Significance
A successful HIV-1 vaccine should generate an immune response capable of neutralizing the enormous diversity of globally circulating viruses. Here, we report the discovery and characterization of two clade C recombinant envelope glycoprotein trimers with native-like structural and antigenic properties, including epitopes for all known classes of broadly neutralizing antibodies (bnAbs). Together with previously described trimers from other clades, these two new trimers will aid in immunization strategies designed to induce bnAbs to HIV-1.
Keywords: HIV-1 envelope, neutralizing antibodies, antigenic diversity, clade C trimers, HIV immunogens
Abstract
A key challenge in the quest toward an HIV-1 vaccine is design of immunogens that can generate a broadly neutralizing antibody (bnAb) response against the enormous sequence diversity of the HIV-1 envelope glycoprotein (Env). We previously demonstrated that a recombinant, soluble, fully cleaved SOSIP.664 trimer based on the clade A BG505 sequence is a faithful antigenic and structural mimic of the native trimer in its prefusion conformation. Here, we sought clade C native-like trimers with comparable properties. We identified DU422 and ZM197M SOSIP.664 trimers as being appropriately thermostable (Tm of 63.4 °C and 62.7 °C, respectively) and predominantly native-like, as determined by negative-stain electron microscopy (EM). Size exclusion chromatography, ELISA, and surface plasmon resonance further showed that these trimers properly display epitopes for all of the major bnAb classes, including quaternary-dependent, trimer-apex (e.g., PGT145) and gp120/gp41 interface (e.g., PGT151) epitopes. A cryo-EM reconstruction of the ZM197M SOSIP.664 trimer complexed with VRC01 Fab against the CD4 binding site at subnanometer resolution revealed a striking overall similarity to its BG505 counterpart with expected local conformational differences in the gp120 V1, V2, and V4 loops. These stable clade C trimers contribute additional diversity to the pool of native-like Env immunogens as key components of strategies to induce bnAbs to HIV-1.
Vaccines have successfully stopped or limited infections by preventing pathogen transmission and have thereby contributed toward eradication of deadly diseases (1). Nonetheless, successful vaccination against highly variable pathogens, such as enveloped RNA viruses, remains a daunting task. For example, vaccination against influenza virus is only modestly effective because constant antigenic drift necessitates yearly updates of the vaccine (2). HIV (HIV-1) is also an RNA virus with a high mutation rate that creates extensive sequence diversity and facilitates immune evasion (3, 4). During HIV-1 infection, immune responses are often narrowly focused against a small subset of viruses (5). The development of broadly neutralizing antibodies (bnAbs) is relatively rare (5–15% of infections), and usually only arises after antigenic stimulation by constantly evolving viral variants over the course of 1–3 y or longer (6, 7). A vaccine that confers protection against the global diversity of HIV-1 is highly challenging and, as yet, an unachieved goal (8).
Neutralizing antibodies (nAbs) are a correlate of protection for almost all successful vaccines (9, 10). The envelope glycoprotein (Env) is the only HIV-1–encoded surface protein and, as such, constitutes the sole bnAb target. A major problem in vaccine development is the intrinsic metastability of Env that is essential for its membrane-fusion function (11). To mediate cell entry, the Env trimer must undergo a series of complex, receptor-triggered conformational changes within and between its noncovalently associated subunits, gp120 and gp41, to progress from prefusion to fusion-active to postfusion forms (12). How to create stable recombinant Env proteins in a prefusion conformation relevant for bnAb recognition and elicitation has been an area of intensive research.
We have previously described a cleaved, stabilized Env trimer from a clade A isolate, termed BG505 SOSIP.664 (13). This SOSIP trimer represents the paradigm for display of all known bNAb epitopes (except the membrane proximal external region, MPER, which was omitted due to its hydrophobicity), with minimal exposure of most non-nAb epitopes (14–16). This BG505 trimer has been extensively characterized and validated structurally in complexes with at least one member of each main bnAb class [trimer apex, PG9 (13); N332-epitope, PGT122 (17, 18); CD4 binding site, PGV04 (19); and cleaved gp41/gp120 interface, PGT151 (20)]. This Env trimer was recently shown to elicit strong, but narrow, nAb responses in animals against the autologous, neutralization-resistant (tier-2) virus (21).
It is unlikely that the BG505 SOSIP.664 trimer by itself will achieve the goal of inducing bnAbs against heterologous tier-2 viruses (21). That task may be beyond the capabilities of any single Env protein. One possible strategy to drive a broader response is to use a diverse set of trimers derived from different HIV-1 isolates/clades, analogous to the trivalent/tetravalent seasonal influenza vaccines. The challenge an HIV vaccine faces is much more onerous, however, because of the far greater global sequence diversity of HIV-1 (3).
We recently described the production and in vitro characterization of another native-like trimer, B41 SOSIP.664 (clade B) (16) to complement its clade A counterpart. As an immunogen, the B41 trimer also generated a strong, but narrow, tier-2 autologous nAb response (21). Here, we sought to identify additional trimers with comparable properties but based on clade C sequences. Clade C is responsible for over 50% of all new infections worldwide, predominantly in sub-Saharan Africa—the epicenter of the AIDS pandemic (22). Neutralization profiles of clade C viruses suggest their trimers may differ subtly from other clades. For example, despite possessing all putative N-linked glycans associated with the 2G12 epitope, many clade C viruses are resistant to this bnAb (23). Thus, clade C trimers may have atypical structural or glycosylation properties.
Here, we screened a panel of 15 clade C env sequences to seek SOSIP.664 trimers with high yield, good thermal stability, native-like antigenicity (display of bnAb epitopes, occlusion of non-nAb epitopes), and native-like structure. We found two trimers, DU422 and ZM197M, with these desired properties. Cryo-electron microscopy (cryo-EM) of ZM197M SOSIP.664 in complex with VRC01 Fab confirmed that the trimers are native-like and provided valuable insights into the distinct neutralization profiles of clade C viruses. These high-quality clade C trimers can now contribute to vaccine strategies aimed at inducing a bnAb response.
Results
Expression and Thermostability of Clade C SOSIP.664 Trimers.
Fifteen clade C env sequences were engineered to incorporate SOSIP modifications, as well as N-linked glycans, to optimize bnAb epitopes (SI Appendix, SI Materials and Methods). The proteins were purified on a Galanthus nivalis lectin (GNL) column to isolate Env, followed by size exclusion chromatography (SEC) to purify trimers. In this initial screening, we used GNL and not the 2G12 bnAb column routinely used for clade A and B trimers because clade C Env proteins are often unreactive with 2G12 and the epitope cannot be reconstituted simply by knocking in N-linked glycan sites associated with 2G12 reactivity (24).
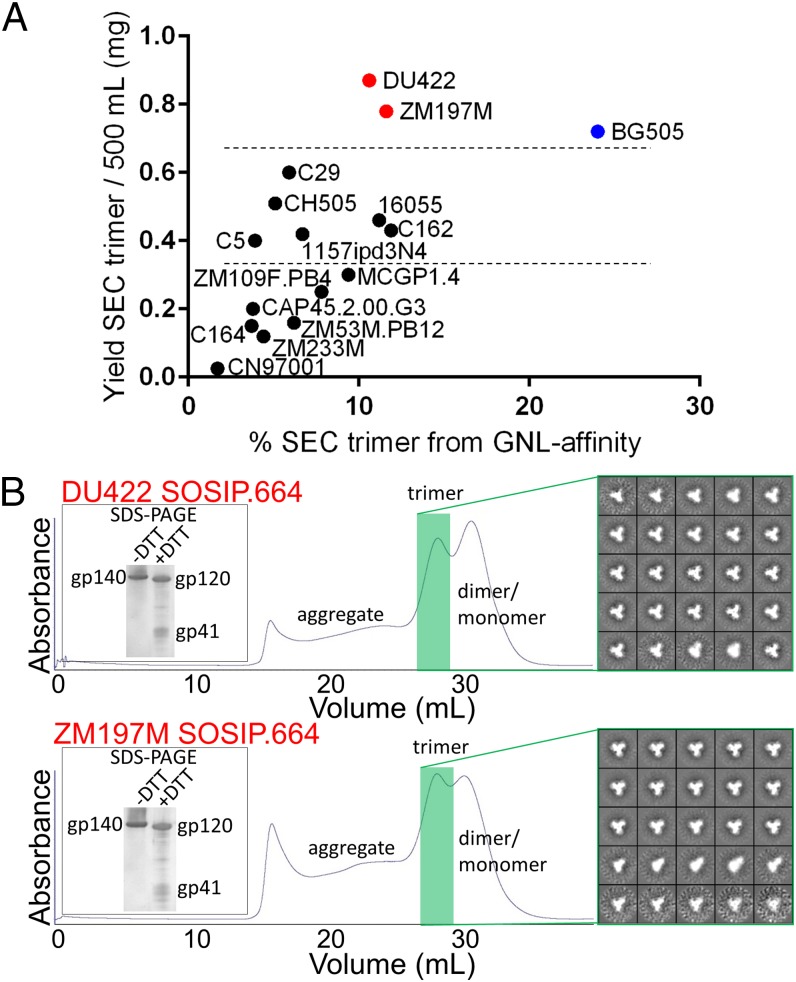
The percentage of Env proteins that formed trimers was assessed from SEC profiles based on elution volumes that corresponded to a molar mass of three gp140 protomers (Fig. 1A). All 15 clade C SOSIP.664 constructs formed trimers, but at yields that varied widely (∼50–900 µg per 500 mL culture). The DU422 and ZM197M trimer yields (∼900 and ∼800 µg, respectively) were comparable to BG505 trimers purified by 2G12/SEC (∼750 µg) (Fig. 1A), despite their relatively low propensity to form trimers (11% and 12% of total GNL-purified protein, respectively), and a corresponding increase in unwanted gp140 dimers/monomers and aggregates (Fig. 1B). The trimer yields therefore reflect a high level of overall Env expression, rather than unusually efficient trimer formation (Fig. 1 A and B). We note that GNL purification might be associated with more contaminants than positive selection on 2G12-affinity columns and, therefore, reduce the proportional yield of trimers. After SEC purification, the DU422 and ZM197M SOSIP.664 trimers adopted predominantly native-like conformations when analyzed by negative-stain electron microscopy (NS-EM) (Fig. 1B), in contrast to aberrant structures, as for uncleaved, non-SOSIP Env trimer constructs (25).
Fig. 1.
Expression of clade C SOSIP.664 trimers. (A) Expression yields in 293S cells and post-SEC recovery of 15 clade C trimers purified by GNL-affinity chromatography. The clade A BG505 trimer produced in this same cell line and purified by 2G12-affinity chromatography is shown in blue for comparison. Dotted lines separate the different constructs into tiers based on expression levels. (B) SEC profiles for the GNL-affinity purified DU422 and ZM197M samples. The shaded green area indicates trimers. The DU422 and ZM197M trimers are efficiently cleaved (Left Inset, showing dissociation of gp140 into gp120 and gp41 subunits in SDS/PAGE upon addition of DTT) and predominantly adopt a native-like Env conformation as assessed by NS-EM (Right Inset, showing representative 2D-class averages of the trimers).
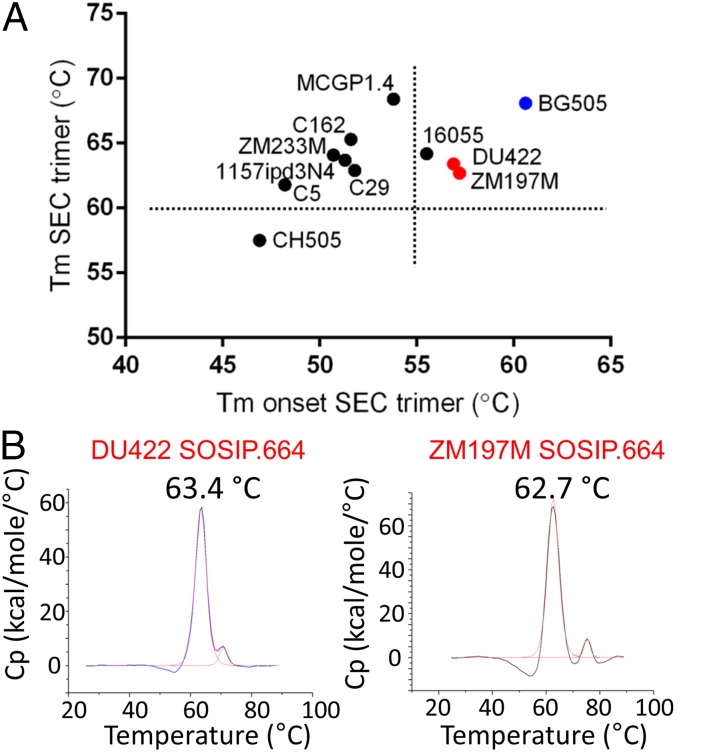
As high stability is a desirable characteristic of native-like trimers, we used differential scanning calorimetry (DSC) to determine the thermostability of the ten SEC-purified clade C SOSIP.664 trimers that could be produced in sufficient quantity (>250 µg/500 mL culture). Two DSC parameters are particularly relevant: melting temperature (Tm) and onset temperature of melting (Tonset). We hypothesized that trimers with high values for both parameters would preferentially adopt a fully closed, prefusion conformation and, hence, have optimal antigenic and immunogenic properties (16, 26). The Tm values for the 10 trimers spanned a wide range, 57.4 °C–68.4 °C, and Tonset was also variable, 46.9 °C–57.2 °C (Fig. 2A). No clade C trimer had an overall thermostability profile as good as BG505 SOSIP.664 (Tm = 68.1 °C; Tonset = 60.9 °C). The most thermostable trimers, judged by both criteria, were 16055 (Tm = 64.2 °C; Tonset = 55.5 °C), DU422 (Tm = 63.4 °C; Tonset = 56.9 °C) and ZM197M (Tm = 62.7 °C; Tonset = 57.2 °C) (Fig. 2 A and B). The MCGP1.4 trimer also had a high Tm (68.4 °C), but its low Tonset (<55 °C) is a sign of instability, perhaps due to structural or glycan heterogeneity. Based on good trimer yields and relatively high thermostability, we selected DU422 and ZM197M for further characterization.
Fig. 2.
Thermostability of clade C SOSIP.664 trimers. (A) Summary of Tm and Tonset of the 10 most efficiently 293S cell-expressed, GNL/SEC-purified clade C trimers. The clade A BG505 trimer is shown in blue for comparison. Dotted lines separate the thermostability data into four quadrants, where the Upper Right quadrant represents the most thermostable trimers with both high Tm and Tonset values. (B) DSC analysis of the DU422 and ZM197M SOSIP.664 trimers reveal sharp melting profiles with Tm values of 63.4 °C and 62.7 °C, respectively.
DU422 and ZM197M SOSIP.664 Trimers Analyzed by NS-EM.
GNL/SEC-purified DU422 and ZM197M SOSIP.664 trimers were further analyzed by NS-EM. Overall, ∼97% and ∼92% of these trimers, respectively, are in a native-like conformation, with few irregularly shaped, nonnative forms (SI Appendix, Fig. S1), similar to BG505 (27). The prefusion trimers can be subdivided into closed and partially open forms, the latter involving an increase in mobility of the gp120 variable loops at the trimer apex (16). Categorized this way, the closed trimer proportions were ∼81% and ∼83% for DU422 and ZM197M, respectively, with a minority subpopulation (∼10–15% in both cases) in partially open forms. The NS-EM analysis also shows that the clade C trimers differ subtly from 2G12/SEC-purified BG505 trimers, for which ∼100% are in the closed, prefusion form (SI Appendix, Fig. S1), but are more reminiscent of B41 trimers, albeit with a lower proportion of partially open forms (10–15%, compared with ∼50–75%) (SI Appendix, Fig. S1) (16). That >90% of the DU422 and ZM197M trimers are in a native-like, prefusion conformation makes them candidates for both structural and immunogenicity studies.
Antigenicity of the DU422 and ZM197M SOSIP.664 Trimers.
We used ELISA to derive antibody-binding EC50 values using GNL/SEC-purified, D7324-tagged trimers produced in 293F cells (i.e., wild-type glycosylation) for comparison with neutralization IC50 endpoints derived using sequence-matched (i.e., glycans knocked in) Env-pseudotyped viruses in a TZM-bl cell assay. The DU422 and ZM197M viruses have tier-2 and tier-1B neutralization classifications, respectively (28), which we also confirmed for the DU422* and ZM197M* glycan knock-in variants. The antibodies (n = 21) spanned a broad range of nAb and non-nAb epitopes. Only a weak correlation between antibody reactivity in ELISA and virus neutralization was observed for DU422 and ZM197M SOSIP.664-D7324 trimers purified by GNL/SEC (R = 0.53 and R = 0.32, respectively, SI Appendix, Fig. S2). This weaker correlation than expected was surprising given that these GNL/SEC-purified clade C trimers coelute with the trimer-specific PGT145 Fab in SEC and are only weakly reactive with non-nAb 19b (stoichiometry, n = 0.2) using isothermal titration calorimetry (ITC) (SI Appendix, Fig. S3 and Table S1). Together, these studies imply that the GNL/SEC-purified DU422 and ZM197M SOSIP.664 preparations do contain native-like trimers, but also other forms of Env with undesirable antigenic properties. As GNL/SEC-purified DU422 and ZM197M trimers were strongly reactive with PGT145 (SI Appendix, Fig. S2), the native-like trimer population can be purified using a PGT145-affinity column (SI Appendix, Fig. S4).
As 2G12 was reactive with GNL/SEC-purified trimers in ELISA, albeit weakly (SI Appendix, Fig. S2), we tested whether native-like DU422 and ZM197M trimers could be purified via 2G12 affinity and SEC. Purification of high-quality, native-like DU422 and ZM197M SOSIP.664-D7324 trimers was successful and enabled a direct comparison with BG505 and B41 trimers purified in the same way. We note that 2G12/SEC resulted in slightly lower yields compared with GNL/SEC purification (SI Appendix, Fig. S4).
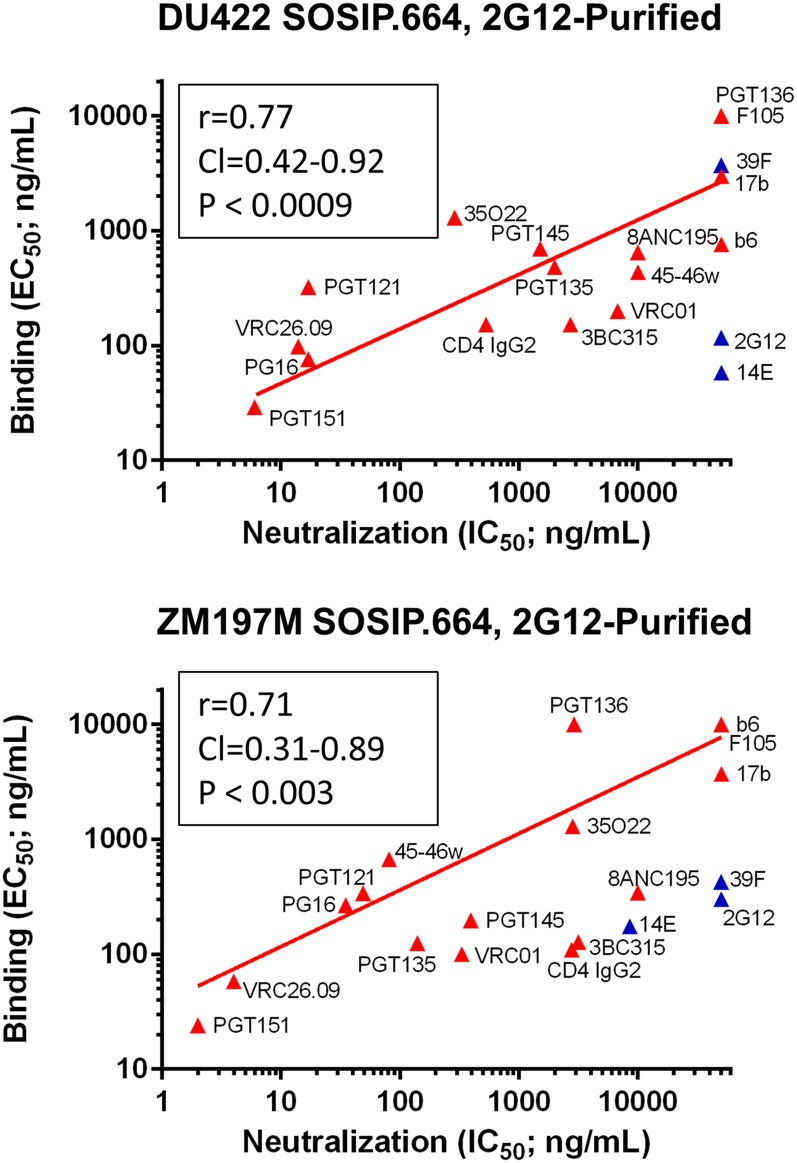
When DU422 and ZM197M SOSIP.664-D7324 trimers were purified by 2G12/SEC, the correlation between their antigenicity (EC50) and virus neutralization (IC50) was much stronger (Fig. 3). The antigenicity vs. neutralization (with 2G12, 14E, and 39F omitted) Spearman correlation coefficients of R = 0.77 and R = 0.71 for DU422 and ZM197M SOSIP.664-D7324 trimers (n = 16 antibodies) are a little lower than for BG505 (R = 0.88 with n = 45 antibodies) (14), but slightly higher than for B41 (R = 0.69 with n = 19 antibodies) (16) (SI Appendix, Table S2). Compared with GNL, purification by 2G12 selects not only for native-like trimers that have a higher affinity for the PGT151 and VRC01 bnAbs, but also a lower affinity for the 17b non-nAb to a CD4-induced epitope (Fig. 3 and SI Appendix, Fig. S2). Surface plasmon resonance (SPR) analysis corroborated the trends observed in ELISA for bnAb binding to the clade C trimers (SI Appendix, Fig. S5). We conclude that 2G12/SEC-purified DU422 and ZM197M SOSIP.664 trimers are good antigenic mimics of the corresponding trimers on Env-pseudotyped viruses, whereas GNL columns are problematic for purifying the highest quality trimers—a conclusion only attainable from detailed NS-EM and antigenicity vs. correlation analyses.
Fig. 3.
Antigenicity of the near-native DU422 and ZM197M SOSIP.664 trimers. The ELISA-derived antigenicity vs. neutralization analysis reveals a positive Spearman correlation (Inset) for trimers expressed in 293F cells and purified by 2G12-affinity chromatography and SEC. The bnAbs and non-nAbs represent a wide set of epitopes. The binding and neutralization values are compared with clade A BG505 and clade B B41 SOSIP.664 trimers in SI Appendix, Table S2. Data points are colored red to indicate their inclusion in the correlation analysis, and blue their omission. The V3 non-nAbs 14E and 39F were excluded from the correlation analysis, as immobilization of SOSIP.664-D7324 trimers on ELISA plates exacerbates the exposure of V3 epitopes in a way not reflected by other techniques (14). 2G12 was also excluded from the correlation analysis because the column yields a subset of trimers that bind this bnAb with higher than average affinity, which does not apply in neutralization assays (14).
Cryo-EM Reconstruction of the ZM197M Trimer in Complex with VRC01 Fab at 9.6-Å Resolution.
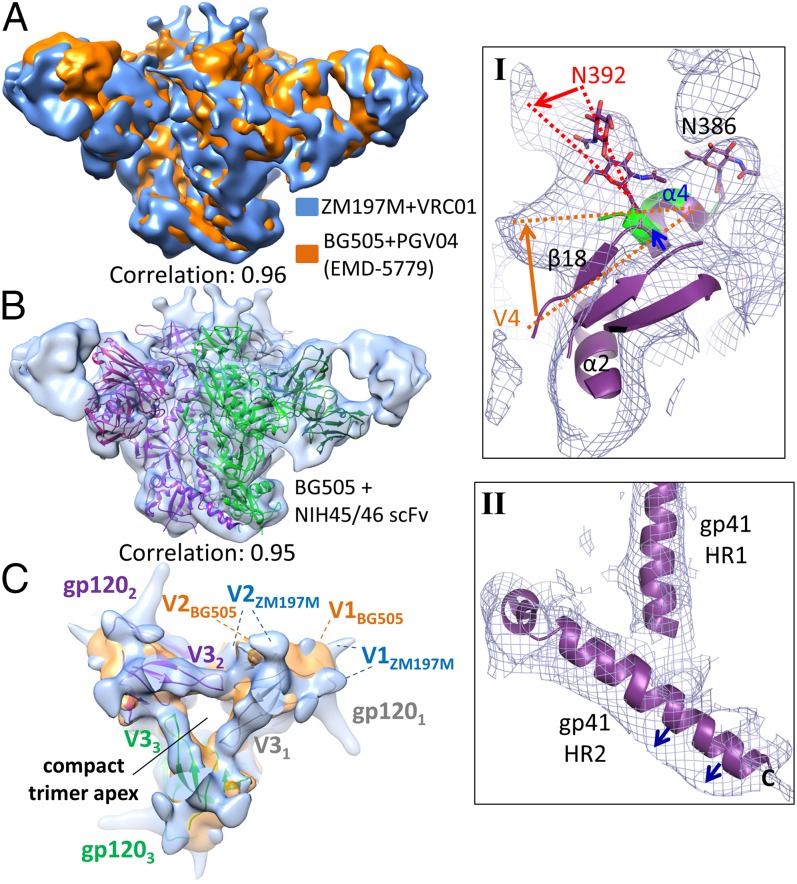
To gain insights into the structure of a clade C trimer, we purified (by SEC) a complex of the ZM197M SOSIP.664 trimer with VRC01 Fab, and obtained a 9.6-Å reconstruction by single-particle cryo-EM (Fig. 4 and SI Appendix, Fig. S6). The clade C and clade A trimer (EMD-5779) reconstructions were highly similar, with a correlation of 0.96 when both maps were low-passed filtered to 9.6 Å (Fig. 4A). Moreover, there was a high degree of overall similarity (correlation of 0.95) with the 4.4-Å BG505 SOSIP.664 trimer + NIH45-46 single-chain Fv (scFv) X-ray structure (Fig. 4B). The apex of the ZM197M trimer has an almost identical closed arrangement to its BG505 counterpart, with gp120 V1, V2, and V3 elements participating in the trimer interface (Fig. 4C) (17–19). Similarly, the three coiled-coil gp41 HR1 helices provide the central platform upon which the gp120 monomers sit. Thus, at this resolution, the clade A BG505 and clade C ZM197M SOSIP.664 trimers are in almost identical overall configurations.
Fig. 4.
Cryo-EM reconstruction of the ZM197M SOSIP.664 trimer in complex with VRC01 Fab at 9.6-Å resolution. (A) Superposition of the clade C ZM197M SOSIP.664 trimer + VRC01 Fab reconstruction (light blue surface) at 9.6-Å resolution vs. the previously reported 5.8-Å clade A BG505 SOSIP.664 trimer + PGV04 Fab reconstruction (EMD-5779, orange surface, low-passed filtered to 9.6-Å resolution) reveals a high degree of structural similarity [Chimera (41) “map-in-map” fitting correlation of 0.96]. Side views of the complexes are shown, with the Fab at the extremities (Left and Right at the 2- and 10-o’clock positions) and the trimer located at the Center (Top, gp120 and Bottom, gp41). (B) Fitting the 4.4-Å resolution clade A BG505 SOSIP.664 trimer + NIH 45/46 scFv crystal structure (shown as secondary structure cartoon and colored by protomer; PDB 5D9Q) into the 9.6-Å clade C ZM197M SOSIP.664 trimer + VRC01 Fab reconstruction also shows a high degree of structural similarity [Chimera (41) “fit-in-map” correlation of 0.95]. Nonetheless, subtle local differences are apparent. (Inset I) α4/β18/V4 elements, including N392 but not N386, appear slightly shifted (blue, orange, and red arrows) in the ZM197M trimer reconstruction from their conformation in the BG505 trimer (purple secondary structure cartoon, with N-linked glycans as sticks) and are more akin to the conformation in the clade C ZM109F.PB4 gp120 core X-ray structure (green secondary structure cartoon, Protein Data Bank ID: 3TIH). (Inset II) The cryo-EM reconstruction shows subtle differences (blue arrows) in the relative position of gp41 HR2, but not HR1, between the clade A BG505 and clade C ZM197M trimers. (C) View down the trimer apex of the superposed reconstructions and crystal structure, highlighting conformational mobility in the highly variable portions of the gp120 V1 and V2 loops. Images were rendered with UCSF Chimera (41) and PyMOL (43).
To assess possible local structural differences between the two trimers, we performed a per-residue map correlation of the 4.4-Å BG505 trimer + NIH45-46 scFv X-ray structure (PDB 5D9Q) into the ZM197M trimer + VRC01 Fab 9.6-Å reconstruction (SI Appendix, Fig. S7). One region with poor density fits is gp120 V1, whether the BG505 trimer was in its PGT122-bound or -unbound conformation (17, 19). These observations are consistent with the ZM197M V1 region being 20-residues longer than its BG505 counterpart, with four extra N-linked glycosylation sites (∼2.3 kDa/complex biantennary glycan), leading to an additional ∼8–10 kDa of mass per gp120 (SI Appendix, Fig. S8). The V1 region is not fully resolved in the cryo-EM reconstruction (Fig. 4C). Another difference between ZM197M and BG505 precedes and includes the V4 loop (Fig. 4B, Inset I and SI Appendix, Fig. S7). Indeed, even at nanometer resolution, the density indicates a slight, but discernible shift in the conformation of gp120 α4/β18/V4 (residues 389–397) for ZM197M compared with BG505, and more akin to the deglycosylated, unliganded gp120 core of the clade C strain ZM109F.PB4 (Fig. 4, Inset I) (29). Whereas the ZM197M N386 glycan density is similar to that in the BG505 trimer, the N392 glycan appears slightly shifted, possibly due to movement in α4/β18/V4 (Fig. 4, Inset I).
The density fit analysis also revealed potential differences in the gp120 C3 β14/β15 elements (SI Appendix, Fig. S7). Little density is present for this region in the ZM197M reconstruction, indicative of high flexibility. The C terminus of gp41 heptad repeat 2 (HR2) (Fig. 4, Inset II and SI Appendix, Fig. S7) appears shifted relative to BG505. Two distinct orientations of the HR2 helix were observed in X-ray and cryo-EM structures of the BG505 SOSIP.664 trimer, and this region was found to be intrinsically flexible in hydrogen/deuterium exchange mass spectrometry experiments (17, 19, 30). It remains to be determined whether the HR2 differences observed here are indicative of true conformational variations between BG505 and ZM197M Env, or instead reflect a higher flexibility for this region in all SOSIP.664 constructs, which are truncated before the gp41 MPER. We also note that these cryo-EM and X-ray structures of Env trimers have been obtained in complex with different ligands, which could subtly affect the trimer conformation.
Discussion
A daunting challenge in HIV-1 vaccine research is to elicit an immune response capable of neutralizing the enormous worldwide diversity of circulating strains. Among strategies to induce bnAbs is the use of multiple, native-like trimers based on diverse sequences, such as env genes from different clades, in sequential or simultaneous immunization protocols. We have already described clade A BG505 and clade B B41 SOSIP.664 trimers, which are excellent antigenic mimics of the corresponding Env spikes (14, 16). Here, we identify and characterize two new trimers of comparably high quality, based on the DU422 and ZM197M clade C viruses.
We selected these two SOSIP.664 trimers after screening 15 candidates to find those that expressed efficiently and were thermally stable. Four of the SOSIP-modified env genes produced GNL/SEC-purified trimers in yields >0.5 mg per 500 mL culture. Furthermore, three of the 10 trimers assessed by DSC had an appropriate stability profile with melting temperatures >60 °C and onsets of melting >55 °C. The Env sequence properties and characteristics that correlate with high-level expression of stable SOSIP.664 trimers are still poorly understood. Combining expression and stability data with structural insights, as described here, should facilitate the rational design of mutation strategies to improve trimer yield, stability, and antigenicity.
Both DU422 and ZM197M env genes were derived from viruses isolated early postinfection: DU422 at 8 wk (31) and ZM197M at 15 wk (32). We therefore note that the five most structurally and antigenically native-like SOSIP.664 trimers that have been identified and characterized to date (clade A BG505, clade B B41 and clade C DU422, ZM197M, and 16055) (15), are all derived from env molecular clones isolated from acute and early infections with viruses that use the CCR5 coreceptor for entry, and that have tier 1B (ZM197M) or tier 2 (BG505, B41, DU422, and 16055) neutralization phenotypes (28). These observations, albeit from a small sample size, lead us to hypothesize that env genes from neutralization-resistant viruses isolated during acute infection may be preferable substrates for expressing soluble, stable, fully native-like SOSIP.664 trimers. It may also be noted that, despite multiple attempts, we have not yet identified fully native trimers from chronic and/or highly neutralization-sensitive viruses of any clade. However, we also found that CAP45 and CH505 clade C sequences derived from acute/resistant viruses did not yield high quality SOSIP.664 trimers efficiently. Clearly, much remains to be learned about the structural consequences of Env sequence variation at the trimer level. Our emerging knowledge of how to further stabilize trimers by targeted sequence changes should also prove fruitful.
The antigenicity vs. neutralization Spearman correlation analyses of the clade A (14), B (16), and C SOSIP.664 trimers reveal that, when properly purified, they mimic the phenotype of the corresponding Env spike on the virus. We now have clear evidence that the purification method substantially influences the properties and antigenicity of native-like trimers. Although GNL columns permit rapid screening of constructs, they were markedly inferior to 2G12-affinity purification for isolation of native-like, antigenically appropriate DU422 and ZM197M SOSIP.664 trimers. Nonnative forms of trimers, and other Env contaminants, can be removed by negative selection with a non-nAb (15) or by positive selection with a bnAb, which we consider to be a superior method (16). The recent improvements in purification strategies will further aid in isolating the most antigenically and structurally native SOSIP.664 trimers from the heterogeneous population of Env proteins that are initially expressed. Rapid identification of suitable native-like Env trimers with high yield and high thermostability using screening platforms analogous to the one described here will be essential to reap the benefits of optimized purification protocols.
The ZM197M SOSIP.664 trimer displays the epitopes recognized by most classes of bnAbs. The ZM197M cryo-EM reconstruction reveals that the VRC01 class of bnAbs binds in the same conserved disposition as on BG505 and that the gp120 variable loops (e.g., V1, V2, and V4) are conformationally different. The N392 glycan, which is important to the 2G12 epitope (33), is slightly shifted on the clade C trimer compared with its position on the clade A trimer. Thus, we hypothesize that the observed differences in the relative disposition of the outer domain glycans at the base of V4 between the ZM197M and BG505 trimers, together with structural differences in neighboring V1, might help explain why 2G12 neutralizes clade C viruses poorly, even when all of the interacting glycans that nominally constitute its epitope are present.
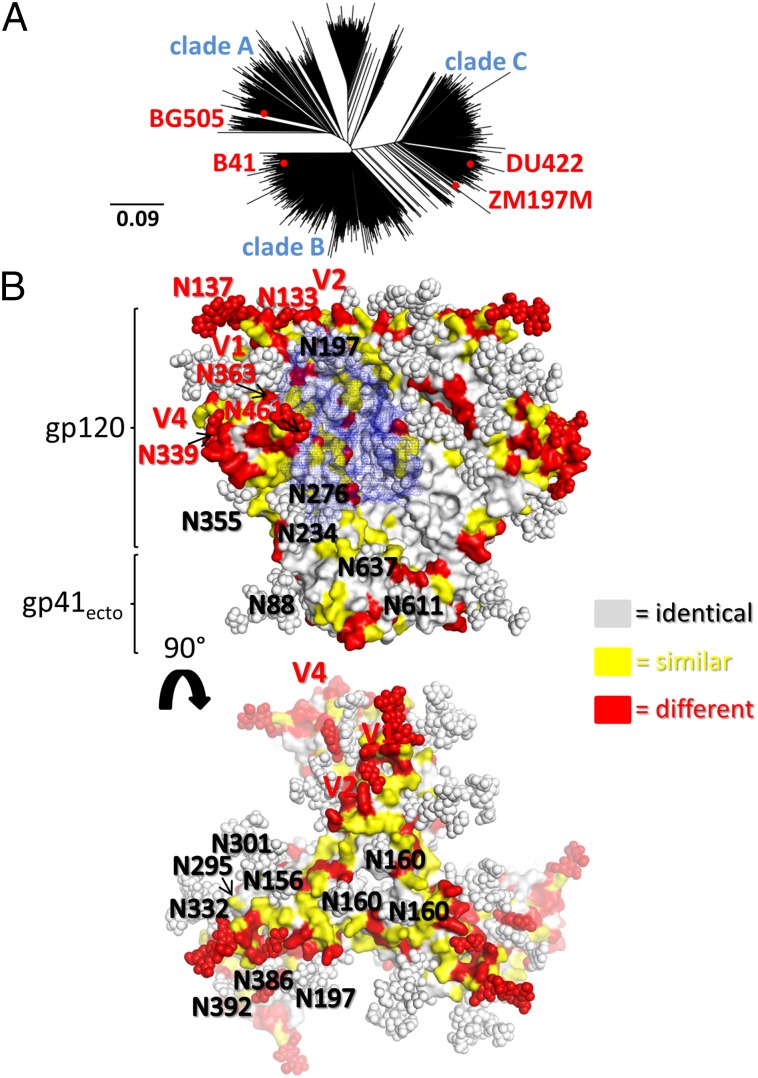
Our identification of antigenically and structurally near-native SOSIP.664 trimers based on sequences from four different strains (BG505, B41, DU422, and ZM197M), three different subtypes (A, B, and C), and with a combined pairwise identity of 77.3% (Fig. 5A and SI Appendix, Fig. S8), should facilitate the development of immunization strategies aimed at inducing bnAbs. For example, knowledge of the specific regions of high sequence and structure conservation among the different trimers may allow immunogens to be designed that direct antibody responses to conserved bnAb epitopes and thereby counter strain and subtype variability (Fig. 5B). Regions of high sequence diversity are often proximal to more conserved sites, such as the CD4 binding site or the trimer apex that contains a tripod arrangement of N160 glycans (Fig. 5B). The DU422 and ZM197M trimers will also be an additional resource for isolating bnAbs from infected individuals (34), as well as valuable reagents for future, higher resolution structural studies, alone and in comparison with the BG505 trimer structures (17–19).
Fig. 5.
The two clade C SOSIP.664 trimers provide added antigenic diversity to the growing arsenal of soluble, cleaved, native-like HIV-1 Env antigens. (A) Phylogenetic tree representation of the sequence divergence between four clade A, B, and C virus sequences that yield native-like recombinant trimers (red dots) among natural HIV-1 Env diversity. The tree was rendered with Geneious (44) using the 2013 precompiled alignment of 4,785 HIV-1 Env sequences downloaded from the Los Alamos National Laboratory HIV sequence database. (B) The four trimer sequences are mapped onto the BG505 SOSIP.664 X-ray structure. Gray surfaces represent positions with identical amino acids among the four sequences, whereas yellow and red surfaces indicate similar or different residues, respectively. N-linked glycans are shown as spheres. The sequence alignment on which this analysis is based, which also delineates the length of variable loops, is shown in SI Appendix, Fig. S8. The blue mesh highlights the CD4 binding site as recognized by VRC01 (blue mesh). Images were produced with PyMOL (43).
Materials and Methods
SOSIP.664-modified env genes were expressed as secreted proteins and purified by either GNL-, 2G12- or PGT145-affinity chromatography, followed by SEC, as previously described (13, 16, 35). Melting temperatures were measured by DSC, and trimer morphology classifications were performed by NS-EM using Appion (36, 37) and MSA/MRA (38) software packages (16). Trimer antigenicity was assessed by ITC, SEC, ELISA, and SPR. For ELISA and SPR, the DU422 and ZM197M SOSIP.664-D7324 trimers were made as described elsewhere (14). Neutralization titers against the DU422* and ZM197M* sequence-matched (i.e., glycan knock-in) Env-pseudotyped viruses were determined in a TZM-bl assay (14). Cryo-EM reconstructions of the ZM197M SOSIP.664 trimer + VRC01 Fab were performed using CTFFind3 (39), DoG Picker (36, 37), MRA/MSA (38), and Relion (40). Fitting of maps and X-ray models into the EM reconstruction was carried out in University of California San Francisco (UCSF) Chimera (41), and subsequent structural analysis were performed in Coot (42) and PyMOL (43). For more details, see SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank James Robinson, John Mascola, Peter Kwong, Mark Connors, and Michel Nussenzweig for donating reagents and antibodies directly or through the AIDS reagents reference program; Celia Labranche and David Montefiori for information on the neutralization tier categorization of the DU422 and ZM197M Env-pseudotyped viruses and their glycan knock-in variants; Robyn Stanfield for insightful discussions; Kevin de los Reyes for technical assistance; and to other members of the HIVRAD team at Weill Medical College of Cornell University, Academic Medical Center, University of Amsterdam, and The Scripps Research Institute. This work was supported by NIH Grants P01 AI082362, P01 AI110657, R01 AI084817, R37 AI36082, UM1 AI100663, the International AIDS Vaccine Initiative, and the Bill and Melinda Gates Foundation (The Collaboration for AIDS Vaccine Discovery). R.W.S. is a recipient of a Vidi grant from the Netherlands Organization for Scientific Research and a starting investigator grant from the European Research Council (ERC-StG-2011–280829-SHEV). J.H.L. is a recipient of a fellowship grant from the California HIV/AIDS Research Program. This is manuscript number 29066 from The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The reconstruction data reported in this paper have been deposited in the Electron Microscopy Data Bank, www.emdatabank.org (EMDB ID code EMD-3059).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507793112/-/DCSupplemental.
References
- 1.Plotkin S. History of vaccination. Proc Natl Acad Sci USA. 2014;111(34):12283–12287. doi: 10.1073/pnas.1400472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: A systematic review and meta-analysis. Lancet Infect Dis. 2012;12(1):36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 3.Korber B, et al. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 4.Julien JP, Lee PS, Wilson IA. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol Rev. 2012;250(1):180–198. doi: 10.1111/imr.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: Clues for vaccine development. Nat Rev Immunol. 2010;10(1):11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overbaugh J, Morris L. The antibody response against HIV-1. Cold Spring Harb Perspect Med. 2012;2(1):a007039. doi: 10.1101/cshperspect.a007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derdeyn CA, Moore PL, Morris L. Development of broadly neutralizing antibodies from autologous neutralizing antibody responses in HIV infection. Curr Opin HIV AIDS. 2014;9(3):210–216. doi: 10.1097/COH.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koff WC, et al. Accelerating next-generation vaccine development for global disease prevention. Science. 2013;340(6136):1232910. doi: 10.1126/science.1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plotkin SA. Vaccines: Correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47(3):401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 10.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol. 2011;12(6):509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munro JB, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346(6210):759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallo SA, et al. The HIV Env-mediated fusion reaction. Biochim Biophys Acta. 2003;1614(1):36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 13.Julien JP, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci USA. 2013;110(11):4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guenaga J, et al. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog. 2015;11(1):e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pugach P, et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol. 2015;89(6):3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pancera M, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514(7523):455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blattner C, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40(5):669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders RW, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349(6244):aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geretti AM. HIV-1 subtypes: Epidemiology and significance for HIV management. Curr Opin Infect Dis. 2006;19(1):1–7. doi: 10.1097/01.qco.0000200293.45532.68. [DOI] [PubMed] [Google Scholar]
- 23.Bures R, et al. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J Virol. 2002;76(5):2233–2244. doi: 10.1128/jvi.76.5.2233-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray ES, Moore PL, Pantophlet RA, Morris L. N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. J Virol. 2007;81(19):10769–10776. doi: 10.1128/JVI.01106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ringe RP, et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci USA. 2013;110(45):18256–18261. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guttman M, et al. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun. 2015;6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seaman MS, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84(3):1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon YD, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci USA. 2012;109(15):5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guttman M, et al. CD4-induced activation in a soluble HIV-1 Env trimer. Structure. 2014;22(7):974–984. doi: 10.1016/j.str.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Harmelen J, et al. Characterization of full-length HIV type 1 subtype C sequences from South Africa. AIDS Res Hum Retroviruses. 2001;17(16):1527–1531. doi: 10.1089/08892220152644232. [DOI] [PubMed] [Google Scholar]
- 32.Li M, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80(23):11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murin CD, et al. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J Virol. 2014;88(17):10177–10188. doi: 10.1128/JVI.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sok D, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci USA. 2014;111(49):17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong L, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20(7):796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166(2):205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166(1):95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogura T, Iwasaki K, Sato C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J Struct Biol. 2003;143(3):185–200. doi: 10.1016/j.jsb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrödinger LLC. 2010. The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.6.
- 44.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.