recombinant E-cad/α, E-cadΔ70/α, and E-cadΔ70/β/α were incubated at room temperature in 0.25 µg/µl sequence-grade trypsin (Roche) in 20 mM Tris, pH 8.0, 150 mM NaCl, and 1 mM DTT. Reactions were stopped with 2× Laemmli buffer at the indicated times, subjected to SDS-PAGE and CBB staining, and analyzed using a LICOR Odyssey imaging system (LI-COR Biosciences).
Dynamic light scattering measurements
Purified monomeric and homodimeric E-cadΔ70/α from S200 were independently concentrated and a DynaPro Plate Reader II (Wyatt Technology) was used at 25°C to assess the change in hydrodynamic radius (Rh).
Native-PAGE
IEC/SEC purified proteins were concentrated to indicated concentrations in Hepes buffer. Before Native-PAGE, samples were diluted to a total concentration of either 1.5 µM (for CBB staining) or 150 nM (for Western blotting) in ice-cold Native-PAGE Sample buffer (Thermo Fisher Scientific). Samples were separated by Native-PAGE on 4–16% Bis-Tris Native-PAGE gels (Thermo Fisher Scientific), and either fixed in 40% methanol, 10% acetic acid for ∼2 h, followed by CBB staining, or transferred onto PVDF membranes for immunostaining with rabbit–αE-catenin and mouse–β-catenin antibodies. After immunostaining, membranes were processed with Alexa Fluor–conjugated secondary antibodies and analyzed using a LICOR Odyssey imaging system (LI-COR Biosciences).
Coimmunoprecipitation
L cells or HEK293 cells transfected with both HA- and Myc-tagged E-cadΔ70/α or E-cadΔ70/β/α were lysed in chilled lysis buffer (1% Triton X-100, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, and protease inhibitors [Roche]). Lysates were cleared by centrifugation and incubated with protein G–Sepharose beads (GE Healthcare) precoupled to anti-HA antibodies for 2 h. Beads were washed four times with chilled lysis buffer, and proteins were eluted with Laemmli buffer) and separated by SDS-PAGE. Upon transfer onto PVDF (GE Healthcare), membranes were immunostained with HA and Myc antibodies, Alexa Fluor–conjugated secondary antibodies, and analyzed using a LICOR Odyssey imaging system (LI-COR Biosciences). To examine coimmunoprecipitation of proteins in low calcium conditions, transfected HEK293 cells were replated in low calcium media (5 µM) 2 h before lysis and processed as described in this section.
Actin co-sedimentation assays
Chicken muscle G-actin (Cytoskeleton, Inc.) was incubated in polymerization buffer (20 mM Tris, pH 8.0, 100 mM KCl, 2 mM MgCl2, 0.5 mM ATP, 1 mM EGTA, and 1 mM DTT) for 1 h at room temperature to polymerize filaments. Gel-filtered proteins were diluted in reaction buffer (20 mM Tris pH 8.0, 150 mM NaCl, 2 mM MgCl2, 0.5 mM ATP, 1 mM EGTA, and 1 mM DTT) in the absence or presence of 2 µM F-actin, and incubated for 30 min at room temperature. Samples were centrifuged at 4°C for 20 min at 100,000 RPM in a TLA 120.1 rotor and pellets were resuspended in Laemmli buffer. The input and pelleted protein were separated by SDS-PAGE followed by CBB staining and analyzed using a LICOR Odyssey imaging system (LI-COR Biosciences).
Actin polymerization assays
Actin was prepared from chicken pectoral muscle as previously described (Spudich and Watt, 1971) and further purified by SEC gel filtration. A 10% solution of pyrene-labeled rabbit muscle G-actin (Cytoskeleton, Inc.) was prepared in buffer containing 2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, and 0.5 mM DTT, incubated on ice for 1 h, and centrifuged at 4°C for 30 min at 14,000 RPM to remove aggregates. Before each experiment, Ca2+ was exchanged for Mg2+ by adding 10% Mg-exchange buffer (50 mM MgCl2, 0.2 mM EGTA) and incubating 2 min at room temperature. To initiate polymerization, polymerization buffer was added to a final concentration of 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole HCl, pH 7.0, and, where indicated, bovine Arp2/3-complex and human GST-nWASp-VCA (Cytoskeleton, Inc.) were added to a final concentration of 50 and 200 nM, respectively. E-cadΔ70/α monomer and homodimer were added to a final concentration of 8 µM. Pyrene fluorescence (365 nm excitation, 407 nm emission) was measured every 10 s, with a 10–20 s delay after addition of polymerization buffer, using a Tecan Infinite M1000 plate reader (Tecan) until fluorescence levels reached a stable value (∼1,000 s).
Isothermal titration calorimetry
ITC experiments were performed on a VP-ITC calorimeter (Microcal; GE Healthcare). A total of 31 aliquots of 9 µl β-catenin at 60 µM were injected into the cell that contained 6 µM of E-cadΔ70/α monomer, E-cadΔ70/α homodimer, or E-cad/α monomer, in 20 mM Hepes, pH 8.0, 150 mM NaCl, and 1 mM DTT at 25°C. Heat change was measured for 240 s between injections. Injection of β-catenin into buffer was used for baseline subtraction, and data were analyzed with Microcal Origin software.
Whole-cell lysate and cell fractionation
L cells were cultured for 2 d in DMEM with 1 g/l sodium bicarbonate, 4.5 g/l glucose, 10% FBS (Atlas Biologicals), 5% penicillin, streptomycin, and kanamycin, and 1 µM dexamethasone to induce expression of E-cadherin (Angres et al., 1996). L cells were transfected with GFP-empty vector, E-cadherin-GFP, E-cadΔ70/α, or E-cadΔ70/β/α using Lipofectamine 3000 (Thermo Fisher Scientific) according to manufacturer’s protocol. For whole-cell lysate, 48 h after transfection cells were washed 1× in PBS, lysed in Triton X-100 buffer (25 mM Hepes, 100 mM NaCl, 1 mM EDTA, 10% [vol/vol] glycerol, and 1% [vol/vol] Triton X-100), and normalized using a BCA Protein Assay kit (Thermo Fisher Scientific). Cell lysates were transferred to a 1.5-ml tube and boiled for 10 min at 100°C, and the same amount of total protein was loaded on an 4–20% Criterion TGX Gel (Bio-Rad Laboratories). Gels were transferred onto nitrocellulose (GE Healthcare) and blocked in 1% milk and 2% BSA in Tris-buffered saline, and probed with indicated antibodies. Cell fractionation was performed as previously described (Benjamin et al., 2010).
Immunofluorescence microscopy
L cells transfected with chimeric constructs were fixed in 4% paraformaldehyde for 20 min, blocked in 0.05% saponin, 10% FBS, 50 mM NH4Cl2, and stained with indicated primary antibodies and, subsequently, secondary Alexa Fluor–conjugated antibodies. Coverslips were mounted using Aqua Polymount (Polysciences, Inc.) and imaged on a Zeiss Axiovert 200 with a plan-apochromat 63× 1.4 NA and 100× 1.4 NA objective. Images were acquired using an AxioCam mRM camera and AxioVision Rel. 4.6 software (Carl Zeiss) and adjusted for brightness and contrast using ImageJ software (National Institutes of Health). For proteasomal inhibitor, 48 h after transfection cells, were incubated with 25 µM MG132 (Sigma-Aldrich) for 6 h before fixing.
Flow cytometry
L cells were transfected with GFP-empty vector, E-cadherin-GFP, E-cadΔ70/α, or E-cadΔ70/β/α using Lipofectamine 3000 (Thermo Fisher Scientific) according to manufacturer’s protocol. Transfected and untransfected control L cells were gently scraped off using ice-cold PBS with 10% FBS (Atlas Biologicals) to maintain surface protein expression levels. Cells were incubated with the RR1 antibody for 1 h, washed three times in PBS + 10% FBS, incubated with DαM568 for 40 min, washed three times, resuspended in 1 ml PBS + 10% FBS, and immediately analyzed for fluorescence intensity using a FACSCalibur Flow Cytometer (BD).
E-cadherin-Fc adhesion assay
For E-cadherin–dependent adhesion of L cells to E-cadherin-Fc, Nunc 96 MicroWell Plates - MaxiSorp (Thermo Fisher Scientific) were coated with 1 µg/well of anti–human IgG (Fc Specific) antibody (I2136; Sigma-Aldrich), blocked with 1% bovine serum albumin, and incubated with 5 µg/ml E-cadherin-Fc that was purified from HEK293 cells (as described in Drees et al., 2005b). Transfected cells were trypsinized, washed in low-calcium DMEM, and allowed to recover surface proteins for 1 h in suspension in low-calcium DMEM containing 10 mM Hepes, pH 7.4, and 0.5% BSA at 37°C with gentle rolling. Subsequently, 1.25 × 105 cells were plated per well in the absence or presence of 1 mM CaCl2. Adhesion was allowed to proceed for 60 min at 37°C, and unbound cells were discarded by washing with HBS (20 mM Hepes, pH 7.4, 137 mM NaCl, and 3 mM KCl) in the absence or presence of 1 mM CaCl2. Adherent cells were lysed and subjected to a luciferase assay using a dual luciferase assay kit (Promega) according to the manufacturer’s protocol.
Hanging drop adhesion assay
L cells were transfected with GFP-empty vector, E-cadherin-GFP, E-cadΔ70/α, or E-cadΔ70/β/α using Lipofectamine 3000 (Thermo Fisher Scientific) according to manufacturer’s protocol. 48 h after transfection, cells were trypsinized using 20 mM Hepes, pH 7.4, 137 mM NaCl, 3 mM KCl, 200 mM CaCl2, 400 mg/l trypsin. Cells were resuspended at 2 × 106 cells/ml in DMEM with 1 g/l sodium bicarbonate, 4.5 g/l glucose, 10% FBS (Atlas Biologicals), and 20-µl drops of cell suspension were pipetted onto 35-mm culture dish lids, and inverted onto dishes filled with 2 ml media to prevent evaporation. At each time point, drops were triturated 20 times through a pipet, fixed with 4% PFA, and each drop was spread onto a glass slide. The entire coverslip was analyzed with an inverted microscope with a 40× 1.3 NA oil objective, and the same total number of cells for each condition was counted. The percent adhesion was calculated by dividing the number of cells in clusters of four or more cells by the total number of cells and the value normalized to E-cadherin.
Online supplemental material
Fig. S1 shows in vitro characterization of E-cadherin/αE-catenin chimeras. Fig. S2 shows cytosolic expression of αE-catenin in L cells, and hanging drop cell–cell adhesion assay. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201411080/DC1.
Supplementary Material
Acknowledgments
We thank Andre Muller and Kathleen Siemers for technical assistance and the Nelson laboratory for support.
This work was supported by a National Institutes of Health (NIH) CMB Training Grant T32-GM007276 (J.M. Bianchini), National Science Foundation GRFP DGE-114747 (J.M. Bianchini), NIH F32 GM096609 (K.N. Kitt), KWF Fellowship BUIT2012-5373 (M. Gloerich), NWO Rubicon Fellowship 82511015 (M. Gloerich), NIH GM35527 (W.J. Nelson), and UO1GM094663 (W.J. Nelson and W.I. Weiss).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ABD
- actin-binding domain
- AJ
- adherens junction
- CBD
- catenin-binding domain
- CCC
- cadherin–catenin complex
- E-cad/α
- E-cadherin/α-catenin chimera
- E-cadΔ70
- E-cadherin lacking the 70 C-terminal amino acids
- E-cadΔ70/β/α
- E-cadΔ70/β-catenin residues 118-151/αE-catenin
- IEC
- ion exchange chromatography
- ITC
- isothermal titration calorimetry
- SEC
- size exclusion chromatography
- TM
- transmembrane
References
- Abe K., and Takeichi M.. 2008. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc. Natl. Acad. Sci. USA. 105:13–19. 10.1073/pnas.0710504105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angres B., Barth A., and Nelson W.J.. 1996. Mechanism for transition from initial to stable cell–cell adhesion: kinetic analysis of E-cadherin-mediated adhesion using a quantitative adhesion assay. J. Cell Biol. 134:549–557. 10.1083/jcb.134.2.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin J.M., Kwiatkowski A.V., Yang C., Korobova F., Pokutta S., Svitkina T., Weis W.I., and Nelson W.J.. 2010. αE-catenin regulates actin dynamics independently of cadherin-mediated cell–cell adhesion. J. Cell Biol. 189:339–352. 10.1083/jcb.200910041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley C.D., Tan J., Anderson K.L., Hanein D., Volkmann N., Weis W.I., Nelson W.J., and Dunn A.R.. 2014. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 346:1254211 10.1126/science.1254211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.T., Stewart D.B., and Nelson W.J.. 1999. Coupling assembly of the E-cadherin/beta-catenin complex to efficient endoplasmic reticulum exit and basal-lateral membrane targeting of E-cadherin in polarized MDCK cells. J. Cell Biol. 144:687–699. 10.1083/jcb.144.4.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.J., Huber A.H., and Weis W.I.. 2006. Thermodynamics of beta-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J. Biol. Chem. 281:1027–1038. 10.1074/jbc.M511338200 [DOI] [PubMed] [Google Scholar]
- Collinet C., and Lecuit T.. 2013. Stability and dynamics of cell-cell junctions. Prog. Mol. Biol. Transl. Sci. 116:25–47. 10.1016/B978-0-12-394311-8.00002-9 [DOI] [PubMed] [Google Scholar]
- Dartsch N., Schulte D., Hägerling R., Kiefer F., and Vestweber D.. 2014. Fusing VE-cadherin to α-catenin impairs fetal liver hematopoiesis and lymph but not blood vessel formation. Mol. Cell. Biol. 34:1634–1648. 10.1128/MCB.01526-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R., Sarpal R., Ishiyama N., Pellikka M., Ikura M., and Tepass U.. 2013. Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat. Cell Biol. 15:261–273. 10.1038/ncb2685 [DOI] [PubMed] [Google Scholar]
- Drees F., Pokutta S., Yamada S., Nelson W.J., and Weis W.I.. 2005a Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 123:903–915. 10.1016/j.cell.2005.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees F., Reilein A., and Nelson W.J.. 2005b Cell-adhesion assays: fabrication of an E-cadherin substratum and isolation of lateral and Basal membrane patches. Methods Mol. Biol. 294:303–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi C.J., Wong E., and Gumbiner B.M.. 2001. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J. Cell Biol. 153:1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B.M. 1996. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 84:345–357. 10.1016/S0092-8674(00)81279-9 [DOI] [PubMed] [Google Scholar]
- Hansen S.D., Kwiatkowski A.V., Ouyang C.Y., Liu H., Pokutta S., Watkins S.C., Volkmann N., Hanein D., Weis W.I., Mullins R.D., and Nelson W.J.. 2013. αE-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Mol. Biol. Cell. 24:3710–3720. 10.1091/mbc.E13-07-0388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinck L., Näthke I.S., Papkoff J., and Nelson W.J.. 1994. Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J. Cell Biol. 125:1327–1340. 10.1083/jcb.125.6.1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Troyanovsky R.B., and Troyanovsky S.M.. 2013. Binding to F-actin guides cadherin cluster assembly, stability, and movement. J. Cell Biol. 201:131–143. 10.1083/jcb.201211054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura Y., Itoh M., Maeno Y., Tsukita S., and Nagafuchi A.. 1999. Functional domains of alpha-catenin required for the strong state of cadherin-based cell adhesion. J. Cell Biol. 144:1311–1322. 10.1083/jcb.144.6.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak A., Pasolli H.A., and Fuchs E.. 2004. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 6:21–30. 10.1038/ncb1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslov E.R., Maupin P., Pradhan D., Morrow J.S., and Rimm D.L.. 1997. Alpha-catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with beta-catenin. J. Biol. Chem. 272:27301–27306. 10.1074/jbc.272.43.27301 [DOI] [PubMed] [Google Scholar]
- Küppers V., Vestweber D., and Schulte D.. 2013. Locking endothelial junctions blocks leukocyte extravasation, but not in all tissues. Tissue Barriers. 1:e23805 10.4161/tisb.23805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski A.V., Maiden S.L., Pokutta S., Choi H.J., Benjamin J.M., Lynch A.M., Nelson W.J., Weis W.I., and Hardin J.. 2010. In vitro and in vivo reconstitution of the cadherin-catenin-actin complex from Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 107:14591–14596. 10.1073/pnas.1007349107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden S.L., and Hardin J.. 2011. The secret life of α-catenin: moonlighting in morphogenesis. J. Cell Biol. 195:543–552. 10.1083/jcb.201103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs J.A., Napolitano E.W., Murphy-Erdosh C., Mays R.W., Reichardt L.F., and Nelson W.J.. 1993. Distinguishing roles of the membrane-cytoskeleton and cadherin mediated cell–cell adhesion in generating different Na+,K(+)-ATPase distributions in polarized epithelia. J. Cell Biol. 123:149–164. 10.1083/jcb.123.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P.W., Pokutta S., Ghosh A., Almo S.C., Weis W.I., Nelson W.J., and Kwiatkowski A.V.. 2013. Danio rerio αE-catenin is a monomeric F-actin binding protein with distinct properties from Mus musculus αE-catenin. J. Biol. Chem. 288:22324–22332. 10.1074/jbc.M113.458406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi A., Ishihara S., and Tsukita S.. 1994. The roles of catenins in the cadherin-mediated cell adhesion: functional analysis of E-cadherin-alpha catenin fusion molecules. J. Cell Biol. 127:235–245. 10.1083/jcb.127.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näthke I.S., Hinck L., Swedlow J.R., Papkoff J., and Nelson W.J.. 1994. Defining interactions and distributions of cadherin and catenin complexes in polarized epithelial cells. J. Cell Biol. 125:1341–1352. 10.1083/jcb.125.6.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K., Zhang J., Fukuhara S., Kunimoto S., Yoshimura M., and Mochizuki N.. 2010. Vascular endothelial-cadherin stabilizes at cell-cell junctions by anchoring to circumferential actin bundles through alpha- and beta-catenins in cyclic AMP-Epac-Rap1 signal-activated endothelial cells. Mol. Biol. Cell. 21:584–596. 10.1091/mbc.E09-07-0580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M. 1998. Identification of the region of α-catenin that plays an essential role in cadherin-mediated cell adhesion. J. Biol. Chem. 273:29524–29529. 10.1074/jbc.273.45.29524 [DOI] [PubMed] [Google Scholar]
- Ozawa M., and Kemler R.. 1998. Altered cell adhesion activity by pervanadate due to the dissociation of α-catenin from the E-cadherin·catenin complex. J. Biol. Chem. 273:6166–6170. 10.1074/jbc.273.11.6166 [DOI] [PubMed] [Google Scholar]
- Ozono K., Komiya S., Shimamura K., Ito T., and Nagafuchi A.. 2011. Defining the roles of α-catenin in cell adhesion and cytoskeleton organization: isolation of F9 cells completely lacking cadherin-catenin complex. Cell Struct. Funct. 36:131–143. 10.1247/csf.11009 [DOI] [PubMed] [Google Scholar]
- Pacquelet A., and Rørth P.. 2005. Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. J. Cell Biol. 170:803–812. 10.1083/jcb.200506131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S., and Weis W.I.. 2000. Structure of the dimerization and beta-catenin-binding region of alpha-catenin. Mol. Cell. 5:533–543. 10.1016/S1097-2765(00)80447-5 [DOI] [PubMed] [Google Scholar]
- Pokutta S., and Weis W.I.. 2007. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu. Rev. Cell Dev. Biol. 23:237–261. 10.1146/annurev.cellbio.22.010305.104241 [DOI] [PubMed] [Google Scholar]
- Pokutta S., Choi H.J., Ahlsen G., Hansen S.D., and Weis W.I.. 2014. Structural and thermodynamic characterization of cadherin·β-catenin·α-catenin complex formation. J. Biol. Chem. 289:13589–13601. 10.1074/jbc.M114.554709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsioen B., Gloerich M., Ritsma L., Rehmann H., Bos J.L., and Jalink K.. 2009. Direct spatial control of Epac1 by cyclic AMP. Mol. Cell. Biol. 29:2521–2531. 10.1128/MCB.01630-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Capaldo C., Gumbiner B.M., and Macara I.G.. 2005. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J. Cell Biol. 171:1061–1071. 10.1083/jcb.200506094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimm D.L., Koslov E.R., Kebriaei P., Cianci C.D., and Morrow J.S.. 1995. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. USA. 92:8813–8817. 10.1073/pnas.92.19.8813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarpal R., Pellikka M., Patel R.R., Hui F.Y., Godt D., and Tepass U.. 2012. Mutational analysis supports a core role for Drosophila α-catenin in adherens junction function. J. Cell Sci. 125:233–245. 10.1242/jcs.096644 [DOI] [PubMed] [Google Scholar]
- Schulte D., Küppers V., Dartsch N., Broermann A., Li H., Zarbock A., Kamenyeva O., Kiefer F., Khandoga A., Massberg S., and Vestweber D.. 2011. Stabilizing the VE-cadherin-catenin complex blocks leukocyte extravasation and vascular permeability. EMBO J. 30:4157–4170. 10.1038/emboj.2011.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih W., and Yamada S.. 2012. N-cadherin-mediated cell-cell adhesion promotes cell migration in a three-dimensional matrix. J. Cell Sci. 125:3661–3670. 10.1242/jcs.103861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., and Watt S.. 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871. [PubMed] [Google Scholar]
- Thomas W.A., Boscher C., Chu Y.S., Cuvelier D., Martinez-Rico C., Seddiki R., Heysch J., Ladoux B., Thiery J.P., Mege R.M., and Dufour S.. 2013. α-Catenin and vinculin cooperate to promote high E-cadherin-based adhesion strength. J. Biol. Chem. 288:4957–4969. 10.1074/jbc.M112.403774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss F., Le Duc Q., Van Der Horst S., Tabdili H., Van Der Krogt G., Wang N., Rehmann H., Huveneers S., Leckband D.E., and De Rooij J.. 2012. Vinculin-dependent Cadherin mechanosensing regulates efficient epithelial barrier formation. Biol. Open. 1:1128–1140. 10.1242/bio.20122428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Degenstein L., Wise B., and Fuchs E.. 2001. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 104:605–617. 10.1016/S0092-8674(01)00246-X [DOI] [PubMed] [Google Scholar]
- Watabe-Uchida M., Uchida N., Imamura Y., Nagafuchi A., Fujimoto K., Uemura T., Vermeulen S., van Roy F., Adamson E.D., and Takeichi M.. 1998. alpha-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J. Cell Biol. 142:847–857. 10.1083/jcb.142.3.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E.E., Kroemker M., Rüdiger A.H., Jockusch B.M., and Rüdiger M.. 1998. Vinculin is part of the cadherin–catenin junctional complex: complex formation between alpha-catenin and vinculin. J. Cell Biol. 141:755–764. 10.1083/jcb.141.3.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M.J., Nagelkerken B., Mertens A.E., Rees-Bakker H.A., Briaire-de Bruijn I.H., and Litvinov S.V.. 2003. Expression of Ep-CAM shifts the state of cadherin-mediated adhesions from strong to weak. Exp. Cell Res. 285:50–58. 10.1016/S0014-4827(02)00045-9 [DOI] [PubMed] [Google Scholar]
- Yamada S., Pokutta S., Drees F., Weis W.I., and Nelson W.J.. 2005. Deconstructing the cadherin-catenin-actin complex. Cell. 123:889–901. 10.1016/j.cell.2005.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.