Abstract
Objective
Persistent infection with oncogenic human papillomavirus (HPV) is known to be the necessary cause of cervical cancer and a majority of vulvar cancers. Persistent HPV infections must evade host immune responses, including cytokines released by activated T-helper (Th) cells. In this study, we investigated the risk of cervical and vulvar cancer associated with common genetic variations in 560 tagging single-nucleotide polymorphisms (SNPs) in candidate cytokine genes.
Methods
The study included 399 invasive squamous cell carcinomas (SCC) and 502 in situ or invasive adenocarcinomas (AC) of the cervix; 357 in situ or invasive vulvar SCC; and 1,109 controls from the Seattle-area case-control studies of HPV-related cancers. Logistic regression was used to estimate odds ratios (OR) and 95% confidence intervals (CI) using a log additive model, with adjustment for multiple testing.
Results
Statistically significant risks were observed for HPV16-containing SCC of the cervix with the variant allele rs879576 in IL17RA and rs2229094 in TNF [OR, 95% CI and multiple-testing corrected p: 1.91(1.30-2.79), p= 0.018 and 0.61 (0.45-0.83), p= 0.02, respectively). We also observed significantly increased risk of HPV-positive vulvar cancers associated with variant alleles in CSF2 (rs25882 and rs27438, 26-28% increased risk) and IL-12B (rs2569254 and rs3181225, 40-41% increased risk) genes.
Conclusions
We found that variation in several Th-cytokine genes are significantly associated with cervical and vulvar cancer risk. The strong association between these HPV-related cancers and common variation in cytokine genes in the Th1 and Th17 pathways may be important for development of new therapies.
Keywords: Cervical cancer, Vulvar cancer, T-helper 1 pathways, T-helper 2 pathway, T-helper 17 pathway, genetic variation
Introduction
Although the burden of carcinoma of the cervix has decreased considerably in countries with wide-spread screening, it remains the third most commonly occurring cancer and fourth most common cause of cancer deaths among women worldwide.[1] Persistent infection with oncogenic human papillomavirus (HPV) has been established as the necessary cause for the development of cervical cancer and a large proportion of HPV-related cancers at other anogenital sites, including vulvar cancer.[2] In contrast with the overall decreased incidence of cervical cancer, there has been a steady rise in the incidence of cervical adenocarcinoma (AC) and squamous cell carcinoma (SCC) of the vulva.[3, 4] HPV infections alone are not sufficient for neoplastic progression to these cancers, as transient HPV infections are extremely common in the general population and relatively few women infected with HPV progress to cancer.[5, 6] The mechanism of clearance of transient infections is complex: it involves HPV antigen presentation to CD4 and CD8 T cells, leading to activated T cells that proliferate into armed effector T cells. [7] In the small group of women who progress from HPV persistent infection to HPV-related cancer, HPV evades detection mainly through activation of the adaptive immune system.[8]
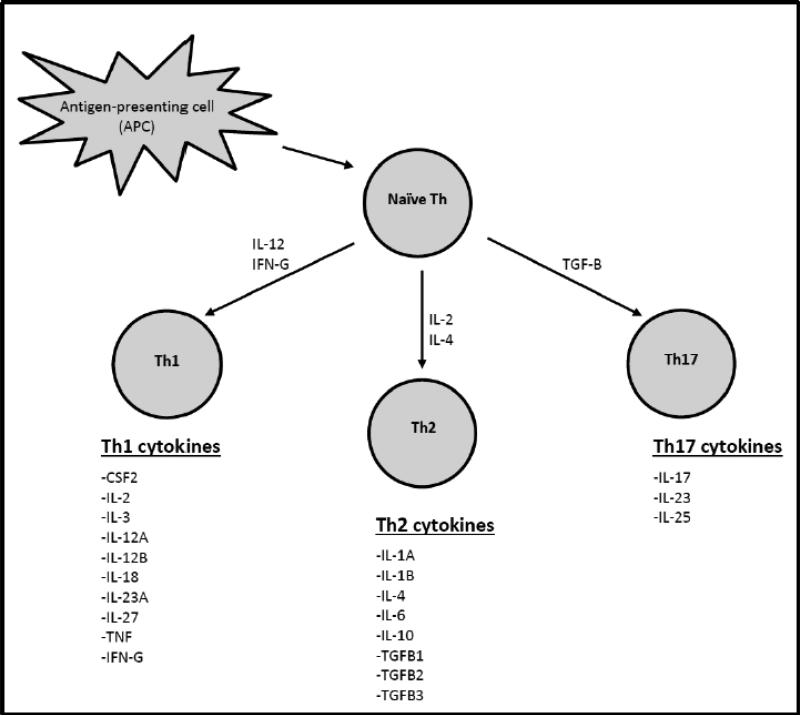
Evidence of the role of adaptive immunity comes from studies that assess T-cells generated in response to HPV infection and lesion development.[9, 10] Those studies suggest that T helper (Th) cells are necessary for production of HPV–specific antibody response and in aiding the development of cytotoxic T lymphocytes. Th responses that result in predominantly Th1-type cytokines favor HPV clearance (Figure 1), whereas activation and terminal differentiation of Th2 cells leads to higher levels of circulating pro-inflammatory cytokines and fewer HPV-specific T-cell responses.[9, 10] Th17 cells are induced by TGFB and produce IL-17-related pro-inflammatory cytokines that act to suppress HPV response.[11]
Figure 1. Cytokines comprising the Th1, Th2 and Th17 immune pathways.
Naïve T-cells may react to interferon-gamma (IFN-G) or interleukin (IL)-12 to differentiate into Th1 cells that clear intracellular pathogens including viruses, or they may respond to IL-4 and IL-2 to evolve into Th2 cells that promote humoral immunity; or transforming growth factor-beta (TGF-β) and IL-6 to promote a Th17 response [Ref: Zhu et. al; Cell Research (2010) 20:4-12.].
Further evidence of the role of immunity in HPV-related cancers comes from natural history studies, which demonstrated that most young women clear the virus within 1-2 years of infection.[12] In contrast, however, a high burden of HPV-related cancers has been observed in several immunosuppressed populations, suggesting that cancer is more likely to develop among individuals with compromised adaptive immune responses.[13] HPV-associated malignancies occur in excess among patients with HIV and AIDS, probably due to reduced HPV clearance among HIV/AIDS patients with low CD4 T-helper cell counts.[13] Incidence of HPV-related cancers is also greatly increased following solid organ transplantation, and these risks increase with length of time on immune suppression.[14] In a rare genetic disease, individuals with WHIM syndrome (OMIM #193670) have mutations in a chemokine receptor, CXCR4, which leads to impaired chemotaxis of leukocytes and inability to clear HPV infections.[15] These studies suggest that the adaptive immune responses are critical to HPV progression to cancer.
In prior publications involving single genes (or a group of genes) within immune response pathways, our research group has found several statistically significant associations with cervical cancer subtypes.[16-19] In this paper, we conducted a comprehensive analysis of genes within the three primary immune response pathways: Th1, Th2 and Th17, and their association with SCC of the cervix, AC of the cervix and vulvar cancers. We focused on subsets, such as HPV16 positive squamous cell and adenocarcinoma of the cervix to determine if HPV16, the most prevalent cause of cervical cancer, was associated with the candidate genes across histologic types. We investigated whether genetic polymorphisms in cytokine genes involved in adaptive immunity, specifically genes important in the Th1, Th2 and Th17 immune response pathway, are associated with cervical and vulvar cancer risk in a population-based case-control study in western Washington state, in the northwest US.
Methods
Study population and data collection
The present analysis utilized data from participants within a large population-based case-control study aimed at understanding the role of host and environmental factors in anogenital cancers. The recruitment procedures, response rates, and details of sample collection have been described elsewhere,[20-22] and are briefly summarized here. Eligible cases were women 18-74 years of age diagnosed between 1986 and 2004 with incident cervical or vulvar cancers (invasive SCC of the cervix, ICDO codes 8010-8081; invasive or in situ AC, ICDO codes 8140-8480; and invasive or in situ SCC of the vulva, ICDO codes 8010, 8070-8077, 8081) while residing in Western Washington state. Case women were identified through the Cancer Surveillance System, a population-based cancer registry that is part of the NCI Surveillance Epidemiology and End Results (SEER) program.[23] Controls were residents of the same region as the cases and were selected through random digit telephone dialing. The controls were frequency matched to the distribution of age at diagnosis for the case groups, in 5-year age intervals. The current report is restricted to white females (91.3%) that had reported at least one sexual partner (99.3%). This study was approved by the Institutional Review Board at the Fred Hutchinson Cancer Research Center, Seattle.
Laboratory Methods
Participants provided peripheral blood samples at the time of interview, from which DNA was extracted. A total of 667 tagging single nucleotide polymorphisms (tagSNPs) were selected from 52 genes in the Th1, Th2 and Th17 families of cytokines (pairwise r2≥ 0.80 used to delineate highly correlated SNPs), with MAF > 5% used for each gene. We used the Snagger algorithm applied to the CEU HapMap population and the Seattle SNPs Variation Discovery Resource to select tagging SNPs.[24] For most groups of correlated SNPs, one tagSNP was selected, however, wherever necessary, we selected multiple SNPs per group to guard against missed coverage due to genotyping failure. For each gene or gene region, the chromosomal location for choosing tagSNPs extended 4kb beyond the 3’ and the 5’ ends to capture variation that may influence gene function.
Genotyping and quality control measures were followed as reported previously.[17, 19] Briefly, Illumina Golden Gate platform (Illumina Inc., San Diego, CA) was used to type the 667 SNPs. Although two SNPs had a Hardy-Weinberg equilibrium (HWE) p-value below 0.05, none had a HWE p-value below 0. 00004 (0.05/1288, total number of passing SNPs assayed on the platform, including those not in this study) and hence none of the SNPs were excluded based on HWE. A subset of the 94 failed SNPs on the Illumina platform was assayed using the KBioscience allele-specific PCR system (KBioscience, Hoddesdon, UK) or TaqMan assays (Life Technologies, Grand Island, NY), leading to a total of 560 SNPs, across all three platforms, included for analyses. Data quality control was achieved through visual inspection of the signal intensity plots by two independent readers. Any SNPs with call frequency below 0.9 were excluded. Genotype data were also evaluated for completeness. There was 1 SNP on the Illumina platform and 8 SNPs on the Taqman platform with more than 5% missing data. Two out of these nine SNPs (IL-10RA rs2229113 and IL-12B rs2569254) were significantly associated with AC of the cervix, as noted in the results.
Our group has previously published on the risk of cervical and vulvar cancers and genetic variation in some of the genes included in the present analyses (IL-2, IL-10, IL-12, TNF and their associated receptors in relation to risk of SCC and AC of cervix as well as SCC of vulva; CSF2, IL-3, IL4, IL13 and associated receptors in relation to risk of SCC of cervix).[16-19] Associations described in prior publications from this dataset are clearly marked as previously published in the tables. These data are presented again for comparison to results restricted to HPV16-positive subgroups, and the new analyses are the focus of this report. New to this manuscript are the following genes and associated receptors (IFNG, IFNG-R1, IFNG-R2, IL17A, IL17B, IL17F, IL17RA, IL17RB, IL18, IL18BP, IL18R1, IL18RAP, IL1A, IL1B, IL1R1-IL1RL2, IL1R2, IL1RL1, IL1RN, IL23A, IL23R, IL25[IL17E], IL27, IL27RA, IL2RA, IL6, IL6R, TGFB2, TGFB1, TGFB2, TGFB3, TGFBR1, TGFBR2).
Presence and type of HPV DNA in paraffin-embedded tumor tissue from biopsies or surgical specimens was previously assessed for cervical and vulvar tumors. We used polymerase chain reaction (PCR) methods to detect HPV DNA in tumors as previously described.[20, 25] Briefly, PCR was performed using MY09/MY11 L1 consensus primers and HPV 16 and HPV18 E6 primers. The identity of the PCR products was confirmed by Southern hybridization, and the L1 consensus products were typed by restriction fragment analysis. Additionally, coamplification of a 536-bp or a 268-bp fragment of the human β-globin gene was conducted to assess specimen adequacy for presence of human DNA. There were HPV DNA PCR results available for 1006 (80%) of the 1258 case subjects included in this analysis.
Statistical analysis
The main analyses comprised of 901 cervical cancers (44% SCC (n=399) and 56% AC (n=502)), 357 cases of HPV-positive vulvar SCC, and each of these case groups was compared with 1109 population-based controls. In secondary analyses, we examined subgroups that had HPV16 positive tumors. We found 158 (158/399, 39.6%) of the cervical SCC tumors had HPV16 in their tumor tissue. A lesser proportion of AC (136/502, 27.1%) were observed to contain HPV16. 85.33% of vulvar cancers were HPV16 positive in our data, hence we looked at all HPV-positive cancers for vulvar SCC. The demographic characteristics between these various subgroups were tested for statistically significant differences using ANOVA or chi-square tests, as appropriate.
Unconditional logistic regression analysis was used to estimate per-allele odds ratios (OR) and 95% confidence intervals (95% CI) for the association of each SNP with all the case groups using a log additive model. Age-adjusted risk estimates were obtained. For SNPs with very low counts of the homozygous variant (less than 5) in either the case or control group, genotypes were modeled assuming a dominant model. P-values less than 0.05 were considered to be significant. The Holm step-down procedure was applied to correct for multiple comparisons on a per-gene basis for each cancer phenotype, and the corrected values reported. All statistical analyses were performed using Stata version 13 (StataCorp, College Station, TX).
Results
Selected demographic characteristics of study cases and controls are presented in Table 1. The mean ages of women presenting with different sub-types of cervical cancers were different (pANOVA<0.001); women presenting with AC of cervix were relatively younger (38.4 years) that those presenting with SCC of cervix (42.6 years). Vulvar carcinoma cases were older (47.2 years) compared to controls (45.6 years) and cervical cancer cases. Compared with control subjects, women with SCC of the cervix and vulva were less educated (pchi-sq<0.001) and more likely to have more than 5 lifetime sexual partners (pchi-sq<0.001). Women with vulvar carcinoma were more likely to be current smokers (60.8%) than both controls (20.7%) and cervical cancer cases (27.3%) (pchi-sq<0.001). Smoking habits and educational attainment for women with AC of the cervix was more similar to controls than to women with SCC of the cervix.
Table 1.
Selected characteristics of women with cervical & vulvar cancers, and controls, Seattle-Puget Sound Region, 1986-2004
| Characteristic | Controls N = 1109(%) | Cases |
||||
|---|---|---|---|---|---|---|
| Squamous cell carcinoma (SCC) |
Adenocarcinoma (AC) |
|||||
| SCC of the cervix | HPV16+ SCCa of the cervix | Vulvar HPV+ SCC | AC of the cervix | HPV16+ ACb of the cervix | ||
| N = 399(%) | N= 158 (%) | N = 357(%) | N = 502(%) | N = 136 (%) | ||
| Age | 45.6 (14.3) | 42.6 (11.7) | 41.9 (11.3) | 47.2 (13.4) | 38.4 (10.3) | 37.8 (10.5) |
| Smoking status | ||||||
| Never | 576 (51.9) | 150 (37.6) | 54 (34.2) | 64 (17.9) | 266 (53.0) | 63 (46.3) |
| Former | 303 (27.3) | 109 (27.3) | 38 (24.1) | 76 (21.3) | 130 (25.9) | 40 (29.4) |
| Current | 230 (20.7) | 140 (35.1) | 66 (41.8) | 217 (60.8) | 106 (21.1) | 33 (24.3) |
| Lifetime sexual partners | ||||||
| 1 | 295 (26.6) | 37 (9.3) | 14 (8.9) | 24 (6.7) | 58 (11.6) | 14 (10.3) |
| 2-4 | 344 (31.0) | 122 (30.6) | 52 (32.9) | 74 (20.7) | 134 (26.7) | 39 (28.7) |
| 5+ | 470 (42.4) | 240 (60.2) | 92 (58.2) | 259 (72.6) | 310 (61.8) | 83 (61.0) |
| Education | ||||||
| High school or less | 327 (29.5) | 157 (39.4) | 67 (42.4) | 150 (42.0) | 126 (25.1) | 32 (23.5) |
| College or more | 782 (70.5) | 242 (60.7) | 91 (57.6) | 207 (58.0) | 376 (74.9) | 104 (76.5) |
118 (29.6%) of squamous cell carcinomas did not have a HPV result
134 (26.7%) of adenocarcinomas did not have a HPV result
c168 (31.8%) of vulvar cancers did not have a HPV result
Of the 667 genotyped SNPs, 94 failed to amplify and 13 were found to be monomorphic and were excluded from further analysis. The results for the remaining 560 SNPs are described by pathway. Of the 318 SNPs from the Th1 pathway genes and their receptors that were evaluated, 64 SNPs were significantly associated with either cervical or HPV-positive vulvar cancers or both (Table 2) before correction for multiple testing, and 13 SNPs within CSF2, IL-12B and TNF genes remained significant after correction for multiple testing. Results not previously published include a missense C/T variant in CSF2 gene (rs25882) that displayed a significant association with HPV-positive vulvar cancers (OR 1.28, 95% CI 1.04-1.57; corrected p= 0.03), and an A/G variant in the intergenic SNP rs27438 for CSF2 gene (OR 1.26, 95% CI 1.03-1.54; corrected p=0.02). A 40% increased risk of HPV-positive vulvar cancers was also observed for minor allele-containing genotypes of two intronic SNPs in IL-12B gene: rs2569254 (OR 1.41, 95% CI 1.13-1.76; corrected p= 0.03) and rs3181225 (OR 1.40, 95% CI 1.13-1.73; corrected p=0.02). Several tagSNPs in the TNF-gene were also found to be associated with cervical and vulvar cancer risk even after multiple test correction, three of which have not been previously reported by our group. The minor allele (G) of rs2229094 missense variant in the TNF-gene was significantly associated with a decreased risk of HPV16 positive cervical cancer (OR 0.61, 95% CI 0.45-0.83; corrected p= 0.02). We also found a 23% decreased risk for cervical adenocarcinomas associated with the minor allele (A) of rs915654 in the TNF-gene (OR 0.77, 95% CI 0.65-0.91; corrected p = 0.02). Finally, a 65% increased risk of HPV-positive vulvar cancer was seen with the minor allele (A) of rs769178 in the TNF region (OR 1.65, 95% CI 1.20-2.26; corrected p= 0.01).
Table 2.
Statistically significant associations between tag SNP's in the Th1 pathway and cervical and vulvar cancer risk, Seattle-Puget Sound Region, 1986-2004
| Gene | Site | Cancer type | SNP | Functiond | Allele | Age-adjusted OR (95% CI) | p-value |
|---|---|---|---|---|---|---|---|
| CSF2a | Vulva | SCC (HPV+) | rs25882 | missense | C/T | 1.28(1.04,1.57) | 0.0187** |
| rs27438 | intergenic | A/G | 1.26(1.03,1.54) | 0.0268** | |||
| CSF2RBa,e | Cervix | SCC (all subtypes) | rs16997517 | missense | C/T | 0.37(0.17,0.82) | 0.0146 |
| Cervix | AC (all subtypes) | rs131840 | coding sequence | C/T | 0.79(0.63,0.97) | 0.0273 | |
| rs738149 | intronic | A/G | 0.79(0.65,0.95) | 0.0131 | |||
| Vulva | SCC (HPV+) | rs16997517 | missense | C/T | 0.32(0.14,0.76) | 0.0092 | |
| rs738149 | intronic | A/G | 1.22(1.00,1.48) | 0.0457 | |||
| rs16845 | missense | C/G | 0.64(0.42,0.98) | 0.0413 | |||
| IL1B | Cervix | AC (all subtypes) | rs1071676 | UTR-3 | C/G | 0.80(0.66,0.97) | 0.0212 |
| rs1143634 | coding sequence | C/T | 0.81(0.67,0.98) | 0.027 | |||
| IL1R1-IL1RL2 | Cervix | SCC (all subtypes) | rs3917273 | intronic | A/T | 0.81(0.69,0.97) | 0.0179 |
| Cervix | AC (all subtypes) | rs7583215 | intronic | C/T | 1.31(1.06,1.61) | 0.0122 | |
| IL1R2 | Cervix | AC (HPV16 only) | rs2072475 | intronic | A/G | 0.57(0.36,0.90) | 0.0168 |
| rs2072476 | intronic | A/G | 0.51(0.29,0.89) | 0.0186 | |||
| rs719250 | intronic | A/G | 0.51(0.28,0.90) | 0.0216 | |||
| Cervix | AC (all subtypes) | rs2072475 | intronic | A/G | 0.74(0.59,0.93) | 0.0095 | |
| IL1RN | Cervix | SCC (all subtypes) | rs9005 | UTR-3 | A/G | 0.80(0.66,0.97) | 0.0209 |
| IL18 | Cervix | SCC (HPV16 only) | rs187238 | nearGene-5 | C/G | 0.75(0.56,1.00) | 0.0486 |
| rs1946519 | nearGene-5 | A/C | 0.75(0.58,0.97) | 0.0305 | |||
| rs5744222 | nearGene-5 | A/C | 1.32(1.01,1.72) | 0.0401 | |||
| IL18BP | Cervix | AC (HPV16 only) | rs1892919 | intronic | C/T | 1.71(1.03,2.82) | 0.037 |
| IL12Ab,f | Vulva | SCC (HPV+) | rs2243154 | intronic | A/G | 1.37(1.04,1.81) | 0.0254 |
| IL12Bb,f | Cervix | AC (all subtypes) | rs10052709 | intronic | C/G | 0.72(0.56,0.93) | 0.0103 |
| rs2569254c | intronic | C/T | 1.23(1.01,1.50) | 0.0438 | |||
| rs3181225 | intergenic | C/T | 1.26(1.04,1.53) | 0.0202 | |||
| rs3213119 | missense | G/T | 0.52(0.29,0.94) | 0.0294 | |||
| Vulva | SCC (HPV+) | rs2569254c | intronic | C/T | 1.41(1.13,1.76) | 0.0024** | |
| rs3181225 | intergenic | C/T | 1.40(1.13,1.73) | 0.0021** | |||
| rs6870828 | intergenic | C/T | 1.23(1.03,1.47) | 0.0218b | |||
| IL12RB2b,f | Cervix | AC (all subtypes) | rs2307153 | missense | A/G | 1.78(1.05,3.00) | 0.0314 |
| IL2RAb,g | Cervix | SCC (all subtypes) | rs2228150 | coding sequence | A/G | 1.91(1.22,2.99) | 0.0047 |
| rs3134883 | intronic | C/T | 1.20(1.00,1.44) | 0.0462 | |||
| Cervix | SCC (HPV16 only) | rs2228150 | coding sequence | A/G | 2.11(1.15,3.86) | 0.0152 | |
| rs10905656 | intronic | A/C | 0.71(0.55,0.92) | 0.0106 | |||
| rs12722516 | intronic | C/T | 1.96(1.11,3.45) | 0.0202 | |||
| rs6602379 | intronic | A/G | 0.71(0.54,0.94) | 0.0174 | |||
| rs7072398 | intronic | A/G | 0.74(0.58,0.95) | 0.0196 | |||
| rs7910961 | intronic | C/T | 0.68(0.51,0.90) | 0.0066 | |||
| g | Cervix | AC (all subtypes) | rs12722588 | intronic | A/G | 1.41(1.03,1.93) | 0.033 |
| rs1323658 | intronic | 0.36(0.15,0.84) | 0.0185 | ||||
| rs2104286 | intronic | A/G | 1.33(1.01,1.76) | 0.0457 | |||
| Cervix | AC (HPV16 only) | rs10905669 | intronic | C/T | 1.24(1.01,1.51) | 0.0351 | |
| rs12722486 | intronic | A/G | 0.59(0.37,0.94) | 0.0274 | |||
| IL2RBb | Cervix | AC (HPV16 only) | rs3218253 | intronic | C/T | 1.34(1.01,1.77) | 0.0435 |
| IL3a | Vulva | SCC (HPV+) | rs181781 | nearGene-5 | A/G | 1.43(1.07,1.91) | 0.0142 |
| rs31481 | intronic | A/G | 1.33(1.07,1.66) | 0.0119 | |||
| rs40401 | missense | C/T | 1.29(1.05,1.58) | 0.0133 | |||
| IFNG | Cervix | AC (all subtypes) | rs2069716 | intronic | A/G | 0.66(0.45,0.97) | 0.0368 |
| TNFb,h | Cervix | SCC (all subtypes) | rs1052248 | nearGene-3 | A/T | 0.74(0.61,0.89) | 0.0019** |
| rs1799964 | nearGene-5 | C/T | 0.65(0.52,0.81) | 0.0001** | |||
| rs2009658 | nearGene-5 | C/G | 0.63(0.49,0.81) | 0.0003** | |||
| rs2229094 | missense | C/T | 0.66(0.54,0.81) | 0.0001** | |||
| rs2239704 | UTR-5 | G/T | 1.31(1.10,1.55) | 0.0019** | |||
| rs2256965 | intronic | A/C/T | 1.22(1.04,1.44) | 0.0174 | |||
| rs3093662 | intronic | A/G | 0.69(0.48,0.98) | 0.0381 | |||
| rs915654 | nearGene-5 | A/T | 0.81(0.68,0.97) | 0.0192 | |||
| Cervix | SCC (HPV16 only) | rs1052248 | nearGene-3 | A/T | 0.70(0.53,0.93) | 0.0149 | |
| rs1799964 | nearGene-5 | C/T | 0.65(0.47,0.90) | 0.0091 | |||
| rs2009658 | nearGene-5 | C/G | 0.57(0.38,0.87) | 0.0085 | |||
| rs2229094 | missense | C/T | 0.61(0.45,0.83) | 0.0017** | |||
| rs2239704 | UTR-5 | G/T | 1.35(1.05,1.72) | 0.0179 | |||
| rs915654 | nearGene-5 | A/T | 0.74(0.57,0.97) | 0.0288 | |||
| h | Cervix | AC (all subtypes) | rs1799964 | nearGene-5 | C/T | 0.80(0.66,0.97) | 0.0261 |
| rs2229094 | missense | C/T | 0.80(0.67,0.96) | 0.0172 | |||
| rs2239704 | UTR-5 | G/T | 1.30(1.10,1.52) | 0.0016** | |||
| rs2256965 | intronic | A/C/T | 1.20(1.03,1.41) | 0.0199 | |||
| rs915654 | nearGene-5 | A/T | 0.77(0.65,0.91) | 0.0019** | |||
| Cervix | AC (HPV16 only) | rs2256965 | intronic | A/C/T | 1.34(1.04,1.73) | 0.0241 | |
| rs915654 | nearGene-5 | A/T | 0.75(0.57,0.99) | 0.0455 | |||
| h | Vulva | SCC (HPV+) | rs1052248 | nearGene-3 | A/T | 0.74(0.60,0.90) | 0.0028** |
| rs1799964 | nearGene-5 | C/T | 0.77(0.62,0.96) | 0.021 | |||
| rs2009658 | nearGene-5 | C/G | 0.71(0.54,0.93) | 0.0137 | |||
| rs2229094 | missense | C/T | 0.71(0.58,0.88) | 0.0013** | |||
| rs2239704 | UTR-5 | G/T | 1.58(1.33,1.88) | <0.0001** | |||
| rs2256965 | intronic | A/C/T | 1.49(1.25,1.77) | <0.0001** | |||
| rs915654 | nearGene-5 | A/T | 0.65(0.53,0.78) | <0.0001** | |||
| rs1800610 | intronic | C/T | 1.52(1.11,2.08) | 0.0093 | |||
| rs769178 | intergenic | A/C | 1.65(1.20,2.26) | 0.0019** | |||
| TNFRSF1Ab,h | Cervix | SCC (all subtypes) | rs4149584 | missense | A/C/G/T | 0.44(0.20,0.99) | 0.0481 |
| Cervix | SCC (HPV16 only) | rs1800693 | intronic | A/G | 1.33(1.04,1.70) | 0.0242 | |
| rs4149575 | intronic | A/C | 0.55(0.32,0.95) | 0.0322 | |||
| rs767455 | coding sequence | C/T | 1.30(1.01,1.66) | 0.0386 | |||
| TNFRSF1Bb | Cervix | AC (HPV16 only) | rs1061631 | UTR-3 | A/G | 1.50(1.11,2.03) | 0.0078 |
| rs1148458 | intronic | C/T | 1.62(1.01,2.60) | 0.0469 | |||
| rs976881 | intronic | A/G | 1.38(1.06,1.81) | 0.0183 | |||
| h | Vulva | SCC (HPV+) | rs1061631 | UTR-3 | A/G | 1.24(1.01,1.53) | 0.0443 |
| rs976881 | intronic | A/G | 1.21(1.01,1.45) | 0.0377 | |||
| rs17882988 | intronic | A/G | 1.26(1.00,1.58) | 0.0466 | |||
| rs499646 | intronic | C/T | 0.62(0.39,0.97) | 0.0365 | |||
| rs652625 | nearGene-5 | A/T | 0.65(0.43,0.99) | 0.0463 |
Highlighted portions indicate previously published results from our study population; references are noted for prior reports.
Significant after correction for multiple testing using Holm's stepdown procedure
Results for squamous cell carcinoma of the cervix previously published by our group
Results for squamous cell carcinoma (all), adenocarconoma (all) and HPV+ vulvar cancer previously published by our group
rs2569254 had 6.1% genotype data
intronic denotes that the SNP is located in the intronic region of the gene (except in the first 2 or last 2 bases), intergenic denotes that the SNP is located in between two genes, nearGene-3 denotes the SNP is located near the 3’ end, nearGene-5 denotes the SNP is located near the 5’ end, UTR-3 denotes that the SNP is located in the untranslated 3’ region, UTR-5 denotes that the SNP is located in the untranslated 5’ region, missense denotes that the SNP is present in the coding region and causes a change in the coded peptide, and coding sequence denotes that the SNP is present in the coding region and does not cause a change in the peptide coded.
See reference 17 (Johnson et. al.)
See reference 18 (Hussain et. al.)
See reference 19 (Hussain et. al.)
See reference 16 (Bodelon et. al.)
We tested 245 SNPs in Th2 genes for their associations with the five case groups for cervical and vulvar cancers. There were 22 significant associations between single SNPs and the five case groups. Except for the significant associations with IL-10 and its associated receptors that we previously reported, none had been previously looked at by our group. However, none of these associations remained statistically significant after correction for multiple testing (Table 3).
Table 3.
Statistically significant associations between tag SNP's in the Th2 pathway and cervical and vulvar cancer risk, Seattle-Puget Sound Region, 1986-2004
| Gene | Site | Cancer type | SNP | Function | Allele | Age-adjusted OR (95% CI) | p-value |
|---|---|---|---|---|---|---|---|
| IL4R | Vulva | SCC (HPV+) | rs3024668 | nearGene-5 | A/G | 0.59(0.37,0.95) | 0.0304 |
| IL6 | Cervix | SCC (all subtypes) | rs2069840 | Intronic | C/G | 1.25(1.05,1.49) | 0.0137 |
| Cervix | SCC (HPV16 only) | rs2069840 | Intronic | C/G | 1.32(1.02,1.70) | 0.0327 | |
| rs2069837 | Intronic | A/G | 0.42(0.21,0.81) | 0.0101 | |||
| IL10a | Cervix | SCC (HPV16 only) | rs3024509 | Intronic | C/T | 0.49(0.26,0.93) | 0.0293 |
| Cervix | AC (HPV16 only) | rs3024509 | Intronic | C/T | 0.48(0.24,0.95) | 0.0337 | |
| rs1800894 | nearGene-5 | A/G | 0.34(0.12,0.94) | 0.0376 | |||
| IL10RAa,c | Cervix | AC (all subtypes) | rs2229113b | Missense | A/G | 1.21(1.02,1.43) | 0.0304 |
| rs2256111 | coding sequence | A/G | 1.19(1.01,1.39) | 0.0326 | |||
| rs2512143 | Intronic | A/G | 1.23(1.05,1.45) | 0.0129 | |||
| rs2512148 | Intergenic | A/C | 1.22(1.04,1.44) | 0.0172 | |||
| IL10RBa | Cervix | AC (HPV16 only) | rs2843701 | Intronic | C/T | 0.75(0.57,0.98) | 0.0327 |
| TGFB1 | Cervix | SCC (all subtypes) | rs11466338 | Intronic | A/G | 1.48(1.05,2.09) | 0.0243 |
| TGFB2 | Cervix | SCC (HPV16 only) | rs17047804 | Intronic | C/T | 1.55(1.06,2.26) | 0.0237 |
| rs2000220 | Intronic | A/G | 1.29(1.01,1.65) | 0.0407 | |||
| Cervix | AC (all subtypes) | rs4846476 | Intronic | C/G | 0.81(0.67,0.99) | 0.0396 | |
| TGFB3 | Cervix | SCC (all subtypes) | rs3917148 | Intronic | A/C | 1.36(1.00,1.85) | 0.0494 |
| Vulva | SCC (HPV+) | rs3917200 | Intronic | C/T | 1.44(1.03,2.01) | 0.0347 | |
| TGFBR1 | Cervix | SCC (all subtypes) | rs10512263 | Intronic | C/T | 0.66(0.46,0.94) | 0.0226 |
| Cervix | SCC (HPV16 only) | rs10512263 | Intronic | C/T | 0.51(0.29,0.91) | 0.0236 | |
| TGFBR2 | Cervix | AC (HPV16 only) | rs9858487 | Intronic | C/G | 1.69(1.00,2.83) | 0.0481 |
| Vulva | SCC (HPV+) | rs3087465 | nearGene-5 | A/G | 1.23(1.00,1.51) | 0.0451 |
Highlighted portions indicate previously published results from our study population.
Results for squamous cell carcinoma (all), adenocarconoma (all) and HPV+ vulvar cancer previously published by our group
rs2229113 had 5.1% missing genotype data
See reference 18 (Hussain et. al.)
For Th17, we tested 69 SNPs in the IL-17, IL-23 and IL-25 gene families and their receptors to evaluate the association between variation in these gene families and risk for cervical and vulvar cancers. We observed that 14 SNPs showed a significant association with cervical or vulvar cancers before multiple testing correction (Table 4). Only one remained significant after correction for multiple testing: the (T) allele of rs879576 in the coding sequence of the IL-17RA gene was associated with HPV16 positive SCC of the cervix (OR 1.91, 95% CI 1.30-2.79; corrected p= 0.018). The full results for genotype associations of all the 560 SNPs typed for candidate genes within the three Th groups can be found in Supplementary Tables 1-5.
Table 4.
Statistically significant associations between tag SNP's in the Th17 pathway and cervical and vulvar cancer risk, Seattle-Puget Sound Region, 1986-2004
| Gene | Site | Cancer type | SNP | Function | Allele | Age-adjusted OR (95% CI) | p-value |
|---|---|---|---|---|---|---|---|
| IL17A | Cervix | SCC (all subtypes) | rs8193036 | nearGene-5 | C/T | 0.80(0.66,0.97) | 0.0231 |
| Cervix | SCC (HPV16 only) | rs8193036 | nearGene-5 | C/T | 0.74(0.55,1.00) | 0.0473 | |
| IL17F | Cervix | SCC (all subtypes) | rs11465549 | intronic | A/G | 1.72(1.07,2.77) | 0.0265 |
| Cervix | AC (all subtypes) | rs12201582 | intronic | A/C | 1.26(1.03,1.54) | 0.0219 | |
| Cervix | AC (HPV16 only) | rs11465549 | intronic | A/C | 2.09(1.04,4.22) | 0.0395 | |
| Vulva | SCC (HPV+) | rs11465549 | intronic | 1.63(1.00,2.65) | 0.0489 | ||
| IL17RA | Cervix | SCC (all subtypes) | rs2241049 | intronic coding | A/G | 0.83(0.70,0.99) | 0.0419 |
| rs879576 | sequence | C/T | 1.38(1.05,1.83) | 0.0225 | |||
| Cervix | SCC (HPV16 only) | rs5992628 | UTR-3 | G/T | 0.77(0.60,0.99) | 0.0408 | |
| rs879574 | intronic coding | A/T | 1.59(1.09,2.34) | 0.0172 | |||
| rs879576 | sequence | C/T | 1.91(1.30,2.79) | 0.001** | |||
| Cervix | AC (all subtypes) | rs4819956 | intergenic coding | A/C | 1.27(1.07,1.50) | 0.0065 | |
| rs879576 | sequence | C/T | 1.31(1.03,1.67) | 0.0289 | |||
| Cervix | AC (HPV16 only) | rs17807076 | intronic | C/T | 0.64(0.47,0.88) | 0.0065 | |
| rs4819956 | intergenic coding | A/C | 1.37(1.04,1.80) | 0.026 | |||
| rs879576 | sequence | C/T | 1.57(1.03,2.41) | 0.0361 | |||
| Vulva | SCC (HPV+) | rs879574 | intronic | A/T | 1.31(1.01,1.70) | 0.0402 | |
| IL23R | Cervix | AC (all subtypes) | rs6588250 | intronic | G/T | 1.21(1.01,1.44) | 0.0386 |
| rs7528924 | intronic | A/G | 1.23(1.03,1.48) | 0.0258 | |||
| rs790633 | intronic | C/T | 1.21(1.03,1.44) | 0.0243 | |||
| Cervix | AC (HPV16 only) | rs6693831 | intronic | C/T | 1.42(1.06,1.89) | 0.0182 | |
| Vulva | SCC (HPV+) | rs10889675 | intronic | A/C | 1.39(1.09,1.78) | 0.008 | |
| rs6693831 | intronic | C/T | 1.30(1.07,1.58) | 0.0084 |
Highlighted portions indicate previously published results from our study population.
Significant after correction for multiple testing using Holm's stepdown procedure
Discussion
In this population-based case-control study, we assessed genetic variation in Th1, Th2 and Th17 immune response associated with HPV-related cervical and vulvar cancers. To our knowledge, this is one of the first investigations of genetic variation in the Th17 genes and cervical and vulvar cancer risk. We also observed associations between SNPs within genes in the Th1 (CSF2, IL-12B and TNF) pathway among subgroups of cervical and vulvar cancer that remained statistically significant even after multiple test correction.
Results from two genome-wide association studies (GWAS) for susceptibility loci for cervical cancer have been recently published.[26, 27] One Swedish study identified three novel independent loci within the major histocompatibility complex (MHC) region contributing to susceptibility to cervical cancers.[27] MHC class I and II molecules play a critical role in antigen presentation to the T-cells during immune response and, therefore, variations in these molecules that are encoded in DNA may affect immune activation against HPV-infected cells.[28] The GWAS results are consistent with the results of a comprehensive study of class I and II alleles in the HLA region for SCC[29] and AC[30] of the cervix by our group. A different GWAS, conducted in Chinese women, also identified two new loci in the 4q12 and 17q12 regions.[26] The most significant SNP in the 17q12 region (rs8067378) is located near the Gasdermin B (GSDMB) gene which is regulated by TGF-β signaling.[31] Variation in the TGF-β genes was related to cervical and vulvar risk in this study, and are a triggering factor for the Th17 immune response pathway.[32] Together, these studies support further evaluating the role of immune response pathways in relation to cervical and vulvar cancers. Although the two GWAS studies identified loci that are related to immune response, they may have missed identifying important variants due to agnostic approaches that rely on high thresholds for significance. Hence, there is some merit to employing candidate gene approaches that may be helpful in identifying some of these missed associations.
Findings of our study not previously reported included statistically significant association (after correction) for the synonymous (T) allele in SNP rs879576 in the IL17RA gene, belonging to the Th17 immune response pathway, with an increased risk for HPV16-positive SCC of the cervix (OR 1.91, 95% CI 1.30-2.79; corrected p= 0.018). In another candidate study, Quan et. al. evaluated the risk of cervical SCC associated with just two Th17 pathway SNPs: one in the IL-17A (rs2275913) and the other in IL-17F (rs763780) gene. They reported an increased risk with the IL-17A gene polymorphism but not with the IL-17F SNP.[33] Genetic variation in Th17 genes has been previously associated with cancers of the breast and the stomach[34, 35], but the exact role of IL-17 and its receptors in the development of cervical cancer has not yet been explored in detail. Our results suggest that genetic variation in the Th17 pathway contributes to cervical carcinogenesis, and that variation in IL17RA warrants replication in other populations as it was associated with HPV16-positive squamous cell cancer and adenocarcinoma of the cervix and HPV-positive vulvar cancer. If supported by functional evaluation, this observation may lead to new approaches to immunotherapy for HPV-related cancers.
Previous candidate gene studies have also suggested that genetic polymorphisms in Th1 and Th2 genes may affect susceptibility to HPV infections.[36] Recently, a meta-analysis involving 58 genetic variants in 25 genes, including IFN-G, IL-1B, IL-10 and TNF genes in Th1 and Th2 pathways, reported significant associations for risk of cervical cancer.[37] A moderately strong association (OR>1.5) was observed for specific polymorphisms in the IL-1B and TNF genes.[37] Although, these particular SNPs were not evaluated in our study, we did find significant associations with other SNPs in these genes as presented in Table 2. In another study, Chen et. al.[38] evaluated the risk associated with 3 candidate polymorphisms in the IL-12A and IL-12B genes and found IL-12A rs568408 and IL-12B rs3212227 to be significantly associated with cervical cancers in Chinese women. That study had limited power (400 cases; 400 controls) and did not correct for multiple comparisons. In our study, we did not find any significant associations between the IL-12B rs3212227 (or rs3212220 which is in linkage disequilibrium with it; r2= 0.95) and cervical cancer risk, although the direction of our results was similar to those from Chen et. al. A Swedish study also utilized the candidate gene approach to test associations between some SNPs in IL-4R, IL-6 and IL-10, and risk of cervical cancer, and found significant associations with a SNP (rs1800797) in the IL-6 gene without multiple testing correction.[39] Although we did not test for rs1800797, we evaluated rs1800795 which is in linkage disequilibrium (r2 = 0.97) with rs1800797 and did not find statistically significant associations with cervical or vulvar cancer. Our results, however, were similar in magnitude and direction to the Swedish study. Finally, a Taiwanese study evaluated five candidate polymorphisms in IL-18 gene and risk of cervical SCC.[40] Similar to our findings, they did not find statistically significant association between SNPs in IL-18 gene with cervical SCC risk.
There are some limitations to this study. As there were five case groups and 560 SNP evaluated in this study, some of the findings may be attributable to chance. We tried to minimize this possibility by correcting for multiple comparisons and focusing our interpretation on findings that remained statistically significant after correction. As our study was restricted to white women, our results may not be applicable to other racial/ethnic population groups. Further, restriction to white women may not have removed all potential confounding by population stratification. Replication of our results is required in larger studies with higher statistical power, especially for the novel associations within the Th17 pathway. Our study has notable strengths including the population-based recruitment of cases and controls, HPV testing of tumor samples, the ability to focus on HPV16 related tumors, and the tagSNP approach allowed for efficient but representative coverage of each of the selected candidate genes.
In conclusion, our study provides some evidence that common genetic variation in the immune response pathways, particularly Th1 and Th17, may be important in the development and progression of HPV-related cervical and vulvar carcinomas. While these findings are interesting and may point to real signals in the genes studied, there is a possibility that these associations may be proxy for other genes that are in linkage disequilibrium with the candidate genes we examined. Therefore, further studies that involve deeper sequencing or functional evaluation are necessary to assess the impact of genetic variability in immune response pathways in cervical and vulvar cancer etiology.
Supplementary Material
Highlights.
Genetic variation in acquired immune response to human papillomavirus (HPV) may contribute to clearance or progression.
Variation in Th1/Th17 pathway genes were associated with cervical and vulvar cancers, especially with HPV16 positive tumors.
Novel findings are presented for IL17 and its ligands and their association with cervical and vulvar cancers.
Acknowledgements
This work was supported by grants from the National Cancer Institute: R01 CA112512-0, P01 CA42792and R25 CA094880.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors do not have any potential conflicts of interest to declare.
REFERENCES
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–5. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 3.Wang SS, Sherman ME, Hildesheim A, Lacey JV, Jr., Devesa S. Cervical adenocarcinoma and squamous cell carcinoma incidence trends among white women and black women in the United States for 1976-2000. Cancer. 2004;100:1035–44. doi: 10.1002/cncr.20064. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Simard EP, Dorell C, Noone AM, Markowitz LE, Kohler B, et al. Annual Report to the Nation on the Status of Cancer, 1975-2009, featuring the burden and trends in human papillomavirus(HPV)-associated cancers and HPV vaccination coverage levels. Journal of the National Cancer Institute. 2013;105:175–201. doi: 10.1093/jnci/djs491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 6.Scott M, Nakagawa M, Moscicki AB. Cell-mediated immune response to human papillomavirus infection. Clin Diagn Lab Immunol. 2001;8:209–20. doi: 10.1128/CDLI.8.2.209-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanley M. Immune responses to human papillomavirus. Vaccine. 2006;24(Suppl 1):S16–22. doi: 10.1016/j.vaccine.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Moscicki AB, Schiffman M, Kjaer S, Villa LL. Chapter 5: Updating the natural history of HPV and anogenital cancer. Vaccine. 2006;24(Suppl 3):S3/42–51. doi: 10.1016/j.vaccine.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Kemp TJ, Hildesheim A, Garcia-Pineres A, Williams MC, Shearer GM, Rodriguez AC, et al. Elevated systemic levels of inflammatory cytokines in older women with persistent cervical human papillomavirus infection. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:1954–9. doi: 10.1158/1055-9965.EPI-10-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong A, van Poelgeest MI, van der Hulst JM, Drijfhout JW, Fleuren GJ, Melief CJ, et al. Human papillomavirus type 16-positive cervical cancer is associated with impaired CD4+ T-cell immunity against early antigens E2 and E6. Cancer Res. 2004;64:5449–55. doi: 10.1158/0008-5472.CAN-04-0831. [DOI] [PubMed] [Google Scholar]
- 11.Gosmann C, Mattarollo SR, Bridge JA, Frazer IH, Blumenthal A. IL-17 suppresses immune effector functions in human papillomavirus-associated epithelial hyperplasia. J Immunol. 2014;193:2248–57. doi: 10.4049/jimmunol.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodman CB, Collins S, Winter H, Bailey A, Ellis J, Prior P, et al. Natural history of cervical human papillomavirus infection in young women: a longitudinal cohort study. Lancet. 2001;357:1831–6. doi: 10.1016/S0140-6736(00)04956-4. [DOI] [PubMed] [Google Scholar]
- 13.Chaturvedi AK, Madeleine MM, Biggar RJ, Engels EA. Risk of human papillomavirus-associated cancers among persons with AIDS. Journal of the National Cancer Institute. 2009;101:1120–30. doi: 10.1093/jnci/djp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madeleine MM, Finch JL, Lynch CF, Goodman MT, Engels EA. HPV-related cancers after solid organ transplantation in the United States. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013;13:3202–9. doi: 10.1111/ajt.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai T, Malech HL. WHIM syndrome: congenital immune deficiency disease. Current opinion in hematology. 2009;16:20–6. doi: 10.1097/MOH.0b013e32831ac557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodelon C, Madeleine MM, Johnson LG, Du Q, Galloway DA, Malkki M, et al. Genetic variation in the TLR and NF-kappaB pathways and cervical and vulvar cancer risk: a population-based case-control study. International journal of cancer Journal international du cancer. 2014;134:437–44. doi: 10.1002/ijc.28364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson LG, Schwartz SM, Malkki M, Du Q, Petersdorf EW, Galloway DA, et al. Risk of cervical cancer associated with allergies and polymorphisms in genes in the chromosome 5 cytokine cluster. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20:199–207. doi: 10.1158/1055-9965.EPI-10-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain SK, Madeleine MM, Johnson LG, Du Q, Galloway DA, Daling JR, et al. Nucleotide variation in IL-10 and IL-12 and their receptors and cervical and vulvar cancer risk: a hybrid case-parent triad and case-control study. International journal of cancer Journal international du cancer. 2013;133:201–13. doi: 10.1002/ijc.28000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain SK, Madeleine MM, Johnson LG, Du Q, Malkki M, Wilkerson HW, et al. Cervical and vulvar cancer risk in relation to the joint effects of cigarette smoking and genetic variation in interleukin 2. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:1790–9. doi: 10.1158/1055-9965.EPI-07-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daling JR, Madeleine MM, McKnight B, Carter JJ, Wipf GC, Ashley R, et al. The relationship of human papillomavirus-related cervical tumors to cigarette smoking, oral contraceptive use, and prior herpes simplex virus type 2 infection. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1996;5:541–8. [PubMed] [Google Scholar]
- 21.Madeleine MM, Daling JR, Schwartz SM, Shera K, McKnight B, Carter JJ, et al. Human papillomavirus and long-term oral contraceptive use increase the risk of adenocarcinoma in situ of the cervix. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2001;10:171–7. [PubMed] [Google Scholar]
- 22.Madeleine MM, Daling JR, Carter JJ, Wipf GC, Schwartz SM, McKnight B, et al. Cofactors with human papillomavirus in a population-based study of vulvar cancer. Journal of the National Cancer Institute. 1997;89:1516–23. doi: 10.1093/jnci/89.20.1516. [DOI] [PubMed] [Google Scholar]
- 23.Hankey BF, Ries LA, Edwards BK. The surveillance, epidemiology, and end results program: a national resource. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1999;8:1117–21. [PubMed] [Google Scholar]
- 24.Edlund CK, Lee WH, Li D, Van Den Berg DJ, Conti DV. Snagger: a user-friendly program for incorporating additional information for tagSNP selection. BMC bioinformatics. 2008;9:174. doi: 10.1186/1471-2105-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter JJ, Madeleine MM, Shera K, Schwartz SM, Cushing-Haugen KL, Wipf GC, et al. Human papillomavirus 16 and 18 L1 serology compared across anogenital cancer sites. Cancer Res. 2001;61:1934–40. [PubMed] [Google Scholar]
- 26.Shi Y, Li L, Hu Z, Li S, Wang S, Liu J, et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nature genetics. 2013;45:918–22. doi: 10.1038/ng.2687. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, Juko-Pecirep I, Hammer J, Ivansson E, Enroth S, Gustavsson I, et al. Genome-wide association study of susceptibility loci for cervical cancer. Journal of the National Cancer Institute. 2013;105:624–33. doi: 10.1093/jnci/djt051. [DOI] [PubMed] [Google Scholar]
- 28.Mota F, Rayment N, Chong S, Singer A, Chain B. The antigen-presenting environment in normal and human papillomavirus (HPV)-related premalignant cervical epithelium. Clinical and experimental immunology. 1999;116:33–40. doi: 10.1046/j.1365-2249.1999.00826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madeleine MM, Johnson LG, Smith AG, Hansen JA, Nisperos BB, Li S, et al. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res. 2008;68:3532–9. doi: 10.1158/0008-5472.CAN-07-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Safaeian M, Johnson LG, Yu K, Wang SS, Gravitt PE, Hansen JA, et al. Human Leukocyte Antigen Class I and II Alleles and Cervical Adenocarcinoma. Frontiers in oncology. 2014;4:119. doi: 10.3389/fonc.2014.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeki N, Kim DH, Usui T, Aoyagi K, Tatsuta T, Aoki K, et al. GASDERMIN, suppressed frequently in gastric cancer, is a target of LMO1 in TGF-beta-dependent apoptotic signalling. Oncogene. 2007;26:6488–98. doi: 10.1038/sj.onc.1210475. [DOI] [PubMed] [Google Scholar]
- 32.Ghilardi N, Ouyang W. Targeting the development and effector functions of TH17 cells. Seminars in immunology. 2007;19:383–93. doi: 10.1016/j.smim.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 33.Quan Y, Zhou B, Wang Y, Duan R, Wang K, Gao Q, et al. Association between IL17 polymorphisms and risk of cervical cancer in Chinese women. Clinical & developmental immunology. 2012;2012:258293. doi: 10.1155/2012/258293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata T, Tahara T, Hirata I, Arisawa T. Genetic polymorphism of interleukin-17A and -17F genes in gastric carcinogenesis. Human immunology. 2009;70:547–51. doi: 10.1016/j.humimm.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Jiang Y, Zhang Y, Wang Y, Huang S, Wang Z, et al. Association analysis of IL-17A and IL-17F polymorphisms in Chinese Han women with breast cancer. PloS one. 2012;7:e34400. doi: 10.1371/journal.pone.0034400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hildesheim A, Wang SS. Host and viral genetics and risk of cervical cancer: a review. Virus research. 2002;89:229–40. doi: 10.1016/s0168-1702(02)00191-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Zhang L, Tian C, Yang L, Wang Z. Genetic variants and risk of cervical cancer: epidemiological evidence, meta-analysis and research review. BJOG : an international journal of obstetrics and gynaecology. 2014;121:664–74. doi: 10.1111/1471-0528.12638. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Han S, Wang S, Zhou X, Zhang M, Dong J, et al. Interactions of IL-12A and IL-12B polymorphisms on the risk of cervical cancer in Chinese women. Clin Cancer Res. 2009;15:400–5. doi: 10.1158/1078-0432.CCR-08-1829. [DOI] [PubMed] [Google Scholar]
- 39.Castro FA, Haimila K, Sareneva I, Schmitt M, Lorenzo J, Kunkel N, et al. Association of HLA-DRB1, interleukin-6 and cyclin D1 polymorphisms with cervical cancer in the Swedish population--a candidate gene approach. International journal of cancer Journal international du cancer. 2009;125:1851–8. doi: 10.1002/ijc.24529. [DOI] [PubMed] [Google Scholar]
- 40.Yang YC, Chang TY, Chen TC, Chang SC, Lin WS, Lee YJ. Genetic variants in interleukin-18 gene and risk for cervical squamous cell carcinoma. Human immunology. 2013;74:882–7. doi: 10.1016/j.humimm.2013.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.