Abstract
Objectives
To identify angiogenic biomarkers associated with tumor angiogenesis and clinical outcome in high-grade serous ovarian cancer (HGSC).
Methods
51 HGSC samples were analyzed using Affymetrix HG-U133A microarray. Microvessel density (MVD) counts were determined using CD31and CD105. Association between mRNA expression levels and overall survival were assessed using rank score statistic. Effect size was estimated as a hazard ratio (HR) under a proportional hazards model. The Storey q-value method was used to account for multiple testing within the false-discovery rate (FDR) framework. Publicly available databases including TCGA and GSE were used for external confirmation.
Results
Thirty-one angiogenic-related genes were significantly associated with survival (q ≤ 0.05). Of these 31 genes, 4 were also associated with outcome in the TCGA data: AKT1 (q=0.02; TCGA p= 0.01, HR=0.8), CD44 (q= 0.003; TCGA p=0.05, HR=0.9), EPHB2 (q= 0.01; TCGA p=0.05, HR=1.2), and ERBB2 (q= 0.02; TCGA p= 0.05, HR=1.2). While 5 were associated with outcome in the GSE database: FLT1 (q= 0.03; GSE26712 p=0.01, HR=3.1); PF4 (q= 0.02; GSE26712 p=0.01, HR=3.0), NRP1 (q= 0.02; GSE26712 p < 0.04, HR>1.4), COL4A3 (q= 0.04; GSE26712 p= 0.03, HR=1.3), ANGPTL3 (q= 0.02; GSE14764 p=0.02, HR=1.5). High AKT1 and CD44 were associated with longer survival. In contrast, high expression of EPHB2, ERBB2, FLT1; PF4, NRP1, COL4A3, and ANGPTL3 were associated with shorter survival. CD105-MVD and CD31-MVD were not significantly associated with angiogenic gene expression.
Conclusions
Thirty-one angiogenic-related genes were associated with survival in advanced HGSC and nine of these genes were confirmed in independent publicly available databases.
Keywords: High-grade serous ovarian carcinoma, biomarkers, angiogenesis, overall survival, gene expression
INTRODUCTION
Epithelial ovarian cancer (EOC) is considered the most lethal gynecologic malignancy and is the fifth leading cause of cancer mortality among women in the United States, resulting in 15,000 deaths annually.[1] High-grade serous ovarian cancer (HGSC) is an aggressive ovarian cancer that is associated with a worse clinical outcome compared to other EOCs. Even with a complete clinical response, more than 90% of the patients with advanced disease will develop a recurrence, resulting in the median survival of 3–4 years.[2, 3] Despite surgical advances and novel anti-angiogenic targeted therapies most women with HGSCs will eventually die from their disease.
Angiogenesis is a critical component of solid tumor growth and metastasis. A high degree of tumor angiogenesis has been shown to be a prognostic factor for a variety of solid tumors including EOC.[4] The regulation of angiogenesis in EOC has not been completely elucidated. However, high serum and tumor levels of a potent pro-angiogenic factor, vascular endothelial growth factor (VEGF), have been associated with worse survival.[5] Recently several anti-angiogenic agents that target VEGF have been evaluated in women with EOC, including those with HGSC, and have demonstrated an improvement in survival.[6] For example bevacizumab (Bev) has been shown to prolong OS in select patients with suboptimally debulked stage III disease and stage IV disease, [7] and cediranib (AZD 2171) is an oral inhibitor of VEGF signaling and has shown increase OS in women with ‘platinum-sensitive’ disease.[8]
However, these agents have significant side effects, are expensive, and have not been shown to be cost-effective.[1] Furthermore, despite initial anti-tumor activity cancers eventually develop resistance to VEGF-blockade. The mechanism of resistance to VEGF inhibitors may be secondary to many factors. For example, one hypothesis maintains that multiple alternate pro-angiogenic pathways overcome VEGF inhibition leading to drug resistance.[9] To effectively overcome anti-VEGF resistance, novel alternate targets must be elucidated in order to develop innovative anti-angiogenic therapies. In addition, there is a need to develop biomarkers to rationally direct anti-angiogenic-specific therapy, improve outcome, and minimize toxicity and cost.
In this study we sought to investigate: i) the association between angiogenic gene expression and overall survival (OS) in HGSC, and ii) the correlation between angiogenic gene expression and MVD using CD105, a marker of activated endothelial cells, and CD31, a non-specific endothelial cell marker. We hypothesized that our results could: a) improve our understanding of tumor angiogenesis regulation; b) identify quantifiable and reproducible angiogenic biomarkers to direct therapy in HGSC; and c) identify candidate genes that may be exploited to develop novel anti-angiogenic therapies.
METHODS
Study population
RNA microarray data from 51 patients with chemotherapy-naïve advanced HGSC (stage III A, B, C and IV) were evaluated. Patients were treated at Duke University Medical Center between 1988 and 2001 under a Duke Institutional Review Board (IRB) approved protocol. Patients enrolled in the study were surgically staged and those with stage III/IV disease all received platin-based combination chemotherapy.[3] The original study had an extreme phenotype design, deliberating genetically profiling tumors from women who had either long or short overall survival (OS). Long-term survival was defined as OS ≥ 7 years vs. short-term survival defined as OS ≤ 3 years.[3] The study population had long-term surveillance of more than 10 years. All genomic data and samples were linked to clinical data.
Microarray analysis
The ovarian cancer specimens were collected and snap-frozen at initial surgery. All pretreatment biopsies were confirmed to have at least 60% invasive disease throughout the core sample before RNA harvesting. RNA was prepared, probes generated, and used for hybridization to Affymetrix HG-U133A GeneChip arrays as described previously in detail.[3]
Immunohistochemistry analysis of MVD
Immunohistochemistry (IHC) was performed for the MVD analysis using CD31 and CD105 markers as previously described.[4] Briefly, five micrometer sections of formalin-fixed, paraffin-embedded tissue were stained using monoclonal antibodies to CD31 (JC70A; DakoCytomation, Glostrup, Denmark) and CD105 (SN6h; DakoCytomation, Carpinteria, CA). For negative control, mouse IgG was used. The 4plus™ Immunoperoxidase Detection System (Biocare Medical, Concord, CA) and Catalyzed Signal Amplification System (DakoCytomation, Carpinteria, CA) were used for CD31 and CD105 staining, respectively. The slides were scanned at low magnification (x40) to identify areas with the greatest MVD (hotspots) for the CD31 or 105 staining signals. Three MVD hotspots were selected by a board certified anatomic pathologist, (SMB), who was blinded to the clinical variables. For each hotspot, a single HPF (x400) was selected and all capillaries expressing CD31 or CD105 were counted. For quantification, an average count from the three MVD hotspot counts was calculated.[4]
Statistical Analysis and validation of microarray results
One hundred and forty-five angiogenic candidate genes based on literature review, known genes in angiogenic pathways (e.g. VEGF signaling pathway), and anti angiogenic therapeutic targets (e.g. VEGF inhibitors, TIE-TEK pathway inhibitors) were evaluated. A panel of 285 probe, sets linked to HUGO Gene Nomenclature Committee (HGNC) gene symbols using annotation data from public databases and a chip annotation file from the Bioconductor package was derived.[10] Affymetrix HG-U133A arrays were pre-processed using the Robust Multi-chip Averaging (RMA) algorithm.[11]
Six publicly available data sets were used to externally confirm of our results. The six external data sets included the TCGA plus five other data sets queried from GEO as referenced below with their data set accession numbers. Information was not aggregated across the six studies. TCGA mRNA data were retrieved from the Cancer Genomic Data Server (CGDS) through the Computational Biology Center Portal (cBio): http://www.cbioportal.org/. The cgdsr extension package (cran.rproject.org/web/packages/cgdsr/) was used to execute the retrieval. In addition to the TCGA database the 31 candidate genes panel was elevated in in 11 publicly available databases as described in the 2012 Bentink el al. paper.[12] Five of the databases (GSE26712 [13], GSE18520 [14], GSE14764 [15], GSE 17260 [16], and GSE 19161 [17]) included information on women with advanced HGSC and the number of cases in these databases match the 2012 Bentink el al. paper.[12] (Supplemental data, Gene Expression Omnibus (GEO) data selection).
The associations between gene expression of each probe set and OS were assessed using a rank score test.[18] Corresponding effect size was quantitatively assessed using a hazard ratio (HR) assuming proportional hazards. Conditional inference trees were used to find optimal cutpoints.[19] The log rank test was used to evaluate the association between clinical and demographic baseline covariates (age, stage and grade) and overall survival. Different cutpoints for Duke, TCGA and GSE data were used to avoid the batch effects. Multiplicity was addressed within the False Discovery Rate (FDR) framework. FDR adjusted P-values (Q-values) were calculated using the method by Storey.[18] The results were not adjusted for multiple testing in the external databases. The Kendall’s tau correlation test was used to evaluate the association between gene expression and MVD. All analyses were performed using the R Statistical Environment and extension packages from CRAN and the Bioconductor project.[18]
The Duke survival data is presented as box plots instead of Kaplan–Meier (KM) plots and HRs due to the extreme phenotype design. Specifically, the 51 samples from the Duke study were pre-selected using a biased sampling approach by selecting patients who had an OS of ≤ 3 years or ≥ 7 years (short vs. long overall survival). Thus the application of the KM method to estimate the survival distribution conditional on the gene expression value was not deemed appropriate, given bias consideration and non-random sampling design. Lin et. al. proposed a method to derive a proper likelihood accounting for biased sampling designs.[20] However, access to the sufficient clinical data was not available to employ such an approach in the current study. Consequently, the distribution of the gene expression was estimated conditional on surviving less than three years and compared to that of the conditional distribution of surviving at least seven years. In the external cohorts, the KM method and Cox model, under the implicit assumption that the sampling was not biased, were used for analysis.
RESULTS
In an adjusted analysis the results demonstrated an association between differential angiogenic gene expression and survival in HGSC. Specifically, forty-three features (Affymetrix probe sets) linked to 31 angiogenic-associated genes were significantly associated with OS (FDR adjusted p-value (q) ≤ 0.05). Stage of the tumor (III vs. IV) was significantly associated with OS (p-value= 0.0042), but patient’s age was not significantly associated with OS (p-value= 0.68) (Table 1). From the 31 genes 4 genes were associated with OS in TCGA, and 5 genes were associated with OS in the GSE database (Tables 2 and 3). The 4 genes in TCGA exhibiting level of significance and concordant directions of effect were: i) v-akt murine thymoma viral oncogene homolog 1 (AKT1), ii) CD44 molecule (CD44), iii) EPH receptor B2 (EPHB2) and, iv) v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (ERBB2/HER2) (Figure 1 and 2). In the GSE database, 5 common genes were identified: i) Fms-like tyrosine kinase 1 (FLT1, also known as VEGFR1), ii) platelet factor 4 (PF4), iii) neuropilin 1 (NRP1), iv) collagen, type IV, alpha-3 (COL4A3), and v) angiopoietin-like 3 (ANGPTL3) (Figure 3). High expression of AKT1 and CD44 were associated with longer survival. In contrast, high expression of EPHB2, ERBB2, FLT1; PF4, NRP1, COL4A3, and ANGPTL3 were associated with shorter survival.
Table 1.
Analysis of assosiation between the 51 patients’ age and tumor stage with overall survival (OS)
| Duke | p-value | |||
|---|---|---|---|---|
| Short Survival (OS ≤ 3 years) | Long Survival (OS ≥ 7 years) | |||
| Stage | III | 23 | 23 | 0.004 |
| IV | 5 | 0 | ||
| Age | 59 (37–79) | 62 (33–78) | 0.68 | |
The stage of the tumor (III vs. IV) was significantly associated with patients OS (p-value= 0.004). However, there were only 5 patients in the stage IV group and none of the patients were in the long survival category. Age of the patients at the time of diagnosis was not significantly associated with OS (p-values= 0.68).
Table 2.
Results summary for associations between angiogenic gene expression and overall survival for TCGA confirmed genes.
| Gene Symbol |
Probe ID | Duke OS p-value |
Duke OS q- value |
Duke Low Expression |
Duke High Expression |
TCGA OS p-value |
TCGA HR |
TCGA Low Expression |
TCGA High Expression |
|---|---|---|---|---|---|---|---|---|---|
| AKT1 | 207163_s_at | 0.002 | 0.02 | 29 | 22 | 0.01 | 0.8 | 190 | 39 |
| CD44 | 212063_at | 0.00002 | 0.003 | 32 | 19 | 0.05 | 0.9 | 77 | 152 |
| EPHB2 | 210651_s_at | 0.0004 | 0.01 | 21 | 30 | 0.05 | 1.2 | 215 | 14 |
| ERBB2 | 210930_s_at | 0.003 | 0.02 | 17 | 34 | 0.05 | 1.2 | 44 | 185 |
HR: Hazard Ratio; OS: Overall Survival; Q-value = adjusted P-value; TCGA data: N=229; Duke Data: N=51.
Conditional inference trees were used to find optimal cutpoints. Based on cutpoints, the TCGA and Duke “low expression” columns represents the number of patients in the “low expression group” and “high expression” columns represents the number of patients in the “high expression group”.
Table 3.
Results summary for associations between angiogenic gene expression and overall survival for GSE confirmed genes.
| Gene Symbol |
Probe ID | Duke OS p-value |
Duke OS q-value |
Duke Low Expression |
Duke High Expression |
GSE OS p-value |
GSE HR |
GSE Low Expression |
GSE High Expression |
|---|---|---|---|---|---|---|---|---|---|
| ANGPTL3** | 219803_at | 0.002 | 0.02 | 23 | 28 | 0.02 | 1.5 | 29 | 34 |
| COL4A3* | 216896_at | 0.01 | 0.04 | 12 | 39 | 0.03 | 1.3 | 61 | 124 |
| FLT1* | 210287_s_at | 0.005 | 0.03 | 19 | 32 | 0.01 | 3.1 | 31 | 154 |
| NRP1* | 210615_at | 0.003 | 0.02 | 13 | 38 | 0.01 | 1.7 | 154 | 31 |
| PF4* | 206390_x_at | 0.003 | 0.02 | 34 | 17 | 0.01 | 3.0 | 24 | 161 |
HR: Hazard Ratio; OS: Overall Survival; Q-value = adjusted P-value;
Denotes GSE 26712: N=185;
Denotes GSE 14764: N= 63.
Conditional inference trees were used to find optimal cutpoints. Based on cutpoints, the GSE and Duke “low expression” columns represents the number of patients in the “low expression group” and “high expression” columns represents the number of patients in the “high expression group”.
Figure 1. Association between AKT1 and CD44 gene expression levels and overall survival (OS) in HGSC.
The Duke data revealed that HGSC patients whose tumors demonstrated high AKT1 and CD44 gene expression were more likely to have a long survival (OS ≥ 7 years) compared to those women whose tumors had low AKT1 and CD44 gene expression (OS ≤ 3 years, AKT1 q-value= 0.02, CD44 q-value= 0.003) (A and C, respectfully). In the TCGA database, higher AKT1 and CD44 gene expression were also associated with longer OS. (B) The median OS for HGSC patients with high AKT1 expression was 58.1 months compared to 39.9 months in patients with lower AKT1 expression (AKT1 p-value= 0.01, HR= 0.8). (D) Similarly, the median OS for HGSC patients with high CD44 expression was 47.7 months compared to 35.8 months in patients with low CD44 expression levels (CD44 p-value= 0.05, HR= 0.9). Conditional inference trees were used to find optimal cutpoints. Based on cutpoints, the TCGA and Duke “low expression” columns represents the number of patients in the “low expression group” and “high expression” columns represents the number of patients in the “high expression group”. The Duke results are shown in box plots. The distribution of the gene expression, y-axis, was estimated conditional on surviving less than three years and compared to that of the conditional distribution of surviving at least seven years. In a boxplot, the box spans the interquartile range (IQR) of the data set and the line within the box denotes the median. Potential outliers (extreme values) are represented as separate points beyond the data set “whiskers”. The whiskers illustrate the spread of the data.
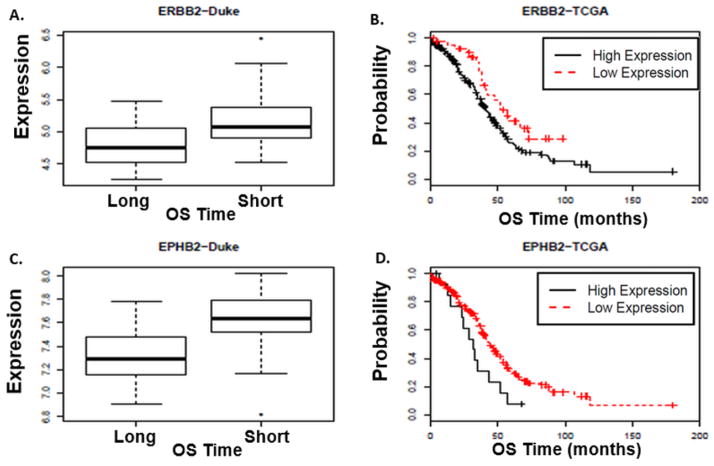
Figure 2. Association between ERBB2 and EPHB2 gene expression levels and overall survival (OS) in HGSC.
The Duke data revealed that HGSC patients whose tumors expressed high ERBB2 and EPHB2 gene expression were more likely to have a short survival (OS ≤ 3 years) compared to those women with low ERBB2 and EPHB2 expression (ERBB2 q-value= 0.02; EPHB2 q-value= 0.01) (A and C, respectfully). In the TCGA database, higher ERBB2 and EPHB2 expression levels were also associated with shorter OS. (B) The median OS for HGSC patients with high ERBB2 expression was 32 months compared to 44.5 months in patients with lower ERBB2 expression (ERBB2 p-value= 0.05, HR= 1.2). (D) The median OS for HGSC patients with high EPHB2 expression was 41.4 months compared to 52 months in patients with lower EPHB2 expression (EPHB2 p-value= 0.05, HR= 1.2). Conditional inference trees were used to find optimal cutpoints. Based on cutpoints, the TCGA and Duke “low expression” columns represents the number of patients in the “low expression group” and “high expression” columns represents the number of patients in the “high expression group”. The Duke results are shown in box plots. The distribution of the gene expression, y-axis, was estimated conditional on surviving less than three years and compared to that of the conditional distribution of surviving at least seven years. In a boxplot, the box spans the interquartile range (IQR) of the data set and the line within the box denotes the median. Potential outliers (extreme values) are represented as separate points beyond the data set “whiskers”. The whiskers illustrate the spread of the data.
Figure 3. Confirming gene expression and OS results in the GSE database.
Higher expression levels of ANGPTL3, COL4A3, FLT1, PF4 and NRP1 were associated with shorter OS in the Duke dataset and the GSE (GSE26712 and GSE14764). (A) ANGPTL3: q-value= 0.02; GSE14764 p-value= 0.02, HR= 1.5, median survival of 43 months (high expression), median survival for low expression has not been reached. (B) FLT1: q-value= 0.03; GSE26712 p-value= 0.01, HR= 3.1, median survival of 34 months (high expression) versus 64 months (low expression); (C) NRP1: q-value = 0.02; GSE26712 p-value < 0.04, HR>1.4, median survival of 20 months (high expression) versus 43 months (low expression); (D) COL4A3: q-value= 0.02; GSE26712 p-value= 0.03, HR=1.3, median survival of 33 months (high expression) versus 44 months (low expression); (E) PF4: q-value= 0.02; GSE26712 p-value= 0.01, HR= 3.0, median survival of 33 months (high expression) versus 68.5 months (low expression). Conditional inference trees were used to find optimal cutpoints. Based on cutpoints, the GSE and Duke “low expression” columns represents the number of patients in the “low expression group” and “high expression” columns represents the number of patients in the “high expression group”. The Duke results are shown in box plots. The distribution of the gene expression, y-axis, was estimated conditional on surviving less than three years and compared to that of the conditional distribution of surviving at least seven years. In a boxplot, the box spans the interquartile range (IQR) of the data set and the line within the box denotes the median. Potential outliers (extreme values) are represented as separate points beyond the data set “whiskers”. The whiskers illustrate the spread of the data.
The MVD counts based on CD105 and CD31 staining had median values of 30 and 33, respectively. After adjusting for multiple comparisons only one gene, the solute carrier family 12 (potassium/chloride transporter), member 6 (SLC12A6), revealed a significant association with MVD-CD31 counts. (CD31p-value= 0.0002, q-value= 0.04). There was no significant association between SLC12A6 expression and survival (OS p-value= 0.19, OS q-value= 0.2). However, the unadjusted analysis revealed associations between several biologically relevant genes, MVD, and survival. In the unadjusted analysis CD105-MVD and CD31-MVD were positively associated with several angiogenic gene features including vascular endothelial growth factor receptor-2 (KDR or VEGF-R2) (CD31 p-value= 0.004, q-value= 0.3), insulin growth factor (IGF)-1 (CD31 p-value= 0.01, q-value= 0.3), and Notch (Drosophila) homolog 4 (NOTCH4) (CD31 p-value= 0.01, q-value= 0.3; CD105 p-value= 0.01, q-value= 0.3). Cullin 7 (CUL7) gene expression levels were associated with both CD105 and CD31 (CD105 p-value= 0.001, q-value= 0.1; CD31 p-value= 0.02 q-value= 0.5 respectively) and OS (q-value= 0.01). CD44, and Coagulation Factor III (F3) expression levels were associated with CD105 MVD (CD44/CD105 p-value= 0.005, q-value= 0.22; for F3/CD105 p-value= 0.04, q-value= 0.7), and survival (CD44, q-value= 0.05; F3, q-value= 0.02). Complete MVD analysis results, and representative IHC staining images for CD31 and CD105 are provided in the supplemental data (Table 1, Table 2. and Figure 1).
Discussion
Our data demonstrated that 31 angiogenic related genes were differentially expressed in women with HGSC who had either short or long-term survival. In addition, 9 of these genes (AKT1, CD44, EPHB2, ERBB2, FLT1, PF4, NRP1, COL4A3, and ANGPTL3) were associated with survival in publicly available databases supporting our initial results. However, none of the genes were associated with survival in both the TCGA and GSE databases. All of the 9 genes have a role in endothelial cell signaling, activation, migration, proliferation, and/or sprouting angiogenesis. In addition, COL4A3, is involved in basement membrane development and may play an important role in extracellular matrix remodeling during angiogenesis.
In our study, women with high AKT1 and CD44, expression had longer survival. CD44 is a cell-surface glycoprotein involved in cell-cell interactions, cell adhesion and migration. CD44 has been implicated as a surface marker in cancer stem cells (CSC) and cancer initiating cells (CIC). Those cells are considered rare but highly tumorigenic and mediate tumor metastasis. In ovarian cancer, a subpopulation of the highly neoplastic progenitors of EOC are derived from CD44+CD117+ cells.[21] CD44+ has been implicated as a marker of CIC in a variety of other cancer.[21] Similar to our findings, in glioblastoma patients higher levels of CD44 expression have been reported to be associated with better survival, although higher expression of CD44 is associated with more aggressive tumor types.[22]
AKT is a serine/threonine kinase with three highly homologous members: AKT1, AKT2, AKT3. The AKT family regulates critical biological processes such as angiogenesis, apoptosis, proliferation, metabolism, cell survival, tissue invasion and tumor formation.[23] There is ongoing effort to target the AKT pathway for cancer therapy and cancer prevention.[24] Increased AKT1 kinase activity has been reported in about 40% of ovarian cancers and the PI3K/AKT pathway is activated in approximately 70% of EOC.[25, 26] The association of higher expression levels of AKT1 and longer OS was surprising because prior data suggests a worse survival for patients with higher levels of AKT1 levels.[25] However, newly published data indicated that activation of TEK might activate downstream signaling through the AKT and endothelial nitric oxide synthase 3 (NOS3 also known as eNOS) pathways. The result was the reduction of tumor metastasis, progression, and growth.[27] Our data in HGSC indicated that higher expression levels of TEK were more likely seen in women with longer OS (data not shown), which may account for our findings regarding AKT and survival. Specifically, TEK overexpression may induce increased AKT expression, but more importantly is associated with a less aggressive phenotype that is driving survival outcomes. Furthermore, we did not distinguish between total AKT and the activated phosphorylated form which may also account for our findings.[23]
Higher expression levels of ANGPTL3 were associated with shorter survival, a finding that was also found in the GSE14764 database. ANGPTL3 is a secreted protein and belongs to the angiopoietin-like family of secreted factors, involved in angiogenesis, endothelial cell adhesion, and migration.[28] Evaluation of the role of ANGPTL3 in HGSC warrants further investigation.
High EphB2 expression was associated with shorter OS. Ephrin (Eph) receptors comprise the largest subgroup of the receptor tyrosine kinase (RTK) family. RTKs and their ligands are involved in processes such as angiogenesis, cell division, migration, metabolism, survival, proliferation, and differentiation.[29] The Eph receptors and their ligands, including EphB2, have been reported to be potential prognostic and diagnostic biomarkers in various cancers.[29] Overexpression of EphB2 receptor in ovarian cancer has been associated with poorer survival which is in concordance with our results.[30]
In addition, our results indicated that overexpression of two important genes in the VEGF pathway, NRP1 and FLT, were associated with reduced OS. VEGF ligands can bind and activate three structurally similar VEGF receptors: VEGFR1 (FLT1), VEGFR2 (KDR) and VEGFR3 (FLT4). VEGF ligands including FLT1 are crucial in biological processes such as endothelial cell survival, proliferation, and migration.[31, 32] In our analysis, overexpression of FLT1 was significantly associated with worse OS, a result which was seen in the external databases (Table 3). The role of VEGF as a major factor in normal and pathological angiogenesis processes has been well established and supported the use of VEGF-targeted anti-angiogenic treatment in various malignancies.[31] The NRP1 and NRP2 receptors can act as co-receptors for VEGF–VEGFR complexes; binding promotes endothelial cell signaling and subsequently endothelial cell migration and angiogenesis. NRP1 forms complex with both FLT1 and KDR. VEGF signals can be mediated via neuropilins in the absence of FLT1 and KDR.[31] Overexpression of NRP1 has been reported in a variety of solid tumors including ovarian and breast carcinomas.[31, 32]
High COL4A3 expression was significantly associated with worse OS in HGSC patients. COL4A3 belongs to the type IV collagen family, which is the main component of basement membranes (BMs) surrounding blood vessels. In order for the malignant cells to metastasize they must penetrate through the epithelial basement membrane (EBM) and the BM that surrounds blood vessels. Overexpression of COL4A3 has been implicated to be associated with tumor size, higher grade, metastasis, and invasion in several malignancies including ovarian cancers.[33, 34]
Similarly, higher PF4 expression was seen in HGSC patients with shorter survival. PF4 is a chemokine that has been reported to possess several biological functions and is a marker of platelet activation. Thrombocytosis (platelet count of >450,000 mm3) is associated with advanced disease and shorter survival.[35] PF4 also functions as an inhibitor to endothelial cell migration, proliferation, and angiogenesis.[36] The association between PF4 and worse survival is most likely secondary to PF4 association with paraneoplastic thrombocytosis rather than its anti-angiogenic properties.
Lastly, high ERBB2 expression was associated with worse survival. ERBB2/HER2 is a well-known transmembrane receptor tyrosine kinases. ERBB2 mediates cell survival, angiogenesis, differentiation, and growth, and as an established oncogene is targeted in cancer therapy in the form of monoclonal antibody against ERBB2. In ovarian cancer elevated levels of ERBB2 are associated with worse survival which is in agreement with our findings.[37] However, anti-HER2 therapy with trastuzumab has minimal activity in women with ovarian and primary peritoneal cancer patients whose tumors demonstrated 2+ or 3+ ERBB2/HER2 overexpression.[38] The clinical utility of this marker has not been promising given the low frequency of overexpression in ovarian cancer and the lack of clinical efficacy with anti-HER2 strategies in ovarian cancer. However, newer strategies that conjugate a potent cytotoxic agent to the monoclonal antibody trastuzumab and target delivery to ERBB2 cells may have activity in select patients with HGSC.
Several authors have reported an “angiogenic” gene expression signature in ovarian cancer. TCGA gene expression data suggest there are 4 subtypes of HGSC: immunoreactive, differentiated, proliferative, and mesenchymal).[39] However, there was no significant difference in survival between the different subtypes. Gottfried and colleagues independently validated the TCGA findings in 175 HGSC samples and confirmed the existence and identity of the four subtypes.[40] In contrast to TCGA data, they found survival differences in univariate (HR= 2.4, 95% CI 1.5–4.1, p-value= 0.007) and multivariate analysis (HR= 2.3, 95% CI 1.3–4.0, p-value= 0.003) after accounting for age, stage, grade, and postoperative residual tumor.[40] Furthermore, Bentink et al reported that an angiogenic gene expression signature predicted for a novel subtype of serous ovarian cancer.[12] Their angiogenic gene signature was associated with worse survival when evaluated in 1,090 patients with advanced HGSC from a collection of 10 previously published studies, (including patients from an ovarian cancer study at Duke).[12] Specifically, women with advanced HGSC whose cancers expressed the angiogenic gene expression signature had a 1.3-fold increased risk of death compared to those who did not have the angiogenic subtype.[12] Their angiogenic signature was evaluated in the TCGA subtypes and correlated with the mensenchymal and immunoreactive groups, with 58% of the angiogenic signature tumors falling in the mesenchymal group.[12] Our data plus the findings from Gottfried et al. and Bentink et al.[12], supports the presence of an angiogenic HGSC subtype that may be useful to identify women with a worse prognosis and could be utilized as a biomarker to direct anti-angiogenic therapy.
While the degree of tumor angiogenesis, as measured by MVD, has been shown to be a prognostic factor in EOC, we did not find an association between MVD and OS after adjusting for multiple testing. However, CD31-MVD was associated with several critical angiogenic genes such as KDR, NOTCH4, IGF-1, CUL7, CD44 and F3 in the unadjusted analysis.
The strengths of our survival analysis study were the following: long term surveillance, available clinical data, and robust statistical analysis. However, none of the subjects in our cohort were treated with anti-angiogenic therapy. Future research is planned to evaluate the expression of angiogenic genes in women treated with anti-angiogenic agents. Additional limitations include those inherent to the TCGA database; the short clinical follow-up and the variety of multiple platforms utilized to determine gene expression. Analysis of the transcriptome via microarray also has inherent limitations, as it does not account for the multiple layers of regulation present in the process of transcription, translation, and protein function. An additional limitation is that the biopsy specimens had 60% tumor and may not be an accurate representation of the tumor microenvironment. Therefore, gene expression profiles specific to the tumor vasculature can be masked.
In conclusion, our findings suggest that several angiogenic-related genes are differentially expressed and associated with survival in women with HGSC. Our current findings while promising are considered preliminary and merit further investigation.
Supplementary Material
Highlights.
Angiogenic gene expression was differentially regulated in short vs. long-term high-grade serous ovarian cancer survivors.
Candidate angiogenic genes were associated with survival in independent databases.
Candidate angiogenic biomarkers may represent novel targets and rationally direct anti-angiogenic therapy.
Acknowledgments
Acknowledgement for financial support: NIH/NCI R25 training fellowship-5R25CA126938 (Integrating Population and Basic Science in Cancer Research), a Gynecological Oncology Group Ovarian Cancer Research Fund Young Investigator Award, and philanthropic funding for ovarian cancer research. As Duke Cancer Institute members, we acknowledge support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: P30 CA014236), specifically the Duke Cancer Institute’s Bioinformatics Shared Resource.
Footnotes
Conflict of interest statement:
The authors wish to report that there are no conflicts of interest with the exception of Dr. Angeles Alvarez Secord who has received research funding from Sanofi-Aventis, Genentech, Bristol-Myers Squibb, Boehringer Ingelheim, Eisai, Incyte, Endocyte, Astex, Amgen, Morphotek, and GlaxoSmithKline Corp; and has served as a consultant for Genentech, Bristol-Myers Squibb, GlaxoSmithKline Corp, and Sanofi-Aventis. In addition Drs. Angeles Alvarez Secord, Sharareh Siamakpour-Reihani, Kouros Owzar, Mark Dewhirst and Chen Jiang have a pending patent (61/825,189 filed on May 20, 2013 entitled “Differential Expression of Angiogenic Genes in Invasive High-Grade Serous Carcinomas”).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burger RA, Brady MF, Bookman MA, Fleming GF, Monk BJ, Huang H, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365:2473–83. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 2.Berchuck A, Iversen ES, Luo J, Clarke JP, Horne H, Levine DA, et al. Microarray analysis of early stage serous ovarian cancers shows profiles predictive of favorable outcome. Clin Cancer Res. 2009;15:2448–55. doi: 10.1158/1078-0432.CCR-08-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berchuck A, Iversen ES, Lancaster JM, Pittman J, Luo J, Lee P, et al. Patterns of Gene Expression That Characterize Long-term Survival in Advanced Stage Serous Ovarian Cancers. Clin Cancer Res. 2005;11:3686–96. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 4.Rubatt JM, Darcy KM, Hutson A, Bean SM, Havrilesky LJ, Grace LA, et al. Independent prognostic relevance of microvessel density in advanced epithelial ovarian cancer and associations between CD31, CD105, p53 status, and angiogenic marker expression: A Gynecologic Oncology Group study. Gynecol Oncol. 2009;112:469–74. doi: 10.1016/j.ygyno.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 5.Frumovitz M, Sood AK. Vascular endothelial growth factor (VEGF) pathway as a therapeutic target in gynecologic malignancies. Gynecol Oncol. 2007;104:768–78. doi: 10.1016/j.ygyno.2006.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teoh D, Secord AA. Antiangiogenic agents in combination with chemotherapy for the treatment of epithelial ovarian cancer. Int J Gynecol Cancer. 2012;22:348–59. doi: 10.1097/IGC.0b013e31823c6efd. [DOI] [PubMed] [Google Scholar]
- 7.Perren TJ, Swart AM, Pfisterer J, Ledermann JA, Pujade-Lauraine E, Kristensen G, et al. A Phase 3 Trial of Bevacizumab in Ovarian Cancer. N Engl J Med. 2011;365:2484–96. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 8.Raja FA, Perren TJ, Embleton A, Rustin GJS, Jayson G, Swart AM, et al. Randomised Double-Blind Phase Iii Trial of Cediranib (Azd 2171) in Relapsed Platinum Sensitive Ovarian Cancer: Results of the Icon6 Trial. Int J Gynecol Cancer. 2013;23 [Google Scholar]
- 9.Loges S, Schmidt T, Carmeliet P. Mechanisms of resistance to anti-angiogenic therapy and development of third-generation anti-angiogenic drug candidates. Genes Cancer. 2010;1:12–25. doi: 10.1177/1947601909356574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 12.Bentink S, Haibe-Kains B, Risch T, Fan JB, Hirsch MS, Holton K, et al. Angiogenic mRNA and microRNA gene expression signature predicts a novel subtype of serous ovarian cancer. Plos One. 2012;7:e30269. doi: 10.1371/journal.pone.0030269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–86. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mok SC, Bonome T, Vathipadiekal V, Bell A, Johnson ME, Wong KK, et al. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated glycoprotein 2. Cancer Cell. 2009;16:521–32. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denkert C, Budczies J, Darb-Esfahani S, Gyorffy B, Sehouli J, Konsgen D, et al. A prognostic gene expression index in ovarian cancer - validation across different independent data sets. J Pathol. 2009;218:273–80. doi: 10.1002/path.2547. [DOI] [PubMed] [Google Scholar]
- 16.Yoshihara K, Tajima A, Yahata T, Kodama S, Fujiwara H, Suzuki M, et al. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. Plos One. 2010;5:e9615. doi: 10.1371/journal.pone.0009615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–9. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owzar K, Barry WT, Jung SH. Statistical Considerations for Analysis of Microarray Experiments. Cts-Clin Transl Sci. 2011;4:466–77. doi: 10.1111/j.1752-8062.2011.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: A conditional inference framework. J Comput Graph Stat. 2006;15:651–74. [Google Scholar]
- 20.Lin DY, Zeng D, Tang ZZ. Quantitative trait analysis in sequencing studies under trait-dependent sampling. Proc Natl Acad Sci U S A. 2013;110:12247–52. doi: 10.1073/pnas.1221713110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–67. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 22.Wei KC, Huang CY, Chen PY, Feng LY, Wu TW, Chen SM, et al. Evaluation of the prognostic value of CD44 in glioblastoma multiforme. Anticancer Res. 2010;30:253–9. [PubMed] [Google Scholar]
- 23.Crowell JA, Steele VE, Fay JR. Targeting the AKT protein kinase for cancer chemoprevention. Mol Cancer Ther. 2007;6:2139–48. doi: 10.1158/1535-7163.MCT-07-0120. [DOI] [PubMed] [Google Scholar]
- 24.Bellacosa A, Kumar CC, Cristofano AD, Testa JR. Activation of AKT Kinases in Cancer: Implications for Therapeutic Targeting. In: George FVW, George K, editors. Adv Cancer Res. Academic Press; 2005. pp. 29–86. [DOI] [PubMed] [Google Scholar]
- 25.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–28. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun M, Wang G, Paciga JE, Feldman RI, Yuan ZQ, Ma XL, et al. AKT1/PKB alpha kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am J Pathol. 2001;159:431–7. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goel S, Gupta N, Walcott BP, Snuderl M, Kesler CT, Kirkpatrick ND, et al. Effects of Vascular-Endothelial Protein Tyrosine Phosphatase Inhibition on Breast Cancer Vasculature and Metastatic Progression. J Natl Cancer Inst. 2013;105:1188–201. doi: 10.1093/jnci/djt164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camenisch G, Pisabarro MT, Sherman D, Kowalski J, Nagel M, Hass P, et al. ANGPTL3 stimulates endothelial cell adhesion and migration via integrin alpha vbeta 3 and induces blood vessel formation in vivo. J Biol Chem. 2002;277:17281–90. doi: 10.1074/jbc.M109768200. [DOI] [PubMed] [Google Scholar]
- 29.Himanen J-P, Saha N, Nikolov DB. Cell–cell signaling via Eph receptors and ephrins. Curr Opin Cell Biol. 2007;19:534–42. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Q, Suo Z, Kristensen GB, Baekelandt M, Nesland JM. The prognostic impact of EphB2/B4 expression on patients with advanced ovarian carcinoma. Gynecol Oncol. 2006;102:15–21. doi: 10.1016/j.ygyno.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 31.Ellis LM, Hicklin DJ. VEGF-targeted therapy: mechanisms of anti-tumour activity. Nat Rev Cancer. 2008;8:579–91. doi: 10.1038/nrc2403. [DOI] [PubMed] [Google Scholar]
- 32.Ellis LM. The role of neuropilins in cancer. Mol Cancer Ther. 2006;5:1099–107. doi: 10.1158/1535-7163.MCT-05-0538. [DOI] [PubMed] [Google Scholar]
- 33.Georgiou GK, Igglezou M, Sainis I, Vareli K, Batsis H, Briasoulis E, et al. Impact of breast cancer surgery on angiogenesis circulating biomarkers: a prospective longitudinal study. World journal of surgical oncology. 2013;11:213. doi: 10.1186/1477-7819-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kauppila S, Saarela J, Stenback F, Risteli J, Kauppila A, Risteli L. Expression of mRNAs for type I and type III procollagens in serous ovarian cystadenomas and cystadenocarcinomas. Am J Pathol. 1996;148:539–48. [PMC free article] [PubMed] [Google Scholar]
- 35.Stone RL, Nick AM, McNeish IA, Balkwill F, Han HD, Bottsford-Miller J, et al. Paraneoplastic thrombocytosis in ovarian cancer. N Engl J Med. 2012;366:610–8. doi: 10.1056/NEJMoa1110352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bikfalvi A. Recent developments in the inhibition of angiogenesis: examples from studies on platelet factor-4 and the VEGF/VEGFR system. Biochem Pharmacol. 2004;68:1017–21. doi: 10.1016/j.bcp.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 37.Zhao D, Zhang F, Zhang W, He J, Zhao Y, Sun J. Prognostic role of hormone receptors in ovarian cancer: a systematic review and meta-analysis. Int J Gynecol Cancer. 2013;23:25–33. doi: 10.1097/IGC.0b013e3182788466. [DOI] [PubMed] [Google Scholar]
- 38.Bookman MA, Darcy KM, Clarke-Pearson D, Boothby RA, Horowitz IR. Evaluation of monoclonal humanized anti-HER2 antibody, trastuzumab, in patients with recurrent or refractory ovarian or primary peritoneal carcinoma with overexpression of HER2: a phase II trial of the Gynecologic Oncology Group. J Clin Oncol. 2003;21:283–90. doi: 10.1200/JCO.2003.10.104. [DOI] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottfried E, Konecny CW, Winterhoff Boris, Dering Judy, Ginther Charles, Chen Hsiao-Wang, Hamidi Habib, Podratz Karl C, Cliby William, Dowdy Sean Christopher, Haluska Paul, Hartmann Lynn C, Kalli Kimberly, Goode Ellen L, Slamon Dennis J David Geffen School of Medicine at University of California, Los Angeles, Los Angeles, CA; Mayo Clinic, Rochester, MN; 1. Division of Gynecologic Surgery, Department of Obstetrics and Gynecology, Mayo Clinic, Rochester, Rochester, MN; University of California, Los Angeles, School of Medicine/Translational Oncology Research Laboratory, Los Angeles, CA; University of California, Los Angeles, Translational Oncology Research Laboratory, Santa Monica, CA; Mayo Medical Center, Rochester, MN. . Prognostic relevance of gene signatures in high-grade serous ovarian carcinoma. 2013 ASCO Annual Meeting. J Clin Oncol. 2013;31(suppl):abstr 5509. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.