Summary
Objectives
Controlled terminologies and their dependent artefacts provide a consensual understanding of a domain while reducing ambiguities and enabling reasoning. However, the evolution of a domain’s knowledge directly impacts these terminologies and generates inconsistencies in the underlying biomedical information systems. In this article, we review existing work addressing the dynamic aspect of terminologies as well as their effects on mappings and semantic annotations.
Methods
We investigate approaches related to the identification, characterization and propagation of changes in terminologies, mappings and semantic annotations including techniques to update their content.
Results and conclusion
Based on the explored issues and existing methods, we outline open research challenges requiring investigation in the near future.
Keywords: Knowledge management, knowledge representation, ontology evolution, biomedical ontologies
1 Introduction
Biomedical terminologies are the foundation for several applications necessary to overcome semantic interoperability issues [1]. To this end, the use of standard vocabularies is key to harmonizing biomedical information and facilitating its exploitation. Well known examples include the National Library of Medicine, which uses MeSH [2] to index scientific publications, or the adoption of ICD [3] terms for encoding medical diagnosis. However, the increasing quantity and size of biomedical controlled terminologies forces ICT (Information and Communication Technologies) systems and domain experts to use a combination of controlled terminologies in order to enable the better sharing and exchange of biomedical information. For example, information encoded with MeSH and information encoded with ICD can only be properly used if the relationships between these terminologies are explicitly defined. This is the role of semantic correspondences (or mappings) which are established between concepts of different terminologies in order to specify their relationship [4].
The biomedical domain is highly dynamic by nature. For instance, 50% of the body of (documented) medical knowledge has been reviewed in the last decade [5] and this has led to frequent publication releases of terminologies [6]. This evolution directly impacts the precision of existing mappings [7] and semantic annotations, and indirectly impacts the performance of ICT systems which rely on these mappings to retrieve information and support decisions with accuracy. In a digital era where more and more knowledge is modelled, linked and shared via computers, there is a real need to provide ways of managing domain knowledge evolution which should: (1) detect the evolution; (2) evaluate the consequences of the evolution on dependent terminologies and mappings; and (3) (semi-) automatically update them to reflect the evolution.
Current evolution approaches for the maintenance of biomedical terminologies are essentially based on manual interventions [8]. This is a labour-intensive task (e.g., SNOMED CT [9] is revised twice a year by hundreds of members), which is both expensive (e.g., SNOMED CT managers spend ~9 million USD per year for maintenance and improvement1) and error-prone (e.g., between 2002 and 2008, SNOMED CT was “cleaned” in more than 20000 concepts because they were duplicated, outdated, or ambiguous2). In order to assist the work of domain experts, more automatic methods and tools to support management tasks in highly dynamic domains (like biomedicine) are required. Limited research presenting solutions to part of this management problem has been published and, to the best of our knowledge, no published work addresses the complete evolution process taking into account the impact of terminology evolution on dependent artefacts.
In this article, we thoroughly review existing techniques concerned with maintaining the validity, from a semantic point of view, of mappings and annotations affected by changes in their associated controlled terminologies. We devote particular attention to the identification and characterization of changes that occur when one version of the terminology evolves to the next. This article also addresses the selection of the appropriate implementation strategy to propagate the terminology changes.
This research presents relevant and recently published approaches that deal with controlled terminology evolution (section 2), mapping maintenance (section 3), and semantic annotation maintenance (section 4). We summarize the analysis of trends and open challenges in section 5 followed by the conclusion in section 6.
2 Controlled Terminologies Evolution
The complexity and size of current biomedical knowledge is reflected in the huge variety of controlled terminologies available today. Repositories such as Bioportal [10] OBO foundry [11] HeTop [12] or UMLS offer controlled vocabularies with services to exploit them. If the number of terminologies is constantly growing within these repositories, then the number of versions for each of these resources will follow the same trajectory. This fact highlights that knowledge engineers and domain experts in charge of the design and maintenance of these terminologies are aware that their content must faithfully follow the progress of domain knowledge over time. As a consequence, ontological elements, like concepts, are added, removed or revised from one version to another in order to reflect new findings in the domain.
To reveal the dynamic aspect of biomedical terminologies, Dos Reis et al. have quantified the number of concepts of SNOMED CT (SCT) that are modified from one version to the next [13]. About 10% (on average) of the total number of concepts present some changes in their description. This represents about 30000 concepts. Other relevant examples are the Gene Ontology [14] (GO) and NCI thesaurus [15] (NCIt), which have more than doubled over the past decade. By virtue of the size of existing terminologies and the fast evolution of the biomedical domain, modifications of their content can be heavily variable in terms of nature and complexity. The work of Hartung et al. [16] pointed out the stability of regions in evolving ontologies. This showed that some components of ontologies are more “stable” than others, i.e., stable parts change very little and these changes do not impact the underlying semantic-based applications. In this general context, it is hard to deny the importance of the terminology evolution problem where the management of dynamic domain knowledge provides a real challenge for semantic-enabled biomedical applications.
We need to consider two major issues during the evolution process: (1) the identification and characterization of changes that occur as one version of the terminology evolves to the next; and (2) the evaluation of the impact of these changes. Ontologies then refer to formal terminologies.
As a huge majority of ICT systems simply use biomedical terminologies rather than designing and constructing new ones, they need to react when new releases of the selected terminologies are published. To this end, theoretical approaches that define changes by analysing the underlying knowledge model used to represent the resource (OWL, OBO, database schemas, etc.) collide with a more pragmatic approach of the problem which focuses on changes that are observed in practice and that lead to an abstraction of the knowledge model (e.g., exploiting linguistic or syntactic aspects).
One traditional (but efficient) method for change identification in ontologies consists of analysing the logs [17] describing the implemented changes. However, this source of information remains rarely available for several reasons which include: (1) the absence of standards to express changes; (2) the complexity of classifying changes; and (3) sometimes the huge amount of changes to report. Taking this fact into account, a family of approaches can be used to detect the evolution of ontologies by automatically comparing their successive versions; a process known as Diff problem identification. State-of-the-art software tools like PROMPTDIFF [18] or COnto-Diff [19] can be used to recognize the differences that exist between two successive versions of the same ontologies resulting from a set of ontology changes. They mostly differ in the complexity of the implemented Diff algorithm and, more interestingly, in the way they define and describe detectable changes (i.e., different classification approaches).
Changes are classified into a set of types of change that can then transform a terminology from one version to another. They range from very simple ones (atomic types), such as the addition or removal of concepts [20], to more complex change types such as the concepts of “merging” or “splitting” [19] which can be seen as a composition of atomic change types (e.g., we can interpret splitting as the removal of a concept and the addition of several others). The empirical observations revealed by Dos Reis et al. [13] have shown that, except for duplicated concepts, concept merging rarely happens in standard biomedical terminologies (knowledge rarely becomes more general). On the contrary, domain knowledge is becoming more and more precise, therefore accelerating concept splitting.
Changes can also affect the structure of a terminology (e.g., the taxonomical hierarchy) without affecting a concept’s description. For this purpose, change patterns have been explored to characterize complex change types and evolution scenarios to minimize the impact of the evolution, and to ensure consistency in the evolution of ontologies [21]. Change patterns can vary according to several elements: (1) the knowledge representation model used to express the ontology [22] (e.g., the addition of OWL classes or Object properties), (2) the structural aspects induced by its semantics [19] e.g., moving part of the ontology), and (3) the formalization [23] (e.g., completion of the ontology by inferring implicitly expressed relationships between concepts). Some change patterns also rely on model-independent features like linguistics [24] (e.g., the way concept terms or labels are expressed), or on specific constraints of the domain [25] (e.g., a naming convention to express genes and proteins in GO).
Another problematic task refers to the evaluation of the impact of changes. If neglected, it can jeopardize the consistencies of the underlying information systems and, in turn, affect medical decisions. As shown in the work of Abgaz et al. [26] the various change types express a different impact on terminology evolution. The approach conducted by Shaban-Nejad & Haarslev [27] addressed this issue using learning agents to reduce the intervention of human experts in the evolution process. Pesquita & Couto [28] exploited machine learning techniques on a significant number of ontology versions in an effort to predict which part of the ontology would evolve in the future in order to better understand it and foresee the potential effects of changes.
Restricting changes to sub-ontologies in order to further control the impact of evolution has also been investigated by Sari et al. [29]. Although logic-based approaches have provided interesting properties to simulate the impact of ontology changes, the lack of formalization in biomedical terminologies precludes applying such techniques. The reference methodology provided by Stojanovic [30] for ontology evolution points out the necessity to have a broader vision of the problem. This research not only advocated an analysis of the impacts to the ontology itself, but also considered propagating them to dependent artefacts. With respect to the biomedical domain, two major artefacts predominate: mappings established between different controlled terminologies and semantic annotations usually associated with documents. Because of the critical role played by them in biomedical information systems, their maintenance over time deserves particular attention.
3 Mapping Maintenance
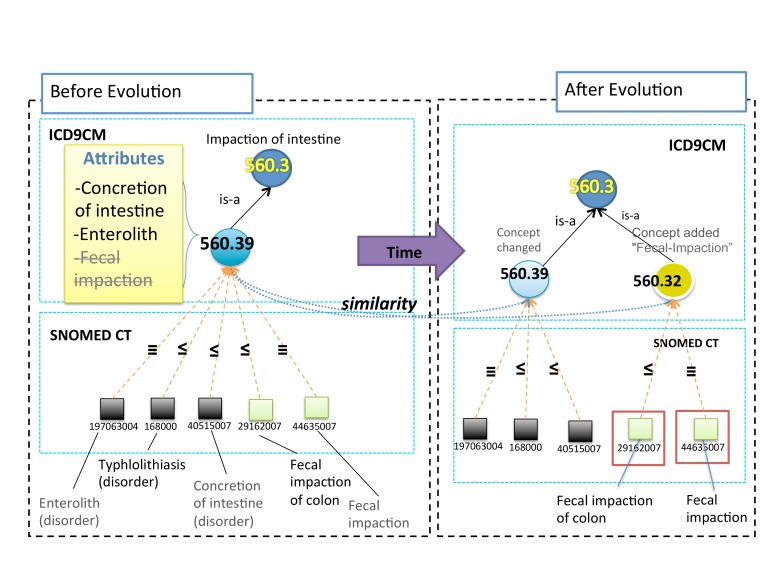
Mapping provides links between elements from different controlled terminologies, making it possible to use them consistently in a combined way. Based on mappings and inference mechanisms, documents semantically annotated with a given terminology can be queried using the vocabulary of a different one [31]. However, when one of these terminologies evolves, mappings can be affected and even invalidated. Figure 1 illustrates the evolution of the concept 560.39 (ICD) where after the evolution of one concept attribute, which becomes the label of a new concept (560.32), two mappings follow this attribute as a consequence. In 2013, Salvadores et al. [32] counted about 9.8 million cross-ontology and mappings accessible via the BioPortal application, and determined that this number was constantly increasing. However, despite the importance of the problem, the maintenance of mappings has not been thoroughly investigated during the past few years.
Fig. 1.
Example of a split of a concept from ICD and the evolution of the associated mappings between ICD and SCT
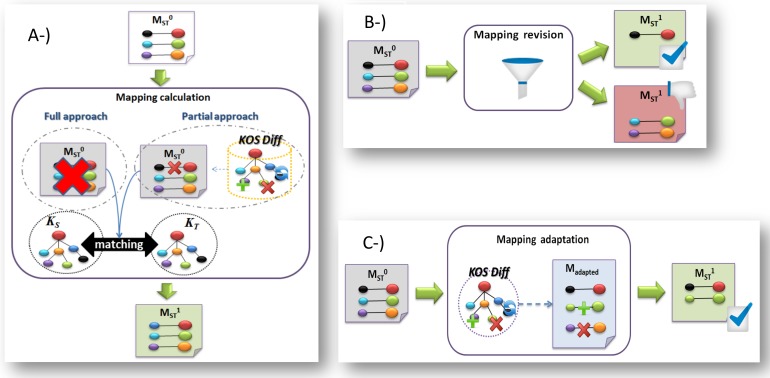
From our research, we highlight three distinct families of approaches for the (semi-automatic) maintenance of mappings currently found in the literature [33]:
The first family (recalculation) relies on an unsophisticated way to maintain mappings that consist in deleting existing ones and realigning terminologies that have evolved over time (cf. Figure 2 part A). This is what has been proposed by Gal et al. [34] where database schemas are realigned once they have been modified. Nevertheless, the size of biomedical controlled terminologies and the necessity of manual validation of all new mappings (if a certain degree of quality has to be ensured) make this type of technique difficult to apply. For instance, there are around 100.000 mappings between SCT and ICD. Therefore, an optimization of the above approach involves considering only part of the mapping set (i.e., only those that are potentially invalid due to terminology changes) and not the whole set [35]. This reduces the maintenance time, but still requires significant validation efforts. For example, approximately 10.000 mappings between the SCT and ICD were affected by the evolution of the involved concepts (from 2009 to 2011).
Fig. 2.
Families of mapping maintenance approaches
The second family (revision) refers to logic-based approaches (cf. Figure 2 part B). This is a two-step process for detecting invalid mappings and repairing (or debugging) them. The former step can be performed using various techniques borrowed from different paradigms: database techniques aiming at periodically comparing the results of queries [36, 37] and deducing invalid mappings when the queries produce erroneous results. For deduction, software engineering techniques and fault-tolerance [38] or logic-based reasoning approaches [39] have been investigated. Although detecting broken mappings is useful to prepare the phase of modifying mappings during maintenance, it is still insufficient to repair mappings and then keep them up-to-date. This is the goal of the second step where ontologies and reasoners with formal approaches are frequently used in order to detect and correct inconsistencies [40-42].
The third family (adaptation) refers to types of approaches that aim to exploit information gained from the analysis of terminology evolution in order to adapt the mappings (cf. Figure 2 part C). To this end, various methods have been investigated such as mapping composition, synchronization of ontologies, change impact minimization, and change propagation.
The mapping composition principle suggests composing various mappings to generate new ones. This technique requires a composition function to combine existing mappings with information coming from the terminology evolution [43]. The techniques of synchronizations [44] were proposed to cope with the heterogeneity of models like databases and ontologies. These methods require strong collaboration between managers of different ontologies to propagate changes from one ontology to the other. Complementary to the previous approaches, the change impact minimization is usually implemented exploiting the logic aspect in ontology [45] (e.g., ontological statements), but it can hardly be applied to the biomedical domain because of the lack of formalization in existing biomedical terminologies.
Change propagation approaches emphasize the way of adapting mappings and the techniques to which information from an evolved ontology can be used to facilitate the maintenance effort [46]. For instance, Martins & Silva [47] studied the consequences (for mappings) of deleting concepts in ontologies, while Dos Reis et al. [48] considered a wider range of defined mapping adaptation actions based on empirical observations to preserve the completeness of the mapping set. The latter approach either aims at determining appropriate concepts or at modifying the semantic relationship of mappings instead of deleting invalid mappings. However, the lack of information documenting the evolution of ontologies (see previous section) limits the use of this approach.
An alternative is to rely on the outcome of Diff tools [48, 49] to detect change patterns and use them to adapt the mappings. For example, an approach implemented by Gross et al. [49] utilizes the COnto-Diff tool to identify ontology changes and then applies mapping adaptation handlers to adapt the mappings. The work presented by Dos Reis et al. [13] analyses the impact of the evolution of SCT on mappings. This shows that correlations between changes affecting concepts and modifications in their associated mappings can exist. These correlations are nontrivial because the same change type may or may not affect associated mappings.
Regarding these correlations, deeper analyses were conducted involving two levels of granularity: (1) the whole concept [7, 13] and (2) its attributes [48]. In the former, the authors characterized changes in an ontology concepts’ descriptions that led to changes in mapping. In the latter, the authors relied on two elements: (a) the identification of specific conceptual information (i.e., subset of the concept’s attributes that change) with changes in the mappings; and (b) the characterization of the changes. Lessons learned from the investigation of Dos Reis et al. [50] highlights that even if biomedical controlled terminologies are mapped in their entirety, mappings are usually defined based on partial conceptual information (attributes’ values).
The findings of the analysis of these diThe findings of the analysis of these dimensions resulted in a method to identify relevant attributes used to support mapping maintenance [24]. The authors observed the changes in these relevant attributes and their impact on the mapping evolution. Thus, with the perspective of mapping maintenance in mind, only changes affecting such relevant concept attributes play a role in adapting affected mappings. However, statements regarding relevant attributes are never kept during the alignment process (when creating mappings), and therefore prevents its reuse for maintenance purposes. In other words, the ontological elements that justify a mapping are not documented when concepts are matched. This is why novel techniques are required to analyse established mappings to detect the relevant attributes [24]. Finally, as pointed out by Hussain et al. [51], it is of utmost relevance to clearly document the alignment process outcome and associate them with mappings as metadata [52] to enhance the quality and usability of the available mappings. This can further support the maintenance of mappings.
4 Semantic Annotation Maintenance
In this section, we explore the impact of the evolution of controlled terminologies and mappings on artefacts that rely on modelled knowledge. Semantic annotation is a typical case. Usually, semantic annotations denote termino-ontological elements including concept labels, attributes, comments, descriptions and relationship symbols that are associated with or attached to a document (texts, multimedia, webpages, etc.). They provide additional information (meta-data) about its content meaning, in order to make its semantics explicit for humans and software applications. The goal is to convert syntactic structures into knowledge structures. Figure 3 shows an example of text annotation, where elements from the text (light blue) are linked to concepts (dark blue) from the Gene Regulation Ontology. In this example, we can observe the identification and annotation of events (green) relating to the elements of the text.
Fig. 3.
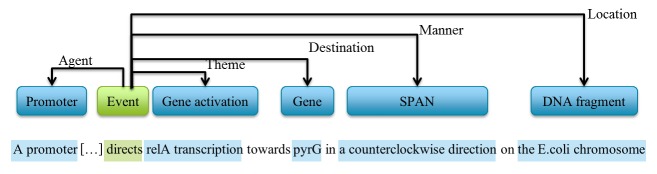
Example from GREC showing a single event annotated based on Gene Regulation Ontology [53].
These annotations facilitate the automatic retrieval and exploitation of relevant information by representing the relationships between real-world entities (Electronic health record - EHRs, genes, publications, etc.) and concepts. However, the dynamic nature of biomedical knowledge leads to frequent revisions of the content of terminologies and, sometimes, to the modification in the content of annotated documents. There is a risk that these changes invalidate existing annotations. To reduce this risk, those responsible for the maintenance of the documents need to address the annotation maintenance problem. Considering the huge quantity of documents produced in the biomedical domain, their maintenance is a labour-intensive task requiring the support of ICT tools. Automatic or semi-automatic annotation maintenance methods have not been deeply studied. For instance, the evolution of annotation in biomedical documents based on biomedical terminologies (e.g., SCT, NCIt, etc.) has rarely been analysed [54, 55].
The most relevant research is focused on the biological domain, in particular on GO annotated documents. Gross et al. [56], analysed the way that GO annotations evolved in the two protein databases Ensembl [57] and Swiss-Prot [58]. The obtained results indicated that a huge number of annotations changed over time, and they may potentially result from the propagation of changes in the underlying ontology, or in the protein descriptions themselves. Similarly, Gillis & Pavlidis [59] and Skunca et al. [60] observed a limited stability of GO-related annotations without explaining why this happens. Gross et al. [61] analysed changes in GO, and statistically evaluated these changes to determine potential influences on the established annotations. Despite these results, what exactly influenced the evolution of annotations or how to repair inconsistencies needs further research. Park et al. [62] analysed existing GO annotations based on the most recent version of GO. These authors identified semantic inconsistencies in the annotations, and they propose a correction method based on the hierarchical structure of the GO graph as well as on the tree structure from the NCBI taxonomy [63].
Outside of the biomedical domain (e.g., XML data management and linked data), the problem of annotation maintenance remains understudied. In the context of XML data management, Köpke & Eder [64] proposed the use of ontology change logs for the detection of out-dated semantic annotations attached to XML schemas. Their approach can identify structural and semantic invalidations, but do not provide mechanisms to correct the annotations.
Maynard et al. [65] considered two different aspects for semantic annotation evolution: (1) propagating meta-data changes to dependent annotations (annotation migration); and (2) analysing changes in the annotations to adapt ontologies. These authors proposed automatic actions only for simple change types, such as the addition or deletion of classes. Volz et al. [66] proposed a protocol to periodically verify whether linked open data sources have changed and if existing links have become invalid. This study only considered addition and deletion of concepts, but not complex changes. The DSNotify framework [67] aims to identify broken links and then notify users to manually correct them, but change detection is limited to additions and deletions of concepts, e.g., to delete broken links between entities. Similarly, Rogozan [68] developed a framework for maintaining semantic annotations while the underlying ontology evolves.
Annotations are classified into non-impacted ones and those that need automatic or manual modifications. Existing approaches rely on a change tracker by logging all performed ontology changes in a specific workbench, which is not feasible for large biomedical terminologies. Furthermore, this requires a significant amount of human intervention for complex changes. In the Semantic Web context, Luong et al. [69, 70] investigated annotation evolution and explored a rule-based approach to detect and correct annotation inconsistencies. This approach converts ontologies to RDF(S) files and detects annotations affected by their evolution, as well as potentially inconsistent annotations using CORESE [71]. Afterwards, inconsistent annotations are detected and corrected. This work focuses on expressive and small-sized ontologies and can hardly be applied to large biomedical ones, because the implemented reasoning techniques require the power of description logics (not always used in biomedical controlled terminologies) to decide on the validity of the annotations.
In summary, existing approaches to annotation evolution mostly handle simple changes (like concept addition and deletion), and only consider small domain ontologies assuming that the annotations are modifiable, which is not always the case, e.g., for EHRs. Therefore, they do not sufficiently address the problem of maintaining semantic annotations in biomedical and clinical use cases
5 Trends and Open Challenges
The efficiency of software applications exploring biomedical data depends on the ability of ICT systems to interpret and transform this data into knowledge. However, systems can quickly be overwhelmed by the ever-increasing amount of data regularly produced. Dealing with large volumes of data raises several challenges: access time, accuracy, completeness, heterogeneity, interpretation, dependency, and data handling methodologies. These challenges become even more complicated with the tendency of using Big Data analyses to support decision makers, pushing towards the redefinition of classical methods for sharing, retrieving, and reasoning over large and heterogeneous data and knowledge sources. It includes a new level of complexity by considering the evolution of existing knowledge-based systems and their maintenance tasks.
To cope with these challenges, we define four complementary trends based on our research observations: (1) Model enrichment, where relevant meta-data is added to explain how/when/why the elements of knowledge exist; (2) Analysis improvement, where novel methods exploit the available data to produce improved knowledge; (3) Knowledge modularization, where existing knowledge is extracted and/or composed to focus on specific end-users’ needs; and (4) Interfaces for model transcription, which transforms information described in formal languages into information described in human friendly languages (like texts, images, etc.).
Several of these challenges can be observed in the development and evolution of standard biomedical terminologies. Indeed, huge sums of money and human effort have been invested in it; the size of the terminologies is continuously increasing as well as the complexity of their representation. This directly impacts the work of thousands of people responsible for generating biomedical data. Typically, the first trend (model enrichment) is observed when designers consider that terminologies are dynamic rather than static objects, and then they enrich the terminology accordingly to include relevant elements for the evolution process, reducing time and maintenance costs. For example, in the definition of ICD-11, designers are considering the work performed by the Semantic Web community about evolution [72]. In fact, the new ICD-11 model will allow a better integration with SCT and will include information that can potentially be used for mapping maintenance tasks [73].
The second trend (analysis improvement) refers to the most advanced one. The focus is on the design and implementation of automatic data analysis tools. This has been developed to cope with missing relevant conceptual information (addressed by trend 1) and the necessity of automatically identifying this information. Rule-based techniques are frequently applied, but the difficulty in harmonizing the rules (defined by experts) or maintaining those rules [74] leads to an increasing use of other techniques. Huge quantities of data favours machine-learning techniques [28]; but the heterogeneity of this data restrains their applicability. New approaches combining these two methods are possible alternatives. For instance, rule-based techniques can be used to select documents on which the learning will be applied.
The third trend (knowledge modularization) consists of implementing modularization techniques for achieving at least one of two objectives: (1) reducing the size of the terminologies, facilitating thus their maintenance and reuse [75], and (2) expressing different points of view about the same knowledge (each point of view can be one module). The former is usually applied to systems that have a limited (low) processing capacity, whereas large controlled terminologies can result in long processing times or create memory overflow issues (e.g., embedded and mobile applications). The latter addresses problems of completeness, dependencies or interpretation of knowledge. For instance, blood can be defined according to a biological, chemical, or clinical point of view. When an overall view is necessary, we need to integrate these “different” meanings. Modularization techniques have been studied to allow this integration or to provide for downsizing. A new potential application of modularization techniques stands for the management of terminology evolution. In this scenario, stable and unstable parts of the ontology can be described in different modules. Hence, the maintenance impact might be limited to specific (unstable) modules.
Finally, the involvement of domain experts in the terminology evolution process creates its own set of issues since domain experts are usually health professionals who are generally unfamiliar with logic-based knowledge representation and technical aspects. We propose the fourth trend to cope with this limitation. For example, this trend requires methods of verbalisation (transformation into natural language text) or illustration (transformation into images) of controlled terminologies [76, 77]. Adequate software applications are needed to further support domain experts in their analyses and to properly interpret their feedback by modifying the out-dated terminologies in an automatic and safe manner.
Despite the limited quantity of published literature regarding the maintenance of mappings and semantic annotations, we strongly suggest that the previously presented trends might provide a means to support and address these problems. We note the additional necessity of defining standard ways of documenting their creation and modification. This limitation has been highlighted in the survey described in section 3 and 4, forcing existing methods to make (sometimes strong) assumptions on the way mappings and annotations have been defined in order to propose analyses and adaptation approaches.
The validation of changes also remains a real problem for Standard Development Organisations. As for ontology evolution, a new generation of intelligent tools is required to improve the interaction between ICT systems and domain experts with the goal of limiting human efforts without compromising the quality of the overall validation and evolution processes.
6 Conclusion
The effective evolution of biomedical terminologies remains a complex problem that involves the identification and characterization of changes, including their impact on existing dependent artefacts (e.g., semantic mappings and annotations), as well as on software applications and other ontology consumers. In this article, we introduced the major problems associated with the management of dynamic, knowledge-based systems. We also provided a systematic literature research on the current state-of-the-art regarding the evolution process of controlled terminologies, including ontologies, as well as mappings and semantic annotations. An analysis of more than 25 distinct approaches covering the three topics discussed here has resulted in the identification of requirements for addressing the management of knowledge evolution, both from an ICT point of view as well as from the perspective of research trends and challenges. From this aspect, we described four trends: Model enrichment (meta-data to improve the explanation of described knowledge); Analysis improvement (combination of machine learning and rule-based techniques); Knowledge modularization (regrouping specific parts of the knowledge); and Interfaces for the transcription of models (verbalisation or illustration of formalisms for end-users).
Acknowledgments
Part of this research has been funded by the Fonds National de la Recherche Luxembourg (FNR) and São Paulo Research Foundation (FAPESP) (Grant #2014/14890-0).
Footnotes
1 http://ihtsdo.org/about-ihtsdo/faq (What would it cost to develop and maintain a new Clinical Terminology based on a global uptake?)
2 http://ihtsdo.org/about-ihtsdo/faq (How can I help to improve SNOMED CT?)
References
- 1.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998;37:394-403. [PMC free article] [PubMed] [Google Scholar]
- 2.Medical Subject Headings. www.nlm.nih.gov/mesh/meshhome.html, Last accessed in 11/11/2014.
- 3.International Classification of Diseases v9. www.cdc.gov/nchs/icd/icd9cm.htm, Last accessed in 11/11/2014.
- 4.Euzenat J, Shvaiko P. Ontology Matching. Heidelberg (DE): Springer-Verlag; 2007. [Google Scholar]
- 5.Baneyx A, Charlet J. Évaluation, évolution et maintenance d’une ontologie en médecine: état des lieux et expérimentation. [Evaluation, evolution and maintenance of a medical ontology: current status and experimentation] Information-Interaction-Intelligence, Special issue; 2006. [Google Scholar]
- 6.Oliver DE, Shahar Y, Shortliffe EH, Musen MA. Representation of change in controlled medical terminologies. Artif Intell Med 1999;15:53-76. [DOI] [PubMed] [Google Scholar]
- 7.Dos Reis JC, Pruski C, Silveira MD, Reynaud-Delaître C. Understanding Semantic Mapping Evolution by Observing Changes in Biomedical Ontologies. J Biomed Inform 2014. February;47:71-82. [DOI] [PubMed] [Google Scholar]
- 8.Mortensen JM, Minty EP, Januszyk M, Sweeney TE, Rector AL, Noy NF, et al. Using the wisdom of the crowds to find critical errors in biomedical ontologies: a study of SNOMED CT. J Am Med Inform Assoc 2015. May;22(3):640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Health Terminology Standards Development Organisation: Systematized Nomenclature of Medicine - Clinical Terms. http://www.ihtsdo.org/snomed-ct, Last accessed in 11/11/2014.
- 10.Noy NF, Shah NH, Whetzel PL, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic acids research. 2009; 37: 1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 2007;25:1251-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosjean J, Merabti T, Dahamna B, Kergourlay I, Thirion B, Soualmia LF, et al. Health multi-terminology portal: a semantic added-value for patient safety. Stud Health Technol Inform 2011;166:129-38. [PubMed] [Google Scholar]
- 13.Dos Reis JC, Pruski C, Da Silveira M, Reynaud-Delaître C. Understanding Semantic Mappings Evolution by Observing Changes in Biomedical Ontologies. J Biomed Inform 2014;47:71-82. [DOI] [PubMed] [Google Scholar]
- 14.Gene Ontology Consortium. http://geneontology.org/, Last accessed in 11/11/2014.
- 15.NCI Thesaurus. http://ncit.nci.nih.gov/, Last accessed in 11/11/2014.
- 16.Hartung M, Gross A, Kirsten T, Rahm E. Discovering Evolving Regions in Life Science Ontologies. 7th Int Conference on Data Integration in the Life Sciences (DILS). Springer; 2010. p. 19-34. [Google Scholar]
- 17.Plessers P, De Troyer O. Ontology Change Detection Using a Version Log. Proceedings of the 4th International Semantic Web Conference. Galway, Ireland; 2005. p. 578-92. [Google Scholar]
- 18.Noy NF, Musen MA. Promptdiff: a fixed-point algorithm for comparing ontology versions. Eighteenth national conference on Artificial intelligence. Edmonton, Alberta, Canada: American Association for Artificial Intelligence; 2002. p. 744-50. [Google Scholar]
- 19.Hartung M, Gross A, Rahm E. COnto-Diff: Generation of Complex Evolution Mappings for Life Science Ontologies. J Biomed Inform 2013;46:15-32. [DOI] [PubMed] [Google Scholar]
- 20.Hartung M, Kirsten T, Gross A, Rahm E. OnEX: Exploring changes in life science ontologies. BMC Bioinformatics 2009;10:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaban-Nejad A, Haarslev V. Bio-medical Ontologies Maintenance and Change Management. Amandeep S. Sidhu and Dillon TS, (eds.). Biomedical Data and Applications. Springer; 2009. p. 143-68. [Google Scholar]
- 22.Djedidi R, Aufaure M-A. OWL Change Management Patterns. The 5th Workshop on Semantic Web Applications and Perspectives (SWAP2008). Rome, Italy; 2008. p. 1-3. [Google Scholar]
- 23.Dragisic Z, Lambrix P, Wei-Kleiner F. Completing the is-a structure of biomedical ontologies. Data Integration in the Life Sciences. Lisbon, Portugal: Springer; 2014. p. 66-80. [Google Scholar]
- 24.Dinh D, Dos Reis JC, Pruski C, Da Silveira M, Reynaud-Delaître C. Identifying relevant concept attributes to support mapping maintenance under ontology evolution. Web Semantics: Science, Services and Agents on the World Wide Web 2014. 29:53-66. [Google Scholar]
- 25.Ogren PV, Cohen KB, Acquaah-Mensah GK, Eberlein J, Hunter L. The compositional structure of Gene Ontology terms. Pacific Symposium on Biocomputing; 2004. p. 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abgaz YM, Javed M, Pahl C. Analyzing Impacts of Change Operations in Evolving Ontologies. 2nd Joint Workshop on Knowledge Evolution and Ontology Dynamics EvoDyn; 2012. [Google Scholar]
- 27.Shaban-Nejad A, Haarslev V. Incremental biomedical ontology change management through learning agents. Agent and Multi-Agent Systems: Technologies and Applications. Springer; 2008. p. 526-35. [Google Scholar]
- 28.Pesquita C, Couto FM. Predicting the extension of biomedical ontologies. PLoS Comput Biol 2012;8(9):e1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sari AK, Rahayu W, Bhatt M. An approach for sub-ontology evolution in a distributed health care enterprise. Information Systems 2013;38:727-44. [Google Scholar]
- 30.Stojanovic L, Maedche A, Motik B, Stojanovic N. User-driven ontology evolution management. In European Conf Knowledge Eng and Management (EKAW 2002). Springer-Verlag; 2002. p. 285-300. [Google Scholar]
- 31.Darmoni SJ, Leroy JP, Baudic F, Douyère M, Piot J, Thirion B. CISMeF: a structured health resource guide. Methods Inf Med 2000;39:30-5. [PubMed] [Google Scholar]
- 32.Salvadores M, Alexander PR, Musen MA, Noy NF. Bioportal as a dataset of linked biomedical ontologies and terminologies in RDF. Semantic Web 2013;4:277-84. [PMC free article] [PubMed] [Google Scholar]
- 33.Dos Reis JC, Pruski C, Reynaud-Delaître C. State-of-the-art on mapping maintenance and challenges towards a fully automatic approach. Expert Systems with Applications 2015;42:1465-78. [Google Scholar]
- 34.Gal A, Anaby-tavor A, Trombetta A, Montesi D. A framework for modeling and evaluating automatic semantic reconciliation. The VLDB Journal 2005;14:50-67. [Google Scholar]
- 35.Zurawski M, Smaill A, Robertson D. Bounded Ontological Consistency for Scalable Dynamic Knowledge Infrastructures. Domingue J, Anutariya C, editors. ASWC. LNCS ed.: Springer; 2008. p. 212-26. [Google Scholar]
- 36.Mccann R, Alshebli B, Le Q, Nguyen H, Vu L, Doan A. Mapping Maintenance for Data Integration Systems. VLDB ‘05 Proceedings of the 31st International Conference on very large data bases; 2005. p. 1018-29. [Google Scholar]
- 37.Colazzo D, Sartiani C. Detection of corrupted schema mappings in XML data integration systems. ACM Transactions on Internet Technology 2009;9:1-53. [Google Scholar]
- 38.Abdul-Rahman Mawlood-Yunis. Fault-tolerant Semantic Mappings Among Heterogeneous and Distributed Local Ontologies. Proceedings of the 2nd international workshop on Ontologies and information systems for the semantic web; 2008. p. 31-8. [Google Scholar]
- 39.Li A, Miao J, Jia Y. Research on Broken Mappings Detecting Method Based on Fuzzy Aggregation Operators in Deep Web Integration Environment. 2010 International Conference on E-Business and E-Government. IEEE; 2010. p. 125-8. [Google Scholar]
- 40.Meilicke C, Stuckenschmidt H, Tamilin A. Reasoning Support for Mapping Revision. Journal of Logic and Computation 2008;19:807-29. [Google Scholar]
- 41.Nikitina N, Rudolph S, Glimm B. Reasoning-supported interactive revision of knowledge bases. The twenty-second international joint conference on artificial intelligence, IJCAI’11. AAAI Press; 2011. p. 1027-32. [Google Scholar]
- 42.Jimenez-Ruiz E, Grau BC. Logmap: Logic-based and scalable ontology matching. The Semantic Web--ISWC 2011. Spinger; 2011. p. 273-88. [Google Scholar]
- 43.Fagin R, Kolaitis PG, Popa L, Tan W-C. Schema Mapping Evolution Through Composition and Inversion. Data-Centric Systems and Applications. Springer-Verlag; 2011. p. 191-222. [Google Scholar]
- 44.An Y, Hu X, Song I-Y. Maintaining Mappings between Conceptual Models and Relational Schemas. Journal of Database Management 2010;21:36-68. [Google Scholar]
- 45.Tang F, Tang R. Minimizing Influence of Ontology Evolution In Ontology-based Data Access System. IEEE International Conference on Progress in Informatics and Computing (PIC); 2010. p. 10-4. [Google Scholar]
- 46.Maedche A, Motik B, Silva N, Volz R. MAFRA - A MApping FRAmework for Distributed Ontologies in the Semantic Web. Knowledge engineering and knowledge management: ontologies and the semantic web. Springer; 2002. p. 235-50. [Google Scholar]
- 47.Martins H, Silva N. A User-Driven and a Semantic-Based Ontology Mapping Evolution Approach. 11th International Conference on Enterprise Information Systems. Milan; 2009. p. 6-10. [Google Scholar]
- 48.Dos Reis JC, Dinh D, Pruski C, Da Silveira M, Reynaud-Delaître C. Mapping Adaptation Actions for the Automatic Reconciliation of Dynamic Ontologies. ACM International Conference on Information and Knowledge Management (CIKM 2013). San Francisco: ACM; 2013. p. 599-608. [Google Scholar]
- 49.Gross A, Dos Reis JC, Hartung M, Pruski C, Rahm E. Semi-Automatic Adaptation of Mappings between Life Science Ontologies. Data Integration in the Life Sciences (DILS 2013). Montreal, Canada: Springer-Verlag; 2013. p. 90-104. [Google Scholar]
- 50.Dos Reis JC, Pruski C, Da Silveira M, Reynaud-Delaître C. Characterizing Semantic Mappings Adaptation via Biomedical KOS Evolution: A Case Study Investigating SNOMED CT and ICD. AMIA 2013 Annual Symposium. Washington DC (USA); 2013. p. 333-42. [PMC free article] [PubMed] [Google Scholar]
- 51.Hussain S, Sun H, Sinaci AA, Erturkmen GB, Mead C, Gray AJ, et al. A Framework for Evaluating and Utilizing Medical Terminology Mappings. e-Health - For Continuity of Care - Proceedings of MIE2014, the 25th European Medical Informatics Conference. Istanbul, Turkey: IOS Press; 2014. p. 594-8. [PubMed] [Google Scholar]
- 52.Dos Reis JC, Da Silveira M, Dinh D, Pruski C, Reynaud-Delaitre C. Requirements for Implementing Mapping Adaptation Systems. WETICE Conference (WETICE), 2014 IEEE 23rd International; 2014. p. 405-10. [Google Scholar]
- 53.Ananiadou S, Pyysalo S, Tsujii JI, Kell DB. Event extraction for systems biology by text mining the literature. Trends Biotechnol 2010;28:381-90. [DOI] [PubMed] [Google Scholar]
- 54.Roberts A, Gaizauskas R, Hepple M, Demetriou G, Guo Y, Roberts I, et al. A semantically annotated corpus of clinical texts. Biomed Inform 2009;42:950-66. [DOI] [PubMed] [Google Scholar]
- 55.Mowery D, Jordan P, Wiebe J, Harkema H, Dowling J, Chapman WW. Semantic Annotation of Clinical Events for Generating a Problem List. AMIA Annu Symp Proc 2013. p. 1032-41. [PMC free article] [PubMed] [Google Scholar]
- 56.Gross A, Hartung M, Kirsten T, Rahm E. Estimating the Quality of Ontology-based Annotations by Considering Evolutionary Changes. 6th Data Integration in the Life Sciences (DILS); 2009. p. 71-87. [Google Scholar]
- 57.Ensembl genome databases. http://www.ensembl.org/index.html, Last accessed in 11/11/2014.
- 58.UniProtKB / Swiss-Prot Database. http://www.uniprot.org/uniprot/?query=*&fil=, Last accessed in 11/11/2014.
- 59.Gillis J, Pavlidis P. Assessing identity, redundancy and confounds in gene ontology annotations over time. Bioinformatics 2013;29:476-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skunca N, Altenhoff A, Dessimoz C. Quality of computationally inferred gene ontology annotations. PLoS Computl Biol 2012;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gross A, Hartung M, Prüfer K, Kelso J, Rahm E. Impact of Ontology Evolution on Functional Analyses. Bioinformatics 2012;28:2671-7. [DOI] [PubMed] [Google Scholar]
- 62.Park YR, Kim J, Lee HW, Yoon YJ, Kim JH. GOChase-II: correcting semantic inconsistencies from Gene Ontology-based annotations for gene products. BMC Bioinformatics 2011;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.National Center for Biotechnology Information Taxonomy. http://www.ncbi.nlm.nih.gov/taxonomy, Last accessed in 11/11/2014.
- 64.Köpke J, Eder J. Semantic Invalidation of Annotations Due to Ontology Evolution. OTM Conferences. Hersonissos, Crete, Greece: Springer; 2011. p. 763-80. [Google Scholar]
- 65.Maynard D, Peters W, d’Aquin M, Sabou M. Change Management for Metadata Evolution. Proceedings of the International Workshop on Ontology Dynamics (IWOD-07); 2007. p. 27--40. [Google Scholar]
- 66.Volz J, Bizer C, Gaedke M, Kobilarov G. Discovering and maintaining links on the web of data. International Semantic Web Conference (ISWC). Washington, DC. Berlin, Heidelberg: Springer; 2009. p. 650-65. [Google Scholar]
- 67.Popitsch N, Haslhofer B. DSNotify–A solution for event detection and link maintenance in dynamic datasets. Web Semantics: Science, Services and Agents on the World Wide Web 2011;9:266-83. [Google Scholar]
- 68.Rogozan D. Gestion de l’évolution d’une ontologie : méthodes et outils pour un référencement sémantique évolutif fondé sur une analyse des changements entre versions d’une ontologie. [Managing ontology evolution: methods and tools for an evolving semantic referring based on an analysis of ontology version changes] PhD Thesis. Rouen: INSA; 2008. [Google Scholar]
- 69.Luong P-H, Dieng-Kuntz R. A Rule-based Approach for Semantic Annotation Evolution. Computational Intelligence 2007;23:320-38. [Google Scholar]
- 70.Luong P-h, Dieng-Kuntz R, Boucher A. Managing semantic annotations evolution in the CosWEM system. Proc 3rd National Symp on Research, Development and Application of Information and Communication Technology,. 2006, p. 215-23. [Google Scholar]
- 71.Corby O, Dieng-Kuntz R, Faron-Zucker C. Querying the Semantic Web with the CORESE Search Engine. 16th European Conference on Artificial Intelligence (ECAI’2004). Valencia, Spain: IOS Press, 2004, p. 705-9. [Google Scholar]
- 72.Tudorache T, Nyulas CI, Noy NF, Musen MA. Using Semantic Web in ICD-11: Three Years Down the Road. The Semantic Web--ISWC 2013; 2013. p. 195-211. [Google Scholar]
- 73.Rodrigues JM, Schulz S, Rector A, Spackman K, Üstün B, Chute CG, et al. Do we need a Common Ontology between ICD 11 and SNOMED CT to ensure Seamless Re-Use and Semantic Interoperability? Stud Health Technol Inform 2013;192:343-6. [PubMed] [Google Scholar]
- 74.Musen MA, Middleton B, Greenes RA. Clinical decision-support systems. Biomedical informatics. Springer; 2014. p. 643-74. [Google Scholar]
- 75.Burgun A. Desiderata for domain reference ontologies in biomedicine. J Biomed Inform 2006;39:307-13. [DOI] [PubMed] [Google Scholar]
- 76.Ben Abacha A, Da Silveira M, Pruski C. Medical Ontology Validation through Question Answering. Artificial Intelligence in Medicine. Murcia, Spain: Springer; 2013. p. 196-205. [Google Scholar]
- 77.Liang S, Stevens R, Scott D, Rector A. Automatic Verbalisation of SNOMED Classes Using OntoVerbal. Peleg M, Lavra N, Combi C, editors. Artificial Intelligence in Medicine. Berlin, Heidelberg: Springer; 2011. p. 338-42. [Google Scholar]