GIV/Girdin, a nonreceptor GEF, directly links integrins to activation of trimeric G proteins to promote the acquisition of proinvasive traits in cancer cells.
Abstract
Signal transduction via integrins and G protein–coupled receptors is critical to control cell behavior. These two receptor classes have been traditionally believed to trigger distinct and independent signaling cascades in response to extracellular cues. Here, we report a novel mechanism of integrin signaling that requires activation of the trimeric G protein Gαi by the nonreceptor guanine nucleotide exchange factor (GEF) GIV (also known as Girdin), a metastasis-associated protein. We demonstrate that GIV enhances integrin-dependent cell responses upon extracellular matrix stimulation and makes tumor cells more invasive. These responses include remodeling of the actin cytoskeleton and PI3K-dependent signaling, resulting in enhanced haptotaxis and invasion. We show that both GIV and its substrate Gαi3 are recruited to active integrin complexes and that tumor cells engineered to express GEF-deficient GIV fail to transduce integrin signals into proinvasive responses via a Gβγ-PI3K axis. Our discoveries delineate a novel mechanism by which integrin signaling is rewired during metastasis to result in increased tumor invasiveness.
Introduction
Integrins are heterodimeric receptors that mediate adhesion to the ECM. Upon activation, integrins recruit intracellular proteins involved in cytoskeletal remodeling and signal transduction, leading to the regulation of multiple aspects of cell behavior (Miranti and Brugge, 2002; Legate et al., 2009). Consequently, dysregulation of integrin function gives rise to different pathologies. In cancer, integrins play a critical role in metastasis by promoting cell migration and invasion (Guo and Giancotti, 2004; Desgrosellier and Cheresh, 2010; Huttenlocher and Horwitz, 2011). However, the molecular mechanisms for this remain poorly understood. An important feature of integrins is that they transmit signals bidirectionally (Hynes, 2002). In the so-called “inside-out” signaling, integrins sense signals from the interior of the cells to modulate their extracellular adhesive function. Upon adhesion to extracellular substrates, they trigger “outside-in” signaling, which is initiated by the association of cytoskeletal and signaling molecules to the tails of the β subunit of integrins. Some key initial signaling events are the activation of tyrosine kinases (e.g., focal adhesion kinase and Src) and phosphatidylinositol-3-kinase (PI3K), followed by the activation of a cascade of kinases and small G proteins of the Rho family (Legate et al., 2009).
Integrin signaling is also intertwined with other signaling pathways. The best-studied case is the cross talk between receptor tyrosine kinase (RTK) signaling and outside-in integrin signaling (Plopper et al., 1995; Miyamoto et al., 1996; Sundberg and Rubin, 1996). It is also well established that the activation of trimeric G proteins by G protein–coupled receptors (GPCRs), another major class of surface receptors, regulates inside-out integrin signaling (Offermanns, 2006; Abram and Lowell, 2009; Shen et al., 2012). In contrast, trimeric G proteins and integrins in the outside-in mode have been traditionally considered to trigger distinct and independent signaling cascades (Shen et al., 2012). This idea has been recently put into question by the discovery that the trimeric G protein Gα13 regulates integrin outside-in signaling. More specifically, active Gα13 directly binds integrin β subunits upon ligand binding and is required for activation of downstream signaling (Gong et al., 2010; Shen et al., 2013). Interestingly, regulation of integrin-mediated cell spreading by active Gα13 occurred in the absence of GPCR stimulation (Gong et al., 2010), suggesting that G protein activation could be achieved by an alternative mechanism. These recent findings indicate that trimeric G proteins may have a role in integrin outside-in signaling more important than previously appreciated, and also raise the question of how trimeric G protein activation is achieved upon integrin activation.
Recent discoveries have challenged the classical view of trimeric G protein activation as an exclusive function of GPCRs. In the classic paradigm, GPCRs are guanine nucleotide exchange factors (GEFs) that activate trimeric G proteins via GDP–GTP exchange (Gilman, 1987). However, it has become evident that some nonreceptor proteins can also exert GEF activity toward trimeric G proteins (Cismowski et al., 1999, 2000; Tall et al., 2003; Sato et al., 2006). We have contributed to the recent characterization of one of these nonreceptor GEFs called GIV (also known as Gα-interacting, vesicle-associated protein or Girdin or KIAA1212; Garcia-Marcos et al., 2015). GIV is a large multidomain protein reported to bind to RTKs (Lin et al., 2014), phosphoinositides (Enomoto et al., 2005), actin fibers (Enomoto et al., 2005), microtubules (Simpson et al., 2005), and dynamin (Simpson et al., 2005; Weng et al., 2014), among others (Anai et al., 2005; Kim et al., 2009). A unique feature that differentiates GIV from other nonreceptor GEFs is that it bears a defined motif of 20–30 aa that is necessary and sufficient to activate G proteins (Garcia-Marcos et al., 2009, 2012). The specific disruption of this motif revealed that G protein activation by GIV is a key function required for signaling downstream of both GPCRs and RTKs in different settings (Garcia-Marcos et al., 2015).
GIV’s role in signaling has been most studied in cancer cells, in which GIV’s GEF activity promotes a set of pro-metastatic features including activation of the PI3K–Akt pathway, actin cytoskeleton remodeling, and cell motility (Garcia-Marcos et al., 2009, 2012). Importantly, GIV expression correlates with metastatic potential in cell lines of different origins (e.g., breast, colon, pancreas, etc.; Ghosh et al., 2010; Garcia-Marcos et al., 2011b; Dunkel et al., 2012) and is required for metastasis in mice (Jiang et al., 2008). GIV expression in primary tumors also correlates with metastasis and predicts patient death in multiple cancers of epithelial origin (Garcia-Marcos et al., 2011b; Ling et al., 2011; Dunkel et al., 2012; Liu et al., 2012a; Shibata et al., 2013; Zhao et al., 2013; Song et al., 2014; Wang et al., 2014), making it a bona fide metastasis-related protein. Thus, G protein activation by GIV’s GEF motif has emerged as a molecular function that underlies the pathological significance of GIV in cancer progression.
Despite these recent advances, all the studies to date on the role of GIV in cancer cell biology have neglected the importance of tumor cell–ECM interactions (Bissell and Radisky, 2001). Here, we studied the role of GIV in controlling the behavior of cancer cells in response to the ECM. We describe a novel signaling mechanism by which GIV transmits cues from activated integrins via direct activation of trimeric G proteins, which results in the acquisition of proinvasive phenotypes in cancer cells. These findings not only define a previously unknown mechanism of trimeric G protein signaling triggered by integrins but also provide new insights into how integrin signaling is rewired during cancer progression.
Results
GIV is required for the acquisition of an invasive growth pattern in 3D cultures of tumor cells
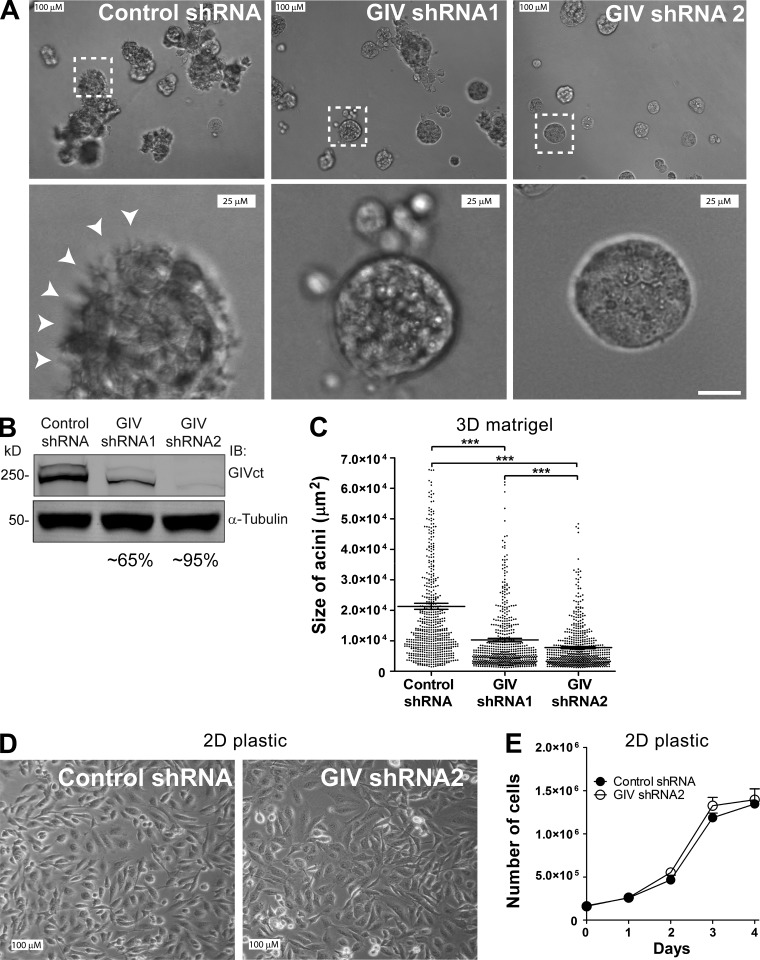
To investigate the role of GIV in controlling tumor cell behavior in the context of interactions with the ECM, we generated MDA-MB-231 cell lines stably depleted of endogenous GIV and tested them in 3D Matrigel tissue cultures (Fig. 1, A and B). 3D Matrigel cultures are well-established models that account for tumor cell interactions with the ECM and recapitulate many of the behavioral features of cancer cells in tumors in situ (Weaver et al., 1997; Wang et al., 1998; Debnath and Brugge, 2005). We chose highly invasive MDA-MB-231 breast cancer cells because they have been characterized previously in this experimental system (Wang et al., 2002; Park et al., 2006; Liu et al., 2012b) and because they express high levels of GIV (Ghosh et al., 2010; Garcia-Marcos et al., 2011b), which is required for their metastatic dissemination in vivo (Jiang et al., 2008). Consistent with previous reports (Wang et al., 2002; Park et al., 2006; Liu et al., 2012b), we found that MDA-MB-231 control cells form large and disorganized acini lacking a defined lumen and frequently display stellate-shaped cells protruding toward the matrix (Fig. 1 A). Depletion of GIV leads to a dramatic morphological change: cells form smaller and more organized acini with defined edges, albeit still lacking a lumen (Fig. 1 A). These observations are consistent with a transition from an invasive tumor phenotype to a less aggressive state upon GIV depletion (Wang et al., 2002; Park et al., 2006; Liu et al., 2012b). Quantification of the size of the acini (Fig. 1 C) revealed an ∼60 and ∼75% reduction for cells expressing GIV shRNA1 and GIV shRNA2, respectively, which parallels the extent of GIV depletion observed for each of the shRNA sequences (Fig. 1 B). In contrast, GIV depletion did not affect the overall cell morphology or growth under standard tissue culture conditions on plastic dishes (Fig. 1, D and E). Therefore, although changes in acini morphology and size observed upon GIV depletion can be caused by alterations of one or multiple processes (cell growth, sprouting, matrix degradation, etc.), these results indicate that they are specific to an alteration in the sensing of the ECM. To rule out that these observations are restricted to MDA-MB-231, we performed identical experiments with an unrelated cell line, i.e., COLO357-FG. Like MDA-MB-231, these invasive pancreatic adenocarcinoma cells express high levels of GIV (Garcia-Marcos et al., 2011b). We found that depletion of GIV in COLO357-FG also reduces acini size in 3D Matrigel cultures without affecting cell growth under standard culture conditions (Fig. S1). Collectively, these results suggest that GIV favors the acquisition of invasive traits in tumor cells in response to the ECM.
Figure 1.
GIV is required for the invasive growth pattern of MDA-MB-231 cells in 3D cultures. (A–C) GIV depletion impairs the proinvasive growth phenotype of MDA-MB-231 cells in Matrigel cultures. (A) MDA-MB-231 cells stably depleted of GIV by two independent shRNA sequences (GIV shRNA1 and GIV shRNA2) or expressing a control shRNA sequence were seeded on Matrigel and pictured by DIC microscopy after 7 d. Bottom panels correspond to a magnified view of areas inside the white boxes drawn on the upper panels. White arrowheads highlight stellate-shaped cells protruding from the acini toward the matrix. (B) The efficiency of GIV depletion (∼65% for shRNA1 and ∼95% for shRNA2 cells) was determined by immunoblotting (IB) using the indicated antibodies. (C) Quantification of the acini size (n = 3; 200 acini per experiment). Each dot is the size of one acini, and the horizontal line is the mean ± SEM (***, P < 0.001). (D and E) GIV depletion does not alter MDA-MB-231 cell morphology or growth on plastic. MDA-MB-231 cells stably depleted of GIV by expression of GIV shRNA2 or expressing a control shRNA were seeded on plastic dishes and grown in complete media for 4 d. A representative field of these cells was pictured by DIC microscopy (D), and cells were counted every day using a hemocytometer (E). Results are depicted as mean ± SEM (error bars; n = 3).
GIV promotes Akt activation upon integrin stimulation
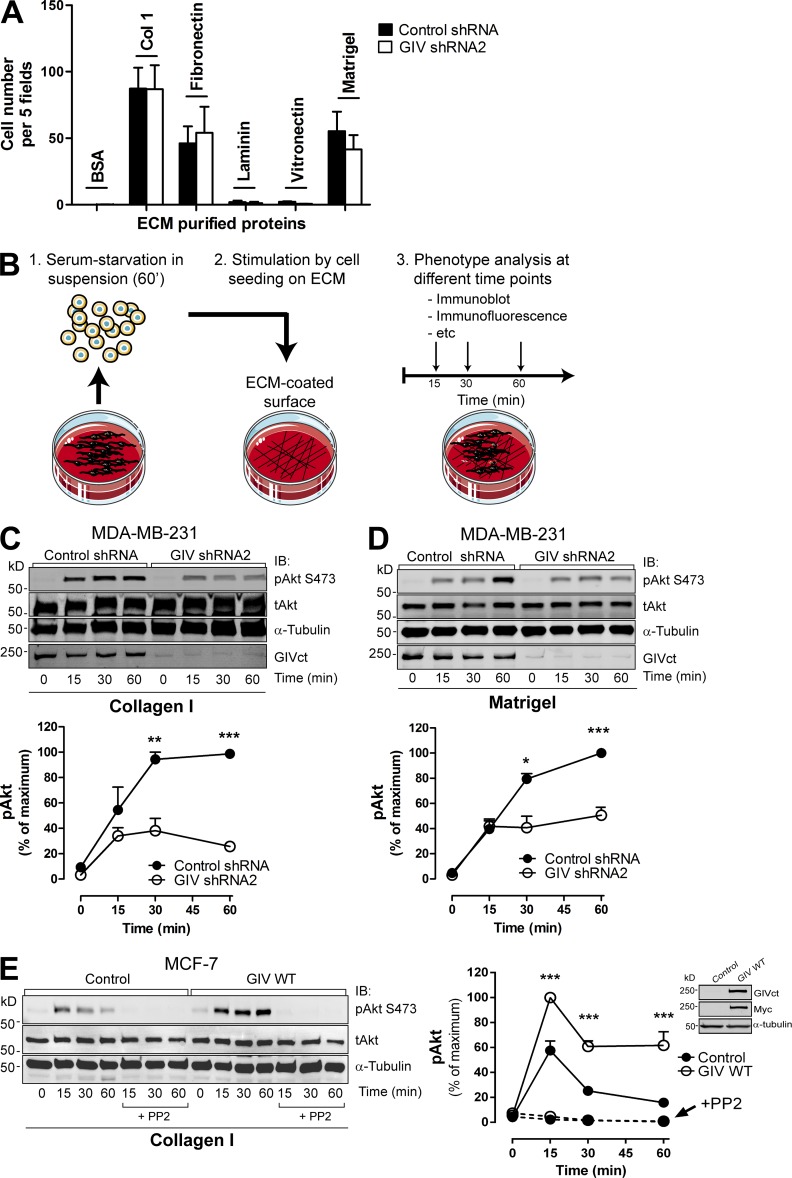
The morphological change of MDA-MB-231 cells in 3D cultures upon GIV depletion closely resembles that described by others upon integrin blockade under the same experimental conditions (Weaver et al., 1997; Wang et al., 2002; Park et al., 2006). For this reason, we next investigated if GIV regulates integrin function. First, we measured cell adhesion to different integrin substrates of the ECM, i.e., collagen I, fibronectin, vitronectin, and laminin. We also tested cell adhesion to Matrigel, which is a complex mixture of different ECM components. Consistent with previous observations (Liu et al., 2012b), MDA-MB-231 cell adhesion was highest to collagen I, followed by fibronectin and Matrigel, and much lower to vitronectin and laminin (Fig. 2 A). GIV-depleted cells showed no difference in adhesion compared with controls (Fig. 2 A), indicating that binding of integrins to these substrates is not significantly affected by GIV.
Figure 2.
GIV promotes Akt activation upon integrin stimulation. (A) GIV depletion does not affect MDA-MB-231 cell adhesion to different ECM substrates. MDA-MB-231 control shRNA and GIV shRNA2 cells were seeded on plates coated with collagen I, fibronectin, laminin, vitronectin, Matrigel, or BSA (as negative control), and cell adhesion was determined 1 h later as described in Materials and methods. Results are depicted as mean ± SEM (error bars; n = 3). (B) Schematic representation of the protocol followed to monitor ECM-specific cell stimulation. Cells were lifted, kept in suspension for 1 h in serum-free media, and seeded on surfaces coated with different ECM components in the absence of serum. Cells were harvested at different time points after seeding for subsequent analyses. Under these conditions, the only stimulus for the cells is mediated through binding to the ECM. (C and D) GIV depletion impairs Akt activation upon integrin activation in MDA-MB-231 cells. MDA-MB-231 control shRNA and GIV shRNA2 cells were stimulated by collagen I (C) or Matrigel (D), as described in B. (C and D; top) Representative immunoblots for the time course of Akt activation (as measured by levels of pAkt) upon ECM stimulation in MDA-MB-231cells. (C and D; bottom) Quantification of Akt activation (as described in Materials and methods). Error bars represent mean ± SEM (n = 3; **, P < 0.01; ***, P < 0.001). (E) Exogenous GIV expression in MCF-7 cells is sufficient to enhance Akt activation in response to collagen I stimulation. MCF-7 cells stably expressing GIV (GIV WT) or an empty plasmid (Control) were stimulated with collagen I as described in B. Some cells were preincubated with PP2 (as indicated). (Left) Representative immunoblots for the time course of Akt activation (as measured by levels of pAkt) upon collagen I stimulation in MCF-7 cells. (Right) Quantification of Akt activation (as described in Materials and methods). Results are depicted as mean ± SEM (error bars; n = 3–7; ***, P < 0.001). (Inset) Expression of exogenous GIV in MCF-7 cells was verified by immunoblotting (IB) using the indicated antibodies.
The results described above indicate that the loss of invasive growth pattern in 3D Matrigel cultures (Fig. 1) is probably not a consequence of defective substrate recognition by integrins. Next, we tested if instead GIV could be required to mediate the signaling responses triggered by integrins upon ECM stimulation. For this we used an experimental protocol (Fig. 2 B) in which cells were sequentially (a) serum starved in suspension, (b) plated on ECM-coated surfaces under serum-free conditions, and (c) analyzed by immunoblotting for activation of Akt. Under these conditions, the only external stimulus is attachment to the ECM proteins. We chose Akt as readout because previous reports have demonstrated that GIV is required for PI3K-dependent activation of Akt in response to other stimuli (Anai et al., 2005; Garcia-Marcos et al., 2009; Kim et al., 2009; Lopez-Sanchez et al., 2014) and because PI3K-Akt is a major signaling pathway also activated by integrins (Legate et al., 2009). In fact, we found that Akt is activated (as determined by phosphorylation of S473, pAkt) during the course of 1 h after MDA-MB-231 adhesion to collagen I. This response is integrin specific because it was not reproduced upon adhesion to the nonintegrin substrate poly-l-lysine (Fig. S2 A) and was inhibited by integrin-blocking antibodies (Fig. S2 B). We found that GIV depletion decreases Akt activation 50–60% in MDA-MB-231 cells stimulated with either collagen I (Fig. 2 C) or Matrigel (Fig. 2 D). Activation of focal adhesion kinase, another integrin-signaling mediator, was also impaired by GIV depletion in the same experiments (not depicted). The defect in Akt activation was not rescued by Mn2+ (Fig. S2 D), indicating that GIV depletion affects outside-in integrin signaling rather than impairing the ability of integrins to adopt an active conformation. This was further confirmed by immunostaining with an antibody that specifically recognizes the active conformation of β1 integrins (not depicted). Depletion of GIV in two other invasive cancer cell lines (i.e., COLO357-FG and HeLa) revealed that Akt activation in response to collagen I is also impaired (Fig. S3), which rules out that the effects observed in MDA-MB-231 cells are cell type specific. In conclusion, these results demonstrate that GIV is required for efficient Akt activation in response to integrin activation by different substrates and in different cell types.
Next, we investigated if expression of GIV is sufficient to enhance integrin-dependent responses. For this we used MCF-7 cells as a model because they are nonmetastatic breast cancer cells that express very low levels of endogenous GIV (Ghosh et al., 2010; Garcia-Marcos et al., 2011b). MCF-7 cells display specific Akt activation in response to collagen I stimulation but not upon binding to poly-l-lysine (Fig. S2 E). We found that MCF-7 cells stably expressing GIV display enhanced Akt activation in response to collagen I stimulation (approximately twofold) compared with control MCF-7 cells (Fig. 2 E). GIV-expressing MCF-7 cells failed to attach to collagen I in the presence of integrin-blocking antibodies (Fig. S2 F), and Akt responses were completely blunted upon inhibition of Src (Fig. 2 E), same as with control MCF-7 cells, which indicates that GIV-dependent signaling relies on canonical integrin-signaling activation mechanisms. Collectively, our results demonstrate that GIV enhances integrin-dependent Akt signaling in tumor cells.
GIV depletion impairs cytoskeletal rearrangements promoted by integrins
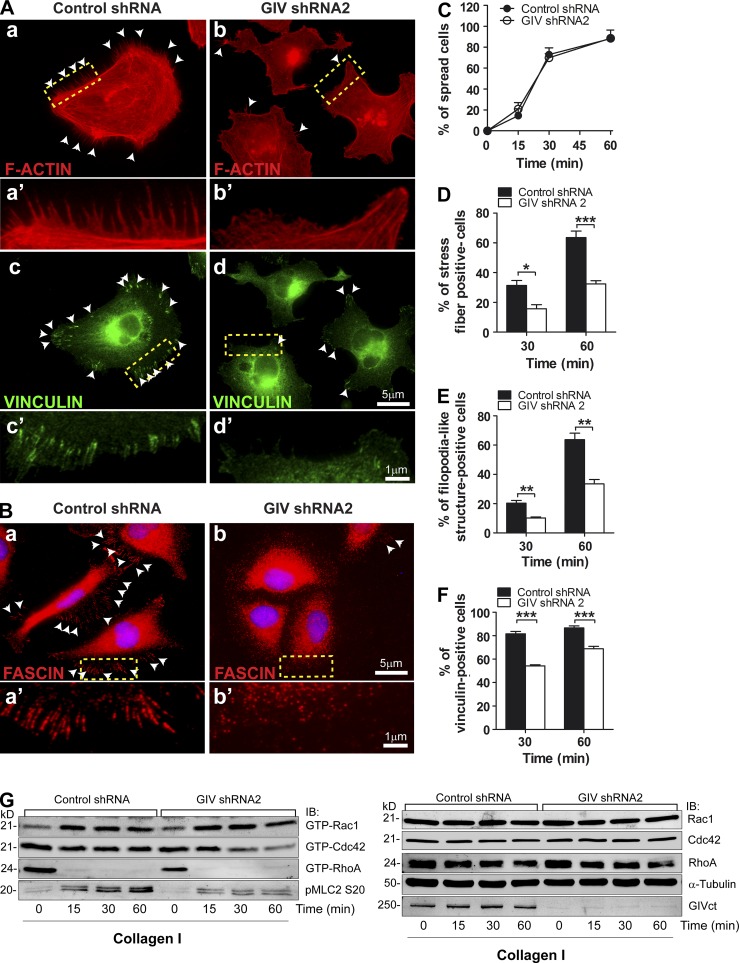
Integrin signaling is closely intertwined with the remodeling of the actin cytoskeleton (Miranti and Brugge, 2002; Legate et al., 2009). Next, we analyzed different cytoskeletal markers by immunofluorescence in MDA-MB-231 cells stimulated by attachment to collagen I as described above. GIV-depleted MDA-MB-231 cells displayed impaired formation of F-actin stress fibers (Fig. 3, A and D) and filopodia-like structures (Fig. 3, A and E, which were confirmed to be filopodia in Fig. 3 B by fascin staining) at 30 and 60 min after collagen I stimulation compared with controls, despite showing no defect in cell spreading (Fig. 3 C). GIV depletion also impaired the formation of mature focal adhesions (as determined by vinculin staining; Fig. 3, A and F). To gain further insight into how GIV affects the remodeling of the cytoskeleton, we measured the activation levels of Rac1, Cdc42, and RhoA, three critical regulators of actin remodeling, upon collagen I stimulation. Because active RhoA levels dropped below the limit of detection after attachment to collagen I (consistent with observations by others: Ren et al., 1999; Danen et al., 2002; Bhadriraju et al., 2007), we monitored MLC2 phosphorylation at Ser20 (pMLC2) as a surrogate for RhoA activity. We found that GIV-depleted cells have lower levels of active RhoA-pMLC2 and Cdc42 than control cells when they are attached to collagen I, whereas levels of Rac1 activity are unchanged (Fig. 3 G). These results are in keeping with the actin cytoskeleton features observed in GIV-depleted cells, i.e., cells spread (no difference in Rac1) but have defective formation of stress fibers (reduced RhoA/pMLC2) and filopodia (reduced Cdc42). Collectively, our results indicate that GIV is required to trigger actin remodeling upon integrin stimulation.
Figure 3.
GIV depletion impairs actin cytoskeleton remodeling upon integrin stimulation. (A and B) MDA-MB-231 shRNA control (left) and GIV shRNA2 (right) cells were stimulated with collagen I as described in Fig. 2 B, except that the collagen I–coated surfaces were glass coverslips and that cells were fixed and stained for F-actin (phalloidin, red) and vinculin (green) in A and fascin (red) in B. Representative pictures of the cells 60 min after seeding are shown. A magnified view of the boxed areas in A (a, b, c, and d) and B (a and b) is shown in A (a′, b′, c′, and d′) and B (a′ and b′). White arrowheads highlight filopodia-like structures in (A, a and b, and B, a and b) and focal adhesions (defined as vinculin-positive structures) in A (c and d). The percentage of cells displaying stress fibers (D), filopodia-like structures (E), and focal adhesions (F) at 30 and 60 min as well as the proportion of spread cells (C) at 15, 30, and 60 min was calculated as described in Materials and methods. Results are depicted as mean ± SEM (error bars; n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.01). (G) MDA-MB-231 shRNA control and GIV shRNA2 cells were stimulated with collagen I as described in Fig. 2 B, and Rac1, Cdc42, and RhoA activity (GTP-Rac1, GTP-Cdc42, and GTP-RhoA) were determined using a pulldown assay as described in Materials and methods. MLC2 phosphorylation was determined by immunoblotting (pMLC2). Representative of at least four experiments.
GIV depletion impairs integrin-mediated cell migration and invasion
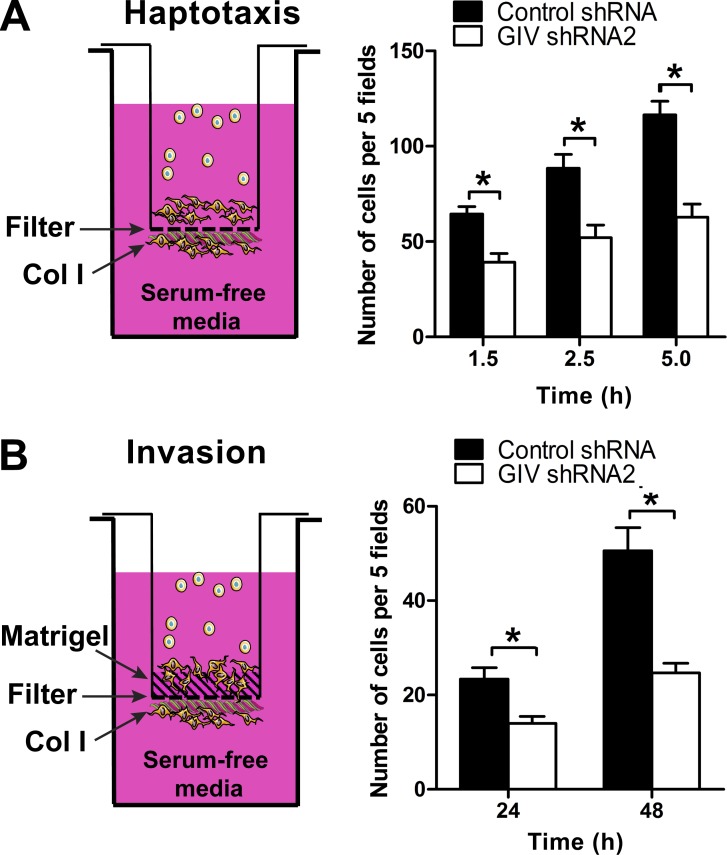
We reasoned that impaired integrin signaling and cytoskeletal remodeling provoked by GIV depletion would also have a deleterious effect on tumor cell phenotypes associated with metastasis, like cell migration and invasion. Previous work has established that GIV is required for enhancing signaling and cell migration of tumor cells in response to soluble factors that activate GPCRs or RTKs (Garcia-Marcos et al., 2015), and we confirmed that this is the case in our experimental conditions with MDA-MB-231 cells (Fig. S4). To test if GIV also controls proinvasive tumor cell phenotypes in response to integrin stimulation, we measured collagen I–driven cell migration (i.e., haptotaxis) by using a modified Boyden chamber assay in which the underside of the filter was coated with collagen I and serum-starved cells were placed on the upper compartment (Fig. 4 A). The lower compartment contained serum-free media so that cell migration was driven exclusively by collagen I. This migration was integrin specific because it was inhibited by incubation of the cells with a β1 integrin–blocking antibody (Fig. S2 C) and it was not observed in control experiments without collagen I coating (not depicted). We found that GIV-depleted MDA-MB-231 cells showed impaired haptotactic cell migration (∼50%) compared with controls (Fig. 4 A). Conversely, exogenous expression of GIV in noninvasive MCF-7 cells enhanced (approximately twofold) the rate of migration in the same assay (see Fig. 7 below). Next, we measured cell invasion. The design for the invasion experiments was the same as for haptotaxis except that the upper side of the filter was coated with a layer of Matrigel as a barrier for the cells (Fig. 4 B). The results also showed an ∼50% reduction in the invasion index in GIV-depleted MDA-MB-231 cells compared with controls (Fig. 4 B), indicating that GIV is required for the pro-metastatic behavior of tumor cells in response to collagen I stimulation.
Figure 4.
GIV is required for efficient haptotaxis and invasion of MDA-MB-231 cells in response to collagen I. (A) The underside of filters in Boyden chambers were coated with collagen I, and serum-starved MDA-MB-231 control shRNA and GIV shRNA2 cells were added to the upper compartment. The lower compartment was filled with serum-free media, and the number of migrated cells on the underside of the filter was counted at different time points (1.5, 2.5, and 5 h). The number of migrated cells was determined as described in Materials and methods. Results are depicted as mean ± SEM (error bars; n = 3; *, P < 0.05). (B) Invasion was determined as in A, except that the upper side of the filter was coated with Matrigel and that the number of cells was counted at 24 and 48 h. The number of cells that invaded through the Matrigel and reached the underside of the filter was determined as described in Materials and methods. Results are depicted as mean ± SEM (error bars; n = 3; *, P < 0.05). No significant cell migration or invasion was observed in controls not stimulated with collagen I (number of cells was <5% of collagen I–stimulated conditions; not depicted).
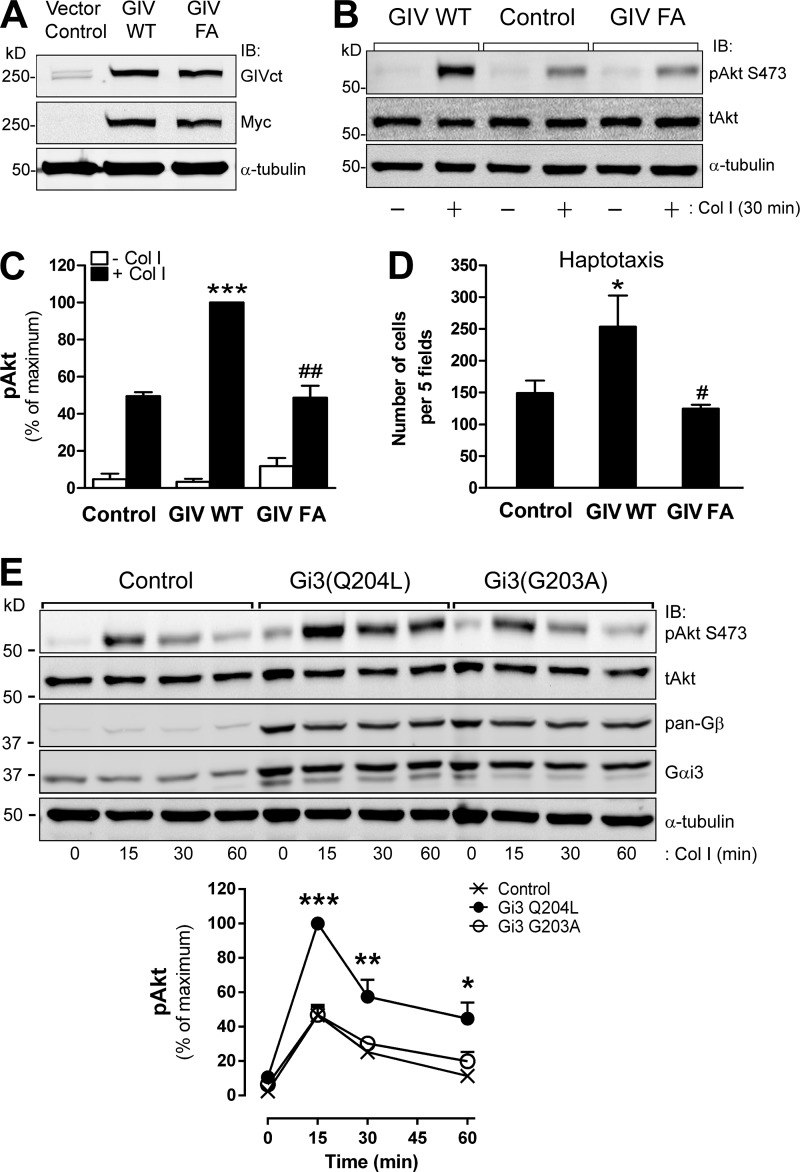
Figure 7.
GIV’s GEF activity is sufficient to enhance integrin-dependent Akt signaling and cell migration. (A) MCF-7 cells stably expressing GIV WT or GIV FA were generated as described in Materials and methods and immunoblotted (IB) with the indicated antibodies. (B and C) Expression of GIV WT, but not the GEF-deficient GIV F1685A (FA) mutant, in MCF-7 cells enhances Akt activation in response to collagen I. MCF-7 cells were stimulated with collagen I for 30 min as described in Fig. 2 B. A representative result is shown in B, and the quantification (n = 3; results are depicted as mean ± SEM (error bars); ***, P < 0.01, compared with control cells; ##, P < 0.01, compared with GIV WT cells) is shown in C. Equivalent results were obtained with cells stimulated with collagen I for 60 min (not depicted). (D) Expression of GIV WT, but not the GEF-deficient GIV F1685A (FA) mutant, in MCF-7 cells enhances haptotaxis. Cells were treated as described in B, except that they were used in haptotaxis assays (as described in Materials and methods). Results indicate mean ± SEM (error bars; *, P < 0.05, compared with control cells; #, P < 0.05, compared with GIV WT cells). (E) Expression of constitutively active but not constitutively inactive Gi3 in MCF-7 cells enhances Akt activation in response to collagen I stimulation. MCF-7 cells were transfected with Gβ1-YC, Gγ2-YN, and Gαi3 mutants Q204L (active) or G203A (inactive), seeded on collagen I for different times as described in Fig. 2 B and analyzed by immunoblotting. (Top) Representative immunoblots for the time course of Akt activation (pAkt) upon collagen I stimulation in MCF-7 cells. (Bottom) Quantification of Akt activation (n = 3; Results indicate mean ± SEM [error bars]; *, P < 0.05; **, P < 0.01, ***, P < 0.001).
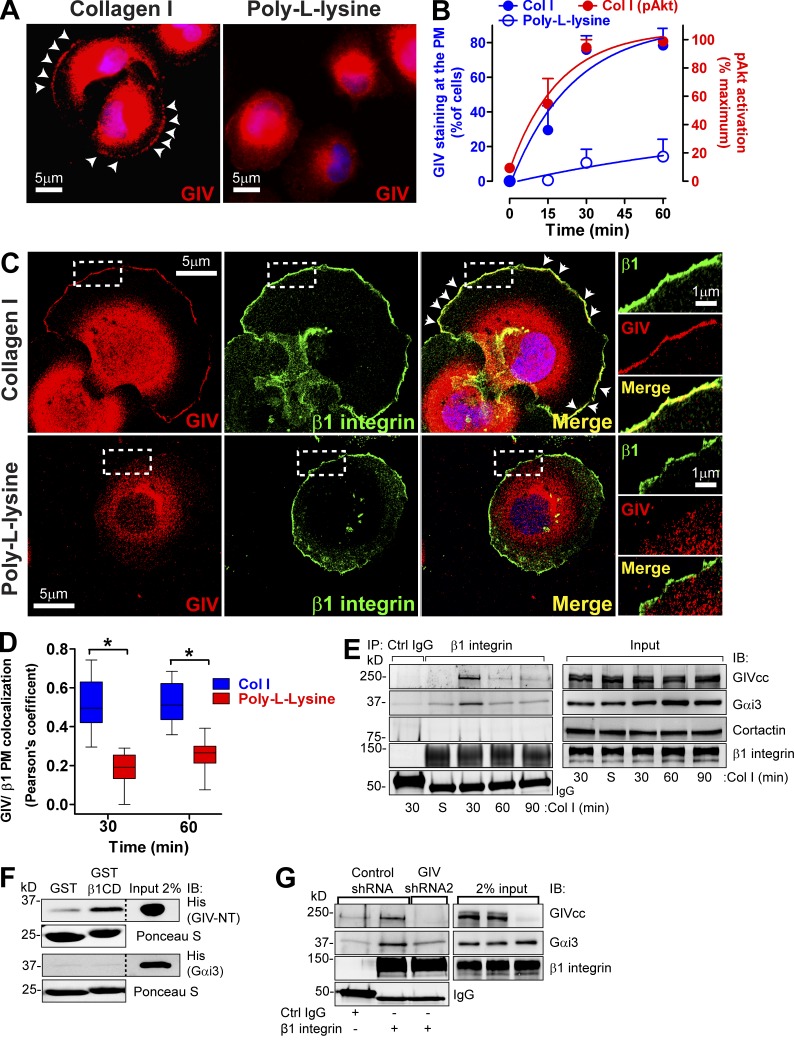
GIV is recruited to the plasma membrane (PM) upon ECM stimulation and colocalizes with integrins
We investigated the localization of GIV upon cell attachment and found that it is recruited to the PM of MDA-MB-231 cells upon binding to collagen I but not upon binding to the nonintegrin substrate poly-l-lysine (Fig. 5 A). The kinetics of GIV recruitment to the PM coincided with the kinetics of Akt activation (Fig. 5 B), suggesting a functional link between the two events. We reasoned that after recruitment to the PM, GIV would colocalize with β1 integrins (a subunit required to form collagen I–binding integrins). We found that this is indeed the case because GIV and β1 integrins colocalize at the PM (Pearson’s coefficient of ∼0.5) after 30 and 60 min of collagen I stimulation (Fig. 5, C and D), but not in cells binding to poly-l-lysine (Pearson’s coefficient of ∼0.2). GIV colocalization with β1 integrins is not caused by nonspecific membrane redundancy at the cell periphery because GIV is not detected at the PM of cells binding to poly-l-lysine, despite the fact that these cells do show β1-integrin staining at the cell periphery (Fig. 5 C). We observed marginal colocalization between GIV and vinculin, and it occurred only at the cell periphery (Fig. S5 A), suggesting that the majority of GIV does not associate with mature focal adhesions. To investigate if GIV associates with nascent focal adhesions, we used a previously validated approach (Kuo et al., 2011; Schiller et al., 2013) consisting of blocking focal adhesion maturation by inhibition of myosin II with blebbistatin. We found that GIV colocalizes with paxillin, a marker of nascent adhesions under these conditions (Kuo et al., 2011), and β1 integrins at the cell periphery (Fig. S5 B), suggesting that it associates with nascent focal adhesions. To further explore the association of GIV with integrin-rich structures involved in the invasive properties of tumor cells, we asked if it was present in invadosomes, which are integrin-rich adhesive structures required for ECM degradation and invasion (Albiges-Rizo et al., 2009; Murphy and Courtneidge, 2011). In NIH3T3-Src* cells, invadosomes have a distinct ring-shaped morphology and are enriched in β1 integrins, F-actin, and cortactin (Seals et al., 2005; Murphy and Courtneidge, 2011). We found that GIV is frequently localized in invadosomes in these cells (Fig. S5 C). Collectively, these results indicate that GIV can associate with different types of integrin-based subcellular structures involved in adhesion and invasion.
Figure 5.
GIV is recruited to β1 integrins at the PM upon collagen I stimulation. (A and B) GIV is recruited to the cell periphery when cells attach to collagen I but not to poly-l-lysine. Control MDA-MB-231 cells were seeded on collagen I or the nonintegrin substrate poly-l-lysine following the protocol described in Fig. 2 B, except that the coated surfaces were glass coverslips and that cells were fixed and stained for GIV (red) or DNA (DAPI; blue). Representative pictures of the cells 60 min after seeding are shown in A. B shows the quantification of percentage of cells displaying GIV staining at the PM (blue; left axis) upon attachment to collagen I (filled circles) or poly-l-lysine (open circles). Results are depicted as mean ± SEM (error bars; n = 3–4). The quantification of Akt activation shown in Fig. 2 C is plotted here in red (right axis) for comparison. (C and D) GIV colocalizes with β1 integrin at the PM in cells attaching to collagen I but not to poly-l-lysine. Cells were treated as described in A but costained for GIV (red) and β1 integrin (green) and imaged by confocal microscopy. Representative pictures of cells 60 min after seeding are shown in C, and the quantification of three independent experiments are shown in D. White arrowheads indicate colocalization, and the boxed areas are shown enlarged on the right. Colocalization at the PM was quantified as described in Materials and methods and shown as box and whiskers plots (midline; median, box; 25–75%, whiskers; min-max range; n = 3; 8–10 cells/ experiment; *, P < 0.05). (E) GIV and Gαi3 coimmunoprecipitates with β1 integrins upon collagen I stimulation. Control MDA-MB-231 cells were seeded on collagen I for 30, 60, and 90 min as described in Fig. 2 B. Lysates of collagen I–attached cells or cells in suspension (S) were immunoprecipitated (IP) with β1-integrin antibodies (AIIB2) as described in Materials and methods. IPs (left) and lysates (right) were immunoblotted with the indicated antibodies. (F) GIV, but not Gαi3, directly binds to the cytoplasmic domain of β1 integrin (β1CD). Binding of purified His-GIV-NT (1–256) or His-Gαi3 to GST-β1CD was determined in pulldown assays as described in Materials and methods. (G) GIV depletion decreases Gαi3 coimmunoprecipitation with β1 integrins upon collagen I stimulation. MDA-MB-231 scr shRNA or GIV shRNA2 cells were seeded on collagen I for 30 min and immunoprecipitated as described in E.
GIV binds to integrins upon collagen I stimulation and is required to recruit Gαi3 to active integrins
To investigate if GIV and integrins are part of the same molecular complex, we performed coimmunoprecipitations from MDA-MB-231 cells either acutely stimulated by collagen I at different times of adhesion or unstimulated cells (maintained in suspension). We found that GIV coimmunoprecipitates with β1 integrins only upon collagen I stimulation (Fig. 5 E). This association is transient and peaks 30 min after cell adhesion, which is in agreement with the association of GIV with nascent adhesions described above. Interestingly, we found that Gαi3, a substrate for GIV’s GEF activity, also coimmunoprecipitates with β1 integrins and that the kinetics of the interaction is the same as for GIV (Fig. 5 E). On the other hand, the cytoskeletal protein cortactin was undetectable in β1 integrin immunoprecipitations, indicating that GIV and Gαi3 interactions are specific.
To test if GIV and/or Gαi3 bind directly to integrins, we performed GST pulldown experiments with recombinant proteins purified from bacteria. We found that the purified N-terminal domain of GIV (aa 1–256) but not Gαi3 bound to the GST-fused cytoplasmic tail of β1 integrins (Fig. 5 F). Collectively with the coimmunoprecipitation results, these findings suggested that Gαi3 could associate indirectly with integrin complexes in cells via GIV. We found that this is the case because Gαi3 did not coimmunoprecipitate with β1 integrins in GIV-depleted MDA-MB-231 cells (Fig. 5 G). Collectively, these results indicate that GIV associates with active integrins via direct interaction and serves as a linker molecule to recruit Gαi3 to integrin-signaling complexes.
GIV’s GEF activity is required for enhancing integrin-dependent Akt activation and cell migration
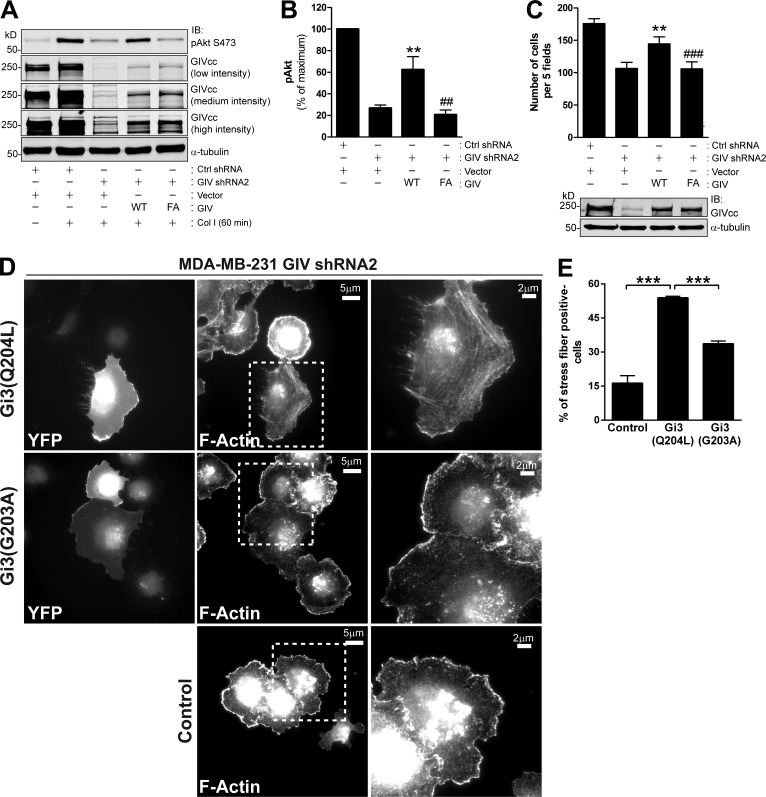
G protein activation by GIV has been previously reported to regulate tumor cell behavior (Garcia-Marcos et al., 2009, 2012; Ghosh et al., 2010). This and the fact that both GIV and Gαi3 are present in integrin-based complexes (Fig. 5) prompted us to investigate the importance of GIV’s GEF activity in regulating integrin-dependent responses. For this, we reintroduced GIV wild type (WT) or the GIV F1685A (FA) mutant into MDA-MB-231 cells depleted of endogenous GIV (i.e., GIV shRNA2). F1685A is a previously characterized mutant (Garcia-Marcos et al., 2009, 2012) that specifically disrupts GIV’s GEF activity. The introduction of GIV WT partially restored GIV’s expression and rescued defective Akt activation (Fig. 6, A and B) and cell migration (Fig. 6 C) observed in GIV-depleted cells in response to collagen I. Importantly, expression of the GIV FA mutant at levels identical to those of GIV WT did not rescue the defects observed in GIV-depleted cells (Fig. 6, A–C), demonstrating that GIV’s GEF activity is required for efficient Akt activation and cell migration in response to collagen I stimulation.
Figure 6.
GIV’s GEF activity is required for enhancing integrin-dependent Akt activation and cell migration. (A and B) Expression of GIV WT, but not the GEF-deficient GIV F1685A (FA) mutant, in MDA-MB-231 cells depleted of endogenous GIV rescues the impaired Akt activation response to collagen I. MDA-MB-231 control shRNA and GIV shRNA2 cells were transduced with lentiviral particles carrying an empty vector, GIV WT, or GIV FA as indicated in Materials and methods. Cells were seeded on collagen I for 60 min as described in Fig. 2 B and analyzed by immunoblotting. A representative result is shown in A, and the quantification of three independent results (results are depicted as mean ± SEM [error bars]; **, P < 0.01, compared with GIV-depleted cells; ##, P < 0.01, compared with cells expressing exogenous GIV WT) is shown in B. Equivalent results were obtained when Akt activation was measured upon collagen I stimulation for 30 min (not depicted). (C) Expression of GIV WT, but not the GEF-deficient GIV F1685A (FA) mutant, in MDA-MB-231 cells depleted of endogenous GIV rescues impaired haptotaxis. Cells were treated as described in A and B, except that they were used in haptotaxis assays (as described in Materials and methods). Results are depicted as mean ± SEM (error bars; n = 3; **, P < 0.05, compared with GIV-depleted cells; ###, P < 0.05, compared with cells expressing exogenous GIV WT). GIV immunoblot of a representative experiment is shown on the bottom. (D and E) Expression of constitutively active but not constitutively inactive Gi3 rescues actin cytoskeleton remodeling defects of GIV-depleted cells upon collagen I stimulation. MDA-MB-231 GIV shRNA2 cells were transfected with Gβ1-YC, Gγ2-YN, and Gαi3 mutants Q204L (active) or G203A (inactive), seeded on collagen I for 30 min as described in Fig. 2 B and analyzed by fluorescence microscopy for YFP expression or F-actin. The boxed areas of transfected cells (as determined by YFP expression) are shown enlarged on the right. A representative result is shown in D, and the quantification (n = 3; results are depicted as mean ± SEM (error bars); ***, P < 0.001) is shown in E.
To further validate that G protein activation by GIV is important for integrin-dependent cell responses, we investigated if Gi3 mutants that render the G protein constitutively active could rescue the cytoskeletal defects that occur upon GIV depletion (Fig. 3). For this, we cotransfected fluorescently tagged Gβ1γ2 with constitutively active (Q204L) or constitutively inactive (G203) Gαi3 mutants (Hermouet et al., 1991; Ghosh et al., 2008) in GIV-depleted MDA-MB-231 cells. Transfected cells were visualized by the YFP fluorescence of Gβ1γ2 dimers. We found that expression of constitutively active Gi3 (Q204L) rescued the defects in actin cytoskeleton remodeling observed in GIV-depleted MDA-MB-231 cells upon collagen I stimulation more efficiently than constitutively inactive Gi3 (G203A) (Fig. 6, D and E). These results indicate that G protein activation is sufficient to restore defective integrin-dependent cell responses upon GIV depletion, suggesting that other functions of GIV are not as critical as or are dependent on its GEF activity.
As an alternative approach to assess the importance of GIV’s GEF activity, we generated stable MCF-7 cells (which are naturally GIV-deficient; Ghosh et al., 2010; Garcia-Marcos et al., 2011b) expressing exogenous GIV WT and GIV FA (Fig. 7 A). The expression of GIV WT not only enhanced Akt activation upon collagen I stimulation (Fig. 7, B and C, as already shown in Fig. 2 E) but also significantly enhanced cell migration (Fig. 7 D). As in the case of MDA-MB-231 cells, none of these phenotypes was reproduced by expression of GIV FA, which supports the conclusion that GIV’s GEF activity promotes efficient integrin-mediated signaling and cell migration. Furthermore, constitutively active Gi3 (Q204L), but not constitutively inactive Gi3 (G203A), enhanced Akt activation in response to collagen I stimulation (Fig. 7 E), indicating that G protein activation phenocopies the effect of GIV expression in MCF-7 cells.
GEF-dependent enhancement of integrin signaling occurs via Gβγ-PI3K
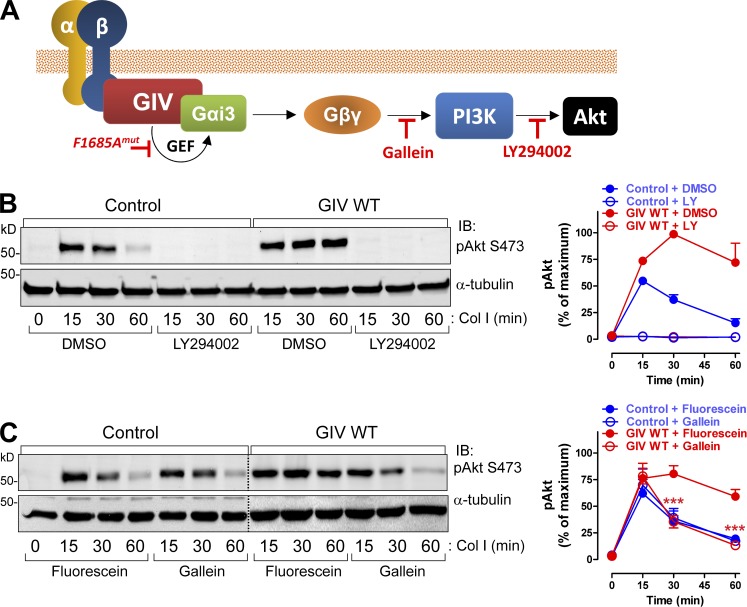
We took advantage of the integrin-signaling gain-of-function observed in MCF-7 cells upon GIV expression to further dissect the mechanism by which GIV’s GEF activity potentiates integrin responses. We hypothesized that Akt signaling enhancement by GIV would be achieved via Gβγ-dependent activation of PI3K (see model, Fig. 8 A). This is because GIV activates Gαi, and it is known that Gαi activation triggers the release of “free” Gβγ subunits (Smrcka, 2008), which in turn activate several effectors including different PI3K isoforms (Dbouk et al., 2012; Vadas et al., 2013). We found that pharmacological inhibition of PI3K with LY294002 blunted collagen I–stimulated Akt activation in both MCF-7 control and MCF-7 GIV WT cells (Fig. 8 B). This indicates that both the native response and the GIV-enhanced response occur via PI3K. On the other hand, gallein, a small molecule that blocks Gβγ binding to PI3K (Bonacci et al., 2006), did not alter the activation of Akt in response to collagen I stimulation in MCF-7 control cells (Fig. 8 C). In MCF-7 GIV WT cells, gallein reduced the Akt response to the same levels of activation observed in MCF-7 control cells (Fig. 8 C), indicating that the GIV-mediated enhancement occurs via Gβγ. Collectively, these results show that GIV promotes a GEF-dependent enhancement of Akt signaling in response to integrin stimulation, and that such enhancement proceeds via a G protein–signaling axis (Gβγ-PI3K) not used in the native response of MCF-7 cells.
Figure 8.
GIV’s GEF activity enhances integrin-dependent signaling via a Gβγ-PI3K-Akt axis. (A) Cartoon depicting a putative GIV-Gβγ-PI3K axis linking integrin stimulation to Akt activation and inhibitors used in subsequent experiments. (B) GIV-induced enhancement of Akt signaling requires PI3K. MCF-7 cells stably expressing vector control or GIV WT were preincubated with the PI3K inhibitor LY294002 or vehicle (DMSO) for 60 min and stimulated with collagen I as described in Fig. 2 B. A representative result is shown on the left, and the quantification (n = 3; results are depicted as mean ± SEM [error bars]) is shown on the right. Blue, MCF-7 control; red, MCF-7 GIV WT; filled circles, DMSO; open circles, LY294002. (C) GIV-induced enhancement of Akt signaling requires signaling via free Gβγ. MCF-7 cells stably expressing vector control or GIV WT were preincubated with the Gβγ inhibitor gallein or its inactive analogue fluorescein for 60 min and stimulated with collagen I as described in Fig. 2 B. A representative result is shown on the left, and the quantification (n = 3; results are depicted as mean ± SEM [error bars]; ***, P < 0.001, gallein vs. fluorescein in GIV WT cells) is shown on the right. Blue, MCF-7 control; red, MCF-7 GIV WT; filled circles, fluorescein; open circles, gallein.
Discussion
Summary and model
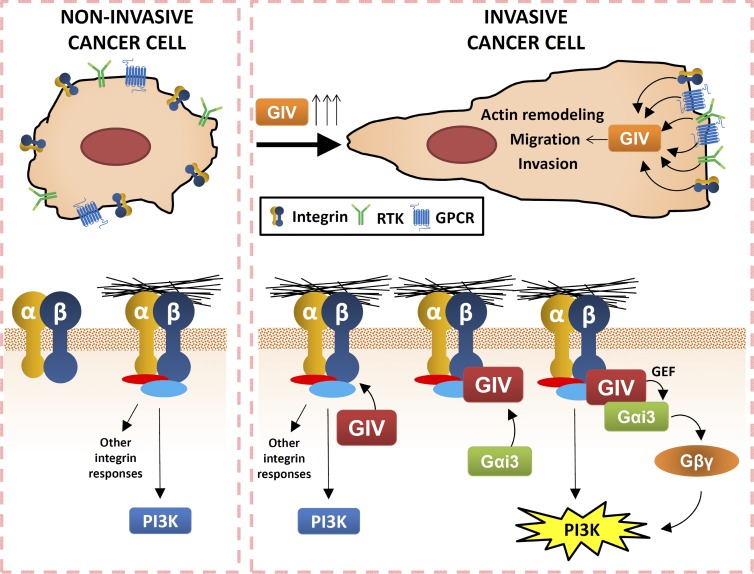
In this study, we describe a new molecular mechanism by which integrins promote the proinvasive behavior of tumor cells. Our results support a model (summarized in Fig. 9) in which integrin signaling is rewired in invasive cancer cells via activation of trimeric G proteins by the nonreceptor protein GIV. Previous work by us (Ghosh et al., 2008, 2010; Garcia-Marcos et al., 2011b) and others (Ling et al., 2011; Dunkel et al., 2012; Liu et al., 2012a; Shibata et al., 2013; Zhao et al., 2013; Song et al., 2014; Wang et al., 2014) has established that GIV expression is up-regulated in invasive tumor cell lines and metastatic tumors, and that GIV is required for metastasis in mouse models (Jiang et al., 2008). Here, we show that without GIV, invasive cancer cells display impaired actin remodeling, motility, and invasiveness in response to integrin stimulation, whereas expression of GIV in noninvasive cancer cells enhances integrin signaling and haptotaxis. Using a combination of genetic and pharmacological manipulations, we show that GIV potentiates integrin-dependent cell responses via activation of trimeric G protein signaling. We found that GIV binds directly to integrin cytoplasmic tails upon ECM stimulation and is required to recruit Gαi3 to active integrin complexes. Subsequently, GIV’s GEF activity triggers Gαi activation to transduce integrin signals into proinvasive responses via a Gβγ-PI3K axis. In summary, our findings establish a previously unappreciated link between integrin and trimeric G protein signaling that promotes proinvasive behavior in cancer cells.
Figure 9.
Proposed model for how GIV regulates integrin signaling in cancer cells. Invasive cancer cells express more GIV than noninvasive cancer cells or nontransformed cells (Ghosh et al., 2010; Garcia-Marcos et al., 2011b), which leads to amplification of signals from GPCRs, RTKs, and integrins (this work). This amplification of signals from integrins (as well as from the other receptors) promotes the remodeling of the actin cytoskeleton, cell motility, and invasion. Mechanistically, GIV binds directly to integrin cytoplasmic tails upon ECM stimulation and is required to recruit Gαi3 to active integrin complexes. Activation of Gαi by GIV triggers the release of free Gβγ, which in turn activates PI3K. This GIV–Gαi–Gβγ–PI3K axis works in parallel to previously described mechanisms of PI3K activation by integrins (Legate et al., 2009), resulting in the enhanced response observed in invasive cancer cells with up-regulated GIV.
Integrins trigger trimeric G protein signaling through a nonreceptor GEF
It is well documented that trimeric G protein signaling activated by GPCRs can alter the adhesive properties of integrins through “inside-out” signaling (Offermanns, 2006; Shen et al., 2012). However, the response of integrins to extracellular cues (“outside-in” signaling) has been traditionally considered to occur through a cascade of events independent from trimeric G proteins and GPCRs. Our findings reported here challenge this traditional view. Recent work by others also indicates that trimeric G protein signaling and integrin signaling intertwine. More specifically, two elegant studies from Du and colleagues (Gong et al., 2010; Shen et al., 2013) have provided evidence indicating that active Gα13 is required for integrin signaling during platelet aggregation. Although they showed that some integrin responses, like cell spreading, were controlled by Gα13 in the absence of direct GPCR stimulation (Gong et al., 2010), the mechanism by which the G protein became activated upon integrin stimulation was not elucidated. Here we propose that activation of trimeric G proteins in response to integrin stimulation can be achieved by a nonreceptor GEF like GIV. Questions that remain open are whether other nonreceptor GEFs can regulate integrin signaling in different contexts, and whether G proteins different from Gαi (the main substrate for GIV’s GEF activity; Garcia-Marcos et al., 2009) can also be regulated by nonreceptor GEFs upon integrin stimulation. For example, the nonreceptor GEF Ric-8 has been recently shown to regulate Concertina, the Drosophila homologue of Gα13, during gastrulation (Peters and Rogers, 2013), but a possible role in integrin signaling has not been explored yet.
GIV assembles an alternative signaling pathway that enhances integrin-dependent invasiveness
Our results not only indicate that GIV is required for enhancing integrin-dependent responses in invasive cells but also that it does so by assembling an alternative signaling cascade. Our combination of genetic and pharmacological manipulations in cells indicates that GIV’s GEF activity is necessary and sufficient to activate trimeric G proteins in response to integrin stimulation, which leads to an enhancement of a Gβγ-PI3K–dependent mechanism not used by noninvasive cells lacking GIV. This assembly of a parallel G protein–signaling pathway that potentiates integrin signaling results in a gain of invasive properties. It remains to be investigated if other integrin-signaling events modulated by GIV like RhoA/pMLC2 and Cdc42 activation are also dependent on its GEF activity. This is likely because Gβγ activates several Rho GEFs specific for RhoA and/or Cdc42 (Blomquist et al., 2000; Niu et al., 2003; Ueda et al., 2008; Wang et al., 2009).
From a molecular standpoint, our results show that GIV (a) can directly bind to the purified cytoplasmic tail of β1 integrins, (b) associates with integrins in cells only after ECM stimulation, and (c) is required for the efficient recruitment of Gαi3 to active integrins. Some of these results are consistent with results by others in the literature. For example, a recent proteomic study (Schiller et al., 2013) identified GIV (named “ccdc88a” in that paper) as a β3 integrin tail-binding protein. More recently, the Takahashi group has reported that GIV coimmunoprecipitates with β1 integrins in a different cell type, and that this occurs via direct binding of the N-terminal region of GIV to the cytoplasmic tail of integrins (Weng et al., 2014), same as we report here. The fact that Gαi3 did not show direct binding to β1 integrin tails in vitro but associated with active integrins in a GIV-dependent manner in cells suggests the following model: GIV would first associate with integrins via direct binding of its N-terminal region to the cytoplasmic domain of integrins, and it would subsequently recruit Gαi3 to integrin complexes via its G protein–binding domain, which is located in the C-terminal region. Because bacterially expressed GIV has GEF activity in vitro without additional modifications (Garcia-Marcos et al., 2010), it is possible that its recruitment to active integrins at the PM in close proximity to its substrate Gαi3 is sufficient to trigger activation.
From a spatiotemporal standpoint, these signaling events seem to occur at an early step of integrin signaling. First, our results indicate that GIV and Gαi3 associate with integrins only transiently at early time points after integrin stimulation. Moreover, GIV probably associates with integrins in nascent but not in mature focal adhesions because it colocalized poorly with markers like vinculin or paxillin. The association with nascent adhesions was further supported by colocalization with paxillin and integrins under conditions in which focal adhesion maturation is blocked. However, we cannot rule out that a pool of GIV associates with mature focal adhesions, but it cannot be efficiently detected by currently available antibodies.
Integrins have proven to be challenging therapeutic targets in cancer because of their roles in normal physiology and because of functional redundancy of different integrin types (Cox et al., 2010; Goodman and Picard, 2012). Current efforts are shifting toward targeting downstream events in integrin signaling (Cox et al., 2010). We propose that blocking GIV is an attractive candidate because of its disease-specific function in integrin signaling and because of the tractability of the molecular interface with its target Gαi (Garcia-Marcos et al., 2012). However, it will be important to address some issues not investigated in the current work, like the mechanistic details of how Gβγ-PI3K signaling is linked to the proinvasive behavior of cancer cells. Although it is tempting to speculate that it is mediated by PIP3-dependent mechanisms, other Gβγ effectors (Smrcka, 2008) or G protein–independent functions of GIV (Lin et al., 2011) may also contribute.
GIV is a convergence-signaling platform for both soluble ligands and immobilized ECM substrates
Our current work expands the range of extracellular cues and receptor types for which GIV serves as signal-transmission platform. Previous work has established that GIV functions downstream of receptors of different classes like GPCRs, RTKs, or TGFβR (Garcia-Marcos et al., 2015), which bind predominantly soluble ligands. It has been shown in many cases that signaling and cell migration triggered by soluble ligands acting on these receptors require trimeric G protein activation by GIV. Our current work expands the role of GIV and its GEF activity in signal transduction to integrins, another major class of surface receptors that responds to immobilized ECM ligands instead of soluble factors. GIV’s ability to enhance responses to both soluble ligands and ECM substrates in the same cell suggests that it works as a node for integrating different physicochemical inputs from the microenvironment. In this regard, an interesting possibility not yet addressed is whether GIV contributes to the synergism and cooperation observed between integrins and other receptors like RTKs (Plopper et al., 1995; Miyamoto et al., 1996; Sundberg and Rubin, 1996). In any case, the ability of GIV to work downstream of multiple receptors is a feature shared with other important signaling nodes in cancer, like for example Src, and it is likely to underlie its role in cancer progression toward metastasis, a process governed by complex cell–environment interactions.
Materials and methods
Reagents and antibodies
Unless otherwise indicated, all chemical reagents were obtained from Sigma-Aldrich or Thermo Fisher Scientific. Escherichia coli DH5α strain was purchased from New England Biolabs, Inc. Pfu ultra DNA polymerase was purchased from Agilent Technologies. Matrigel Growth Factor reduced, rat tail collagen I, human fibronectin, and mouse laminin were purchased from BD. Human vitronectin, LY294002, poly-l-Lysine (P-8920), PP2 (P-0042), and insulin (I9278) were obtained from Sigma-Aldrich. Gallein and fluorescein were purchased from TCI. (S)-(-)-blebbistatin was purchased from Santa Cruz Biotechnology, Inc. Pierce Protein G agarose beads were obtained from Thermo Fisher Scientific. Puromycin and blasticidin were purchased from Gold Biotechnology. Paraformaldehyde 16% was purchased from Alfa Aesar.
Mouse monoclonal antibodies raised against vinculin (V9264), poly-His tag (H1029), and α-tubulin (T6074) were obtained from Sigma-Aldrich. Mouse monoclonal antibody raised against integrin β1 (clone AIIB2) was obtained from the Developmental Studies Hybridoma Bank (The University of Iowa) created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health and maintained at The University of Iowa, Department of Biology. Mouse monoclonal antibody raised against fascin was obtained from Thermo Fisher Scientific. Mouse monoclonal antibodies raised against paxillin, cortactin, and Cdc42 were obtained from BD. Rabbit polyclonal antibody raised against phospho-myosin light chain 2 (S20; catalog no. 2480) was purchased from Abcam. Generation of rabbit serum (GIVcc) against the coiled-coil domain of rat GIV (aa 1174–1399) was described previously (Le-Niculescu et al., 2005). Rabbit polyclonal antibodies raised against an epitope in the C terminus of GIV (GIVct; T-13), Gαi3 (C-10), pan-Gβ (M-14), and mouse monoclonal antibodies raised against β1 integrins (P5D2) and total Akt (B-1) were purchased from Santa Cruz Biotechnology, Inc. Rabbit antibodies raised against β1 integrins (clone D2E5), phospho-Akt (S473; clone D9E), RhoA (clone 67B9), and mouse antibodies for Myc-tag (9B11) were obtained from Cell Signaling Technology. Goat anti–rabbit and goat anti–mouse Alexa Fluor 680 or IRDye 800 F(ab′)2 used as secondary antibodies for immunoblotting were from Invitrogen and Li-Cor Biosciences, respectively. DAPI, goat anti–rabbit Alexa Fluor 594–coupled antibody, and Alexa Fluor 594–coupled phalloidin (catalog no. A12381) were purchased from Invitrogen. Goat anti–mouse Alexa Fluor 488–coupled antibody was from The Jackson Laboratory.
Plasmids
shRNA sequences were designed using two previously validated GIV-targeting sequences (GIV shRNA1: GAAGGAGAGGCAACTGGAT [Kitamura et al., 2008]; GIV shRNA2: AAGAAGGCTTAGGCAGGAATT [Enomoto et al., 2005; Ghosh et al., 2008]) or a nontargeting sequence (control shRNA: GGATTGAGATCAGAAGATAGC, corresponding to GIV shRNA2 scrambled), and inserted into the AgeI/EcoRI sites of pLKO.1-puro (Addgene). Full-length human GIV with a 3xFLAG tag and a silent mutation to confer resistance to the GIV shRNA2 sequence was amplified from a previously described construct (Garcia-Marcos et al., 2012) and inserted into the XhoI/NotI sites of a pLVX-puro plasmid with an expanded multicloning site. The 3xFLAG was excised by digestion with BamHI/NotI and replaced by a 2xMyc tag, and an additional silent mutation to confer resistance to GIV shRNA1 was inserted by mutagenesis. The entire RNAi-resistant GIV-2xMyc cassette was cut with SpeI/XbaI and ligated into the SalI/XbaI sites of pLENTI-CMV-Blast (Addgene). Point mutations were introduced using the Quikchange II kit (Agilent Technologies), and all constructs were verified by DNA sequencing. The N-terminal domain (aa 1–256) of GIV was cloned into the NcoI/NdeI sites of pET28b plasmid to encode for His-tagged GIV-NT (1–256). Cloning of rat Gαi3 WT, Q204L, and G203A in the mammalian expression plasmid pcDNA3 was described previously (Ghosh et al., 2008). Bacterial expression plasmid pGEX4T-1-β1 integrin cytoplasmic domain (β1CD) was provided by X. Du (University of Illinois at Chicago, Chicago, IL). Bacterial expression plasmids pGEX4T-1-PBD and pGEX4T-1-RBD were provided by L. Dubrez (Institut National de la Santé et de la Recherche Médicale UMR866, Dijon, France). Mammalian expression plasmids pcDNA3.1-Venus 155–239 Gβ1 (Gβ1-YC) and pcDNA3.1-Venus 1–155 Gγ2 (Gγ2-YN; Hollins et al., 2009) were provided by N. Lambert (Georgia Regents University, Augusta, GA).
Cell culture, transfections, and generation of stable cells lines
MDA-MB-231, MCF-7, COLO357-FG ( provided by M. Bouvet, University of California, San Diego, La Jolla, CA), HeLa, and NIH3T3-Src* (provided by S. Courtneidge, Sanford-Burnham Institute, La Jolla, CA) cells were grown in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 1% l-glutamine (37°C, 5% CO2). Stable cell lines were generated by lentiviral transduction followed by selection with the appropriate antibiotics. Lentivirus packaging was performed in HEK-293FT cells (R700-07; Invitrogen) by cotransfection of the lentiviral plasmid of interest with the packaging plasmid pSPAX2 and a VSV-G encoding plasmid at a 1:1:0.5 ratio. Approximately 6 h after transfection the media was changed, and ∼42 h later, lentivirus-containing media were collected, centrifuged at 500 g for 3 min, and filtered through 0.45 µm. In some cases (i.e., GIV-containing plasmids), the lentivirus-containing media was concentrated approximately fivefold with a Pierce Concentrator (9K MWCO/7 mL; Thermo Fisher Scientific) to increase the virus titer.
MDA-MB-231 cell lines stably expressing GIV shRNA sequences were generated by incubation with supernatants of pLKO.1-puro–packaged lentivirus (mixed 1:1 with fresh media) for 2 d, followed by selection with 1 µg/ml puromycin. All surviving clones were pooled and maintained in the presence of 1 µg/ml puromycin. MDA-MB-231 cell lines stably expressing GIV shRNA2 or control shRNA sequences were used as the starting point for the reconstitution experiments with RNAi-resistant GIV. In brief, pLENTI-GIV plasmids bearing silent mutations to confer resistance to the GIV shRNA sequence expressed in these cells were used to generate lentiviral particles as described above, except that concentrated supernatants were used and that selection was performed in the presence of 7.5 µg/ml blasticidin. Because from initial experiments we noticed that GIV expression in these cell lines decayed rapidly after selection, we performed all of our experiments in freshly transduced cells soon after selection. In brief, 2 d after incubation with lentiviral supernatants, cells were transferred to p6 dishes, and 6 h later selection was started with 7.5 µg/ml blasticidin (in addition to 1 µg/ml puromycin). Once they reached confluency (∼2 d), they were transferred to p10 dishes and used for experiments immediately right after reaching confluency again (∼2 d). MCF-7 cell lines stably expressing GIV-coding plasmids were generated similarly except that concentrated pLVX-puro–packaged lentiviruses were used. MCF-7 cell lines stably transduced with these plasmids were incubated overnight in the presence of 10 mM sodium butyrate before all experiments. COLO357-FG and HeLa cells stably depleted of GIV were generated exactly as described for MDA-MB-231 cells.
Cell transfection was performed using Lipofectamine LTX Reagent with PLUS Reagent (Invitrogen) for DNA plasmids according to the manufacturer’s protocols. In brief, 650,000 MDA-MB-231 cells were reverse transfected in a 6-well plate with pcDNA3.1-Gαi3/pcDNA3.1-Gβ1-YC/pcDNA3.1-Gγ2-YN (1:0.75:0.75 µg). 750,000 MCF-7 cells were plated overnight in p6 dishes before transfection of pcDNA3.1-Gαi3/pcDNA3.1-Gβ1-YC/pcDNA3.1-Gγ2-YN (1:0.75:0.75 µg). For both cell lines, media was changed 6 h after transfection, and the cells were used for experiments 24–48 h after transfection.
3D Matrigel culture
This assay was performed exactly as described previously (Debnath et al., 2003). In brief, 40 µl of ice-cold Matrigel (growth factor reduced) was spread as a thick layer on the bottom of Lab-Tek II 8-chamber slides (Thermo Fisher Scientific) and allowed to solidify at 37°C. Approximately 5 × 103 MDA-MB-231 or COLO375-FG cells were seeded on the Matrigel-coated chambers containing complete medium supplemented with 2% Matrigel. Media were changed every other day, and cells were analyzed 7 d after seeding by phase-contrast microscopy with a fluorescence microscope (10× objective; Axio Observer Z1; Carl Zeiss) equipped with a camera (C10600/Orca-R2; Hamamatsu Photonics).The size of 200 acini (in µm2) was quantified in each independent experiment using a measuring tool of Axiovision 4.8.1 software (Carl Zeiss). All individual images were processed using ImageJ software and assembled for presentation using Photoshop and Illustrator software (both Adobe).
Cell adhesion assay
This assay was performed as described previously (Liu et al., 2012b), with minor modifications. In brief, the bottom of 24-well plates (Corning) was coated overnight at 4°C with BSA, collagen I, fibronectin, vitronectin, laminin, or Matrigel (1 µg/cm2). Plates were then washed twice with PBS and incubated with PBS supplemented with 2% BSA at 37°C for 1 h to block nonspecific adhesion sites. MDA-MB-231 control shRNA or GIV shRNA2 cell lines were washed with PBS and detached by incubation in PBS supplemented with 20 mM EDTA, pH 7.4, at 37°C (∼6 min) and dispersed as single cells. MDA-MB-231 cells were washed three times in serum-free DMEM (450 g, 2 min) and resuspended in the same medium at a concentration of 3 × 105 cells/ml. 1.25 × 105 cells/well in serum-free DMEM were seeded in triplicates and incubated for 1 h at 37°C. For some experiments (Fig. S2), MCF-7 cells were treated as described above, except that they were incubated with the β1 integrin–blocking antibody AIIB2 (2, 4, and 8 µg/ml) or control IgG (8 µg/ml) for 15 min at 37°C with constant tumbling before seeding. Medium was aspirated and unattached cells were removed by washing three times with PBS. Attached cells were fixed with 3% paraformaldehyde for 30 min and stained with 0.5% crystal violet for 1 h. The number of cells in five randomly chosen fields was counted in each triplicate and averaged for each independent experiment.
Cell stimulation by ECM attachment and other treatments
The protocol for cell stimulation by ECM attachment was performed as described previously (Leyme et al., 2012), with modifications. Tissue culture dishes (p6) and/or glass coverslips were coated with collagen I dissolved in 0.02N acetic acid (1.6 µg/cm2) or Matrigel dissolved in serum-free DMEM (1 µg/cm2) overnight at 4°C and washed twice with PBS right before the experiments. In some cases, plates were coated with poly-l-lysine by incubation for 5 min at room temperature (5 µg/cm2), washed twice with PBS, and dried for 15 min right before use. Cells at 90% confluency were washed with PBS and detached by incubation in PBS supplemented with 20 mM EDTA, pH 7.4, for ∼6 min at 37°C. Cells were washed three times with 10 ml of serum-free DMEM by cycles of centrifugation (450 g, 2 min) and resuspension. Washed cells at a final concentration of 3 × 105 cells/ml were maintained in suspension in serum-free media for 1 h at room temperature, and subsequently seeded on ECM-coated plastic dishes and/or glass coverslips (6 × 105 and 1.8 × 106 cells for p6 and p10 dishes, respectively). Stimulations were stopped at different time points (typically 15, 30, and 60 min) by washing with cold PBS and either freezing the plastic dishes at −20°C (for subsequent immunoblot analyses) or fixing the coverslips in 3% PFA for 30 min (for subsequent fluorescence microscopy analyses). The time 0 min in the immunoblot analyses corresponds to an aliquot of the cells in suspension, which was pelleted and frozen at −20°C right before seeding. For some experiments, 10 µM LY294002, 10 µM gallein, 10 µM fluorescein, 10 µM PP2, or 50 µM blebbistatin (or the equivalent volume of DMSO) was added to the media during the 1-h incubation of the cells in suspension and maintained during the time course of attachment to collagen I. For experiments with integrin-blocking antibodies, the β1 integrin–blocking antibody P5D2 (10 µg/ml) was added to cells in suspension for 15 min at 37°C with constant tumbling before seeding, and the same concentration of antibody was maintained in the media during the adhesion to collagen I. For experiments with insulin stimulation, cells were cultured in DMEM plus 0.5% FBS overnight before stimulation with 100 nM insulin for 10 min. Cell stimulation was stopped by washing with ice-cold PBS followed by harvesting in lysis buffer.
Cell lysis and quantitative immunoblotting
Cells frozen at −20°C on coated tissue culture dishes (as described above) were harvested on ice with lysis buffer (20 mM Hepes, pH 7.2, 5 mM Mg(CH3COO)2, 125 mM K(CH3COO), 0.4% [vol:vol] Triton X-100, 1 mM DTT, 10 mM β-glycerophosphate, and 0.5 mM Na3VO4 supplemented with a protease inhibitor cocktail [Sigma-Aldrich]) and cleared (14,000 g, 10 min) before use. Proteins were quantified by Bradford (Bio-Rad Laboratories), and samples were boiled in Laemmli sample buffer for 5 min. Proteins were separated by SDS-PAGE and transferred to PVDF membranes (EMD Millipore). Membranes were blocked with PBS supplemented with 5% nonfat milk or BSA (for antibodies against phosphorylated proteins) before sequential incubation with primary and secondary antibodies. Infrared imaging and quantification of Western blots were performed according to the manufacturer’s protocols using an Odyssey Infrared Imaging System (Li-Cor Biosciences). Akt activation was determined by calculating the phospho-Akt (pAkt)/α-tubulin ratio and normalizing it to the maximum activation in each experiment (percentage of maximum). Primary antibodies were diluted as follows: GIVcc (rabbit serum), 1:1,000; GIVct, 1:500; Gαi3, 1:250; panGβ, 1:250; pAkt (S473), 1:1,000; total Akt, 1:250; β1 integrins (clone D2E5), 1:250; Myc, 1:1,000; cortactin, 1:1,000; Rac1, 1:250; RhoA, 1:250; pMLC2 (S20), 1:250; α-tubulin, 1:2,500. All Odyssey images were processed using ImageJ software and assembled for presentation using Photoshop and Illustrator software (Adobe).
Immunofluorescence microscopy
This assay was performed as described previously (Garcia-Marcos et al., 2011a). MDA-MB-231 cells were seeded on coverslips coated with collagen I or poly-l-lysine for different times (typically 30 and 60 min). Cells were fixed with 3% paraformaldehyde for 30 min, permeabilized and blocked in PBS containing 10% normal goat serum and 0.1% Triton X-100 for 30 min, and then sequentially incubated with primary and secondary antibodies for 1 h at room temperature. The mounting media used was ProLong Diamond Antifade reagent (Life Technologies). Antibody dilutions were as follows: GIVct, 1:50; β1 integrin (P5D2), 1:200; vinculin, 1:200; paxillin, 1:50; secondary antibodies, 1:300. The same procedure was followed for fascin (1:50) and cortactin (1:50) staining, except that cells were fixed with absolute methanol at −20°C for 10 min. F-actin was visualized with Alexa Fluor 594–conjugated phalloidin and DNA stained with DAPI.
For the phenotypic analysis of MDA-MB-231 cells (i.e., assessment of stress fiber, filopodia-like structure and focal adhesion formation, cell spreading, and GIV recruitment to the PM), images were acquired at room temperature with a fluorescence microscope (Axio Observer Z1; Carl Zeiss) equipped with a digital camera (C10600/ORCA-R2; Hamamatsu Photonics) and AxioVision 4.8.1 software (Carl Zeiss) using a 63× oil-immersion objective (NA 1.4). At least 100 cells were analyzed in each individual experiment for the quantitative assessment of phenotypes. Cells showing prominent actin filaments across cell bodies were considered “stress fiber positive.” For the experiments investigating the rescue of stress-fiber defects in GIV-depleted cells by Gi3 mutants, only YFP-positive cells were counted. Cells displaying more than 20 vinculin-stained structures (presumably focal adhesions) or more than 20 filopodia-like structures were considered “vinculin positive” or “filopodia-like structure positive” cells, respectively. Cell spreading was assessed by counting the number of cells presenting a flat and extended morphology (as opposed to rounded cells) during the attachment to the substrate.
For colocalization studies, images were acquired at room temperature with a laser scanning confocal microscope (Axiovert 200 LSM510; Carl Zeiss) equipped with a laser diode (405 nm), argon (488), and 2 HeNe (543 and 633) lasers and controlled by its proprietary software (Carl Zeiss). Images were taken using a 63× oil-immersion objective (NA 1.4). Regions of interest were drawn around the PM region of 8–10 cells per experiment using ImageJ, and the Pearson’s correlation coefficient was determined to assess the extent of colocalization. All individual images were assembled for presentation using Photoshop and Illustrator software (both Adobe).
NIH3T3-Scr* cells were seeded on glass coverslips and cultured for 24 h in complete media before fixation and imaging as described above.
Rac1, Cdc42, and RhoA activation assay
This assay was performed as described previously (Benard and Bokoch, 2002). In brief, MDA-MB-231 cells were seeded on collagen I as described in Cell stimulation by ECM attachment and other treatments. Stimulations were stopped at different time points (15, 30, and 60 min) by washing with ice-cold PBS before lysis (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 5 mM MgCl2, 1% [wt:vol] NP-40, and 5% [vol:vol] glycerol supplemented with a protease inhibitor cocktail [Sigma-Aldrich]). After ∼5 min in ice, cell lysates were clarified by centrifugation (10,000 g, 5 min, 4°C) and quickly frozen in liquid nitrogen. Approximately 20 µg GST-PBD or GST-RBD was immobilized on 20 µl of glutathione agarose beads by incubation of bacterial lysates expressing the GST-fused proteins for 1 h at 4°C with rotation. Approximately 500 µg of total protein from the MDA-MB-231 cell lysates was incubated with immobilized GST-PBD or GST-RBD for 45 min at 4°C with constant tumbling. Beads were washed four times with wash buffer (25 mM Tris-HCl, pH 7.5, 40 mM NaCl, 30 mM MgCl2, and 1 mM DTT supplemented with a protease inhibitor cocktail [Sigma-Aldrich]), and resin-bound proteins were eluted by boiling in Laemmli sample buffer, separated by SDS-PAGE and immunoblotted with Rac1, Cdc42, and RhoA antibodies.
Haptotaxis and invasion assays
Haptotaxis experiments were performed using a modified Boyden chamber assay as described previously (Thibault et al., 2007; Abair et al., 2008; Liu et al., 2012b). In brief, the bottom side of the membrane filters (8-µm pores, 24-well format; Greiner Bio-One) was coated with 1.5 µg/cm2 collagen I at 4°C overnight, washed twice in PBS, and then blocked with BSA 2% for 2 h at 37°C. Cells were prepared exactly as described in Cell stimulation by ECM attachment and other treatments. The upper chamber was filled with serum-free media containing 5 × 104 MDA-MB-231 cells or 105 MCF-7 cells (300 µl total), whereas the lower chamber was filled with 500 µl of serum-free medium. MDA-MB-231 cells were incubated for 1.5, 2.5, and 5 h, and MCF-7 cells were incubated for 16 and 24 h at 37°C. For some experiments (Fig. S2), MDA-MB-231 cells were treated as described above, except that they were incubated with the integrin-blocking antibody AIIB2 (10 µg/ml) for 15 min at 37°C before seeding. For chemotaxis cell migration assays (Fig. S4), MDA-MB-231 cells were treated as described above, except that the filters were not coated with collagen I and the lower chamber was filled with 500 µl DMEM plus 10% FBS or DMEM plus 100 nM insulin.
Filters were fixed in 3% paraformaldehyde for 30 min at room temperature, stained with crystal violet for 1 h, and washed three times in PBS. Cells on the upper side of the filters were removed with cotton-tipped swabs, and cells on the bottom side of the filters were counted as follows. Images were acquired with a fluorescence microscope (Axio Observer Z1; Carl Zeiss) using a 20 or 40× objective for MCF-7 or MDA-MB-231 cells, respectively. The microscope was equipped with a digital camera (C10600/ORCA-R2; Hamamatsu Photonics) and AxioVision 4.8.1 software (Carl Zeiss). Contrast-phase pictures were captured in five different fields of each insert, and cells were counted. The results are presented as the average number of migrating cells in two replicates per experiment in three independent experiments.
Invasion experiments were performed exactly as described above, except that the upper side of the filters was coated with a layer of Matrigel (BD) and that invading cells were counted after 24 and 48 h. No significant migration or invasion was observed in controls in which the bottom side of the filters was not coated with collagen I but blocked with 2% BSA (not depicted).
Immunoprecipitation
Protein G agarose beads (Thermo Fisher Scientific) were blocked with 5% BSA for 2 h at room temperature, washed, and incubated with either 4 µg of the β1-integrin mouse antibody AIIB2 or mouse IgG control (Santa Cruz Biotechnology, Inc.) for 90 min at 4°C with rotation. Beads were then washed four times with IP buffer (10 mM Pipes, pH 6.8, 300 mM sucrose, 50 mM NaCl, 3 mM MgCl2, 0.5% Triton X-100, and 0.5 mM Na3VO4 supplemented with a protease inhibitor cocktail [Sigma-Aldrich]). MDA-MB-231 cells were prepared essentially as described above in Cell stimulation by ECM attachment and other treatments. MDA-MB-231 cells were maintained in suspension or plated for different time points (30, 60, and 90 min) on tissue culture dishes coated with collagen I. Cell lysates (∼1–2 mg) prepared in IP buffer were incubated with IgG-coupled protein G–agarose beads for 4 h at 4°C. After four washes with IP buffer, immunoprecipitates were boiled in Laemmli sample buffer, and proteins were separated by SDS-PAGE and immunoblotted with GIVcc, cortactin, integrin β1 (D2E5), and Gαi3 antibodies.
Protein purification
GST, GST-fused β1CD, rat His-Gαi3 WT, and His-GIV-NT were purified as described previously by Garcia-Marcos et al. (2010). Plasmids were transformed in E. coli strain BL21(DE3), and protein expression was induced overnight at 23°C with 1 mM 1-isopropyl-β-d-thiogalactopyranoside. Pelleted bacteria from 1 liter of culture were resuspended in 10 ml of GST-lysis buffer (25 mm Tris-HCl, pH 7.5, 20 mM NaCl, 1 mM EDTA, 20% [vol/vol] glycerol, and 1% [vol/vol] Triton X-100 supplemented with protease inhibitor cocktail [1 µM leupeptin, 2.5 µM pestatin, 0.2 µM aprotinin, and 1 mM PMSF]) or His-lysis buffer (50 mM NaH2PO4, pH 7.4, 300 mM NaCl, 10 mM imidazole, and 1% [vol/vol] Triton X-100 supplemented with protease inhibitor cocktail [1 µM leupeptin, 2.5 µM pestatin, 0.2 µM aprotinin, and 1 mM PMSF]) for GST- or His-fused proteins, respectively. After sonication (4 × 20 s, 1 min between cycles), lysates were centrifuged at 12,000 g at 4°C for 20 min. Solubilized proteins were affinity purified on glutathione-agarose beads or HisPur Cobalt Resin (Thermo Fisher Scientific). Proteins were eluted, dialyzed overnight against phosphate-buffered saline, and stored at −80°C. His-Gαi3 was buffer exchanged into G protein storage buffer (20 mM Tris-HCl, pH 7.4, 20 mM NaCl, 1 mM MgCl2, 1 mM DTT, 10 µM GDP, and 5% [vol/vol] glycerol) before storage at −80°C.
In vitro protein-binding assay
20–25 µg GST or GST-β1CD was immobilized on glutathione agarose beads for 1 h at room temperature in PBS. Beads were resuspended in binding buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.4% [vol:vol] NP-40, 10 mM MgCl2, 5 mM EDTA, 2 mM DTT, and 30 µM GDP) and incubated overnight at 4°C with constant tumbling in the presence of 10 µg His-GIV-NT or His-Gαi3. Beads were washed (four times) with 1 ml wash buffer (4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 0.1% [vol/vol] Tween 20, 10 mm MgCl2, 5 mm EDTA, 1 mm DTT, and 30 µM GDP), proteins were eluted by boiling in Laemmli sample buffer, and proteins were separated by SDS-PAGE and immunoblotted with anti-His antibodies.
Statistical analysis
Each experiment was performed at least three times. Data shown are expressed as mean ± SEM or as one representative experiment. Statistical significance between various conditions was assessed with Student’s t test. P < 0.05 was considered significant.
Online supplemental material
Fig. S1 shows that GIV depletion impairs the growth of COLO357-FG cells on Matrigel but not on plastic. Fig. S2 shows controls validating the integrin dependency of different responses of MDA-MB-231 and MCF-7 cells. Fig. S3 shows that GIV depletion impairs Akt activation in response to collagen stimulation in COLO357-FG and HeLa cells. Fig. S4 shows that GIV depletion impairs MDA-MB-231 cell migration and Akt activation in response to soluble factors. Fig. S5 shows the localization GIV in different integrin-based subcellular structures. The online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506041/DC1.
Supplementary Material
Acknowledgments
We thank K. Kandror, B. Ritter, and V. Trinkaus-Randall (BU) for access to microscopes and Laura Macias and Miriam Lopez-Hinojosa for their technical help.
This work was supported by National Institutes of Health (grants GM108733 and GM112631), American Cancer Society (grant RSG-13-362-01-TBE), and the Elsa U. Pardee Foundation (to M. Garcia-Marcos).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- GEF
- guanine nucleotide exchange factor
- GPCR
- G protein–coupled receptor
- PI3K
- phosphatidylinositol-3-kinase
- PM
- plasma membrane
- RTK
- receptor tyrosine kinase
- WT
- wild type
References
- Abair T.D., Sundaramoorthy M., Chen D., Heino J., Ivaska J., Hudson B.G., Sanders C.R., Pozzi A., and Zent R.. 2008. Cross-talk between integrins alpha1beta1 and alpha2beta1 in renal epithelial cells. Exp. Cell Res. 314:3593–3604. 10.1016/j.yexcr.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abram C.L., and Lowell C.A.. 2009. The ins and outs of leukocyte integrin signaling. Annu. Rev. Immunol. 27:339–362. 10.1146/annurev.immunol.021908.132554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiges-Rizo C., Destaing O., Fourcade B., Planus E., and Block M.R.. 2009. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J. Cell Sci. 122:3037–3049. 10.1242/jcs.052704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anai M., Shojima N., Katagiri H., Ogihara T., Sakoda H., Onishi Y., Ono H., Fujishiro M., Fukushima Y., Horike N., et al. . 2005. A novel protein kinase B (PKB)/AKT-binding protein enhances PKB kinase activity and regulates DNA synthesis. J. Biol. Chem. 280:18525–18535. 10.1074/jbc.M500586200 [DOI] [PubMed] [Google Scholar]
- Benard V., and Bokoch G.M.. 2002. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 345:349–359. 10.1016/S0076-6879(02)45028-8 [DOI] [PubMed] [Google Scholar]
- Bhadriraju K., Yang M., Alom Ruiz S., Pirone D., Tan J., and Chen C.S.. 2007. Activation of ROCK by RhoA is regulated by cell adhesion, shape, and cytoskeletal tension. Exp. Cell Res. 313:3616–3623. 10.1016/j.yexcr.2007.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell M.J., and Radisky D.. 2001. Putting tumours in context. Nat. Rev. Cancer. 1:46–54. 10.1038/35094059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomquist A., Schwörer G., Schablowski H., Psoma A., Lehnen M., Jakobs K.H., and Rümenapp U.. 2000. Identification and characterization of a novel Rho-specific guanine nucleotide exchange factor. Biochem. J. 352:319–325. 10.1042/bj3520319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacci T.M., Mathews J.L., Yuan C., Lehmann D.M., Malik S., Wu D., Font J.L., Bidlack J.M., and Smrcka A.V.. 2006. Differential targeting of Gbetagamma-subunit signaling with small molecules. Science. 312:443–446. 10.1126/science.1120378 [DOI] [PubMed] [Google Scholar]
- Cismowski M.J., Takesono A., Ma C., Lizano J.S., Xie X., Fuernkranz H., Lanier S.M., and Duzic E.. 1999. Genetic screens in yeast to identify mammalian nonreceptor modulators of G-protein signaling. Nat. Biotechnol. 17:878–883. 10.1038/12867 [DOI] [PubMed] [Google Scholar]
- Cismowski M.J., Ma C., Ribas C., Xie X., Spruyt M., Lizano J.S., Lanier S.M., and Duzic E.. 2000. Activation of heterotrimeric G-protein signaling by a ras-related protein. Implications for signal integration. J. Biol. Chem. 275:23421–23424. 10.1074/jbc.C000322200 [DOI] [PubMed] [Google Scholar]
- Cox D., Brennan M., and Moran N.. 2010. Integrins as therapeutic targets: lessons and opportunities. Nat. Rev. Drug Discov. 9:804–820. 10.1038/nrd3266 [DOI] [PubMed] [Google Scholar]
- Danen E.H., Sonneveld P., Brakebusch C., Fassler R., and Sonnenberg A.. 2002. The fibronectin-binding integrins alpha5beta1 and alphavbeta3 differentially modulate RhoA-GTP loading, organization of cell matrix adhesions, and fibronectin fibrillogenesis. J. Cell Biol. 159:1071–1086. 10.1083/jcb.200205014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dbouk H.A., Vadas O., Shymanets A., Burke J.E., Salamon R.S., Khalil B.D., Barrett M.O., Waldo G.L., Surve C., Hsueh C., et al. . 2012. G protein-coupled receptor-mediated activation of p110β by Gβγ is required for cellular transformation and invasiveness. Sci. Signal. 5:ra89 10.1126/scisignal.2003264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J., and Brugge J.S.. 2005. Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer. 5:675–688. 10.1038/nrc1695 [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., and Brugge J.S.. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 30:256–268. 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- Desgrosellier J.S., and Cheresh D.A.. 2010. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 10:9–22. 10.1038/nrc2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkel Y., Ong A., Notani D., Mittal Y., Lam M., Mi X., and Ghosh P.. 2012. STAT3 protein up-regulates Gα-interacting vesicle-associated protein (GIV)/Girdin expression, and GIV enhances STAT3 activation in a positive feedback loop during wound healing and tumor invasion/metastasis. J. Biol. Chem. 287:41667–41683. 10.1074/jbc.M112.390781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A., Murakami H., Asai N., Morone N., Watanabe T., Kawai K., Murakumo Y., Usukura J., Kaibuchi K., and Takahashi M.. 2005. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev. Cell. 9:389–402. 10.1016/j.devcel.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ghosh P., and Farquhar M.G.. 2009. GIV is a nonreceptor GEF for Gαi with a unique motif that regulates Akt signaling. Proc. Natl. Acad. Sci. USA. 106:3178–3183. 10.1073/pnas.0900294106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ghosh P., Ear J., and Farquhar M.G.. 2010. A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration. J. Biol. Chem. 285:12765–12777. 10.1074/jbc.M109.045161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ear J., Farquhar M.G., and Ghosh P.. 2011a A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol. Biol. Cell. 22:673–686. 10.1091/mbc.E10-08-0738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M., Jung B.H., Ear J., Cabrera B., Carethers J.M., and Ghosh P.. 2011b Expression of GIV/Girdin, a metastasis-related protein, predicts patient survival in colon cancer. FASEB J. 25:590–599. 10.1096/fj.10-167304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M., Kietrsunthorn P.S., Pavlova Y., Adia M.A., Ghosh P., and Farquhar M.G.. 2012. Functional characterization of the guanine nucleotide exchange factor (GEF) motif of GIV protein reveals a threshold effect in signaling. Proc. Natl. Acad. Sci. USA. 109:1961–1966. 10.1073/pnas.1120538109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ghosh P., and Farquhar M.G.. 2015. GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. J. Biol. Chem. 290:6697–6704. 10.1074/jbc.R114.613414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Garcia-Marcos M., Bornheimer S.J., and Farquhar M.G.. 2008. Activation of Galphai3 triggers cell migration via regulation of GIV. J. Cell Biol. 182:381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Beas A.O., Bornheimer S.J., Garcia-Marcos M., Forry E.P., Johannson C., Ear J., Jung B.H., Cabrera B., Carethers J.M., and Farquhar M.G.. 2010. A Galphai-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Mol. Biol. Cell. 21:2338–2354. 10.1091/mbc.E10-01-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman A.G. 1987. G proteins: Transducers of receptor-generated signals. Annu. Rev. Biochem. 56:615–649. 10.1146/annurev.bi.56.070187.003151 [DOI] [PubMed] [Google Scholar]
- Gong H., Shen B., Flevaris P., Chow C., Lam S.C., Voyno-Yasenetskaya T.A., Kozasa T., and Du X.. 2010. G protein subunit Galpha13 binds to integrin alphaIIbbeta3 and mediates integrin “outside-in” signaling. Science. 327:340–343. 10.1126/science.1174779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S.L., and Picard M.. 2012. Integrins as therapeutic targets. Trends Pharmacol. Sci. 33:405–412. 10.1016/j.tips.2012.04.002 [DOI] [PubMed] [Google Scholar]
- Guo W., and Giancotti F.G.. 2004. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 5:816–826. 10.1038/nrm1490 [DOI] [PubMed] [Google Scholar]
- Hermouet S., Merendino J.J. Jr., Gutkind J.S., and Spiegel A.M.. 1991. Activating and inactivating mutations of the alpha subunit of Gi2 protein have opposite effects on proliferation of NIH 3T3 cells. Proc. Natl. Acad. Sci. USA. 88:10455–10459. 10.1073/pnas.88.23.10455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollins B., Kuravi S., Digby G.J., and Lambert N.A.. 2009. The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell. Signal. 21:1015–1021. 10.1016/j.cellsig.2009.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher A., and Horwitz A.R.. 2011. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 3:a005074 10.1101/cshperspect.a005074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: Bidirectional, allosteric signaling machines. Cell. 110:673–687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Jiang P., Enomoto A., Jijiwa M., Kato T., Hasegawa T., Ishida M., Sato T., Asai N., Murakumo Y., and Takahashi M.. 2008. An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer Res. 68:1310–1318. 10.1158/0008-5472.CAN-07-5111 [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Duan X., Liu C.Y., Jang M.H., Guo J.U., Pow-anpongkul N., Kang E., Song H., and Ming G.L.. 2009. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 63:761–773. 10.1016/j.neuron.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T., Asai N., Enomoto A., Maeda K., Kato T., Ishida M., Jiang P., Watanabe T., Usukura J., Kondo T., et al. . 2008. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nat. Cell Biol. 10:329–337. 10.1038/ncb1695 [DOI] [PubMed] [Google Scholar]
- Kuo J.C., Han X., Hsiao C.T., Yates J.R. III, and Waterman C.M.. 2011. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13:383–393. 10.1038/ncb2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K.R., Wickström S.A., and Fässler R.. 2009. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 23:397–418. 10.1101/gad.1758709 [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H., Niesman I., Fischer T., DeVries L., and Farquhar M.G.. 2005. Identification and characterization of GIV, a novel Galpha i/s-interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J. Biol. Chem. 280:22012–22020. 10.1074/jbc.M501833200 [DOI] [PubMed] [Google Scholar]
- Leyme A., Bourd-Boittin K., Bonnier D., Falconer A., Arlot-Bonnemains Y., and Théret N.. 2012. Identification of ILK as a new partner of the ADAM12 disintegrin and metalloprotease in cell adhesion and survival. Mol. Biol. Cell. 23:3461–3472. 10.1091/mbc.E11-11-0918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Ear J., Pavlova Y., Mittal Y., Kufareva I., Ghassemian M., Abagyan R., Garcia-Marcos M., and Ghosh P.. 2011. Tyrosine phosphorylation of the Gα-interacting protein GIV promotes activation of phosphoinositide 3-kinase during cell migration. Sci. Signal. 4:ra64 10.1126/scisignal.2002049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Ear J., Midde K., Lopez-Sanchez I., Aznar N., Garcia-Marcos M., Kufareva I., Abagyan R., and Ghosh P.. 2014. Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol. Biol. Cell. 25:3654–3671. 10.1091/mbc.E14-05-0978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y., Jiang P., Cui S.P., Ren Y.L., Zhu S.N., Yang J.P., Du J., Zhang Y., Liu J.Y., and Zhang B.. 2011. Clinical implications for girdin protein expression in breast cancer. Cancer Invest. 29:405–410. 10.3109/07357907.2011.568568 [DOI] [PubMed] [Google Scholar]
- Liu C., Zhang Y., Xu H., Zhang R., Li H., Lu P., and Jin F.. 2012a Girdin protein: A new potential distant metastasis predictor of breast cancer. Med. Oncol. 29:1554–1560. 10.1007/s12032-011-0087-6 [DOI] [PubMed] [Google Scholar]
- Liu H., Radisky D.C., Yang D., Xu R., Radisky E.S., Bissell M.J., and Bishop J.M.. 2012b MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat. Cell Biol. 14:567–574. 10.1038/ncb2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I., Dunkel Y., Roh Y.S., Mittal Y., De Minicis S., Muranyi A., Singh S., Shanmugam K., Aroonsakool N., Murray F., et al. . 2014. GIV/Girdin is a central hub for profibrogenic signalling networks during liver fibrosis. Nat. Commun. 5:4451 10.1038/ncomms5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranti C.K., and Brugge J.S.. 2002. Sensing the environment: a historical perspective on integrin signal transduction. Nat. Cell Biol. 4:E83–E90. 10.1038/ncb0402-e83 [DOI] [PubMed] [Google Scholar]
- Miyamoto S., Teramoto H., Gutkind J.S., and Yamada K.M.. 1996. Integrins can collaborate with growth factors for phosphorylation of receptor tyrosine kinases and MAP kinase activation: roles of integrin aggregation and occupancy of receptors. J. Cell Biol. 135:1633–1642. 10.1083/jcb.135.6.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.A., and Courtneidge S.A.. 2011. The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Profirovic J., Pan H., Vaiskunaite R., and Voyno-Yasenetskaya T.. 2003. G Protein betagamma subunits stimulate p114RhoGEF, a guanine nucleotide exchange factor for RhoA and Rac1: Regulation of cell shape and reactive oxygen species production. Circ. Res. 93:848–856. 10.1161/01.RES.0000097607.14733.0C [DOI] [PubMed] [Google Scholar]
- Offermanns S. 2006. Activation of platelet function through G protein-coupled receptors. Circ. Res. 99:1293–1304. 10.1161/01.RES.0000251742.71301.16 [DOI] [PubMed] [Google Scholar]
- Park C.C., Zhang H., Pallavicini M., Gray J.W., Baehner F., Park C.J., and Bissell M.J.. 2006. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Res. 66:1526–1535. 10.1158/0008-5472.CAN-05-3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K.A., and Rogers S.L.. 2013. Drosophila Ric-8 interacts with the Gα12/13 subunit, Concertina, during activation of the Folded gastrulation pathway. Mol. Biol. Cell. 24:3460–3471. 10.1091/mbc.E12-11-0813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plopper G.E., McNamee H.P., Dike L.E., Bojanowski K., and Ingber D.E.. 1995. Convergence of integrin and growth factor receptor signaling pathways within the focal adhesion complex. Mol. Biol. Cell. 6:1349–1365. 10.1091/mbc.6.10.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X.D., Kiosses W.B., and Schwartz M.A.. 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18:578–585. 10.1093/emboj/18.3.578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Blumer J.B., Simon V., and Lanier S.M.. 2006. Accessory proteins for G proteins: Partners in signaling. Annu. Rev. Pharmacol. Toxicol. 46:151–187. 10.1146/annurev.pharmtox.46.120604.141115 [DOI] [PubMed] [Google Scholar]
- Schiller H.B., Hermann M.R., Polleux J., Vignaud T., Zanivan S., Friedel C.C., Sun Z., Raducanu A., Gottschalk K.E., Théry M., et al. . 2013. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15:625–636. 10.1038/ncb2747 [DOI] [PubMed] [Google Scholar]
- Seals D.F., Azucena E.F. Jr., Pass I., Tesfay L., Gordon R., Woodrow M., Resau J.H., and Courtneidge S.A.. 2005. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 7:155–165. 10.1016/j.ccr.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Shen B., Delaney M.K., and Du X.. 2012. Inside-out, outside-in, and inside-outside-in: G protein signaling in integrin-mediated cell adhesion, spreading, and retraction. Curr. Opin. Cell Biol. 24:600–606. 10.1016/j.ceb.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Zhao X., O’Brien K.A., Stojanovic-Terpo A., Delaney M.K., Kim K., Cho J., Lam S.C., and Du X.. 2013. A directional switch of integrin signalling and a new anti-thrombotic strategy. Nature. 503:131–135. 10.1038/nature12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Matsuo Y., Shamoto T., Hirokawa T., Tsuboi K., Takahashi H., Ishiguro H., Kimura M., Takeyama H., and Inagaki H.. 2013. Girdin, a regulator of cell motility, is a potential prognostic marker for esophageal squamous cell carcinoma. Oncol. Rep. 29:2127–2132. [DOI] [PubMed] [Google Scholar]
- Simpson F., Martin S., Evans T.M., Kerr M., James D.E., Parton R.G., Teasdale R.D., and Wicking C.. 2005. A novel hook-related protein family and the characterization of hook-related protein 1. Traffic. 6:442–458. 10.1111/j.1600-0854.2005.00289.x [DOI] [PubMed] [Google Scholar]
- Smrcka A.V. 2008. G protein βγ subunits: Central mediators of G protein-coupled receptor signaling. Cell. Mol. Life Sci. 65:2191–2214. 10.1007/s00018-008-8006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.Y., Jiang P., Li N., Wang F.H., and Luo J.. 2014. Clinical significance of Girdin expression detected by immunohistochemistry in non-small cell lung cancer. Oncol Lett. 7:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg C., and Rubin K.. 1996. Stimulation of beta1 integrins on fibroblasts induces PDGF independent tyrosine phosphorylation of PDGF beta-receptors. J. Cell Biol. 132:741–752. 10.1083/jcb.132.4.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall G.G., Krumins A.M., and Gilman A.G.. 2003. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J. Biol. Chem. 278:8356–8362. 10.1074/jbc.M211862200 [DOI] [PubMed] [Google Scholar]
- Thibault M.M., Hoemann C.D., and Buschmann M.D.. 2007. Fibronectin, vitronectin, and collagen I induce chemotaxis and haptotaxis of human and rabbit mesenchymal stem cells in a standardized transmembrane assay. Stem Cells Dev. 16:489–502. 10.1089/scd.2006.0100 [DOI] [PubMed] [Google Scholar]
- Ueda H., Nagae R., Kozawa M., Morishita R., Kimura S., Nagase T., Ohara O., Yoshida S., and Asano T.. 2008. Heterotrimeric G protein betagamma subunits stimulate FLJ00018, a guanine nucleotide exchange factor for Rac1 and Cdc42. J. Biol. Chem. 283:1946–1953. 10.1074/jbc.M707037200 [DOI] [PubMed] [Google Scholar]
- Vadas O., Dbouk H.A., Shymanets A., Perisic O., Burke J.E., Abi Saab W.F., Khalil B.D., Harteneck C., Bresnick A.R., Nürnberg B., et al. . 2013. Molecular determinants of PI3Kγ-mediated activation downstream of G-protein-coupled receptors (GPCRs). Proc. Natl. Acad. Sci. USA. 110:18862–18867. 10.1073/pnas.1304801110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Weaver V.M., Petersen O.W., Larabell C.A., Dedhar S., Briand P., Lupu R., and Bissell M.J.. 1998. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: A different perspective in epithelial biology. Proc. Natl. Acad. Sci. USA. 95:14821–14826. 10.1073/pnas.95.25.14821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Hansen R.K., Radisky D., Yoneda T., Barcellos-Hoff M.H., Petersen O.W., Turley E.A., and Bissell M.J.. 2002. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J. Natl. Cancer Inst. 94:1494–1503. 10.1093/jnci/94.19.1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Kumamoto Y., Wang P., Gan X., Lehmann D., Smrcka A.V., Cohn L., Iwasaki A., Li L., and Wu D.. 2009. Regulation of immature dendritic cell migration by RhoA guanine nucleotide exchange factor Arhgef5. J. Biol. Chem. 284:28599–28606. 10.1074/jbc.M109.047282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Lin J., Li L., and Wang Y.. 2014. Expression and clinical significance of girdin in gastric cancer. Mol. Clin. Oncol. 2:425–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver V.M., Petersen O.W., Wang F., Larabell C.A., Briand P., Damsky C., and Bissell M.J.. 1997. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 137:231–245. 10.1083/jcb.137.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L., Enomoto A., Miyoshi H., Takahashi K., Asai N., Morone N., Jiang P., An J., Kato T., Kuroda K., et al. . 2014. Regulation of cargo-selective endocytosis by dynamin 2 GTPase-activating protein girdin. EMBO J. 33:2098–2112. 10.15252/embj.201488289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Ma S., Liu Q., and Liang P.. 2013. Clinical implications of Girdin protein expression in glioma. ScientificWorldJournal. 2013:986073 10.1155/2013/986073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.