Abstract
Microsporidia are highly divergent fungi that are obligate intracellular pathogens of a wide range of host organisms. Here we review recent findings from the genome sequences of mosquito-infecting microsporidian species Edhazardia aedis and Vavraia culicis, which show large differences in genome size, although similar numbers of predicted genes. We also show a video of E. aedis polar tube firing, which is the dramatic mechanism used by microsporidia to deliver the germ cell (sporoplasm) into the host cell to initiate intracellular infection.
Keywords: Polar tube, microsporidia, Edhazardia aedis, Vavraia culicis, mosquito, genome sequence, Aedes aegypti
1. Introduction
The phylum Microsporidia contains over 1400 species of obligate intracellular pathogens that infect a wide range of hosts, from invertebrates to mammals (Vavra and Lukes, 2013). Microsporidia were originally proposed to be protozoans or ‘ancient eukaryotes’, but with a growing database of genome sequence information it has become clear they are most closely related to fungi. Recent phylogenomic analyses have placed them together with the Cryptomycota as the earliest branching clade in the fungal kingdom (James et al., 2013). Microsporidia have dramatic mechanisms of invasion into their host cells with a polar tube infection apparatus that is used to deliver a cell wall-deficient ‘sporoplasm’ into the host cell (Xu and Weiss, 2005). This dramatic polar tube ‘firing’ has been described for several species of microsporidia and previous studies have described which conditions will induce germination for various species. While the dynamics of polar tube firing have been described before (Frixione et al., 1997), this report represents the first video publication of this dramatic event specific to the phylum Microsporidia. In particular, we show a video of polar tube firing of the mosquito-infecting species Edhazardia aedis, which has recently been subjected to genome analysis, together with another mosquito-infecting species Vavraia culicis (Desjardins et al., 2015). By describing these recent genomic and transcriptomic findings, together with a video of the most distinctive feature of microsporidia, we aim to facilitate understanding and increase exposure for these ubiquitous, but poorly understood parasites.
2. Genomic and transcriptomic analysis of mosquito-infecting microsporidian species E. aedis and V. culicis
For more than 100 years microsporidia have been studied in mosquitoes because they are excellent model systems to investigate applied studies such as use for biocontrol or in basic studies to resolve complex developmental cycles and host-pathogen relationships (Becnel et al., 2005). Two microsporidian species in particular have been studied for their ability to infect mosquitoes: E. aedis, which is a specialist species that specifically infects the yellow fever mosquito Aedes aegypti, and V. culicis, which is a generalist species that infects a wide range of mosquito species including important Anopheles spp. that vector malaria. Recent genomic and transcriptomic analysis has demonstrated interesting differences and similarities between these species. Surprisingly, the E. aedis genome is 51.3 Mb, which almost 10-fold larger than the 6.1 Mb V. culicis genome (Table 1). As such, the E. aedis genome represents the largest microsporidian genome sequenced to date. Previously, the largest genome reported was 25 Mb (Corradi et al., 2009). Microsporidian genomes are famous for their reduction and compaction, with the human-infecting microsporidian species Encephalitozoon intestinalis having the smallest known eukaryotic genome at 2.3 Mb. The increase in E. aedis genome size is not due to repetitive sequence, but rather due to expansion of AT-rich intergenic regions, perhaps suggesting additional regulation of gene expression. Although the E. aedis genome is much larger than the genomes of V. culicis and other microsporidian species, it has only about a 1.5-fold increase in gene content with 4190 predicted genes. This is in comparison to 2773 predicted genes in V. culicis, which is similar to the number of predicted genes in microsporidian species that infect humans, insects and nematodes (Table 1). These findings are similar to other microsporidian genomes, where an increase in genome size does not appear to be accompanied by a similar increase in magnitude of the predicted proteome size. Interestingly, the increased gene content in E. aedis appears not to be due to retention of genes from the last common ancestor shared with true fungi, but rather due to species-specific expansion of genes, which has been observed in the genomes of other microsporidian species.
Table 1.
Genome size and predicted gene number for several microsporidian species
| Species | Host | Genome size (Mb) | Predicted gene number | Reference |
|---|---|---|---|---|
| Edhazardia aedis | yellow fever mosquito Aedes | 51.3 | 4190 | (Desjardins et al., 2015) |
| Vavraia culicis | wide range of mosquito species | 6.1 | 2773 | (Desjardins et al., 2015) |
| Enterocytozoon bieneusi | humans | ~6 | 3804 | (Akiyoshi et al., 2009) |
| Encephalitozoon intestinalis | humans | 2.3 | 1833 | (Corradi et al., 2010) |
| Encephalitozoon cuniculi | humans | 2.9 | 1999 | (Katinka et al., 2001) |
| Nosema ceranae | honey bees | 7.86 | 2614 | (Cornman et al., 2009) |
| Spraguea lophii | fish | 6.2 to 7.3 | 2573 | (Campbell et al., 2013) |
| Trachipleistophora hominis | humans | 8.5 – 11.6 | 3266 | (Heinz et al., 2012) |
| Nematocida parisii | Caenorhabditis nematodes | 4.1 | 2661 | (Cuomo et al., 2012) |
| Nematocida sp1 | Caenorhabditis nematodes | 4.7 | 2770 | (Cuomo et al., 2012) |
In addition to genome sequencing of these two microsporidian species, transcriptomic analysis was performed on various stages of the microsporidian life cycle (Desjardins et al., 2015). In the simplest overview, the microsporidian life cycle begins with the horizontally transmissible spore form, which fires its polar tube in the midgut to invade host cells, where it replicates intracellularly and eventually differentiates back into spores that escape back into the environment. Different microsporidian species have more complex versions of this cycle, involving horizontal or vertical transmission, and sometimes multiple forms of replicative cells, which we do not discuss here because of space constraints. In general, transcriptomic analysis of the spores of E. aedis and V. culicis indicated similar gene expression, with primarily expression of genes that encode ribosomal proteins and Hsp70 domain proteins, indicating a focus of these spores on protein production and protein folding. In contrast, there were very distinct expression patterns in the replicative forms of E. aedis and V. culicis, with genes upregulated in E. aedis enriched for Gene Ontology (GO) terms for growth, carbohydrate metabolism and DNA replication, as well as genes of unknown function that are predicted to encode secreted proteins. Genes upregulated in the replicative form of V. culicis were enriched for GO terms for protein modification and trafficking, whereas there was not an enrichment in genes predicted to encode secreted proteins. These differences may reflect the specialist vs. generalist lifestyles of E. aedis and V. culicis, with the specialist E. aedis involved in a host/pathogen arms race with its mosquito host Ae aegypti.
3. Kinetics of polar tube firing in E. aedis
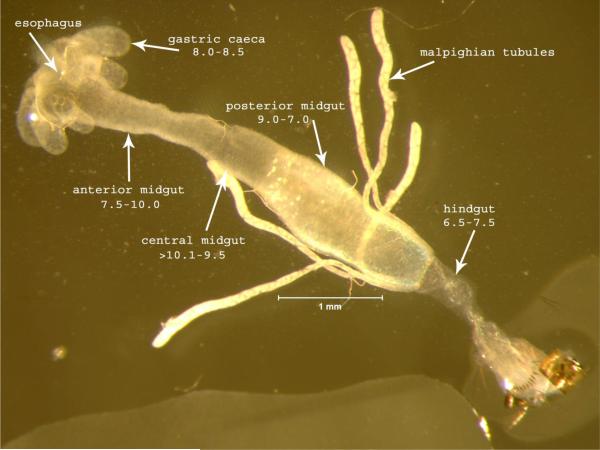
One of the most distinctive features of microsporidia is the polar tube infection apparatus. Traditionally this structure has been called the polar filament when coiled within the spore, and then has been referred to as a polar tube outside the spore after it everts. For simplicity, here we will refer to it as the polar tube. The polar tube is coiled inside of the transmissible spore form until it receives a stimulus, at which point it fires dramatically and everts outside of the spore, in an event called germination. This tube is thought to pierce a host cell and then inject a single microsporidian parasite directly into that cell, although there is evidence that invasion can also occur through phagocytosis ((Leitch et al., 2005) and references therein). There are a variety of stimuli that have been shown to induce polar tube firing, with different conditions used for different species. Polar tube firing characterization has been performed with mosquito-infecting microsporidia because of the size of spores and ease of germination in vitro. Previous studies have characterized in detail the conditions that will induce Edhazardia germination (Undeen and Becnel, 1992). In particular, this event is influenced by both the type of cation present and the pH, with the optimal germination conditions identified to be 0.1M KCl at pH10.5-11, although germination can also be triggered with other monovalent ions. Here we show the dynamics of this polar tube firing in E. aedis spores treated with 0.1M KCl pH10.5-11 (Video File 1). Interestingly, the digestive tract of the mosquito has a pH range that should facilitate firing in the anterior-central midgut, where E. aedis commonly germinates to infect gastric caecal cells (Figure 1). While the underlying mechanisms of this dramatic event remain poorly understood, the firing can be divided into several distinct phases, including 1) activation, 2) increase in intrasporal osmotic pressure, 3) eversion of the polar tube outside the spore, and 4) passage of sporoplasm through the polar tube. A clear event shown in the last frames of this video is the expanding posterior vacuole that appears to push the spore contents into the everted polar tube. This video should provide a useful resource for presentations and analysis of this dramatic polar tube firing event that characterizes the microsporidia.
Figure. Ae. aegypti digestive gut.
Dissected alimentary canal from a larva of Aedes aegypti indicating the pH shifts in each region from anterior to posterior. Spores enter through the esophagus and are carried to the midgut where they encounter digestive enzymes and pH shifts that contribute to initiating the germination process.
4. Conclusions
Microsporidia are morphologically quite distinct from fungi but share a number of traits including spore formation, chitin as a component of the spore wall, the presence of trehalose, and a closed mitosis (Vavra and Lukes, 2013). These characters are not unique to the fungi and microsporidia (some occur in a number of protist groups) but a more definitive link between the two groups has been made with genome sequencing. Several phylogenomic comparisons between fungi and microsporidia have concluded that the microsporidia can best be considered the earliest diverging branch of fungi or their sister group (Vavra and Lukes, 2013). The new findings in genome sequence and transcriptional analysis for these two divergent microsporidian species from mosquitoes provides additional information to understand core microsporidian genes and pathways as well as specific adaptations to different hosts by both generalist and specialist species.
The environmental spores of E. aedis demonstrated in this video contain an injection apparatus composed of a polar filament, polaroplast and posterior vacuole. It is generally thought that germination is an osmotic event with various stimuli (pH shifts, ion concentrations etc.) that results in tremendous internal pressure (for E. aedis ~40 atmospheres). The spores rupture at the apex and the filament is expelled and everted to become a tube through which the spore contents are forced out, likely by the expanding posterior vacuole, and released at the tip (Video File 1). There remain many unanswered questions including the dynamics and factors that cause spore germination, as well as the very basic aspects of the manner in which the polar tube penetrates/enters the host cell. The recently acquired genome information coupled with functional studies should shed light on this intriguing invasion process common to microsporidia.
Supplementary Material
Highlights.
Review of genome sequences for two microsporidian species that infect mosquitoes
Largest genome size reported to date for microsporidia, which characteristically have small genomes
Video of the dynamics of polar tube firing in mosquito-infecting microsporidia
Acknowledgements
E.R.T. acknowledges support from NIAID R01 AI087528. J.J.B. acknowledges support from the USDA-Agricultural Research Service and Neil Sanscrainte for video editing.
Abbreviations
- Mb
megabase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyoshi DE, Morrison HG, Lei S, Feng X, Zhang Q, Corradi N, Mayanja H, Tumwine JK, Keeling PJ, Weiss LM, et al. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS pathogens. 2009;5:e1000261. doi: 10.1371/journal.ppat.1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel JJ, White SE, Shapiro AM. Review of microsporidia-mosquito relationships: from the simple to the complex. Folia Parasitol (Praha) 2005;52:41–50. [PubMed] [Google Scholar]
- Campbell SE, Williams TA, Yousuf A, Soanes DM, Paszkiewicz KH, Williams BA. The genome of Spraguea lophii and the basis of host-microsporidian interactions. PLoS genetics. 2013;9:e1003676. doi: 10.1371/journal.pgen.1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Chen YP, Schatz MC, Street C, Zhao Y, Desany B, Egholm M, Hutchison S, Pettis JS, Lipkin WI, et al. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS pathogens. 2009;5:e1000466. doi: 10.1371/journal.ppat.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, Haag KL, Pombert JF, Ebert D, Keeling PJ. Draft genome sequence of the Daphnia pathogen Octosporea bayeri: insights into the gene content of a large microsporidian genome and a model for host-parasite interactions. Genome biology. 2009;10:R106. doi: 10.1186/gb-2009-10-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, Pombert JF, Farinelli L, Didier ES, Keeling PJ. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat Commun. 2010;1:77. doi: 10.1038/ncomms1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, Desjardins CA, Bakowski MA, Goldberg J, Ma AT, Becnel JJ, Didier ES, Fan L, Heiman DI, Levin JZ, et al. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 2012;22:2478–2488. doi: 10.1101/gr.142802.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Sanscrainte ND, Goldberg JM, Heiman D, Young S, Zeng Q, Madhani HD, Becnel JJ, Cuomo CA. Contrasting host-pathogen interactions and genome evolution in two generalist and specialist microsporidian pathogens of mosquitoes. Nat Commun. 2015;6:7121. doi: 10.1038/ncomms8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frixione E, Ruiz L, Cerbon J, Undeen AH. Germination of Nosema algerae (Microspora) spores: conditional inhibition by D2O, ethanol and Hg2+ suggests dependence of water influxupon membrane hydration and specific transmembrane pathways. The Journal of eukaryotic microbiology. 1997;44:109–116. doi: 10.1111/j.1550-7408.1997.tb05946.x. [DOI] [PubMed] [Google Scholar]
- Heinz E, Williams TA, Nakjang S, Noel CJ, Swan DC, Goldberg AV, Harris SR, Weinmaier T, Markert S, Becher D, et al. The genome of the obligate intracellular parasite Trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution. PLoS pathogens. 2012;8:e1002979. doi: 10.1371/journal.ppat.1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, Pelin A, Bonen L, Ahrendt S, Sain D, Corradi N, Stajich JE. Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr Biol. 2013;23:1548–1553. doi: 10.1016/j.cub.2013.06.057. [DOI] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Leitch GJ, Ward TL, Shaw AP, Newman G. Apical spore phagocytosis is not a significant route of infection of differentiated enterocytes by Encephalitozoon intestinalis. Infection and immunity. 2005;73:7697–7704. doi: 10.1128/IAI.73.11.7697-7704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undeen AH, Becnel JJ. Longevity and Germination of Edhzardia aedis (Microspora: Amblyosporidae) Spores. Biocontrol Science and Technology. 1992;2:247–256. [Google Scholar]
- Vavra J, Lukes J. Microsporidia and 'the art of living together'. Advances in parasitology. 2013;82:253–319. doi: 10.1016/B978-0-12-407706-5.00004-6. [DOI] [PubMed] [Google Scholar]
- Xu Y, Weiss LM. The microsporidian polar tube: a highly specialised invasion organelle. Int J Parasitol. 2005;35:941–953. doi: 10.1016/j.ijpara.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.