Significance
Loading of Argonautes with the correct strand of the pre-miRNA duplex and disposal of the other strand are essential steps in microRNA biogenesis. Here we report characterization of the protein and microRNA populations associated with mutant ALG-1 Argonautes that are defective in transitioning from microRNA processing to target repression. We show that mutant Argonaute erroneously associates with the normally discarded microRNA* strands, signifying a role for Argonaute ALG-1 in microRNA strand selection. Accumulation of microRNA* is dependent on the microRNA identity, suggesting that specific microRNA features allow wild-type Argonautes to distinguish among different microRNAs. These findings are relevant to understanding Argonaute roles in microRNA biogenesis and, more broadly, to the functions of microRNAs in development and disease.
Keywords: Argonaute, ALG-1, microRNA, microRNA*, passenger
Abstract
MicroRNAs are regulators of gene expression whose functions are critical for normal development and physiology. We have previously characterized mutations in a Caenorhabditis elegans microRNA-specific Argonaute ALG-1 (Argonaute-like gene) that are antimorphic [alg-1(anti)]. alg-1(anti) mutants have dramatically stronger microRNA-related phenotypes than animals with a complete loss of ALG-1. ALG-1(anti) miRISC (microRNA induced silencing complex) fails to undergo a functional transition from microRNA processing to target repression. To better understand this transition, we characterized the small RNA and protein populations associated with ALG-1(anti) complexes in vivo. We extensively characterized proteins associated with wild-type and mutant ALG-1 and found that the mutant ALG-1(anti) protein fails to interact with numerous miRISC cofactors, including proteins known to be necessary for target repression. In addition, alg-1(anti) mutants dramatically overaccumulated microRNA* (passenger) strands, and immunoprecipitated ALG-1(anti) complexes contained nonstoichiometric yields of mature microRNA and microRNA* strands, with some microRNA* strands present in the ALG-1(anti) Argonaute far in excess of the corresponding mature microRNAs. We show complex and microRNA-specific defects in microRNA strand selection and microRNA* strand disposal. For certain microRNAs (for example mir-58), microRNA guide strand selection by ALG-1(anti) appeared normal, but microRNA* strand release was inefficient. For other microRNAs (such as mir-2), both the microRNA and microRNA* strands were selected as guide by ALG-1(anti), indicating a defect in normal specificity of the strand choice. Our results suggest that wild-type ALG-1 complexes recognize structural features of particular microRNAs in the context of conducting the strand selection and microRNA* ejection steps of miRISC maturation.
MicroRNAs are small noncoding RNAs that, as part of a microRNA induced silencing complex (miRISC), bind to complementary sites in the 3′UTR of target messenger RNAs and cause translational repression or degradation of the target mRNA. microRNAs are predicted to posttranscriptionally regulate as much as 60% of mammalian protein coding genes (1), making them powerful regulators of physiological and developmental processes. Faithful microRNA biogenesis and programming of miRISC with the appropriate microRNA and accessory protein factors are essential for proper posttranscriptional gene regulation by microRNAs.
Mature ∼22-nt single-stranded microRNAs are generated and loaded into specialized Argonaute proteins [ALG-1 (Argonaute-like gene) and ALG-2 in Caenorhabditis elegans] through a series of enzymatic and RNA-protein assembly steps. First, the primary microRNA transcript is processed by the Drosha/Pasha microprocessor complex into a hairpin precursor and exported into the cytoplasm, where the RNAse III enzyme Dicer cleaves the precursor to generate a duplex, consisting of two short strands of RNA, corresponding to the “5p” and “3p” strands of the precursor hairpin. The microRNA duplex is then bound by Argonaute, such that one of the duplex strands (the eventual guide strand, or “miR” strand) becomes stably associated with Argonaute and the other strand (referred to as the passenger strand, or “miR*” strand) becomes discarded and degraded.
It is generally thought that the microRNA duplex is transferred to Argonaute in the context of a miRISC loading complex (miRLC), which contains Argonaute associated with Dicer (2, 3). The loading of the Argonaute with the duplex is also assisted by chaperone proteins Hsp70/Hsp90 (4–6).
Critical to proper miRISC assembly and subsequent microRNA-mediated gene repression is the accurate and consistent selection of which strand of the 5p/3p duplex will be loaded into Argonaute as the guide microRNA. This process of guide strand selection involves establishing the proper orientation of the duplex within the miRLC, followed by ejection and disposal of the passenger strand (7). For most microRNAs, guide strand selection is highly asymmetric and specific, so that either the 5p or the 3p accumulate in dramatic excess over the other (8, 9). This specificity of guide strand choice has been shown to be associated with certain features of the 5p::3p duplex: (i) The presence and configuration of centrally located mismatches for Drosophila microRNAs (10–12), (ii) the identity of the 5′ nucleotide of the guide microRNA (13–15), and (iii) the relative thermodynamic stability of the two ends of the miR::miR* duplex (16–18), which has a dramatic impact on strand choice in both flies and mammals. It should be noted that the relative thermodynamic stability of the duplex ends may be a primary determinant of strand selection for perfectly paired duplexes of siRNAs, but may be less important for the mismatched duplexes of most microRNAs (10, 11, 16, 17). Additionally, in C. elegans, microRNAs that produce 5p guides were reported to have similar thermodynamic stabilities of duplex ends as microRNAs that produce 3p guides (9), suggesting that at least in C. elegans other duplex features may be more important for strand selection (9). Features, such as microRNA duplex sequence composition and position of mismatches, could instead be major contributors to strand selection (19–21).
Although guide strand selection appears to be highly asymmetric for most microRNAs, for a small subset of microRNAs substantial quantities of both 5p and 3p strands can accumulate, and in at least some cases there is evidence for functional roles for both strands (22–25). It’s possible that the relative loading of 5p and 3p strands as the guide could be regulated by upstream signals acting via Argonaute or other components of the miRLC. How C. elegans Dicer DCR-1 may contribute mechanistically to the specificity of microRNA loading and guide strand selection is unknown. Mammalian Dicer-associated proteins TRBP/PACT contribute to strand selection (26), and R2D2, a key component of Drosophila miRLC, is dispensable for miR strand selection (27). Interestingly, mammalian Dicer itself may be dispensable for asymmetric miRISC duplex loading, at least under certain circumstances (28, 29). The Argonaute protein itself may have the capacity to distinguish among specific microRNAs to determine the guide/miR vs. passenger/miR* fates of the 5p and 3p strands. Indeed, a very recent report has suggested a direct role for mAGO2 in miR strand choice (30). All these considerations emphasize the importance of acquiring a better understanding of the regulation of microRNA guide strand choice by Argonaute complexes in vivo.
We reported previously novel antimorphic alleles of the C. elegans Argonaute gene alg-1 that broadly impair the functions of many microRNAs, apparently by sequestering microRNAs into immature and ineffectual miRLC complexes (31). The mutant ALG-1(anti) proteins exhibit an increased association with the miRLC component Dicer DCR-1 in vivo, and a decreased association with the miRISC effector AIN-1 (ALG-1 interacting protein-1). We proposed that ALG-1(anti)–containing complexes associate with microRNAs, but fail to properly mature from the Dicer-containing miRLCs to the effector miRISCs, thereby sequestering microRNAs in immature (and nonfunctional) RISC complexes (31). To better understand both the protein and the microRNA dynamics associated with this miRLC-to-miRISC maturation step, we further characterized the ALG-1(anti)–associated proteins and microRNAs using high-throughput proteomics and small RNAseq. Our results support the idea that alg-1(anti) mutants are defective in transitioning from biogenesis (miRLC) to effector (miRISC) status and identify conserved Argonaute-interacting proteins that may be specific to either miRLC or miRISC complexes. Moreover, our small RNAseq analysis shows that alg-1(anti) mutants accumulate miR* strands at levels dramatically greater than the wild-type, indicating that the ALG-1(anti) mutant proteins are defective in microRNA strand selection and miR* strand disposal. We show that the biogenesis of different microRNAs can be impacted very differently by ALG-1(anti). For example, in the case of mir-58, miR* strands are retained by the ALG-1(anti) Argonaute apparently as part of the miR::miR* duplex, suggesting defective miR* strand release. In contrast, for mir-2, the ALG-1(anti) complexes contained far more miR* strands than miR, indicating that miR-2 biogenesis suffers from defects in the strand selection step of miRISC maturation. These findings support a model wherein microRNA biogenesis and miRISC maturation involve critical roles for Argonaute both in recognizing features that define specific microRNAs and in exercising microRNA-specific programs of guide strand selection and miR* strand disposal.
Materials and Methods
C. elegans Culture and Genetics.
C. elegans culture was performed using standard nematode growth conditions (32), except the HB101 Escherichia coli strain was used as the food source. All strains were grown at 20 °C. Because of their strong heterochronic phenotypes, alg-1(anti) mutant animals often burst through the vulva during the L4-adult molt (31). Therefore, alg-1(anti) mutations were maintained in a lin-31(n1053) genetic background that impairs vulva development and thereby suppresses the bursting phenotype of alg-1 mutants, while leaving their heterochronic phenotypes intact. The adult-specific col-19::gfp reporter transgene is also present in all of the strains. Therefore, the alg-1(anti) strains used here contain lin-31;col-19::gfp, and in all experiments the wild-type controls were lin-31(n1053);col-19::gfp.
Northern Blotting.
Total RNA was isolated from mixed population of animals using TRIzol reagent (Life Technologies), and Northern blots were performed as previously described (33, 34). For oligo probe sequences please see SI Materials and Methods.
Firefly microRNA Quantifications.
FirePlex microRNA assays were performed as previously described (31). For experiments involving 2′O-methyl oligonucleotide-mediated pull-down, equivalent fractions of input material and supernatant were tested using the FirePlex assays to quantitatively assess microRNA abundance in the starting material and microRNA depletion from the supernatant by the 2′-O-methylated oligonucleotide.
Small RNA Library Preparation and High-Throughput Sequencing.
Small RNA cDNA libraries were prepared as previously described (35) and sequenced on the Illumina GAIIx instrument or the NextSeq500 (for L2 staged samples) using standard manufacturer’s protocols.
Computational Analysis of cDNA Library Sequence Data.
Detailed description of cDNA library sequence data analysis can be found in SI Materials and Methods.
ALG-1 Immunoprecipitation and Western Blot Analysis.
Detailed description of antibodies, lysate preparation, immunoprecipitation, and Western blot analysis can be found in SI Materials and Methods.
2′O-Methyl Oligo Pull-Downs.
The 2′O-methyl oligo pull-downs from extracts of whole worms were performed as described previously (36). For mass spectrometry, each sample contained 20 mg of total protein. Sequences of the 2′-O-methylated, biotinylated oligonucleotides can be found in SI Materials and Methods.
Mass Spectrometry Analysis of ALG-1 Immunopurified Complexes and 2′-O-Methyl Oligonucleotide Pulldown Complexes and Computational Analysis of Proteomic Data.
For a detailed description of mass spectrometry analysis and computational analysis of ALG-1 associated proteomes, as well as data analysis of miR-58 and miR-58* associated proteins, see SI Materials and Methods.
SI Materials and Methods
Oligo Probes for Northern Blotting Analysis.
The starfire oligo probes were obtained from IDT: mir-80* (5′-GTTCAGAATCATGTCGAAAGCT/3StarFire/-3′), mir-58* (5′-GATGAGATGCGAAGAGTAGGGCA/3StarFire/-3′), and mir-77* (5′-ATTTCCTCAGAGCACAACCATC/3StarFire/-3′).
ALG-1 IP, and Western Blot Analysis.
For small RNAseq and Western blot analysis of the constituents of ALG-1 complexes, preparation of whole-protein lysates of worm populations, and ALG-1 IPs were performed as previously described using a custom ALG-1–specific polyclonal antibody (Anaspec) (31). Lysates were divided into two equal parts: one for ALG-1 IP, and one for RNA preparation to quantitatively profile microRNAs in the starting material. Subsequently, half of the immunoprecipitated sample was used to assess the efficiency of IP by Western blotting and the other half was used for RNA preparation to assess the profile of ALG-1–associated microRNAs. Each experiment was carried out in duplicate or triplicate, using populations of worms from independent cultures.
To assess ALG-1, AIN-1, and DCR-1 protein levels, total protein was extracted from mixed-stage animals, as previously described (47), except RNase inhibitor (Invitrogen) was added to the lysis buffer. Western blotting for ALG-1, AIN-1, and DCR-1 was performed as previously described using custom antibodies (31).
For mass spectrometric analysis of material coimmunoprecipitated with ALG-1 from worm extracts, IPs were performed using a cross-linked bead/anti–ALG-1 antibody matrix. For each IP experiment, 200 µg of ALG-1 affinity-purified polyclonal antibody was covalently linked to 20 µL of Sepharose A beads using dimethylpimelimidate.
The 2′O-Methyl Oligo Sequences.
The following 2′-O-methylated, biotinylated oligos were used in the pull-down experiments:
Against miR-58: 5′-CAUCAUUGCCGUACUGAACGAUCUCAAGUC-3′
Against miR-58*: 5′-CUAGGAUGAGAUGCGAAGAGUAGGGCACAUU-3′
Against miR-2: 5′-AUUCAGCACAUCAAAGCUGGCUGUGAUAUUCCA-3′
Against miR-2*: 5′-ACUUUACAUCAACCACCGCUUUGAUGUCCAA-3′
Against miR-52: 5′-AUAAGAGCACGGAAACAUAUGUACGGGUGUUGAU-3′
Against miR-52*: 5′-UCAUUGCUACCCUUUCAUUGUAACGUGUCCUU-3′
Scrambled control: 5′-CAUCACGUACGCGGAAUACUUCGAAAUGUC-3′
Deep-Sequencing and Analysis of cDNA Library Sequence Data.
Deep-sequencing was performed on cDNA libraries made from total RNA and RNA immunoprecipitated with ALG-1 from mixed-staged populations of three strains: three biological replicates from wild-type animals and two biological replicates each from alg-1(ma192) and alg-1(ma202) mutant animals. In addition, deep-sequencing was performed on cDNA libraries made from L2-staged total RNA in two biological replicates from wild-type and alg-1(ma202) animals and one biological replicate of alg-1(ma192).
Sequencing files in FastQ formats were processed using the Fastx toolkit to remove the adapter sequences and split the sequence reads into libraries according to the barcode sequences. Reads shorter than 17 nt were removed. Reads with identical sequences were combined and the combined count was saved in Fasta files. Reads were than aligned to the Caenorhabditis elegans genome (WormBase release WS215) using bowtie (48) with arguments, -v 3 -f -B 1 -a–best –strata. Alignments were then filtered based on the length of the read and the number of mismatches as follows: for sequence lengths 17, 18–19, 20–24, or >24: zero, one, two, or three mismatches were allowed, respectively. Annotations of tRNAs, rRNAs, piRNAs, and microRNAs were obtained from WormBase (release WS215) and miRBase (49) (Release 20). Passenger strand microRNA sequences that were not annotated previously were manually curated according to the deep-sequencing data. An in-house developed code was used to analyze the mapping results. To assign read counts to the microRNA sequences we considered all reads that mapped to a pre-microRNA sequence within −5 to +5 nucleotides of the annotated mature microRNA start. For all of the other small RNA species, we included all reads that map within the annotated region. The counts of all microRNA sequences were normalized to the total microRNA reads and presented as parts per million. Sequence data files are available in the GEO database under the accession number GSE72659.
Mass Spectrometry Analysis of ALG-1–Immunopurified Complexes and 2′-O-Methyl Oligonucleotide Pull-Down Complexes.
ALG-1–associated immunopurified protein complexes or 2′-O-methyl–purified protein complexes, were dissolved in digestion buffer (100 mM Tris⋅HCl, pH 8.5, 8M urea), reduced, alkylated, and digested by sequential addition of lys-C and trypsin proteases, as previously described (50, 51). ALG-1 samples were analyzed on a LTQ-orbitrap (ThermoScinetific), as described previously (51), whereas peptide digests prepared from 2′-O-methyl–purified samples were desalted and fractionated online using a 50-µM inner diameter fritted fused silica capillary column with a 5-µM pulled electrospray tip and packed in-house with 15 cm of Luna C18(2) 3-µM reversed-phase particles. The gradient was delivered by an easy-nLC 1,000 ultrahigh-pressure liquid chromatography system (Thermo Scientific). MS/MS spectra were collected on a Q-Exactive mass spectrometer (Thermo Scientific) (52, 53). Data analysis was performed using the ProLuCID and DTASelect2 algorithms as implemented in the Integrated Proteomics Pipeline-IP2 (Integrated Proteomics Applications) (54–56). Protein and peptide identifications were filtered using DTASelect and the filtering parameter required at least two unique peptides per protein and a peptide-level false-positive rate of less than 5%, as estimated by a decoy database strategy (57). Normalized spectral abundance factor (NSAF) values were obtained as described and multiplied by 105 to improve readability (58).
Computational Analysis and Comparison of ALG-1(wt)– and ALG-1(anti)–Associated Proteomes.
ALG-1 IP followed by mass spectrometry was conducted in three biological replicates from wild-type, alg-1(ma192), and alg-1(null), and one biological replicate from alg-1(ma202) animals. Proteins identified in this data were first filtered by removing proteins with marginal signals according to the following criteria: (i) spectral counts less than four across all samples in an experiment, or (ii) NSAF values of 0 in alg-1(ma202), and two or more replicates of wild-type and alg-1(ma192).
After application of the above filters, the data were analyzed to identify bona fide coimmunoprecipitating interactors with ALG-1 as follows: NSAF values from wild-type, alg-1(ma192), and alg-1(ma202) were divided by NSAF values obtained from the control alg-1(null) to derive an “experimental/control ratio”; separate experimental/control ratios were calculated for each of the three replicates for each genotype [with the exception of alg-1(ma202)]. In this experimental/control ratio calculation, [alg-1(null)] NSAF values equal to 0 were replaced by 1. A protein was considered to be a bona fide interacting partner according to the following criteria: (i) the experimental/control ratio was greater than 1 in all replicates and, (ii) its average experimental/control ratio was greater than 4. For proteins that satisfied the above criteria for interacting partners, the background-corrected NSAF values shown in Dataset S1 were calculated as follows: control NSAF values were subtracted from raw wild-type and alg-1(ma192) NSAF values, and the resulting corrected values were averaged across three replicates. For proteins that did not satisfy the above criteria for interacting partners, Dataset S1 shows their NSAF values set to 0. Proteomic mass spectrometry data have been deposited to the ProteomeXchange Consortium via the MassIVE partner repository with the data set identifier PXD002835.
microRNA Pull-Down MudPIT Data Analysis.
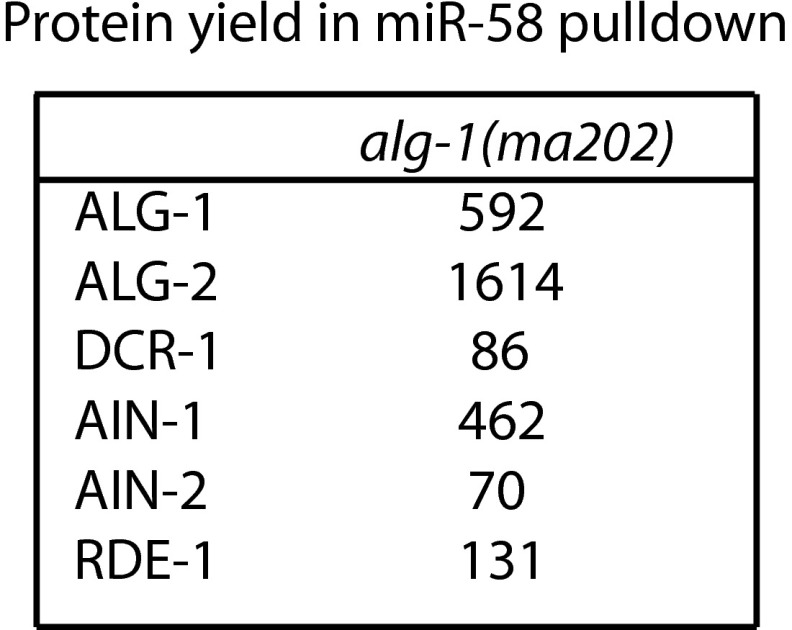
microRNA pull-down followed by mass spectrometry was conducted in two biological replicates from two strains, wild-type and alg-1(ma192), using three different oligos: oligo against miR-58 guide, oligo against miR-58*, and a scrambled oligo control. In addition, microRNA pull-down was performed in one biological replicate from alg-1(ma202) using oligo against miR-58 guide and a scrambled oligo control. Analysis of data from these miR and miR* pull-down experiments was restricted to ALG-1, ALG-2, DCR-1, AIN-1, AIN-2, RDE-1, and is reported in Fig. 4B and Fig. S5. Proteins were scored as bona fide interactors with the oligo-targeted microRNA as follows. First, NSAF obtained from microRNA pull-down was divided by the NSAF obtained from the scrambled control to derive an experimental/control ratio. If this experimental/control ratio was greater than 4 in all replicates, the protein was considered to be a specific interactor. NSAF of nonspecific interactors are reported as 0 in Fig. 4B. In addition to standard NSAF value calculations for ALG-1 and ALG-2, which take into account shared peptides, we calculated NSAFunique values using only peptides that unambiguously mapped to either ALG-1 or ALG-2 proteins (by discarding the peptides shared between the two proteins). Background-corrected NSAF values were calculated as follows: scrambled control NSAF values were subtracted from raw mir-58 guide and passenger NSAF values or NSAFunique values, and the resulting corrected values were averaged across the two replicates. Protein yield per microRNA family pull-down was calculated by normalizing background corrected NSAF values (or NSAFunique values) to the amount of miR-58, miR-80, and miR-81 present in total RNA samples (input material) and to the amount of these microRNAs depleted by anti–miR-58 oligo.
Fig. 4.
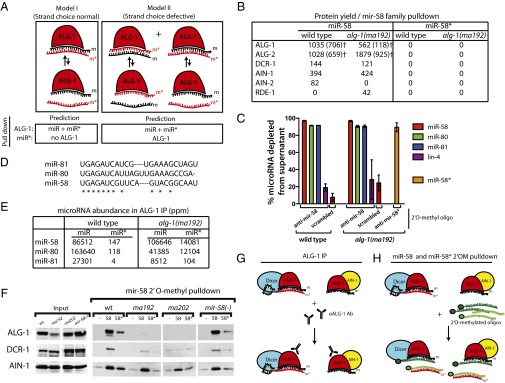
2′O-methyl oligonucleotide pull-downs from wild-type and alg-1(anti) mutants suggest that miR-58* accumulation in ALG-1 is a result of failure of ALG-1(anti) to release it from the duplex. (A) Two models for miR* microRNA accumulation in alg-1(anti) mutant animals. Model I: ALG-1(anti) complexes retain both microRNA strands primarily in a bound duplex. Model II: ALG-1(anti) complexes contain single strands of miR and miR* microRNAs because of a loss in the ability of the protein to differentiate between the two strands. Predictions of the outcome for IP and pull-down experiments are shown directly below each model. (B) Protein yield of miRISC components and other small RNA related factors from mir-58 family microRNA pull-down as determined by MudPIT proteomics and normalized to the amount of RNA depleted by the 2′O-methyl pull-down. †ALG-1 and ALG-2 protein yield as determined based on normalized spectral abundance factor (NSAF)unique (SI Materials and Methods). (C) Efficiency of the miR-58 and miR-58* strand pull-downs presented as percent microRNA depleted from the supernatant in samples shown in B. (D) Sequence alignments between members of the mir-58 family of microRNAs. (E) miR-58 family microRNA and miR* abundances in wild-type ALG-1 and ALG-1(anti) IP. (F) Western blot analysis of mir-58 microRNA family pull-downs from extracts of alg-1(anti) and wild-type animals showing association of ALG-1, DCR-1, and AIN-1 with mir-58 family microRNAs (-, scrambled oligo control; 58, oligo against miR-58; 58*, oligo against miR-58*). (G and H) Summary model for the miR-58* strand and related proteins accumulation as observed in alg-1(anti) mutants by (G) ALG-1(anti) IP and (H) miR-58 and miR-58* 2′O-methyl pull-downs (m, miR; m*, miR*).
Fig. S5.
Protein yield (represented as normalized NSAF values) of miRISC components and other small RNA related factors from mir-58 family microRNA pull-down from alg-1(ma202) mutant animals as determined by MudPIT proteomics.
Classification of miRNAs into Classes.
miRNAs that fulfilled the following two conditions: (i) read counts higher than 40 ppm in IP samples of wild-type, alg-1(ma192), and alg-1(ma202), and (ii) miR*/miR < 0.2 in wild-type IP were classified into four classes based on their average miR*/miR ratios in the ALG-1(ma202) IP (Table 1).
Table 1.
microRNAs classified according to their miR*/miR ratio in input or ALG-1 IPs from alg-1(anti) mutants and wild-type
| Class | mir | Wild-type | alg-1(ma192) | alg-1(ma202) | Fold-change in miR*/miR ratios between ALG-1(ma202) IP and ALG-1(wt) IP | |||||||||
| Average microRNA abundance (ppm) | Average miR*/miR ratio | Average microRNA abundance (ppm) | Average miR*/miR ratio | Average microRNA abundance (ppm) | Average miR*/miR ratio | |||||||||
| Input | IP | Input | IP | Input | IP | Input | IP | Input | IP | Input | IP | |||
| Class I: Asymmetric (IP miR*/miR > 1) | mir-244 | 335 | 336 | 0.149 | 0.068 | 1,644 | 467 | 0.762 | 0.937 | 132 | 52 | 21.024 | 21.700 | 321 |
| mir-87 | 297 | 327 | 0.102 | 0.005 | 258 | 181 | 2.400 | 1.081 | 158 | 94 | 13.566 | 9.205 | 1,942 | |
| mir-2 | 518 | 966 | 0.091 | 0.020 | 1,308 | 3,748 | 3.520 | 0.484 | 140 | 442 | 30.950 | 4.261 | 212 | |
| mir-86 | 3,622 | 4,019 | 0.006 | 0.002 | 3,073 | 6,598 | 0.216 | 0.112 | 1,206 | 1,407 | 5.593 | 2.496 | 1,005 | |
| mir-252 | 310 | 1,187 | 0.006 | 0.002 | 129 | 1,618 | 4.769 | 0.526 | 240 | 1,024 | 3.349 | 1.565 | 818 | |
| mir-241 | 2,460 | 2,454 | 0.016 | 0.007 | 746 | 470 | 0.460 | 0.491 | 524 | 262 | 1.206 | 1.559 | 218 | |
| mir-45 | 2,269 | 1,868 | 0.014 | 0.006 | 6,359 | 2,322 | 0.010 | 0.014 | 10,737 | 6,100 | 1.298 | 1.127 | 183 | |
| Class II: Asymmetric (input miR*/miR > 1) | mir-235 | 998 | 5,019 | 0.022 | 0.003 | 821 | 6,086 | 2.879 | 0.416 | 453 | 2,528 | 11.603 | 0.487 | 192 |
| mir-90 | 2,323 | 13,937 | 0.054 | 0.002 | 4,580 | 11,900 | 0.316 | 0.022 | 1,572 | 8,970 | 3.301 | 0.256 | 105 | |
| mir-54 | 20,260 | 26,474 | 0.012 | 0.003 | 11,576 | 30,654 | 0.140 | 0.017 | 4,804 | 14,626 | 1.818 | 0.330 | 112 | |
| mir-786 | 329 | 314 | 0.139 | 0.051 | 2,067 | 2,586 | 0.149 | 0.037 | 167 | 118 | 1.188 | 0.373 | 7 | |
| mir-79 | 3,644 | 2,092 | 0.011 | 0.012 | 15,606 | 12,644 | 0.238 | 0.159 | 8,892 | 6,286 | 1.024 | 0.607 | 51 | |
| Class III: Accumulated | mir-793 | 843 | 710 | 0.001 | 0.000 | 688 | 439 | 0.104 | 0.040 | 613 | 447 | 0.350 | 0.214 | 492 |
| mir-77 | 34,022 | 14,471 | 0.002 | 0.001 | 19,458 | 18,383 | 0.114 | 0.063 | 53,246 | 27,746 | 0.190 | 0.467 | 463 | |
| mir-67 | 4,987 | 4,783 | 0.002 | 0.001 | 3,705 | 4,158 | 0.197 | 0.197 | 3,536 | 5,558 | 0.533 | 0.336 | 457 | |
| mir-82 | 23,791 | 24,879 | 0.001 | 0.000 | 7,637 | 5,013 | 0.065 | 0.010 | 18,774 | 19,543 | 0.133 | 0.057 | 213 | |
| mir-64 | 16,189 | 18,788 | 0.000 | 0.000 | 3,522 | 8,989 | 0.077 | 0.008 | 13,263 | 16,721 | 0.087 | 0.027 | 170 | |
| mir-1 | 68,107 | 82,018 | 0.002 | 0.000 | 13,608 | 6,031 | 0.183 | 0.065 | 29,357 | 26,087 | 0.202 | 0.068 | 160 | |
| mir-43 | 1,365 | 580 | 0.012 | 0.006 | 3,638 | 791 | 0.232 | 0.291 | 5,604 | 1,702 | 0.716 | 0.833 | 136 | |
| mir-50 | 9,314 | 10,552 | 0.004 | 0.001 | 2,792 | 1,126 | 0.151 | 0.017 | 698 | 659 | 0.564 | 0.087 | 134 | |
| mir-52 | 61,157 | 45,748 | 0.001 | 0.000 | 112,055 | 286,661 | 0.131 | 0.017 | 35,070 | 49,065 | 0.312 | 0.057 | 131 | |
| mir-792 | 217 | 186 | 0.003 | 0.002 | 111 | 119 | 0.071 | 0.082 | 53 | 50 | 0.286 | 0.237 | 125 | |
| lin-4 | 28,721 | 34,049 | 0.000 | 0.000 | 12,529 | 3,248 | 0.438 | 0.083 | 9,432 | 5,610 | 0.051 | 0.003 | 114 | |
| mir-248 | 267 | 314 | 0.001 | 0.000 | 122 | 84 | 0.104 | 0.033 | 61 | 49 | 0.066 | 0.028 | 113 | |
| mir-58 | 99,377 | 86,512 | 0.003 | 0.002 | 186,421 | 106,646 | 0.066 | 0.162 | 75,670 | 48,452 | 0.119 | 0.205 | 112 | |
| mir-38 | 4,656 | 2,615 | 0.007 | 0.001 | 12,010 | 1,343 | 0.025 | 0.014 | 6,628 | 1,940 | 0.142 | 0.129 | 95 | |
| mir-1829c | 458 | 385 | 0.007 | 0.001 | 568 | 718 | 0.011 | 0.017 | 138 | 113 | 0.024 | 0.067 | 92 | |
| mir-84 | 7,428 | 8,996 | 0.004 | 0.001 | 5,116 | 3,399 | 0.097 | 0.029 | 11,952 | 14,754 | 0.184 | 0.063 | 92 | |
| mir-81 | 24,466 | 27,301 | 0.001 | 0.000 | 7,395 | 8,512 | 0.190 | 0.013 | 15,353 | 28,579 | 0.142 | 0.012 | 84 | |
| mir-74 | 3,845 | 2,478 | 0.001 | 0.000 | 6,947 | 2,253 | 0.005 | 0.002 | 2,180 | 1,301 | 0.043 | 0.009 | 79 | |
| mir-71 | 54,306 | 58,135 | 0.009 | 0.003 | 34,269 | 18,007 | 0.354 | 0.206 | 29,816 | 28,479 | 0.688 | 0.248 | 78 | |
| mir-55 | 39,420 | 39,856 | 0.001 | 0.000 | 37,619 | 77,519 | 0.047 | 0.002 | 19,635 | 35,531 | 0.059 | 0.006 | 75 | |
| Class IV: Unaffected | mir-259 | 1,321 | 1,470 | 0.005 | 0.004 | 1,013 | 2,108 | 0.130 | 0.020 | 544 | 816 | 0.072 | 0.018 | 4 |
| mir-250 | 6,847 | 1,897 | 0.013 | 0.002 | 10,518 | 2,811 | 0.007 | 0.005 | 24,762 | 9,243 | 0.010 | 0.009 | 4 | |
| mir-1829a | 71 | 136 | 0.284 | 0.030 | 49 | 92 | 1.107 | 0.108 | 43 | 74 | 0.506 | 0.096 | 3 | |
| mir-356b | 53 | 96 | 0.047 | 0.021 | 24 | 60 | 0.431 | 0.141 | 45 | 114 | 0.291 | 0.064 | 3 | |
| mir-59 | 105 | 53 | 0.010 | 0.016 | 564 | 182 | 0.028 | 0.032 | 529 | 151 | 0.010 | 0.047 | 3 | |
| mir-63 | 2,915 | 2,920 | 0.034 | 0.017 | 1,520 | 999 | 0.035 | 0.013 | 3,177 | 4,966 | 0.105 | 0.041 | 2 | |
| mir-61 | 1,652 | 848 | 0.003 | 0.002 | 2,693 | 1,119 | 0.008 | 0.005 | 1,637 | 1,117 | 0.013 | 0.005 | 2 | |
| mir-36 | 1,492 | 468 | 0.105 | 0.027 | 9,510 | 2,694 | 0.047 | 0.027 | 13,420 | 6,710 | 0.056 | 0.057 | 2 | |
| mir-230 | 355 | 285 | 0.202 | 0.093 | 409 | 328 | 0.195 | 0.089 | 466 | 762 | 0.612 | 0.190 | 2 | |
| mir-2214 | 189 | 84 | 0.021 | 0.112 | 403 | 148 | 0.056 | 0.202 | 617 | 151 | 0.050 | 0.220 | 2 | |
| mir-4816 | 97 | 57 | 0.073 | 0.021 | 124 | 54 | 0.210 | 0.038 | 133 | 112 | 0.202 | 0.037 | 2 | |
| mir-75 | 3,038 | 2,426 | 0.044 | 0.026 | 4,394 | 5,836 | 0.107 | 0.072 | 29,063 | 89,083 | 0.117 | 0.044 | 2 | |
| mir-65 | 30,192 | 20,108 | 0.000 | 0.000 | 5,417 | 4,792 | 0.005 | 0.001 | 24,775 | 43,527 | 0.001 | 0.000 | 2 | |
| mir-62 | 486 | 485 | 0.244 | 0.018 | 1,397 | 478 | 0.123 | 0.027 | 1,070 | 928 | 0.075 | 0.021 | 1 | |
| mir-791 | 47 | 364 | 0.797 | 0.117 | 37 | 422 | 2.076 | 0.076 | 45 | 221 | 0.694 | 0.053 | 0 | |
| mir-34 | 3,106 | 3,459 | 0.084 | 0.054 | 2,950 | 1,253 | 0.233 | 0.096 | 4,709 | 1,753 | 0.149 | 0.023 | 0 | |
Class I: Asymmetric. MicroRNAs that had a miR*/miR ratio >1 in ALG-1(ma202) IP. Class II: Asymmetric in input. MicroRNAs that had a miR*/miR ratio >1 in input for the ALG-1(ma202) IP. Class III: Accumulated. 20 microRNAs representative of the microRNAs whose miR*/miR ratio was significantly increased in ALG-1(ma202) IP compared with wild-type ALG-1 IP. See Dataset S4 for a complete list. Class IV: Unaffected. MicroRNAs whose miR*/miR ratio changed ≤4 between wild-type ALG-1 IP and ALG-1(ma202) IP. ppm, parts per million.
miRNA Duplex Features Calculation.
Precursor arm origin of the guide miRNA, 5′ identity of the guide and passenger strands were determined based on the miRNA annotation in miRBase (48) and by considering the most abundant miRNA isoforms in deep-sequencing data. Duplex energies (ΔG) were calculated on the full miRNA sequences of both guide and passenger miRNAs using RNAduplex (59). Duplex end energies were calculated on the terminal two nucleotides form each side of the duplex (after removing the nucleotides that are overhanged), similar to refs. 11 and 16, using RNAduplex (59). Differences in the duplex end energies (ΔΔG ends) are equal to guide 5′ end duplex energy minus passenger 5′ end duplex energy.
Seed energies were calculated on nucleotides 2–7 from the 5′ end of the guide sequence and nucleotides 2–7 from the 5′ end of the star sequence. Differences in seed energies (ΔΔG seed) are equal to guide 5′ end seed duplex energy minus passenger 5′ end seed duplex energy.
Results
alg-1(anti) Mutant Phenotypes Correlate with a Shift in ALG-1–Associated Protein Complexes.
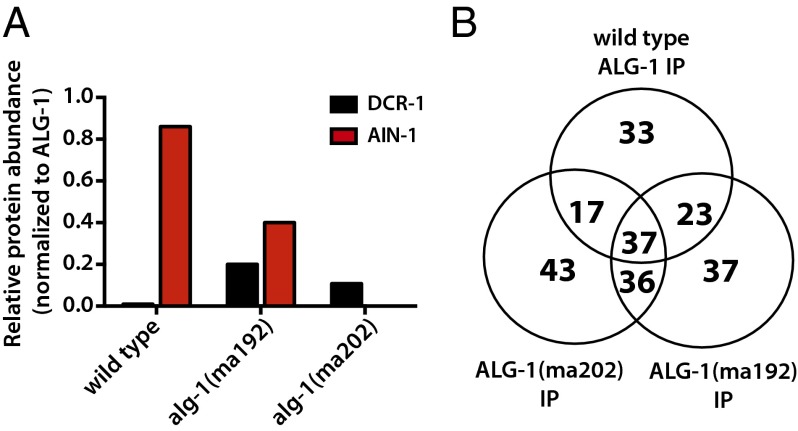
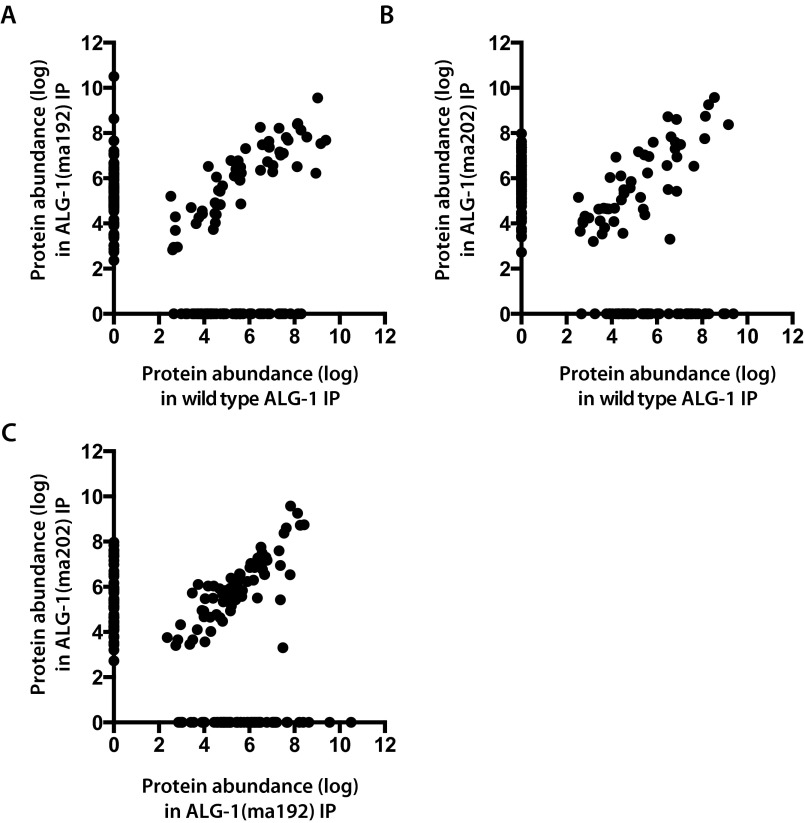
We reported previously that ALG-1(anti) protein showed a reduced interaction with the miRISC effector protein AIN-1 compared with wild-type ALG-1, and an increased association with Dicer, DCR-1 (31), suggesting an inability of ALG-1(anti)–containing miRISC to mature from processing complexes to effector complexes. To determine if interactions of ALG-1 with other proteins were affected by the alg-1(anti) mutations, we conducted ALG-1 IP from wild-type and alg-1(anti) animals followed by MudPIT (Multidimensional Protein Identification Technology) analysis to determine the composition of the associating complexes. The MudPIT data confirmed the results of our previous Western blot experiments, of a reduced ALG-1(anti) association with AIN-1 and an increased association with DCR-1 (Fig. 1A and Dataset S1), and also revealed a dramatic shift in the overall composition of the ALG-1(anti)–associated complexes (Datasets S1–S3). For example, 33 proteins were detected only in the immunoprecipitation (IP) of wild-type ALG-1, with 37 and 43 proteins detected only in the IP of ALG-1(ma192) and ALG-1(ma202), respectively. (Fig. 1B, Fig. S1, and Dataset S2). Consistent with the idea that ALG-1(anti) pre-miRISC complexes are defective in maturation from a miRISC biogenesis stage to the mature effector miRISC complex, we observed that the ALG-1(anti) IP recovered an increased yield, compared with wild-type, of certain hsp-60 and hsp-70 family proteins and other putative chaperones (Datasets S1–S3), and a reduced yield of several known miRISC effector components, including the poly-A binding proteins PAB-1 and PAB-2 (Dataset S1), which have been shown to play critical roles in target repression (37 and 38; reviewed in ref. 39). This trend was seen across the two alg-1(anti) mutants examined: alg-1(ma192) and alg-1(ma202) (Fig. 1 and Dataset S1), which affect the PIWI and MID domains of the protein, respectively (31). ALG-1(ma192) and ALG-1(ma202) coimmunoprecipitated with overlapping yet nonidentical sets of proteins (Fig. 1, Fig. S1, and Dataset S1), consistent with the hypothesis that the two mutations may not have identical effects on the ability of ALG-1 to interact with various partners.
Fig. 1.
Wild-type ALG-1 and ALG-1(anti) associate with distinct but overlapping protein populations. ALG-1 was immunoprecipitated from extracts of wild-type or alg-1(anti) animals using anti–ALG-1 antisera, and proteins were quantified by MudPIT proteomics, as described in SI Materials and Methods. (A) Association of AIN-1 with ALG-1(ma192) and ALG-1(ma202) is reduced compared with wild-type ALG-1, whereas association of DCR-1 with ALG-1(ma192) and ALG-1(ma202) is increased compared with wild-type ALG-1. AIN-1 and DCR-1 abundances in IPs are normalized to the abundance of ALG-1. (B) Venn diagram representation of the wild-type ALG-1–, ALG-1(ma192)–, and ALG-1(ma202)–associated proteins.
Fig. S1.
Scatterplots showing protein abundances (expressed as background-corrected NSAF values) for proteins detected by MudPIT proteomics in ALG-1 immunopreciptates from wild-type or alg-1(anti) extracts. (A) Wild-type ALG-1 vs. ALG-1(ma192). (B) Wild-type ALG-1 vs. ALG-1(ma202). (C) ALG-1(ma192) vs. ALG-1(ma202).
It should be noted that the proteins coimmunoprecipitated with ALG-1 in these experiments could include factors that directly interact with ALG-1 or that indirectly associate with miRISC by binding to target mRNAs, because our IP experiments were performed in the absence of RNase treatment.
alg-1(anti) Animals Accumulate miR* Strands and Precursor Hairpin Loops.
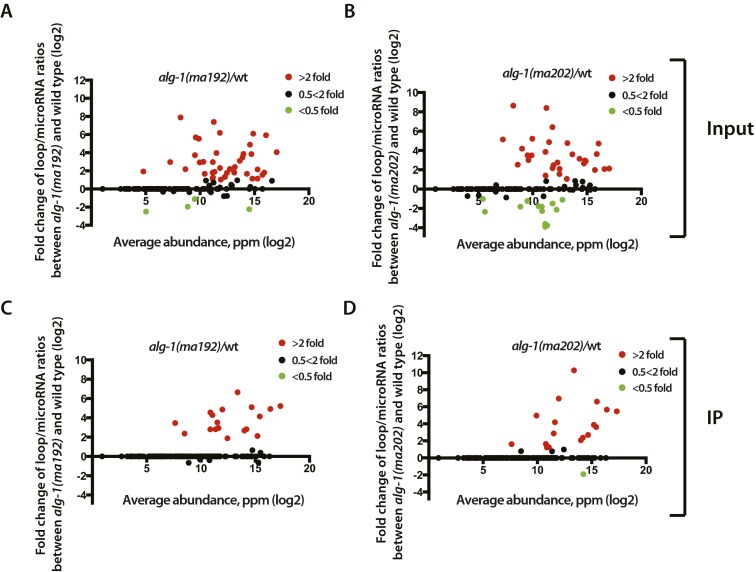
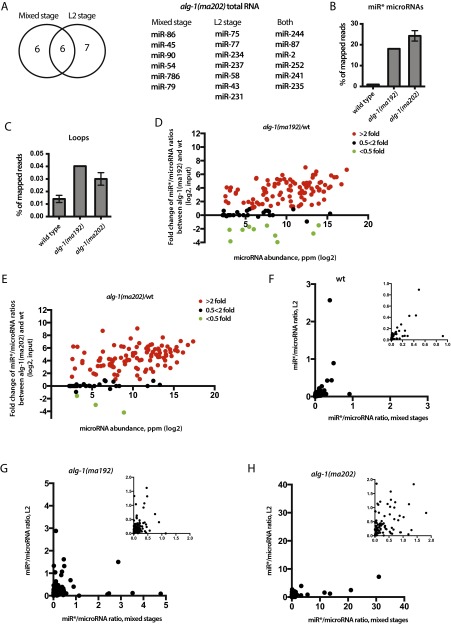
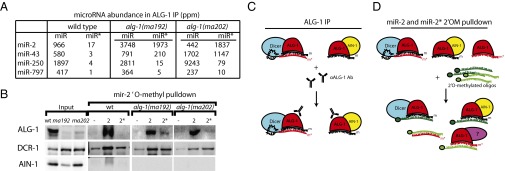
Given that ALG-1(anti) miRLC exhibits an apparent defect in maturing from a Dicer-containing configuration to the effector miRISC configuration, and considering that miR* strand release is expected to be associated with that maturation process, we tested whether alg-1(anti) mutants accumulate abnormal levels of miR* strands. Indeed, Northern blot analysis reveals a dramatic accumulation of the miR* strands for mir-80, mir-58, and mir-77 in alg-1(anti) mutants (Fig. 2A). Deep-sequencing of small RNA cDNA libraries from wild-type and alg-1(anti) mixed-staged animals confirmed that miR* strand accumulation occurs in alg-1(anti) mutants for most microRNAs, where in many cases the miR* strand increased many-fold in alg-1(anti) compared with wild-type (Fig. 2B). Moreover, alg-1(anti) mutants exhibited an increase (threefold, on average) in accumulation of the loop biproduct of the precursor microRNA processing (Fig. 2C and Fig. S2). MiR* strand accumulation occurred for most microRNAs regardless of the normal wild-type microRNA abundance (Fig. 2 D and E). For some microRNAs, such as mir-2 and mir-244, alg-1(anti) mutants showed an astounding miR*/miR ratio of 20–30, meaning that the passenger strand accumulated to levels 20- to 30-times greater than its microRNA counterpart (Table 1 and Dataset S4). This finding was in stark contrast to the wild-type, where the miR*/miR ratio was always less than one, and for most microRNAs rarely exceeded 0.02 (Table 1 and Dataset S4). Interestingly, the alg-1(ma202) allele affects miR* accumulation to a greater extent than does alg-1(ma192) (Fig. 2, Table 1, and Dataset S4), with little correlation between the profiles of miR* accumulation between the two alleles (Fig. S3). This finding is not surprising considering that alg-1(ma192) and alg-1(ma202) affect distinct domains of ALG-1.
Fig. 2.
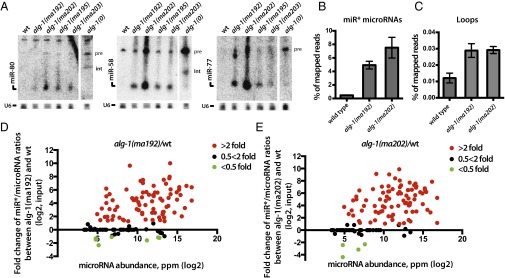
alg-1(anti) mutants show an accumulation of miR* strands in total RNA. (A) Northern blot analysis of total RNA extracted from wild-type and alg-1 mutants using probes against miR* strands (pre, precursor microRNA; int, intermediate species of microRNA processing presumably containing the passenger strand and the loop). (B and C) Deep-sequencing analysis of small RNA populations from wild-type, alg-1(ma192) and alg-1(ma202) mutant animals shows an increase in miR* populations (B) and loop accumulation (C) in the alg-1 mutants compared with wild-type. (D and E) Scatterplots showing fold-change in individual miR*/miR ratios between alg-1(anti) and wild-type total RNA. Increased miR*/miR ratio is seen in alg-1(ma192) (D) and alg-1(ma202) (E) mutants and is independent of microRNA abundance (ppm). Red dots represent microRNAs with an increased miR*/miR ratio in the alg-1 mutant over wild-type of at least twofold; green dots represent miRNAs with a decreased miR*/miR ratio in the alg-1 mutant compared with wild-type of at least twofold. Black dots represent no change in miR*/miR ratio in the alg-1 mutants compared with wild-type.
Fig. S2.
ALG-1(anti) associate with a greater amount of precursor microRNA processing byproducts; loops compared with wild-type ALG-1. (A and B) Scatterplots showing fold-change in individual loop/miR ratios between alg-1(anti) and wild-type total RNA. Increased loop/miR ratio is seen in alg-1(ma192) (A) and alg-1(ma202) (B) mutants and is independent of microRNA abundance. (C and D) Scatterplots showing fold-change in individual loop/miR ratios in material coimmunoprecipitated with ALG-1 from alg-1(anti) and wild-type. Increased loop/miR ratio is seen in alg-1(ma192) (C) and alg-1(ma202) (D) mutants and is independent of microRNA abundance. Red dots represent miRNAs with an increased loop/miR ratio in the alg-1 mutant over wild-type by at least twofold; green dots represent miRNAs with a decreased loop/miR ratio in the alg-1 mutant compared with wild-type by at least twofold. Black dots represent no change in loop/miR ratio in alg-1 mutants compared with wild-type.
Fig. S3.
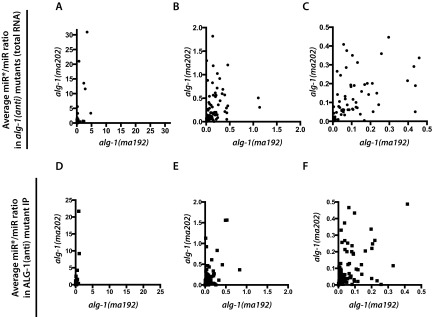
Average miR*/miR ratio in alg-1(ma192) vs. alg-1(ma202) mutant total RNA (A–C) and ALG-1(ma192) vs. ALG-1(ma202) IP (D–F). B and C how progressive zooming of the graph in A. E and F show progressing zooming of the graph in D.
To assess whether the alg-1(anti) miR*/miR ratio phenotype may be affected by developmental stage, we sequenced small RNA cDNA libraries from second larval-stage (L2) populations of wild-type and alg-1(anti) animals. L2 stage alg-1(anti) mutants were found to accumulate miR* strands many-fold compared with wild-type (Fig. S4 and Dataset S4), similarly to mixed-staged populations. Interestingly, microRNAs that showed a miR*/miR ratio of >1 in total RNA samples formed overlapping yet distinct groups in mixed-stage vs. L2 animals (Fig. S4). The differences observed could reflect differential microRNA tissue specificity and abundance at various stages of development. Importantly, however, a subset of microRNAs exhibited consistent miR*/miR ratios in both L2 and mixed-animals populations (Fig. S4 and Dataset S4).
Fig. S4.
alg-1(anti) second larval (L2)-stage animals accumulate miR* strands. (A) miRNAs that showed asymmetry (miR*/miR >1) in the total RNA of alg-1(ma202) mixed-stage populations overlapped with miRNAs that showed asymmetry in the total RNA of alg-1(ma202) L2 stage animals. (B and C) Deep-sequencing analysis of cDNA libraries from small RNA populations shows an increase in miR* (B) and loop (C) levels in alg-1(ma192) and alg-1(ma202) animals compared with wild-type. (D and E) Scatterplots showing fold-change in individual miR*/miR ratios between alg-1(anti) and wild-type total RNA. Increased miR*/miR ratio is seen in alg-1(ma192) (D) and alg-1(ma202) (E) mutants and is independent of microRNA abundance. Red dots represent microRNAs with an increased miR*/miR ratio in the alg-1 mutant over wild-type of at least twofold; green dots represent miRNAs with a decreased miR*/miR ratio in the alg-1 mutant compared with wild-type of at least twofold. Black dots represent no change in miR*/miR ratio in the alg-1 mutants compared with wild-type. (F–H) Scatterplots showing miR*/miR ratios observed in mixed-stage vs. L2-staged populations in wild-type (F), alg-1(ma192) (G), and alg-1(ma202) (H) animals. F–H, Insets show zoomed-in graphs.
Immunoprecipitated ALG-1(anti) Complexes Show an Increased Association with miR* Strands and Exhibit a Reversed Strand Bias for Some microRNAs.
Because we observed the passenger microRNA strand accumulation in the alg-1(anti) animals, we hypothesized that these passenger strands may be bound to the ALG-1(anti) Argonaute. To address this question, we immunoprecipitated ALG-1 from wild-type and mutant mixed-staged animals and cloned and sequenced the associated small RNAs. The immunoprecipitated material was enriched for microRNA populations compared with the input (Fig. 3A) and was correspondingly de-enriched for other RNA populations (Fig. 3B). We found that immunoprecipitated ALG-1(anti) associated with greater quantities of miR* strands compared with the wild-type ALG-1 (Fig. 3C). In many cases, the miR*/miR ratios were clearly enriched in ALG-1 (ma202) IP compared with input (for example, for mir-77) (Table 1 and Dataset S4), supporting a specific association of those passenger strands with the ALG-1(anti) Argonaute. Interestingly, the immunoprecipitated ALG-1(anti) also showed an increased association with loop biproducts of precursor microRNA processing (Fig. 3D), suggesting that ALG-1(anti) miRISCs may be failing to completely eject both miR* and loop products after Dicer cleavage.
Fig. 3.
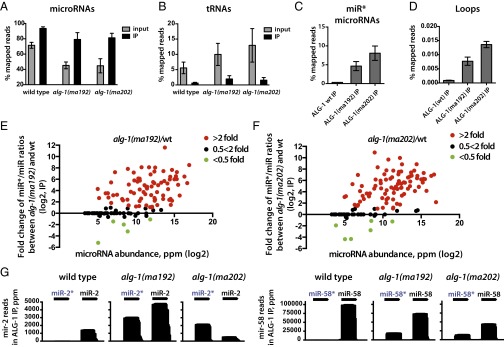
ALG-1(anti) Argonaute associates with miR* strands to a much greater degree than does wild-type ALG-1. (A and B) microRNAs are enriched (A), and tRNAs are de-enriched (B) in the ALG-1 IP compared with input. (C and D) ALG-1(anti) associates with a greater amount of miR* strands (C) and precursor microRNA loops (D) compared with the wild-type ALG-1. (E and F) Scatterplots showing fold-change in individual miR*/miR ratios in material coimmunoprecipitates with ALG-1 from alg-1(anti) and wild-type. Increased miR*/miR ratio for microRNAs associated with ALG-1 is exhibited for alg-1(ma192) (E) and alg-1(ma202) (F) mutants and is independent of microRNA abundance. Red dots represent miRNAs with an increased miR*/miR ratio in the alg-1 mutant over wild-type by at least twofold; green dots represent miRNAs with a decreased miR*/miR ratio in the alg-1 mutant compared with wild-type of at least twofold. Black dots represent no change in miR*/miR ratio in alg-1 mutants compared with wild-type. (G) Histograms showing miR-2 and miR-2*, as well as miR-58 and miR-58* strand association with immunoprecipitated wild-type and mutant ALG-1 protein.
MiR* strand accumulation in ALG-1(anti) complexes appeared to occur for the majority of microRNAs regardless of their normal abundance (Fig. 3 E and F, Table 1, and Dataset S4), including very highly abundant microRNAs, such as mir-58 (Fig. 3G and Table 1). Depending on the microRNA, the ratio of miR* to miR in ALG-1(anti) complexes could vary across a wide range, and for most microRNAs the two strands associated with ALG-1(anti) nonstoichiometrically (Table 1 and Dataset S4). Notably, for some microRNAs, the miR* strand accumulated in ALG-1(anti) complexes to a greater extent than the corresponding miR strand (Table 1). For example, for mir-2, the miR-2* strand associated with ALG-1(anti) several-fold more than the miR-2 strand (Fig. 3G and Table 1). These cases of miR* strand accumulation in excess to miR strand seem to reflect that ALG-1(anti) proteins are defective in not only miR* strand disposal, but also in the normal specificity of the miR guide strand choice (Fig. 4A and Table 1).
The 2′O-Methyl Oligo-Mediated Pull-Down of mir-58 Family and mir-2 microRNAs from Wild-Type and alg-1(anti) Differentially Enriches for miRISC Components.
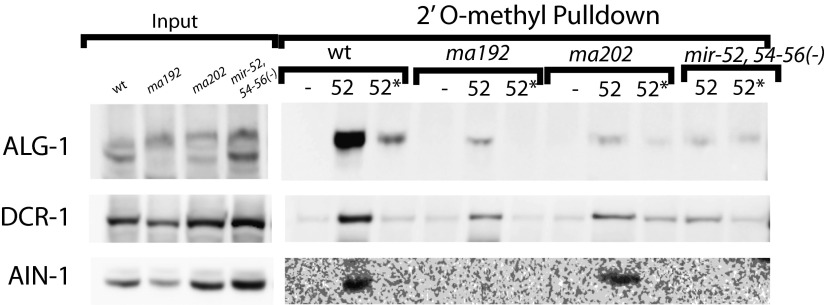
As noted above, the accumulation of miR* strands in ALG-1(anti) immunoprecipitates could reflect failure to release miR* in the context of normal strand loading choice (Fig. 4A, model I) or an additional defect in strand choice (Fig. 4A, model II). According to model I, we expect to recover ALG-1(anti) (and perhaps other miRISC components) preferentially using an anti-miR oligo, but not with an anti-miR* oligo. In contrast, model II predicts that ALG-1(anti) and the associated miRISC components should be recovered both by anti-miR pull-down and by anti-miR* pull-down. To distinguish between these models for mir-58 family microRNAs, we examined complexes recovered from wild-type and alg-1(ma192) lysates by 2′O-methyl oligo-mediated pull-down followed by MudPIT analysis of the associated complexes. We observed that known miRISC components were recovered by pull-down using an anti–miR-58 oligo but were not detected after pull-down of miR-58* (Fig. 4B). Both the miR-58* and three members of the mir-58 family microRNAs were efficiently depleted from the extract by 2′O-methyl pull-down (Fig. 4C), presumably because of the sequence homology between the related microRNAs (Fig. 4D). Therefore, we do not believe that the low signals for miRISC proteins in the anti–miR-58* pull-down (Fig. 4B) can be explained by inefficient base pairing of passenger with anti–miR-58* oligo. Rather, we interpret this result to support model I, where mir-58 family microRNA passenger strand accumulation reflects retention of the passenger strand in duplexes with an ALG-loaded guide strand. This explains the recovery of both miR-58* and miR-58 by IP of ALG-1(anti) (Fig. 4 E and G) and the asymmetry of miRISC component recovery in pull-down of miR-58 vs. miR-58* (Fig. 4 B and H). We believe that the efficient recovery of both strands by oligo pull-down reflects efficient competition of the 2′-O-methyl oligos for their cognate microRNA strands in extracts under the condition of the pull-down experiment.
Incidentally, Western blot analysis of the material recovered by the 2′O-methyl pull-down of mir-58 family guide strands (Fig. 4F) is qualitatively consistent with the MudPIT proteomic analysis of the same samples (Fig. 4B). Both the Western blot and MudPIT showed reduced association of ALG-1(anti) with mir-58 family microRNAs. However, the signal for ALG-1(anti) appeared more reduced as detected by the Western blot compared with the wild-type than the MudPIT data indicate (Fig. 4 B and F and Fig. S5). This finding could in part reflect the fact that the protein abundances of the miRISC components and other factors, as determined by MudPIT analysis, were normalized to the amount of the microRNA present in the starting material, as well as the efficiency of the pull-down (Fig. 4B). Furthermore, if ALG-1 and ALG-2 protein yields (Fig. 4B) were recalculated using only peptides that were unique to each Argonaute (discarding the shared peptides; see Materials and Methods), the difference between ALG-1 abundance in wild-type vs. alg-1(anti) animal pull-downs increases from twofold to ∼sixfold (Fig. 4B), thus becoming more consistent with the Western blot data. However, it remains possible that ALG-1(anti) detection on a Western blot by the anti–ALG-1 antibody is hampered for unknown technical reasons. Another MudPIT/Western blot discrepancy, the presence of ALG-1 in the miR-58*-associated material as detected by Western blotting in wild-type (Fig. 4F), could be a result of the more stringent washing of the 2′-O-methyl pull-down material in preparation for the MudPIT analysis.
Curiously, although the miR-58 family miR strand pull-down from alg-1(anti) animals yielded a decreased recovery of ALG-1 compared with the wild-type ALG-1, an increased amount of ALG-2 was recovered, in addition to the RDE-1 Argonaute (Fig. 4B and Fig. S5). This finding suggests a possible reapportioning of some of the mir-58 family microRNAs in alg-1(anti) mutants that normally associate with ALG-1 into ALG-2 and perhaps other Argonautes. Consistent with this scenario, we did not detect a decrease in association of mir-58 family microRNAs with the miRISC effector protein AIN-1 (Fig. 4 B and F and Fig. S5), suggesting that at least a portion of the microRNAs must be in miRISCs that contain AIN-1. The observed AIN-1 and DCR-1 signal (Fig. 4F) is likely to at least in part reflect complexes of miR-58 with ALG-2. Similar observations were made for the mir-52 family microRNA pull-downs (Fig. S6). Interestingly, mir-58 family microRNA association with AIN-2 was not observed in alg-1(ma192) mutant animals (Fig. 4B), suggesting that AIN-2 may differ from its homolog AIN-1 with respect to its interaction with Argonautes ALG-1 and ALG-2.
Fig. S6.
Western blot analysis of mir-52 microRNA family pull-downs from extracts of alg-1(anti) and wild-type animals showing association of ALG-1, DCR-1, and AIN-1 with mir-52 family microRNAs (-, scrambled oligo control pull-down; 52, miR-52 pull-down; 52*, miR-52* pull-down).
To understand the basis for miR-2* strand accumulation (Figs. 3G, 4A, and 5A) we conducted 2′O-methyl pull-down experiments from wild-type and alg-1(anti) animals using oligos complementary to miR-2 and miR-2* strands. Pull-down against miR-2 recovered ALG-1 protein from both wild-type and alg-1(anti) animals (Fig. 5B). Surprisingly, we found that pull-downs with an oligo complementary to the miR-2* also recovered ALG-1(anti) protein (Fig. 5B). This finding suggests that at least a subpopulation of miR-2* strands are associated with the mutant ALG-1(anti) as single strands, indicating that ALG-1(anti) is defective in proper strand selection for mir-2 (model II in Fig. 4A). This cannot be the only defect in miR-2::ALG-1(anti) association, because the quantity of ALG-1(anti) detected in the anti-miR-2* pull-down is less than what is associated with the miR-2 strand, even though miR-2* is in excess to miR-2 (Fig. 5A). This finding indicates that perhaps miR-2* strand accumulation in ALG-1(anti) complexes reflects a combination of two defects: its inefficient ejection from the duplex (model I in Fig. 4A) as well as erroneous miR-2* incorporation into the ALG-1(anti) Argonaute as guide (Fig. 4A, model II, and Fig. 5 C and D). However, miR-2 complementary 2′-O-methylated oligo cross-reacts with other members of the mir-2 family, given the sequence overlaps between family members, whereas miR-2* complementary oligo is expected to specifically recover miR-2*. These differential specificities of oligos may also contribute to the differing amount of ALG-1 observed as associated with both the mir-2 family microRNAs and miR-2* alone (Fig. 5 A and B).
Fig. 5.
2′-O-methyl oligonucleotide pull-downs from wild-type and alg-1(anti) mutants suggest that miR-2* accumulation in ALG-1 is in part a result of inappropriate loading into the ALG-1(anti) Argonaute. (A) mir-2 family microRNA and miR* abundances in wild-type ALG-1 and ALG-1(anti) IP. (B) Western blot analysis of miR-2 and miR-2* pull-downs from extracts of alg-1(anti) and wild-type animals showing association of ALG-1, DCR-1, and AIN-1 with mir-58 family microRNAs (-, scrambled oligo control; 2, oligo against miR-2; 2*, oligo against miR-2*). (C and D) Summary model for the miR-2* strand and related proteins accumulation as observed in alg-1(anti) mutants by (C) ALG-1(anti) IP and (D) miR-2 and miR-2* 2′O-methyl pull-downs.
The Set of microRNAs Exhibiting Grossly Abnormal miR* Loading by ALG-1(anti) Are Not Easily Distinguished by Features of Their microRNA Duplex Structures.
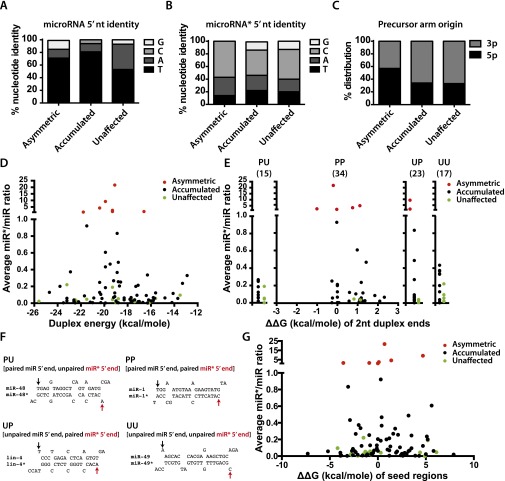
None of class I asymmetric microRNAs (Table 1) are particularly enriched (relative to the other classes; see Table 1) for microRNAs that preferentially associate with ALG-1 compared with ALG-2 (40), suggesting that a simple bias of microRNAs to preferentially load into one Argonaute vs. another cannot explain the asymmetric miR* loading into ALG-1(anti). In addition, class I asymmetric microRNAs do not have a 5′ nucleotide bias for either miR or miR* (Fig. S7 A and B). Similarly, no 5p vs. 3p precursor arm bias was detected for asymmetric microRNAs (class I) compared with other classes (Table 1 and Fig. S7C). MicroRNAs with highly ALG-1(anti)–enriched miR* strands (class I in Table 1) also showed similar distribution of duplex and duplex end energies compared with those microRNAs whose miR* strands were not enriched in ALG-1 IP (classes II, III, and IV in Table 1, and Fig. S7 D and E). Pairing (or lack thereof) of the 5′ nucleotide of miR or miR* also does not explain the differential incorporation of some miR* strands into ALG-1(anti) (Fig. S7 E and F). Similarly, our analysis of differences between duplex seed energies on the 5′ end of the guide vs. 5′ end of the miR* (ΔΔG seed) did not distinguish the asymmetrically loaded microRNAs from the other classes (Fig. S7G).
Fig. S7.
microRNA feature analysis of microRNAs with a miR*/miR ratio of >1 in ALG-1(ma202) IP (termed asymmetric microRNAs) (class I in Table 1); classes II and III, microRNAs that showed an increase of miR*/microRNA ratio in ALG-1(ma202) compared with wild-type ALG-1 (accumulated) (Table 1); and the microRNAs whose miR*/microRNA ratio was not affected (termed “unaffected”) (class IV in Table 1). (A) The identity of the 5′ nucleotide of the guide microRNA. (B) The identity of the 5′ nucleotide of the miR* microRNA. (C) Precursor arm origin. (D) Duplex energies comparison between the asymmetric (class I in Table 1), accumulated (classes II and III in Table 1), and unaffected (class IV in Table 1) microRNA populations. (E) Differences in the end-pairing and duplex end energies (ΔΔG) of the three microRNA populations. PP, paired 5′ nucleotide of the microRNA, paired 5′ nucleotide of the miR*; PU, paired 5′ nucleotide of the microRNA, unpaired 5′ nucleotide of the miR*; UP, unpaired 5′ nucleotide, paired 5′ nucleotide of the miR*. Number in parenthesis represents the total number of microRNAs within each group. ΔΔG(kcal/mole) of two nucleotides at each end of duplex is plotted as a function of miR*/microRNA ratio for the microRNA duplexes that had paired nucleotides at each duplex end. (F) MicroRNA duplex examples of the four groups (PU, PP, UP, UU) shown in E. (G) ΔΔG(kcal/mole) of seed energies (ΔG of miR seed minus ΔG of miR* seed) is plotted as a function of miR*/miR ratio for the microRNA duplexes.
Discussion
Here we report detailed molecular characterizations of the previously genetically described antimorphic alg-1 mutations (31). These mutations were shown to have broad effects on microRNA function and are more detrimental than complete loss of ALG-1, yet do not significantly affect Dicer processing of microRNA precursors or mature guide microRNA accumulation (31). The strong phenotypes caused by ALG-1(anti) could be explained by the observation that ALG-1(anti) complexes contain microRNAs, but poorly associate with the miRISC effector AIN-1, and hence sequester microRNAs in ineffective complexes (31). Previous findings also showed that ALG-1(anti) complexes are more enriched for Dicer than are wild-type ALG-1 complexes, further indicating that ALG-1(anti) proteins could be defective in transition from miRISC biogenesis to miRISC function (31).
In this study, we aimed to better understand the effects of these alg-1(anti) mutations on ALG-1 function through comprehensive analysis of the profile of proteins and microRNAs associated with ALG-1(anti) complexes in vivo. MudPIT mass spectometry characterization of ALG-1(anti)–associated proteins revealed a shift in the profile of protein interactors compared with the wild-type that is consistent with defective miRISC maturation by ALG-1(anti). Furthermore, we show that for most microRNAs the ALG-1(anti) complexes inappropriately retain the miR* strand, the strand of the pre-microRNA duplex that is ordinarily, in the wild-type, chosen as the passenger strand and hence disposed of. In some cases, the miR* strand accumulates in dramatic excess to the microRNA strand, revealing defects in the specificity of guide strand choice.
These results suggest that the normal function of ALG-1 includes direct or indirect roles in (i) the specificity of the microRNA strand selection, and (ii) the disposal of the miR* strands. Importantly, we observe that the effects of ALG-1(anti) on microRNA strand selection and miR* disposal can differ widely—qualitatively and quantitatively—depending on the particular microRNA. This finding suggests that wild-type ALG-1 Argonaute normally functions as an active component of complexes that recognize specific microRNA precursors in such a way as to determine the specificity of microRNA strand selection and the efficiency of miR* strand disposal.
ALG-1(anti) Complex Composition Is Consistent with Stalled miRLC Complexes.
Our MudPIT analysis of the ALG-1 coimmunoprecipitates from wild-type and alg-1(anti) mutant animals revealed a shift in the profile of ALG-1–associated proteins (Fig. 1, Fig. S1, and Dataset S1). This shift includes the detection of protein populations associated with ALG-1(anti) that were not detected in wild-type ALG-1 complexes, and an absence of proteins that were detected only in wild-type complexes. This finding is consistent with the model that both ALG-1(ma192) and ALG-1(ma202)-containing complexes are defective in properly maturing into functional miRISC. These MudPIT data support and expand our previous findings from Western blot analyses of Dicer and AIN-1 association with ALG-1(anti) (31). By MudPIT, ALG-1(anti) are observed to associate more with DCR-1 and less with AIN-1, than does wild-type ALG-1 (Fig. 1A and Dataset S1). The MudPIT data further show that ALG-1(anti) exhibit a reduced association with a set of additional proteins known to be required for microRNA target translational repression and target degradation, including poly(A)-binding proteins PAB-1 and PAB-2. At the same time, ALG-1(ma192) IPs contained an increase in heat-shock family proteins that have been previously shown to assist in the loading of the Argonautes (4, 6). Therefore, this observation further supports our model that ALG-1(anti)–containing complexes are stalled in a pre-miRISC form, perhaps as the miRLC.
Our deep-sequencing of cDNA libraries prepared from ALG-1(anti) immunoprecipitated complexes further supports the interpretation of ALG-1(anti) complexes as being stalled in a miRLC form. Notable in this regard is our finding that both miR* and loop sequences, which are excised by Dicer from microRNA precursors and usually ejected before miRISC maturation, are enriched in ALG-1 IP from alg-1(anti) worms compared with the wild-type (Fig. 3).
microRNAs Can Shift from ALG-1 to Other Argonautes in alg-1(anti) Animals.
MudPIT analysis of complexes recovered by oligonucleotide-mediated pull-down of mir-58 family microRNAs from alg-1(anti) worm extracts revealed an increase in the amount of ALG-2 recovered (Fig. 4B). This increase could reflect an up-regulation of ALG-2 protein abundance in the alg-1(anti) background (for example, via a hypothetical compensatory regulation of ALG-2). We showed previously that alg-2 mRNA levels were not detectably increased in alg-1(anti) animals, but we were not able to determine ALG-2 protein abundance because of a lack of ALG-2–specific antiserum (31). Alternatively, inability of ALG-1(anti) Argonautes to appropriately load miR-58 may result in a release of the mir-58 duplex from ALG-1(anti) and subsequent take up by the available microRNA competent Argonautes such as ALG-2 and RDE-1, according to a previously proposed “duplex sorting” model (11).
Interestingly, oligonucleotide-mediated pull-down of the miR-58 family guide also showed a relatively minor, but nevertheless striking, shift of these microRNAs to RDE-1 in alg-1(anti) mutants (Fig. 4B and Fig. S5). RDE-1 has been previously shown to associate with multiple classes of small RNAs, including microRNAs (41). It is possible that RDE-1 may function as a natural reservoir for microRNAs that are, under certain circumstances, excluded from ALG-1 and ALG-2. It is worth noting that if there were a broad shift in microRNAs from ALG-1(anti) to other Argonauts, then that could result in a general mitigation of the alg-1(anti) phenotypes by providing some measure of functional replacement of the defective ALG-1(anti) protein.
ALG-1(anti) Causes Defects in Guide Strand Choice and Passenger Strand Disposal.
Ordinarily, for the majority of microRNA precursors, one strand of the duplex is specifically selected to associate with Argonaute in the guide strand configuration, whereas the other strand is ejected and subsequently degraded. Therefore, in the wild-type, the miR*/miR ratio is exceedingly small for most microRNAs, with few exceptions, such as mir-30 in mammals (42) and mir-47 in C. elegans (Dataset S4). A striking aspect of our findings is that the usual asymmetry of miR vs. miR* accumulation is dramatically altered in ALG-1(anti) complexes (Table 1). In ALG-1(ma202) complexes, 75 microRNAs exhibited a 4- to ∼2,000-fold increase in the ratio of miR* to miR compared with the wild-type (Table 1 and Dataset S4).
Importantly, for certain microRNAs, the miR* strand accumulated in ALG-1(anti) complexes to levels in excess to their miR strand counterparts [for example, miR-2* in alg-1(ma202)]. In such cases of miR* excess, at least a portion of the miR*, must be loaded into ALG-1(anti) in the guide strand configuration. This interpretation is supported by our finding that oligonucleotide pull-down of miR-2* from alg-1(ma202) extracts yielded ALG-1, as detected by Western Blot (Fig. 5). Thus, for at least some microRNAs (class I in Table 1), ALG-1(anti) complexes fail to execute the proper specificity of guide strand choice.
For those microRNAs whose miR* strand did accumulate in ALG-1(anti) complexes, but not in excess to the microRNA strand (classes II and III in Table 1), the underlying defect could be a defective guide strand choice selection, although less potent than for the class I microRNAs. Alternatively, for some of these class II/III microRNAs, miR* strand accumulation could reflect simply a miR* strand release defect in the context of normal strand choice. Pull-down of miR-58* strands in alg-1(anti) did not appreciably recover ALG-1, ALG-2, AIN-1/2, or DCR-1 (Fig. 4 B and F), whereas pull-down of the miR-58 strand did recover these known Argonaute complex components (Fig. 4 B and F). Thus, unlike miR-2, where oligonucleotide pull-down of the miR* strand did yield ALG-1 (Fig. 5), the miR-58* strand seems to be not directly associated with ALG-1 but in a duplex with ALG-bound miR-58 microRNA. This finding indicates that for mir-58 and likely many other microRNAs exhibiting miR* strand accumulation, ALG-1(anti) causes a miR* strand release defect (Fig. 4A, model I).
Defective passenger strand release by ALG-1(anti) suggests an active role for ALG-1 in the passenger release process. This role may represent a direct activity of ALG-1, such as cleavage of passenger strand by ALG-1 slicer activity. However, recombinant ALG-1(anti) protein seems to retain slicer activity by in vitro assays (31). Moreover, many microRNA precursors contain mismatched bases in the center of their precursor helix, precluding passenger strand slicing (43). Thus, excess miR* accumulation in ALG-1(anti) Argonautes likely reflects passenger release by slicing-independent mechanisms.
It is curious that for certain microRNAs we observed high levels of miR* strands in whole-worm extracts, yet the miR* strands of those particular microRNAs were not efficiently recovered by in ALG-1 immunoprecipitation (Fig. 4B and Table 1). This finding suggests that many miR* strands are either not stably associated with ALG-1 under the conditions of IP, or they are associated with other complexes of unknown composition.
Throughout our characterizations of the alg-1(anti) mutants we observed quantitative and qualitative differences between alg-1(ma192) and alg-1(ma202) alleles in how these two mutant ALG-1 proteins affect miR* strand accumulation for specific microRNAs. alg-1(ma202) animals appear to accumulate miR* strands to a greater degree than do alg-1(ma192) mutants, both in total RNA samples and in ALG-1 IP (Figs. 2 and 3, Table 1, and Fig. S3). alg-1(ma192) and alg-1(ma202) alleles affect two distinct domains of ALG-1 (PIWI and MID, respectively) and may differentially affect ALG-1 interactions with specific microRNAs and protein cofactors. It is intriguing to consider that the two mutated forms of ALG-1 Argonaute both disrupt miRLC to miRISC maturation but impose distinct effects on the ability of ALG-1 to interact with specific microRNA and protein cofactors.
Distinct Effects of ALG-1(anti) on Different microRNAs.
Our data indicate at least two distinct effects of ALG-1(anti) on microRNA biogenesis that are exhibited to varying degrees by different microRNAs: loss of specificity of guide strand selection (for example, mir-2) (Table 1), and defective passenger strand release (for example: mir-58) (Table 1). At this point we cannot estimate how many microRNAs in classes I and II/III could be defective in only passenger strand release (such as mir-58), or only guide strand selection, or could be affected by compound defects in both processes.
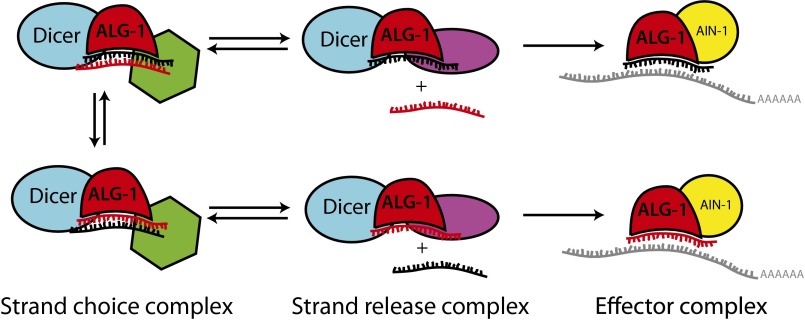
We speculate that differing effects of ALG-1(anti) on miR strand selection and miR* disposal for distinct microRNAs could reflect how ALG-1 recognizes sequence-specific or duplex structure-specific features characteristic of each pre-microRNA in the context of miRLC. Presumably, for all microRNAs, guide strand selection involves a sampling by the miRLC of both orientations of the duplex (with either miR or miR* sampled as the potential guide), with the specificity of the guide strand choice determined by mechanisms that couple passenger release selectively to a specific duplex orientation (Fig. 6). There is evidence that microRNA passenger strand selection by Argonaute may involve conformational changes in the Argonaute::pre-mir complex that could be triggered by structural features characteristic of one duplex orientation, and that subsequently favor passenger strand release (19). It is possible that ALG-1(anti) mutant proteins are primarily defective in key miRLC conformational changes that couple miR* or passenger strand release to a specific duplex orientation for specific microRNAs. According to this model, microRNAs, such as mir-2 and other class I asymmetric microRNAs, correspond to cases where strand selection depends on a conformational change that is defective in ALG-1(anti). On the other hand, microRNAs (such as mir-58) that display proper strand selection in alg-1(anti) mutants, but poor miR* strand release, are those for which strand selection is determined before the defective conformational change.
Fig. 6.
A model representing factors that influence the orientation of microRNA duplex loading.
Interestingly, the class of asymmetric microRNAs, whose miR* strands were more abundant in the ALG-1(anti) than their miR strands (class I in Table 1), is enriched for microRNAs with paired duplex ends at both the 5′ and the 3′ ends (Fig. S7E), with all of the asymmetric microRNA duplexes being paired at the 3′ end. At the same time, the majority of the unaffected microRNAs had an unpaired 3′ nucleotide (Fig. S7E). This observation is intriguing but not predictive of asymmetric loading, as that difference alone cannot explain the disparity between miR* Argonaute loading for class I microRNAs compared with all other microRNAs. It is possible that our inability to identify microRNA duplex features as predictors of asymmetric loading into ALG-1(anti) was hampered by the small sample size of class I microRNAs in this study (Table 1). Therefore, we do not preclude the possibility that certain features or some combination thereof may in fact influence strand loading in alg-1(anti) mutants. However, our data raise an intriguing possibility that additional microRNA-extrinsic mechanisms may help determine which microRNA strand becomes loaded and functional.
It’s interesting to consider that different rules governing guide strand selection and passenger disposal may apply to different microRNAs. Given the differences between protein cofactors associated with ALG-1 in wild-type and alg-1(anti) mutant animals, one attractive possibility is that Argonaute cofactors may be what imparts the context/microRNA specificity of strand selection and allows for modulation of strand choice (Fig. 6). We suggest that duplex orientation is sampled by the miRLC and that loading of the duplex into the Argonaute in correct orientation depends on several properties, both intrinsic and extrinsic to the microRNA duplex itself. We propose that features of the duplex structure, conformation of the Argonaute protein itself, and the presence of microRNA-specific protein cofactors (shown in green and purple in Fig. 6) may all be important for the appropriate orientation of the duplex during miRLC loading.
Given the evidence that passenger strand may be functional in certain circumstances (23, 44–46), the ability to modulate the choice between guide and passenger strands must be highly regulated. Such regulation could be indispensable in developmental and pathological contexts in which the specificity of guide/passenger strand selection is critical. Further study is required to develop a fuller understanding of how microRNA strand choice is executed and regulated by Argonautes and their associated cofactors.
Supplementary Material
Acknowledgments
We thank the members of the V.R.A. and the Mello laboratories for helpful discussions, especially Weifeng Gu for help with initial high-throughput sequencing data analysis; and the University of Massachusetts Medical School Molecular Biology Core for performing the high-throughput sequencing. This work was supported in part by a Tara Bean postdoctoral fellowship (to A.Y.Z.); a Leukemia and Lymphoma Society postdoctoral fellowship (to I.V.-L.); and National Institutes of Health Grants R01GM089778 (to J.A.W.) and R01GM34028 (to V.R.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GEO database (accession no. GSE72659). Proteomic mass spectrometry data have been deposited to the ProteomeXchange Consortium via the MassIVE partner repository (data set identifier PXD002835).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506576112/-/DCSupplemental.
References
- 1.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maniataki E, De Planell Saguer MDA, Mourelatos Z. Immunoprecipitation of microRNPs and directional cloning of microRNAs. Methods Mol Biol. 2005;309:283–294. doi: 10.1385/1-59259-935-4:283. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Jin D-Y, McManus MT, Mourelatos Z. Precursor microRNA-programmed silencing complex assembly pathways in mammals. Mol Cell. 2012;46(4):507–517. doi: 10.1016/j.molcel.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwasaki S, et al. Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex. Nature. 2015;521(7553):533–536. doi: 10.1038/nature14254. [DOI] [PubMed] [Google Scholar]
- 5.Martinez NJ, Chang H-M, Borrajo J de R, Gregory RI. The co-chaperones Fkbp4/5 control Argonaute2 expression and facilitate RISC assembly. RNA. 2013;19(11):1583–1593. doi: 10.1261/rna.040790.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwasaki S, et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39(2):292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Meister G. Argonaute proteins: Functional insights and emerging roles. Nat Rev Genet. 2013;14(7):447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 8.Miki TS, Rüegger S, Gaidatzis D, Stadler MB, Großhans H. Engineering of a conditional allele reveals multiple roles of XRN2 in Caenorhabditis elegans development and substrate specificity in microRNA turnover. Nucleic Acids Res. 2014;42(6):4056–4067. doi: 10.1093/nar/gkt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warf MB, Johnson WE, Bass BL. Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA. 2011;17(4):563–577. doi: 10.1261/rna.2432311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36(3):431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czech B, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36(3):445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16(1):43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seitz H, Tushir JS, Zamore PD. A 5′-uridine amplifies miRNA/miRNA* asymmetry in Drosophila by promoting RNA-induced silencing complex formation. Silence. 2011;2:4. doi: 10.1186/1758-907X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465(7299):818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 15.Mi S, et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133(1):116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 17.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 18.Krol J, et al. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem. 2004;279(40):42230–42239. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 19.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16(9):953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 20.Yoda M, et al. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17(1):17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu HY, et al. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, et al. The expression of miR-30a* and miR-30e* is associated with a dualistic model for grading ovarian papillary serious carcinoma. Int J Oncol. 2014;44(6):1904–1914. doi: 10.3892/ijo.2014.2359. [DOI] [PubMed] [Google Scholar]
- 23.Bang C, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124(5):2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J-S, et al. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17(2):312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamura K, et al. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15(4):354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson RC, et al. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell. 2015;57(3):397–407. doi: 10.1016/j.molcel.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishida KM, et al. Roles of R2D2, a cytoplasmic D2 body component, in the endogenous siRNA pathway in Drosophila. Mol Cell. 2013;49(4):680–691. doi: 10.1016/j.molcel.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 28.Betancur JG, Tomari Y. Dicer is dispensable for asymmetric RISC loading in mammals. RNA. 2012;18(1):24–30. doi: 10.1261/rna.029785.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki HI, et al. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat Struct Mol Biol. 2015;22(7):512–521. doi: 10.1038/nsmb.3050. [DOI] [PubMed] [Google Scholar]
- 31.Zinovyeva AY, Bouasker S, Simard MJ, Hammell CM, Ambros V. Mutations in conserved residues of the C. elegans microRNA Argonaute ALG-1 identify separable functions in ALG-1 miRISC loading and target repression. PLoS Genet. 2014;10(4):e1004286. doi: 10.1371/journal.pgen.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12(4):589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 35.Gu W, Claycomb J, Batista P, Mello C, Conte D. In: Methods in Molecular Biology. Hobman TC, Duchaine TF, editors. Humana Press; Totowa, NJ: 2011. pp. 251–280. [DOI] [PubMed] [Google Scholar]
- 36.Jannot G, Vasquez-Rifo A, Simard MJ. In: Methods in Molecular Biology. Hobman TC, Duchaine TF, editors. Humana Press; Totowa, NJ: 2011. pp. 233–249. [DOI] [PubMed] [Google Scholar]
- 37.Huntzinger E, et al. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 2013;41(2):978–994. doi: 10.1093/nar/gks1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuzuoglu-Öztürk D, Huntzinger E, Schmidt S, Izaurralde E. The Caenorhabditis elegans GW182 protein AIN-1 interacts with PAB-1 and subunits of the PAN2-PAN3 and CCR4-NOT deadenylase complexes. Nucleic Acids Res. 2012;40(12):5651–5665. doi: 10.1093/nar/gks218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun JE, Huntzinger E, Izaurralde E. A molecular link between miRISCs and deadenylases provides new insight into the mechanism of gene silencing by microRNAs. Cold Spring Harb Perspect Biol. 2012;4(12):4. doi: 10.1101/cshperspect.a012328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasquez-Rifo A, et al. Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS One. 2012;7(3):e33750. doi: 10.1371/journal.pone.0033750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corrêa RL, Steiner FA, Berezikov E, Ketting RF. MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet. 2010;6(4):e1000903. doi: 10.1371/journal.pgen.1000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouasker S, Simard MJ. The slicing activity of miRNA-specific Argonautes is essential for the miRNA pathway in C. elegans. Nucleic Acids Res. 2012;40(20):10452–10462. doi: 10.1093/nar/gks748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozomara A, Hunt S, Ninova M, Griffiths-Jones S, Ronshaugen M. Target repression induced by endogenous microRNAs: Large differences, small effects. PLoS One. 2014;9(8):e104286. doi: 10.1371/journal.pone.0104286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji H, et al. Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLoS One. 2014;9(10):e110314. doi: 10.1371/journal.pone.0110314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Almeida MI, et al. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142(4):886–896.e9. doi: 10.1053/j.gastro.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou Y, et al. Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science. 2013;340(6130):372–376. doi: 10.1126/science.1231321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser P, Wohlschlegel J. Identification of ubiquitination sites and determination of ubiquitin-chain architectures by mass spectrometry. Methods Enzymol. 2005;399:266–277. doi: 10.1016/S0076-6879(05)99018-6. [DOI] [PubMed] [Google Scholar]
- 51.Wohlschlegel JA. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol. 2009;497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- 52.Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J Proteome Res. 2012;11(6):3487–3497. doi: 10.1021/pr3000249. [DOI] [PubMed] [Google Scholar]
- 53.Michalski A, et al. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10(9):M111.011015. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cociorva D, Tabb DL, Yates JR. Validation of tandem mass spectrometry database search results using DTASelect. Curr Protoc Bioinformatics. 2006;Chapter 13:Unit 4. doi: 10.1002/0471250953.bi1304s16. [DOI] [PubMed] [Google Scholar]
- 55.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu T, et al. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics 2006 , 5(10 suppl):S174. [Google Scholar]
- 57.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 58.Florens L, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40(4):303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.