Significance
Coral reefs form marine-biodiversity hotspots of enormous ecological, economic, and aesthetic importance that rely energetically on a functional symbiosis between the coral animal and a photosynthetic alga. The ongoing decline of corals worldwide due to anthropogenic influences, including global warming, ocean acidification, and pollution, heightens the need for an experimentally tractable model system to elucidate the molecular and cellular biology underlying the symbiosis and its susceptibility or resilience to stress. The small sea anemone Aiptasia is such a system, and our analysis of its genome provides a foundation for research in this area and has revealed numerous features of interest in relation to the evolution and function of the symbiotic relationship.
Keywords: coral reefs, endosymbiosis, horizontal gene transfer, dinoflagellate, pattern-recognition receptors
Abstract
The most diverse marine ecosystems, coral reefs, depend upon a functional symbiosis between a cnidarian animal host (the coral) and intracellular photosynthetic dinoflagellate algae. The molecular and cellular mechanisms underlying this endosymbiosis are not well understood, in part because of the difficulties of experimental work with corals. The small sea anemone Aiptasia provides a tractable laboratory model for investigating these mechanisms. Here we report on the assembly and analysis of the Aiptasia genome, which will provide a foundation for future studies and has revealed several features that may be key to understanding the evolution and function of the endosymbiosis. These features include genomic rearrangements and taxonomically restricted genes that may be functionally related to the symbiosis, aspects of host dependence on alga-derived nutrients, a novel and expanded cnidarian-specific family of putative pattern-recognition receptors that might be involved in the animal–algal interactions, and extensive lineage-specific horizontal gene transfer. Extensive integration of genes of prokaryotic origin, including genes for antimicrobial peptides, presumably reflects an intimate association of the animal–algal pair also with its prokaryotic microbiome.
Coral reefs form marine-biodiversity hotspots that are of enormous ecological, economic, and aesthetic importance. Coral growth and reef deposition are based energetically on the endosymbiosis between the cnidarian animal hosts and photosynthetic dinoflagellate algae of the genus Symbiodinium, which live in vesicles within the gastrodermal (gut) cells of the animal and typically supply ≥90% of its total energy, while the host provides the algae with a sheltered environment and the inorganic nutrients needed for photosynthesis and growth (1). This tight metabolic coupling allows the holobiont (i.e., the animal host and its microbial symbionts) to thrive in nutrient-poor waters. Although the ecology of coral reefs has been studied intensively, the molecular and cellular mechanisms underlying the critical endosymbiosis remain poorly understood (2). As coral reefs face an ongoing and increasing threat from anthropogenic environmental change (3), new insights into these mechanisms are of critical importance to understanding the resilience and adaptability of coral reefs and thus to the planning of conservation strategies (4).
Aiptasia is a globally distributed sea anemone that harbors endosymbiotic Symbiodinium like its Class Anthozoa relatives the stony corals (Fig. 1 and SI Appendix, Fig. S1A) (4, 5). Aiptasia has a range of polyp sizes convenient for experimentation and is easily grown in laboratory culture, where it reproduces both asexually (so that large clonal populations can be obtained) and sexually (allowing experiments on larvae and potentially genetic studies), and it can be maintained indefinitely in an aposymbiotic (dinoflagellate-free) state and reinfected by a variety of Symbiodinium strains (6, 7). These characteristics make Aiptasia a highly attractive model system for studies of the molecular and cellular basis of the cnidarian–dinoflagellate endosymbiosis (2, 4). To provide a solid platform for research on Aiptasia, we have sequenced and analyzed its genome. The results have already provided important insights into several aspects of the evolution and function of the symbiotic system.
Fig. 1.
Phylogenetic position and different symbiotic states of Aiptasia. (A) Partial phylogenetic tree (see SI Appendix, SI Materials and Methods and Fig. S1A for details) shows Aiptasia grouped with other anthozoans among the cnidarians. Numbers on nodes denote bootstrap values. (B–D) An aposymbiotic Aiptasia polyp (B) and symbiotic polyps viewed under white light (C) or by fluorescence microscopy to visualize the red chlorophyll autofluorescence of the endosymbiotic Symbiodinium algae (D).
Results and Discussion
Genome Size and Assembly.
We used flow cytometry to estimate the haploid genome size of Aiptasia, obtaining a value of ∼260 Mb (SI Appendix, Fig. S2 A, 1–3). This value is smaller than those reported previously for three other cnidarians (SI Appendix, Table S1), including two other anthozoans, the anemone Nematostella vectensis (450 Mb) (8) and the stony coral Acropora digitifera (420 Mb) (9). However, we estimated a genome size of ∼329 Mb for N. vectensis by flow cytometry (SI Appendix, Fig. S2 A, 4–6), and a previous independent estimate by densitometry of Feulgen-stained embryonic nuclei gave an even lower value of ∼220 Mb (10).
To generate a genome sequence, we used DNA from a clonal population of aposymbiotic anemones to obtain ∼140× coverage by Illumina short-read sequencing plus additional sequence from long-insert mate-pair libraries (SI Appendix, Table S2). From these sequences we assembled a draft genome of ∼258 Mb comprised of 5,065 scaffolds and 29,410 contigs, with half of the genome found in scaffolds longer than 440 kb and in contigs longer than 14.9 kb (Table 1 and SI Appendix, Table S3). Using ab initio prediction informed by a reference transcriptome covering several developmental and symbiotic states (SI Appendix, Tables S4 and S5 and Fig. S1C), we identified 29,269 gene models, of which 26,658 appear to be complete (Table 1) and 27,469 (including 25,370 of the seemingly complete genes) were supported by RNA evidence (SI Appendix, Table S6). Of the 29,269 gene models, 26,162 (89%) have identifiable homologs in other eukaryotes and 22,993 have annotations in the UniProt database (SI Appendix, Table S6 and Dataset S1.1). The Aiptasia genome assembly and gene predictions compare well to those in the other sequenced cnidarians (SI Appendix, Table S1), and this comparison also suggests that the smaller genome size of Aiptasia reflects largely the smaller sizes of introns and their lower frequency (as indicated by a larger mean exon size) in Aiptasia (SI Appendix, Table S1). Indeed, as expected, the diversity of molecular functions represented in the predicted Aiptasia protein set is comparable to those in other cnidarians and in other “basal” metazoans (SI Appendix, Fig. S2 B and C) (11).
Table 1.
Statistics for the Aiptasia genome assembly and gene annotation
| Parameter | Value |
| Genome size, by measurement of DNA content* | 260 Mb |
| Total size of genome assembly, in 5,065 scaffolds | 258 Mb |
| Total contig size, in 29,410 contigs | 213 Mb |
| Scaffold N50 | 440 kb |
| Longest scaffold | 2,344 kb |
| Contig N50 | 14.9 kb |
| Longest contig | 228 kb |
| Number of gene models | 29,269 |
| Number of apparently complete gene models† | 26,658 |
| Number of predicted proteins with recognizable (E-value ≤1e-5) homologs in other eukaryotes‡ | 26,162 |
| Mean predicted protein length | 517 amino acids |
| Repeat content | 26% |
See also SI Appendix, Tables S1, S3, and S6. kb, kilobase; Mb, megabase pairs; N50, 50% of the assembly is in scaffolds or contigs longer than this value.
Gene models with both predicted start and stop codons.
See SI Appendix, Table S6 for details.
Repeat Content and Evolution, Synteny, and Fast-Evolving Genes.
The Aiptasia genome contains ∼26% repeated sequences, intermediate between what has been reported for other cnidarians (SI Appendix, Table S1) and similar to the estimates for other basal metazoans such as Capitella teleta (31%) and Lottia gigantea (20%) (11, 12). Of the repeated sequences, ∼36% can be assigned to previously known repetitive elements (SI Appendix, Table S7) that are in multiple categories including a variety of known transposable elements (TEs). We found at least four classes of TEs that showed an expansion at a Jukes–Cantor distance of 0.4–0.5 (Fig. 2A and SI Appendix, Fig. S3 A and B). This ancient TE expansion aligns with the time of divergence of Aiptasia from Verrilactis paguri and Sagartia elegans, two anemone species that are not symbiotic with dinoflagellates, based on Nei–Gojobori synonymous-substitution rates in the cytochrome c oxidase III gene (COX3) (Fig. 2A). Such burst-like expansions of TEs have been reported to reflect ancient population bottlenecks (13) and/or speciation events, as well as major increases in genome size, such as that found in Hydra (14). Remarkably, an ancient expansion of TEs in the stony coral A. digitifera resembles that in Aiptasia, whereas the more closely related N. vectensis (which is not symbiotic with dinoflagellates) appears not to have had such an expansion (SI Appendix, Fig. S3A). The existence of similar repeat expansions in Aiptasia and A. digitifera is intriguing, and it remains to be determined if these genomic rearrangements were functionally associated with some common event affecting their symbiotic lifestyles.
Fig. 2.
Genomic rearrangements in Aiptasia. (A) Correlation between a period of speciation and a period of high TE activity. (Top) Approximate times of divergence of Aiptasia from the anemone species S. elegans, V. paguri, and Bartholomea annulata based on Nei–Gojobori synonymous substitution rates in the COX3 gene (SI Appendix, SI Materials and Methods), which correspond to the Jukes–Cantor distances used to estimate the times of past TE activity. Aiptasia and B. annulata are symbiotic with dinoflagellates; V. paguri and S. elegans are not. (Bottom) The dynamics of seven distinct TE classes, plotted as the cumulative percentage of the genome covered by each class (ordinate) at given Jukes–Cantor substitution distances [abscissa; calculated based on the nucleotide differences between the individual genomic TEs and the consensus sequence for the corresponding TE family (SI Appendix, SI Materials and Methods)]. (B) Arrangements of HOX gene clusters in three anthozoans. N. vectensis and A. digitifera genes are named as described previously (15); Aiptasia genes are named based on the sequence similarities as shown in SI Appendix, Fig. S3C. Arrows indicate directions of transcription. Blue, other homeobox genes; green, HOX and HOX-related genes.
Despite the potential genome-rearranging effects of such TE activity (11), a total of 3,377 Aiptasia genes are found in synteny blocks of 3–33 genes (mean of 4.8) that are shared with one or more other metazoans (SI Appendix, SI Materials and Methods). This is similar to previously reported findings with other animals (11). About half of these synteny blocks, containing 1,727 genes, are shared only with one or more other cnidarians. Interestingly, although many of the Aiptasia HOX genes are in clusters, as in other metazoans, the detailed organization of the clusters is distinct even from that in other anthozoans, leaving the ancestral organization of the HOX cluster uncertain (15, 16) (Fig. 2B and Dataset S1.2).
To seek further insights into Aiptasia evolution and symbiosis, we identified the gene families with elevated rates of amino acid substitution relative to other cnidarians (SI Appendix, SI Materials and Methods). Among 2,478 gene families, 143 (containing 165 genes) were found to show accelerated evolution (Dataset S1.3), but analysis of the functions ascribed to these families did not reveal obvious clues to the molecular basis of the endosymbiotic relationship.
Taxonomically Restricted Genes.
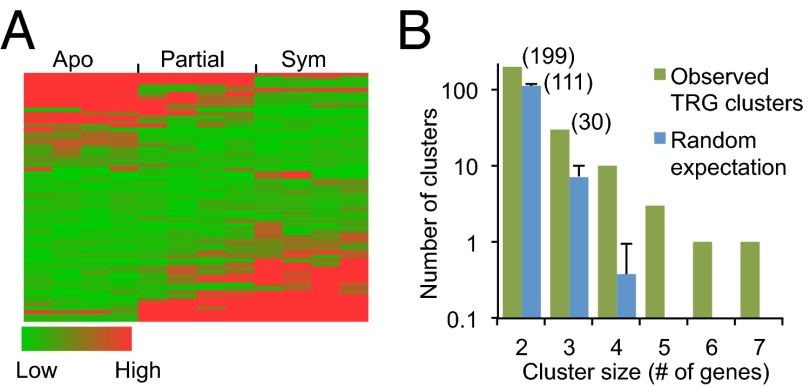
We found 3,107 genes whose predicted products had no discernible homologs in other organisms (SI Appendix, Table S6). Of these, 1,946 were supported by transcriptomic evidence. Such taxonomically restricted genes (TRGs) are thought to play important roles in the creation of evolutionary novelties and morphological diversity within the taxonomic group (17, 18). Although the TRG products lack similarity to known proteins from which putative functions could be derived, we found that many of the TRGs were expressed differentially in different states of symbiosis (Fig. 3A and SI Appendix, Fig. S4A) and/or between adults and larvae (SI Appendix, Fig. S4A), suggesting that at least some are real genes with specific functions, possibly in Aiptasia-specific aspects of endosymbiosis or development.
Fig. 3.
Differential expression and nonrandom chromosomal clustering of taxonomically restricted genes (TRGs) in Aiptasia. (A) Heat map of FPKM expression values (SI Appendix, SI Materials and Methods) for 63 putative TRGs with expression-level changes of ≥eightfold (up or down) between partially (Partial) or fully (Sym) infected anemones and anemones without dinoflagellates (Apo). (B) To evaluate genomic clustering, the observed numbers of clusters of two or more putative TRGs without intervening genes were compared with the expectations based on a random distribution of such genes in the genome (SI Appendix, SI Materials and Methods). Error bars indicate SDs; numbers in parentheses indicate the actual numbers of clusters of those sizes.
To search further for homologies of the TRGs, a Hidden-Markov-Model analysis was conducted (SI Appendix, SI Materials and Methods), identifying 52 putatively new protein domains. These domains were found in 122 of the putative TRGs, including 14 cases in which the two genes encoding the same seemingly new domain were immediately adjacent to each other or separated by a single gene, suggesting recent gene duplication. In addition, these putatively new domains were found in 145 Aiptasia proteins that were not classified as TRG products (Dataset S1.4). Also of interest was that the putative TRGs were not randomly distributed in the genome, with about one-third being present in clusters of two to seven with no interspersed genes (Fig. 3B). The genes in these clusters were not coexpressed (SI Appendix, Fig. S4B), and the biological reason for the nonrandom genomic arrangement remains unclear.
Metabolic Exchanges Between the Partners.
The cnidarian–dinoflagellate symbiosis involves an intimate metabolic exchange between the partners, but many of the details remain obscure. In particular, the respective contributions to animal metabolism from ingested food, synthesis by Symbiodinium, synthesis by the prokaryotic microbiome, and synthesis by the animal cells themselves remain unclear in many cases. The Aiptasia genome sequence should help to elucidate these matters, as the following examples illustrate. First, BLASTP (protein basic local alignment search tool) searches of the gene models confirm previous transcriptomic evidence (19) that anthozoans, like other animals, lack the ability to synthesize 12 of the 20 common amino acids from central-pathway intermediates (SI Appendix, Table S8). The missing enzymes include two that are essential for the synthesis of the sulfur-containing amino acids, but the genome also confirms that Aiptasia, like other animals including N. vectensis (SI Appendix, Table S8) and several corals (9), contains a gene for a cystathionine-β-synthase, so that it should be able to synthesize cysteine from methionine (19). Thus, the reported absence of such a gene in several Acropora species (9) remains a puzzle. Although labeling studies have indicated that various essential amino acids are transported from the dinoflagellate to the host (20), it remains unclear if this supply is adequate for the animals’ needs or whether additional supplies from ingested food are also necessary.
Second, analysis of the genome sequence confirmed the presence of five genes (19) encoding Npc2-type sterol-transport proteins and identified a sixth such gene. Interestingly, five of these genes occur in two clusters that probably reflect their evolutionary origins by tandem duplication (SI Appendix, Fig. S5A and Dataset S1.5). The two genes encoding proteins with typical cholesterol-binding sites (NPC2A and the newly identified NPC2F) are closely linked and have three introns apiece, whereas three of the four genes encoding proteins that presumably cannot bind cholesterol (19) are present in a second cluster and have either zero or one intron. The latter genes may represent an anthozoan-specific expansion of the family (19), and both differential expression (19, 21) and protein localization (22) data suggest that the product of one of these genes (NPC2D) is involved in transport of dinoflagellate-synthesized sterols to the host cytosol in symbiotic anemones. Such transport may provide the host with the bulk sterols that it needs for membrane formation, and indeed analysis of the predicted protein sets of Aiptasia, N. vectensis, and A. digitifera indicates that anthozoans (like Drosophila, but unlike both mammals and Symbiodinium) are unable to synthesize sterols from glycolytic intermediates (SI Appendix, Fig. S5B and Dataset S1.6). However, most dinoflagellate-synthesized sterols are structurally distinct from the bulk of those found in the cnidarian hosts (23–26), and it remains unclear whether the dinoflagellate and host can transport and biochemically convert the dinoflagellate-produced sterols in sufficient quantity to fulfill the host’s needs in the absence of a supply also from ingested food. As suggested previously (19), an intriguing alternative would be that the transported sterols serve a role in host recognition of the algal symbionts.
Interactions of the Host with Algal Symbionts and Other Microbes.
A cnidarian host must distinguish among potential symbionts, pathogens, and particles of food. Moreover, a given host can establish symbiosis with some strains of Symbiodinium but not others (6, 7), and there is good evidence from Hydra that cnidarian hosts also actively shape the assembly of a specific and beneficial prokaryotic microbiome (27, 28). Although understanding of microbiome assembly in corals and anemones is much less complete, recent studies suggest that processes similar to those in Hydra are involved (29–32). Such discriminations must be accomplished in the absence of an adaptive immune system and presumably depend on innate immunity mechanisms that involve the recognition of microbial cell-surface molecules by host pattern-recognition receptors (PRRs) (2, 33). Indeed, there is evidence that the recognition of compatible Symbiodinium types depends on the specific binding of algal cell-surface glycans by host lectins (34).
Thus, it was of interest that among the significantly enriched protein domains in the Aiptasia genome (SI Appendix, Fig. S6A) was the fibrinogen domain, which acts as a carbohydrate-binding moiety in secreted vertebrate PRRs that triggers the innate immunity cascade of the lectin-complement pathway (35). Searching the Pfam annotation (SI Appendix, SI Materials and Methods) revealed 116 Aiptasia proteins containing fibrinogen domains, of which 13 also contain an N-terminal collagen domain as in the bilaterian ficolins (35). Strikingly, not only has this protein family undergone apparently independent expansions in the symbiotic anemone and stony-coral lineages (Fig. 4A), but most family members (including all identified to date in Aiptasia) also contain two or three Ig domains lying between the collagen and fibrinogen domains (Fig. 4A). As proteins with this tripartite domain structure appear to be absent from bilaterians and have not previously been described, we have named them Cnidarian ficolin-like proteins (CniFLs), and we speculate that the Ig domains may contribute to their ability to recognize a variety of microbial surface patterns with high specificity. Interestingly, a similar expansion of proteins containing both Ig-superfamily and fibrinogen domains (FREPs) has been identified in the snail Biomphalaria glabrata, in which the Ig domains are not only expanded in the genome but further diversified through point mutations and somatic recombination of the germ-line source sequences (36). Even though the Ig domain diversity in CniFLs and FREPs falls far short of that in the antibody system of vertebrates, our findings further support the hypothesis that the pattern-recognition capabilities of invertebrate innate immunity systems are more flexible than once thought.
Fig. 4.
Cnidarian ficolin-like proteins (CniFLs), a newly recognized family of putative PRRs. (A) Maximum-likelihood phylogenetic tree of all CniFLs identified in Aiptasia and stony corals (A. digitifera and Stylophora pistillata) and of all canonical ficolins (Fcns) found in stony corals and the anemone N. vectensis (Dataset S1.7). A human and a mouse ficolin (one of three and two, respectively, in those species) were also included as an out-group. The tree is based on an alignment of a 113-amino-acid sequence (with gaps removed) that spans portions of the collagen and fibrinogen domains. Most of the identified CniFLs contain three central Ig domains, but five (indicated by an asterisk) contain only two. Numbers on nodes denote bootstrap values. (B) Components of the lectin-complement pathway that are encoded in the Aiptasia genome (based on KEGG analysis; SI Appendix, SI Materials and Methods) and may be involved in signaling downstream of the CniFLs. BF, complement factor B; C2, C3, and C3b, complement components 2 and 3 and the cleavage product of C3; C4BP, complement component 4 binding protein; CR, complement receptor; HF1, complement factor H; MASP, mannose-binding-lectin–associated serine protease. Complement component C4 was not unequivocally identified in the currently available sequence but is included here for the sake of completeness (Dataset S1.8).
Both analogy to the function of vertebrate PRRs and previous studies of complement components in cnidarians (37, 38) suggest that the Aiptasia CniFLs might function through the complement pathway, and indeed we were able to identify putative orthologs of most components of this pathway in the Aiptasia predicted protein set (Fig. 4B and Dataset S1.8). Accordingly, a CniFL-complement pathway might be involved in the recognition of compatible Symbiodinium types, in shaping other aspects of the host microbiome, or both. Consistent with the former possibility, we note (i) that the CniFLs have so far been found only in symbiotic cnidarians (Aiptasia and two corals, but not N. vectensis) and (ii) that the Aiptasia CniFLs were almost all up-regulated—in some cases dramatically so—in aposymbiotic relative to symbiotic anemones (SI Appendix, Table S9). This up-regulation might reflect a role in recognition and uptake of compatible Symbiodinium cells by animals that lack them, with down-regulation following the successful establishment of symbiosis.
Toll-like receptors (TLRs) and Interleukin-like receptors (ILRs) are additional PRR classes; both recognize extracellular microbial patterns and signal through the intracellular Toll/Interleukin-receptor (TIR) domain. Consistent with a previous transcriptome study (39), the current Aiptasia genome assembly has not revealed any canonical TLR with both TIR and extracellular leucine-rich-repeat (LRR) domains, but it does contain four ILR homologs with both a TIR domain and one to three extracellular Ig domains, as well as two proteins with a TIR domain only (SI Appendix, Table S10 and Fig. S6B), which may partner with an unknown extracellular PRR. Consistent with previous studies (39, 40), many other predicted components of the TLR/ILR signaling pathways were identified using the Kyoto Encyclopedia of Genes and Genomes (KEGG)-annotated protein sets of Aiptasia, N. vectensis, and A. digitifera (SI Appendix, Fig. S6C and Dataset S1.9), suggesting that these pathways are functional in anthozoans. The close similarity between the Aiptasia and N. vectensis TLR/ILR protein repertoires and the apparently lineage-specific expansion of these proteins in A. digitifera (SI Appendix, Table S10 and Fig. S6B), but not in Aiptasia, suggest that these pathways are not involved in the interaction between host and Symbiodinium. Instead, as suggested by studies in Hydra (41), these proteins might function in shaping the host-associated prokaryotic microbiome. The microbiome might be more complex in corals than in anemones, or coral and anemone hosts might rely on different sets of mechanisms to shape their microbiomes.
Microbes that evade the extracellular recognition systems and invade the animal cell can be recognized by intracellular “nucleotide-binding and oligomerization domain” (NOD)-like receptors (NLRs), which trigger defensive responses overlapping with those of the TLRs and ILRs (27) (SI Appendix, Fig. S6 C and D). NLRs are characterized by a central NACHT or NB-ARC domain and are greatly expanded and diversified in cnidarians (42, 43). A search of the Aiptasia Pfam annotation revealed 86 proteins containing NACHT domains and 22 proteins containing NB-ARC domains, numbers similar to those reported for N. vectensis but much smaller than those reported for both Hydra magnipapillata and A. digitifera. It is not clear what could explain these substantial lineage-specific differences. As reported previously for H. magnipapillata (42), KEGG-based analysis revealed the presence in Aiptasia and other anthozoans of homologs of many components of the vertebrate NLR-triggered pathways but, surprisingly, not of the centrally important “receptor-interacting protein kinases” RIPK1 and RIPK2 (SI Appendix, Fig. S6D and Dataset S1.10). This result suggests that cnidarians have a distinct mechanism for the processing of signals resulting from NLR recognition of microbial patterns.
Evidence for Extensive Horizontal Gene Transfer.
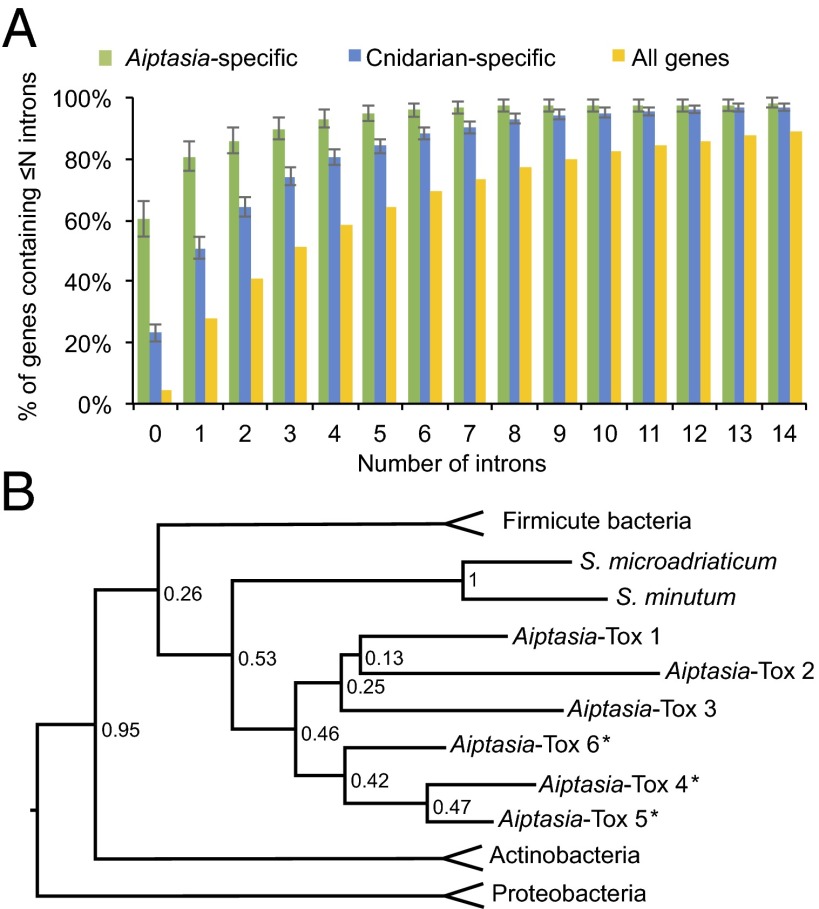
The associations between cnidarians and their endosymbiotic dinoflagellates and prokaryotic microbiomes have evolved over millions of years, making it likely that horizontal gene transfer (HGT) has occurred (44). To explore this possibility, we searched the predicted Aiptasia protein set for cases in which the best alignment was to a nonmetazoan rather than a metazoan sequence (SI Appendix, SI Materials and Methods). This search identified 275 HGT candidates that are specific to Aiptasia (SI Appendix, Fig. S7 A, Left) and an additional 548 “cnidarian-specific” cases in which the Aiptasia protein has a hit in one or more other cnidarians, but not in other metazoans, so that the gene is likely to have been transferred from a nonmetazoan source to a basal cnidarian (SI Appendix, Fig. S7 A, Right and Dataset S1.11). Although the HGT candidates have a variety of apparent sources, genes of putative bacterial origin predominate (SI Appendix, Fig. S7A). Consistent with origins in bacterial sequences (and/or elements arising by reverse transcription) and a subsequent gradual accumulation of introns, both the Aiptasia-specific and cnidarian-specific HGT candidates have, on average, fewer introns than the overall Aiptasia gene set (Fig. 5A). Similar patterns have been observed for HGT candidates in H. magnipapillata, N. vectensis, and the rotifer Adineta vaga (14, 45, 46). As the Aiptasia-specific HGT candidates also have, on average, fewer introns than the cnidarian-specific HGT candidates (Fig. 5A), the Aiptasia lineage has presumably continued to acquire new genes by horizontal transfer since its split from the other cnidarian lineages examined here (rather than these genes having been lost in the other lineages).
Fig. 5.
Evidence for horizontal gene transfer (HGT) in Aiptasia. (A) Cumulative-distribution plot of intron numbers in Aiptasia-specific (green) and cnidarian-specific (blue) HGT candidates and in all 29,269 Aiptasia gene models (yellow). Because of the smaller sample sizes of the first two categories (275 and 823, respectively), 95% binomial-proportion confidence intervals (SI Appendix, SI Materials and Methods) are shown for them. (B) Maximum-likelihood phylogenetic tree for the Aiptasia Tox-Art-HYD1 domains apparently derived by intraspecific duplications after an HGT event. For simplicity, the numerous and varied bacterial species whose Tox-Art-HYD1 domains form the outer branches of the tree are not shown individually (SI Appendix, Fig. S7E). Numbers on nodes denote bootstrap values. An asterisk represents the three proteins in one genomic region.
Among the HGT candidates were 29 (17 Aiptasia-specific and 12 cnidarian-specific) whose best alignment was to a Symbiodinium protein (SI Appendix, Fig. S7A), suggesting that they were transferred from Symbiodinium or another dinoflagellate(s) into cnidarian hosts (or possibly the reverse). [Because the databases used (SI Appendix, SI Materials and Methods) contained sequences from Symbiodinium but not other dinoflagellates, there is ambiguity about the dinoflagellate(s) involved.] To test this possibility further, we constructed maximum-likelihood phylogenetic trees for 4 of the 12 cnidarian-specific proteins and sequences representing the full phylogenetic diversity in the UniProt database (SI Appendix, SI Materials and Methods). In each case, the Aiptasia protein and its homologs in other cnidarians and Symbiodinium form a distinct clade that is itself embedded in a larger clade containing almost entirely proteins from nonmetazoans (SI Appendix, Fig. S7B), supporting the hypothesis of a dinoflagellate-to-cnidarian HGT (or vice versa).
One of the 12 cnidarian-specific genes has already been identified in previous studies as a probable case of HGT (9, 47). In N. vectensis, A. digitifera, and several dinoflagellates, a single gene was found to encode a 3-dehydroquinate synthase–like domain fused to an O-methyltransferase–like domain; these domains appear to catalyze consecutive steps in the synthesis of UV-protective mycosporine amino acids (48). Both domains from these dinoflagellate and cnidarian fusion proteins cluster in phylogenetic trees with each other and with the corresponding domains found in several cyanobacteria (9, 47) (SI Appendix, Fig. S7 C and D), although the domains are encoded in distinct but adjacent genes in those cyanobacteria (49). Indeed, our search of 89 cyanobacterial genomes (https://img.jgi.doe.gov/) for gene models containing a 3-dehydroquinate synthase domain (as annotated by Pfam) revealed 119 genes, none of which contained a fused O-methyltransferase domain. We also found that both domains of the predicted Aiptasia fusion protein cluster in phylogenetic trees as described previously for the N. vectensis and A. digitifera proteins and that Symbiodinium microadriaticum (Clade A1) also contains a gene encoding both domains with similar sequences (SI Appendix, Fig. S7 C and D). As the same fusion-protein gene is present in both dinoflagellates and cnidarians and the cnidarian sequences of both domains are more similar to those in dinoflagellates than to those in cyanobacteria, it seems likely that the gene fusion occurred just once and simultaneously with the transfer from a cyanobacterium into a basal dinoflagellate (49), with a subsequent transfer into one or more cnidarian ancestors from a dinoflagellate(s) that was potentially symbiotic (providing the intimate association that would facilitate the gene transfer). On this model, the presence of the fusion gene in N. vectensis can be explained most readily by the hypothesis that the modern species had an ancestor with a symbiotic dinoflagellate, a possibility that is supported by the fact that at least 9 of the 11 additional cnidarian-specific HGT candidates whose best alignment was to Symbiodinium are present in N. vectensis.
Among the 17 Aiptasia-specific HGT candidates whose best alignment was to a Symbiodinium protein, seven had domains that were annotated through the Pfam database. Three of these annotations were to the bacterial Tox-Art-HYD1 domain of ∼100 amino acids, which is known to act as an ADP ribosyltransferase toxin in several bacterial pathogens (50, 51) but has not previously been described in eukaryotic genomes (although one crustacean sequence is annotated in the Pfam database). In addition to the three genes found in the HGT screen, we found three more Aiptasia genes that contain Tox-Art-HYD1 domains based on Pfam annotation. Moreover, three of these six Tox-Art-HYD1 genes are present in one genomic region of ∼64 kb without intervening genes, and these three encode proteins that are particularly closely related in sequence (Fig. 5B and SI Appendix, Fig. S7E). Thus, it appears that Tox-Art-HYD1 domain-containing genes have undergone intraspecific duplications in Aiptasia after the initial HGT event.
Key residues of the Tox-Art-HYD1 domain are conserved in Aiptasia and Symbiodinium (SI Appendix, Fig. S7E), and in a phylogenetic analysis, the Aiptasia and Symbiodinium domains formed a distinct clade embedded within a large clade of bacterial sequences (Fig. 5B), supporting the hypothesis of a Symbiodinium–Aiptasia HGT event following an initial transfer of a bacterial gene into one partner or the other. Bacterial toxins containing Tox-Art-HYD1 domains are thought to function in interspecific conflicts (51), and the Aiptasia and Symbiodinium proteins may be used similarly to combat pathogens and/or shape the composition of the host prokaryotic microbiome, much as species-specific antimicrobial peptides in Hydra appear to help shape its microbiome (52).
Conclusions
Recent years have brought dramatically increased appreciation of the importance of understanding the molecular, cellular, and organismal bases of mutualistic host-microbe symbioses. Here we have reported on the assembly and analysis of the genome sequence and predicted protein sets of the sea anemone Aiptasia. Although differences are likely to exist between different symbiotic anthozoans, our analyses have revealed a variety of conserved features that should help to illuminate the evolution of the symbiotic lifestyle and provide the basis for the continued development of Aiptasia as a critically needed model for coral-dinoflagellate endosymbiosis, which underlies one of the most important marine ecosystems, coral reefs.
Materials and Methods
For genome sequencing and assembly, DNA from anemones of the clonal Aiptasia strain CC7 was used for the preparation of Illumina short- and long-insert paired-end sequencing libraries. The reference transcriptome was sequenced and assembled using RNA derived from different developmental and symbiotic states of adult anemones from Aiptasia strain CC7 and larval crosses of Aiptasia strains CC7 and H2. SI Appendix details the sequencing, assembly, annotation, and genome analysis.
Supplementary Material
Acknowledgments
We thank John Parkinson for help on the Symbiodinium phylogeny and Cory J. Krediet and Ved Chirayath for providing pictures. Research reported in this publication was supported by the King Abdullah University of Science and Technology and by Grant 2629 from the Gordon and Betty Moore Foundation. L.Y.E. and E.M.L. were supported by National Science Foundation Graduate Research Fellowships, a Stanford Graduate Fellowship, and National Institutes of Health Training Grant 5 T32 HG000044. E.A.H. and A.G. were supported by funding from the Deutsche Forschungsgemeinschaft (GU 1128/3-1 to A.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper are available at aiptasia.reefgenomics.org and have been deposited in the NCBI database (accession no. PRJNA261862).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513318112/-/DCSupplemental.
References
- 1.Dubinsky Z, Stambler N. Coral Reefs: An Ecosystem in Transition. Springer Science & Business Media; Berlin: 2010. [Google Scholar]
- 2.Davy SK, Allemand D, Weis VM. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev. 2012;76(2):229–261. doi: 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoegh-Guldberg O, et al. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318(5857):1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- 4.Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR. Cell biology in model systems as the key to understanding corals. Trends Ecol Evol. 2008;23(7):369–376. doi: 10.1016/j.tree.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Thornhill DJ, Xiang Y, Pettay DT, Zhong M, Santos SR. Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol Ecol. 2013;22(17):4499–4515. doi: 10.1111/mec.12416. [DOI] [PubMed] [Google Scholar]
- 6.Schoenberg DA, Trench RK. 1980. Genetic variation in Symbiodinium (=Gymnodinium) microadriaticum freudenthal, and specificity in its symbiosis with marine invertebrates. III. Specificity and infectivity of Symbiodinium microadriaticum. P Roy Soc B-Biol Sci 207(1169):445–460.
- 7.Hambleton EA, Guse A, Pringle JR. Similar specificities of symbiont uptake by adults and larvae in an anemone model system for coral biology. J Exp Biol. 2014;217(Pt 9):1613–1619. doi: 10.1242/jeb.095679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317(5834):86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 9.Shinzato C, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476(7360):320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- 10.Gregory TR. 2015 Animal Genome Size Database. Available at genomesize.com/
- 11.Simakov O, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493(7433):526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453(7198):1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 13.Lynch M, Walsh B. The Origins of Genome Architecture. Sinauer Associates, Sunderland; MA: 2007. [Google Scholar]
- 14.Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464(7288):592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chourrout D, et al. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature. 2006;442(7103):684–687. doi: 10.1038/nature04863. [DOI] [PubMed] [Google Scholar]
- 16.DuBuc TQ, Ryan JF, Shinzato C, Satoh N, Martindale MQ. Coral comparative genomics reveal expanded Hox cluster in the cnidarian-bilaterian ancestor. Integr Comp Biol. 2012;52(6):835–841. doi: 10.1093/icb/ics098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12(10):692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 18.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25(9):404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Lehnert EM, et al. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3 (Bethesda) 2014;4(2):277–295. doi: 10.1534/g3.113.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Douglas A. Essential amino acid synthesis and nitrogen recycling in an alga–invertebrate symbiosis. Mar Biol. 1999;135(2):219–222. [Google Scholar]
- 21.Ganot P, et al. Adaptations to endosymbiosis in a cnidarian-dinoflagellate association: Differential gene expression and specific gene duplications. PLoS Genet. 2011;7(7):e1002187. doi: 10.1371/journal.pgen.1002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dani V, Ganot P, Priouzeau F, Furla P, Sabourault C. Are Niemann-Pick type C proteins key players in cnidarian-dinoflagellate endosymbioses? Mol Ecol. 2014;23(18):4527–4540. doi: 10.1111/mec.12876. [DOI] [PubMed] [Google Scholar]
- 23.Giner JL. Biosynthesis of marine sterol side chains. Chem Rev. 1993;93(5):1735–1752. [Google Scholar]
- 24.Withers NW, Kokke WC, Fenical W, Djerassi C. Sterol patterns of cultured zooxanthellae isolated from marine invertebrates: Synthesis of gorgosterol and 23-desmethylgorgosterol by aposymbiotic algae. Proc Natl Acad Sci USA. 1982;79(12):3764–3768. doi: 10.1073/pnas.79.12.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kokke WCMC, Fenical W, Bohlin L, Djerassi C. Sterol synthesis by cultured zooxanthellae; implications concerning sterol metabolism in the host-symbiont association in caribbean gorgonians. Comp Biochem Physiol B. 1981;68(2):281–287. [Google Scholar]
- 26.Yamashiro H, Oku H, Higa H, Chinen I, Sakai K. Composition of lipids, fatty acids and sterols in Okinawan corals. Comp Biochem Physiol B. 1999;122(4):397–407. [Google Scholar]
- 27.Bosch TC. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu Rev Microbiol. 2013;67:499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- 28.Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA. 2007;104(32):13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gochfeld DJ, Aeby GS. Antibacterial chemical defenses in Hawaiian corals provide possible protection from disease. Mar Ecol Prog Ser. 2008;362:119–128. [Google Scholar]
- 30.Bayer T, et al. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol. 2013;79(15):4759–4762. doi: 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidal-Dupiol J, et al. Innate immune responses of a scleractinian coral to vibriosis. J Biol Chem. 2011;286(25):22688–22698. doi: 10.1074/jbc.M110.216358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fusetani N, Toyoda T, Asai N, Matsunaga S, Maruyama T. Montiporic acids A and B, cytotoxic and antimicrobial polyacetylene carboxylic acids from eggs of the scleractinian coral Montipora digitata. J Nat Prod. 1996;59(8):796–797. doi: 10.1021/np9604036. [DOI] [PubMed] [Google Scholar]
- 33.Kvennefors EC, Leggat W, Hoegh-Guldberg O, Degnan BM, Barnes AC. An ancient and variable mannose-binding lectin from the coral Acropora millepora binds both pathogens and symbionts. Dev Comp Immunol. 2008;32(12):1582–1592. doi: 10.1016/j.dci.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Wood-Charlson EM, Hollingsworth LL, Krupp DA, Weis VM. Lectin/glycan interactions play a role in recognition in a coral/dinoflagellate symbiosis. Cell Microbiol. 2006;8(12):1985–1993. doi: 10.1111/j.1462-5822.2006.00765.x. [DOI] [PubMed] [Google Scholar]
- 35.Fujita T. Evolution of the lectin-complement pathway and its role in innate immunity. Nat Rev Immunol. 2002;2(5):346–353. doi: 10.1038/nri800. [DOI] [PubMed] [Google Scholar]
- 36.Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. 2004;305(5681):251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- 37.Kvennefors EC, et al. Analysis of evolutionarily conserved innate immune components in coral links immunity and symbiosis. Dev Comp Immunol. 2010;34(11):1219–1229. doi: 10.1016/j.dci.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 38.Kimura A, Sakaguchi E, Nonaka M. Multi-component complement system of Cnidaria: C3, Bf, and MASP genes expressed in the endodermal tissues of a sea anemone, Nematostella vectensis. Immunobiology. 2009;214(3):165–178. doi: 10.1016/j.imbio.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Poole AZ, Weis VM. TIR-domain-containing protein repertoire of nine anthozoan species reveals coral-specific expansions and uncharacterized proteins. Dev Comp Immunol. 2014;46(2):480–488. doi: 10.1016/j.dci.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Miller DJ, et al. The innate immune repertoire in cnidaria--Ancestral complexity and stochastic gene loss. Genome Biol. 2007;8(4):R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franzenburg S, et al. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci USA. 2012;109(47):19374–19379. doi: 10.1073/pnas.1213110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange C, et al. Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol Biol Evol. 2011;28(5):1687–1702. doi: 10.1093/molbev/msq349. [DOI] [PubMed] [Google Scholar]
- 43.Hamada M, et al. The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Mol Biol Evol. 2013;30(1):167–176. doi: 10.1093/molbev/mss213. [DOI] [PubMed] [Google Scholar]
- 44.Dunning Hotopp JC. Horizontal gene transfer between bacteria and animals. Trends Genet. 2011;27(4):157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320(5880):1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 46.Artamonova II, Mushegian AR. Genome sequence analysis indicates that the model eukaryote Nematostella vectensis harbors bacterial consorts. Appl Environ Microbiol. 2013;79(22):6868–6873. doi: 10.1128/AEM.01635-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starcevic A, et al. Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins. Proc Natl Acad Sci USA. 2008;105(7):2533–2537. doi: 10.1073/pnas.0707388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balskus EP, Walsh CT. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science. 2010;329(5999):1653–1656. doi: 10.1126/science.1193637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waller RF, Slamovits CH, Keeling PJ. Lateral gene transfer of a multigene region from cyanobacteria to dinoflagellates resulting in a novel plastid-targeted fusion protein. Mol Biol Evol. 2006;23(7):1437–1443. doi: 10.1093/molbev/msl008. [DOI] [PubMed] [Google Scholar]
- 50.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng Q, Barbieri JT. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu Rev Microbiol. 2008;62:271–288. doi: 10.1146/annurev.micro.62.081307.162848. [DOI] [PubMed] [Google Scholar]
- 52.Franzenburg S, et al. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci USA. 2013;110(39):E3730–E3738. doi: 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.