Significance
In this work, we took a proteome-centric view to analyze the cell division and lifespan asymmetry between mother and daughter cells in budding yeast. Using a flow cytometry-based, high-throughput approach, we quantified the partitioning of the proteome and identified 74 mother-enriched and 60 daughter-enriched proteins. Functional analysis of these proteins suggests mechanisms of asymmetric partitioning at an organelle/suborganelle level. We found that mother-enriched proteins are much more likely to becoming aging factors than those proteins chosen at random. The proposed mechanism, as supported by our single-cell observations, is that these proteins accumulate in old mother cells to a high level that becomes lifespan-limiting. Our work sheds new light on the mechanisms of asymmetric cell division and aging.
Keywords: aging, asymmetric cell division, proteome
Abstract
Budding yeast divides asymmetrically, giving rise to a mother cell that progressively ages and a daughter cell with full lifespan. It is generally assumed that mother cells retain damaged, lifespan limiting materials (“aging factors”) through asymmetric division. However, the identity of these aging factors and the mechanisms through which they limit lifespan remain poorly understood. Using a flow cytometry-based, high-throughput approach, we quantified the asymmetric partitioning of the yeast proteome between mother and daughter cells during cell division, discovering 74 mother-enriched and 60 daughter-enriched proteins. While daughter-enriched proteins are biased toward those needed for bud construction and genome maintenance, mother-enriched proteins are biased towards those localized in the plasma membrane and vacuole. Deletion of 23 of the 74 mother-enriched proteins leads to lifespan extension, a fraction that is about six times that of the genes picked randomly from the genome. Among these lifespan-extending genes, three are involved in endosomal sorting/endosome to vacuole transport, and three are nitrogen source transporters. Tracking the dynamic expression of specific mother-enriched proteins revealed that their concentration steadily increases in the mother cells as they age, but is kept relatively low in the daughter cells via asymmetric distribution. Our results suggest that some mother-enriched proteins may increase to a concentration that becomes deleterious and lifespan-limiting in aged cells, possibly by upsetting homeostasis or leading to aberrant signaling. Our study provides a comprehensive resource for analyzing asymmetric cell division and aging in yeast, which should also be valuable for understanding similar phenomena in other organisms.
Cellular aging and asymmetric cell division are intimately linked. In budding yeast, asymmetric cell division yields a mother cell and a daughter cell that are easily distinguishable under the microscope. Tracking the fate of the mother lineage led to the discovery that individual mother cells have a finite replicative lifespan, defined by the number of daughters a mother cell produced before senescence (1). It is known that although the mother cell ages with each division, their daughters retain the same full lifespan independent of the age of the mother at least until the last few mother cell divisions (2, 3). Thus, the asymmetry in cell division leads to asymmetry of aging.
Even in single-celled organisms in which cell division is seemingly morphologically symmetric, such as fission yeast or Escherichia coli, asymmetric partitioning of cellular contents can still occur and have a differential impact on the aging/death fate of the two offspring (4–8). Asymmetric cell division is also a general phenomenon in mammalian cells (e.g., during development or in mitotically active tissues), where cell division typically leads to two cells with distinct fates, often with different replicative potential. It has been argued on theoretical grounds that asymmetric cell division may be favored by natural evolution (9–11); when the accumulation of lifespan-limiting damage outpaces the dilution by symmetric cell division, keeping the damage to one of the two offspring via asymmetric partitioning is a general strategy to avoid population senescence.
It is generally assumed that budding yeast mother cells retain damaged/lifespan-limiting materials (referred to as “aging factors” hereafter), allowing their daughter cells to reset the clock. Indeed, a number of potential aging factors have been reported to accumulate preferentially in mother cells through asymmetric partitioning. One example is extrachromosomal rDNA circles, which are known to be a limiting factor for lifespan (12, 13) and are retained in mother cells (14–16). Protein aggregates, carbonylated proteins, and reactive oxygen species have also been reported to distribute asymmetrically between old mothers and their daughters (17–22). Preferential retention of membrane transporters in the mother cells has also been associated with lifespan asymmetry (23, 24). These observations support the general notion that mother cells retain aging factors to themselves, enabling their daughters to rejuvenate. However, a global view of the identities of asymmetrically partitioned aging factors and the mechanism through which they influence lifespan is still lacking.
In this work, we took a proteome-centric approach to explore cell division asymmetry and its connection to lifespan asymmetry in budding yeast. Using high-throughput proteomics based on a green fluorescent protein (GFP) library and mother cell labeling, we quantified the asymmetric partitioning of the proteome between mother and daughter cells and systematically identified mother-enriched and daughter-enriched proteins. Functional analyses of these proteins suggest that macrostructures are one basis for asymmetric partition. We found that mother-enriched proteins tend to accumulate in mother cells over time and that the deletion mutants are much more likely to extend lifespan than genes chosen at random, arguing that it is the high concentration that limits lifespan. These observations provide a consistent picture of how asymmetric partitioning of the proteome influences lifespan asymmetry, and serve as a starting point for generating new hypotheses on the mechanism of asymmetry and aging.
Results
Identification of Mother- and Daughter-Enriched Proteins by Quantifying the Asymmetric Partitioning of the Yeast Proteome.
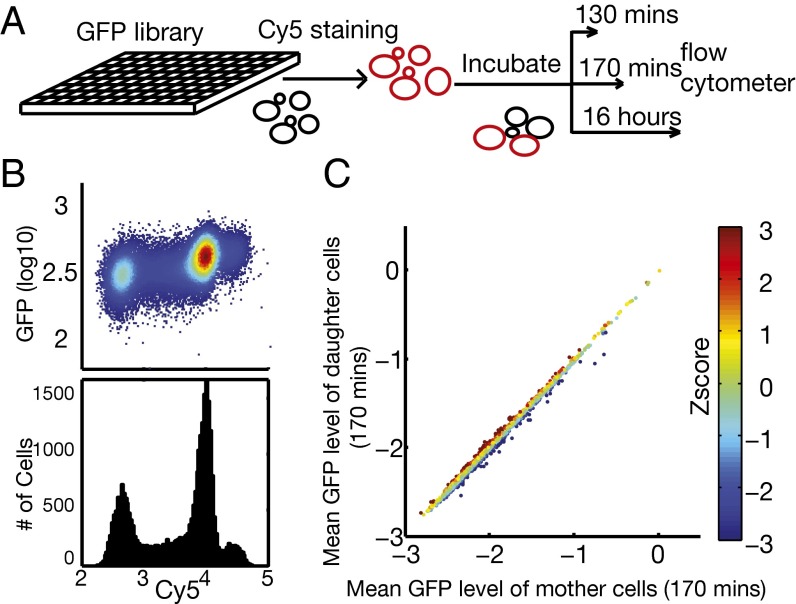
To systematically identify proteins that are asymmetrically segregated, we have developed a high-throughput approach to measure quantitatively the partition of the whole proteome between mother and daughter cells during cell division. To measure the partition of a GFP-tagged protein from the yeast GFP tagging library, we labeled mother cells with Cy5 and let the population grow for about one generation. The resulting cell population was then subjected to flow cytometry analysis for both the Cy5 and GFP signals (Fig. 1A). Because the daughter cells synthesize a new cell membrane and cell wall, they do not inherit the surface dye, and thus can be distinguished from the Cy5-positive mothers (Fig. 1B). Measuring the GFP signal normalized by the cell size (using the side-scatter data) then gives the intensity of the protein in the mother and daughter cell populations. We systematically screened the GFP-tagging library (25) in 96-well plates using a BD Biosciences LSR II flow cytometer with a high-throughput sampler (HTS). Fig. 1C shows the average intensity in mother vs. daughter cells for all ∼4,000 GFP-tagged strains. The majority of the proteins fall on a straight line on the diagonal, with the abundance spanning three orders of magnitude. This result indicates that most of the proteins are evenly (or symmetrically) distributed, with the intensity in the mother cells the same as the intensity in the daughter cells; therefore the total amount of protein is proportional to the average cell volume of the two populations.
Fig. 1.
High-throughput screening for proteins asymmetrically distributed between mother and daughter cells. (A) Schematic of the experimental procedure. S. cerevisiae strains from the GFP tagging library were grown in 96-well plates to exponential phase and then stained with Cy5 dye (cells in red). Newly budded daughter cells after the initial staining carry little Cy5 dye (cells in black) because they do not inherit the cell wall from their mothers. (B) Example of the flow cytometry data collected from one well at 170 min. Mother and daughter populations are clearly visible from the 2D density plot of Cy5 vs. GFP signals (Upper) and the histogram of Cy5 signal (Lower). (C) Identification of mother- or daughter-enriched proteins. The log10-transformed mean GFP intensity (normalized by cell volume) of the mother population is plotted against the log10-transformed mean GFP intensity of the daughter population (170-min data), with each dot representing one tagged strain. The straight line is the linear fit (slope = 0.985, intercept = −0.034). The deviation of the GFP intensity of the daughter population from the fitted line was converted to a z-score (indicted by the color scale) to measure the asymmetry, where negative or positive z-scores indicate enrichment in mother or daughter cells, respectively.
However, there is a small subset of proteins that deviate from the diagonal; they are either enriched in mother cells (below the diagonal) or enriched in the daughter cells (above the diagonal), and therefore asymmetrically distributed. We quantified the deviation from the diagonal by a z-score, defined as the vertical distance from the diagonal normalized by the standard deviation calculated from all of the strains (Methods). Mother-enriched proteins have a negative z-score, whereas daughter-enriched proteins have a positive z-score (Fig. 1C).
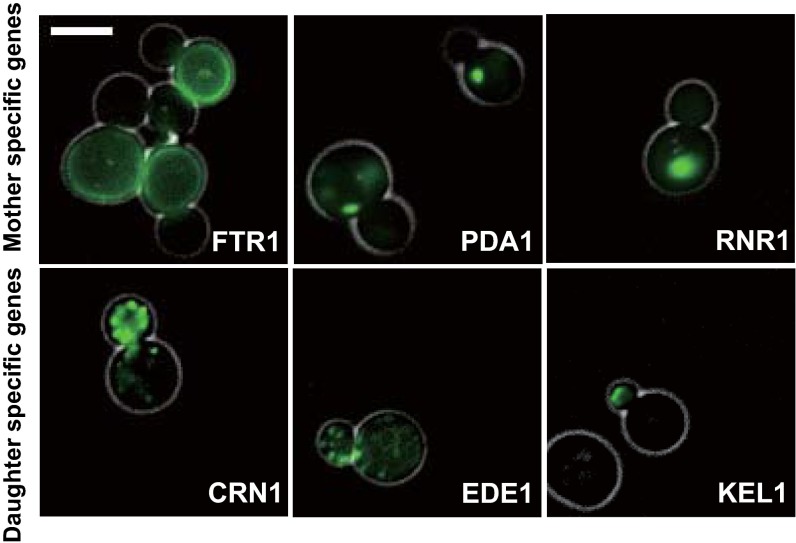
We verified some of these asymmetrically partitioned proteins directly by microscopy and found that there is a clearly visible difference between mother and daughter cells for high z-score proteins (Fig. S1 and images of ref. 25). However, it is difficult to identify asymmetrically partitioned proteins with moderate z-scores from fluorescent images. Our method based on mother cell labeling and flow cytometry is much more sensitive and allows better quantification because a large number of cells (103 to 104) are measured for each GFP-tagged strain.
Fig. S1.
Example images of cells with GFP-tagged mother-enriched (Upper) or daughter-enriched (Lower) proteins. (Scale bar: 5 μm.)
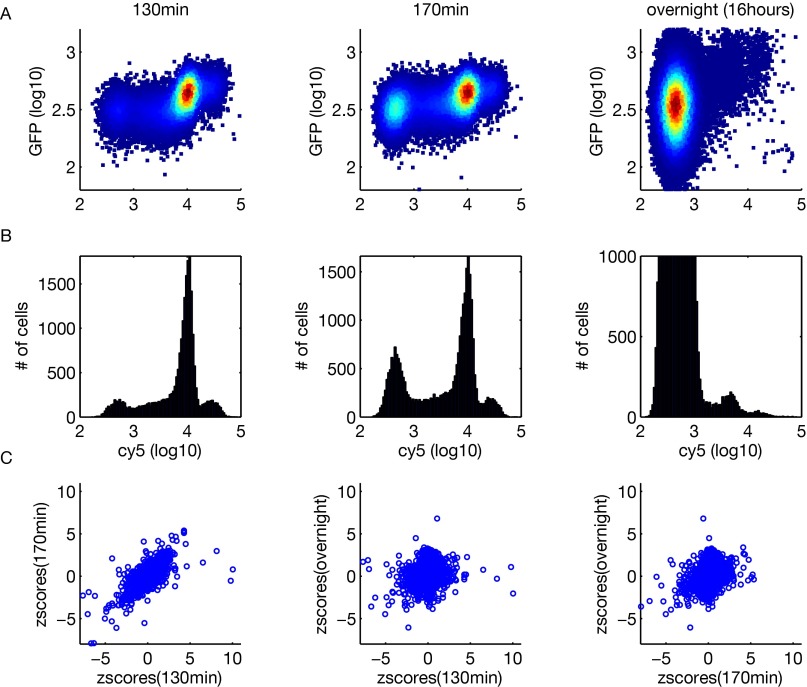
To follow the dynamics of proteome partitioning, we performed experiments at three different time points after the original cell population was labeled with Cy5: 130 min, 170 min, and overnight (about 16 h). The first two time points compare mother and daughter cells with different degrees of maturation (the unlabeled daughter population becomes obvious around 130 min), whereas the third time point gives a comparison between labeled middle-aged mother cells (∼10 generations old after overnight growth) and an unlabeled population that is a mixture of daughter cells and mother cells at different ages. A negative z-score for the first two time points indicates a protein enriched in mother cells, whereas a negative z-score for the third time point indicates that the protein is enriched in the middle-aged mother cells (∼10 generations old) compared with the general mixed-cell population. In general, the magnitude of the z-scores (thus, the asymmetry) decreases over time as the unlabeled cell population develops into more mature daughter cells and eventually becomes mixed with both mother and daughter cells (Fig. S2 and Dataset S1).
Fig. S2.
Typical flow cytometry data from the three time points (130 min, 170 min, and 16 h overnight) and correlations of z-scores. (A) Two-dimensional density plots of Cy5 vs. GFP signals. (B) Histograms of the Cy5 signal. (C) Pairwise scatter plots of z-scores from the three time points.
Functional Bias of Mother- and Daughter-Enriched Proteins Suggests Specific Mechanisms for Asymmetry.
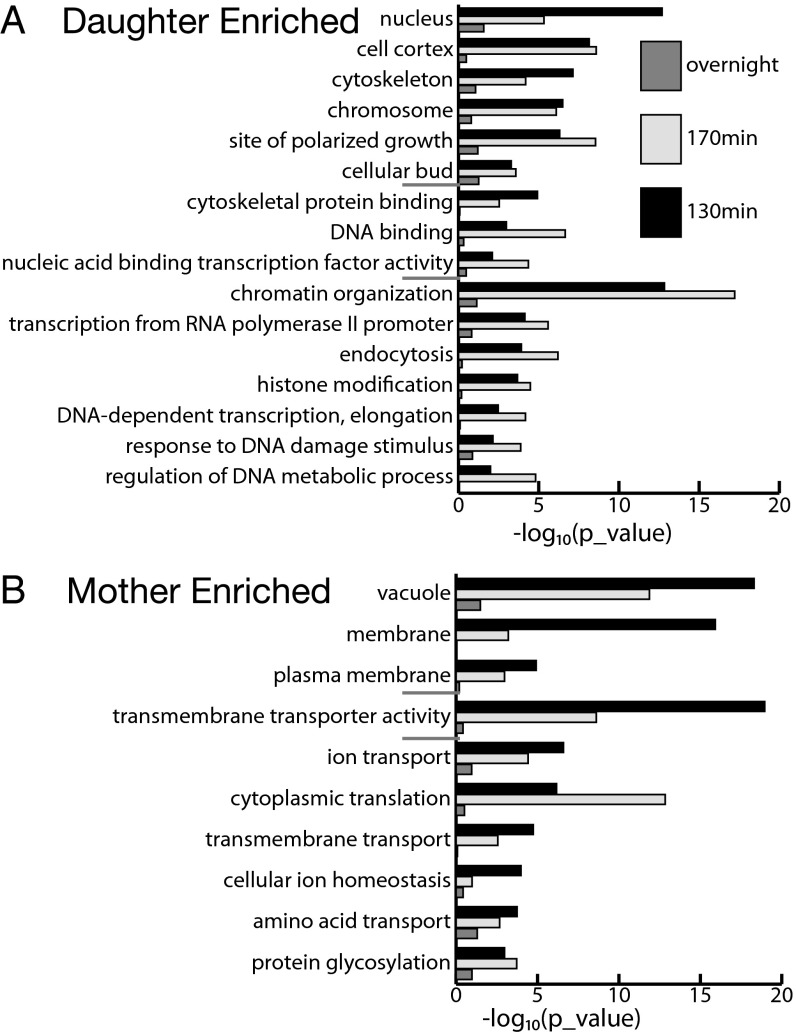
To analyze the biological function of asymmetrically partitioned proteins, we first performed gene ontology (GO) analysis. For each time point, we ranked genes by their z-scores from the most negative to the most positive and performed rank sum test for genes in each GO category associated with a specific molecular function, biological process, or cellular component. GO categories that ranked at the top tend to have more negative z-scores and are preferentially retained in mother cells, whereas GO categories ranked at the bottom are enriched in daughter cells. A number of GO categories are significantly enriched in either the mother or daughter cells (Fig. 2). With a few exceptions, the statistical significance of these categories is generally highest at the 130-min time point, decreases at 170 min, and diminishes overnight, indicating that the asymmetry is highest between mother cells and newborn daughter cells.
Fig. 2.
GO analysis of mother/daughter-enriched proteins. At each of the three time points, proteins are rank-ordered by their z-scores, and a rank sum test was performed for each of the GO-slim categories. GO categories with proteins ranked toward positive z-scores are daughter-enriched (A), whereas GO categories with proteins ranked toward negative z-scores are mother-enriched (B). P values for the significant GO categories associated with a specific cellular component, molecular function, or biological process are shown in the upper, middle, and lower portions of the graphs (separated by short gray lines).
Several clear functional themes emerge from the GO analysis. For daughter-enriched proteins, there is a strong bias toward certain cellular components, such as the cell cortex, cytoskeleton, site of polarized growth, and biological processes like endocytosis (Fig. 2A). Many of these proteins are known to be preferentially distributed to the bud neck, inside the bud, or at the bud tip (Dataset S2 provides a list of the daughter-enriched proteins discussed below). For example, the nuclear protein encoded by the LOC1 gene is required for the daughter-specific localization of the ASH1 mRNA, and CRN1 encodes a cortical actin cytoskeletal component that regulates actin patch assembly. This analysis indicates that proteins needed for the emergence, construction, and scission of the bud are enriched in daughter cells, possibility through active transport processes. Another theme for daughter-enriched proteins is reflected by those proteins localized at chromosomes or in the nucleus that function in chromatin organization. Examples are HST2, which encodes a cytoplasmic NAD(+)-dependent protein deacetylase and a member of the SIR2 family, and HMO1, a chromatin-associated high-mobility group family member involved in genome maintenance. This functional theme suggests that maintenance of chromatin in a silenced and repressive state at certain loci is important for the newborn daughter cells.
Examination of GO categories enriched in mother cells revealed an interesting pattern suggestive of organelle-based asymmetry. Among highly significant GO categories are the vacuole and plasma membrane (cellular components) and transmembrane transporters (molecular function), many of them involved in ion and amino acid transport (Fig. 2B). Because newly budded daughter cells do not inherit the plasma membrane from their mothers, it is expected that many proteins in the plasma membrane of the daughter cells need to be synthesized anew, and thus will appear depleted in the young daughter cells relative to the mother cells (or, equivalently, mother-enriched). Somewhat unexpectedly, the most significant GO category is the vacuole, because a large number of proteins localized to the vacuole or vacuolar membranes are enriched in mother cells (rank sum test, P < 10−17; Dataset S3 provides a list of the mother-enriched proteins discussed below). This enrichment suggests that, quantitatively, the vacuole is unevenly distributed, although it is known that a daughter does inherit the vacuole from its mother in an active process (26, 27).
To analyze specific asymmetrically partitioned proteins, we defined a set of mother-enriched and daughter-enriched proteins based on their z-scores at the three time points. For mother-enriched proteins, we picked those proteins with strong negative z-scores at any of the three time points (z < −3.0) or with robust negative z-scores across time points (z < −1.0 for all three time points). Seventy-four genes are mother-enriched by these criteria. Similarly, 60 proteins are found to be daughter-enriched, defined by z > 3.0 at any time point or z > 1.0 across all three time points. These criteria ensure that the probability of including a random gene in either one of the lists is less than 0.008, assuming that the z-scores are independent and normally distributed.
GO enrichment analysis of the mother-enriched and daughter-enriched proteins gave similar results to the rank sum test. For example, the membrane and vacuole are the two most significantly enriched categories for the mother-enriched proteins, with 37 of 74 proteins and 18 of 74 proteins annotated as localized to the membrane and vacuole, respectively (P = 2.2 * 10−11 and P = 2 * 10−8, respectively). For daughter-enriched proteins, the cell periphery (17 of 60 proteins; P = 3 * 10−4) and site of polarized growth (10 of 60 proteins; P = 2 * 10−3) are the two most significant categories.
In addition to the plasma membrane and vacuole, mother-enriched proteins are found in other organelles. However, these organelles did not score as significantly in the GO analysis because most of the proteins localized in these organelles are symmetrically distributed. This observation suggests that proteins localized in symmetrically partitioned organelles can still be preferentially retained in the mother cell. For example, although most of the mitochondrially localized proteins are symmetrically distributed, several are found enriched in the mother cells, including the cytochrome C protein Cyc1, pyruvate dehydrogenase protein Pda1, the TCA cycle enzymes Idh2 and Kgd1, and the ATPase components Atp1 and Atp2. Interestingly, three of the six proteins (Pda1, Kgd1, and Atp1) are known to be associated with the mitochondrial nucleoid (28). It has been shown that the actively replicating mitochondrial nucleoid is associated with the endoplasmic reticulum (ER)/mitochondria junction (29). It is plausible that the part of the mitochondria tethered to the ER may not be partitioned symmetrically. Thus, asymmetry can be caused by asymmetric partitioning of a substructure of an organelle that is symmetrically partitioned overall.
Mother-Enriched Proteins Are More Likely to be Lifespan-Limiting.
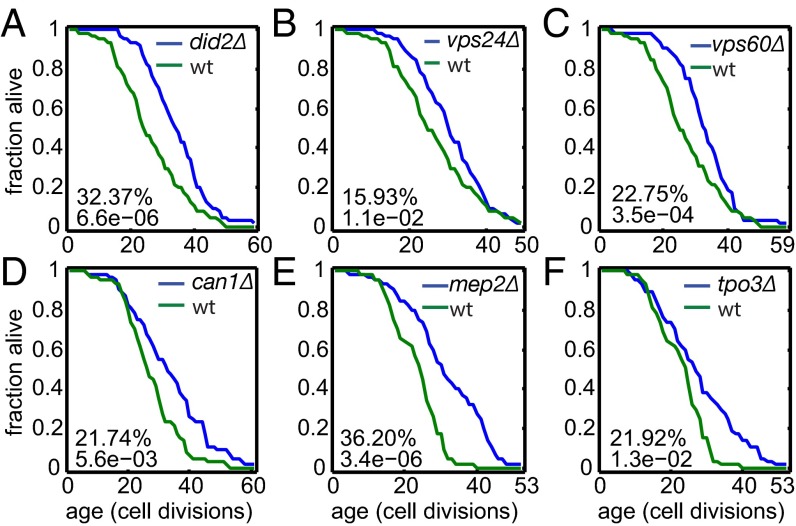
To analyze the relationship between cell division asymmetry and lifespan asymmetry, we focused on mother-enriched proteins. We reasoned that proteins preferentially retained in mother cells are more likely to be aging factors because their accumulation in the mother cells may be deleterious, eventually becoming lifespan-limiting. To test this hypothesis, we first examined the overlap between this set of genes and 228 long-lived mutants from a previous genomics screen (30) and found 13 that were in common [enrichment: P < 8.6 × 10−6]. We then measured the lifespan of the deletion mutants of all 74 mother-enriched proteins. Strikingly, we found that deletion of 23 of the 74 genes extends lifespan (Table 1); this fraction is approximately sixfold the frequency of lifespan extension observed in viable genome-wide gene deletions.
Table 1.
Mother-enriched lifespan-limiting proteins
| Name | Mean lifespan | WT lifespan | Percent increase |
| Did2 (1) | 31.6 (125) | 27.2 (125) | 16.2 (0.0006) |
| Vps24 (1) | 29.7 (125) | 27.4 (125) | 8.4 (0.0348) |
| Vps60 (1) | 32.2 (125) | 27.5 (125) | 17.1 (0.0002) |
| Can1 (2) | 35.6 (165) | 25 (225) | 42.4 (<0.0001) |
| Mep2 (2) | 30.6 (125) | 25 (125) | 22.4 (<0.0001) |
| Tpo3 (2) | 28.8 (125) | 24.8 (125) | 16.1 (0.0013) |
| Itr1 (2) | 29.6 (125) | 27.3 (125) | 8.4 (0.0257) |
| Fet3 (2) | 27.9 (50) | 24.2 (50) | 15.3 (0.0325) |
| Yro2 (2) | 32.3 (145) | 27.2 (185) | 18.8 (<0.0001) |
| Fcy2 (2) | 28.9 (125) | 25.6 (165) | 12.9 (0.0015) |
| Scw4 (2) | 27.8 (45) | 23 (45) | 20.9 (0.0142) |
| Cyc1 (3) | 29.3 (125) | 25.6 (125) | 14.5 (0.0015) |
| Idh2 (3) | 30.8 (735) | 26.8 (904) | 14.9 (<0.0001) |
| Fmp42 (3) | 31.5 (45) | 24.2 (45) | 30.2 (0.0022) |
| Pml39 (4) | 29.2 (45) | 25.2 (45) | 15.9 (0.0234) |
| Rai1 (4) | 35.3 (45) | 23.5 (45) | 50.2 (<0.0001) |
| Pgm2 (5) | 31.3 (125) | 27.2 (125) | 15.1 (0.0011) |
| Ypt6 (5) | 30.7 (205) | 27 (245) | 13.7 (0.007) |
| Rpl7a (5) | 32.1 (172) | 26 (208) | 23.5 (<0.0001) |
| Gsy1 (5) | 30.7 (125) | 25.7 (125) | 19.5 (<0.0001) |
| Yeh1 (5) | 30.4 (45) | 26.8 (65) | 13.4 (0.0143) |
| YEL020C (5) | 29 (45) | 25.3 (45) | 14.6 (0.0468) |
| YER128W (5) | 30 (45) | 25 (45) | 20 (0.0036) |
Column 1 shows the protein name (grouped based on related function or similar subcellular localization). 1, ESCRT; 2, transporters/cell periphery; 3, mitochondrial; 4, nuclear; 5, others. Columns 2 and 3 show the mean lifespan (number of cells dissected) for the deletion mutant and the WT control. Column 4 shows the percent increase in mean lifespan (P values).
There are clear functional themes among these newly discovered lifespan-extending genes. Besides two genes involved in mitochondrial energy generation and several localized to the vacuole, we found three genes involved in endosomal sorting and endosome-to-vacuole transport, including VPS24, encoding one of the four subunits of the endosomal sorting complex required for transport III (ESCRT-III) complex; DID2, encoding a class E protein of the vacuolar protein-sorting (Vps) pathway that binds Vps4p and directs it to dissociate from the ESCRT-III complex; and VPS60, encoding a protein involved in late endosome-to-vacuole transport (Fig. 3 A–C). Interestingly, another component of the ESCRT-III complex, SNF7, is also mother-enriched. However, deletion of SNF7 leads to shortened lifespan. Snf7 is a core component of the ESCRT-III complex, suggesting that reduced but not abolished activity of the ESCRT-III complex extends lifespan. To our knowledge, our study represents the first time that genes involved in endosomal sorting/endosome-to-vacuole transport have been implicated in lifespan regulation, although it was known previously that ESCRT complexes are tightly linked to metabolic regulation and target of rapamycin (TOR) activity (31).
Fig. 3.
Examples of mother-enriched proteins, the deletion of which extends lifespan. Shown are lifespan curves for the deletion strains of three genes involved in endosomal sorting/endosome-to-vacuole transport (A–C) and three nitrogen source transporters (D–F). The percent lifespan increase relative to the wild-type (wt) control and the P value based on a rank sum test are indicated.
As another functional theme, we found that deletion of three genes that encode various transporters for nitrogen sources extends lifespan. These genes are MEP2, encoding an ammonia transporter; CAN1, encoding an arginine transporter; and TPO3, encoding a polyamine transporter (Fig. 3 D–F). The strong enrichment of transporters for nitrogen sources suggests there might be a common mechanism. It is possible that the unregulated uptake of nitrogen sources may lead to the overproduction of metabolic intermediates that become lifespan-limiting in aged cells.
Mother-Enriched and Lifespan-Limiting Proteins Tend to Accumulate over Time in Aged Mother Cells.
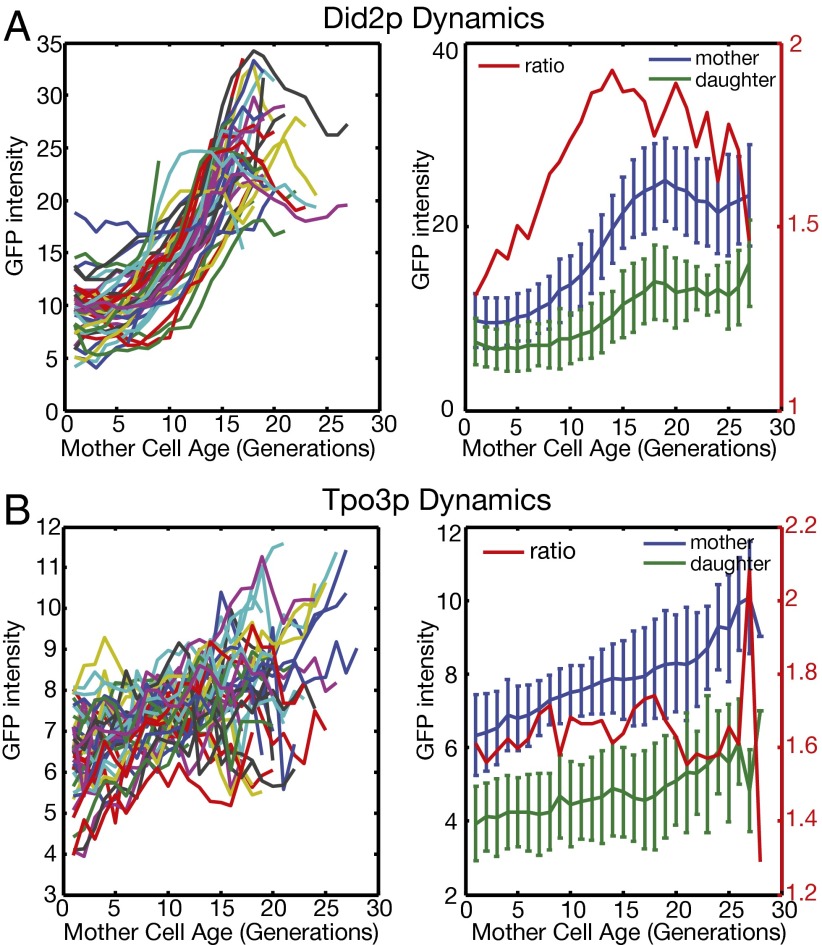
To connect the asymmetric division in young cells to the aging phenotype in old cells, we tracked how these proteins change over time in single mother cells throughout their lifespan, using a recently developed microfluidic device (32, 33). We analyzed Did2p and Tpo3p, which represent a protein involved in endosomal sorting/endosome-to-vacuole transport and a nitrogen source transporter, respectively.
We followed the intensity of the proteins in single mother cells and their daughter cells as a function of the mother cell age (Fig. 4). A common trend is that the protein level in the mother cells increases steadily as they age. As the protein level increases in the mother cell, the daughter cells also inherit more from the mother. However, there is a clear asymmetry between mother and daughter cells; this asymmetry keeps the protein level in the daughter cells low even when they are born from old mother cells. For Did2p, daughters born from old mother cells aged around 20 generations have a protein level comparable to middle-aged mothers of about 10 generations. For Tpo3p, daughter cells (regardless of the age of their mothers) always have a level lower than the level of the young mother cells. It is known that the lifespan of daughter cells is independent of the age of their mother cells, except for the daughters produced in the last few cell divisions of the mother (3). A possible scenario is that these proteins become lifespan-limiting only when they reach the same level as in the old mother cells.
Fig. 4.
Dynamic expression of mother-enriched proteins in single cells as a function of mother cell age. Mother cells were tracked using a microfluidic device and a time-lapsed microscope. GFP intensities of C-terminal–tagged Did2p (A) and Tpo3p (B) were measured in individual mother cells as well as in their daughters. (Left) GFP intensity in individual mother cells, where each colored line represents one cell. (Right) Mean and SD of the GFP intensity in mother cells and in daughter cells produced by mother cells at a given age. The red line is the ratio of the means of the mother and daughter cells.
Discussion
One major mechanism for lifespan asymmetry between yeast mother and daughter cells is asymmetric division of cellular contents. Using a novel high-throughput approach, we have quantified the asymmetric partitioning of the yeast proteome between mother and daughter cells and identified proteins that are enriched in mother or daughter cells. These proteins are interesting candidates for the future investigation of the mechanisms of asymmetric cell division and aging.
How can a protein asymmetrically partition between mother and daughter cells? For a typically sized protein (∼400 amino acids) freely floating in cytosol, the time it takes to diffuse throughout the cell (a few microns in size) is a few seconds; thus, the concentration can quickly equilibrate between the mother and the bud, and it is difficult to develop asymmetry. Consistent with this notion, we observed that the majority of proteins are symmetrically distributed, with equal concentrations in mother and daughter cells. However, we did observe a subset of proteins that are preferentially distributed to one of the two cell types. Interestingly, mother-specific proteins are not more likely to form complexes [based on the protein complex data from the Saccharomyces Genome Database (SGD), downloads.yeastgenome.org/curation/literature/interaction_data.tab], further contraindicating diffusion constraints as a major mechanism of asymmetry.
Instead, analysis of the mother- and daughter-enriched proteins suggested two scenarios for their asymmetric partitioning: (i) active transport and (ii) confinement within an organelle or a substructure of an organelle that is not symmetrically distributed. Proteins preferentially distributed to daughter cells are highly enriched for those proteins needed for the construction of the bud, many of which are delivered to bud neck and bud through active transport processes. Mother-specific proteins are highly enriched for those proteins localized to membranes or in the vacuole, suggesting that the vacuole is preferentially retained in the mother cell. This result is consistent with the observation that the vacuole in the mother cell keeps increasing in size as the cell ages, and eventually reaches a high-volume fraction of the cell. Quantitative analysis also revealed a faster than linear scaling of the vacuole size with the cell volume, consistent with a rapid increase of vacuole size as mother cells become older and bigger (27).
We have observed preferential retention of a number of mitochondrial proteins (e.g., Kgd1, Pda1, Atp1) in the mother cells, even though the majority of mitochondrial proteins are symmetrically distributed. It is known that the mitochondria-to-cell volume ratio of daughter cells is very close to, and even slightly higher than, the mitochondria-to-cell volume ratio of their mother cells (34). Therefore, the preferential retention of these mitochondrial proteins cannot be explained by the preferential distribution of mitochondria in mother cells. Kgd1, Pda1, and Atp1 are physically associated with the mitochondrial nucleoid and are likely bifunctional enzymes involved in both energy metabolism and mitochondrial genome maintenance (28). Previous studies have shown that the actively replicating mitochondrial nucleoid is associated with the mitochondria/ER junction (29). We thus conjecture that a substructure of mitochondria that is tethered to the ER is not equally transmitted to the daughter cell. Consistent with this conjecture is a recent observation showing that aggregated proteins in the ER are captured by mitochondria and preferentially retained in the mother cells (18).
How does asymmetric protein partitioning connect with lifespan asymmetry? Our data show that mother-enriched proteins are more likely to have a lifespan extension phenotype when deleted, suggesting that they are more likely to become lifespan-limiting in old cells. Dynamic tracking of these proteins in single mother cells showed a steady increase to a high level in the old cells, whereas the daughter cells keep a relatively low level through asymmetry, arguing that the high concentration may be deleterious in old cells.
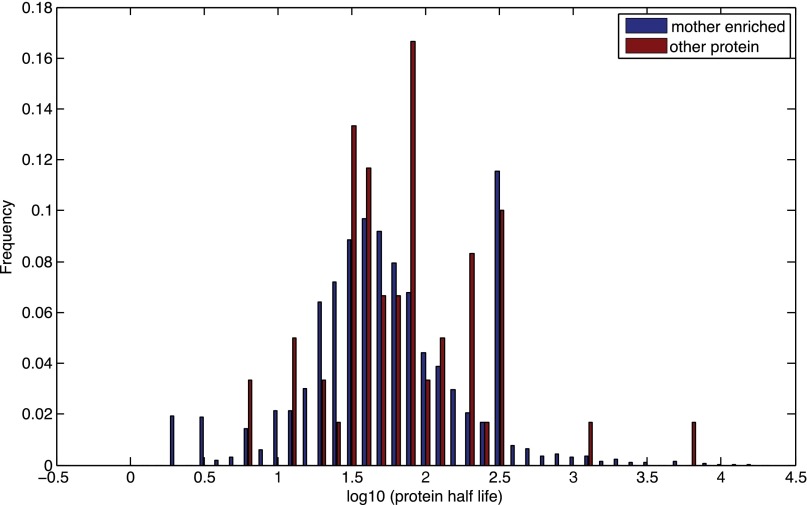
Thayer et al. (35) recently used isotope labeling and total proteome MS to identify long-lived asymmetrically retained proteins (LARPs). They identified several full-length proteins (Mrh1, Pma1, Sur7, Thr1, and Hsp26) and many protein fragments. We found that Pma1 is significantly enriched in mother cells (z-score of −4.5 for the 170 min), but is filtered out due to a small number of cells (Methods). Mrh1 is also in our list of mother-enriched proteins, but the deletion mutant is short-lived. Unlike the LARPs, which are long-lived proteins, the mother-enriched proteins we identified have a normal half-life (Fig. S3), and hence normal turnover. This observation suggests that the lifespan-limiting effect is not due to old and nonfunctional proteins; rather, it is the high concentration in mothers that interferes with cell homeostasis and/or leads to aberrant cell signaling, limiting lifespan.
Fig. S3.
Mother-enriched proteins have a normal t1/2. Shown is the histogram of the t1/2s of the set of mother-enriched proteins and the t1/2s of other proteins. The two distributions are not significantly different (P = 0.125 based on the rank sum test).
Are there fitness benefits of the preferential retention of proteins in the mother cell? Theoretical studies suggest that asymmetric segregation may evolve as a general strategy to cope with cellular damage (9–11), conferring a fitness benefit at the population level by producing rejuvenated offspring at the expense of an aging parent. Such asymmetric segregation may operate at the level of the organelle, where the daughter cell synthesizes an organelle de novo or grows it from a seed inherited from the mother. Mother-specific retention may also confer a fitness benefit by ensuring that certain proteins (e.g., those proteins not needed for, or even inhibitory to, the early development of the bud) are expressed just in time in the daughter cell, and that their level of expression does not become too high in the daughter/young mother cells. Interestingly, we found that none of 74 mother-enriched proteins are essential, which is a significant depletion from the genome-wide average of about 18%.
The observed asymmetric distribution of many vacuolar proteins and the rapid increase in vacuole size as the mother cell ages suggest that vacuoles in old mother cells might be a major source of aging factors limiting lifespan. Interestingly, it was observed that the spores do not inherit the vacuole from the mother during meiosis (36), and sporulation was found to rejuvenate the gametes generated from old cells (37). Three of the 23 proteins we identified (Did2, Vps24, and Vps60) are involved in endosomal sorting and endosome-to-vacuole transport via the multivesicular body (MVB) pathway. It is known that the MVB pathway is strongly linked to the regulation of cellular metabolism, and mutants of ESCRT complexes exhibit decreased TOR activity (31, 38). Interestingly, chemical-genetic profiling in yeast revealed that DID2 and VPS60 deletion mutants show strong sensitivity to rapamycin treatment (39), adding to the evidence that these genes positively influence TOR activity. Thus, one possible scenario is that accumulated proteins involved in endosomal sorting and endosome-to-vacuole transport eventually lead to aberrant TOR signaling that limits the lifespan. Consistent with this scenario, our dynamic tracking of the protein Did2 showed that a decrease of the concentration later in life in a subset of cells correlates with a longer lifespan (Fig. 4A).
In this study, we have focused mainly on mother-enriched proteins. However, our genome-wide screening also produced a comprehensive set of daughter-enriched proteins. These proteins may be important for bud development, or may act as rejuvenating factors necessary for the reset of the lifespan. Data from our genome-wide screening indicate that the deletion mutants are enriched for those mutants with an increased lifespan (Dataset S4). For example, deletion of CRN1 and LOC1, involved in construction of the bud- and daughter-specific mRNA localization, significantly extends lifespan; the latter is consistent with a previous finding (40). The enrichment of lifespan-extending mutants is consistent with the antagonistic pleiotropy hypothesis that genes beneficial to early development may become detrimental later in life (41). This hypothesis has been explored in other species (e.g., in worms), where a strong overlap between longevity genes and genes important for development was observed (42). For potential rejuvenating factors, we expect that the deletion will shorten lifespan. We did found a number of mutants with a significantly shortened lifespan, among them SRS2 and HMO1, which are involved in DNA repair and genome maintenance.
In summary, we have used a high-throughput approach to identify asymmetrically distributed proteins between mother and daughter cells during division. Proteins enriched in mother cells also tend to be lifespan-limiting, suggesting that their accumulation during mother cell aging is restrictive to continued cell division. We also identified a set of daughter-enriched proteins with lifespan phenotypes. These findings set the stage for a comprehensive understanding of cell division asymmetry and replicative aging in yeast, and point to a number of new mechanisms that may modulate aging in more complex eukaryotes.
Methods
All strains of Saccharomyces cerevisiae used in high-throughput flow cytometry screening were taken from the GFP tagging library (25). Deletion strains used for lifespan analysis were derived from BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). Yeast cultures were grown in selective minimal media containing 2% (mass/vol) glucose.
Replicative lifespan analysis and single-cell tracking using the microfluidic device were performed as previously described (33, 43, 44).
Protocols for cell staining and flow cytometry experiments and FACS data analysis and z-score calculation are described in SI Methods.
SI Methods
Cell Staining and Flow Cytometry.
Yeast cells were cultured in 96-well plates in a 30 °C shaker overnight and were then diluted to OD600 = 0.05–0.075 and grown for 6 h. The cells (180 μL per well) were spun down (3,000 rpm, 5 min, Beckman Coulter Allegra 6 Centrifuge), washed with 0.01 M PBS, and then stained with Cy5 dye (one pack of Cy5 dissolved in 2 mL of PBS, Cy5 Post-Labeling Reactive Dye Pack; Amersham Bioscience) for 10 min. After staining, cells were washed twice with PBS and resuspended in culture media. Stained cells were then divided into three new 96-well plates and grown in a 30 °C shaker to three different time points [130 min, 170 min, and overnight (16 h)] before being subjected to flow cytometry analysis. For the overnight plates, cell density was diluted to OD600 = 0.0012–0.0015. A BD Biosciences LSR II flow cytometer and HTS were used to measure the GFP and Cy5 signals [by the areas of fluorescein isothiocyanate (FITC) channel (510 nm) and allophycocyanin (APC) channel (650 nm), respectively].
FACS Data Analysis.
For the 130-min and 170-min time points, each well was sampled for 10 s using customized flow cytometer data collection software. It was observed that the GFP signal occasionally became unstable in a small time interval during the 10-s time window. To remove the unstable signals, we divided the 10-s time window into 200 bins, calculated the mean of the log10 (FITC-height) signal for each bin, and used the distribution of the means from the 200 bins to remove the extreme outliers (5 standard deviations above or below the average). After the time window filtering, we further removed the cells having the top/bottom 0.1% forward-scattering area signal or the top/bottom 0.2% of the area of side-scattering (SSCA) signal to filter out cell debris or aggregates. We also removed any event with extremely high Cy5 signals (>105, given our staining protocol and the flow cytometer settings) to get rid of small staining particles not dissolved or washed away. For the overnight time point, we applied a slightly different filtering strategy. Time window filtering was not necessary because data collected by the BD Biosciences software Diva did not suffer from the problem of unstable signals. We did not filter cells based on their forward-scattering and side-scattering signals to avoid removing aged mother cells, because they tend to have larger sizes. We removed cells with a Cy5 signal greater than 105 for the same reason described above.
Selecting Mother and Daughter Populations.
For the 170-min time point, the distribution of the Cy5 signal is typically bimodal, with low and high Cy5 peaks corresponding to the daughter and mother populations (Fig. 1B). To select the two populations automatically, we first used an intermediate Cy5 level [log10(Cy5_h) = 3.3 based on the observations of many wells] in the valley of the distribution to separate the cells into two groups. We then refined the selection by locating the positions of the peaks of the log10(Cy5_h) signal (Pd and Pm for the daughter and mother populations, respectively) and used the two windows (Pd − 0.1, Pd + 0.1) and (Pm − 0.1, Pm + 0.1) to select daughter cells and mother cells for the final GFP signal calculation (changing the bin width has little effect on the final results).
For the 130-min time point, it was sometimes difficult to determine the peak position of the daughter population due to the relatively small number of daughter cells. In that case, we used the gating intervals defined by the corresponding well (the same strain and the same initial staining) at the 170-min time point to select the daughter and mother populations.
For the overnight time point, because the fraction of the initially labeled mother cells becomes very small after 16 h (∼10−3), we used a different strategy to separate them from the unlabeled population (which is mixed with daughter cells and mother cells of different ages). We aggregated the cells from all 96 wells on each plate and selected those cells having Cy5 values in the top 1% (more than 104 cells) so as to enrich for the labeled mother cells. The distribution of the Cy5 values of this subset of the cells generally has two peaks, representing the labeled middle-aged mother cells and a small fraction of the unlabeled population. We then determined the positions of the peaks and found the valley that separated them. This valley position in the Cy5 axis was then used to separate the labeled and unlabeled populations for each of the 96 wells on the same plate.
To ensure good statistics, we also marked wells with a small number of cells and excluded them from the subsequent analysis. For the 130-min and 170-min time points, we excluded the wells with fewer than 8,000 total cells, or with the number of mother/daughter cells with less than 100 total cells. For the overnight data, we excluded the wells with fewer than 80,000 unlabeled cells or fewer than 200 labeled mother cells. The strains in the excluded wells are reported as “no data” (empty wells in Dataset S1) for the given time point.
Z-Score Calculation.
We first normalized the GFP signal of a cell by its size: , where FITCA and SSCA are the areas of the GFP and the side-scattering signals. For each population, we calculated the mean of the normalized GFP signal and took the log transform: , where the is the number of cells of the ith population (i = m or d, for mother or daughter, respectively) in the jth well.
To calculate the z-scores for the 130-min and 170-min time points, we first performed a linear regression, , for all of the wells. Using a and b derived from the regression, we calculated the expected GFP value for the daughter cell population from the jth well, given the measured value of the mother population, : . We then took the difference between the measured and expected GFP values for the daughter population, , and converted it to a z-score, , where is the SD of the distribution of from all of the wells.
For the overnight data, we performed a similar linear regression, , for the labeled (l) mother population and the unlabeled (u) mixed population. We used the unlabeled population to calculate the predicted value for the labeled population, because the former has a much larger number of cells, and hence less noise. We then took the difference between the predicted and measured values for the labeled mother cell population, , and converted it to a z-score.
For wells with replicated experiments, the averaged z-scores from the multiple measurements were reported.
Replicative Lifespan Analysis.
Yeast replicative life span (RLS) assays were performed as previously described (43, 44). In short, virgin daughter cells were isolated from each strain and then allowed to grow into mother cells, whereas their corresponding daughters were microdissected and counted until the mother cell could no longer divide. Statistical significance was determined by calculating P values using the Wilcoxon rank sum test (45, 46).
Microfluidic Experiments and Time-Lapsed Microscopy.
Microfluidic devices for the experiments were fabricated as previously described (33). After mold fabrication, polydimethylsiloxane (PDMS; parts A and B in a 10:1 ratio) was poured into the mold and allowed to cure at 80 °C. Access holes to the channels were punched in the PDMS, and the final chip was sealed to a cover glass slide after treating with plasma.
For time-lapsed microscopy, cells were prepared as previously described (33). Temperature of the microfluidic chip was kept at 30 °C with a stage top incubator (INU-TIZHB-F1, Tokai Hit Co., Ltd., Fujinomiya-shi, Japan). Images were collected with epi-fluorescence microscopy using a Nikon Ti-E inverted microscope equipped with the objective lens Plan Apo VC 100×/1.40 Oil DIC N2 or Plan Fluor 40× DIC M N2, the motorized XY stage and the Perfect-Focus System (Nikon Co., Tokyo). Images were acquired with an CoolSNAP HQ2 camera (1,392 × 1,040, 6.45 μm, Photometrics, Tucson, AZ) and Lambda SC shutter controllers (Sutter Instruments, Novato, CA). NIS Elements AR v3.2 (Nikon Co., Tokyo) was used to automate image acquisition and microscope control. The microfluidic device was mounted on the microscope by a customized holder. For Did2p, images (bright field and GFP) were taken for 40 h with a 10-min interval. For Tpo3p, images were taken for 40 h with a 10-min interval for the bright field and a 50-min interval for the GFP channel. Images were analyzed using ImageJ (NIH). Mean GFP intensity in the whole cell was measured for Did2p. For each generation of a cell, more than five images were captured, and the mean intensity from these images was reported as the intensity for that generation. For Tpo3, the total intensity from a cell is normalized by its surface area.
For the images of mother- or daughter-enriched proteins, cells were cultured overnight in synthetic defined media (SD) media, diluted to OD600 = 0.1, and then grown for 4 h. GFP/bright-field images were taken using a Nikon Ti-E time-lapsed microscope.
Supplementary Material
Acknowledgments
We thank Cynthia Kenyon, Changhui Deng, and Ke Zou for helpful discussions. This research was supported by NIH Grant AG043080, by a Packard Fellowship in Science and Engineering (to H.L.), by NIH Training Grant T32AG000266 (to M.A.M.), by National Natural Science Foundation of China (NSFC) Grant 11434001, by Ministry of Science and Technology of China (MOST) Grant 2012AA02A702, and by NIH Center for Systems and Synthetic Biology Grant P50 GM081879.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506054112/-/DCSupplemental.
References
- 1.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183(4677):1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 2.Jazwinski SM. An experimental system for the molecular analysis of the aging process: The budding yeast Saccharomyces cerevisiae. J Gerontol. 1990;45(3):B68–B74. doi: 10.1093/geronj/45.3.b68. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy BK, Austriaco NR, Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127(6 Pt 2):1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coelho M, et al. Fission yeast does not age under favorable conditions, but does so after stress. Curr Biol. 2013;23(19):1844–1852. doi: 10.1016/j.cub.2013.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker MG, Walmsley RM. Replicative ageing in the fission yeast Schizosaccharomyces pombe. Yeast. 1999;15(14):1511–1518. doi: 10.1002/(sici)1097-0061(199910)15:14<1511::aid-yea482>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 6.Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3(2):e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105(8):3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, et al. Robust growth of Escherichia coli. Curr Biol. 2010;20(12):1099–1103. doi: 10.1016/j.cub.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ackermann M, Chao L, Bergstrom CT, Doebeli M. On the evolutionary origin of aging. Aging Cell. 2007;6(2):235–244. doi: 10.1111/j.1474-9726.2007.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans SN, Steinsaltz D. Damage segregation at fissioning may increase growth rates: A superprocess model. Theor Popul Biol. 2007;71(4):473–490. doi: 10.1016/j.tpb.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watve M, Parab S, Jogdand P, Keni S. Aging may be a conditional strategic choice and not an inevitable outcome for bacteria. Proc Natl Acad Sci USA. 2006;103(40):14831–14835. doi: 10.1073/pnas.0606499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell. 1997;91(7):1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 13.Defossez PA, et al. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3(4):447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 14.Denoth-Lippuner A, Krzyzanowski MK, Stober C, Barral Y. Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. eLife. 2014;3:3. doi: 10.7554/eLife.03790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454(7205):728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 16.Gehlen LR, et al. Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Curr Biol. 2011;21(1):25–33. doi: 10.1016/j.cub.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 17.McFaline-Figueroa JR, et al. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 2011;10(5):885–895. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou C, et al. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 2014;159(3):530–542. doi: 10.1016/j.cell.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou C, et al. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell. 2011;147(5):1186–1196. doi: 10.1016/j.cell.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140(2):257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Erjavec N, Larsson L, Grantham J, Nyström T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21(19):2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299(5613):1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 23.Eldakak A, et al. Asymmetrically inherited multidrug resistance transporters are recessive determinants in cellular replicative ageing. Nat Cell Biol. 2010;12(8):799–805. doi: 10.1038/ncb2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson KA, Hughes AL, Gottschling DE. Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. eLife. 2014;3:e03504. doi: 10.7554/eLife.03504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 26.Weisman LS. Yeast vacuole inheritance and dynamics. Annu Rev Genet. 2003;37:435–460. doi: 10.1146/annurev.genet.37.050203.103207. [DOI] [PubMed] [Google Scholar]
- 27.Chan YH, Marshall WF. Organelle size scaling of the budding yeast vacuole is tuned by membrane trafficking rates. Biophys J. 2014;106(9):1986–1996. doi: 10.1016/j.bpj.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6(11):815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 29.Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J Cell Biol. 2003;163(3):503–510. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormick MA, et al. A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metabolism. 2015 doi: 10.1016/j.cmet.2015.09.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones CB, et al. Regulation of membrane protein degradation by starvation-response pathways. Traffic. 2012;13(3):468–482. doi: 10.1111/j.1600-0854.2011.01314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Z, et al. Molecular phenotyping of aging in single yeast cells using a novel microfluidic device. Aging Cell. 2012;11(4):599–606. doi: 10.1111/j.1474-9726.2012.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Single cell analysis of yeast replicative aging using a new generation of microfluidic device. PLoS One. 2012;7(11):e48275. doi: 10.1371/journal.pone.0048275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rafelski SM, et al. Mitochondrial network size scaling in budding yeast. Science. 2012;338(6108):822–824. doi: 10.1126/science.1225720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thayer NH, et al. Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc Natl Acad Sci USA. 2014;111(39):14019–14026. doi: 10.1073/pnas.1416079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roeder AD, Shaw JM. Vacuole partitioning during meiotic division in yeast. Genetics. 1996;144(2):445–458. doi: 10.1093/genetics/144.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unal E, Kinde B, Amon A. Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science. 2011;332(6037):1554–1557. doi: 10.1126/science.1204349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babst M, Odorizzi G. The balance of protein expression and degradation: An ESCRTs point of view. Curr Opin Cell Biol. 2013;25(4):489–494. doi: 10.1016/j.ceb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long RM, et al. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J Cell Biol. 2001;153(2):307–318. doi: 10.1083/jcb.153.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams G. Pleiotropy, natural selection, and evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 42.Chen D, Pan KZ, Palter JE, Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6(4):525–533. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2(9):E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. J Vis Exp. 2009;25(28):e1209. doi: 10.3791/1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilcoxin F. Probability tables for individual comparisons by ranking methods. Biometrics. 1947;3(3):119–122. [PubMed] [Google Scholar]
- 46.Wilcoxon F. Individual comparisons of grouped data by ranking methods. J Econ Entomol. 1946;39:269. doi: 10.1093/jee/39.2.269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.