Abstract
Epigenetic inactivation of tumor-related genes is an important characteristic in the pathology of human cancers, including melanomagenesis. We analyzed the epigenetic inactivation of Claudin 11 (CLDN11) in malignant melanoma (MM) of the skin, including six melanoma cell lines, 39 primary melanoma, 41 metastases of MM and 52 nevus cell nevi (NCN). CLDN11 promoter hypermethylation was found in 19 out of 39 (49%) of the primary MM and in 21 out of 41 (51%) of the MM metastases, but only in eight out of 52 (15%) of NCN (p = 0.001 and p = 0.0003, respectively). Moreover, a significant increase in the methylation level of CLDN11 from primary melanomas to MM metastases was revealed (p = 0.003). Methylation of CLDN11 was significantly more frequent in skin metastases (79%) compared to brain metastases (31%; p = 0.007). CLDN11 methylation was also found in five out of six MM cell lines (83%) and its promoter hypermethylation correlated with a reduced expression. Treatment of MM cell lines with a DNA methylation inhibitor reactivated CLDN11 transcription by its promoter demethylation. In summary, CLDN11 proved to be an epigenetically inactivated tumor related gene in melanomagenesis, and analysis of CLDN11 methylation level represents a potential tool for assisting in the discrimination between malignant melanoma and nevus cell nevi.
Keywords: malignant melanoma, Claudin 11, tumor suppressor gene, epigenetics, DNA methylation
1. Introduction
Malignant melanoma is a malignant skin cancer showing a rising incidence worldwide [1]. Several molecular pathways have been found altered in melanocytic tumors including the MAPK pathway, the p16INK4a/RB pathway and the Hippo/Ras Association Domain Family (RASSF) pathway [2,3,4]. Aberrant regulation of these pathways is accomplished through inactivation of tumor suppressor genes (e.g., RASSF10) and activation of proto-oncogenes (e.g., BRAF) [5,6]. Apart from mutation, the epigenetic silencing of tumor suppressor genes is a frequent and fundamental event in the pathogenesis of cancer, including melanomagenesis [7,8]. This inactivation is achieved by hypermethylation of CpG island promoters in malignant melanoma. In this context, methylation markers may serve as important tools to distinguish between benign lesions and aggressive tumors. Recently, it has been suggested that Claudin 11 (CLDN11) could be a useful epigenetic biomarker for identifying malignant melanoma [9]. CLDN11 is a member of the claudin family that encodes integral membrane proteins and is involved in the formation of the paracellular tight junction seal in tissues [10,11]. Thus CLDN11 harbors a Claudin_2/PMP22 domain (Figure 1a) that is also found in the peripheral myelin protein PMP22 and the epithelial membrane proteins (e.g., EMP1) [12]. So far, 27 members of the CLDN family (CLDN1 to 27) have been identified in the human genome [13]. Expressional analysis suggests that several claudin genes exhibit decreased transcript levels in cancer. However, CLDN3, CLDN4 and CLDN7 levels are elevated in certain tumor entities [10]. For CLDN11 it has been reported that it is silenced in gastric cancer by promoter hypermethylation and its inactivation is associated with invasiveness of this cancer [14]. A genome-wide analysis has identified the methylation of CLDN11 in primary cutaneous melanoma [15]. However, the epigenetic regulation (e.g., expression) in melanoma has not been analyzed.
Figure 1.
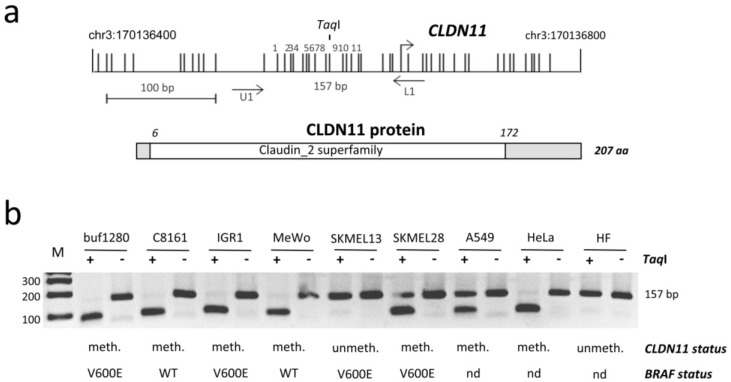
Epigenetic regulation of Claudin 11 (CLDN11) in malignant melanoma. (a) Structure of the CLDN11 CpG island promoter on chromosome 3 and the CLDN11 protein (207 aa). Arrows mark transcriptional (+1) start site for CLDN11. Vertical lines indicate CpGs. The 157 bp PCR product with respective primers and the TaqI site are depicted. The conserved Claudin_2 superfamily domain of CLDN11 is marked; (b) Methylation analysis of CLDN11 by COBRA. Bisulfite-treated DNA from MM cell lines (buf1280, C8161, IGR1, MeWo, SKMEL13 and SKMEL28), lung cancer A549, HeLa and human fibroblasts (HF) was amplified, digested with TaqI (+) or mock digested (−) and resolved on 2% gels with a 100 bp marker (M). The methylation status of CLDN11 (meth./methylated and unmeth./unmethylated) and BRAF status (WT/wild type, V600E/Codon 600 mutation and nd/not determined) are indicated.
The aim of our study was to illuminate the epigenetic inactivation of CLDN11 in malignant melanomas (MM) in more detail. Here, we report a significant increase in the methylation level of CLDN11 in MM metastases compared to primary MM and nevus cell nevi.
2. Results
2.1. Epigenetic Inactivation of CLDN11 in Malignant Melanoma
Recently hypermethylation of Claudin 11 (CLDN11) has been reported in primary melanomas [9], however its epigenetic regulation was not analyzed in detail. The schematic promoter region of CLDN11 and according CpGs are shown in Figure 1a. The promoter lies within a CpG island of 1644 bp on chromosome 3q26.2 from position 170′136′243 to 170′137′886 (UCSC genome browser). To reveal the epigenetic status of CLDN11 in malignant melanoma (MM) cell lines, we have analyzed its aberrant methylation in buf1280, C8161, IGR1, MeWo, SKMEL13, SKMEL28, lung cancer (A549), cervix cancer (HeLa) and human fibroblast (HF) by COBRA (Figure 1b). Fragmentation of the PCR product by TaqI indicates an underlying methylated CLDN11 promoter. In five MM cell lines (buf1280, C8161, IGR1, MeWo, SKMEL28) hypermethylation of CLDN11 was detected (Figure 1b). CLDN11 was unmethylated in normal human fibroblast (HF) and melanoma cell line SKMEL13. Methylation of CLDN11 was also observed in A549 and HeLa cancer cells (Figure 1b). Previously, we analyzed the BRAF mutational status in MM cell lines [5]. There was no obvious correlation between CLDN11 methylation and BRAF mutation status in MM cell lines (Figure 1b).
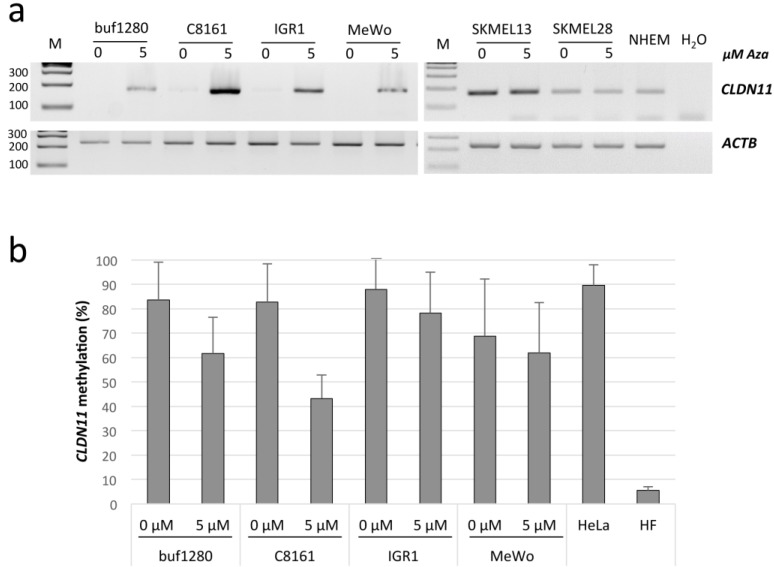
Subsequently, we analyzed the expression of CLDN11 in six MM cell lines and normal human epidermal melanocytes (NHEM) by RT-PCR (Figure 2a). CLDN11 mRNA levels were reduced in buf1280, C8161, IGR1 and MeWo compared to NHEM (Figure 2a). Treatment of these four cell lines with 5-aza-2′-deoxycytidine (Aza), a substance that inhibits DNA methylation, resulted in increased CLDN11 expression (Figure 2a). In SKMEL13 cells, which harbor an unmethylated promoter, CLDN11 expression was observerd in untreated cells (Figure 2a). In SKMEL28 cells with a partially methylated CLDN11 promoter, there was no induction of CLDN11 expression after Aza treatment. To analyze the impact of Aza treatment on DNA methylation, quantitative bisulfite sequencing was performed (Figure 2b). For all four MM cell lines that exhibited elevated CLDN11 expression after Aza treatment, a demethylation of CLDN11 was detected (Figure 2). Especially in C8161 which exhibit a high re-expression of CLDN11 (Figure 2a), a strong demethylation (2-fold reduction in methylation level) of the CLDN11 promoter region was observed after Aza treatment (Figure 2b).
Figure 2.
Epigenetic reactivation of CLDN11 in malignant melanoma. (a) RNA expression of CLDN11 in MM cell lines and normal human epidermal melanocytes (NHEM). MM cell lines (buf1280, C8161, IGR1, MeWo, SKMEL13 and SKMEL28) were treated for four days with 5 µM of 5-aza-2′-deoxycytidine (Aza). RNA was isolated and analyzed by RT-PCR. Products for CLDN11 (167 bp) and a 100 bp ladder (M) were resolved on 2% gel. Expression of ACTB (225 bp) was determined as a control for RNA integrity; (b) Methylation of CLDN11 in Aza-treated MM cell lines. DNA was isolated and analyzed by quantitative bisulfite pyrosequencing. 11 CpGs within the PCR products obtained from the indicated MM cell lines, HeLa and human fibroblasts (HF) were analyzed. The mean frequency of CpG methylation is indicated.
2.2. CLDN11 Promoter Hypermethylation Occurs Frequently in Melanoma, Is a Rare Event in Nevi
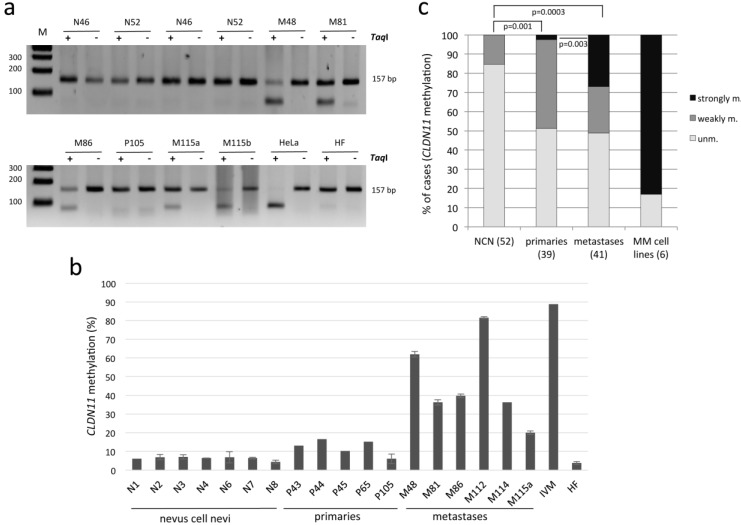
Subsequently, we analyzed the methylation of CLDN11 in primary MM, MM metastasis and NCN by COBRA and bisulfite pyrosequencing (Figure 3 and Table 1). In 21 out of 41 (51%) metastases (e.g., M48/lymph node and M81/skin metastasis), the promoter of CLDN11 was methylated and therefore restriction products were detected (Figure 3a and Table 1). Methylation of CLDN11 was frequently found in 15 out of 19 (79%) skin metastases of MM, but only in five out of 16 (31%) brain metastases of MM (p = 0.007; Table 1). Methylation levels of 11 CpGs (Figure 1a) were quantified by pyrosequencing and rated unmethylated (<10%), weakly methylated (10% to 20% methylation) and strongly methylated (>20%; Figure 3b,c). The background was under 10% in all samples that were negative in COBRA (Figure 3 and data not shown). None of the 52 (0%) NCN exhibited strong methylation of CLDN11 and only 15% (8/52) NCN showed weak CLDN11 methylation. In primary melanoma a significantly higher frequency of CLDN11 methylation (49%; 19/39) was revealed (p = 0.001). Interestingly, primary melanoma exhibited lower methylation levels compared to metastases (Table 1, Figure 3b,c). We observed an increase in methylation levels with significantly stronger methylated metastases compared to primaries (27% vs. 3%; p = 0.003, respectively; Figure 3c and Table 1). In summary, our data suggest that hypermethylation of CLDN11 occurs frequently in primary MM and metastases, however is rarely found in NCN (p = 0.001 and p = 0.0003, respectively).
Figure 3.
Methylation of CLDN11 in nevus cell nevi (N), primary malignant melanoma (P) and melanoma metastases (M). (a) Methylation of CLDN11 was analyzed by COBRA. PCR products (157 bp) from bisulfite-treated DNA were digested with TaqI (+) or mock digested (−) and resolved on 2% gel. DNA from HeLa and fibroblast (HF) was utilized as methylated and unmethylated control. Sizes of a 100 bp ladder (M) are marked; (b) Methylation analysis of CLDN11 by bisulfite pyrosequencing. 11 CpGs within PCR products generated from nevus cell nevi (N), primary MM (P/primaries) and MM metastases (M) and in vitro methylated DNA (IVM) were analyzed in technical replicates. M48 is a lymph node metastasis and all other metastases originated from skin; (c) Comparison between methylation of MM (primaries, metastases) and NCN. Bar charts indicate percentage of cases with unmethylated (unm. <10%), weakly methylated (10% to 20%) and strongly methylated (>20%) CLDN11. Significance was calculated with the two-tailed Fisher exact probability test.
Table 1.
Summary of CLDN11 methylation (n).
| Case | Unmethylated a | Weakly Methylated b | Strongly Methylated c |
|---|---|---|---|
| Nevus cell nevus | |||
| dysplastic (14) | 86% (12) | 14% (2) | 0% |
| Non-dysplastic (38) | 84% (32) | 16% (6) | 0% |
| Total (52) | 85% (44) | 15% (8) | 0% |
| Primary melanoma (39) | 51% (20) | 46% (18) | 3% (1) |
| Melanoma metastasis | |||
| Skin (19) | 21% (4) | 37% (7) | 42% (8) |
| Brain (16) | 68.75% (11) | 18.75% (3) | 12.5% (2) |
| Lymph node (4) | 75% (3) | 0% | 25% (1) |
| Others (2) | 100% (2) | 0% | 0% |
| Total (41) | 49% (20) | 24% (10) | 27% (11) |
| MM cell lines (6) | 17% (1) | 0% | 83% (5) |
a <10%; b 10%–20%; c >20% methylation.
3. Discussion
The Claudin gene family consists of 27 members, which encode membrane proteins of the paracellular tight junction. Claudin 11 (CLDN11) was recently identified as a member that is hypermethylated in human cancers including malignant melanoma (MM) [9,14]. Here we confirm that CLDN11 is frequently hypermethylated in cutaneous MM, including primaries and metastases (49% and 51% methylation, respectively). Additionally to previous studies, we have performed quantitative methylation analysis and revealed significantly increased methylation levels of MM metastases especially in skin metastases (Figure 3 and Table 1). Moreover, we show that hypermethylation of CLDN11 correlates with inactivation of its expression and that inhibition of DNA methyltransferase epigenetically reactivated CLDN11 transcription (Figure 2). Gao et al. have reported a similar frequency of 48% methylation for primary melanoma, but 73% for metastatic melanoma [9]. However, we show that the location of metastases correlates significantly with the methylation frequency of CLDN11 (Table 1). Especially skin metastases exhibit significantly higher methylation frequency compared to brain metastases (79% compared to 31%, respectively). It is tempting to speculate that CLDN11 methylation levels in primary MM contribute to differences in metastatic capacity of melanomas. Thus it will be interesting to analyze the functional consequences of CLDN11 inactivation for invasiveness potential of melanomas in more detail. Interestingly, the RASSF6 tumor suppressor gene exhibited its highest methylation frequency in melanoma brain metastases [16]. Since most melanomas are driven by an activating mutation at codon 600 of BRAF, we also analyzed its mutational status and CLDN11 methylation in MM cell lines (Figure 1b). There was no direct or inverse correlation between both events. It has been reported that BRAF mutations are found with similar frequencies in brain metastases (48%) and skin metastases (53%) [17]. We revealed 66% of BRAF mutation and 83% of CLDN11 methylation in MM cell lines (Figure 1b). To date, the methylation rate of CLDN11 had not been analyzed in MM cell lines.
Moreover, we utilized bisulfite pyrosequencing, a method that provides quantitative data on the methylation levels of CLDN11 in different skin samples. Here we report significantly elevated methylation levels in metastatic MM compared to primary cancers (Figure 3c), thus the frequency and level of CLDN11 methylation increases with the malignancy of melanoma. Previously, we have analyzed RASSF10 methylation in MM and we reported frequent methylation in melanoma, although RASSF10 methylation was not found in non-dysplastic nevi [6]. Here, we observed a weak CLDN11 methylation in few non-dysplastic and at a similar frequency (15%) in dysplastic NCN (Table 1). Gao et al. reported only 3% methylation of CLDN11 methylation in dysplastic NCN [9]. However they have utilized methylation specific PCR and analyzed a region within the 1st Exon (+100 to +300) [9]. Since we have analyzed a region upstream of the transcriptional start site (Figure 1a), the difference in methylation could be attributed to gradual spreading of DNA methylation from the borders of the CLDN11 CpG island. This progressive epigenetic inactivation event has been reported previously for human mammary epithelial cells during stress induced senescence for RASSF1A [18]. Considering DNA methylation as an epigenetic biomarker for MM the region of CLDN11 where methylation occurs during melanomagenesis should be utilized, since low methylation levels were also observed in NCN.
Additionally, we have also analyzed the epigenetic regulation of CLDN11 in MM cell lines (Figure 1). The level and frequency of CLDN11 hypermethylation is increased compared to primary tissues (Figure 3c). Moreover, we found that aberrant CLDN11 promoter methylation correlates with its transcriptional silencing in four MM cell lines (Figure 2). Inhibition of DNA methyltransferases by 5-aza-2′-deoxycytidine reactivates CLND11 expression through promoter demethylation in these cell lines. These observations suggest that aberrant DNA methylation has an important impact on CLDN11 expression, which had not been analyzed previously in MM.
Aberrant epigenetic regulation of other Claudin members has also frequently been reported in human cancers. CLDN1 methylation was detected in colon cancer and has been found in breast cancer [19,20]. For CLDN3 it has been observed that it methylation occurs in esophageal and hepatocellular carcinoma [21,22]. Methylation of CLDN4 and CLDN5 has been reported in bladder and pancreatic cancer, respectively [23,24]. Moreover, hypermethylation of CLDN6, CLND7 and CLDN15 have also been observed in different cancer entities [25,26,27]. Thus hypermethylation of distinct Claudins were frequently reported in human cancers and it will be interesting to analyze the methylation of several Claudin family members (e.g., CLDN1 and CLDN15) in cutaneous melanoma.
4. Experimental Section
4.1. Tissue and Cell Lines
Primary tissues and cancer cell lines were previously published [6]. All patients signed informed consent at initial clinical investigation. The study was approved by local ethic committees (University of Heidelberg, Heidelberg, Germany). Primary Normal Human Epidermal Melanocytes (NHEM) obtained from PromoCell (Heidelberg, Germany). All cell lines were cultured in humidified atmosphere (37 °C) with 5% CO2 and 1xPenicillin/Streptomycin in according medium.
4.2. Methylation Analysis
DNA was isolated by phenol-chloroform extraction and then bisulfite treated prior to COBRA analysis and pyrosequencing [28]. Methylation analyses were performed in technical replicates. Bisulfite treated DNA (150 ng) was used for PCR with primer CLDN11BSU1 (TTTTGGGGTTATTTTGTTTTTTTTTA) and 5′-biotinylated primer CLDN11BioL1 (AAAACAACAACRCTACTAAACAAC). Products were digested with 0.5 µL TaqI (Fermentas GmbH, St. Leon-Rot, Germany) for 1 h at 65 °C and resolved on 2% TBE gel. Methylation status was quantified utilizing the primer CLDN11Seq1 (ATTTTGTTTTTTTTTAYGTTTTTTTT) and PyroMark Q24 (Qiagen, Hilden, Germany). Eleven CpGs are included in the analyzed region of CLDN11 and mean methylation was calculated (Figure 1a). For in vitro methylation of genomic DNA we used CpG methyltransferase (M. SssI, NEB, Frankfurt, Germany).
4.3. Expression Analysis
RNA was isolated using the Isol-RNA lysis procedure (5 Prime, Hamburg, Germany). RNA was DNase (Fermentas GmbH, St. Leon-Rot, Germany) digested and then reversely transcribed [29]. RT-PCR was performed with primers: CLDN11RTF2: CCCACCTGCCGCAAGCTGGA, CLDN11RTR2: GGCAGACCCAGGACCGAGGC, ßACTFW: CCTTCCTTCCTGGGCATGGAGTC, ßACTRW: CGGAGTACTTGCGCTCAGGAGGA.
4.4. Statistical Evaluation
Categorical variables were plotted into contingency tables and evaluated using Fisher’s exact probability test. All reported p-values are two-sided and considered significant for p < 0.05.
5. Conclusions
In summary, our results show that hypermethylation of CLDN11 promoter occurs frequently in MM, but was rarely found in NCN. This data suggests that CLDN11 may encode a novel melanoma-specific tumor suppressor gene. Further studies are necessary to elaborate the exact tumor suppressor function of CLDN11. Furthermore, quantitative CLDN11 methylation analysis may serve as candidate biomarker tool in melanomagenesis in combination with other markers (e.g., RASSF10), since low CLDN11 methylation levels were also observed in NCN.
Acknowledgments
The work was supported by grants (TRR81, LOEWE) from the DFG and Land Hessen to Reinhard Dammann. These organizations had no involvement in the study design, acquisition, analysis, data interpretation, writing of the manuscript or in the decision to submit the manuscript for publication. We thank Michelle Woods for proofreading the manuscript.
Abbreviations
- CLDN11
Claudin 11
- COBRA
combined bisulfite restriction analysis
- Aza
5-Aza-2′-deoxycytidine
- MM
malignant melanoma
- NCN
nevus cell nevus
- RASSF
Ras Association Domain Family
- ACTB
Beta-Actin
Author Contributions
RHD has created the study. SKW, AMR and PH acquired data. SKW, AMR, PH and RHD controlled analyzed and interpreted data. RHD prepared the manuscript. SKW, AMR, PH and RHD read, corrected and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Dahl C., Guldberg P. The genome and epigenome of malignant melanoma. Apmis. 2007;115:1161–1176. doi: 10.1111/j.1600-0463.2007.apm_855.xml.x. [DOI] [PubMed] [Google Scholar]
- 3.Miller A.J., Mihm M.C., Jr. Melanoma. N. Engl. J. Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 4.Rastetter M., Schagdarsurengin U., Lahtz C., Fiedler E., Marsch W., Dammann R., Helmbold P. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol. Histopathol. 2007;22:1005–1015. doi: 10.14670/HH-22.1005. [DOI] [PubMed] [Google Scholar]
- 5.Lahtz C., Stranzenbach R., Fiedler E., Helmbold P., Dammann R.H. Methylation of pten as a prognostic factor in malignant melanoma of the skin. J. Investig. Dermatol. 2010;130:620–622. doi: 10.1038/jid.2009.226. [DOI] [PubMed] [Google Scholar]
- 6.Helmbold P., Richter A.M., Walesch S., Skorokhod A., Marsch W., Enk A., Dammann R.H. RASSF10 promoter hypermethylation is frequent in malignant melanoma of the skin but uncommon in nevus cell nevi. J. Investig. Dermatol. 2012;132:687–694. doi: 10.1038/jid.2011.380. [DOI] [PubMed] [Google Scholar]
- 7.Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothhammer T., Bosserhoff A.K. Epigenetic events in malignant melanoma. Pigment Cell Res. 2007;20:92–111. doi: 10.1111/j.1600-0749.2007.00367.x. [DOI] [PubMed] [Google Scholar]
- 9.Gao L., van den Hurk K., Moerkerk P.T., Goeman J.J., Beck S., Gruis N.A., van den Oord J.J., Winnepenninckx V.J., van Engeland M., van Doorn R. Promoter cpg island hypermethylation in dysplastic nevus and melanoma: Cldn11 as an epigenetic biomarker for malignancy. J. Investig. Dermatol. 2014;134:2957–2966. doi: 10.1038/jid.2014.270. [DOI] [PubMed] [Google Scholar]
- 10.Hewitt K.J., Agarwal R., Morin P.J. The claudin gene family: Expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Itallie C.M., Anderson J.M. Claudin interactions in and out of the tight junction. Tissue Barriers. 2013;1:e25247. doi: 10.4161/tisb.25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jetten A.M., Suter U. The peripheral myelin protein 22 and epithelial membrane protein family. Progr. Nucleic Acid Res. Mol. Biol. 2000;64:97–129. doi: 10.1016/s0079-6603(00)64003-5. [DOI] [PubMed] [Google Scholar]
- 13.Mineta K., Yamamoto Y., Yamazaki Y., Tanaka H., Tada Y., Saito K., Tamura A., Igarashi M., Endo T., Takeuchi K., et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011;585:606–612. doi: 10.1016/j.febslet.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal R., Mori Y., Cheng Y., Jin Z., Olaru A.V., Hamilton J.P., David S., Selaru F.M., Yang J., Abraham J.M., et al. Silencing of claudin-11 is associated with increased invasiveness of gastric cancer cells. PLoS ONE. 2009;4:e8002. doi: 10.1371/journal.pone.0008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao L., Smit M.A., van den Oord J.J., Goeman J.J., Verdegaal E.M., van der Burg S.H., Stas M., Beck S., Gruis N.A., Tensen C.P., et al. Genome-wide promoter methylation analysis identifies epigenetic silencing of mapk13 in primary cutaneous melanoma. Pigment Cell Melanoma Res. 2013;26:542–554. doi: 10.1111/pcmr.12096. [DOI] [PubMed] [Google Scholar]
- 16.Mezzanotte J.J., Hill V., Schmidt M.L., Shinawi T., Tommasi S., Krex D., Schackert G., Pfeifer G.P., Latif F., Clark G.J. Rassf6 exhibits promoter hypermethylation in metastatic melanoma and inhibits invasion in melanoma cells. Epigenetics. 2014;9:1496–1503. doi: 10.4161/15592294.2014.983361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colombino M., Capone M., Lissia A., Cossu A., Rubino C., de Giorgi V., Massi D., Fonsatti E., Staibano S., Nappi O., et al. Braf/nras mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol. 2012;30:2522–2529. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 18.Strunnikova M., Schagdarsurengin U., Kehlen A., Garbe J.C., Stampfer M.R., Dammann R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the rassf1a promoter. Mol. Cell Biol. 2005;25:3923–3933. doi: 10.1128/MCB.25.10.3923-3933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Cello F., Cope L., Li H., Jeschke J., Wang W., Baylin S.B., Zahnow C.A. Methylation of the claudin 1 promoter is associated with loss of expression in estrogen receptor positive breast cancer. PLoS ONE. 2013;8:e68630. doi: 10.1371/journal.pone.0068630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogoshi K., Hashimoto S., Nakatani Y., Qu W., Oshima K., Tokunaga K., Sugano S., Hattori M., Morishita S., Matsushima K. Genome-wide profiling of DNA methylation in human cancer cells. Genomics. 2011;98:280–287. doi: 10.1016/j.ygeno.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Adams L., Roth M.J., Abnet C.C., Dawsey S.P., Qiao Y.L., Wang G.Q., Wei W.Q., Lu N., Dawsey S.M., Woodson K. Promoter methylation in cytology specimens as an early detection marker for esophageal squamous dysplasia and early esophageal squamous cell carcinoma. Cancer Prev. Res. 2008;1:357–361. doi: 10.1158/1940-6207.CAPR-08-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang L., Yang Y.D., Fu L., Xu W., Liu D., Liang Q., Zhang X., Xu L., Guan X.Y., Wu B., et al. Cldn3 inhibits cancer aggressiveness via wnt-emt signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget. 2014;5:7663–7676. doi: 10.18632/oncotarget.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boireau S., Buchert M., Samuel M.S., Pannequin J., Ryan J.L., Choquet A., Chapuis H., Rebillard X., Avances C., Ernst M., et al. DNA-methylation-dependent alterations of claudin-4 expression in human bladder carcinoma. Carcinogenesis. 2007;28:246–258. doi: 10.1093/carcin/bgl120. [DOI] [PubMed] [Google Scholar]
- 24.Sato N., Fukushima N., Maitra A., Matsubayashi H., Yeo C.J., Cameron J.L., Hruban R.H., Goggins M. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–3742. [PubMed] [Google Scholar]
- 25.Kim B., Kang S., Jeong G., Park S.B., Kim S.J. Identification and comparison of aberrant key regulatory networks in breast, colon, liver, lung, and stomach cancers through methylome database analysis. PLoS ONE. 2014;9:e97818. doi: 10.1371/journal.pone.0097818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kominsky S.L., Argani P., Korz D., Evron E., Raman V., Garrett E., Rein A., Sauter G., Kallioniemi O.P., Sukumar S. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–2033. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 27.Tsunoda S., Smith E., de Young N.J., Wang X., Tian Z.Q., Liu J.F., Jamieson G.G., Drew P.A. Methylation of cldn6, fbn2, rbp1, rbp4, tfpi2, and tmeff2 in esophageal squamous cell carcinoma. Oncol. Rep. 2009;21:1067–1073. doi: 10.3892/or_00000325. [DOI] [PubMed] [Google Scholar]
- 28.Haag T., Herkt C.E., Walesch S.K., Richter A.M., Dammann R.H. The apoptosis associated tyrosine kinase gene is frequently hypermethylated in human cancer and is regulated by epigenetic mechanisms. Genes Cancer. 2014;5:365–374. doi: 10.18632/genesandcancer.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter A.M., Walesch S.K., Wurl P., Taubert H., Dammann R.H. The tumor suppressor rassf10 is upregulated upon contact inhibition and frequently epigenetically silenced in cancer. Oncogenesis. 2012;1:e18. doi: 10.1038/oncsis.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]