Abstract
Successful vaccine development remains a huge challenge for infectious diseases such as malaria, HIV and influenza. As a novel way to present antigenic epitopes to the immune system, we have developed icosahedral self-assembling protein nanoparticles (SAPNs) to serve as a prototypical vaccine platform for infectious diseases. Here we examine some biophysical factors that affect the self-assembly of these nanoparticles, which have as basic building blocks coiled-coil oligomerization domains joined by a short linker region. Relying on in silico computer modeling predictions, we selected five different linker regions from the RCSB protein database that connect oligomerization domains, and then further studied the self-assembly and stability of in vitro produced nanoparticles through biophysical characterization of formed particles. One design in particular, T2i88, revealed excellent self-assembly and homogeneity thus paving the way toward a more optimized nanoparticle for vaccine applications.
From the Clinical Editor
Despite the widespread use of vaccines worldwide, successful development of vaccines against some diseases remains a challenge still. In this article, the authors investigated the physic-chemical and biological properties of icosahedral self-assembling protein nanoparticles (SAPNs), which mimic viral particles, in order to utilize this technology as potential platform for future design of vaccines.
Key words: Protein nanoparticle, Malaria, Vaccine carrier, Protein design, Self-assembly
Graphical Abstract
Using in silico computer modeling predictions, we selected five different linker regions from proteins in the RCSB protein database to connect the coiled-coil oligomerization domains in our nanoparticles, and studied the self-assembly and stability of in vitro produced nanoparticles through biophysical characterization. One design revealed excellent self-assembly and homogeneity thus paving the way toward a more optimized nanoparticle for vaccine applications.
Inspired by the numerous applications of virus capsids, virus-like particles1, 2 and capsid-like protein cages,3, 4, 5, 6 the self-assembling protein nanoparticles (SAPNs) developed in our laboratory aim to mimic the architecture of viruses and their icosahedral symmetry. Virus capsids are often composed of just one single polypeptide chain that self-assembles into a proteinaceous cover thus protecting the genomic material of the virion. In light of this information, our design has a single polypeptide chain as the building block that self-assembles into a nanoparticle. Icosahedrons have 5-fold, 3-fold and 2-fold symmetry axes. To mimic icosahedral viruses, two of these axes were included in our design by creating a fusion protein of a pentameric coiled coil that is covalently linked to a trimeric coiled coil. Modeling showed that the superposition of these two oligomerization domains onto the corresponding symmetry axes of the icosahedron and application of the symmetry elements gives rise to a nanoparticle with icosahedral symmetry. Under the right conditions, identical copies of the protein chain self-assemble to produce the nanoparticle.
In a nanoparticle with T = 1 icosahedral symmetry, 60 protein chains form the 20 triangular faces and every chain occupies an identical environment. The requirement of a minimal number of 60 asymmetric units is related to the number of symmetry-related asymmetric units in an icosahedron. With a relatively small deviation from the exact symmetry of a T = 1 icosahedron, more protein chains can be incorporated in a viral capsid. Certain multiples (1, 3, 4, etc.) of 60 subunits are allowed, leading to capsids with 60, 180, 240, … protein chains. These multiples are referred to as triangulation numbers T = 1, T = 3, T = 4, etc. Thus, in the case of the nanoparticles, it is theoretically possible to accommodate more than one protein subunit in an asymmetric unit to form nanoparticles with T = 3 or T = 4 icosahedral symmetry. However, to achieve this some of the pentameric coiled coils of the fusion protein would have to switch to form a hexamer, which is rather unlikely. According to the viral tiling theory, which includes surface representations in terms of more than one type of building block per particle,7 it is more likely that such larger assemblies are non-quasi-equivalent pentamer-only capsids, like the capsids of polyoma or papilloma viruses. For this reason, we refer to them as T = 3-like icosahedra in the context of this manuscript.
Initially, we used solid-phase peptide synthesis to produce a peptide chain composed of two oligomerization domains with different oligomerization states joined by a short linker segment.8 Biophysical characterization of the synthetic peptide nanoparticles showed that they had a diameter of about 16 nm and that there were 60 peptide chains per nanoparticle, in support of a T = 1 icosahedral model. We also showed that when SAPNs were functionalized with a fragment of the surface protein of severe acute respiratory syndrome (SARS) coronavirus, they were able to elicit conformation-specific antibodies that inhibited infection of cells by SARS viruses in vitro. 9 Likewise, we have made nanoparticles for use as repetitive antigen displays for vaccines against malaria,10, 11, 12, 13 HIV,14 toxoplasmosis15 and influenza.16 They can also be used for the design of other tools for nano-biotechnological applications.17, 18, 19
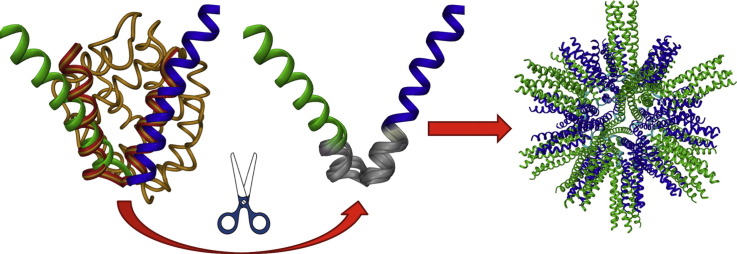
Given the potential of SAPNs as repetitive antigen display systems for the development of vaccines, we next aimed to rationally modify their building blocks in order to affect epitope density. Our goal was to find out what would happen if we modified the short linker region connecting the pentameric and the trimeric oligomerization domains of the fusion protein (Figure 1 ). This part of the fusion protein is of great importance as the packing of chains in the nanoparticle is governed by the flexibility of the linker region (Figure 1). By decreasing the angular flexibility between the pentamer and the trimer, nanoparticles with 60 or more chains could be obtained. Researchers have long been investigating the effect of linker length and composition when creating a fusion protein. In 2002, George and Heringa developed a linker database intended for the rational design of linkers for domain fusion,20 and we have previously noted that one of the factors influencing protein assembly into icosahedral nanoparticles is the length of the linker region that connects the two oligomerization domains.8 Not only the length of the linker but also its composition is crucial. Indeed, Linhult et al. investigated three different linker regions in order to obtain a stable divalent proteinaceous human serum albumin binding ligand under alkaline conditions.21 They concluded that the length and composition of the linker region was important when connecting two functional domains. In the context of protein design and dynamics, Wriggers et al. highlighted the distinction between flexible, glycine-rich linkers and the more rigid, often helical kind.22 Missirlis et al. studied a system of peptide amphiphiles where a bioactive peptide was connected to a hydrophobic segment by a linker.23 By incorporating different linkers, they showed that linker chemistry influenced peptide folding, which in turn impacted the peptide’s biofunction. Arai et al. 24 demonstrated the importance of linker rigidity versus flexibility by separating two fluorescent protein domains (EGFP and EBFP) by various types of linkers. Their research showed that FRET was more efficient in flexible, glycine-rich linkers than rigid, helical ones.
Figure 1.
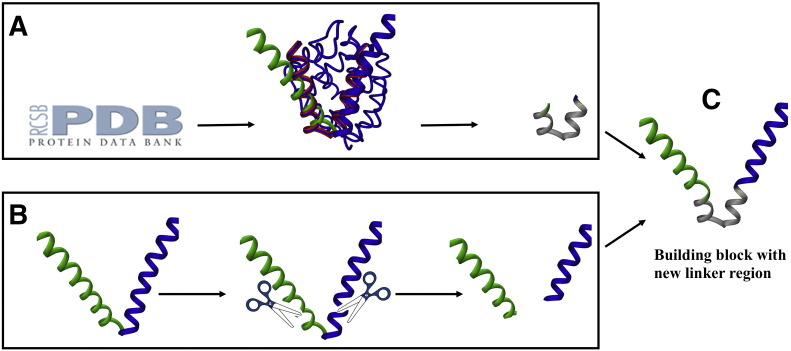
(A) PDB search for protein structures with inter-helical angles similar to the mode of the nanoparticle followed by superposition of the helices of the different PDB structures (blue and red) onto the pentamer (green) and trimer (blue) helices of the monomer of the peptide nanoparticle. The residues with angles similar to the angle between the helices of the pentamer and the trimer in the peptide nanoparticle were selected (gray region). (B) Schematics of the molecular biology strategy for inserting the new linker region: the original construct with pentameric (green) and trimeric (blue) helices is double digested with restriction enzymes ApaI and XhoI. (C) Linker oligonucleotides selected from (A) are ligated into double digested vector (B) using T4 DNA ligase generating new plasmid which codes for the genetically modified single polypeptide chain.
The de novo design of a protein sequence that will result in a folded protein with the required oligomerization domains connected by the perfect angle for icosahedral symmetry is difficult.25, 26 Therefore, in the course of the present work we used advanced database searching algorithms and molecular visualization tools to obtain promising structures from the Protein Database (PDB). The SAPN T81c-8-pf was selected for the subsequent tests. The construct T81c-8-pf is a prototype malaria vaccine described in detail elsewhere.11 It contains a modified tryptophan zipper as a pentameric coiled coil connected by two glycine residues to the trimeric coiled coil, which contains a helper T-cell epitope. At the C-terminus a short spacer sequence GGSG connects the nanoparticle core and the B-cell epitope (NANP)4 from the circumsporozoite protein of Plasmodium falciparum. Our main intention was to understand how the incorporation of different linkers would affect self-assembly of this nanoparticle. To this end, we genetically modified the sequence of the linker region in the construct to include one of the five linker regions obtained from the PDB search. The nanoparticles formed by the three constructs that could be refolded (T1qgt, T2ic8 and T2i88) were biophysically characterized by transmission electron microscopy (TEM) and dynamic light scattering (DLS). Mass measurements by analytical ultracentrifugation (AUC) and scanning transmission electron microscopy (STEM) were performed on the most promising candidate, T2i88 revealing a T = 3-like icosahedral symmetry.
Methods
Structural similarity search
The DEJAVU program was used to search the RCSB protein database for structurally similar proteins with the helix–turn–helix motif in the nanoparticles.27 Different two-helix models in which the helices had the same relative orientation as the structures predicted by molecular modeling experiments placing a pentameric and trimeric coiled coil close to the five-fold and three-fold symmetry axes of an icosahedron, were used as search models. The helices of the search models contained from 10 to 20 amino acids. The results from the different similarity searches made with varying helical lengths, were then analyzed with the graphical program O.27 The five structures (PDB codes 2nr9, 1jil, 1qgt, 2ic8 and 2i88) that were deemed to be the best fits to the expected helix–turn–helix motif, were selected to engineer the five nanoparticle constructs by the approach detailed in Figure 1.
Gene expression and purification
Oligonucleotides (purchased from IdtDNA) coding for the five different linker regions 2nr9, 1jil, 1qgt, 2ic8 and 2i88 were annealed (verified by agarose gel electrophoresis). The final protein sequence of T2i88 was: MGHHHHHHASWKWDGGLVPRGSWQTWNAKWDQWSNDWNAWRSDWQAWKDDWAYWTLTWKYGELYSKLAELERRLEELAKFVAAWTLKAAAVDLELAALRRRLEELARGGSGANANPNANPNANPNANP. In the corresponding constructs T2nr9, T1jil, T1qgt, and T2ic8, the portion in italics (underlined) is replaced by the corresponding linker sequences indicated in Figure 2 . A modified pPEP-T vector that coded for the monomer of the core particle was double digested with restriction enzymes ApaI (New England Biolabs — NEB) and XhoI (NEB) and then the linker oligonucleotides were ligated using T4 DNA ligase (NEB). Plasmids harboring the desired genes were transformed into BL21(DE3)pLysS Escherichia coli expression cells (Novagen). Bacterial growth was carried out at 37 °C in Luria Broth (LB) medium in the presence of ampicillin (200 μg/ml) and chloramphenicol (30 μg/ml). Expression was induced with 1 mM isopropyl β-d-thiogalactopyranoside. After 3 hours of expression, cells were harvested and centrifuged at 4000g for 10 min and frozen at − 80 °C. Purification was done under denaturing conditions (9 M urea). The cell pellet was thawed on ice, resuspended in lysis buffer A at pH 8.0 (9 M urea, 100 mM NaH2PO4, 10 mM Tris and 10 mM β-mercaptoethanol) and lysed by sonication (SONICATOR®3000 Ultrasonic Liquid Processor, cycle of 4 s pulse at 55% amplitude followed by a 6 s rest repeated for a 10 min period). The cell membranes were removed by centrifugation (45 min at 30500g). The supernatant was then incubated with nickel-NTA beads (Qiagen) for one hour so that the protein could be purified by affinity chromatography. Possible DNA contamination was removed by a wash with buffer B at pH 8.0 (9 M urea, 500 mM NaH2PO4, 10 mM Tris, 10 mM imidazole). Protein contaminants were washed from the column using a pH gradient and 10 mM imidazole in the buffers. The first wash was done with lysis buffer A and the second and third washes were done at pH 6.3 and pH 5.9, respectively, with a buffer containing 9 M urea, 100 mM NaH2PO4, 20 mM sodium citrate, 10 mM imidazole and 10 mM β-mercaptoethanol. Elution was achieved at pH 5.2 and pH 4.5 with 9 M urea, 100 mM NaH2PO4, 20 mM sodium citrate, 10 mM imidazole and 10 mM β-mercaptoethanol. Protein purity was verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Following purification, the denatured monomeric polypeptides were refolded using one of the following three methods: (1) dialysis performed in a stepwise manner until 0 M urea was reached (stepwise refolding) (2) dialysis against a buffer with no urea (direct refolding) or (3) concentration (Amicon ultracentrifugal filter devices) and dilution to a buffer with no urea (quick refolding). The proteins were filtered with a 0.1 μm polyvinylidene fluoride membrane filter before and after dialysis (Milipore #SLVV 033 RS).
Figure 2.
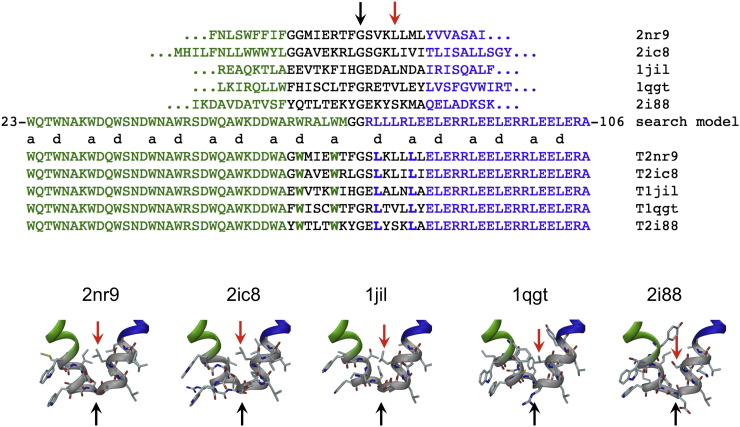
Alignment of the sequences of the PDB structures, the existing design (search model termed “T81c-8-pf”) and the studied designs. The pentamer is shown in green, the trimer is in blue and the critical, modified residues at coiled-coil core positions are shown in boldface. There are some noteworthy trends in the PDB structures: completely conserved glycine in position 61 (black arrow) and either a hydrophobic amino acid (leucine or valine) identical or similar to leucine found in the original design (T81c-8-pf) or a small serine residue (T2i88) at position 65 (red arrow).
Dynamic light scattering
The hydrodynamic diameter of the nanoparticles was obtained with a Malvern Zetasizer Nano S equipped with a 633 nm laser using a 3 mm path length quartz suprasil cell. The measurements were performed at 25 °C and for each protein five scans were collected and the average was recorded.
Transmission electron microscopy (TEM)
Samples at a concentration of 50 μg/ml were negatively stained with 1% uranyl acetate (SPI). Electron micrographs were taken with an FEI Tecnai T12 transmission electron microscope at an accelerating voltage of 80 kV or with a Philips EM 300 transmission electron microscope at an accelerating voltage of 80 kV (the micrographs were scanned at 600 dpi).
Analytical ultracentrifugation
Sedimentation velocity analysis was conducted at 20 °C and 30,000 RPM using interference optics with a Beckman-Coulter XL-I analytical ultracentrifuge. Double sector synthetic boundary cells equipped with sapphire windows were used to match the sample and reference menisci. The rotor was equilibrated under vacuum at 20 °C, and after a period of ~ 1 hour at 20 °C the rotor was accelerated to 30,000 RPM. Interference scans were acquired at 45 second intervals for ~ 5 hours. Extinction coefficients, molecular masses, partial specific volumes and solvent densities were calculated using Sednterp.28 Initial analysis was performed using Sedfit29 to obtain c(s) distributions and DcDt +30, 31, 32 to obtain g(s*) distributions.
Scanning transmission electron microscopy (STEM)
The nanoparticles were at a concentration of 60 μg/ml in 20 mM Hepes pH 7.5, 50 mM NaCl, and 5% glycerol. Glycerol was removed from the undiluted sample using spin columns following the Zeba™ Desalt Spin Column Instructions (Pierce). Afterwards, a 5 μl sample aliquot was adsorbed for 1 min to a glow discharged STEM film (thin carbon film that spans a thick fenestrated carbon layer covering 200-mesh/inch, gold-plated copper grids). The grid was then blotted, washed on 8 drops of quartz double-distilled water, blotting between each step, and plunge frozen in liquid nitrogen. It was freeze-dried at − 80 °C and 5 × 10− 8 Torr overnight in the microscope. Tobacco mosaic virus (TMV) particles (kindly provided by R. Diaz Avalos) were used as a mass standard. These particles were similarly adsorbed to separate STEM grids, washed on 8 drops of 100 mM ammonium bicarbonate, and air-dried.
An HB-5 vacuum generator scanning transmission electron microscope interfaced to a modular computer system (Tietz Video and Image Processing System GmbH, Gauting, Germany) was employed. Series of 512 × 512-pixel, dark-field images were recorded from the unstained sample at an acceleration voltage of 80 kV and a nominal magnification of 200,000 ×. The recording dose was 402 ± 18 electrons/nm2. Selected regions of the sample grid were also repeatedly scanned to determine the beam-induced mass-loss incurred by the nanoparticles. The digital images were evaluated using the program package MASDET.33 In short, projections were selected in circular boxes and the total scattering of each region was calculated. The average background scattering of the carbon support film was then subtracted and the mass calculated. The results were scaled according to the mass-per-length measured for TMV, corrected for beam-induced mass-loss,34 binned into histograms and described by Gaussian curves. The number of particle masses described by each (n) was estimated from the peak area. Nanoparticles with masses in operator-defined ranges centered at the peak maxima were sorted into galleries for inspection.
Results
In order to modify the flexibility of the linker region, we incorporated five different linker sequences into our parent SAPN termed “T81c-8-pf”.11 As the de novo design of a protein sequence that will result in a folded protein with the required oligomerization domains and geometry is difficult25, 26 these were based on the results of a PDB search. We identified five suitable protein structures with the PDB entries 1jil, 2nr9, 1qgt, 2ic8 and 2i88 (Figure 1, A). The linker regions found in these structures originally connected two monomeric helices in the context of a fully folded globular protein. Thus mutations were necessary to convert the monomeric helices into coiled-coil forming helices while keeping them at an angle compatible with the icosahedral symmetry of the nanoparticle. Therefore, in the pentameric domain, in positions a and d in the heptad repeat, we mutated the amino acid to tryptophan. Likewise, in the trimeric domain, positions a and d of the heptad repeat were changed to leucine (Figure 2).
Remarkably, our experimental data show that these de novo designs self-assemble into nanoparticles. The biophysical analyses of the three constructs that expressed well and were soluble upon refolding (T1qgt, T2ic8 and T2i88) are discussed below. In general, three different pHs (6.5, 7.5 and 8.5) each at either high or low ionic strength (50 or 150 mM NaCl) were used as the six starting conditions. Based on the results from the initial screening the refolding conditions were then further optimized.
T1qgt-8-pf: Linker region containing a cysteine residue at position 57 of the heptad repeat
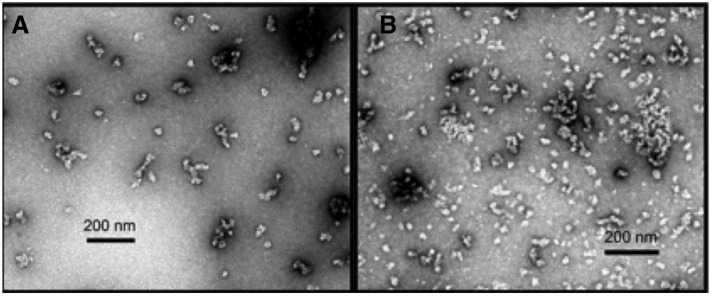
One feature of this design is the presence of the bulky phenylalanine residue in the two-residue linker connecting the pentamer (green) to the trimer (blue) and a cysteine residue at position 57 in the pentamer. In the original PDB structure (human hepatitis B virus capsid) this residue is reduced and does not form a disulfide bond that might stabilize protein structure.20 Likewise for our construct (T1qgt-8-pf) we wanted to prevent the occurrence of disulfide bridges so as to mimic the cysteine’s oxidation state in the original PDB structure. Another advantage of a reduced cysteine for our SAPN, is that aggregation caused by intermolecular disulfide bridges would be prevented. Given this knowledge, we included the reducing agent TCEP in the refolding buffer. The buffer used for stepwise removal of urea was 20 mM Tris pH 8.5, 50 mM NaCl, 5% glycerol and 0.1 mM TCEP (reducing agent). High pH and low NaCl concentration were selected because of our research group’s successful refolding of constructs containing pan-DR epitope under these conditions.11, 12 Despite the fact that refolding was performed under reducing conditions there was substantial aggregation caused by unknown interactions (Figure 3 , A). To corroborate self-assembly of the protein in another way, we used dynamic light scattering (DLS), which is a biophysical technique that measures the size and size distribution of molecules in solution. The DLS graph (Figure 3, B) confirms the results obtained by TEM depicting heterogeneity and aggregation.
Figure 3.
T1qgt-8-pf stepwise refolded at low salt concentration and high pH. (A) TEM image. The particles are very heterogeneous and tend to aggregate. Scale bar, 200 nm. (B) Dynamic light scattering graph. The size distribution by volume shows high polydispersity.
T2ic8-8-pf: Linker region with leucine at position 65 of the heptad repeat
Direct refolding (dialysis from 8 M urea to buffer with no urea) at pH 7.5 was initially attempted with this construct, but resulted in insoluble protein with visible aggregation in the dialysis bag. Therefore, a change in pH in the refolding buffer was deemed necessary. Direct refolding at pH 8.7 resulted in soluble nanoparticles but with some aggregation (Figure 4 , A). Stepwise refolding at pH 8.7 was also investigated and, as shown in Figure 4, B, yielded a similar distribution of single particles and aggregates as the direct refolding scheme. DLS results confirmed the polydispersity found in the TEM images (data not shown).
Figure 4.
TEM images of T2ic8-8-pf under different refolding conditions. (A) Direct refolding with 20 mM Tris pH 8.7, 50 mM NaCl and 5% (v/v) glycerol. Sample concentration was 0.06 mg/ml. (B) Stepwise refolding with same buffer as in (A) with sample concentration of 0.07 mg/ml.
T2i88-8-pf: Linker region with tyrosine in lieu of one of the glycines at position 60
For this construct two refolding schemes were investigated: stepwise and quick refolding. Stepwise refolding was done under different pH values using Hepes (20 mM) as the buffering agent (pKa = 7.55). The pH values studied were: 7.0, 7.5 and 8.5. The salt concentration (50 mM NaCl) and glycerol volume (5%) were kept constant. Biophysical characterization by DLS and TEM (data not shown) revealed that while single nanoparticles were formed they had a slight tendency to aggregate and form what appeared to be beads on a string.
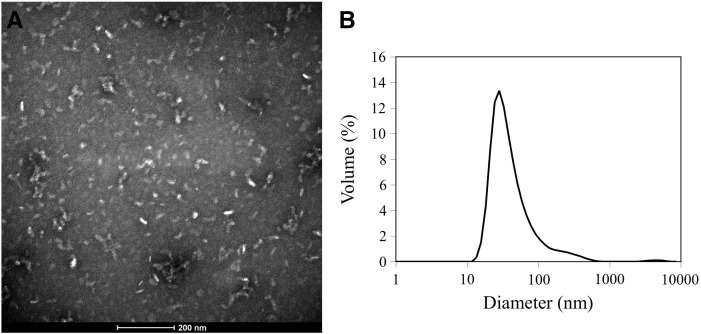
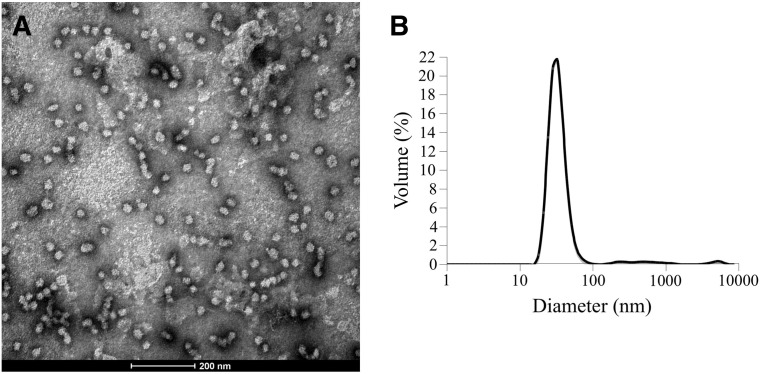
The other refolding scheme that was examined for T2i88-8-pf was quick refolding. As with stepwise refolding the salt concentration (50 mM NaCl) and glycerol volume (5%) remained unchanged. What was varied was the buffering agent and the pH: Mes (pH 6.5) and Hepes (pH 7.0, 7.5 and 8.0). Aggregation tendencies were found at all pH values except pH 7.0 (Figure 5 ). The electron micrograph (Figure 5, A) shows that the polypeptide chains self-assembled into nanoparticles with fairly homogeneous, spherical shapes with diameters ranging from 32 nm to 36 nm. The solution data observed with dynamic light scattering (Figure 5, B) correlate with the TEM data. Clearly neutral pH seemed to work best for the refolding of T2i88-8-pf.
Figure 5.
T2i88-8-pf quick refolded at low salt concentration (50 mM NaCl) and 5% (v/v) glycerol. (A) TEM image. Scale bar is 200 nm. (B) Dynamic light scattering graph. Peak ranges from 32 nm to 36 nm.
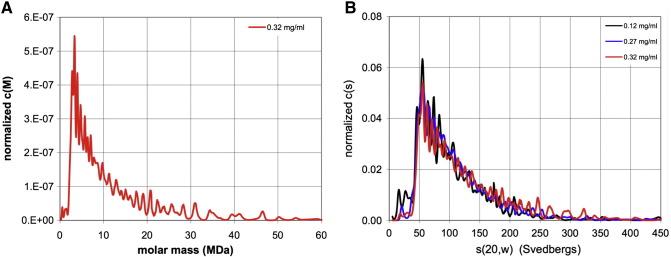
Of all the designs, T2i88-8-pf was the most promising given the biophysical characterization data obtained (Figure 5). To understand more about the nature of this nanoparticle and its self-assembly T2i88-8-pf was analyzed by analytical ultracentrifugation. Surprisingly, the sedimentation velocity analysis showed that there is a polydisperse mixture of species (Figure 6 , A). However, the distribution peaks at 55 S corresponding to a particle with a molecular weight of about 2.5 MDa (Figure 6, B). Given that the single chain has a molecular weight of 14900 Da, the peak at 55 S would correlate with roughly 180 chains per nanoparticle. This would imply a T = 3-like icosahedral symmetry.
Figure 6.
Sedimentation velocity analysis of 0.32 mg/ml T2i88-8-pf stepwise refolded at pH 7.0 (A) Continuous distribution of molecular masses. The molecular masses range from a few hundred thousand daltons to over 50 MDa (B) Continuous distribution of sedimenting species at the concentrations of 0.12 mg/ml (black), 0.27 mg/ml (blue) and 0.32 mg/ml (red), respectively. The distribution peaks at ~ 55 S, with a significant amount of material evident at S values up to ~ 300 S.
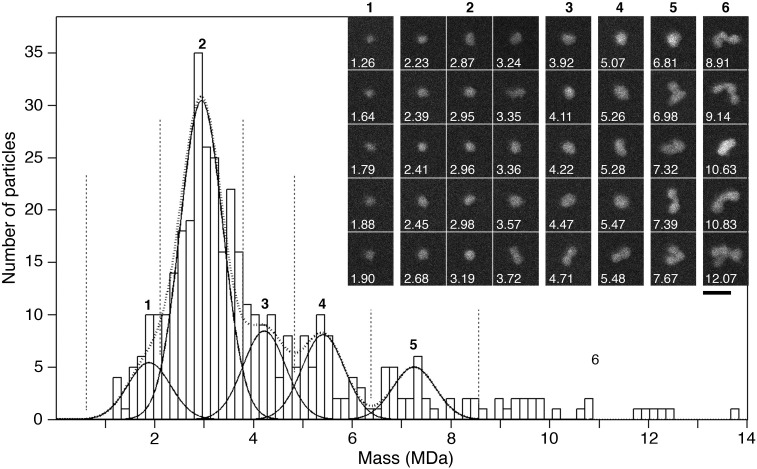
T2i88-8-pf nanoparticles were also analyzed by STEM. According to Müller and Engel,35 the importance of the STEM for mass determination lies in its ability to analyze intact biological structures regardless of their shape and at the same time to provide an image from which the quality and homogeneity of the specimen may be independently assessed. The mass of T2i88-8-pf was determined by analyzing 389 nanoparticles from the dark-field STEM images (Figure 7 ). The masses range from 1.2 to 13.8 MDa. Together they form a histogram that tails off to higher mass and can be described by 5 Gaussians. The major peak is at 2.9 ± 0.4 MDa, which it is compatible with the presence of about 180 peptide chains per particle. Examination of the corresponding images shows that the higher masses frequently arise from clusters of nanoparticles, documenting the type of aggregation that occurs (Figure 7, inset), while particles with masses below about 4.0 MDa have essentially circular projections compatible with a roughly spherical shape.
Figure 7.
Dark-field STEM mass analysis of T2i88-8-pf stepwise refolded at pH 7.0. The histogram displays the masses of 389 imaged nanoparticles calculated from their scattering power and corrected for beam-induced mass loss. Particles with masses in the different regions of the histogram (delimited by dotted lines) are shown in the inset; scale bar: 50 nm. The mass distribution could be described by fitting 5 Gaussian curves of equal width. While peaks 1 (1.9 ± 0.4 MDa, n = 35) and 2 (2.9 ± 0.4 MDa, n = 195) primarily arise from single nanoparticles often with almost circular projections, peaks 3 (4.2 ± 0.4 MDa, n = 54), 4 (5.4 ± 0.4 MDa, n = 47) and 5 (7.2 ± 0.4 MDa, n = 30) and the few higher mass particles seem to be the result of aggregation. According to the mass indicated by peak 2, most of the nanoparticles are composed of 195 ± 27 peptide chains (a single chain has a molecular weight of 14900 Da).
Discussion
Protein engineering is a challenging subject. The “rules” for protein self-assembly are still being investigated.36 Even virus capsid folding, such as that of the hepatitis B core (HBc), is not always well understood.37 Janssens et al. analyzed improvements in the HBc particle assembly for vaccination research.37 Likewise, we have attempted to change the epitope density of the SAPN, which could lead to superior immunological properties.
Examination of the sequences of the five linker designs obtained from the PDB reveals some trends in amino acid usage. The most striking similarity between all of these structures is a completely conserved glycine in position 61 (black arrow in Figure 2) backing up the rationale of our design. At position 65 (red arrow in Figure 2) all of these structures have either a hydrophobic amino acid (leucine or valine) identical or similar to the leucine found in the original design (T81c-8-pf), or a small serine residue (T2i88). Three designs (T2ic8, T2i88 and T2nr9) have bulky residues at position 60, whereas T1qgt has a leucine at position 60 and T1jil has a histidine at this position. T1qgt is unique, as it is the only design with a cysteine at position 57.
T1qgt-8-pf: Linker region containing a cysteine residue at position 57 of the heptad repeat
The aggregation behavior seen in Figure 3 can be regarded as the start of fiber formation. Aili et al. demonstrated that helix–loop–helix polypeptides containing cysteine in the loop region can form four-arm dendrimers by forming a disulfide bridge.38 These dendrimers would be the units that assemble to form fibers. The construct studied here could also be viewed as a helix–loop–helix polypeptide able to form fibers in the same way. However, in our case the situation is more complex as the helices interact with one another as pentamers and trimers, which would then create sheets of aggregation or other structures that are hard to predict. The fibers formed by these would not be similar to those described by Aili et al. Strings of nanoparticles with a length of 200 nm can be observed in the TEM micrographs (Figure 3, A). DLS also shows a peak around 200 nm (Figure 3, B). This is of interest because studies have shown that protein nanofibers can act as self-adjuvanting vaccines in mice39, 40
T2ic8-8-pf: Linker region with leucine at position 65 of the heptad repeat
The aggregation results reported (Figure 4) can be understood in light of the work by Shahbazi et al. on this particular linker region.41 Predicting the formation of hydrogen bonds allows the expected mobility of protein chains to be estimated. Mobility reduction is observed when there is more rigidity due to the formation of hydrogen bonds.41 Shahbazi et al. studied the linker 2ic8 and found that there were many flexible segments, which can result in aggregation without icosahedral structural arrangement, as can be seen from the TEM data in Figure 4.
T2i88-8-pf: Linker region with tyrosine in lieu of one of the glycines at position 60
According to the work of Shahbazi et al. the linker 2i88 is very rigid due to the formation of hydrogen bonds, which should lead to the production of homogeneous nanoparticles. Indeed, this is what we observed in our biophysical characterization (Figure 5). AUC and STEM were carried out to extend this analysis. The results of both of these mass measurement methods support the presence of a prominent species that contains 180 peptide chains per nanoparticle, suggesting the formation of T = 3-like icosahedral particles. Both also indicate a degree of polydispersity, which according to the STEM data may be at least partly due to the association of lower mass species (Figure 7, inset). The images also show that masses below about 3.5 MDa generally arise from particles with essentially circular projections compatible with a roughly spherical shape.
For exact T = 3 icosahedral symmetry the chains of T2i88-8-pf would have to rearrange in such a way that the Trp-zipper becomes a hexameric coiled coil. Given that it is nearly impossible for a Trp-zipper to change to a hexamer we are left with nanoparticles that are only T = 3-like. We are currently further investigating this topic with tessellation theories.7
In summary, this work provides insight into the self-assembly of coiled-coil based nanoparticles which can be used for rational vaccine design. By substituting the linker region between the pentamer and trimer with protein sequences from helix–turn–helix motifs from the protein structural database we were able to show how the linker chemistry affected nanoparticle formation. The flexibility in the linker region allows different inter-helical angles, which influences the self-assembly of the nanoparticles. Considering the potential application of nanoparticles as vaccines, the different angles would allow different epitope densities, which may result in distinct immune responses. The database search yielded a new optimized sequence of the core of the particles that meanwhile has been used to design an improved malaria vaccine candidate compared to the one described in references.11, 12, 13
Footnotes
Support by the NIH/NIGMS (award 1P01GM096971), the NIH/NIDA (award 1DP1DA033524) and the NIH/NIAID (award 5R01AI068761) for this work is gratefully acknowledged. The STEM microscopy was funded by the Maurice E. Müller Foundation of Switzerland and Swiss National Foundation Grant 3100A0-108299 to Andreas Engel and by the Swiss systems biology initiative SystemsX.ch (grant CINA to Andreas Engel and Henning Stahlberg).
Competing interests: PB has an interest in the company Alpha-O Peptides that has patents or patents pending on the technology.
References
- 1.Kushnir N., Streatfield S.J., Yusibov V. Virus-like particles as a highly efficient vaccine platform: Diversity of targets and production systems and advances in clinical development. Vaccine. 2012;31(1):58–83. doi: 10.1016/j.vaccine.2012.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith D.M., Simon J.K., Baker J.R. Applications of nanotechnology for immunology. Nat Rev Immunol. 2013;13(8):592–605. doi: 10.1038/nri3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doll T.A., Raman S., Dey R., Burkhard P. Nanoscale assemblies and their biomedical applications. J R Soc Interface. 2013;10(80):20120740. doi: 10.1098/rsif.2012.0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bozic S., Doles T., Gradisar H., Jerala R. New designed protein assemblies. Curr Opin Chem Biol. 2013;17(6):940–945. doi: 10.1016/j.cbpa.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher J.M., Harniman R.L., Barnes F.R., Boyle A.L., Collins A., Mantell J. Self-assembling cages from coiled-coil peptide modules. Science. 2013;340(6132):595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Der B.S., Kuhlman B. Cages from coils. Nat Biotechnol. 2013;31(9):809–810. doi: 10.1038/nbt.2670. [DOI] [PubMed] [Google Scholar]
- 7.Twarock R. A tiling approach to virus capsid assembly explaining a structural puzzle in virology. J Theor Biol. 2004;226(4):477–482. doi: 10.1016/j.jtbi.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Raman S., Machaidze G., Lustig A., Aebi U., Burkhard P. Structure-based design of peptides that self-assemble into regular polyhedral nanoparticles. Nanomedicine. 2006;2(2):95–102. doi: 10.1016/j.nano.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Pimentel T.A., Yan Z., Jeffers S.A., Holmes K.V., Hodges R.S., Burkhard P. Peptide nanoparticles as novel immunogens: Design and analysis of a prototypic severe acute respiratory syndrome vaccine. Chem Biol Drug Des. 2009;73(1):53–61. doi: 10.1111/j.1747-0285.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaba S.A., Brando C., Guo Q., Mittelholzer C., Raman S., Tropel D. A nonadjuvanted polypeptide nanoparticle vaccine confers long-lasting protection against rodent malaria. J Immunol. 2009;183(11):7268–7277. doi: 10.4049/jimmunol.0901957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaba S.A., McCoy M.E., Doll T.A., Brando C., Guo Q., Dasgupta D. Protective antibody and CD8 + T-cell responses to the Plasmodium falciparum circumsporozoite protein induced by a nanoparticle vaccine. PLoS One. 2012;7(10):1–11. doi: 10.1371/journal.pone.0048304. e48304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Q., Dasgupta D., Doll T.A., Burkhard P., Lanar D.E. Expression, purification and refolding of a self-assembling protein nanoparticle (SAPN) malaria vaccine. Methods. 2013;60(3):242–247. doi: 10.1016/j.ymeth.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCoy M.E., Golden H.E., Doll T.A., Yang Y., Kaba S.A., Zou X. Mechanisms of protective immune responses induced by the Plasmodium falciparum circumsporozoite protein-based, self-assembling protein nanoparticle vaccine. Malar J. 2013;12(1):1–12. doi: 10.1186/1475-2875-12-136. 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahome N., Pfeiffer T., Ambiel I., Yang Y., Keppler O.T., Bosch V. Conformation-specific display of 4E10 and 2F5 epitopes on self-assembling protein nanoparticles as a potential HIV vaccine. Chem Biol Drug Des. 2012;80(3):349–357. doi: 10.1111/j.1747-0285.2012.01423.x. [DOI] [PubMed] [Google Scholar]
- 15.El Bissati K., Zhou Y., Dasgupta D., Cobb D., Dubey J.P., Burkhard P. Effectiveness of a novel immunogenic nanoparticle platform for toxoplasma peptide vaccine in HLA transgenic mice. Vaccine. 2014;32(26):3243–3248. doi: 10.1016/j.vaccine.2014.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babapoor S., Neef T., Mittelholzer C., Girshick T., Garmendia A., Shang H. A novel vaccine using nanoparticle platform to present immunogenic M2e against avian influenza infection. Influenza Res Treat. 2011;2011:1–12. doi: 10.1155/2011/126794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Ringler P., Muller S.A., Burkhard P. Optimizing the refolding conditions of self-assembling polypeptide nanoparticles that serve as repetitive antigen display systems. J Struct Biol. 2012;177(1):168–176. doi: 10.1016/j.jsb.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y., Burkhard P. Encapsulation of gold nanoparticles into self-assembling protein nanoparticles. J Nanobiotechnol. 2012;10:1–11. doi: 10.1186/1477-3155-10-42. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y., Neef T., Mittelholzer C., Garcia Garayoa E., Blauenstein P., Schibli R. The biodistribution of self-assembling protein nanoparticles shows they are promising vaccine platforms. J Nanobiotechnol. 2013;11:1–12. doi: 10.1186/1477-3155-11-36. 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George R.A., Heringa J. An analysis of protein domain linkers: Their classification and role in protein folding. Protein Eng. 2002;15(11):871–879. doi: 10.1093/protein/15.11.871. [DOI] [PubMed] [Google Scholar]
- 21.Linhult M., Gulich S., Graslund T., Nygren P.A., Hober S. Evaluation of different linker regions for multimerization and coupling chemistry for immobilization of a proteinaceous affinity ligand. Protein Eng. 2003;16(12):1147–1152. doi: 10.1093/protein/gzg121. [DOI] [PubMed] [Google Scholar]
- 22.Wriggers W., Chakravarty S., Jennings P.A. Control of protein functional dynamics by peptide linkers. Biopolymers. 2005;80(6):736–746. doi: 10.1002/bip.20291. [DOI] [PubMed] [Google Scholar]
- 23.Missirlis D., Farine M., Kastantin M., Ananthanarayanan B., Neumann T., Tirrell M. Linker chemistry determines secondary structure of p5314-29 in peptide amphiphile micelles. Bioconjug Chem. 2010;21(3):465–475. doi: 10.1021/bc900383m. [DOI] [PubMed] [Google Scholar]
- 24.Arai R., Ueda H., Kitayama A., Kamiya N., Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14(8):529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 25.King N.P., Lai Y.T. Practical approaches to designing novel protein assemblies. Curr Opin Struct Biol. 2013;23(4):632–638. doi: 10.1016/j.sbi.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Lai Y.T., King N.P., Yeates T.O. Principles for designing ordered protein assemblies. Trends Cell Biol. 2012;22(12):653–661. doi: 10.1016/j.tcb.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Laue T.M. Analytical ultracentrifugation in biochemistry and polymer science. R Soc Chem. 1992:90–125. [Google Scholar]
- 29.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philo J.S. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem. 2000;279(2):151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 31.Philo J.S. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354(2):238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 32.Stafford W.F. Boundary analysis in sedimentation transport experiments: A procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 1992;203(2):295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 33.Krzyzanek V., Müller S.A., Engel A., Reichelt R. MASDET — A fast and user-friendly multiplatform software for mass determination by dark-field electron microscopy. J Struct Biol. 2009;165(2):78–87. doi: 10.1016/j.jsb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Müller S.A., Goldie K.N., Burki R., Haring R., Engel A. Factors influencing the precision of quantitative scanning transmission electron microscopy. Ultramicroscopy. 1992;46:317–334. [Google Scholar]
- 35.Müller S.A., Engel A. Structure and mass analysis by scanning transmission electron microscopy. Micron. 2001;32:21–31. doi: 10.1016/s0968-4328(00)00022-6. [DOI] [PubMed] [Google Scholar]
- 36.Papapostolou D., Howorka S. Engineering and exploiting protein assemblies in synthetic biology. Mol BioSyst. 2009;5(7):723–732. doi: 10.1039/b902440a. [DOI] [PubMed] [Google Scholar]
- 37.Janssens M.E., Geysen D., Broos K., De Goeyse I., Robbens J., Van Petegem F. Folding properties of the hepatitis B core as a carrier protein for vaccination research. Amino Acids. 2010;38(5):1617–1626. doi: 10.1007/s00726-009-0365-1. [DOI] [PubMed] [Google Scholar]
- 38.Aili D., Tai F.I., Enander K., Baltzer L., Liedberg B. Self-assembly of fibers and nanorings from disulfide-linked helix–loop–helix polypeptides. Angew Chem. 2008;47(30):5554–5556. doi: 10.1002/anie.200801155. [DOI] [PubMed] [Google Scholar]
- 39.Chen J., Pompano R.R., Santiago F.W., Maillat L., Sciammas R., Sun T. The use of self-adjuvanting nanofiber vaccines to elicit high-affinity B cell responses to peptide antigens without inflammation. Biomaterials. 2013;34(34):8776–8785. doi: 10.1016/j.biomaterials.2013.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hudalla G.A., Modica J.A., Tian Y.F., Rudra J.S., Chong A.S., Sun T. A self-adjuvanting supramolecular vaccine carrying a folded protein antigen. Adv Healthc Mater. 2013;2(8):1114–1119. doi: 10.1002/adhm.201200435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahbazi Z., Pimentel T.A.P.F., Ilies H., Kazerounian K., Burkhard P. Advances in Robot Kinematics: Motion in Man and Machine. Part 3. 2010. A kinematic observation and conjecture for creating stable constructs of a peptide nanoparticle; pp. 203–210. [Google Scholar]