Significance
The apolipoprotein E4 (ApoE4) genotype is the strongest genetic risk factor for developing Alzheimer’s disease (AD). However, the mechanisms that underlie this link between ApoE4 genotype and AD are not well understood. Our data in this paper are the first mechanistic studies to our knowledge that link ApoE4 genotype-specific changes in brain phospholipid homeostasis to ApoE4-increased susceptibility to develop AD. Our studies indicate previously unidentified therapeutic options for the treatment of AD, targeting ApoE4’s pathogenic nature.
Keywords: Alzheimer's disease, apolipoprotein E4, phospholipid, dysregulation, cognitive deficits
Abstract
The apolipoprotein E4 (ApoE4) allele is the strongest genetic risk factor for developing sporadic Alzheimer’s disease (AD). However, the mechanisms underlying the pathogenic nature of ApoE4 are not well understood. In this study, we have found that ApoE proteins are critical determinants of brain phospholipid homeostasis and that the ApoE4 isoform is dysfunctional in this process. We have found that the levels of phosphoinositol biphosphate (PIP2) are reduced in postmortem human brain tissues of ApoE4 carriers, in the brains of ApoE4 knock-in (KI) mice, and in primary neurons expressing ApoE4 alleles compared with those levels in ApoE3 counterparts. These changes are secondary to increased expression of a PIP2-degrading enzyme, the phosphoinositol phosphatase synaptojanin 1 (synj1), in ApoE4 carriers. Genetic reduction of synj1 in ApoE4 KI mouse models restores PIP2 levels and, more important, rescues AD-related cognitive deficits in these mice. Further studies indicate that ApoE4 behaves similar to ApoE null conditions, which fails to degrade synj1 mRNA efficiently, unlike ApoE3 does. These data suggest a loss of function of ApoE4 genotype. Together, our data uncover a previously unidentified mechanism that links ApoE4-induced phospholipid changes to the pathogenic nature of ApoE4 in AD.
The apolipoprotein E4 (ApoE4) allele is a primary genetic risk factor for sporadic Alzheimer’s disease (AD) (1). Although only ∼20% of humans are ApoE4 carriers, these individuals account for up to 65% of all AD cases. Elucidation of the contribution of ApoE4 to AD pathogenesis has been a considerable challenge. The mechanisms that underlie the link between ApoE4 genotype and AD are not yet well understood, but have been suggested to involve reduced clearance of brain Aβ (2, 3). Other Aβ-independent mechanisms have been implicated (4), including alterations in brain membrane lipid composition and metabolism (5–7). For example, evidence suggests that in postmortem AD brains, certain alterations in brain membrane lipid composition are exaggerated by the ApoE4 genotype (5–8). In addition, a recent paper reported changes in serum levels of 10 phospholipids as predicting phenotype conversion from normal aging to either amnestic mild cognitive impairment (MCI) or early AD in 2–3-y intervals with more than 90% accuracy (9). These findings suggest that specific phospholipid homeostasis may play an important role in the pathogenesis of AD, and possibly in ApoE4-increased susceptibility. In the present study, we test the hypotheses that ApoE proteins are critical determinants of brain phospholipid homeostasis and that the ApoE4 isoform is dysfunctional in this process.
Results
Changes in Phosphoinositol Metabolites Correlate with AD Disease Development, and ApoE4 Genotype Exacerbates Phosphoinositol Biphosphate Reduction.
We first analyzed phospholipid composition of human parietal cortex tissues of both ApoE4 carriers and non-ApoE4 subjects (Fig. S1A). There was significant correlation between changes in phosphoinositol metabolites [PI; phosphoinositol phosphate (PIP) and phosphoinositol biphosphate (PIP2)] and AD disease development and progression (Fig. 1A). A specific pattern of changes including reduction of PIP2 and PI levels, as well as a reciprocal elevation in PIP levels, was observed along with disease progression from normal aging to MCI and early AD [clinical dementia rating (CDR) scale from 0 to 1].
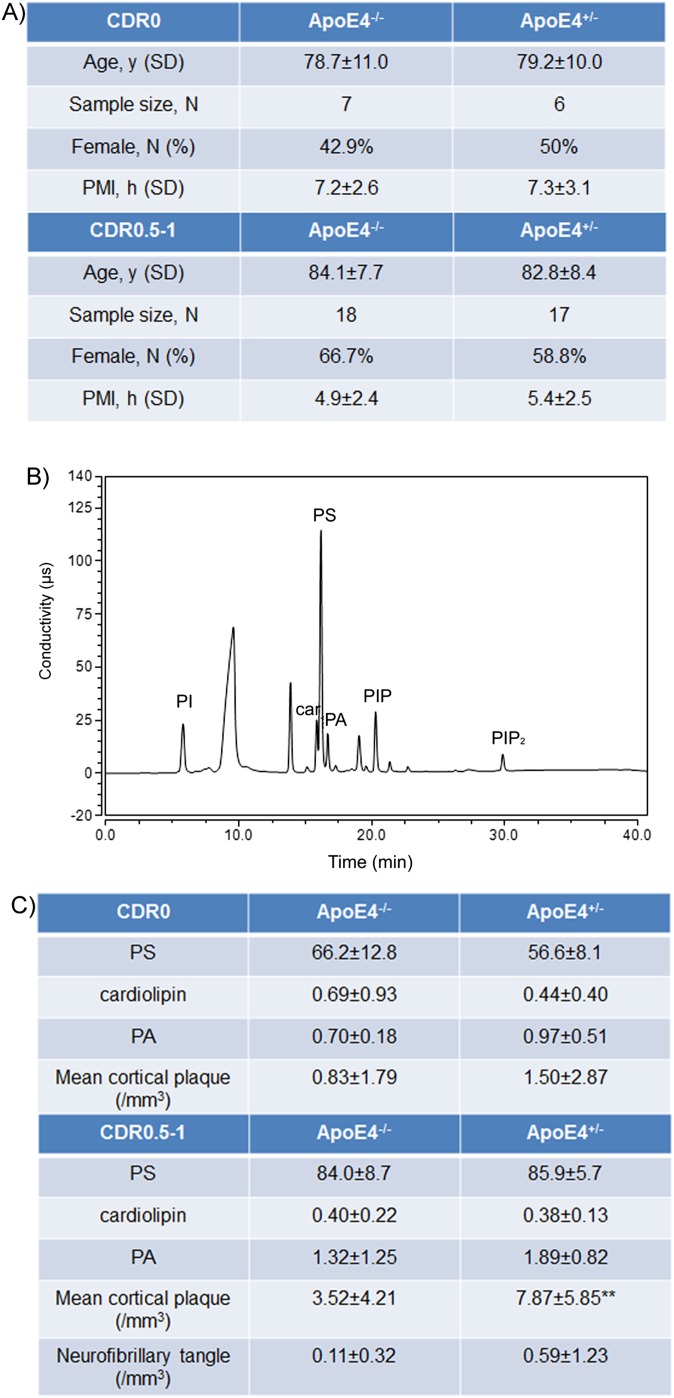
Fig. S1.
Sample information for cohorts of CDR 0 and 0.5–1. (A) The sample size and characteristics of subjects in the CDR 0 and 0.5–1 with ApoE4−/− and ApoE4+/− genotypes are shown (PMI, postmortem interval). The PMI of each sample was less than 8 h for all samples and was comparable between groups. (B) A typical example of HPLC chromatogram showing phospholipid peaks including phosphoinositol (PI), cardiolipin (car), phosphoserine (PS), phosphatidic acid (PA), phosphoinositol phosphate (PIP), and phosphoinositol biphosphate (PIP2). (C) The average values (calculated as the percentage of total lipids measured) of three other phospholipids (PS, cardiolipin, and PA) are shown for ApoE4−/− and ApoE4+/− subjects from the CDR 0 and 0.5–1 cohorts; the mean cortical plaque and neurofibrillary tangle burden are also shown (**P < 0.01 with ANOVA tests).
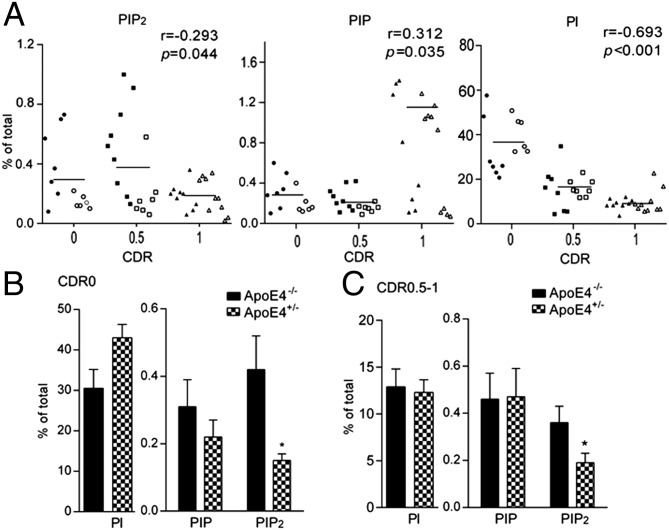
Fig. 1.
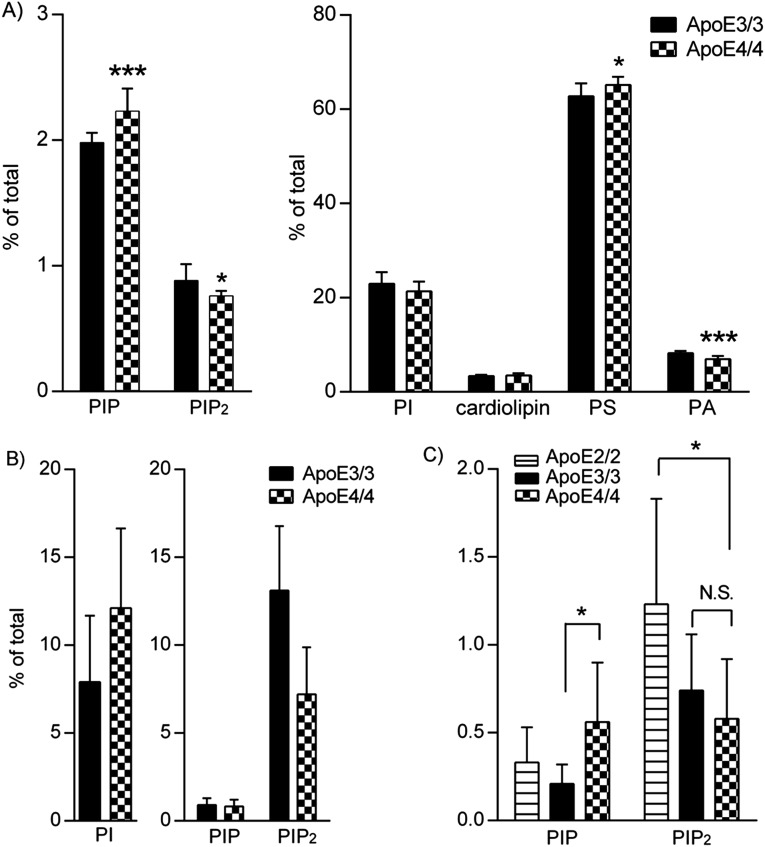
Changes in PI, PIP, and PIP2 correlate with AD disease development and progression. Reductions in PIP2 levels are exacerbated in ApoE4 carriers. (A) Correlation of PI/PIP/PIP2 and AD stages. Closed symbols, ApoE4−/− subjects; open symbols, ApoE4+/− carriers. Phospholipid profile of ApoE4+/− brain samples (B) with CDR 0 and (C) with CDR 0.5–1. *P < 0.05 with ANOVA tests.
Interestingly, ApoE4 genotype specifically exacerbates the changes in PIP2 levels along with disease progression. A reduction of PIP2 was observed in ApoE4+/− tissue in both CDR 0 (Fig. 1B) and CDR 0.5–1 cohorts (Fig. 1C). PIP2 levels were much lower in the ApoE4+/− brain tissues of normal aged subjects (0.15% of total phospholipids measured) compared with ApoE4−/− counterparts (0.42%; P = 0.026). A trend of elevation in substrate PI levels in ApoE4+/− brains was seen but did not achieve statistical significance. No changes were seen in other phospholipid species such as PIP, phosphoserine, phosphatidic acid, and cardiolipin (Fig. S1 B and C). Reduction in PIP2 levels was also observed in patients with early AD with the ApoE4 genotype, using a cohort of brain tissues from patients with a clinical diagnosis of MCI or early AD (CDR ranges from 0.5 to 1; Fig. 1C). Brain PIP2 levels in ApoE4+/− patients (0.19%) were significantly lower than those in ApoE4−/− patients (0.36%; P = 0.036). There were no changes seen in PI or PIP levels in ApoE4+/− versus ApoE4−/− brain samples. Concomitantly, the amounts of amyloid plaque and neurofibrillary tangles in ApoE4+/− brains were much higher than those in ApoE4−/− brains, even in the normal aging population (Fig. S1C), suggesting that ApoE4 effects on amyloid burden may exacerbate its effects on PIP2 reduction, as implicated by a previous report that oligomeric Aβ can reduce PIP2 levels in mouse cortical neuronal cultures (10). It should be noted, however, that in advanced AD stages (a cohort of CDR 3 subjects; Fig. S2 A and B), there was a trend of reduction in PIP2 levels in ApoE4+/− brains compared with ApoE4−/− brains, but it did not achieve statistical significance as a result of large sample variations. The PI, in contrast, was statistically reduced in ApoE4+/− brains compared with ApoE4−/− counterparts at this stage, possibly because of a significant disruption of cell membrane integrity by the ApoE4 genotype at the advanced disease stage.
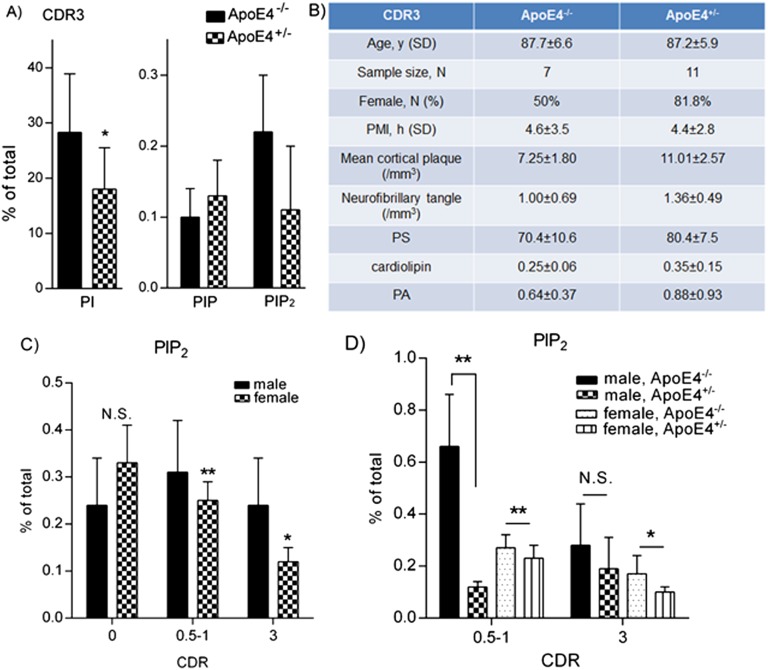
Fig. S2.
Phospholipid analysis of brain samples from cohorts of CDR 0–3. (A) In the cohort of CDR 3, the PI levels were significantly reduced in ApoE4+/− subjects if compared with ApoE4−/− counterparts (18.0% vs. 28.3% with ANOVA tests), whereas PIP2 levels were reduced in ApoE4+/− subjects as well, but did not achieve statistical significance because of large variations within groups (0.11% vs. 0.22% with ANOVA tests). No significant differences were detected in PIP levels between the two groups. (B) In the CDR 3 cohort, there was a higher percentage of female subjects and a tendency of increased plaque burden in the ApoE4+/− group compared with its ApoE4−/− counterparts. Age and PMI are comparable between two groups, and no significant differences were detected in other phospholipid species. (C) The brain PIP2 levels of female subjects were lower than those of male subjects in cohorts of CDR 0.5–1 (0.25% vs. 0.31%; **P = 0.005 with independent-sample t tests), as well as CDR 3 (0.12% vs. 0.24%; *P = 0.048 with independent-sample t tests). No significant differences were seen in brain PIP2 levels of female versus male subjects from the cohort of CDR 0 (0.33% vs. 0.24%; P = 0.484). (D) ApoE4 genotype-associated PIP2-lowering effects can be observed in female subjects from cohorts of CDR 0.5–1 (ApoE4+/− 0.23% vs. ApoE4−/− 0.27%; **P = 0.007 with independent-sample t tests) and CDR 3 (ApoE4+/− 0.10% vs. ApoE4−/− 0.17%; *P = 0.021 with independent-sample t tests), whereas these effects can only be seen in male subjects in the cohort of CDR 0.5–1 (ApoE4+/− 0.12% vs. ApoE4−/− 0.66%; **P = 0.007 with independent-sample t tests).
Interestingly, ANOVA analysis of all samples reveals a main effect of sex on brain PIP2 homeostasis independent of CDR stage and ApoE genotype (P = 0.039). Brain PIP2 levels of female subjects were lower than those of male subjects in cohorts with CDR 0.5–1 and 3 (Fig. S2C). The reduction of PIP2 levels over the course of disease development and progression was more closely correlated in female subjects (Fig. S2C; r = −0.530) than in the overall population (Fig. 1A; r = −0.293). In addition, ApoE4 genotype-associated effects on PIP2 levels can be observed in female subjects from cohorts with CDR 0.5–1 and 3, whereas these effects can only be seen in the cohort of CDR 0.5–1 male subjects (Fig. S2D). Together, these results suggest that sex-specific effects on brain PIP2 homeostasis may, in addition to ApoE4 genotype, synergistically contribute to AD pathogenesis and disease progression.
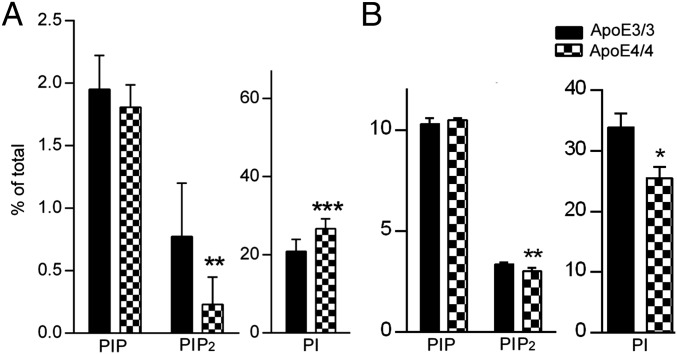
Reduction of PIP2 in ApoE4 human brains was further recapitulated in animal models carrying the human ApoE4 genotype (11–13), as well as in neurons and astrocytes expressing ApoE4. As shown in Fig. 2, the amount of PIP2 was significantly reduced in male ApoE4/4 homozygous KI mouse hippocampal regions at 9 mo of age (0.23%) if compared with ApoE3/3 tissues (0.77%; P = 0.002). PI levels in these mice were elevated (26.72% vs. 20.93% in ApoE3 mice; P < 0.001), similar to what we observed in human brain tissues derived from CDR 0 group (Fig. 1B). No statistically significant changes were seen in levels of other phospholipids. A decrease in PIP2 levels was also seen in 9-mo-old female ApoE4/4 hippocampal brain tissues (Fig. S3A). Trends of PI/PIP2 changes were seen in ApoE4/4 mouse brains at as early as 3 mo of age (whole-brain lipid analysis; Fig. S3B), suggesting the intrinsic effects of ApoE4 on PI metabolism, particularly PIP2 levels, which become more prominent with the ApoE4-induced accelerated aging processes (14–19). Therefore, we speculate that ApoE4-induced PIP2 changes may contribute to ApoE4-induced exacerbation of neurodegenerative processes.
Fig. 2.
Changes in PIP2 levels observed in mouse brain and primary neurons in ApoE4/4 mice. (A) Amounts of PIP2 in hippocampus of male ApoE4/4 homozygous KI mice at 9 mo of age; **P < 0.01, ***P < 0.001 compared with ApoE3/3 male homozygous KI mice with independent-sample t tests. (B) PIP2 levels in primary hippocampal neurons derived from ApoE4/4 or ApoE3/3 homozygous KI mice. *P < 0.05, **P < 0.001 versus ApoE3/3 neurons with independent-sample t tests. The levels of PI were dramatically reduced in ApoE4 neurons (25.45 ± 1.95%) compared with ApoE3 (33.9 ± 2.3%; P = 0.03), suggesting neuron-specific changes in PI/PIP2.
Fig. S3.
Changes in PIP2 levels observed in mouse brain, primary neurons, and astrocytes of ApoE4/4 mice. (A) Amounts of PIP2 in hippocampus of female ApoE4/4 homozygous KI mice at 9 mo of age; *P < 0.05, ***P < 0.001 compared with ApoE3/3 male homozygous KI mice with independent-sample t tests. (B) There were trends toward changes in PI/PIP2 levels in ApoE4/4 mouse brains as early as 3 mo of age (determined by whole-brain lipid analysis). These results suggest an intrinsic effect of ApoE4 on PI metabolism, particularly PIP2, which becomes more prominent with the ApoE4-induced accelerated aging processes. (C) A statistically significant reduction of PIP2 was seen in ApoE4 astrocytes compared with those in ApoE2 or E3 astrocytes (ApoE2 1.23 ± 0.60%, ApoE3 0.74 ± 0.32%, and ApoE4 0.58 ± 0.34%; *P < 0.05 with ANOVA tests).
Moreover, a reduction in PIP2 levels was observed in cultured hippocampal neurons (Fig. 2B) and astrocytes (Fig. S3C) derived from mice expressing human ApoE4 genotype compared with ApoE3 counterparts. A reduction of PIP2 levels was seen in ApoE4 neurons (3.02%) compared with ApoE3 neurons (3.36%; P = 0.008). However, the levels of PI were dramatically reduced in ApoE4 neurons (25.45%) compared with ApoE3 (33.9%; P = 0.03), suggesting neuronal-specific changes in PI/PIP2. A statistically significant reduction of PIP2 was seen in ApoE4 astrocytes compared with in ApoE2 astrocytes, with a lesser decrease of reduction compared with in ApoE3 astrocytes (Fig. S3C; ApoE2, 1.23%; ApoE3, 0.74%; and ApoE4, 0.58%). Together, our data suggest that ApoE4 specifically induces reduction in PIP2 levels.
Expression Levels of a PIP2-Degrading Enzyme, Synaptojanin 1, Are Elevated in ApoE4 Brains and Neurons.
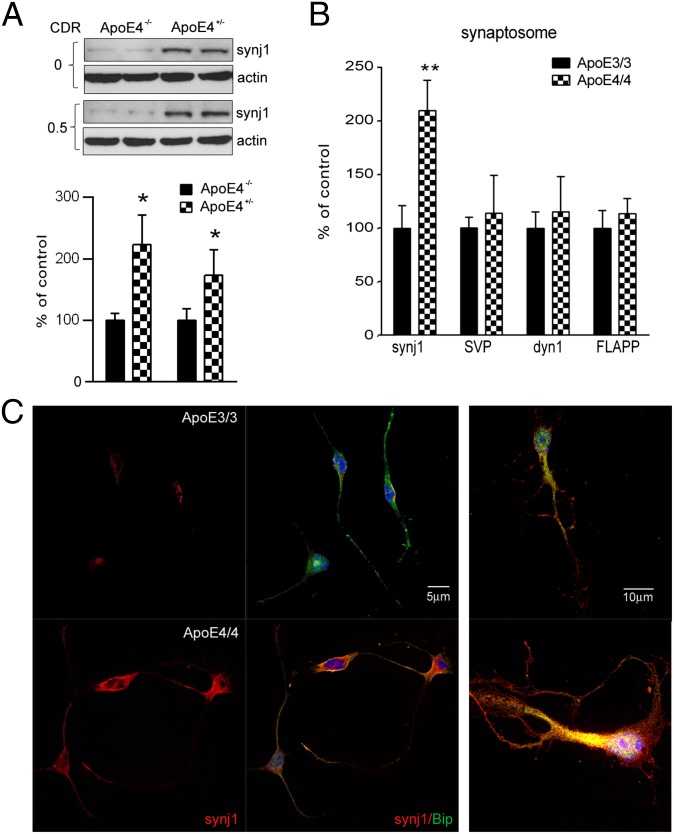
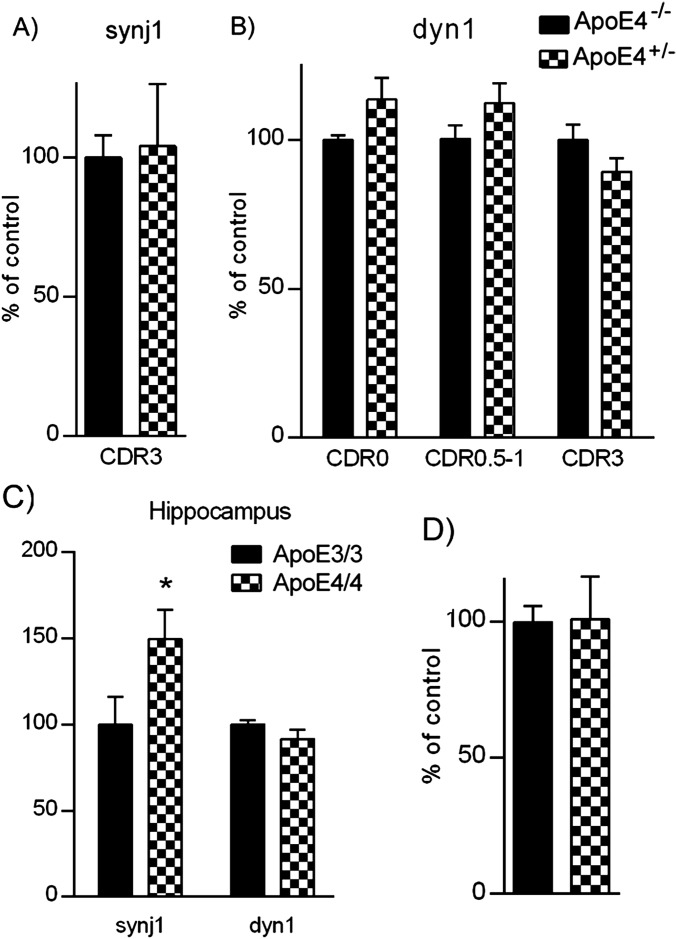
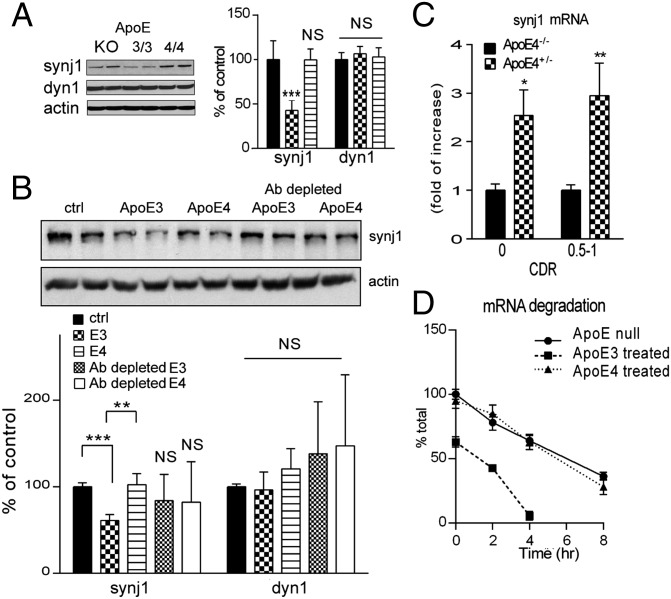
Next, we investigated whether the reduction of PIP2 levels in ApoE4 brain is secondary to increased expression and/or enzymatic activities of the rate-limiting enzyme of brain PIP2 pathway, synaptojanin 1 (synj1). Synj1 is a PIP2-degrading enzyme that is highly enriched in the brain (20, 21). As shown in Fig. 3A, synj1 protein levels in ApoE4+/− subjects were much higher than those in ApoE4−/− subjects. The differences can be seen in both the normal aged cohort (with CDR 0; synj1 levels in ApoE4+/− brains were 223.1% of those in ApoE4−/− brains) and the early AD cohort (with CDR 0.5–1; synj1 levels in ApoE4+/− brains were 173.3% of those in ApoE4−/− brains). However, at the advanced stage (CDR 3), this difference in synj1 protein levels was diminished (Fig. S4A; synj1 levels in ApoE4+/− are 104.1% of levels in ApoE4−/−). There were no differences seen in levels of another endocytic adaptor protein dynamin 1 (dyn1) between ApoE4+/− and ApoE4−/− groups at all stages of AD, suggesting the changes in synj1 are specific (Fig. S4B). Consistently, protein levels of synj1, but not dyn1, in ApoE4/4 mouse neocortical and hippocampal tissues were significantly elevated compared with in ApoE3/3 brains (Fig. S4C). It should be noted that the differences in synj1 expression levels are not yet seen in 3-mo-old ApoE4/4 and E3/3 brains, suggesting a possible compensatory mechanism on regulation of synj1 expression levels at early ages of animals (Fig. S4D).
Fig. 3.
Expression levels of synaptojanin 1 are elevated in ApoE4 brains. (A) The synj1 protein levels in ApoE4+/− brains from CDR 0 and CDR 0.5–1 cohorts. *P < 0.05 versus ApoE4−/− brains with ANOVA tests. (B) Synj1 and dyn1 protein levels in ApoE4/4 mouse hippocampal synaptosomes. *P < 0.05, **P < 0.01 versus ApoE3/3 with independent-sample t tests. (C) Immunofluorescence staining of synj1 (red) and Bip (green) in ApoE4/4 or ApoE3/3 neurons. (Right) Magnified image.
Fig. S4.
Expression levels of synj1 but not dyn1 are elevated in ApoE4 human and mouse brains compared with ApoE3/3 counterparts. (A) At advanced stage (CDR 3), the differences in synj1 protein levels between ApoE4+/− and ApoE4−/− subjects were diminished (synj1 levels in ApoE4+/− were 104.1 ± 22.4% of levels in ApoE4−/−). (B) No differences were seen in levels of another endocytic adaptor protein dyn1 between ApoE4+/− and ApoE4−/− groups at all stages of AD, suggesting the changes in synj1 are specific. (C) The synj1 and dyn1 protein levels in ApoE4/4 male mouse hippocampal tissues are shown. *P < 0.05 versus ApoE3/3. (D) The differences in synj1 expression levels are not yet seen in ApoE4/4 and E3/3 brains at 3 mo of age.
Because synj1 is enriched at the synapse (20, 21), we then analyzed synj1 levels in synaptosomes [validated by a synaptosome marker, synaptic vesicle protein (SVP)] extracted from ApoE4/4 mouse hippocampal brain tissues. As shown in Fig. 3B, the amount of synj1 proteins in ApoE4/4 mouse hippocampal synaptosomes was elevated versus that in ApoE3/3 brains (193.1% of controls; n = 4; P = 0.003; this was more robust compared with changes in ApoE4/4 brain total lysates shown in Fig. S4C). There were no significant changes in dyn1 or total amyloid precursor protein (holoAPP) protein levels in synaptosomes of ApoE4/4 mouse hippocampal brain tissues compared with in ApoE3/3 counterparts. These data suggest that neuronal synj1/PIP2 levels at synaptic terminals are probably more sensitive to changes induced by ApoE isoforms, which could contribute to ApoE4-induced synaptic dysfunction.
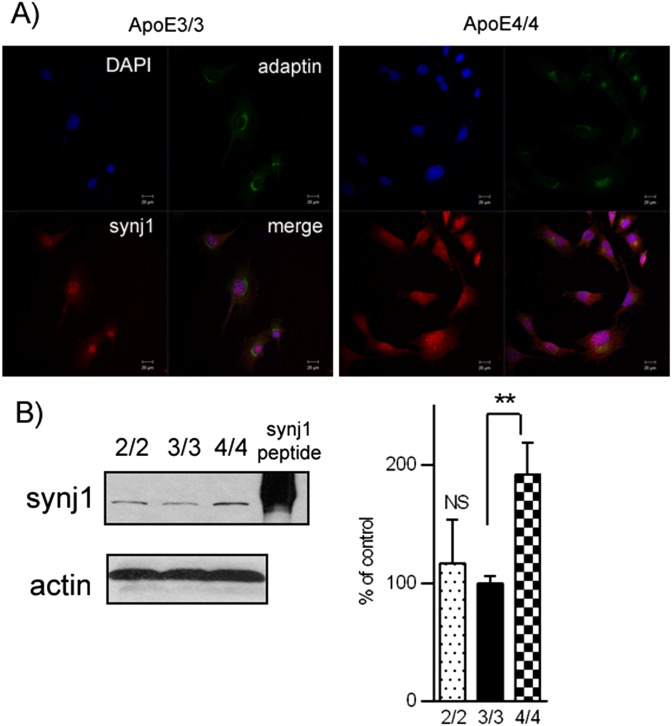
Interestingly, we observed a more prominent increase in synj1 protein levels induced by ApoE4 in neurons (Fig. 3C) than that in astrocytes (Fig. S5A). The amount of synj1 (red fluorescent signals) was increased in ApoE4/4 primary neurons compared with that in ApoE3/3 cells. A magnified image of individual cells showed dramatic differences in synj1 levels at synaptic terminals and neuronal processes, as well as in cell bodies of ApoE4/4 and E3/3 neurons. An endoplasmic reticulum marker, pre-B cell immunoglobulin heavy chain-binding protein Bip, was stained as a control (green fluorescent signals). Similar differences in fluorescent intensity are seen when comparing synj1 levels in ApoE4/4 and E3/3 astrocytes (Fig. S5A). Consistently, the levels of synj1 protein in ApoE4/4 primary neurons were 253.8% of those in ApoE3/3 neurons, whereas synj1 levels in ApoE4/4 astrocytes were 176% of those in ApoE3/3 astrocytes (Fig. S5B). Although the differences in synj1 expression levels in ApoE3/3 versus ApoE4/4 astrocytes were less robust, they were still significant; these changes may play a functional role in differential regulation of astrocyte functions by various ApoE isoforms.
Fig. S5.
Expression levels of synj1 are elevated in ApoE4-expressing astrocytes with their ApoE3 counterparts. (A) In immunofluorescence studies, synj1 levels (red fluorescent signals) were reduced in ApoE4/4 astrocytes relative to ApoE3/3 astrocytes. A Golgi marker, γ-adaptin (labeled as green fluorescent signals) and a nuclear marker DAPI (blue fluorescent signals) were used as controls. (B) The synj1 protein levels in ApoE4/4 astrocytes. **P < 0.01 versus ApoE3/3 astrocytes with independent-sample t tests. Synj1 peptide was used as a loading control.
Loss-of-Function Effects on Synj1 Expression by ApoE4 Genotype.
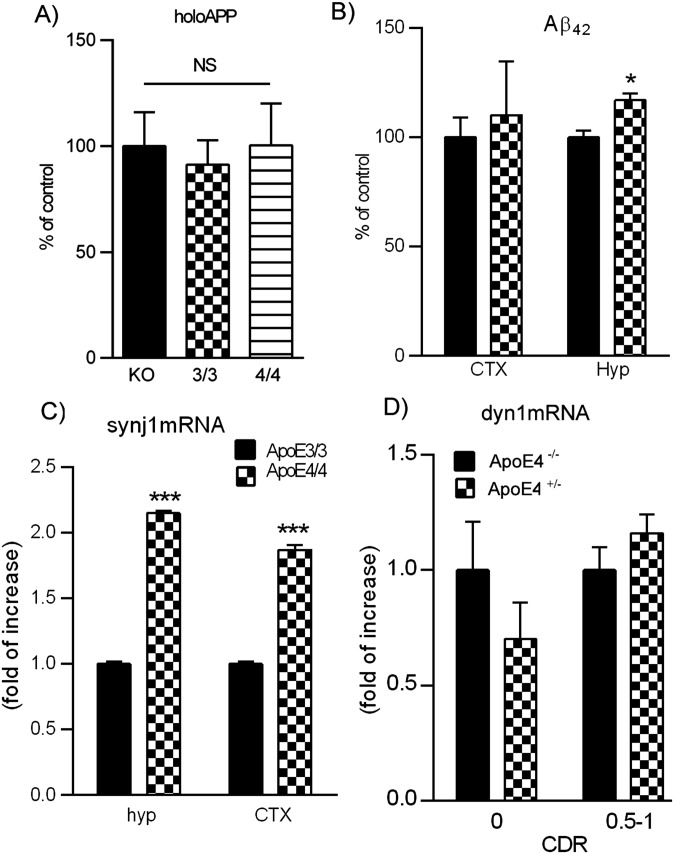
We next investigated whether the differences in synj1 expression levels between ApoE4 versus E3 alleles are secondary to ApoE3-induced down-regulation or ApoE4-induced up-regulation of synj1 expression. First, we compared synj1 levels in ApoE3/3 or ApoE4/4 brains to that of ApoE null mouse brains and found that the levels of synj1 were comparable in ApoE4/4 and ApoE−/− brains, whereas ApoE3/3 brain synj1 levels were much lower (Fig. 4A). These results suggest a loss-of-function effect on synj1 expression by ApoE4 genotype, similar to ApoE knockout conditions. No differences were detected in the amount of dyn1 (Fig. 4A) or holoAPP (Fig. S6A) in ApoE4 versus E3 mouse brain lysates compared with ApoE KO brain lysates. There were modest elevations in endogenous mouse Aβ levels of ApoE4 hippocampal brain lysates compared with ApoE3 counterparts (Fig. S6B). These data are consistent with previous reports that ApoE genotypes do not affect APP processing but instead regulate Aβ degradation (2, 3).
Fig. 4.
Loss-of-function effects on synaptojanin 1 expression by ApoE4 genotype. (A) The synj1 levels were comparable between ApoE4/4 and ApoE−/− mouse brains (E4 99.5 ± 12.3% of controls, with ApoE−/− synj1 levels as 100%), whereas ApoE3/3 mouse brain synj1 levels were much lower (42.8 ± 11.5%; ***P < 0.001 versus ApoE−/− synj1 levels with ANOVA tests). No significant differences were seen in dyn1 levels. (B) Synj1 and dyn1 protein levels in ApoE−/− neurons after exposure to conditioned media containing no ApoE, ApoE3, or ApoE4 with or without immune depletion by an anti-ApoE antibody. E3, 61.1 ± 7.6% of controls; ***P < 0.001 versus ApoE−/− synj1 levels; E4, 102.6 ± 13.3% of controls; **P = 0.01 with ANOVA tests. Antibody-depleted E3 media 83.9 ± 14.2% of controls; P = 0.86; antibody-depleted E4 media 82.3 ± 21.9% of controls; P = 0.60. (C) Synj1 mRNA levels for ApoE4+/− and ApoE4−/− brains from CDR 0 and 0.5–1 cohorts. *P < 0.05, **P < 0.01 versus ApoE4−/− with ANOVA tests. (D) Synj1 mRNA in ApoE−/− neurons exposed to ApoE3 or E4 conditioned media over time after the addition of actinomycin. The slope of synj1 mRNA degradation in ApoE3-treated cells (y = −15.73x + 66.58; R2 = 0.96) was much steeper than that in ApoE4-treated (y = −8.62x + 97.88; R2 = 0.99) or ApoE−/− conditions (y = −7.77x + 96.82; R2 = 0.99).
Fig. S6.
Expression levels of synj1 and other AD-related proteins in ApoE mouse and human brains. (A) The holoAPP levels were comparable between ApoE−/−, ApoE4/4, and ApoE3/3 brain tissues. (B) There was a modest elevation in mouse Aβ42 levels in ApoE4 hippocampal but not neocortical brain lysates compared with ApoE3 counterparts (*P < 0.05 with independent-sample t tests). (C) Synj1 mRNA levels were increased in ApoE4/4 mouse hippocampal and neocortical brain tissues compared with the levels in ApoE3/3 brain tissues (hippocampus: 2.15 ± 0.02 folds of increase; neocortex: 1.87 ± 0.04 fold of increase; ***P < 0.001 with independent-sample t tests). (D) No changes were seen in dyn1 mRNA levels between ApoE4+/− and ApoE4−/− brain tissues of all cohorts.
We next studied the dynamic changes in synj1 expression levels using ApoE null neurons incubated with conditioned media derived from ApoE4 or E3 astrocytes. As shown in Fig. 4B, the baseline synj1 levels in ApoE−/− neurons were rather high, similar to what we observed in ApoE−/− brain tissues (Fig. 4A). Incubation of ApoE−/− neurons with conditioned media derived from ApoE3, but not ApoE4 or ApoE−/− astrocytes, reduced synj1 protein levels (Fig. 4B). The inhibitory effects of ApoE3-containing media on synj1 were almost completely abolished after conditioned media was depleted of ApoE (Fig. 4B). There were no differences in dyn1 levels in neurons treated with various conditions, suggesting the specific regulation of synj1 expression by ApoE isoforms. These results indicate that the ApoE3 isoform suppresses synj1 expression. In contrast, the ApoE4 allele has no effect on synj1, indicating a loss of function for this allele.
The amount of synj1 mRNA was also elevated in ApoE4 human (Fig. 4C) and mouse (Fig. S6C) brains compared with their ApoE3 counterparts, whereas dyn1 mRNA levels were unchanged (Fig. S6D). Conditioned media from ApoE3-expressing astrocytes accelerated the degradation of synj1 mRNA in ApoE−/− neurons (Fig. 4D). The rate of synj1 mRNA degradation, however, was similar in neurons treated with conditioned media from ApoE4 or ApoE−/− astrocyte cultures. These data suggest that ApoE3 down-regulates synj1 expression by promoting degradation of synj1 mRNA. In contrast, ApoE4 lacks this activity and is unable to reduce neuronal synj1 levels.
Synj1 Reduction Can Rescue Cognitive Deficits and Restore PIP2 Homeostasis in ApoE4 KI Mice.
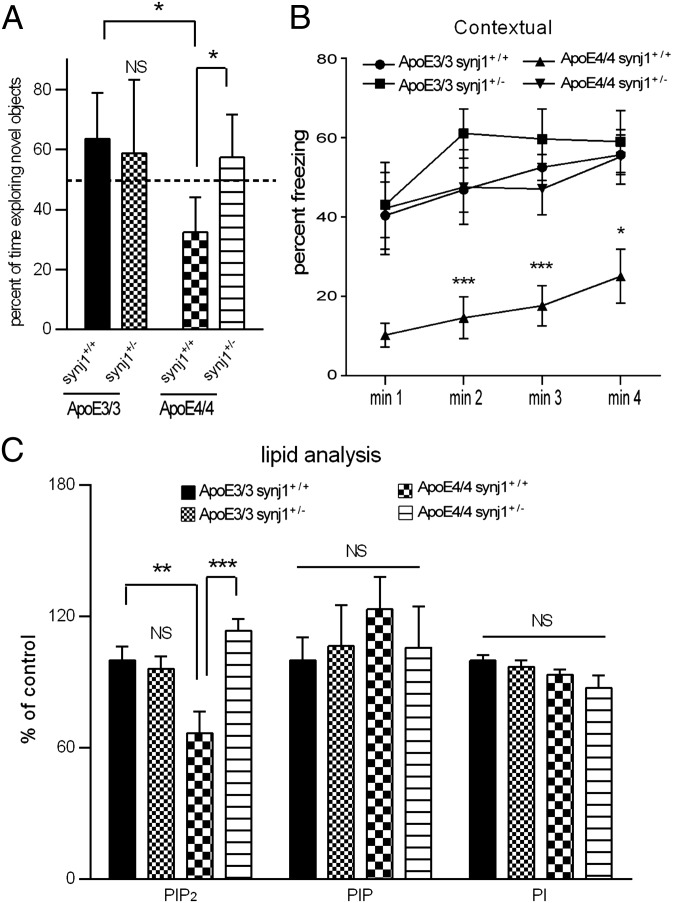
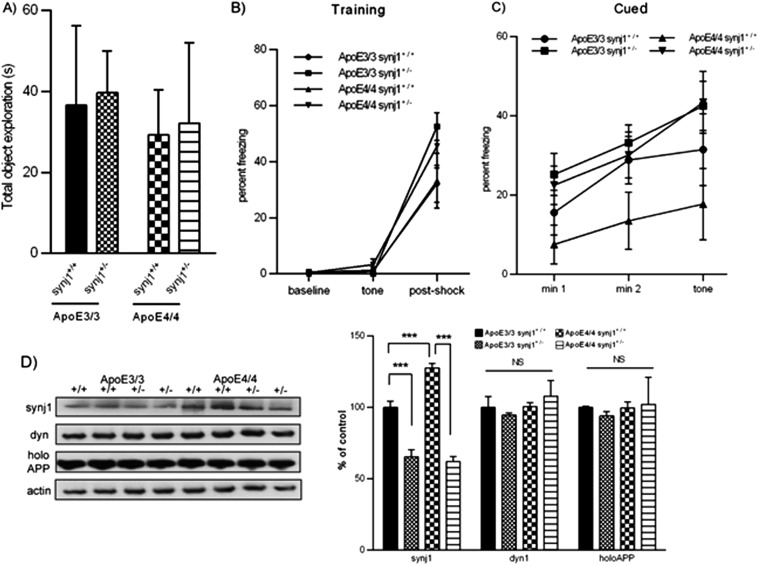
Mice with a human ApoE4 KI background (11–13) spent less time exploring a novel object than ApoE3 KI mice (Fig. 5A), indicating that ApoE4 mice had significant memory impairments including an inability to discriminate novel from familiar objects. This deficit of the ApoE4 KI mice was abolished by genetic knockdown of synj1. The ApoE3+/+ synj1+/− animals behaved similarly to wild-type littermates (ApoE3+/+ synj1+/+). The total amount of time spent in exploring objects (seconds) was comparable among all four genotype groups (Fig. S7A). In the fear conditioning test, all four groups responded similarly in the training phase, showing increased freezing after the foot shock (P < 0.001 baseline vs. postshock; Fig. S7B). On day 2 in the contextual phase, ApoE4+/+ synj1+/+ animals froze less compared with ApoE3+/+ synj1+/+ mice (Fig. 5B), suggesting impaired hippocampal function of ApoE4+/+ synj1+/+ mice. Importantly, this defect was not observed in ApoE4+/+ synj1+/− mice. Furthermore, there were no significant differences in freezing behavior between ApoE3+/+ synj1+/+ and ApoE3+/+ synj1+/− groups. On day 3 in the cued phase, all groups exhibited increased freezing when the tone was presented with the novel context, a result consistent with intact amygdala function (Fig. S7C; P < 0.001 baseline vs. posttone).
Fig. 5.
Reduced synj1 rescues cognitive deficits and restores PIP2 homeostasis in ApoE4 KI mice. (A) The amount of total time spent exploring a novel object (n = 6/group) and (B) the frequency of freezing behavior during the contextual phase of a fear conditioning test (n = 9/group) in four groups of mice: ApoE4+/+ synj1+/+, ApoE4+/+ synj1+/−, ApoE3+/+ synj1+/+, and ApoE3+/+ synj1+/−. *P < 0.05, **P < 0.01, ***P < 0.001 versus ApoE3+/+ synj1+/+ with ANOVA tests. (C) The levels of PI/PIP/PIP2 in mouse hippocampus.
Fig. S7.
Synj1 reduction can rescue cognitive deficits and restore PIP2 homeostasis in ApoE4 KI mice. (A) The total amount of time spent in exploring objects (seconds) was comparable among the four groups (ApoE4+/+ synj1+/+, ApoE4+/+ synj1+/−, ApoE3+/+ synj1+/+, ApoE3+/+ synj1+/−; n = 6/group). (B) All four groups responded similarly in the day 1 training phase of the fear conditioning test, showing increased freezing after the foot shock (P < 0.001 baseline vs. postshock with ANOVA tests; n = 9/group). (C) In the day 3 cued phase of the fear conditioning test, all four groups exhibited increased freezing when the tone was presented with the novel context, a result consistent with intact amygdala function (P < 0.001 baseline vs. posttone with ANOVA tests). In ApoE4+/+ synj1+/+ mice, freezing after the tone was increased to a lesser extent compared with three other groups, but the differences did not achieve statistical significance. (D) The Synj1 haploinsufficiency in ApoE4 mice (ApoE4+/+ synj1+/−) reduced the synj1 levels to those comparable with ApoE3+/+ synj1+/− mouse brains. There were no changes in dyn1 or holoAPP levels with synj1 reduction in either ApoE4 or E3 mice.
The synj1+/− genotype reversed the decreased PIP2 levels in the ApoE4+/+ synj1+/+ mice (Fig. 5C). The amount of synj1 in ApoE3+/+ synj1+/+ mice was significantly lower than that in ApoE4+/+ synj1+/+ mice (Fig. S7D), which is consistent with our prior observations (Figs. 3 and 4). Synj1 haploinsufficiency in ApoE4 mice (ApoE4+/+ synj1+/−) reduced the levels of synj1 to comparable levels in ApoE3+/+ synj1+/− mouse brains. No changes were seen in dyn1 or holoAPP levels with synj1 reduction in ApoE4 or E3 mice. Together, these results suggest that genetic reduction of synj1 in ApoE4 KI mouse models can restore impaired PIP2 homeostasis and rescue AD-related cognitive deficits in these mice.
Discussion
Accumulating evidence indicates that phospholipid regulators including synj1 (22–24), phospholipase D (25), and PI-binding clathrin assembly protein (26) play important roles in pathways leading to neurodegenerative processes such as lipid granule accumulation, endosomal/lysosomal degradation, and regulation of microglia and astrocyte functions. Our data show that ApoE3 increases brain PIP2 levels through promotion of synj1 mRNA degradation and suppression of synj1 expression. In contrast, ApoE4 has lost the ability to regulate synj1 mRNA degradation and protein expression, and thereby ApoE4 reduces brain PIP2 levels. Our findings reveal a previously unidentified molecular mechanism underlying the strong association of ApoE4 with sporadic AD, whereby ApoE4 genotype-specific changes in brain phospholipid homeostasis predispose to AD susceptibility.
Several ApoE functions including Aβ-dependent and Aβ-independent pathways have been proposed for ApoE4-related AD pathogenesis (27). One such pathway may be aberrant metabolism of membrane phospholipids. Recent literature, including data from our laboratory, suggests that PI homeostasis, particularly PIP2, plays an important role in AD (10, 22, 24, 28, 29). For example, one study demonstrated PIP2 reduction in the prefrontal cortex of patients with AD (29). Here, our phospholipid analysis of human brain tissues is the first to our knowledge to demonstrate a specific pattern of changes in PI metabolism (including reduction of PI and PIP2, and a reciprocal increase in PIP) that correlates with disease conversion from normal aging to MCI and early AD. More important, our data suggest that ApoE4 genotype exacerbates the changes in PIP2 levels in particular, along with disease development and progression. It is interesting that reduced PIP2 is found even in normal aged ApoE4 carriers (CDR 0; Fig.1), suggesting that impaired brain phospholipid metabolism precedes the development of overt AD clinical symptoms. Reduced synj1 rescues cognitive deficits in ApoE4 KI mice, which lack amyloid pathology (Fig. 5), suggesting an Aβ-independent mechanism underlying ApoE4-induced cognitive impairments. Sex-specific effects on brain PIP2 homeostasis are also observed in our analysis (Fig. S2 C and D), consistent with recent evidence showing that compared with men, women’s brains are more vulnerable to AD pathological processes, and cognitive functions decline faster (30, *, †).
A recent paper reported changes in serum levels of 10 phospholipids (including PI) as predicting phenotypic conversion to either amnestic MCI or early AD in 2–3 y intervals with more than 90% accuracy (9). Despite the fact that this study was performed in a rather small-sized patient cohort and did not have cerebrospinal fluid analysis of lipidomics or brain imaging studies to correlate with their serum biomarker studies, our findings in addition to this report suggest a potential importance of assessing specific phospholipid profiling as AD biomarkers in predicting conversion and progression of disease. In contrast, the reduction of PI levels has a much stronger correlation with AD disease development and progression, suggesting a putative functional role for this phospholipid in the structure and function of cell membranes, which are most likely disrupted at early stages of AD. However, it should be noted that the changes in PI metabolism are prominently seen in early disease progression (from CDR 0 to 1; Fig. 1), but not much so at the advanced stage (CDR 3; Fig. S2). It is possible that at advanced AD stages, the severity of disrupted membrane integrity may confound any specific patterns of changes in phosphoinositol metabolism and become less indicative for disease progression at later stages.
Our data showing a reduction of PIP2 and a reciprocal increase in substrate PIP/PI levels along AD phenotype conversion suggest a possibility of accelerated PIP2 degradation processes (such as elevated PIP2 degrading enzyme synj1 levels), rather than impaired PIP2 synthesis. PIP2 is a signaling lipid involved in ion channel regulation, exocytosis, endocytosis, actin cytoskeleton rearrangement, and cell signaling (20). PIP2 dephosphorylation at the synapse is mediated primarily by synj1, a type 2 polyphosphate-5-phosphatase encoded by a gene present on human chromosome 21. Synj1 is expressed and enriched at the synapse and has been shown to play a role in endocytosis, presynaptic vesicle recycling, and postsynaptic receptor trafficking (20, 21). A potential link between AD and synj1 was suggested by studies demonstrating that familial AD mutations in the phosphoserine 1 result in a perturbation of PIP2 metabolism. Overexpression of synj1 led to an increase in Aβ production (23). In addition, one of the yeast orthologs of synj1, INP52, was identified in an unbiased genome-wide screen for modifiers of Aβ toxicity (31). We have recently shown that genetically reducing synj1 promoted Aβ degradation through lysosomes and ameliorated memory deficits in an AD transgenic mouse model (22). Here our data are the first, to our knowledge, to show specific changes in PIP2 and synj1 levels induced by ApoE4 genotype. More important, we demonstrate that differential expression in synj1 levels may contribute to ApoE4-induced AD pathological process. We speculate that one pathogenic mechanism of ApoE4 is to perturb neuronal, and in particular synaptic, PIP2 homeostasis by altering synj1 expression at synapse (Fig. 3), which could contribute to the development of ApoE4-induced cognitive deficits and synaptic dysfunction.
Interestingly, the regulation of synj1 expression levels by ApoE isoforms is mediated by differential regulation of synj1 mRNA degradation rates in cells expressing various ApoE genotypes. As shown in Fig. 4, ApoE3 degrades synj1 mRNA rapidly (its half-life is ∼2 h), whereas ApoE4 fails to do so. In contrast, synj1 protein degradation rates are comparable between different ApoE isoforms, given the fact that synj1 protein half-life is usually long (∼16 h). The mRNA stability is often regulated by microRNA binding to a 3′-UTR regions (32). We have performed microRNA array studies and found distinct changes in certain microRNA levels between ApoE4 and ApoE3 samples. These results give credence to the potential involvement of and regulation of synj1 expression by ApoE isoforms through microRNA.
Overall, our studies implicate a novel therapeutic strategy targeted at PIP2 homeostasis to modify the ApoE4 pathogenic phenotypes. As demonstrated by genetic reduction of synj1 in ApoE4 mice, we are able to restore impaired PIP2 homeostasis and rescue cognitive deficits in an ApoE4 mouse model. Therefore, pharmacological interventions to reduce synj1 expression levels and/or phosphatase activities could achieve similar functional outcomes in ApoE4 conditions. Several current therapeutic strategies have been focused on removing or ablating ApoE4, based on the observations that ApoE4 contributes to AD-related neurodegenerative processes and that total absence of ApoE proteins has no significant neurocognitive deficits (33, 34). However, our data suggest that ApoE is critical to maintain brain phospholipid homeostasis and that ApoE4 exerts less of this effect than other isoforms. For example, ApoE4 demonstrates a loss-of-function effect on regulating synj1 mRNA stability and protein expression, similar to ApoE null condition. These data raise a concern for therapeutic strategies directed at reduction of brain ApoE levels, which may not be sufficient to rescue ApoE4-related phospholipid dysregulation because of loss-of-function effects on phospholipid homeostasis induced by ApoE4 isoform.
In summary, our findings represent the first mechanistic studies to our knowledge that link ApoE4 genotype-specific changes in brain phospholipid homeostasis with ApoE4-dependent increased susceptibility to develop AD. These studies may uncover new therapeutic directions for the treatment of sporadic AD.
Methods
Human Brain Sample Selection and Preparation.
Equal amounts of postmortem brain tissues (50 μg by net weight) from parietal cortex regions were obtained from the Icahn School of Medicine at Mount Sinai Brain Bank and used for lipid analysis and mRNA/protein expression studies.
Phospholipid Analysis.
Human or mouse brain samples were used for lipid extraction, followed by the quantification by anion-exchange HPLC, as described previously (22, 35).
Generation of Synj1 Haploinsufficiency Mice with Human ApoE Background.
The human ApoE4 or ApoE3 KI mouse models (12, 13, 36) were mated with heterozygous synj1 null mice (synj1+/−) (37). Double heterozygous F1s were then bred with F0 ApoE4/4+/+ or ApoE3/3+/+ mice to generate offspring that express human ApoE4/4 or E3/3 in the synj1+/− background. Genotypes were determined by PCR amplification, as described (37).
Immunocytochemical Studies.
Hippocampal neurons were obtained from 17-d-old mouse embryos and grown for 7 d in vitro, as described (38), before being fixed and stained for confocal microscopy analysis (LSM510) (39).
Brain and Neuronal Lysate Preparation and Analysis.
Snap-frozen mouse hemibrains or cultured neurons were harvested in lysis buffer (40) and processed via step-wise solubilization (40, 41), followed by SDS/PAGE to determine levels of synj1, dyn1, holoAPP, and CTFs. Levels of Aβ42 were determined by high-sensitive mouse Aβ42 ELISA kits (Wako). Alternatively, homogenates of mouse brains were fractionated via sucrose gradient to collect synaptosomes according to Huttner et al. (42).
mRNA Extraction and Degradation.
Synj1 mRNA synthesis were determined by quantitative real time-PCR. The half-life of synj1 mRNA was determined by treatment of actinomycin at 50 μg/mL for various periods before being subjected to RNA extraction (43, 44).
Behavior Studies.
Six- to 9-mo-old male ApoE4 synj1+/+ and synj1+/− mice were tested with the novel object recognition memory task (45) and fear conditioning studies, as described (46).
Antibodies and Reagents.
The anti-synj1 (mouse human Ab, Novus), anti-Bip and β actin (Santa Cruz), anti-MAB348 and clone 41 (Millipore), anti-mouse and rabbit HRP, Texas-Red or Alexa488 conjugated anti-mouse and rabbit IgG (Vector Laboratories Inc.) were purchased. pAb369 (C-terminal APP antibody) was used to detect human and mouse holo-APP and CTFs (47).
Statistical Analysis.
Levels of synj1, dyn1, and holoAPP were normalized to β-actin levels and expressed as percentage of control. Absolute Aβ42 concentrations were quantitatively determined by ELISA (Wako) and expressed as percentage of control. Independent sample t tests were used to determine significant mean differences (the threshold for significance set at P < 0.05). The ANOVA with post hoc tests were used to determine group differences for multiple comparisons. The Pearson correlation coefficients were measured to determine the linear relationship between two variables. All statistical analysis was performed using SPSS v21.0.
Acknowledgments
We thank Dr. Pietro De Camilli (Yale School of Medicine) for providing synj1 haploinsufficiency mice. Antibody p369 was generously provided by Dr. Paul Greengard. We also thank Dr. William Netzer (The Rockefeller University) and Dr. Michaela Kiernan (Stanford University) for critical reading of the manuscript. This work was supported in whole or in part by funding from the Department of Veteran Affairs Basic Life Science Research & Development CDA (Award CAI-spring 2010); Alzheimer Association (Award NIRP14-304720); Department of Veteran Affairs Rehabilitation Research & Development Small Projects in Rehabilitation Research (Grant 1I21RX001558-01A1); and NIH R01 (Grant 1R01AG048923-01) (to D.C.), NIH R37 (Grant AG017926) and R01 (Grant 2R01AG008200) (to N.K.R.), NIH UO1 (Grant AG046170), P50 (Grant AG005138), R34 (Grant AG049649), and RO1 (Grant NS075685) (to S.G.); as well as Department of Veterans Affairs RR&D National Center for Excellence for the Medical Consequences of Spinal Cord Injury (Grant B9212C) (to C.C.). Confocal microscopy studies were supported by the James J. Peters VA Medical Center Research Core Facility.
Footnotes
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510011112/-/DCSupplemental.
*Yaffe K, et al. Alzheimer's Association International Conference, July 24–28, 2015, Washington, DC, 3121 (abstr).
†Tosun D, et al. Alzheimer's Association International Conference, July 24–28, 2015 Washington, DC, 5556 (abstr).
References
- 1.Mayeux R. Epidemiology of neurodegeneration. Annu Rev Neurosci. 2003;26:81–104. doi: 10.1146/annurev.neuro.26.043002.094919. [DOI] [PubMed] [Google Scholar]
- 2.Castellano JM, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med. 2011;3(89):89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, et al. Differential regulation of amyloid-β endocytic trafficking and lysosomal degradation by apolipoprotein E isoforms. J Biol Chem. 2012;287(53):44593–44601. doi: 10.1074/jbc.M112.420224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holtzman DM, Herz J, Bu G. Apolipoprotein E and apolipoprotein E receptors: Normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2(3):a006312. doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan RB, et al. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J Biol Chem. 2012;287(4):2678–2688. doi: 10.1074/jbc.M111.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanfer JN, Pettegrew JW, Moossy J, McCartney DG. Alterations of selected enzymes of phospholipid metabolism in Alzheimer’s disease brain tissue as compared to non-Alzheimer’s demented controls. Neurochem Res. 1993;18(3):331–334. doi: 10.1007/BF00969091. [DOI] [PubMed] [Google Scholar]
- 7.Pettegrew JW, Panchalingam K, Hamilton RL, McClure RJ. Brain membrane phospholipid alterations in Alzheimer’s disease. Neurochem Res. 2001;26(7):771–782. doi: 10.1023/a:1011603916962. [DOI] [PubMed] [Google Scholar]
- 8.Klunk WE, Panchalingam K, McClure RJ, Stanley JA, Pettegrew JW. Metabolic alterations in postmortem Alzheimer’s disease brain are exaggerated by Apo-E4. Neurobiol Aging. 1998;19(6):511–515. doi: 10.1016/s0197-4580(98)00105-5. [DOI] [PubMed] [Google Scholar]
- 9.Mapstone M, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20(4):415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman DE, et al. Oligomeric amyloid-beta peptide disrupts phosphatidylinositol-4,5-bisphosphate metabolism. Nat Neurosci. 2008;11(5):547–554. doi: 10.1038/nn.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gad H, et al. Fission and uncoating of synaptic clathrin-coated vesicles are perturbed by disruption of interactions with the SH3 domain of endophilin. Neuron. 2000;27(2):301–312. doi: 10.1016/s0896-6273(00)00038-6. [DOI] [PubMed] [Google Scholar]
- 12.Grootendorst J, et al. Human apoE targeted replacement mouse lines: H-apoE4 and h-apoE3 mice differ on spatial memory performance and avoidance behavior. Behav Brain Res. 2005;159(1):1–14. doi: 10.1016/j.bbr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 13.Wang C, et al. Human apoE4-targeted replacement mice display synaptic deficits in the absence of neuropathology. Neurobiol Dis. 2005;18(2):390–398. doi: 10.1016/j.nbd.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Caselli RJ, et al. Longitudinal modeling of frontal cognition in APOE ε4 homozygotes, heterozygotes, and noncarriers. Neurology. 2011;76(16):1383–1388. doi: 10.1212/WNL.0b013e3182167147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caselli RJ, et al. Longitudinal modeling of age-related memory decline and the APOE epsilon4 effect. N Engl J Med. 2009;361(3):255–263. doi: 10.1056/NEJMoa0809437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caselli RJ, et al. Longitudinal changes in cognition and behavior in asymptomatic carriers of the APOE e4 allele. Neurology. 2004;62(11):1990–1995. doi: 10.1212/01.wnl.0000129533.26544.bf. [DOI] [PubMed] [Google Scholar]
- 17.Wolf AB, et al. Apolipoprotein E as a β-amyloid-independent factor in Alzheimer’s disease. Alzheimers Res Ther. 2013;5(5):38. doi: 10.1186/alzrt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caselli RJ. Age-related memory decline and apolipoprotein E e4. Discov Med. 2009;8(41):47–50. [PubMed] [Google Scholar]
- 19.Caselli RJ, et al. Cognitive domain decline in healthy apolipoprotein E epsilon4 homozygotes before the diagnosis of mild cognitive impairment. Arch Neurol. 2007;64(9):1306–1311. doi: 10.1001/archneur.64.9.1306. [DOI] [PubMed] [Google Scholar]
- 20.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 21.McPherson PS, et al. A presynaptic inositol-5-phosphatase. Nature. 1996;379(6563):353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- 22.Zhu L, et al. Reduction of synaptojanin 1 accelerates Aβ clearance and attenuates cognitive deterioration in an Alzheimer mouse model. J Biol Chem. 2013;288(44):32050–32063. doi: 10.1074/jbc.M113.504365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landman N, et al. Presenilin mutations linked to familial Alzheimer’s disease cause an imbalance in phosphatidylinositol 4,5-bisphosphate metabolism. Proc Natl Acad Sci USA. 2006;103(51):19524–19529. doi: 10.1073/pnas.0604954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIntire LB, et al. Reduction of synaptojanin 1 ameliorates synaptic and behavioral impairments in a mouse model of Alzheimer’s disease. J Neurosci. 2012;32(44):15271–15276. doi: 10.1523/JNEUROSCI.2034-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruchaga C, et al. UK Brain Expression Consortium; Alzheimer’s Research UK Consortium Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505(7484):550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat Rev Neurol. 2013;9(2):106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voronov SV, et al. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down’s syndrome. Proc Natl Acad Sci USA. 2008;105(27):9415–9420. doi: 10.1073/pnas.0803756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morel E, et al. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat Commun. 2013;4:2250. doi: 10.1038/ncomms3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heise V, et al. Apolipoprotein E genotype, gender and age modulate connectivity of the hippocampus in healthy adults. Neuroimage. 2014;98:23–30. doi: 10.1016/j.neuroimage.2014.04.081. [DOI] [PubMed] [Google Scholar]
- 31.Treusch S, et al. Functional links between Aβ toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011;334(6060):1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 33.Mak AC, et al. Effects of the absence of apolipoprotein e on lipoproteins, neurocognitive function, and retinal function. JAMA Neurol. 2014;71(10):1228–1236. doi: 10.1001/jamaneurol.2014.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potter H, Wisniewski T. Apolipoprotein e: Essential catalyst of the Alzheimer amyloid cascade. Int J Alzheimers Dis. 2012;2012:489428. doi: 10.1155/2012/489428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nasuhoglu C, et al. Nonradioactive analysis of phosphatidylinositides and other anionic phospholipids by anion-exchange high-performance liquid chromatography with suppressed conductivity detection. Anal Biochem. 2002;301(2):243–254. doi: 10.1006/abio.2001.5489. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez GA, Burns MP, Weeber EJ, Rebeck GW. Young APOE4 targeted replacement mice exhibit poor spatial learning and memory, with reduced dendritic spine density in the medial entorhinal cortex. Learn Mem. 2013;20(5):256–266. doi: 10.1101/lm.030031.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cremona O, et al. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 1999;99(2):179–188. doi: 10.1016/s0092-8674(00)81649-9. [DOI] [PubMed] [Google Scholar]
- 38.Xu H, et al. Estrogen reduces neuronal generation of Alzheimer beta-amyloid peptides. Nat Med. 1998;4(4):447–451. doi: 10.1038/nm0498-447. [DOI] [PubMed] [Google Scholar]
- 39.Berg I, Nilsson KP, Thor S, Hammarström P. Efficient imaging of amyloid deposits in Drosophila models of human amyloidoses. Nat Protoc. 2010;5(5):935–944. doi: 10.1038/nprot.2010.41. [DOI] [PubMed] [Google Scholar]
- 40.Lane RF, et al. Diabetes-associated SorCS1 regulates Alzheimer’s amyloid-beta metabolism: Evidence for involvement of SorL1 and the retromer complex. J Neurosci. 2010;30(39):13110–13115. doi: 10.1523/JNEUROSCI.3872-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawarabayashi T, et al. Age-dependent changes in brain, CSF, and plasma amyloid (beta) protein in the Tg2576 transgenic mouse model of Alzheimer’s disease. J Neurosci. 2001;21(2):372–381. doi: 10.1523/JNEUROSCI.21-02-00372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huttner WB, DeGennaro LJ, Greengard P. Differential phosphorylation of multiple sites in purified protein I by cyclic AMP-dependent and calcium-dependent protein kinases. J Biol Chem. 1981;256(3):1482–1488. [PubMed] [Google Scholar]
- 43.Cuadrado A, García-Fernández LF, Imai T, Okano H, Muñoz A. Regulation of tau RNA maturation by thyroid hormone is mediated by the neural RNA-binding protein musashi-1. Mol Cell Neurosci. 2002;20(2):198–210. doi: 10.1006/mcne.2002.1131. [DOI] [PubMed] [Google Scholar]
- 44.Tani H, Akimitsu N. Genome-wide technology for determining RNA stability in mammalian cells: Historical perspective and recent advantages based on modified nucleotide labeling. RNA Biol. 2012;9(10):1233–1238. doi: 10.4161/rna.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howlett DR, et al. Cognitive correlates of Abeta deposition in male and female mice bearing amyloid precursor protein and presenilin-1 mutant transgenes. Brain Res. 2004;1017(1-2):130–136. doi: 10.1016/j.brainres.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 46.Elder GA, et al. Blast exposure induces post-traumatic stress disorder-related traits in a rat model of mild traumatic brain injury. J Neurotrauma. 2012;29(16):2564–2575. doi: 10.1089/neu.2012.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buxbaum JD, et al. Processing of Alzheimer beta/A4 amyloid precursor protein: Modulation by agents that regulate protein phosphorylation. Proc Natl Acad Sci USA. 1990;87(15):6003–6006. doi: 10.1073/pnas.87.15.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]