Abstract
Background
Zebrafish express five cytochrome P450 1 genes: CYP1A, CYP1B1, CYP1C1, CYP1C2, inducible by aryl hydrocarbon receptor agonists, and CYP1D1, a constitutively expressed CYP1A-like gene. We examined substrate selectivity of CYP1s expressed in yeast.
Methods
CYP1s were expressed in W(R) yeast, engineered to over-express P450 reductase, via pYES/DEST52 and via pYeDP60. Microsomal fractions from transformed yeast were examined for activity with fluorogenic substrates, benzo[a]pyrene and testosterone. Modeling and docking approaches were used to further evaluate sites of oxidation on benzo[a]pyrene and testosterone.
Results
CYP1s expressed in yeast dealkylated ethoxy-, methoxy-, pentoxy- and benzoxy-resorufin (EROD, MROD, PROD, BROD). CYP1A and CYP1C2 had the highest rates of EROD activity, while PROD and BROD activities were low for all five CYP1s. The relative rates of resorufin dealkylation by CYP1C1, CYP1C2 and CYP1D1 expressed via pYeDP60 were highly similar to relative rates obtained with pYES/DEST52-expressed enzymes. CYP1C1 and CYP1C2 dealkylated substituted coumarins and ethoxy-fluorescein-ethylester, while CYP1D1 did not. The CYP1Cs and CYP1D1 co-expressed with epoxide hydrolase oxidized BaP with different rates and product profiles, and all three produced BaP-7,8,9,10-tetrol. The CYP1Cs but not CYP1D1 metabolized testosterone to 6β-OH-testosterone. However, CYP1D1 formed an unidentified testosterone metabolite better than the CYP1Cs. Testosterone and BaP docked to CYP homology models with poses consistent with differing product profiles.
Conclusions
Yeast-expressed zebrafish CYP1s will be useful in determining further functionality with endogenous and xenobiotic compounds.
General significance
Determining the roles of zebrafish CYP1s in physiology and toxicology depends on knowing the substrate selectivity of these enzymes.
1. Introduction
Enzymes in the vertebrate cytochrome P450 1 (CYP1) family metabolize many xenobiotics, including polycyclic aromatic hydrocarbons (PAHs), natural products, and drugs. These metabolic activities have implications for environmental chemical effects in humans and wildlife, including carcinogenesis and other health outcomes [1–4], and for drug therapy in humans [5–8]. CYP1 enzymes also metabolize endogenous regulatory molecules, including steroids and arachidonic acid [2,9–13]. Mammals express three CYP1 genes in two subfamilies, CYP1A (genes CYP1A1 and CYP1A2) and CYP1B (gene CYP1B1), all three of which are inducible by aryl hydrocarbon receptor (AHR) agonists [14]. Other vertebrates (fish, amphibians, birds) have four CYP1 subfamilies, CYP1A and CYP1B, as well as the more recently discovered CYP1C and CYP1D [15–20]. Teleost fish such as zebrafish typically have five CYP1 genes, CYP1A, CYP1B1, CYP1C1, CYP1C2 and CYP1D1 [16,17]. Functional properties of the mammalian CYP1s are rather well known [14], but catalytic functions of the non-mammalian CYP1s by comparison remain poorly characterized.
The zebrafish is a major vertebrate model, yielding insights into mechanisms in normal developmental processes as well as in environmental toxicology and chemically induced diseases [21]. Zebrafish increasingly are used also in drug discovery and toxicity screening [22,23]. Knowledge of the regulation and catalytic functions of the full suite of CYP1s (and indeed all CYPs) in zebrafish is important to strengthen inference from toxicological and pharmacological studies with this model. In teleost fish, four of the five CYP1 genes, i.e., CYP1A, CYP1B1, CYP1C1 and CYP1C2, are inducible by AHR agonists such as 3,3′,4,4′,5-pentachlorobiphenyl (PCB126) and benzo[a]pyrene (BaP) [24,25], although with differing degrees of responsiveness that change over development [24]. Notably, CYP1C2 transcript inducibility diminishes substantially after hatching, and it effectively is not inducible in adult liver [24,26]. In contrast to the other CYP1 genes, zebrafish CYP1D1 has not been found to be inducible by AhR agonists, in adult or developmental stages [17]. CYP1D1 has a gene structure identical to that of CYP1A, but with a single dioxin response element in the 10 kb promoter region, and it appears to be constitutively regulated. There also are pronounced tissue and developmental differences in basal expression of the five CYP1s. For example, in adult zebrafish brain, CYP1D1 is relatively more highly expressed than the other CYP1 genes [17].
Differences in tissue distribution, basal expression levels, and inducibility by AhR agonists suggest that the five zebrafish CYP1s play different roles in vivo, which could as well involve differences in substrate selectivity and catalytic efficiency of these proteins. Enzyme functions of the CYP1As have been studied in several teleost species, including zebrafish (e.g., [27]), yet the substrate selectivity of the multiple CYP1s that occur in teleost species, including zebrafish, still are poorly known. Here we report on catalytic functions of recombinant zebrafish CYP1 enzymes expressed in yeast, with a suite of fluorogenic substrates as well as with a model PAH (BaP) and a model steroid (testosterone). The catalytic functions of the lesser-known CYP1s in fish, the CYP1Cs and CYP1D1, in particular need attention, and we emphasize these enzymes. Activities of the zebrafish CYP1s expressed in Escherichia coli have been examined with a variety of substrates like those we use here, and a different steroid substrate, 17β-estradiol [28,29]. The results provide a foundation for further studies of CYP1s in the zebrafish model.
2. Materials and methods
2.1. Chemicals
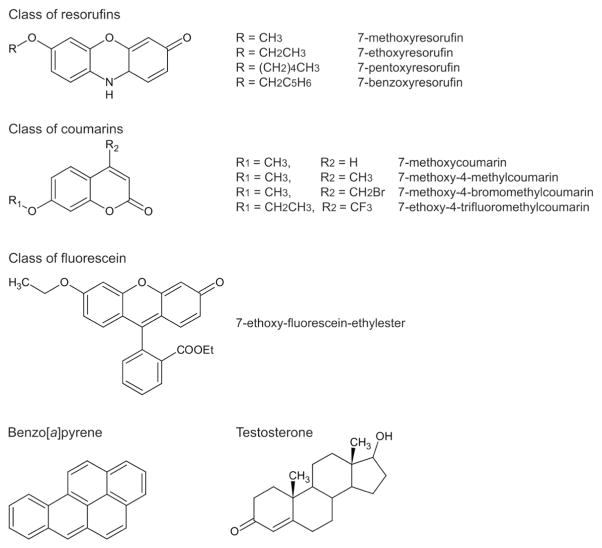
NADPH, benzo[a]pyrene and testosterone were purchased from Sigma-Aldrich (St. Louis, MO). The substituted resorufin substrates, 7-ethoxyresorufin, 7-methoxyresorufin, and 7-pentoxyresorufin and 7-benzyloxyresorufin were from Sigma-Aldrich or from Molecular Probes (Eugene, OR). The substrates 7-ethoxyfluorescein-ethylester (EFEE); 7-methoxy-coumarin; 7-methoxy-4-methyl-coumarin; 7-methoxy-4-bromomethyl-coumarin; 7-methoxy-4-trifluoromethyl-coumarin; and 7-ethoxy-4-trifluoromethyl-coumarin all were from Molecular Probes. Substrate structures are shown in Fig. 1. Other chemicals were from Fisher Scientific or Sigma-Aldrich.
Fig. 1.
Structures of substrates tested for activity with zebrafish CYP1s.
2.2. Heterologous expression
The open reading frames of zebrafish CYP1s were amplified by PCR (using primers shown in Supplemental Table 1), gel-purified using Nucleospin columns, and ligated into an entry vector, either pENTR/D/TOPO® (Invitrogen) or PCRScript (Stratagene), according to instructions.
CYP1s were expressed in the Saccharomyces cerevisiae W(R) strain (MATa; ade2-1; his3-11,15; leu2-3, 112; trp1-1; ura3-1; canR; cyr+), engineered to over-express the yeast microsomal NADPH-cytochrome P450 reductase (POR) [30]. In W(R) yeast transformed with CYP genes, POR activity has been found consistently to be at about 2200 ± 200 nmol cyt c reduced/min/mg of microsomal protein with cells cultivated in a synthetic medium, and 3200 ± 200 nmol cyt c reduced/min/mg of microsomal protein from cells cultivated in a rich medium. In the current work, W(R) cells were cultivated in a rich medium.
Two approaches were taken using different expression vectors. In the first procedure, CYP1s were transferred from the entry vector pENTR/D/TOPO to the destination vector pYES/DEST52/V5-His using the TOPO Cloning Kit (Invitrogen), as per the kit protocol. A pYES/DEST52 plasmid carrying the Arabidopsis β-glucuronidase (gus) gene was used as a positive control for transformation and a negative control for CYP heterologous expression. In the second procedure, CYP1s were excised from the PCRScript entry vector and then ligated into the yeast expression vector pYeDP60 by standard ligation protocols, and an empty vector was used to prepare control yeast. Plasmids for yeast expression were purified from E. coli cultures grown overnight, and were used to transform competent yeast. Yeast cells were made competent using the S.c. EasyComp™ transformation kit according to the manufacturer’s instructions (Invitrogen), or by using the standard lithium acetate procedure [31].
Single pYES/DEST52-transformed yeast colonies were picked from selective plates and used to inoculate 30 mL of SGAI medium [32]. Cultures in SGAI were grown overnight at 28 °C with shaking at 130 rpm, then 10 mL was transferred to a 2-liter flask containing 500 mL of YPGE media [32]. YPGE cultures were grown overnight as above. Yeast cells were induced by the addition of 2% galactose and incubated for various periods of time up to 18 h at 28°–30 °C with shaking at 130 or 140 rpm. Cells were harvested by centrifugation, washed with TES50 buffer (50 mM Tris–HCl, 1 mM EDTA, 0.6 M sorbitol; pH 7.4) and cell pellets were used in whole cell assays or frozen in liquid nitrogen (LN2) and stored at −80 °C for preparation of microsomal fractions used in assay of catalytic activities.
Alternatively, pYEDP60 yeast colonies picked from selective plates were used to inoculate 50 mL of SGAI medium, the cultures were grown overnight at 28 °C with shaking at 160 rpm, then transferred to a 1-liter flask containing 250 mL of YPGE media. The YPGE culture was grown for 48 h at 28 °C with shaking at 140 rpm. Yeast cells were then induced by the addition of 2% galactose for 12 h at 28 °C with shaking at 140 rpm. Cells were harvested by centrifugation, washed with TES buffer (50 mM Tris–HCl, 1 mM EDTA, 0.6 M sorbitol; pH 7.4) and immediately processed for microsome preparation.
2.3. Microsomal fraction preparation
Yeast cells from the first procedure (pYES/DEST52) were suspended in a vacuum degassed TES 50 buffer containing 1.0 mM dithiothreitol (DTT) and protease inhibitor cocktail (1×) (Sigma) and mechanically disrupted using the BeadBeater (BioSpec Products, Inc., Bartlesville, OK). A 25 mL chamber containing glass beads (0.5 mm diameter, amounting to 1/2 of the total volume) and the yeast cell slurry was cooled with an ice water/methanol (20%) cooling jacket. Air remaining in the chamber was displaced by adding a degassed TES 50 buffer. The cells were disrupted by twenty 5-s BeadBeater cycles with 45-s resting (cooling) periods between cycles. This method of disruption has been shown to be effective at maintaining the sample temperature below 8 °C during disruption and results in greater than 90% lysis of yeast cells.
All subsequent steps were carried out at 4 °C. Cell lysate was decanted and the beads remaining in the chamber were washed once with 2 mL of a degassed buffer, which was then added to the cell lysate. The cell lysate was then centrifuged with conditions used before [33] to collect the microsomal fraction. The resulting microsomal pellet was resuspended in 1 to 2 mL of TEDG buffer (50 mM Tris–HCl, 1 mM EDTA, 20% glycerol by volume, 1 mM DTT, pH 7.4) by gentle hand homogenization using a Potter-Elvehjem homogenizer and microsomal suspensions were stored in LN2. An aliquot of suspension was taken for microsomal protein determination using the BCA protein assay (Pierce) with BSA as the standard [34].
Yeast cells from the second procedure (with the pYeDP60 vector) were suspended in a minimal volume of TES buffer (usually 1.5–2.0 mL) and hand-broken using glass microbeads (0.4–0.5-mm diameter) at 4 °C. All subsequent steps were carried out at 4 °C. Microsomal fractions were prepared from disrupted yeast cells using a glycerol–NaCl precipitation procedure as previously described [32]. The resulting microsomal pellet was resuspended in 1 mL of TEG buffer (50 mM Tris–HCl, 1 mM EDTA, 20% glycerol by volume, pH 7.4) as above. An aliquot of the suspension was taken for microsomal protein determination as above, and for CO-reduced spectral measurement of total P450.
Microsomal proteins from pYES/DEST52 yeast cells expressing the CYP1s were resolved electrophoretically and blots probed with antibody to the V5 epitope. As a surrogate for the amounts of P450 expressed, the amounts of V5 tag were determined by ImageJ analysis of the stain density of bands in the immunoblot. In addition, microsomes from pYES/DEST52 yeast expressing CYP1A were stained with antibodies to CYP1A.
2.4. Assay of catalytic activities
Activities with substituted resorufin substrates (ethoxy-, methoxy-, pentoxy- and benzyloxyresorufin) were measured in yeast microsomal fractions, and also in intact yeast cells, with endogenous NADPH. For assay of intact cells, freshly prepared yeast cell pellets were diluted to 200 mg/mL in assay buffer (50 mM Tris, 0.1 M NaCl, pH 7.8). Control samples were boiled yeast and yeast expressing Arabidopsis β-glucuronidase (gus). Reactions were initiated by mixing 1 volume of yeast suspension with 1 volume of 8 μM resorufin substrate in assay buffer (4 μM final concentration of substrate), and incubated at 30 °C. Duplicate 150 μL aliquots were taken at 0, 5, 10, 15 and 30 min after mixing, and placed immediately on ice to stop the reaction. Aliquots of reaction mixtures were centrifuged to remove yeast cells and 100 μL of supernatant fluid was transferred to a 96 well plate and fluorescence was measured and compared with resorufin standards, using a fluorescence plate reader as described previously [35,36]. Activities in pmol min−1 mg−1 whole yeast protein were determined from the linear portion of the resorufin production time course.
Activities of microsomal fractions prepared from transformed yeast cultures with the substituted resorufin substrates were assessed kinetically using a fluorescence plate reader as above, at 30 °C, in a buffer of 50 mM Tris, 0.1 M NaCl, pH 7.8. Activities obtained with CYPs in yeast microsomal preparations were determined from the linear portion of the resorufin production time course, in pmol min−1 mg−1. Activity with substituted resorufins also was measured with a spectrofluorometer, using an excitation wavelength of 530 nm and an emission wavelength of 586 nm. NADPH concentrations were 120 μM and substrate was saturating. Stock solutions were prepared in methanol, and the final concentration of methanol in incubation mixtures was 0.2–0.3%.
Activities with the substrates 7-methoxy-, 7-methoxy-4-methyl-, and 7-methoxy-4-bromomethyl-coumarin were measured fluorometrically using excitation and emission wavelengths at 380 nm and 460 nm, respectively, and with 7-methoxy- and 7-ethoxy-4-trifluoromethyl-coumarin activities were determined with excitation and emission wavelengths set at 385 nm and 502 nm, respectively. For these assays, reactions were in 1 mL of 50 mM Tris–HCl buffer (pH 7.4) containing 1 mM EDTA, substrate at 2 μM (saturating), and 100 μg of yeast microsomal protein; reactions were initiated by adding NADPH to achieve 150 μM. Stock solutions of coumarins were prepared in methanol, except for 7-methoxy-4-bromomethyl-coumarin which was prepared in N,N-dimethylformamide (DMF). The final concentration of MeOH or DMF in incubation mixtures was 0.2%.
Ethoxyfluorescein ethyl ester O-deethylase activities were measured fluorometrically using excitation and emission wavelengths set at 479 nm and 560 nm, respectively. Reactions were carried out in 1 mL of 50 mM Tris–HCl buffer (pH 7.4) containing 1 mM EDTA, NADPH 120 μM, EFEE 1 μM (saturating), and 150 μg of yeast microsomal proteins. The stock solution of EFEE was prepared in methanol, with a final concentration of 0.1% in incubation mixtures.
Saturation of the different fluorogenic substrates was determined by the usual kinetic method that consists of two parallel incubations, one with the presumed saturating concentration of the substrate, the other with half of this concentration. The calculated reaction rates for the two incubations are compared, and if they are of similar value within experimental error, the concentration is said to be saturating.
Rates of benzo[a]pyrene oxidation were determined using a radiometric assay with 14C-BaP, modified from a method described previously [37]. Replicate 100 μL reaction mixtures contained CYP1C1 or CYP1C2 yeast microsomal protein (1–2 mg/mL final concentration), uniformly labeled 14C-BaP 70 μM final concentration in 50 mM Tris buffer, pH 7.4. Reactions were initiated with addition of NADPH (0.25 mM, final concentration), and incubated at 28 °C. Reactions were stopped at various intervals with 0.15 M KOH in DMSO on ice, extracted with hexane, and aliquots of the aqueous phase added to scintillation vials prefilled with 3 mL Scintaverse + 10 μL 0.6 N HCl, and counted with a Beckman LS 6500 scintillation counter. Blank reactions without NADPH were included as controls.
Reaction mixtures for generation of metabolites of benzo[a]pyrene and testosterone contained 50 mM Tris–HCl buffer, 1 mM EDTA, pH 7.4, recombinant zebrafish CYP in 250 or 300 mg of total yeast microsomal protein, yeast-expressed human microsomal epoxide hydrolase in 200 mg total microsomal protein, 0.2 mM NADPH, in a final volume of 0.33 mL. Incubations were at 28 °C. Benzo[a]pyrene was added at a final concentration of 12 μM, from a stock solution in DMF; the final concentration in DMF of the incubation mixtures was 1%. For testosterone metabolism assays, epoxide hydrolase was omitted and the substrate concentration was 120 μM from a stock solution in methanol. The final concentration in methanol of the incubation mixtures was 0.1%. Reactions were stopped at different intervals by adding 5 μL trifluoracetic acid in water (1:1 vol.) and 1 volume of acetonitrile, and the mixture was centrifuged at 10,000 rpm for 10 min to remove membranes and precipitated proteins. An aliquot of the supernatant (10–30 μL) was analyzed by HPLC separation, as below.
2.5. Reverse phase high-throughput HPLC analyses
The BaP and testosterone metabolites were separated and analyzed on an Alliance HT2795 HPLC Waters module connected to both a Waters 2475 fluorescence detector and a Micromass ZQ mass spectrometer with a Spheri-5 RP18 5 μm Brownlee column (4.6 × 100-mm). The column was heated at 40 °C for a better peak resolution. Metabolites were separated using a gradient starting at 90:10 (water:acetonitrile) followed by a linear increase to 0:100 over 10 min and, then, by a plateau at 100% CH3CN for 2 min, and return to initial conditions and hold for 2 min; 14 min of total run length. For identification of metabo-lites from BaP, the fluorescence detection was set at 278 nm for excitation and 407 nm for emission, and chromatographic quantitation was standardized with benzo[ghi]perylene as an internal standard. Testosterone metabolites were identified and quantified by mass spectrometry with electrospray positive ionization. Parameters of ioni-zation were as follows: capillary voltage 3.4 kV, cone voltage 20.0 V, desolvation gas flow 550 L·h−1, desolvation temperature 350 °C, and source temperature 120 °C. Continuous metabolite mass detection was using both full scan spectra by scanning mass range 200–500 amu and a SIR channel (single ion response) set at the precise m/z corresponding to the protonated mass of hydroxytestosterone: [M+H]+ = 305.4. The detected m/z corresponds to M+H since positive electrospray mode is used. The amounts of metabolite were quantified by measuring peak areas from the SIR data. Initial velocities were determined by plotting the peak area vs. time for each metabolite after 5, 10, 15, and 20 min of incubation.
2.6. Homology modeling and docking
Homology models of zebrafish CYP1C1, CYP1C2 and CYP1D1 were constructed using Modeller 9v8 [38], based on the crystal structures of human CYP1A2 [39] and CYP1B1 [40]. The N-terminal membrane anchor regions of the fish CYP1A sequences were truncated to match the sequence of the human structure. Initial CLUSTALX alignments were refined using the salign_2d function of Modeller. Default parameters in Modeller were applied, excluding water molecules and any ions that were part of any of the templates with the exception of the heme and heme iron. Homology modeling was carried out by satisfaction of spatial restraints using the automodel function of Modeller, with very thorough variable target function method (VTFM), thorough molecular dynamics (MD), and two repeat cycles of minimization. One hundred randomly seeded models were generated for each protein. Side chain positions were optimized in a solvated model by molecular dynamics minimization and equilibration using NAMD (NAnoscale Molecular Dynamics freeware) (v2.8) [41]. The P450 heme-cysteine bonds were explicitly patched, and models were solvated in a cube of water to allow periodic boundary condition, charge-neutralized, and minimized for 100 steps prior to relaxation for 2500 steps (5 ps). Models and crystal structures were prepared for docking using AutoDockTools (v1.5.4) [42], with the addition of polar hydrogens and the assignment of Gasteiger charges. Ligand structure and charge minimization was performed with semiempirical methods (PM6 Hamiltonian in Mopac2009 [43,44]). Gasteiger–Marsili partial charges were used in the final docking runs. High throughput docking was performed using Autodock Vina (v1.1.1) [45]. Flexible ligands were docked into models with rigid protein backbones and rigid side chains, performing 100 replicate dockings and retaining a broad range of calculated energies (6 kcal/mol).
3. Results
3.1. Activity of five zebrafish CYP1s expressed via pYES/DEST52
Initially, we examined expression of the five CYP1s in W(R) yeast via the pYES/DEST52/V5-His vector. Immunoblot assay for the V5 tag in microsomal fractions of yeast induced with galactose showed V5 tag expression in each case, confirming successful expression of the CYPs (Fig. 2). Examining the time-course of V5 tag levels, we determined that maximal expression occurred between 12–18 h after addition of galactose (not shown). Immunoblotting of microsomal fractions of CYP1A-transformed yeast with monoclonal antibody 1-12-3 that recognizes zebrafish CYP1A confirmed the yeast expression of CYP1A (Supplemental Fig. 1). Prior immunoblotting [17] had confirmed the expression of CYP1D1.
Fig. 2.
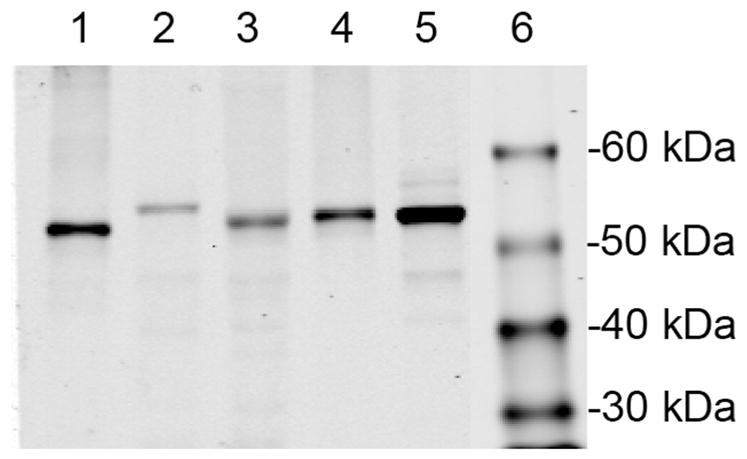
Zebrafish CYP1s expressed via pYES/DEST52 in yeast. Immunodetection with antibodies to the V5-tag. Each lane had 5 μg of microsomal protein loaded: lane 1, CYP1A; lane 2, CYP1B1; lane 3, CYP1C1; lane 4, CYP1C2; lane 5, CYP1D1; and lane 6, mol wt. standards.
Although immunoblot assays for V5 tag or specific CYPs indicated expression of the target proteins, we were unable to detect P450 spectrophotometrically in microsomal fractions of yeast transformed with CYP1s via pYES/DEST52/V5-His. However, screening for enzyme activity in intact cells showed that both EROD and MROD could be detected in yeast transformed with CYP1s, but not in yeast transformed with Arabidopsis β-glucuronidase (not shown).
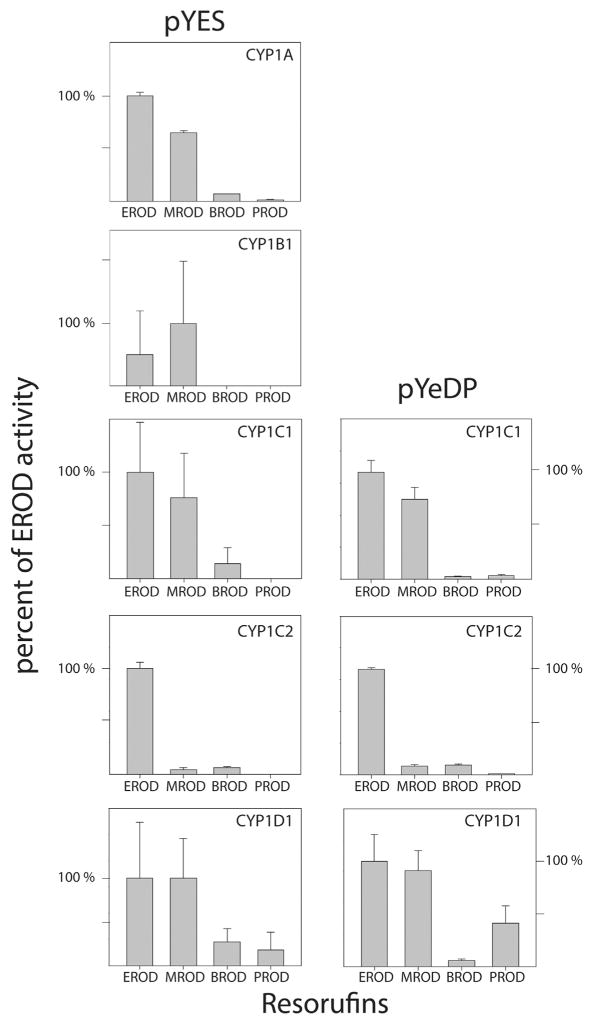
Following the demonstration of functional CYP in intact yeast cells, specific activities with the four substituted resorufins were measured kinetically, at saturating substrate concentrations, using microsomal fractions prepared from CYP1 transformed (pYES/DEST52) yeast strains. Activity with one or more of the resorufin substrates was detected with microsomal fractions from yeast transformed with the zebrafish CYP1s, but not with microsomes from control yeast transformed with Arabidopsis β-glucuronidase, or with CYP1 yeast microsomes that were boiled prior to assay. Microsomal recombinant CYP1A had the highest rates of EROD activity, and MROD activity of CYP1A was approximately 2/3 the rate of EROD at saturating concentrations of substrate (Table 1 and Fig. 3, left panel). The profile of CYP1B1 was different in that MROD seemed to be greater than EROD. The CYP1C1 profile seemed to be similar to CYP1A, while CYP1C2 yeast microsomes had EROD rates that were substantially greater than the MROD, which was less than 10% of the rates with 7-ER (Fig. 3). With all five CYP1s, the rates with 7-BR and 7-PR were less than those with 7-ER or 7-MR (Fig. 3).
Table 1.
Activities of CYP1s expressed in yeast with resorufin and ethoxyfluorescein ethylester.
| Substrate | CYP expressed via pYES/DEST52a
|
CYP expressed via pYeDP60a
|
||||||
|---|---|---|---|---|---|---|---|---|
| 1A | 1B1 | 1C1 | 1C2 | 1D1 | 1C1 | 1C2 | 1D1 | |
| ER d | 89.3 ± 1.9 | 0.2 ± 0.2 | 2.1 ± 0.7 | 46.5 ± 2.0 | 1.1 ± 0.5 | 757 ± 60 | 1301 ± 13 | 35 ± 4 |
| MR | 58.1 ± 0.8 | 0.4 ± 0.1 | 1.6 ± 0.7 | 2.2 ± 0.7 | 1.1 ± 0.0 | 567 ± 69 | 110 ± 17 | 31 ± 3 |
| PR | 1.3 ± 0.3 | NDb | ND | ND | 0.2 ± 0.1 | 27 ± 6 | 11 ± 2 | 14 ± 5 |
| BR | 6.3 ± 0.3 | ND | 0.3 ± 0.3 | 3.0 ± 0.5 | 0.3 ± 0.2 | 20 ± 4 | 124 ± 11 | 2.2 ± 0.2 |
| EFEE | –c | – | – | – | – | 78 ± 4 | 21 ± 8 | ND |
All activities are in pmol substrate oxidized min−1 mg−1 microsomal protein. Each of these values is the mean of four to six different measurements. Boiled microsomes from each transformant and microsomes from yeast expressing Arabidopsis β-glucuronidase exhibited no catalytic activity with any substrate. The final concentration of methanol in incubation mixtures was 0.2–0.3%.
ND is not detected.
Not assayed with CYPs expressed via pYES/DEST52.
Abbreviations are ethoxy-, methoxy-, propyloxy- and benzyloxyresorufin (ER, MR, PR and BR) and 7-ethoxyfluorescein-ethylester (EFEE).
Fig. 3.
Relative rates of microsomal alkoxy-O-dealkylase activities of zebrafish CYP1s expressed in yeast. For each CYP, the activities with 7-ER (EROD) are set at 100%. The enzymes expressed via pYES/DEST52 are in the left column, and for CYP1Cs and CYP1D1 expressed via pYeDP60 are in the right column. The error bars reflect error propagation.
The immunoblot of V5 tag (Fig. 2) indicated that there were differences in amounts of the CYP1 proteins expressed, which would affect the values for activity per mg of yeast microsomal protein. Determining the amounts of V5 tag on the immunoblots allowed us to normalize the levels of catalytic activity to the amounts of protein expressed (Supplemental Table 2). While this normalization did affect the absolute activities, the relative activities of a given enzyme with the various substrates were unchanged (Supplemental Fig. 3). Since the relative amounts of yeast POR do not vary significantly from one microsomal preparation to another [30,46–48], differences in amounts of yeast POR in microsomal fractions here are not likely to contribute to the observed differences in activity between the zebrafish CYP1s.
3.2. Activity of CYP1Cs and CYP1D1 expressed via pYeDP60
Next we focused on characterizing activity of the CYP1Cs and CYP1D1, with a broader set of substrates, using yeast transformed via the pYeDP60 vector. As above, we initially tested intact cells for activity with the substituted resorufins. Cells that had been transformed with an empty vector had no activity, while cells transformed with CYP1C1, CYP1C2 and CYP1D1 via pYeDP60 all showed activity with 7-ER, indicating expression of catalytically active enzyme (data not shown). Subsequent analysis of microsomal preparations from yeast transformed with CYP1C1, CYP1C2, or CYP1D1 showed again that all three enzymes acted on substituted resorufins as substrates (Table 1). The CYP1Cs also acted on EFEE. All activities were NADPH-dependent, as under identical conditions no product formation was observed with NADH (not shown).
The rates of activity of microsomal preparations of yeast transformed with CYP1s via pYeDP60 were greater than rates obtained with microsomes of yeast transformed with the same CYPs via pYES/DEST52 (Table 1). However, the relative specific activities with the four substituted resorufin substrates (normalized to EROD activity in each case) were highly similar for recombinant enzymes from yeast transformed via pYeDP60 and pYES/DEST52 (Fig. 3). This is evident also comparing the EROD to MROD ratios; for example, with CYP1C1 this ratio was 1.3 with the pYES/DEST52-expressed enzyme and with the pYeDP60 expressed enzyme.
Spectrophotometric analysis of microsomal fractions prepared from yeast transformed with CYP1C1 and CYP1C2 via pYeDP60 had CO-bound reduced absorbance at ca 450 nm, consistent with P450 (e.g., illustrated in Fig. 4). Turnover numbers (kcat) for substrate oxidation were calculated with CYP1Cs, using microsomal fractions that had quantifiable P450 levels (Table 2). The kcat for metabolism of the substituted resorufin substrates and EFEE (Table 2) indicate that the yeast expressed CYP1Cs are quite efficient catalysts for these activities. The kcat for EROD catalyzed by CYP1C2 in one transformation was 35 nmol/nmol P450/min, higher than that for human or rodent CYP1A1 expressed in the same yeast strain with the same vector [49]. Although CYP1D1 showed catalytic activity with resorufins, the microsomal P450 content was very low and in most preparations was not detected, consistent with a lesser expression of CYP1D1. The lower activity of yeast-expressed CYP1D1 was reflected also in a higher Km with 7-ER (Table 2).
Fig. 4.
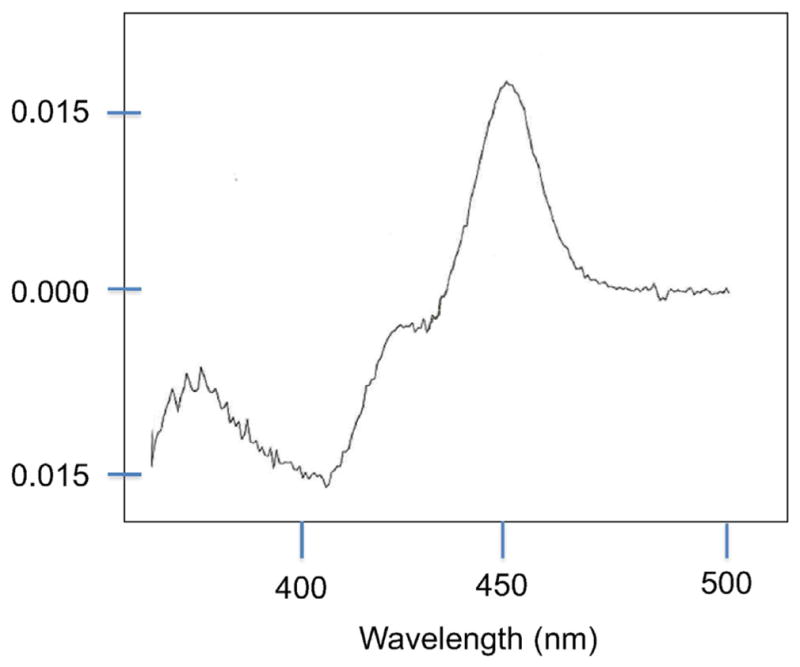
Spectral analysis of recombinant CYP1C1. Reduced, CO difference spectrum obtained with yeast microsomes transformed with CYP1C1 expressed via pYeDP60.
Table 2.
Km and kcat for dealkylation of resorufin substrates.
| Enzyme | Km for ER (nM) | kcat (mol substrate oxidized/mol P450/min)b
|
||||
|---|---|---|---|---|---|---|
| ERc | MR | PR | BR | EFEE | ||
| CYP1C1 | 85 ± 5 | 6.5 ± 0.5 | 4.9 ± 0.6 | 0.27 ± 0.06 | 0.20 ± 0.04 | 0.67 ± 0.03 |
| CYP1C2 | 110 ± 10 | 35.1 ± 0.4 | 3.3 ± 0.5 | 0.30 ± 0.06 | 3.35 ± 0.29 | 0.40 ± 0.15 |
| CYP1D1 | 1000a | – | – | – | – | – |
Estimated from the specific activities expressed per mg of yeast microsomal protein, from yeast transformed via pYeDP60. Each of these values is the mean of four to eight different measurements. The final concentration of methanol was 0.2–0.3% for resorufins and 0.1% for EFEE assays.
The P450 contents of the microsomal samples used for these assays were 0.12 and 0.04 nmol/mg for CYP1C1 and CYPC2, respectively. A P450 spectrum was detected in some CYP1D1 preparations, but not those that had been used in this analysis.
Abbreviations are as in Table 1.
Given that we observed greater specific activities of the pYeDP60-expressed CYP1Cs and CYP1D1 than those expressed via pYES/DEST52, we used these enzymes to assess the activity with additional fluorogenic substrates. In addition to EFEE, we examined 7-methoxy-coumarin, 7-methoxy-4-methyl-coumarin, 7-methoxy-4-bromomethyl-coumarin, 7-methoxy-4-trifluoromethyl-coumarin, and 7-ethoxy-4-trifluoromethyl-coumarin (see Fig. 1). Both CYP1C1 and CYP1C2 were active with all of these substrates. While the activity of pYeDP60-expressed CYP1C2 with 7-ER seemed to exceed the activity of CYP1C1 the reverse appeared to be true with some of the coumarin substrates, particularly MMC, MBC and MFC (Table 3). Although CYP1D1 was weakly active with the resorufin substrates, it had no O-dealkylase activity with any of the coumarins or with the EFEE. However, preliminary studies suggest it may be active in hydroxylation of some of these coumarins (unpublished data).
Table 3.
kcat values with coumarin substrates.
| Enzyme | kcat (mol substrate oxidized/mol P450/min)a
|
||||
|---|---|---|---|---|---|
| MOCb,c | MMCb,c | MBCb,c | MFCb,c | EFCc,d | |
| CYP1C1 | 0.70, 0.85 | 1.35, 1.11 | 0.215, 0.27 | 0.36, 0.43 | 1.51 ± 0.14 |
| CYP1C2 | 0.29, 0.36 | 0.18, 0.22 | 0.04, 0.06 | 0.04, 0.04 | 1.18 ± 0.02 |
| CYP1D1e | 0 | 0 | 0 | 0 | 0 |
kcat values obtained with microsomal preparations from yeast expressing the indicated CYP via pYeDP60, and in which P450 content could be measured. The P450 contents in these preparations were 0.12 and 0.04 nmol/mg for CYP1C1 and CYPC2, respectively. The final concentration of methanol (with all coumarins except MBC) or DMF (MBC assays) was 0.2%.
Abbreviations are: 7-methoxy-coumarin (MOC); 7-methoxy-4-methyl-coumarin (MMC); 7-methoxy-4-bromomethyl-coumarin (MBC); 7-methoxy-4-trifluoromethyl-coumarin (MFC); and 7-ethoxy-4-trifluoromethyl-coumarin (EFC).
These values were obtained from measurements with two independent preparations, and both values are shown.
These values are from measurements with four to eight independent preparations.
No dealkylation activity was detected with CYP1D1 for any of these substrates.
3.3. Benzo[a]pyrene oxidation
Microsomes from the yeast transformed via pYeDP60 were used in radiometric assays to measure the overall activity of CYP1Cs and CYP1D1 with B[a]P. The rates of total B[a]P metabolism with this assay were 0.81 nmol min−1 nmol−1 microsomal P450 for CYP1C1, and 2.26 nmol min−1 nmol−1 P450 for CYP1C2. CYP1D1 metabolism of BaP was not quantifiable with the radiometric assay. However, metabolites of BaP were generated in reactions together with human epoxide hydrolase for all three enzymes. Both CYP1C1 and CYP1C2 formed multiple metabolites of BaP (Fig. 5), including the 9,10-dihydrodiol-BP, 7,8-dihydrodiol-BP, 3-hydroxy-BP and 9-hydroxy-BP derivatives, and the 7,8,9,10-tetrol-BP. The tetrol metabolite observed with zebrafish CYP1s co-eluted with the authentic tetrol produced by human CYP1A1 in the presence of epoxide hydrolase, consistent with identity as the 7β,8α,9α,10β-tetrahydrotetrol derivative.
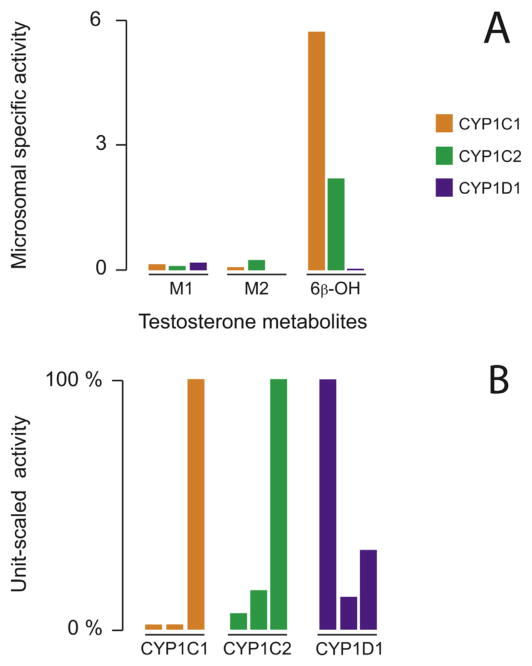
Fig. 5.
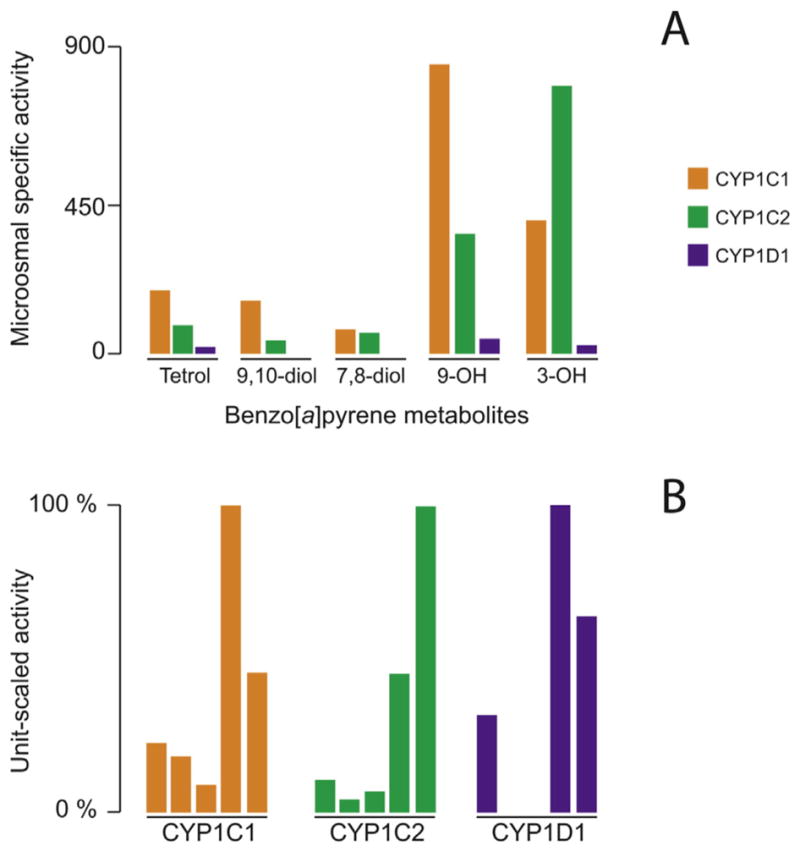
Benzo[a]pyrene metabolite profiles for CYP1Cs and CYP1D1. Panel A, microsomal specific activity calculated for each benzo[a]pyrene metabolite from its peak area (arbitrary units) expressed per min and per mg of yeast microsomal protein. The retention times of the different compounds were tetrol (3.2 min), 9,10-dhydrodiol (4.2 min), 7,8-dihydrodiol (5.6 min), 9-hydroxy (7.9 min), 3-hydroxy (8.3 min), and benzo[a]pyrene (11.0 min) for a total run time of 14 min. Panel B, benzo[a]pyrene metabolite profiles obtained for each CYP enzyme from unit-scaling the specific activities to the highest one. For each CYP enzyme, bars represent from left to right tetrol, 9,10-dihydrodiol, 7,8-dihydrodiol, 9-hydroxy-, and 3-hydroxy-benzo[a]pyrene metabolite.
The ratio of CYP1C2 to CYP1C1 activity calculated from rates of formation of the sum of metabolites of BaP per nmol P450 was 2.40, close to the ratio of rates measured with the radiometric assay, 2.79. CYP1D1 yeast microsomes also formed 7,8,9,10-tetrol-BP and the 9-hydroxy-BP and 3-hydroxy-BP metabolites, but the amounts formed were less than 0.2% of the amounts formed by CYP1C yeast microsomes. BaP-4,5-dihydrodiol and quinones were not examined in this analysis. Finally, BaP was shown to be a competitive inhibitor of EROD activity for the zebrafish enzymes, with a Ki in the range of 100 nM for both CYP1Cs and in the range of 10 μM for CYP1D1. Thus, the affinity of BaP for CYP1D1 appears to be a hundred times lower than that for either of the CYP1Cs (Table 4).
Table 4.
Benzo[a]pyrene interaction with zebrafish CYP1Cs and CYP1D1 expressed in yeast, and metabolite formation.
| Benzo[a]pyrene metaboliteb
|
||||||
|---|---|---|---|---|---|---|
| Kia | Tetrol | 9,10-Diol | 7,8-Diol | 9-OH | 3-OH | |
| CYP1C1 | 0.06 μM | 0.223 | 0.179 | 0.082 | 1.000 | 0.456 |
| CYP1C2 | 0.20 μM | 0.099 | 0.040 | 0.064 | 0.421 | 0.926 |
| CYP1D1 | 12 μM | 0.007 | 0 | 0 | 0.023 | 0.015 |
Competitive inhibition of ethoxyresorufin O-deethylation reaction by benzo[a]pyrene.
Metabolite formation is derived from mass spec (MS) peak areas expressed as arbitrary units min−1 mg−1 microsomal protein, normalized to the highest MS peak area. Expressing the total metabolite formation to microsomal P450 content gave arbitrary units min−1 mg−1 P450 of 16.1 for CYP1C1 and 38.7 for CYP1C2, and a ratio of CYP1C2/CYP1C1 of 2.4.
3.4. Testosterone oxidation
CYP1C1, CYP1C2 and CYP1D1 all were able to oxidize testosterone, forming three metabolites; one that was identified as 6β-OH-testosterone and two that were unidentified (M1 and M2 in Fig. 6). The major metabolite was 6β-OH-testosterone, and was formed in greater amounts by CYP1C1 than CYP1C2 (Fig. 6). CYP1D1 produced very little 6β-hydroxytestosterone, less than 0.5 % of the amount formed by CYP1C1. However, despite being sluggish with testosterone overall, the hydroxylated metabolite M1 was formed by CYP1D1 at nearly twice the rate of the CYP1Cs (per mg of microsomal protein) (Fig. 6).
Fig. 6.
Testosterone metabolite profiles for CYP1Cs and CYP1D1. Panel A, microsomal specific activity calculated for each testosterone metabolite from its peak area (arbitrary units) expressed per min and per mg of yeast microsomal protein. The retention times of the different compounds were M1 (5.3 min), M2 (6.6 min), 6β-hydroxy (7.1 min), and testosterone (8.1 min) for a total run time of 14 min. Panel B, testosterone metabolite profiles obtained for each CYP enzyme from unit-scaling the specific activities to the highest one. For each CYP enzyme, bars represent from left to right, hydroxylated testosterone M1 (unknown), hydroxylated testosterone M2 (unknown), 6β-hydroxy-testosterone.
3.5. Homology modeling and docking
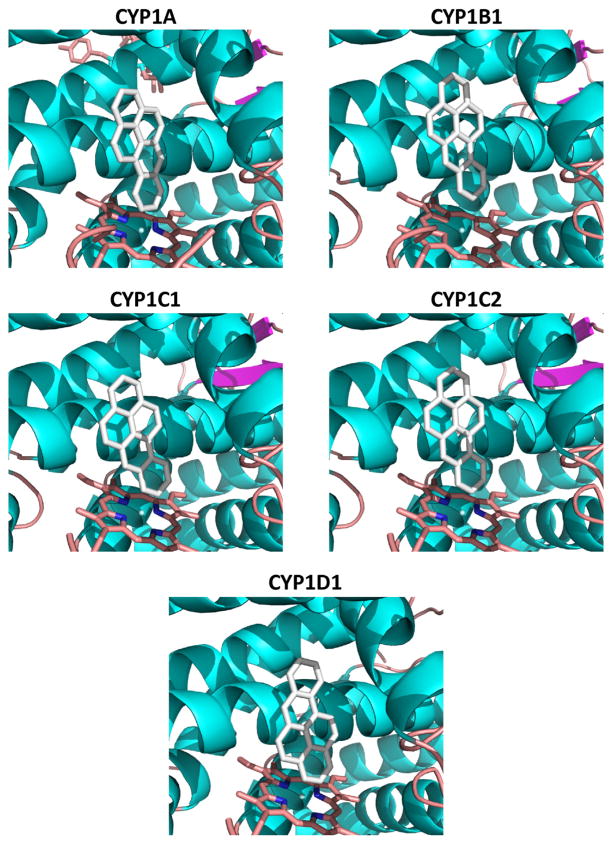
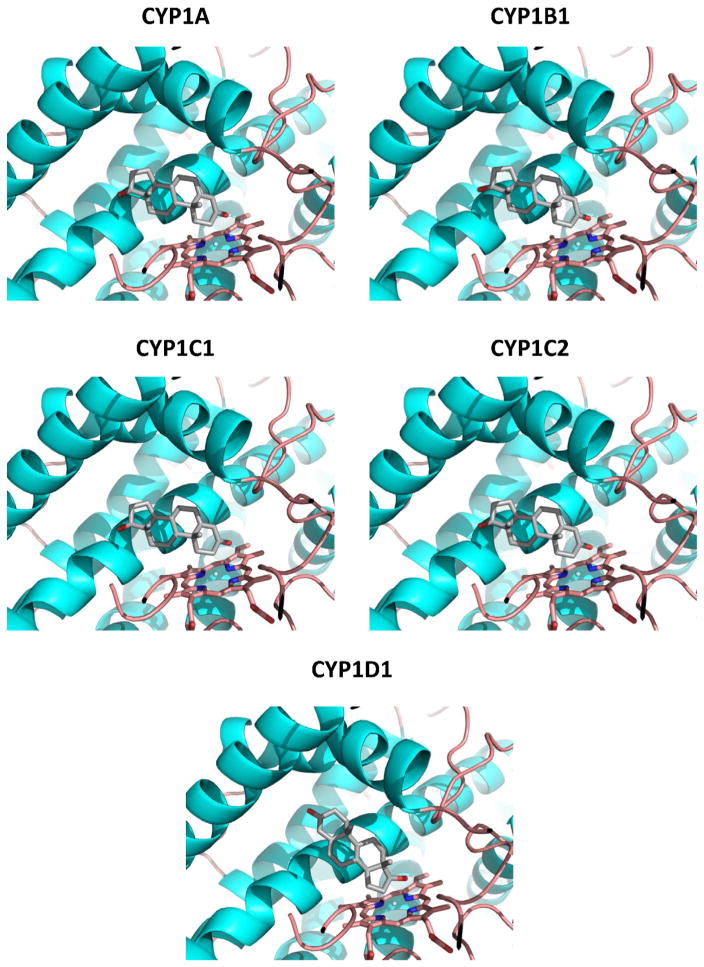
An analysis of docked orientations of potential substrates BaP and testosterone suggests likely sites of oxidation that support the in vitro data with the CYP1Cs and CYP1D1. Although in this study we did not analyze BaP metabolism by zebrafish CYP1A or CYP1B1, others have identified BaP metabolites formed by those enzymes (expressed in E. coli) [29] and we therefore modeled those CYPs and docked BaP to them as well. BaP docked in a family of productive orientations that was planar in CYP1A, CYP1B1, CYP1C1 and CYP1C2, but not in CYP1D1 (Supplemental Fig. 3). The top poses in CYP1A, CYP1B1, CYP1C1 and CYP1C2 were those with the benzoring of BaP oriented toward the heme iron (Fig. 7). BaP docking in CYP1D1 differed from the other CYP1s, with some poses that were out of the plane and not likely productive. The top poses had the 3-carbon positioned toward the heme iron, rather than the benzoring as in the other CYP1s (Fig. 7). Relative energy calculations also show that BaP docking to CYP1D1 was less favored than to the CYP1Cs by an average of 2.6 kcal/mol (Table 5), corresponding to a 100-fold higher calculated KD for CYP1D1.
Fig. 7.
Best single positions for BaP docked in zebrafish CYP1s. Homology models for the five CYP1s were constructed, and substrates docked as described in the Materials and methods section. The benzo-ring is oriented in a productive position for all models except for CYP1D1.
Table 5.
Calculated affinities and relative KD values from docking studies.
| BaP
|
Testosterone
|
|||||
|---|---|---|---|---|---|---|
| Best affinity (kcal/mol) | Mean affinitya | Relative KD | Best affinity (kcal/mol) | Mean affinity | Relative KD | |
| CYP1A | −14.5 | −14.3 | 0.026 | −11.6 | −9.5 | 0.127 |
| CYP1B1 | −14.4 | −14.2 | 0.035 | −10.5 | −8.3 | 1.000 |
| CYP1C1 | −14.1 | −13.7 | 0.072 | −11.0 | −8.9 | 0.363 |
| CYP1C2 | −15.1 | −14.9 | 0.011 | −11.6 | −8.5 | 0.713 |
| CYP1D1 | −12.5 | −12.2 | 1.000 | −10.7 | −9.9 | 0.065 |
Mean of top 5 poses.
Docking of testosterone to the CYP1Cs showed that the top productive poses have the A- and B-rings positioned proximal to the heme iron, which would allow the 6-carbon to potentially be in an oxidizable position (Fig. 8). The docking of testosterone in CYP1D1 differed from the CYP1cs, in that the orientations were more diverse (Supplemental Fig. 4), and the top poses showed the D-ring proximal to the heme. Docking of testosterone to the CYP1A and CYP1B1 models showed orientation similar to that in the CYP1Cs (Fig. 8), allowing a prediction that they would have similar product profiles. Testosterone affinity values were 2–3 kcal/mol lower than for BaP, and both CYP1D1 and CYP1B1 were predicted to have relatively poor binding of testosterone.
Fig. 8.
Best single positions for testosterone docked in zebrafish CYP1s. Homology models for the five CYP1s were constructed, and substrates docked as described in the Materials and methods section. Note that the pose in CYP1D1 is different from top poses in the other CYP1s.
4. Discussion
4.1. Fluorogenic substrates
Zebrafish have five CYP1 genes, CYP1A, CYP1B1, CYP1C1, CYP1C2 and CYP1D1. In these studies we expressed all five CYP1s in W(R) yeast via pYES/DEST52, and also expressed the CYP1Cs and CYP1D1 via pYeDP60. The yeast-expressed recombinant enzymes were variously active with alkyl- or benzyl-substituted resorufin substrates commonly used to characterize CYP1 enzymes. Among the five CYP1s expressed via pYES/DEST52, the highest activity with ER was obtained with CYP1A, consistent with CYP1As in other fish and CYP1A1s in mammals being highly active with this substrate [50–52]. The values we obtained for EROD activity with CYP1A expressed via the pYES/DEST52 ranged from 89–150 pmol min−1 mg−1 yeast microsomal protein, similar to the optimum value of 142 pmol min−1 mg−1 microsomal protein obtained for zebrafish CYP1A expressed in another yeast strain, INVsc1, also using the pYES/DEST52 destination vector [27].
CYP1C1, CYP1C2 and CYP1D1 expressed via the pYeDP60 vector had rates of activity with a given resorufin substrate that were greater than rates obtained with the same enzymes expressed via pYES/DEST52, However, we also observed very similar profiles of relative activity with the resorufin substrates for the enzymes expressed via the different vectors, analyzed in different laboratories. The relative rates of activity with the resorufin substrates we observed with the CYP1s expressed in yeast were in large part similar to the relative activity profiles with these substrates, for the same five zebrafish CYP1s expressed in E. coli [29]. In particular, the substrate profiles for recombinant CYP1As from W(R) yeast and E. coli were essentially the same. The CYP1B1, CYP1Cs and CYP1D1 showed some differences between the yeast and E. coli recombinant enzymes. For example, as before [17] we found that CYP1D1 acted on 7-MR as well as 7-ER, while activity with 7-MR was not detected with CYP1D1 expressed in E. coli [29].
The redox partner for P450 in the W(R) yeast was the native yeast POR, while in the E. coli preparations it was the human POR enzyme [29]. Differences in reductase identity might contribute to differences in activity of a CYP with some substrates, perhaps especially evident at rates that are very low. Differences in specific content of POR in the preparations of E. coli and yeast also could contribute to differences in observed activity of recombinant CYP1s. Nevertheless, the similarities in relative activities between the enzymes expressed in yeast transformed via different vectors, and the similarities between yeast- and E. coli-expressed enzymes, indicate that these relative activities reflect real differences in substrate selectivity for the zebrafish CYP1 enzymes.
Cytochrome P450 was detected spectrophotometrically in microsomal preparations of the yeast expressing CYP1C1, CYP1C2 and CYP1D1 via pYeDP60, but not in yeast transformed via pYES/DEST52. The lack of a spectral P450 signal in the yeast transformed via pYES/DEST52 could occur if there were substantially lower levels of expression with that vector. Weaker expression in the yeast transformed with the pYES/DEST52 vector possibly could result from some interference by the V5 tag. The V5 tag also might interfere with the activity of the expressed enzymes. The fact that the pYeDP60 vector contains two selection markers also might contribute to higher levels of expression with that vector. Apparent differences in expression also could involve some artifact resulting from differences in the method of cell disruption [27], which was more vigorous with the pYES/DEST52 yeast. However, a more likely possibility is that untranslated sequence at the 5′ of the coding sequence between the initiation ATG and the last bp of the GAL promoter is detrimental for translation efficiency of P450-coding sequence [53]. There is an untranslated stretch between the inserted coding sequence and the end of the GAL promoter in pYES/DEST52, but not in CYP1C1, CYP1C2 and CYP1D1 constructs in pYeDP60.
As the spectrophotometric and catalytic assays indicated that the transformation of yeast via pYeDP60 resulted in greater levels of expression of CYP1C1, CYP1C2 and CYP1D1, microsomal preparations from these yeasts were used for further characterization of these CYP1s with additional fluorogenic substrates typically metabolized by CYP1s from mammalian systems [49]. The three pYeDP60-expressed zebrafish CYP1s differed markedly from one another with these substrates. Thus, EFEE and the substituted coumarins all were metabolized more efficiently by CYP1C1 than by CYP1C2, a pattern opposite to that seen for EROD. Notably, the CYP1D1 yeast microsomes showed no activity with any of these substrates. Scornaienchi et al. [29] also assayed the CYP1s expressed in E. coli for activity with a series of substituted coumarins, but only one compound (7-methoxy-4-trifluoromethyl-coumarin) was assayed in common in their series and ours. This compound was not a substrate for O-dealkylation reactions by CYP1D1 expressed in yeast or E. coli. Despite the lack of O-dealkylation activity, some coumarinic compounds appear to be susceptible to hydroxylation by CYP1D1 (unpublished); further examination may suggest a functional significance for CYP1D1.
Ratios of activity with different substrates can reveal whether a novel CYP1 is more like mammalian CYP1A1, CYP1A2, or CYP1B1. Based on the EROD/MROD ratios, CYP1C2 is an “exacerbated” 1A1-type (ratio = 10.6; human CYP1A1 has an EROD/MROD ratio of 2.0, Consistent with that, the kcat for EROD determined with CYP1C2 is higher than the kcat for recombinant human CYP1A1 measured in our studies [54]. The high level of activity with EFEE also suggests that CYP1C2 is a 1A1-type enzyme. In contrast, CYP1C1 and CYP1D1 have EROD/MROD ratios that are more “1A2-type” (ratio = 1.3 and 1.1, respectively; human CYP1A2 has an EROD/MROD ratio of 0.6. CYP1D1 is also a 1A2-type, in that neither human CYP1A2 nor zebrafish CYP1D1 had activity with EFEE. However, CYP1C1, which is more 1A2-type based on its EROD/MROD ratio, has 1A1-type characteristics based on high activity with EFEE.
4.2. Benzo[a]pyrene and testosterone
The rates we obtained with the radiometric assay for overall BaP metabolism by CYP1C1 and CYP1C2 were 0.81 and 2.26 nmol/min/nmol P450, respectively, indicating that CYP1C2 is more efficient at metabolism of this PAH than is CYP1C1. The ratio of CYP1C2 BaP metabolism to CYP1C1 BaP metabolism per nmol P450 in the radiometric assay was 2.79, and a similar CYP1C2/CYP1C1 ratio of 2.4 was obtained from the sum of metabolites formed, when normalized to P450 (see Table 4). The rates of metabolism we measured with the radiometric assay were somewhat greater than the overall rates for BaP metabolism reported for CYP1C1 and CYP1C2 expressed in E. coli, calculated from the sum of amounts of individual metabolites [29]. However, in E. coli as in yeast, CYP1C2 was more active than CYP1C1 with BaP. That CYP1C2 is more efficient than CYP1C1 at metabolism of BaP is consistent with the observation from EROD data that CYP1C2 is catalytically more like mammalian CYP1A1. The amounts of BaP metabolites formed, and the Ki for EROD inhibition, showed that BaP was a poor substrate for the yeast-expressed CYP1D1, observed also with the E. coli-expressed CYP1D1 [29].
Both of the CYP1Cs expressed in yeast were formed the benzoring 7,8- and 9,10-dihydrodiols of BaP. The CYP1Cs and the CYP1A and CYP1B1 expressed in E. coli also formed benzoring dihydrodiols of BaP [29]. The metabolite results are supported by the orientation of BaP docked into homology models of these four zebrafish CYP1s, all of which had top poses with the benzo-ring oriented toward the heme iron. In contrast, the CYP1D1 expressed either in yeast or E. coli did not form the benzo-ring dihydrodiols, and docking to CYP1D1 models showed a very different suite of poses than observed in the other CYP1s.
We also observed formation of BaP-7,8,9,10-tetrol by both CYP1Cs and by CYP1D1. The BaP-7,8,9,10-tetrol reflects formation of the BaP-7,8-diol-9,10-oxides, which include the ultimate carcinogenic derivative of BaP [55]. This suggests that metabolism by the CYP1Cs and CYP1D1 could contribute to activation of BaP to mutagenic or carcinogenic diol-epoxides of BaP, although the lesser affinity of BaP for CYP1D1 suggests that it may play less of a role than the CYP1Cs. It has been known for many years that various fish tissues are able to oxidize BaP to suites of metabolites, favoring oxidation both on the benzo-ring and at the 3-carbon (e.g. [56]). Our results suggest that the CYP1Cs could contribute significantly to overall metabolism of BaP observed with microsomal preparations of organs or cells where these proteins might be expressed abundantly; based on mRNA transcript levels this could include embryos. Determining patterns of metabolism of other PAHs by these CYP1s will be important to assessing their possible contribution to PAH effects in the zebrafish model.
As with BaP, CYP1D1 was much less active at overall metabolism of testosterone than the CYP1Cs. A similar difference was observed for estradiol metabolism by the E. coli expressed enzymes [28]. Thus, CYP1D1 is less active than the CYP1Cs with both androstane- and estrane-derived compounds. While the CYP1Cs both formed 6β-hydroxytestosterone CYP1D1 did not. Rather, in contrast to 6β-hydroxylation, testosterone metabolite 1 was formed more actively by CYP1D1 than the CYP1Cs. A difference between CYP1D1 and the CYP1Cs was evident as well in the orientation of testosterone docked into the homology models. Generally, the CYP3 family enzymes are the major catalysts for testosterone 6β-hydroxylation in fish [57–59]. That does not preclude other enzymes, such as the CYP1Cs, from contributing to this metabolism, particularly in cells or life stages where CYP3As might be expressed weakly relative to these CYP1s. Thus, the oxidation of testosterone by the three CYP1Cs or CYP1D1 could be physiologically relevant in some organs, cell types, or developmental stages.
The contribution of CYP1Cs relative to that of CYP1A or CYP1B1 to overall catalytic activity is yet to be established in most organs in fish. Our studies on inhibition of EROD by a monoclonal antibody to fish CYP1A indicate that, at least in liver of some fish species, about 95% of microsomal EROD activity is due to CYP1A [52], implying little contribution from other enzymes, and likely low level expression of the other CYP1s in liver overall. There could be a high-level expression of the other CYP1s in some cell type(s), but with rare exception (e.g., [60–62]), there is as yet little information on the levels of the CYP1 proteins other than CYP1A in different organs or cell types of fish, including throughout development. Such information can point to the contribution of the various CYP1s to cell and organ responses to xenobiotic or endogenous substrates in fish models such as zebrafish, exemplified by the reported participation of CYP1Cs in TCDD effects in brain vessels [62].
5. Conclusions
The five CYP1s of zebrafish were expressed in yeast, and activities assessed with substituted resorufins. The results with yeast-expressed enzymes were largely similar to results with E. coli expressed zebrafish CYP1s [29], indicating that the recombinant enzymes from both expression systems can give valid information about the functional capabilities of the zebrafish CYP1s. As a general finding CYP1D1 tended to have less activity with most substrates than the other CYP1s. This also was found with E. coli-expressed enzymes [29]. However, the formation of a novel testosterone metabolite suggests that there could be unrecognized biological functions of CYP1D1. It also may be that the biological importance of CYP1D1 is diminishing over evolution, suggested by the loss or pseudogenization of the CYP1D1 gene in some vertebrates [20,63].
Results obtained with docking of BaP and testosterone to homology models of the CYP1s were consistent with the metabolite data on sites of oxidation of these substrates. The approach thus may be used to predict the regioselectivity of metabolism of substrates with multiple sites of oxidation. It is important to note, however, that the docking here does not consider possible differences in access channels that can govern substrate ingress to the active center [64]. Studies are underway to identify activities with other endogenous and xenobiotic substrates, which will be important to inferring the roles of these multiple CYP1s in physiology and toxicology in the zebrafish model. The functions of different CYP1s in vivo will also depend on the relative levels of CYP1 protein expression, including the response to inducers, in different organs and cell types, and at different developmental stages. Important questions remain regarding the extent to which the multiple CYP1s might participate in the toxic effects of AHR agonists, which tend also to be CYP1 substrates or inhibitors. The finding of substantial catalytic function for the CYP1Cs with some substrates, as well as uncoupling of the CYP1Cs by halogenated AHR agonists (unpublished observations), suggests that there could be a complex involvement of multiple CYP1s in effects of AHR agonists in zebrafish. These CYP1s also present an interesting opportunity for further exploring the structural features associated with substrate selectivity among related P450s. The full range of substrates of zebrafish CYP1s is yet to be established, with either the E. coli or yeast expressed enzymes. Determining activities with endogenous and xenobiotic substrates is important to inferring the roles of these multiple CYP1s in health and disease in the zebrafish model.
Supplementary Material
Highlights.
Zebrafish CYP1 enzymes have been compared in two yeast expression systems.
Fluorogenic substrates, benzo[a]pyrene and testosterone were assayed.
The two yeast systems gave similar relative activities with resorufins.
Docking benzo[a]pyrene and testosterone to CYP1 models supports sites of oxidation.
An unknown testosterone hydroxylated metabolite is produced by CYP1D1.
Acknowledgments
This work was supported in part by NIH grants R01ES015912 and P42ES007381 (the Superfund Research Program at Boston University) to JJS, and the Centre National de la Recherche Scientifique, France, by the Japan Society for the Promotion of Science Postdoctoral Fellowship and Postdoctoral Fellowship for Research Abroad to AK (JSPS nos. 194313 and 24820, respectively), and by a Belgian-American Educational Foundation (BAEF) Postdoctoral Fellowship (BL). Additional support (JJS) was provided by NIH grant P01ES021923 and National Science Foundation Grant OCE-1314642 through the Woods Hole Center for Oceans and Human Health. NAMD was developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign.
Abbreviations
- AhR
aryl hydrocarbon receptor
- BaP or BP
benzo[a]pyrene
- BR
7-Benzoxyresorufin
- BROD
7-Benzoxyresorufin O-debenzylation
- CYP or P450
cytochrome P450
- DMF
N,N-dimethylformamide
- DTT
dithiothreitol
- EFC
7-Ethoxy-4-trifluoromethyl-coumarin
- EFEE
7-Ethoxyfluorescein ethylester
- ER
7-Ethoxyresorufin
- EROD
7-Ethoxyresorufin O-deethylation
- LN2
liquid nitrogen
- MBC
7-Methoxy-4-bromomethyl-coumarin
- MD
molecular dynamics
- MFC
7-Methoxy-4-trifluoromethyl-coumarin
- MMC
7-Methoxy-4-methyl-coumarin
- MOC
7-Methoxy-coumarin
- MR
7-Methoxyresorufin
- MROD
7-Methoxyresorufin O-demethylation
- MS
mass spectra
- NAMD
nanoscale molecular dynamics
- PAH
polycyclic aromatic hydrocarbon
- POR
NADPH-cytochrome P450 reductase
- PR
7-Pentoxyresorufin
- PROD
7-Pentoxyresorufin O-depentylation
- TEDG buffer
Tris–HCl 50 mM, EDTA 1 mM, glycerol 20%, DTT 1 mM, pH 7.4
- TEG buffer
Tris–HCl 50 mM, EDTA 1 mM, glycerol 20%, pH 7.4
- TES50 buffer
Tris–HCl 50 mM, EDTA 1 mM, sorbitol 0.6 M pH 7.4
- Tetrol
7,8,9,10-Tetrahydrotetrol-benzo[a]pyrene
- VTFM
variable target function method
- 9,10-diol
9,10-Dihydrodiol-benzo[a]pyrene
- 7,8-diol
7,8-Dihydrodiol-benzo[a]pyrene
- 9-OH
9-Hydroxy-benzo[a]pyrene
- 3-OH
3-Hydroxy-benzo[a]pyrene
- 6β-OH
6b-Hydroxy-testosterone
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagen.2015.07.010.
Footnotes
Transparency document
The Transparency document associated with this article can be found, in online version.
References
- 1.Ioannides C, Lewis DF. Cytochromes P450 in the bioactivation of chemicals. Curr Top Med Chem. 2004;4(16):1767–1788. doi: 10.2174/1568026043387188. [DOI] [PubMed] [Google Scholar]
- 2.Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer. 2006;6(12):947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez FJ, Kimura S. Understanding the role of xenobiotic-metabolism in chemical carcinogenesis using gene knockout mice. Mutat Res. 2001;477(1–2):79–87. doi: 10.1016/s0027-5107(01)00109-9. [DOI] [PubMed] [Google Scholar]
- 4.Kleiner HE, Vulimiri SV, Hatten WB, Reed MJ, Nebert DW, Jefcoate CR, DiGiovanni J. Role of cytochrome p4501 family members in the metabolic activation of polycyclic aromatic hydrocarbons in mouse epidermis. Chem Res Toxicol. 2004;17(12):1667–1674. doi: 10.1021/tx049919c. [DOI] [PubMed] [Google Scholar]
- 5.Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- 6.Guengerich FP. Cytochrome p450 and chemical toxicology. Chem Res Toxicol. 2008;21(1):70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 7.Guengerich FP. Human cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. Kluwer Academic; New York: 2005. pp. 377–463. [Google Scholar]
- 8.Kim D, Guengerich FP. Cytochrome P450 activation of arylamines and heterocyclic amines. Annu Rev Pharmacol Toxicol. 2005;45:27–49. doi: 10.1146/annurev.pharmtox.45.120403.100010. [DOI] [PubMed] [Google Scholar]
- 9.Dragin N, Shi Z, Madan R, Karp CL, Sartor MA, Chen C, Gonzalez FJ, Nebert DW. Phenotype of the Cyp1a1/1a2/1b1−/− triple-knockout mouse. Mol Pharmacol. 2008;73(6):1844–1856. doi: 10.1124/mol.108.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz D, Kisselev P, Chernogolov A, Schunck WH, Roots I. Human CYP1A1 variants lead to differential eicosapentaenoic acid metabolite patterns. Biochem Biophys Res Commun. 2005;336(3):779–783. doi: 10.1016/j.bbrc.2005.08.172. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz D, Kisselev P, Ericksen SS, Szklarz GD, Chernogolov A, Honeck H, Schunck WH, Roots I. Arachidonic and eicosapentaenoic acid metabolism by human CYP1A1: highly stereoselective formation of 17(R),18(S)-epoxyeicosatetraenoic acid. Biochem Pharmacol. 2004;67(8):1445–1457. doi: 10.1016/j.bcp.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 12.Schlezinger JJ, Parker C, Zeldin DC, Stegeman JJ. Arachidonic acid metabolism in the marine fish Stenotomus chrysops (Scup) and the effects of cytochrome P450 1A inducers. Arch Biochem Biophys. 1998;353(2):265–275. doi: 10.1006/abbi.1998.0651. [DOI] [PubMed] [Google Scholar]
- 13.Rifkind AB, Lee C, Chang TK, Waxman DJ. Arachidonic acid metabolism by human cytochrome P450s 2C8, 2C9, 2E1, and 1A2: regioselective oxygenation and evidence for a role for CYP2C enzymes in arachidonic acid epoxygenation in human liver microsomes. Arch Biochem Biophys. 1995;320(2):380–389. doi: 10.1016/0003-9861(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 14.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279(23):23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 15.Uno T, Okamoto S, Masuda S, Imaishi H, Nakamura M, Kanamaru K, Yamagata H, El-Kady MA, Kaminishi Y, Itakura T. Bioconversion by functional P450 1A9 and P450 1C1 of Anguilla japonica. Comp Biochem Physiol C Toxicol Pharmacol. 2008;147(3):278–285. doi: 10.1016/j.cbpc.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Godard CA, Goldstone JV, Said MR, Dickerson RL, Woodin BR, Stegeman JJ. The new vertebrate CYP1C family: cloning of new subfamily members and phylogenetic analysis. Biochem Biophys Res Commun. 2005;331(4):1016–1024. doi: 10.1016/j.bbrc.2005.03.231. [DOI] [PubMed] [Google Scholar]
- 17.Goldstone JV, Jönsson ME, Behrendt L, Woodin BR, Jenny MJ, Nelson DR, Stegeman JJ. Cytochrome P450 1D1: a novel CYP1A-related gene that is not transcriptionally activated by PCB126 or TCDD. Arch Biochem Biophys. 2009;482(1–2):7–16. doi: 10.1016/j.abb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstone JV, Goldstone HM, Morrison AM, Tarrant A, Kern SE, Woodin BR, Stegeman JJ. Cytochrome P450 1 genes in early deuterostomes (tunicates and sea urchins) and vertebrates (chicken and frog): origin and diversification of the CYP1 gene family. Mol Biol Evol. 2007;24(12):2619–2631. doi: 10.1093/molbev/msm200. [DOI] [PubMed] [Google Scholar]
- 19.Jonsson ME, Berg C, Goldstone JV, Stegeman JJ. New CYP1 genes in the frog Xenopus (Silurana) tropicalis: induction patterns and effects of AHR agonists during development. Toxicol Appl Pharmacol. 2010;250(2):170–183. doi: 10.1016/j.taap.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonsson ME, Woodin BR, Stegeman JJ, Brunstrom B. Cytochrome p450 1 genes in birds: evolutionary relationships and transcription profiles in chicken and Japanese quail embryos. PLoS One. 2011;6(12):e28257. doi: 10.1371/journal.pone.0028257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stegeman JJ, Goldstone JV, Hahn ME. Perspectives on zebrafish as a model in environmental toxicology. In: Perry SF, editor. Zebrafish. Academic Press; New York: 2010. pp. 367–439. [Google Scholar]
- 22.Mandrell D, Truong L, Jephson C, Sarker MR, Moore A, Lang C, Simonich MT, Tanguay RL. Automated zebrafish chorion removal and single embryo placement: optimizing throughput of zebrafish developmental toxicity screens. J Lab Autom. 2012;17(1):66–74. doi: 10.1177/2211068211432197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson RT, Macrae CA. Systematic approaches to toxicology in the zebrafish. Annu Rev Pharmacol Toxicol. 2012;52:433–453. doi: 10.1146/annurev-pharmtox-010611-134751. [DOI] [PubMed] [Google Scholar]
- 24.Jönsson ME, Orrego R, Woodin BR, Goldstone JV, Stegeman JJ. Basal and 3,3′,4,4′,5-pentachlorobiphenyl-induced expression of cytochrome P450 1A, 1B and 1C genes in zebrafish. Toxicol Appl Pharmacol. 2007;221(1):29–41. doi: 10.1016/j.taap.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Scheffier BE, Willett KL. CYP1C1 messenger RNA expression is inducible by benzo[a]pyrene in Fundulus heteroclitus embryos and adults. Toxicol Sci. 2006;93(2):331–340. doi: 10.1093/toxsci/kfl072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonsson ME, Jenny MJ, Woodin BR, Hahn ME, Stegeman JJ. Role of AHR2 in the expression of novel cytochrome P450 1 family genes, cell cycle genes, and morphological defects in developing zebra fish exposed to 3,3′,4,4′,5-pentachlorobiphenyl or 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2007;100(1):180–193. doi: 10.1093/toxsci/kfm207. [DOI] [PubMed] [Google Scholar]
- 27.Chung WG, Sen A, Wang-Buhler JL, Yang YH, Lopez N, Merrill GF, Miranda CL, Hu CH, Buhler DR. cDNA-directed expression of a functional zebrafish CYP1A in yeast. Aquat Toxicol. 2004;70(2):111–121. doi: 10.1016/j.aquatox.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Scornaienchi ML, Thornton C, Willett KL, Wilson JY. Cytochrome P450-mediated 17beta-estradiol metabolism in zebrafish (Danio rerio) J Endocrinol. 2010;206(3):317–325. doi: 10.1677/JOE-10-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scornaienchi ML, Thornton C, Willett KL, Wilson JY. Functional differences in the cytochrome P450 1 family enzymes from zebrafish (Danio rerio) using heterologously expressed proteins. Arch Biochem Biophys. 2010;502(1):17–22. doi: 10.1016/j.abb.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truan G, Cullin C, Reisdorf P, Urban P, Pompon D. Enhanced in vivo monooxygenase activities of mammalian P450s in engineered yeast cells producing high levels of NADPH-P450 reductase and human cytochrome b5. Gene. 1993;125(1):49–55. doi: 10.1016/0378-1119(93)90744-n. [DOI] [PubMed] [Google Scholar]
- 31.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 32.Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 33.Urban P, Truan G, Bellamine A, Laine R, Gautier JC, Pompon D. Engineered yeasts simulating P450-dependent metabolisms: tricks, myths and reality. Drug Metabol Drug Interact. 1994;11(3):169–200. doi: 10.1515/dmdi.1994.11.3.169. [DOI] [PubMed] [Google Scholar]
- 34.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 35.Hahn ME, Lamb TM, Schultz ME, Smolowitz RM, Stegeman JJ. Cytochrome-P4501a Induction and Inhibition by 3,3′,4,4′-tetrachlorobiphenyl in an Ah receptor-containing fish hepatoma-cell line (Plhc-1) Aquat Toxicol. 1993;26(3–4):185–208. [Google Scholar]
- 36.Kennedy SW, Lorenzen A, James CA, Collins BT. Ethoxyresorufin-O-deethylase and porphyrin analysis in chicken-embryo hepatocyte cultures with a fluorescence multiwell plate reader. Anal Biochem. 1993;211(1):102–112. doi: 10.1006/abio.1993.1239. [DOI] [PubMed] [Google Scholar]
- 37.Binder RL, Stegeman JJ. Induction of aryl hydrocarbon hydroxylase activity in embryos of estuarine fish. Biochem Pharmacol. 1980;29(6):949–951. doi: 10.1016/0006-2952(80)90226-9. [DOI] [PubMed] [Google Scholar]
- 38.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007;Chapter 2(Unit 2):9. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 39.Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem. 2007;282(19):14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 40.Wang A, Savas U, Stout CD, Johnson EF. Structural characterization of the complex between alpha-naphthoflavone and human cytochrome P450 1B1. J Biol Chem. 2011;286(7):5736–5743. doi: 10.1074/jbc.M110.204420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cosconati S, Forli S, Perryman AL, Harris R, Goodsell DS, Olson AJ. Virtual screening with AutoDock: theory and practice. Expert Opin Drug Discovery. 2011;5(6):597–607. doi: 10.1517/17460441.2010.484460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart JJ. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J Mol Model. 2007;13(12):1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart JJ. MOPAC2009, Stewart Computational Chemistry. Colorado Springs, CO: 2008. [Google Scholar]
- 45.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peyronneau MA, Renaud JP, Jaouen M, Urban P, Cullin C, Pompon D, Mansuy D. Expression in yeast of three allelic cDNAs coding for human liver P-450 3A4. Different stabilities, binding properties and catalytic activities of the yeast-produced enzymes. Eur J Biochem. 1993;218(2):355–361. doi: 10.1111/j.1432-1033.1993.tb18384.x. [DOI] [PubMed] [Google Scholar]
- 47.Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272(31):19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- 48.Urban P, Werck-Reichhart D, Teutsch HG, Durst F, Regnier S, Kazmaier M, Pompon D. Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast. Kinetic and spectral properties of the major plant P450 of the phenylpropanoid pathway. Eur J Biochem. 1994;222(3):843–850. doi: 10.1111/j.1432-1033.1994.tb18931.x. [DOI] [PubMed] [Google Scholar]
- 49.Taly V, Urban P, Truan G, Pompon D. A combinatorial approach to substrate discrimination in the P450 CYP1A subfamily. Biochim Biophys Acta. 2007;1770(3):446–457. doi: 10.1016/j.bbagen.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Bellamine A, Gautier JC, Urban P, Pompon D. Chimeras of the human cytochrome P450 1A family produced in yeast. Accumulation in microsomal membranes, enzyme kinetics and stability. Eur J Biochem. 1994;225(3):1005–1013. doi: 10.1111/j.1432-1033.1994.1005b.x. [DOI] [PubMed] [Google Scholar]
- 51.Burke MD, Prough RA, Mayer RT. Characteristics of a microsomal cytochrome P-448-mediated reaction. Ethoxyresorufin O-de-ethylation. Drug Metab Dispos. 1977;5(1):1–8. [PubMed] [Google Scholar]
- 52.Kloepper-Sams PJ, Park SS, Gelboin HV, Stegeman JJ. Specificity and cross-reactivity of monoclonal and polyclonal antibodies against cytochrome P-450E of the marine fish scup. Arch Biochem Biophys. 1987;253(1):268–278. doi: 10.1016/0003-9861(87)90660-6. [DOI] [PubMed] [Google Scholar]
- 53.Pompon D. cDNA cloning and functional expression in yeast Saccharomyces cerevisiae of beta-naphthoflavone-induced rabbit liver P-450 LM4 and LM6. Eur J Biochem. 1988;177(2):285–293. doi: 10.1111/j.1432-1033.1988.tb14375.x. [DOI] [PubMed] [Google Scholar]
- 54.Gautier JC, Urban P, Beaune P, Pompon D. Engineered yeast cells as model to study coupling between human xenobiotic metabolizing enzymes. Simulation of the two first steps of benzo[a]pyrene activation. Eur J Biochem. 1993;211(1–2):63–72. doi: 10.1111/j.1432-1033.1993.tb19870.x. [DOI] [PubMed] [Google Scholar]
- 55.Ma Q, Lu AY. CYP1A induction and human risk assessment: an evolving tale of in vitro and in vivo studies. Drug Metab Dispos. 2007;35(7):1009–1016. doi: 10.1124/dmd.107.015826. [DOI] [PubMed] [Google Scholar]
- 56.Stegeman JJ, Woodin BR, Binder RL. Patterns of benzo[a]pyrene metabolism by varied species, organs, and developmental stages of fish. Natl Cancer Inst Monogr. 1984;65:371–377. [PubMed] [Google Scholar]
- 57.Klotz AV, Stegeman JJ, Woodin BR, Snowberger EA, Thomas PE, Walsh C. Cytochrome P-450 isozymes from the marine teleost Stenotomus chrysops: their roles in steroid hydroxylation and the influence of cytochrome b5. Arch Biochem Biophys. 1986;249(2):326–338. doi: 10.1016/0003-9861(86)90009-3. [DOI] [PubMed] [Google Scholar]
- 58.Celander M, Buhler DR, Forlin L, Goksoyr A, Miranda CL, Woodin BR, Stegeman JJ. Immunochemical relationships of cytochrome P4503A-like proteins in teleost fish. Fish Physiol Biochem. 1996;15(4):323–332. doi: 10.1007/BF02112359. [DOI] [PubMed] [Google Scholar]
- 59.Lee SJ, Buhler DR. Functional properties of a rainbow trout CYP3A27 expressed by recombinant baculovirus in insect cells. Drug Metab Dispos. 2002;30(12):1406–1412. doi: 10.1124/dmd.30.12.1406. [DOI] [PubMed] [Google Scholar]
- 60.Bugiak B, Weber LP. Hepatic and vascular mRNA expression in adult zebrafish (Danio rerio) following exposure to benzo-a-pyrene and 2,3,7,8-tetrachlorodibenzo-p-dioxin. Aquat Toxicol. 2009;95(4):299–306. doi: 10.1016/j.aquatox.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 61.Wang L, Camus AC, Dong W, Thornton C, Willett KL. Expression of CYP1C1 and CYP1A in Fundulus heteroclitus during PAH-induced carcinogenesis. Aquat Toxicol. 2010;99(4):439–447. doi: 10.1016/j.aquatox.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kubota A, Stegeman JJ, Woodin BR, Iwanaga T, Harano R, Peterson RE, Hiraga T, Teraoka H. Role of zebrafish cytochrome P450 CYP1C genes in the reduced mesen-cephalic vein blood flow caused by activation of AHR2. Toxicol Appl Pharmacol. 2011;253(3):244–252. doi: 10.1016/j.taap.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawai YK, Ikenaka Y, Fujita S, Ishizuka M. The CYP1D subfamily of genes in mammals and other vertebrates. Mamm Genome. 2010;21(5–6):320–329. doi: 10.1007/s00335-010-9263-9. [DOI] [PubMed] [Google Scholar]
- 64.Urban P, Truan G, Pompon D. Access channels to the buried active site control substrate specificity in CYP1A P450 enzymes. Biochim Biophys Acta. 2015;1850(4):696–707. doi: 10.1016/j.bbagen.2014.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.