Abstract
Current prognostic factors are insufficient for precise risk-discrimination in breast cancer patients with low grade breast tumors, which, in disagreement with theoretical prognosis, occasionally form early lymph node metastasis. To identify markers for this group of patients, we employed iTRAQ-2DLC-MS/MS proteomics to 24 lymph node positive and 24 lymph node negative grade 1 luminal A primary breast tumors. Another group of 48 high-grade tumors (luminal B, triple negative, Her-2 subtypes) was also analyzed to investigate marker specificity for grade 1 luminal A tumors. From the total of 4405 proteins identified (FDR<5%), the top 65 differentially expressed together with 30 previously identified and control markers were analyzed also at transcript level. Increased levels of carboxypeptidase B1 (CPB1), PDZ and LIM domain protein 2 (PDLIM2), and ring finger protein 25 (RNF25) were associated specifically with lymph node positive grade 1 tumors, whereas stathmin 1 (STMN1) and thymosin beta 10 (TMSB10) associated with aggressive tumor phenotype also in high grade tumors at both protein and transcript level. For CPB1, these differences were also observed by immunohistochemical analysis on tissue microarrays. Up-regulation of putative biomarkers in lymph node positive (versus negative) luminal A tumors was validated by gene expression analysis of an independent published data set (n = 343) for CPB1 (p = 0.00155), PDLIM2 (p = 0.02027) and RELA (p = 0.00015). Moreover, statistically significant connections with patient survival were identified in another public data set (n = 1678). Our findings indicate unique pro-metastatic mechanisms in grade 1 tumors that can include up-regulation of CPB1, activation of NF-κB pathway and changes in cell survival and cytoskeleton. These putative biomarkers have potential to identify the specific minor subpopulation of breast cancer patients with low grade tumors who are at higher than expected risk of recurrence and who would benefit from more intensive follow-up and may require more personalized therapy.
Breast cancer is the most common form of cancer in women worldwide and distant metastases are the main reasons for patient mortality. Cancer emerges as a consequence of multiple genetic aberrations, whereas metastatic characteristics may be predisposed or acquired during disease development and are governed by a number of genetic and biochemical mechanisms (1, 2). In clinical practice, both traditional and molecular prognostic markers are used for risk-group discrimination and determination of metastatic potential. Traditional prognostic markers in breast cancer involve age at diagnosis, tumor size and grade, lymph node status, and presence of distant metastasis. Tumor size is a potent prognostic factor predicting higher probability of metastatic behavior for larger tumors. More differentiated tumors (e.g. grade 1) have low dissemination potential in general, although less differentiated, more proliferative high grade tumors (e.g. grade 3) form metastases much more frequently. Low grade breast tumor cells spread predominantly via lymph vessels and lymph nodes are therefore the first site of tumor cell dissemination prior to eventual spread into distant organs such as lung or bone (3). Molecular prognostic markers involve hormonal receptors (estrogen receptor (ER)1, progesterone receptor (PR)), Her-2/neu receptor, and expression panels like Oncotype DX and MammaPrint. Also, the American Society for Clinical Oncology (ASCO) has recommended urokinase plasminogen activator (PLAU) and urokinase plasminogen activator inhibitor (SERPINE1) as indicative factors for metastatic potential in breast cancer (4, 5), however their use in clinical practice has not been generally accepted (4).
Currently available markers are not sufficient for precise risk-group or individual assessment specifically in low grade luminal-A tumors, whose general prognosis is very favorable, resulting in treatment by less aggressive adjuvant therapy and no chemotherapy. However, a low percentage of these tumors develop early lymph node metastases. The molecular mechanism of this phenomenon is not known and current clinical practice lacks the means for predicting its occurrence. New knowledge is thus essential for identifying biomarkers that can identify high risk individuals within the predominantly low risk population of patients with low grade breast cancers. These high risk patients should then receive more intensive follow-up and could be considered for more aggressive therapy, which cannot be achieved currently in view of the detrimental effects of therapy on the majority of patients who will not show benefit. In addition, understanding the mechanisms of metastasis of low grade breast cancer may lead to the identification of new therapeutic targets.
Shotgun proteomics with isobaric tags for relative and absolute quantification (iTRAQ) is an established approach for quantification of proteins related to cancer metastasis (6, 7). Moreover, recent developments made to multidimensional liquid chromatography and mass spectrometry, including the FT-Orbitrap detector technology, have significantly advanced the discovery proteomics field (8). We have used this untargeted quantitative approach to identify proteins correlating with lymph node metastasis in low grade breast cancer. A complementary targeted transcriptomics study was performed on the same sample set to identify those proteins that exhibited correlation with gene expression. The combining of protein and transcript level profiles allowed us to interrogate independent large patient data sets for validation and their impact on survival. In addition, we compared G1 and G3 tumors with and without metastasis to see whether the metastatic process is the same or different between these groups, providing fundamental information into the mechanisms of tumor progression in different tumor grades.
EXPERIMENTAL PROCEDURES
Tissue Procurement and Patient Characteristics
Patient informed consent forms along with tissue procurement procedures were approved by the ethics committee of the Masaryk Memorial Cancer Institute (MMCI) (see supplemental File 1 for ethics documents). Tissues were frozen in liquid nitrogen within 20 min after surgical removal and stored at −180 °C in tissue bank of MMCI. A complementary formalin fixed, paraffin embedded tissue block was available for each sample for histological evaluation and immunohistochemical (IHC) analysis. A set of 96 preoperatively untreated breast carcinomas of 11–20 mm maximum diameter (pT1c) was selected for analysis. The sample set characteristics are shown in supplemental File 1 in detail and the study design is summarized in Fig. 1. The sample set included 48 grade 1 tumors positive for both ER and PR, without HER2 amplification; 24 of these had lymph node metastases at the time of operation, selected from a total collection of about 4000 carcinoma cases, as they are quite rare. The matching node negative pT1c grade 1 tumors exhibiting identical profiles of ER, PR, and HER2 were selected randomly from the total collection.
Fig. 1.
Experimental design based on three iTRAQ eight-plex experiments, overall workflow and overview of key data outputs across the study. The design allowed analysis of 24 samples (each pooled from 4 patients' lysates, 24 × 4 = 96 patients involved); these 24 samples are represented by 24 rectangles in three rows corresponding to three iTRAQ eight-plex experiments (E1, E2, E3). Each pooled sample in each of three eight-plex experiments was labeled by one of eight iTRAQ labels (113, 114, 115, 116, 117, 118, 119, 121). Patient's numbers correspond with their data in Supplemental file 1. The colors represent grouping of samples according to tumor grade and lymph node status: Lymph node positive grade 1 (red) were labeled with iTRAQ 113 and 114 labels in all three eight-plex experiments, lymph node negative grade 1 (yellow) were labeled with iTRAQ 115 and 116 labels, lymph node positive grade 3 (all tones of blue) were labeled with iTRAQ 117 and 118 labels and lymph node negative grade 3 (all tones of green) were labeled with iTRAQ 119 and 121 labels. Stratification of the samples according to estrogen receptor (ER), progesterone receptor (PR) and HER2 receptor across the sample set is also shown in the figure: All grade 1 tumors (luminal A subtype) were ER positive, PR positive, and HER2 negative (red and yellow color, depending on lymph node status). The grade 3 tumor group involved subgroups with different ER, PR, and HER2 status: (i) Triple negative (ER, PR, and HER2 negative), (b) luminal B type (ER positive, PR positive or negative, and HER2 negative), (c) luminal B HER2 positive (ER positive, PR positive or negative, and HER2 positive), (g) HER2 positive (and ER, PR negative) and luminal B HER2 positive; all subtypes both lymph node positive and negative. These subgroups are distinguished by different tones of blue (lymph node positive) or green (lymph node negative). Samples with the same color tone belong to the same cancer subtype with the same lymph node status and are thus considered as biological replicates. For more clinical characteristics of individual patients see Supplemental file 1.
To investigate similarities and difference in metastasis biomarkers in high and low grade breast cancers, a second sample set was collected of 48 pT1c grade 3 carcinomas, 24 of them node positive and 24 node negative. These were selected to ensure representation of the following immunophenotypes: triple negative (n = 16); tumors with HER2 amplification (n = 16), of which 8 were ER positive and 8 ER negative; and ER positive tumors without HER2 amplification (n = 16), always half of them with and half without metastases.
For all samples, frozen tumor samples were cut into two pieces used for (1) RNA isolation for confirmation of RNA integrity and for qRT-PCR analyses on TaqMan Low density arrays, and (2) protein isolation for iTRAQ-2DLC-MS/MS analysis. The set of the same 96 tumors was used for proteomics, transcriptomics, and IHC analyses. An independent set of 64 additional grade 1 luminal A breast carcinomas was used for IHC validation of CPB1 protein levels. These samples were preoperatively untreated, ER positive, PR positive, HER2 negative tumors of 11–30 mm maximum diameter (T1c-T2); 43 were lymph node negative and 21 lymph node positive.
RNA Isolation, RNA Integrity Control and Reverse Transcription
After homogenization in a MM301 mechanical homogenizer (Retsch, Haan, Germany) using a metal ball for 2 × 2 min at 25 s−1 in 600 μl of RLT buffer (Qiagen, Hilden, Germany) with 1% β-mercaptoethanol, total RNA was isolated using RNeasy Mini Kit (Qiagen) following the manufacturer's protocol. RNA was eluted with 30 μl of RNase-free water, quantified at 260 nm using NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE) and quality checked by measurement of RNA integrity number (RIN) on Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Samples that did not pass the criterion of RNA quality (RIN>7) were excluded and replaced by other tissues with the same clinicopathological characteristics for the whole study (transcriptomics, proteomics, and IHC). The final sample set is presented in supplemental File 1. Isolated RNA was stored at −80 °C. Total RNA (0.9 μg) was reverse transcribed using H Minus M-MuLV Reverse Transcriptase (Fermentas, Vilnius, Lithuania) according to the manufacturer's protocol with the use of random hexamer primers (Fermentas).
Proteomics Sample Preparation, Pooling, Digestion, and Labeling
One hundred and fifty microliters lysis solution (0.5 m triethylammonium bicarbonate, pH 8.5; 0.05% w/v SDS) was added to each tissue and homogenized in a mechanical homogenizer (Retsch Technology, Haan, Germany) using a metal ball for 2 × 2 mins at 25 s−1. The homogenates were then subjected to needle sonication (Bandelin 2200 Ultrasonic homogenizer, Bandelin, Germany, 30 × 0.1 s pulses at 50 W), kept on ice for 1 h, centrifuged at 14000 × g for 20 min at 4 °C and the total protein in the supernatant quantified using a modified Bradford assay (Bio-Rad, Hercules, CA) according to manufacturer's instruction.
To match the sample set size with the sample capacity of iTRAQ-2DLC-MS/MS approach, sample pooling was performed as outlined in Fig. 1 with details presented in supplemental File 1: 25 μg (total protein) aliquots of tumor lysates from four patients with the same clinicopathological characteristics (tumor grade, lymph node status, ER and HER2 status) were pooled together. The pooled lysates containing 100 μg of protein in 20 μl of 0.5 m triethylammonium bicarbonate, pH 8.5 and 0.05% w/v SDS were subjected to reduction of cysteine S-S bridges by the addition of 2 μl of 50 mm tris-2-carboxyethyl phosphine (TCEP), followed by incubation for 1 h at 60 °C. Cysteines were blocked by adding 1 μl 200 mm methyl methanethiosulfonate in isopropanol and 10 min incubation at room temperature. For trypsin digestion, 6 μl of freshly prepared trypsin (Roche, Mannheim, Germany) solution (500 ng/μl) were added and incubated for 12 h at RT (protein/trypsin ratio ∼1:30). Labeling with iTRAQ eight-plex (ABSCIEX, Darmstadt, Germany) was performed at RT for 2 h according to the manufacturer's instructions (see Fig. 1 for the design of three iTRAQ experiments and supplemental File 1 for the tissue specimens involved). The samples in each eight-plex were then mixed and evaporated in a centrifugal vacuum concentrator to 100 μl final volume.
ZIC-HILIC Peptide Fractionation
A Dionex P680 HPLC Pump with PDA-100 photodiode Array Detector and SeQuant ZIC-pHILIC column 150 × 4.6 mm, 5 μm with a corresponding precolumn (20 × 2.1 mm, 5 μm) were used for peptide fractionation. Mobile phase (A) comprised 0.1% ammonium hydroxide in acetonitrile. Mobile phase (B) comprised 10 mm ammonium formate in water adjusted with ammonium hydroxide to pH 10. The mixture of iTRAQ labeled peptides (100 μl) was diluted with 100 μl ZIC-HILIC mobile phase (A) and centrifuged at 14000 × g 10 min at RT. Sample injection volume was 190 μl. Separation conditions were: isocratic 10% B for 10 min; gradient up to 40% B over 40 min; gradient up to 100% B over 40 min; isocratic 100% B for 10 min and gradient down to 10% B over 5 min. Flow rate was 0.4 ml/min, column temperature 30 °C and UV detection at 280 and 254 nm. Thirteen to fourteen fractions were collected from each fractionation and dried with centrifugal vacuum concentrator.
LC-MS/MS Analysis
All LC-MS/MS experiments were performed by nanoscale reversed phase liquid chromatography (Easy-nLCII, Thermo Fisher Scientific, Bremen, Germany) coupled on-line to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Individual ZIC-HILIC fractions were redissolved in 15 μl 0.1% formic acid (aq) and 12 μl of each fraction was loaded onto a peptide cartridge (CapTrap, Michrom Bioresources, Auburm, CA) using water containing 0.1% formic acid (mobile phase A) at 10 μl/min. Peptide separation was performed on a pulled tip column (15 cm × 100 μm id) containing C18 Reprosil, 5 μm particles (Nikkyo Technos, Tokyo, Japan) using increasing amounts of acetonitrile containing 0.1% formic acid (mobile phase B) at 300 nl/min. Gradient conditions were: 5–40% B (0 to 40 min), 40–80% B (5 min), 80% B held for 4 min, 80–5% B (1 min).
Protein identification and quantification was based on accurate precursor mass measurements and high resolution/accurate fragmentation data. The mass spectrometer was operated in positive ion mode and a data-dependent “Top 10” method was employed. In each cycle a full scan spectrum was acquired in Orbitrap Velos (Thermo Fisher Scientific) (m/z range 400–2000) at a target value of 1 × 106 ions (two microscans) with resolution r = 30.000 at m/z 400 followed by higher energy collision dissociation (HCD) on the 10 most intense ions with a target value of 5 × 104 ions (1 microscan). Fragment ions were measured in the Orbitrap mass analyzer with resolution r = 7500 at m/z 400. The ‘lock mass’ function was enabled for the MS mode, where the background ion at m/z 445.1200 was used as the lock mass ion. General mass spectrometric conditions were as follows: spray voltage, 1.75 kV; no sheath or auxiliary gas flow; S-lens, 60%. FT preview mode was disabled, charge state screening enabled and rejection of singly charged ions enabled. Ion selection thresholds were 500 counts for MS2, isolation width 1.2 Th, HCD normalized collision energy was 42. Dynamic exclusion was employed and ±5 ppm window of the selected m/z was excluded for 30 s.
Proteomics Data Analysis
Protein identification and quantification in the iTRAQ experiments was performed with Proteome Discoverer™ version 1.1 software (Thermo Fisher Scientific, Bremen, Germany) using the Mascot database search algorithm (Mascot server version 2.2.4, Matrixscience, London, UK). The data analysis parameters were as follows: Spectrum properties filter: Peptide mass range: 800–7000 Da. Peak filters: S/N = 3. Input data: Protein database: SwissProt (version 2010_04), enzyme name: Trypsin (cleaving polypeptides at the carboxyl side of lysine or arginine except when either is followed by proline), max. missed cleavage sites 2, taxonomy: Homo sapiens (20279 human protein entries were searched in total). Peptide scoring options: Peptide cut-off score: 10 (default by Proteome Discoverer). Protein scoring options: Use MudPIT Scoring: Yes. Protein relevance threshold: 20. Decoy database search: True. Target FDR 0.05 (as calculated by Proteome Discoverer). Tolerances: 5 ppm precursor mass tolerance and 0.8 Da fragment mass tolerance. Modifications: Dynamic (variable): Phosphorylation (STY), oxidation (M), deamidation (NQ), acetylation (K). Static (fixed): iTRAQ eight-plex (K, N-term), methylthio (C). Quantitation method: iTRAQ eight-plex (Thermo Scientific Instruments). Protein quantification was based on unique peptides (supplemental File 2) with at least three quantitative ratios using statistical analysis described below. Protein grouping function was disabled.
Statistical Analysis of Proteomics Data
Loess and global median normalization was used to process the proteomics data. Data were log2-transformed and analyzed on both peptide and protein level. Statistical significance of observed fold-change ratios was determined by one sample t test. p values were adjusted for multiple hypothesis testing by Benjamini-Hochberg procedure. To select differentially expressed proteins for further validation, three criteria were applied in parallel: (1) Fold change higher than 1.2 for up-regulation or lower than 0.8 for down-regulation, (2) lower limit of the fold change confidence interval above 1.0 for up-regulation and upper limit below 1.0 for down-regulation; (3) FDR adjusted p values < 0.05 were considered as statistically significant for proteins with high and medium number of observations (N>10) and FDR adjusted p values of 0.1 for proteins with low number of observations (N ≤ 10). The rationale for choosing the above criteria was to filter out the false positive protein level changes caused by (1) technical method variability up to 20% (criterion 1), (2) inconsistent protein levels accross biological replicates (criteria 2 and 3), and (3) variability in levels of various peptides representing a single protein (criteria 2 and 3). Higher threshold of FDR adjusted p values (0.1) was allowed to not filter out proteins whose quantification relied on low numbers of peptides; this criterion was chosen based on the HER2/ERBB2 protein change between HER2-positive and HER2-negative carcinomas (P04626, n = 4, FCH = 5.27, p = 0.099, Supplemental file 4, HER2+ versus HER2- sheet and supplemental File 5); HER2 tumor positivity/negativity was independently determined by IHC. All calculations were performed in R 2.10 (9) using packages from Bioconductor (www.bioconductor.org).
qRT-PCR Gene Expression Analysis Using Low Density Arrays
Gene expression analysis of 95 genes (see supplemental File 6 for detailed information on the TaqMan assays used) in 96 primary breast cancer tissues was performed using Low Density Arrays (Micro Fluidic Card System, 384 qRT-PCR reactions/card) on 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Each RNA isolated from an individual tissue piece was analyzed in three analytical replicates that were run independently in three different cards. 100 μl of sample mix containing 200 ng of cDNA and 1× TaqMan Universal PCR Master Mix UNG was used in each loading reservoir. All analysis parameters were electronically provided by the manufacturer together with the custom MicroFluidic Cards. Gene expression was evaluated by the comparative CT method of relative quantification using RQ Manager 1.2 software (Applied Biosystems) with 18S rRNA as an endogenous control. The baseline was established manually to 0.25 for all samples and ΔCT values were exported for external statistical evaluation.
Statistical Analysis of qRT-PCR Data
The comparative Ct method (10) was used to calculate ΔΔCt, r = 2−ΔΔCt and their standard deviations. Discrepancies in observation numbers for each gene were inspected by Fisher exact test. Ct values were then compared by two sample t test, the results were considered significant when p ≤ 0.05.
Correlation Analysis and Hierarchical Clustering
Fisher Z-transformed Pearson's correlation was used as the distance measure between samples coupled with average linkage criterion. Spearman correlation coefficients for each pair of samples in qRT-PCR study were computed. Additionally, qRT-PCR profiles from samples pooled in proteomic experiments were averaged and Spearman correlation coefficients for combined proteomics/qRT-PCR profiles calculated.
Immunohistochemistry (IHC) on Tissue Microarrays (TMA)
IHC was used to evaluate protein levels and staining patterns of selected putative biomarkers in TMAs prepared from formaldehyde fixed, paraffin embedded blocks, taken in parallel with the frozen tissue. Antibodies and protocols are presented in supplemental File 9. The antibody dilutions were optimized at test TMA from breast carcinomas from the same sample set. The paraffin sections were placed on Superfrost Plus slides (Gerhard Menzel GmbH, Braunschweig, Germany), slides were deparaffinized by three changes of xylene (5 min each step), followed by rehydration in 95%, 70%, 50% ethanol (5 min each step) and brought to distilled water. After blocking endogenous peroxidase (3% hydrogen peroxide in phosphate buffered saline (PBS), pH 7.5, for 15 min) antigen retrieval was performed in 10 mm sodium citrate buffer, pH 6.0 for 30 min at 95 °C. The primary antibodies diluted in DAKO antibody diluent (DAKO, Glostrup, Denmark) were incubated overnight at 4 °C followed by detection by DAKO EnVision™/peroxidase kit (cat. No K4007 for anti-mouse and K4011 for anti-rabbit secondary antibody, DAKO) and counterstained with Gill's hematoxylin (Sigma-Aldrich, St. Louis, MO). All slides were scored by the same pathologist who was blinded to other data. Associations between staining intensity of each selected protein and clinicopathological characteristics were assessed by Wilcoxon rank sum test for continuous variables and by Pearson's Chi-squared for categorical variables (Fisher's exact test was used when the number of observations was less than 5). All statistical tests were two-sided. All analyses were performed in R (9).
Analysis of Gene Expression and Connection of Gene Expression With Patient Survival in Independent Published Sample Sets
Publicly available gene expression data set SUPERTAM_HGU133A including data from four studies (all platform Affymetrix Human Genome U133A, 856 samples in total) was downloaded in a log2 normalized form that was used in (11). Samples were classified into 4 breast cancer subtypes using a subtype classification model based on gene modules called SCMOD2 (12) resulting in 348 luminal A samples. Information on lymph node status was available for 343 cases (n = 76 node positive, n = 267 node negative). Association between gene expression and lymph node status within luminal A samples was assessed by Wilcoxon rank sum test (also known as Mann-Whitney test).
Survival analysis was performed using Kaplan-Meier Plotter (http://kmplot.com) for both relapse-free survival (RFS) and distant metastasis free survival (DMFS) involving a microarray data set from 4142 breast cancer tissues (2014 database version) (13). For each gene, the population was split according to upper quartile (based on approximate proportion of lymph node positive patients in luminal A grade 1 group) and 15 years follow up threshold was applied. Each gene was represented by user-defined probe set, Affymetrix IDs were as follows: 205509_at (CPB1), 219165_at (PDLIM2), 218861_at (RNF25), 200783_s_at (STMN1), 217733_s_at (TMSB10), 215411_s_at (TRAF3IP2), 201783_s_at, and 209878_s_at (RELA), 201020_at (YWHAH). The following target group of patients were analyzed: luminal A (data from 1678 patients were available for analysis of RFS and from 918 patients for DMFS), luminal A restricted to grade 1 (n = 228 for RFS and n = 140 for DMFS), luminal A restricted to N0 patients (n = 933 for RFS and n = 546 for DMFS), luminal A restricted to grade 1 N0 patients (n = 159 for RFS and n = 109 for DMFS) and luminal B (n = 989 for RFS and n = 360 for DMFS).
RESULTS
Untargeted Proteomics Screening
To identify metastasis-related proteins in low grade breast cancer, we performed a high-resolution proteomics discovery study on a set of 48 clinicopathologically well characterized small primary grade 1 luminal A (ER+, PR+, HER2-) breast tumors; 24 lymph node positive and 24 lymph node negative. A similar matched set of 48 high grade tumors was used to investigate the selectivity of proteins for the low grade luminal A tumor group and to gain potential insight into common versus distinct metastatic processes during progression of low and high grade tumors and of different breast cancer subtypes. The workflow of the proteomic experiment together with all follow-up studies is shown in Fig. 1. A total of 4405 proteins were identified based on at least one tryptic peptide (FDR<0.05). Protein and peptide identification data together with peptide fractionation chromatograms are available in supplemental File 2. Peptide spectra are accessible through MS-Viewer at http://prospector2.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer, search key access dd7asd2je5. The mass spectrometry raw proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) (14) via the PRIDE partner repository with the data set identifier PXD000029.
The quantitative data at peptide level and protein level are presented in supplemental Files 3 and 4, respectively, where detailed comparisons of protein levels between the key breast tumor characteristics (grade, lymph node status, estrogen and HER2 receptors) are available. Forty-two proteins whose levels correlated with lymph node metastasis either positively or negatively were selected for further verification. An additional 23 proteins connected to metastasis according to the literature and that exhibited dysregulation in other parameters under comparison (Table I) were also involved in the verification process to investigate their correlation with low-grade cancer metastasis.
Table I. Top 65 targets in proteomics study with statistically significant changes (with major focus on proteins related to lymph node positivity of grade 1 breast tumors) that were selected for analysis of gene expression at transcript level (right part of the table). Proteins with low number of observations (N≤10) in the proteomics study are in italics (see Experimental procedures for details). Agreements in statistically significant changes at protein and transcript levels are indicated in bold.
| Protein level | Annotation Acc. No. | Gene symbol | Protein name | Proteomics |
Fold change | Lower limit of CI | Upper limit of CI | p value | Transcriptomics |
Rsd | p value | Protein function |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | R | |||||||||||
| G1: N1–2/N0 | P15086 | CPB1 | Carboxypeptidase B OS = Homo sapiens GN = CPB1 PE = 1 SV = 4 - [CBPB1_HUMAN] | 12 | 4.34 | 2.86 | 6.59 | 0.002 | 6.74 | 5.53 | 0.025 | Protease, Metalloprotease, Select calcium binding protein |
| G1: N1–2/N0 | Q14161 | GIT2 | ARF GTPase-activating protein GIT2 OS = Homo sapiens GN = GIT2 PE = 1 SV = 2 - [GIT2_HUMAN] | 3 | 1.53 | 1.49 | 1.56 | 0.028 | 1.15 | 0.11 | 0.138 | Select regulatory molecule, G-protein modulator, Other G-protein modulator |
| G1: N1–2/N0 | P31947 | SFN | 14–3-3 protein sigma OS = Homo sapiens GN = SFN PE = 1 SV = 1 - [1433S_HUMAN] | 20 | 1.45 | 1.28 | 1.64 | 0.001 | 1.17 | 0.14 | 0.215 | Miscellaneous function, Other miscellaneous function protein |
| G1: N1–2/N0 | Q96BH1 | RNF25 | E3 ubiquitin-protein ligase RNF25 OS = Homo sapiens GN = RNF25 PE = 1 SV = 1 - [RNF25_HUMAN] | 3 | 1.44 | 1.39 | 1.48 | 0.053 | 1.27 | 0.09 | 0.002 | Molecular function unclassified |
| G1: N1–2/N0 | Q9Y592 | CCDC41 | Coiled-coil domain-containing protein 41 OS = Homo sapiens GN = CCDC41 PE = 1 SV = 2 - [CCD41_HUMAN] | 10 | 1.41 | 1.21 | 1.64 | 0.050 | 1.17 | 0.11 | 0.088 | Molecular function unclassified |
| G1: N1–2/N0 | Q9NZT2 | OGFR | Opioid growth factor receptor OS = Homo sapiens GN = OGFR PE = 1 SV = 3 - [OGFR_HUMAN] | 18 | 1.39 | 1.24 | 1.56 | 0.002 | 1.13 | 0.09 | 0.124 | Receptor, Other receptor |
| G1: N1–2/N0 | Q96JB5 | CDK5RAP3 | CDK5 regulatory subunit-associated protein 3 OS = Homo sapiens GN = CDK5RAP3 PE = 1 SV = 2 - [CK5P3_HUMAN] | 10 | 1.33 | 1.14 | 1.55 | 0.082 | 1.02 | 0.10 | 0.877 | Molecular function unclassified |
| G1: N1–2/N0 | P63313 | TMSB10 | Thymosin beta-10 OS = Homo sapiens GN = TMSB10 PE = 1 SV = 2 - [TYB10_HUMAN] | 32 | 1.32 | 1.18 | 1.49 | 0.003 | 1.25 | 0.14 | 0.050 | Cytoskeletal protein, Actin family cytoskeletal protein, Non-motor actin binding protein |
| G1: N1–2/N0 | Q04206 | RELA | Transcription factor p65 OS = Homo sapiens GN = RELA PE = 1 SV = 2 - [TF65_HUMAN] | 18 | 1.30 | 1.16 | 1.47 | 0.020 | 1.22 | 0.09 | 0.012 | Transcription factor, Other transcription factor, Nucleic acid binding |
| G1: N1–2/N0 | Q96JY6 | PDLIM2 | PDZ and LIM domain protein 2 OS = Homo sapiens GN = PDLIM2 PE = 1 SV = 1 - [PDLI2_HUMAN] | 8 | 1.30 | 1.21 | 1.40 | 0.009 | 1.29 | 0.11 | 0.007 | Cytoskeletal protein, Actin family cytoskeletal protein, Non-motor actin binding protein |
| G1: N1–2/N0 | P35442 | THBS2 | Thrombospondin-2 OS = Homo sapiens GN = THBS2 PE = 1 SV = 2 - [TSP2_HUMAN] | 68 | 1.25 | 1.10 | 1.43 | 0.043 | 0.92 | 0.27 | 0.765 | Cell adhesion molecule, Extracellular matrix, Extracellular matrix glycoprotein |
| G1: N1–2/N0 | P16949 | STMN1 | Stathmin OS = Homo sapiens GN = STMN1 PE = 1 SV = 3 - [STMN1_HUMAN] | 18 | 1.24 | 1.15 | 1.33 | 0.002 | 1.28 | 0.16 | 0.052 | Miscellaneous function, Other miscellaneous function protein |
| G1: N1–2/N0 | P67936 | TPM4 | Tropomyosin alpha-4 chain OS = Homo sapiens GN = TPM4 PE = 1 SV = 3 - [TPM4_HUMAN] | 130 | 1.23 | 1.16 | 1.31 | 0.000 | 1.16 | 0.14 | 0.209 | Cytoskeletal protein, Actin family cytoskeletal protein, Non-motor actin binding protein |
| G1: N1–2/N0 | Q04917 | YWHAH | 14–3-3 protein eta OS = Homo sapiens GN = YWHAH PE = 1 SV = 4 - [1433F_HUMAN] | 63 | 1.20 | 1.10 | 1.32 | 0.012 | 1.20 | 0.09 | 0.024 | Miscellaneous function, Other miscellaneous function protein |
| G1: N1–2/N0 | P01042 | KNG1 | Kininogen-1 OS = Homo sapiens GN = KNG1 PE = 1 SV = 2 - [KNG1_HUMAN] | 24 | 0.60 | 0.45 | 0.79 | 0.050 | 1.12 | 0.09 | 0.186 | Signaling molecule, Other signaling molecule, Select regulatory molecule, Protease inhibitor |
| G1: N1–2/N0 | P00747 | PLG | Plasminogen OS = Homo sapiens GN = PLG PE = 1 SV = 2 - [PLMN_HUMAN] | 38 | 0.74 | 0.64 | 0.85 | 0.007 | NA | NA | NA | Protease, Serine protease |
| G1: N1–2/N0 | P06396 | GSN | Gelsolin OS = Homo sapiens GN = GSN PE = 1 SV = 1 - [GELS_HUMAN] | 172 | 0.82 | 0.74 | 0.91 | 0.016 | 1.13 | 0.14 | 0.327 | Select calcium binding protein, Other select calcium binding proteins, Cytoskeletal protein |
| G3: N1–2/N0 | P22676 | CALB2 | Calretinin OS = Homo sapiens GN = CALB2 PE = 1 SV = 2 - [CALB2_HUMAN] | 8 | 2.32 | 1.86 | 2.91 | 0.003 | 2.00 | 0.82 | 0.100 | Select calcium binding protein, Calmodulin related protein |
| G3: N1–2/N0 | Q9Y2M0 | MTMR15 | Coiled-coil domain-containing protein MTMR15 OS = Homo sapiens GN = MTMR15 PE = 2 SV = 4 - [MTMRF_HUMAN] | 4 | 1.66 | 1.47 | 1.86 | 0.034 | 0.93 | 0.12 | 0.584 | Molecular function unclassified |
| G3: N1–2/N0 | P21926 | CD9 | CD9 antigen OS = Homo sapiens GN = CD9 PE = 1 SV = 4 - [CD9_HUMAN] | 9 | 1.43 | 1.22 | 1.68 | 0.026 | 1.23 | 0.23 | 0.261 | Cell adhesion molecule, Other cell adhesion molecule |
| G3: N1–2/N0 | P58107 | EPPK1 | Epiplakin OS = Homo sapiens GN = EPPK1 PE = 1 SV = 1 - [EPIPL_HUMAN] | 59 | 1.32 | 1.21 | 1.44 | 0.000 | 1.26 | 0.37 | 0.433 | Cytoskeletal protein, Actin family cytoskeletal protein, Non-motor actin binding protein |
| G3: N1–2/N0 | Q15063 | POSTN | Periostin OS = Homo sapiens GN = POSTN PE = 1 SV = 2 - [POSTN_HUMAN] | 244 | 1.31 | 1.24 | 1.38 | 0.000 | 1.51 | 0.34 | 0.070 | Cell adhesion molecule, Other cell adhesion molecule |
| G3: N1–2/N0 | Q15582 | TGFBI | Transforming growth factor-beta-induced protein ig-h3 OS = Homo sapiens GN = TGFBI PE = 1 SV = 1 - [BGH3_HUMAN] | 72 | 1.30 | 1.17 | 1.44 | 0.000 | 1.35 | 0.28 | 0.157 | Signaling molecule, Cell adhesion molecule, Other cell adhesion molecule |
| G3: N1–2/N0 | P60903 | S100A10 | Protein S100-A10 OS = Homo sapiens GN = S100A10 PE = 1 SV = 2 - [S10AA_HUMAN] | 16 | 1.28 | 1.17 | 1.39 | 0.001 | 1.16 | 0.15 | 0.256 | Select calcium binding protein, Calmodulin related protein |
| G3: N1–2/N0 | P31949 | S100A11 | Protein S100-A11 OS = Homo sapiens GN = S100A11 PE = 1 SV = 2 - [S10AB_HUMAN] | 74 | 1.26 | 1.19 | 1.33 | 0.000 | 1.19 | 0.18 | 0.279 | Signaling molecule, Select calcium binding protein, Calmodulin related protein |
| G3: N1–2/N0 | O15173 | PGRMC2 | Membrane-associated progesterone receptor component 2 OS = Homo sapiens GN = PGRMC2 PE = 1 SV = 1 - [PGRC2_HUMAN] | 28 | 1.25 | 1.15 | 1.36 | 0.000 | 1.03 | 0.12 | 0.782 | Receptor, Other receptor |
| G3: N1–2/N0 | O43707 | ACTN4 | Alpha-actinin-4 OS = Homo sapiens GN = ACTN4 PE = 1 SV = 2 - [ACTN4_HUMAN] | 167 | 1.25 | 1.21 | 1.29 | 0.000 | 1.04 | 0.13 | 0.767 | Cytoskeletal protein, Actin family cytoskeletal protein, Non-motor actin binding protein |
| G3: N1–2/N0 | P04083 | ANXA1 | Annexin A1 OS = Homo sapiens GN = ANXA1 PE = 1 SV = 2 - [ANXA1_HUMAN] | 111 | 1.24 | 1.18 | 1.30 | 0.000 | 1.28 | 0.23 | 0.175 | Signaling molecule, Select calcium binding protein, Annexin,Transfer/carrier protein |
| G3: N1–2/N0 | P40763 | STAT3 | Signal transducer and activator of transcription 3 OS = Homo sapiens GN = STAT3 PE = 1 SV = 2 - [STAT3_HUMAN] | 38 | 1.24 | 1.14 | 1.35 | 0.000 | 1.25 | 0.16 | 0.086 | Transcription factor, Other transcription factor, Nucleic acid binding |
| G3: N1–2/N0 | Q05682 | CALD1 | Caldesmon OS = Homo sapiens GN = CALD1 PE = 1 SV = 2 - [CALD1_HUMAN] | 134 | 1.24 | 1.18 | 1.31 | 0.000 | 1.13 | 0.27 | 0.624 | Cytoskeletal protein, Actin family cytoskeletal protein, Non-motor actin binding protein |
| G3: N1–2/N0 | P55268 | LAMB2 | Laminin subunit beta-2 OS = Homo sapiens GN = LAMB2 PE = 1 SV = 2 - [LAMB2_HUMAN] | 33 | 1.24 | 1.13 | 1.35 | 0.002 | 1.33 | 0.19 | 0.048 | Extracellular matrix, Extracellular matrix linker protein |
| G3: N1–2/N0 | P05556 | ITGB1 | Integrin beta-1 OS = Homo sapiens GN = ITGB1 PE = 1 SV = 2 - [ITB1_HUMAN] | 29 | 1.22 | 1.14 | 1.30 | 0.000 | 1.28 | 0.14 | 0.034 | Receptor, Other receptor, Cell adhesion molecule |
| N1–2/N0 | P31944 | CASP14 | Caspase-14 OS = Homo sapiens GN = CASP14 PE = 1 SV = 2 - [CASPE_HUMAN] | 10 | 1.74 | 1.32 | 2.29 | 0.064 | 2.30 | 1.41 | 0.179 | Protease, Cysteine protease |
| N1–2/N0 | Q96HF1 | SFRP2 | Secreted frizzled-related protein 2 OS = Homo sapiens GN = SFRP2 PE = 1 SV = 2 - [SFRP2_HUMAN] | 8 | 1.48 | 1.23 | 1.79 | 0.097 | 1.31 | 0.24 | 0.148 | Signaling molecule, Other signaling molecule |
| N1–2/N0 | O43734 | TRAF3IP2 | Adapter protein CIKS OS = Homo sapiens GN = TRAF3IP2 PE = 2 SV = 2 - [CIKS_HUMAN] | 28 | 1.37 | 1.21 | 1.55 | 0.002 | 1.23 | 0.10 | 0.012 | Molecular function unclassified |
| N1–2/N0 | Q14993 | COL19A1 | Collagen alpha-1(XIX) chain OS = Homo sapiens GN = COL19A1 PE = 1 SV = 3 - [COJA1_HUMAN] | 10 | 1.36 | 1.27 | 1.46 | 0.001 | 1.20 | 0.77 | 0.779 | Cell adhesion molecule, Extracellular matrix, Extracellular matrix structural protein |
| N1–2/N0 | P06756 | ITGAV | Integrin alpha-V OS = Homo sapiens GN = ITGAV PE = 1 SV = 2 - [ITAV_HUMAN] | 38 | 1.23 | 1.12 | 1.35 | 0.010 | 1.10 | 0.09 | 0.206 | Cell adhesion molecule, Other cell adhesion molecule |
| N1–2/N0 | P07951 | TPM2 | Tropomyosin beta chain OS = Homo sapiens GN = TPM2 PE = 1 SV = 1 - [TPM2_HUMAN] | 80 | 1.21 | 1.12 | 1.32 | 0.002 | 1.11 | 0.13 | 0.353 | Cytoskeletal protein, Actin family cytoskeletal protein, Non-motor actin binding protein |
| N1–2/N0 | P20962 | PTMS | Parathymosin OS = Homo sapiens GN = PTMS PE = 1 SV = 2 - [PTMS_HUMAN] | 378 | 1.12 | 1.07 | 1.17 | 0.000 | 1.17 | 0.15 | 0.220 | Molecular function unclassified |
| N1–2: G3/G1 | P06702 | S100A9 | Protein S100-A9 OS = Homo sapiens GN = S100A9 PE = 1 SV = 1 - [S10A9_HUMAN] | 30 | 1.83 | 1.43 | 2.34 | 0.001 | 4.27 | 2.15 | 0.006 | Select calcium binding protein, Calmodulin related protein |
| N1–2: G3/G1 | P51153 | RAB13 | Ras-related protein Rab-13 OS = Homo sapiens GN = RAB13 PE = 1 SV = 1 - [RAB13_HUMAN] | 10 | 1.30 | 1.10 | 1.54 | 0.090 | 0.70 | 0.07 | 0.001 | Select regulatory molecule, G-protein, Small GTPase |
| N1–2: G3/G1 | P60709 | ACTB | Actin, cytoplasmic 1 OS = Homo sapiens GN = ACTB PE = 1 SV = 1 - [ACTB_HUMAN] | 4 | 1.21 | 1.14 | 1.28 | 0.052 | 1.045 | 0.120 | 0.701 | Cytoskeletal protein, Actin family cytoskeletal protein, Actin and actin related protein |
| N0: G3/G1 | P08670 | VIM | Vimentin OS = Homo sapiens GN = VIM PE = 1 SV = 4 - [VIME_HUMAN] | 881 | 0.74 | 0.73 | 0.76 | 0.000 | 1.076 | 0.119 | 0.511 | Cytoskeletal protein, Intermediate filament |
| G3/G1 | Q9NZT1 | CALML5 | Calmodulin-like protein 5 OS = Homo sapiens GN = CALML5 PE = 1 SV = 2 - [CALL5_HUMAN] | 116 | 1.69 | 1.37 | 2.07 | 0.000 | 1.44 | 0.46 | 0.248 | Select calcium binding protein, Calmodulin related protein |
| G3/G1 | P23528 | CFL1 | Cofilin-1 OS = Homo sapiens GN = CFL1 PE = 1 SV = 3 - [COF1_HUMAN] | 72 | 1.25 | 1.16 | 1.35 | 0.000 | 1.11 | 0.08 | 0.168 | Cytoskeletal protein, Actin family cytoskeletal protein, Non-motor actin binding protein |
| G3/G1 | Q8NC51 | SERBP1 | Plasminogen activator inhibitor 1 RNA-binding protein OS = Homo sapiens GN = SERBP1 PE = 1 SV = 2 - [PAIRB_HUMAN] | 72 | 1.22 | 1.15 | 1.30 | 0.000 | 1.09 | 0.08 | 0.237 | Nucleic acid binding, Other RNA-binding protein |
| G3/G1 | O95433 | AHSA1 | Activator of 90 kDa heat shock protein ATPase homolog 1 OS = Homo sapiens GN = AHSA1 PE = 1 SV = 1 - [AHSA1_HUMA5N2] | 52 | 1.22 | 1.10 | 1.35 | 0.003 | 1.206 | 0.093 | 0.017 | Molecular function unclassified |
| G3/G1 | P26447 | S100A4 | Protein S100-A4 OS = Homo sapiens GN = S100A4 PE = 1 SV = 1 - [S10A4_HUMAN] | 151 | 1.28 | 1.18 | 1.38 | 0.000 | 1.206 | 0.125 | 0.074 | Select calcium binding protein, Calmodulin related protein |
| G3/G1 | P12830 | CDH1 | Cadherin-1 OS = Homo sapiens GN = CDH1 PE = 1 SV = 3 - [CADH1_HUMAN] | 48 | 0.83 | 0.75 | 0.92 | 0.006 | 1.16 | 0.16 | 0.265 | Cell adhesion molecule, Cadherin |
| G3/G1 | Q13740 | ALCAM | CD166 antigen OS = Homo sapiens GN = ALCAM PE = 1 SV = 2 - [CD166_HUMAN] | 28 | 0.84 | 0.73 | 0.96 | 0.050 | 0.43 | 0.07 | 0.000 | Cell adhesion molecule, CAM family adhesion molecule |
| G3/G1 | P02753 | RBP4 | Retinol-binding protein 4 OS = Homo sapiens GN = RBP4 PE = 1 SV = 3 - [RET4_HUMAN] | 24 | 0.72 | 0.62 | 0.83 | 0.001 | 0.948 | 0.326 | 0.877 | Transfer/carrier protein, Other transfer/carrier protein |
| G3/G1 | Q96NY7 | CLIC6 | Chloride intracellular channel protein 6 OS = Homo sapiens GN = CLIC6 PE = 2 SV = 3 - [CLIC6_HUMAN] | 231 | 0.47 | 0.42 | 0.53 | 0.000 | 0.082 | 0.030 | 0.000 | Ion channel, Voltage-gated ion channel |
| ER+/ER- | O95994 | AGR2 | Anterior gradient protein 2 homolog OS = Homo sapiens GN = AGR2 PE = 1 SV = 1 - [AGR2_HUMAN] | 8 | 3.19 | 1.84 | 5.54 | 0.023 | 25.81 | 17.52 | 0.000 | Miscellaneous function, Surfactant |
| ER+/ER- | P50238 | CRIP1 | Cysteine-rich protein 1 OS = Homo sapiens GN = CRIP1 PE = 1 SV = 3 - [CRIP1_HUMAN] | 5 | 3.07 | 2.31 | 4.06 | 0.010 | 5.29 | 1.30 | 0.000 | Molecular function unclassified |
| ER+/ER- | Q8TD06 | AGR3 | Anterior gradient protein 3 homolog OS = Homo sapiens GN = AGR3 PE = 1 SV = 1 - [AGR3_HUMAN] | 10 | 2.21 | 1.70 | 2.88 | 0.002 | 72.69 | 46.42 | 0.000 | Miscellaneous function, Surfactant |
| ER+/ER- | P35443 | THBS4 | Thrombospondin-4 OS = Homo sapiens GN = THBS4 PE = 1 SV = 2 - [TSP4_HUMAN] | 4 | 2.16 | 1.73 | 2.70 | 0.031 | 3.03 | 1.17 | 0.006 | Cell adhesion molecule, Extracellular matrix, Extracellular matrix glycoprotein |
| ER+/ER- | Q96S44 | TP53RK | TP53-regulating kinase OS = Homo sapiens GN = TP53RK PE = 1 SV = 2 - [PRPK_HUMAN] | 4 | 1.57 | 1.43 | 1.71 | 0.014 | 1.28 | 0.19 | 0.110 | Protease, Metalloprotease |
| ER+/ER- | Q9BTC8 | MTA3 | Metastasis-associated protein MTA3 OS = Homo sapiens GN = MTA3 PE = 1 SV = 2 - [MTA3_HUMAN] | 5 | 1.40 | 1.31 | 1.51 | 0.005 | 1.52 | 0.19 | 0.002 | Transcription factor, Zinc finger transcription factor, Other zinc finger transcription factor |
| ER+/ER- | Q9P2M7 | CGN | Cingulin OS = Homo sapiens GN = CGN PE = 1 SV = 2 - [CING_HUMAN] | 11 | 1.38 | 1.21 | 1.56 | 0.004 | 2.54 | 0.60 | 0.000 | Molecular function unclassified |
| ER+/ER- | Q01995 | TAGLN | Transgelin OS = Homo sapiens GN = TAGLN PE = 1 SV = 4 - [TAGL_HUMAN] | 59 | 1.26 | 1.22 | 1.30 | 0.000 | 0.94 | 0.19 | 0.759 | Cytoskeletal protein, Actin family cytoskeletal protein, Non-motor actin binding protein |
| ER+/ER- | P16070 | CD44 | CD44 antigen OS = Homo sapiens GN = CD44 PE = 1 SV = 2 - [CD44_HUMAN] | 20 | 0.82 | 0.73 | 0.94 | 0.035 | 1.00 | 0.15 | 0.974 | Receptor, Other receptor, Cell adhesion molecule, Cell junction protein |
| HER2+/HER2- | P04626 | ERBB2 | Receptor tyrosine-protein kinase erbB-2 OS = Homo sapiens GN = ERBB2 PE = 1 SV = 1 - [ERBB2_HUMAN] | 4 | 5.27 | 3.12 | 8.89 | 0.099 | 22.866 | 5.402 | 0.000 | Receptor, Protein kinase receptor, Kinase, Protein kinase, Tyrosine protein kinase receptor |
| HER2+/HER2- | Q9UGT4 | SUSD2 | Sushi domain-containing protein 2 OS = Homo sapiens GN = SUSD2 PE = 1 SV = 1 - [SUSD2_HUMAN] | 20 | 2.92 | 1.80 | 4.77 | 0.013 | 7.97 | 3.39 | 0.000 | Molecular function unclassified |
| HER2+/HER2- | Q9UNE0 | EDAR | Tumor necrosis factor receptor superfamily member EDAR OS = Homo sapiens GN = EDAR PE = 1 SV = 1 - [EDAR_HUMAN] | 7 | 0.66 | 0.56 | 0.78 | 0.058 | 0.69 | 0.27 | 0.346 | Receptor, Other receptor |
| HER2+/HER2- | P06703 | S100A6 | Protein S100-A6 OS = Homo sapiens GN = S100A6 PE = 1 SV = 1 - [S10A6_HUMAN] | 20 | 0.76 | 0.68 | 0.84 | 0.004 | 0.73 | 0.16 | 0.170 | Signaling molecule, Growth factor, Select calcium binding protein, Calmodulin related protein |
Targeted Transcriptomics
To further elucidate the mechanisms of protein alterations in low grade breast cancer, we designed a custom TaqMan Low Density Array (Microfluidic card). The arrays were made to monitor (1) 65 gene transcripts selected on the basis of the observed changes at proteome level (shown in Table I), (2) an additional 30 genes related to prometastatic mechanisms according to the literature that were not detected at protein level, or that internally validated the sample set design (see list in supplemental File 6). Some genes within the array were also members of Oncotype DX (15) (MMP11, ESR1, PGR, ERBB2, ACTB) and MammaPrint (16) (TGFB1, STMN1 and MMP9) gene expression arrays. Complete results of transcriptomics experiment are presented in supplemental File 6.
Connecting Proteomics and Transcriptomics Data
The targets that exhibited statistically significant changes at both protein and transcript levels in lymph node positive versus negative grade 1 tumors were then selected. This group involved carboxypeptidase B1 (CPB1), PDZ and LIM domain protein 2 (PDLIM2), ring finger protein 25 (RNF25), NF-κB transcription factor p65 (RELA), 14–3-3η (YWHAH), stathmin 1 (STMN1) and thymosin beta 10 (TMSB10). TRAF3 interacting protein 2 (TRAF3IP2) was up-regulated in lymph node positive versus negative tumors regardless of grade and integrin beta-1 (ITGB1) was up-regulated in grade 3 but not grade 1 tumors with metatasis (see Tables I to III, in bold). Table II also summarizes genes and proteins that exhibited both differential expression and different protein levels between other subgroups of breast tumors that were involved in the study to investigate clinicopathological selectivity of identified targets.
Table III. Top targets related to lymph node metastasis in grade 1 breast tumors and their clinicopathological selectivity. Proteins with low number of observations (N ≤ 10) in the proteomics study are in italics (see Experimental procedures for details). Significant changes in protein levels and expression (according to criteria defined in the Experimental Procedures section) are indicated in bold.
| G1: N1–2/N0 |
G3:N1–2/N0 |
N1–2/N0 |
G3/G1 |
ER+/ER- |
HER2+/HER2- |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fold change (IHC trend) | p value | fold change (IHC trend) | p value | fold change (IHC trend) | p value | fold change (IHC trend) | p value | fold change (IHC trend) | p value | fold change (IHC trend) | p value | ||
| CPB1 | protein | 4.34 | 0.002 | 0.87 | 0.741 | 1.95 | 0.114 | 0.55 | 0.063 | 2.22 | 0.007 | 0.56 | 0.248 |
| transcript | 6.737 | 0.025 | 0.197 | 0.073 | 1.199 | 0.779 | 0.192 | 0.009 | 15.571 | 0.001 | 1.266 | 0.799 | |
| IHC total intensity | ↑ | 0.096 | ∼ | 0.514 | ∼ | 0.607 | ∼ | 0.206 | ↑ | 0.514 | ∼ | 0.722 | |
| IHC histoscore | ↑ | 0.039 | ∼ | 0.475 | ∼ | 0.406 | ∼ | 0.246 | 0.449 | ∼ | 0.589 | ||
| RNF25 | protein | 1.44 | 0.053 | 1.18 | 0.354 | 1.30 | 0.089 | 0.84 | 0.379 | N<3 | N<3 | N<3 | N<3 |
| transcript | 1.267 | 0.002 | 0.870 | 0.218 | 1.050 | 0.512 | 0.763 | 0.000 | 1.418 | 0.002 | 1.168 | 0.160 | |
| IHC tumor cells staining | ∼ | 0.817 | ∼ | 0.766 | ↑ | 0.570 | ∼ | 0.626 | ↓ | 0.286 | ↑ | 0.825 | |
| IHC stromal cell staining | ∼ | 1.000 | ↑ | 0.742 | ∼ | 0.905 | ↓ | 0.521 | ↓ | 0.439 | ↓ | 0.433 | |
| PDLIM2 | protein | 1.30 | 0.009 | 1.06 | 0.866 | 1.18 | 0.415 | 0.71 | 0.008 | 1.42 | 0.208 | 0.65 | 0.060 |
| transcript | 1.287 | 0.007 | 0.851 | 0.215 | 1.047 | 0.571 | 1.038 | 0.645 | 1.159 | 0.258 | 1.056 | 0.671 | |
| STMN1 | protein | 1.24 | 0.002 | 1.02 | 0.720 | 1.12 | 0.012 | 1.14 | 0.039 | 1.01 | 0.949 | 0.77 | 0.036 |
| transcript | 1.283 | 0.050 | 1.116 | 0.608 | 1.197 | 0.211 | 2.042 | 0.000 | 1.250 | 0.296 | 1.086 | 0.705 | |
| IHC total intensity | ↑ | 0.454 | ↓ | 0.879 | ↑ | 0.750 | ↑ | 0.006 | ∼ | 0.879 | ∼ | 0.110 | |
| IHC % of stained cells | ∼ | 0.415 | ∼ | 0.350 | ∼ | 0.327 | ↑ | 0.000 | ∼ | 0.73 | ∼ | 0.246 | |
| IHC membrane staining | ↑ | 0.108 | ↓ | 0.370 | ∼ | 1.000 | ↑ | 0.000 | ∼ | 0.562 | ↓ | 0.210 | |
| IHC stained lymphocytes | ∼ | 1.000 | ↓ | 0.659 | ↓ | 0.434 | ↑ | 0.203 | ↓ | 0.05 | ↓ | 1.000 | |
| TMSB10 | protein | 1.32 | 0.003 | 0.97 | 0.668 | 1.13 | 0.066 | 1.26 | 0.005 | 0.89 | 0.161 | 1.09 | 0.540 |
| transcript | 1.245 | 0.050 | 0.990 | 0.951 | 1.110 | 0.364 | 1.727 | 0.000 | 0.738 | 0.066 | 1.925 | 0.000 | |
| RELA | protein | 1.30 | 0.020 | 1.10 | 0.246 | 1.20 | 0.007 | 0.85 | 0.029 | 1.26 | 0.161 | 0.80 | 0.066 |
| transcript | 1.215 | 0.012 | 1.020 | 0.835 | 1.111 | 0.850 | 1.097 | 0.133 | 1.094 | 0.337 | 1.050 | 0.601 | |
| YWHAH | protein | 1.20 | 0.012 | 1.04 | 0.382 | 1.12 | 0.007 | 0.84 | 0.000 | 1.17 | 0.022 | 0.83 | 0.001 |
| transcript | 1.200 | 0.024 | 0.877 | 0.330 | 1.026 | 0.750 | 1.226 | 0.011 | 1.056 | 0.687 | 1.075 | 0.583 | |
| IHC tumor cells staining | ∼ | 1.000 | ∼ | 0.145 | ∼ | 0.348 | ↑ | 0.002 | ↓ | 0.336 | ∼ | 0.767 | |
| IHC membrane staining in tumor cells | ↓ | 0.246 | ↓ | 0.552 | ↓ | 0.269 | ↑ | 0.060 | ↓ | 0.126 | ∼ | 1.000 | |
| IHC stromal cell staining | ∼ | 0.866 | ∼ | 0.789 | ∼ | 0.788 | ↑ | 0.070 | ∼ | 0.529 | ∼ | 0.840 | |
| TRAF3IP2 | protein | 1.24 | 0.387 | 1.52 | 0.001 | 1.37 | 0.002 | 0.63 | 0.000 | 1.32 | 0.042 | 1.09 | 0.403 |
| transcript | 1.404 | 0.003 | 1.084 | 0.505 | 1.233 | 0.012 | 0.858 | 0.070 | 1.088 | 0.486 | 1.335 | 0.023 | |
| IHC percentage of stained cells | ∼ | 0.928 | ∼ | 0.220 | ∼ | 0.357 | ∼ | 0.494 | ∼ | 0.403 | ∼ | 0.342 | |
| IHC stromal fibroblasts stained | ∼ | 1.000 | ↑ | 0.755 | ∼ | 0.825 | ∼ | 0.825 | ↑ | 0.063 | ↓ | 0.191 | |
Table II. Expression profiles of the top 65 gene products screened at both protein and transcript level and of 30 additional transcripts based on the literature in all breast cancer immnophenotypes involved in the study. Proteins with low number of observations (N ≤ 10) in the proteomics study are in italics (see Experimental procedures for details). Agreements in statistically significant changes at protein and transcript levels are indicated in bold. Up = up-regulation, down = down-regulation. Gene products monitored only at transcript level are underlined.
| G1: N1–2/N0 | |
|---|---|
| Protein | up: CPB1, GIT2, SFN, RNF25, OGFR, TMSB10, RELA, PDLIM2, THBS2, STMN1, TPM4, YWHAH ‖ down: KNG1, CALML5, PLG |
| Transcript | up: RNF25, TRAF3IP2, PPP5C, PDLIM2, RELA, TIMP1, YWHAH, CPB1, RAB13, ITGAV, AGR3, CDH5, TMSB10, STMN1 ‖ down: - |
| G3: N1–2/N0 | |
| Protein | up: CALB2, SFRP2, MTMR15, TRAF3IP2, CD9, COL19A1, EPPK1, POSTN, TGFBI, S100A10, TPM2, S100A11, PGRMC2, ACTN4, ANXA1, STAT3, CALD1, LAMB2, PTMS, ITGB1, TAGLN ‖ down: S100A9 |
| Transcript | up: SERPINE1, ITGB1, THBS2, SPP1, CGN, LAMB2 ‖ down: - |
| N1–2/N0 | |
| Protein | up: SFRP2, TRAF3IP2, COL19A1, RNF25, OGFR, ITGAV, TPM2, TPM4, LAMB2 ‖ down: - |
| Transcript | up: TRAF3IP2, SERPINE1, CFL1, CGN, ITGB1, ANXA1, TPM4, RAB13, POSTN, SPP1 ‖ down: - |
| N1–2: G3/G1 | |
| Protein | up: CALML5, CALB2, S100A9, AHSA1, CFL1, S100A4, ANXA1, ACTB ‖ down: CPB1, THBS4, CLIC6, CCDC41, COL19A1, AGR3, EDAR, PDLIM2, TP53RK, POSTN, TPM2, THBS2, TRAF3IP2, SFN, MTA3, TAGLN, TPM4, YWHAH, LAMB2 |
| Transcript | up: STMN1, MMP9, MMP1, TMSB10, S100A9, EPCAM, CASP14 ‖ down: LAMB2, ESR1, TIMP1, PGR, HGF, CLIC6, ITGAV, ERBB3, MUC1, RNF25, CPB1, AGR3, MTA3, ALCAM, POSTN, RAB13, FLT4, SFRP2, MTMR15, CDH5, PGRMC2, CGN, TRAF3IP2, VEGFC, GIT2, STAT3, AGR2, GSN, THBS10, CCDC41, CRIP1, FLT1, CD44, PPP5C, THBS2, CASP14 |
| N0: G3/G1 | |
| Protein | up: S100A9, TMSB10, SERBP1, S100A4, STMN1 ‖ down: CLIC6, POSTN, CRIP1, TRAF3IP2, COL19A1, TAGLN, EPPK1, TPM2, KNG1, TP53RK, RBP4, THBS4, CALD1, S100A10, ITGB1, ITGAV, VIM, LAMB2, PLG, PTMS, RAB13, GSN, CCDC41, TGFBI, THBS2 |
| Transcript | up: COL19A1, TMSB10, FASLG, STMN1, S100A9, YWHAH, MMP9, MMP1, CXCR4, RELA, S100A4, OGFR, CASP14, PDLIM2 ‖ down: LAMB2, PGR, CLIC6, MUC1, SFRP2, POSTN, STAT3, THBS2, ERBB3, ESR1, CRIP1, ALCAM, TAGLN, CGN, AGR3, GSN, MTA3, THBS4, ITGAV, RAB13, HGF, TPM4, ITGB1, TIMP1, TPM2, AGR2 |
| G3/G1 | |
| Protein | up: S100A9, CALML5, SUSD2, S100A4, TMSB10, CFL1, SERBP1, AHSA1 ‖ down: THBS4, POSTN, COL19A1, CRIP1, CCDC41, TRAF3IP2, AGR3, TP53RK, TPM2, TAGLN, PDLIM2, EPPK1, RBP4, EDAR, THBS2, CALD1, S100A10, ITGAV, LAMB2, MTA3, TPM4 |
| Transcript | up: STMN1, TMSB10, MMP9, MMP1, S100A9, FASLG, COL19A1, CASP14, EPCAM, CXCR4, YWHAH, AHSA1, OGFR, EGFR, MET ‖ down: LAMB2, PGR, CLIC6, ERBB3, ESR1, SFRP2, POSTN, AGR3, TIMP1, ALCAM, ITGAV, HGF, MTA3, STAT3, THBS2, CGN, RAB13, CRIP1, RNF25, GSN, THBS4, FLT4, MTMR15, AGR2, TAGLN, GIT2, VEGFC, CCDC41, TPM4, CPB1, PGRMC2, MMP11, CDH5, CD9, TPM2 |
| ER+/ER- | |
| Protein | up: AGR2, CRIP1, CLIC6, CPB1, AGR3, THBS4, EDAR, COL19A1, CCDC41, TP53RK, PTMS, SUSD2, POSTN, MTA3, ALCAM, CGN, TRAF3IP2, TPM2, TAGLN, EPPK1, TPM4, THBS2 ‖ down: CASP14, S100A9, CALML5, S100A4 |
| Transcript | up: ESR1, CRIP1, AGR3, ERBB3, PGR, AGR2, PPP5C, CGN, CPB1, RNF25, MTA3, ALCAM, MTMR15, PGRMC2, CDH1, THBS4, PTMS, LAMB2, GIT2, OGFR, CDK5RAP3, CLIC6, RAB13 ‖ down: CD82, EGFR, S100A9, S100A10, ANXA1, VIM, MMP1, S100A6, CALD1, CALB2 |
| HER2+/HER2- | |
| Protein | up: ERBB2, CRIP1, SUSD2, KNG1 ‖ down: CLIC6, THBS4, SFN, PDLIM2, EDAR, S100A6, STMN1, PTMS |
| Transcript | up: ERBB2, SUSD2, MUC1, CDH1, TMSB10, TP53RK, S100A11, MMP11, CD9, POSTN, EPPK1, GIT2, KISS1, TRAF3IP2, PGRMC2, CASP14, RAB13, AGR2, MTA3 ‖ down: - |
| TN/LA | |
| Transcript only | up: TMSB10, S100A9, EGFR, MMP1, FASLG, COL19A1, CD82, MMP9, STMN1, EDAR, MET, ANXA1, S100A10, CASP14 ‖ down: ESR1, PGR, CLIC6, MTA3, LAMB2, POSTN, AGR3, CRIP1, ERBB3, AGR2, ERBB2, MTMR15, CPB1, MUC1, ALCAM, THBS2, CGN, ITGAV, RNF25, PGRMC2, FLT4, MMP11, RAB13, THBS4, SFRP2, STAT3, GIT2, VEGFC, FASLG, HGF, TPM4, CD9, PPP5C, TAGLN, TRAF3IP2, CCDC41, TIMP1, TP53RK, SUSD2, GSN, CDH1 |
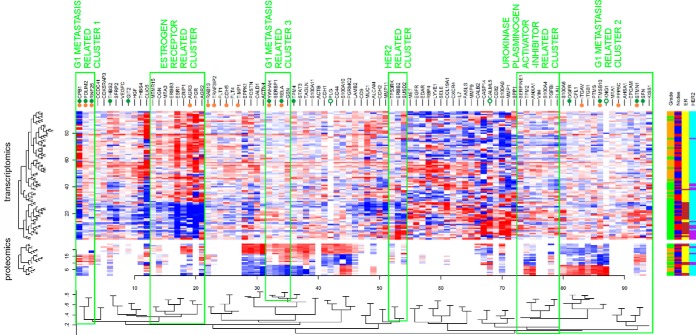
Protein levels and gene expression were also analyzed together using hierarchical clustering (Fig. 2 and supplemental Files 7A–7J). Three clusters of gene products in cold maps related to lymph node metastasis in grade 1 tumors were revealed (Fig. 2): G1 metastasis related cluster 1 involves CPB1, PDLIM2 and RNF25. It neighbors a large cluster of estrogen receptor related gene products composed of ESR1, PGR and anterior gradient proteins 2 and 3 (AGR2 and AGR3). Among the genes within the second lymph-node positivity related cluster in grade 1 tumors (G1 metastasis related cluster 2 in Fig. 2) there are STMN1, TMSB10, and ITGB1. This cluster involves other metastasis-related genes EPCAM, KISS1 and MTA1, and neighbors with a cluster containing PLAU and SERPINE1. Two other, RELA and YWHAH are involved in the third G1 metastasis related cluster 3, together with other metastasis-associated Plasminogen activator inhibitor 1 RNA-binding protein (SERBP1) and gelsolin (GSN) (Fig. 2).
Fig. 2.
Heatmap of expression patterns of proteins and transcripts in connection with tumor grade, lymph node status, ER and HER2 status. Genes marked with full green dots were up-regulated whereas those marked with empty green dots were down-regulated at protein level in G1 N1–2 versus G1 N0 tumors. Genes marked with full orange dots were statistically significantly up-regulated at transcript level in G1 N1–2 versus G1 N0 tumors (no down-regulated transcripts were found here). The genes up-regulated at both levels are considered as core genes of G1 metastasis related clusters 1 (CPB1, PDLIM2, RNF25, associated with lymph node metastasis of grade 1 tumors and had low levels in grade 3 tumors), G1 metastasis related clusters 2 (STMN1, TMSB10 associated with lymph node metastasis of grade 1 tumors and had high levels in grade 3 tumors) and G1 metastasis related clusters 3 (RELA, YWHAH associated with lymph node metastasis of grade 1 tumors with no significant difference between low and high grade tumors). Clusters related to estrogen receptor (ESR1) and HER2 receptor, and PLAU+SERPINE1 genes are also highlighted.
Immunohistochemistry
IHC staining was performed for the top targets from the previous analysis and for which IHC compatible, specific antibodies were available: CPB1, RNF25, STMN1, ITGB1, and YWHAH. Data confirming specificity of antibodies are available in supplemental File 8, including effects of siRNA silencing on protein level using Western blot and protein profiles in breast cancer cell lines. Representative IHC images showing protein staining in low grade breast tumors for CPB1 are shown in Fig. 3, for other proteins see supplemental File 9. Although IHC is by nature a rather semiquantitative approach, the data confirmed key trends observed in proteomics and transcriptomics data: (1) Up-regulation of CPB1 in lymph node positive versus lymph node negative grade 1 tumors, (2) up-regulation of STMN1 in grade 3 versus grade 1 tumors. Data are available in Table III, with further details in supplemental Files 9 and 10. The most promising target, CPB1, was tested in an independent set of grade 1 luminal A tumors (n = 64), see Fig. 3.
Fig. 3.
A, Representative examples of CPB1 IHC in luminal A grade 1 tumors. B, CPB1 levels in lymph node negative and positive luminal A grade 1 tumors in discovery (n = 48) and validation (n = 64) set of patients. The difference in CPB1 levels between lymph node positive and negative tumors was statistically significant in discovery but not in validation set although the trend is visible. Combining of both data sets (n = 112) would however lead to statistical significance at p = 0.02742.
Evaluation of Clinicopathological Selectivity of Gene Products Correlating with Metastasis of Low Grade Tumors
Analysis of clinicopathological selectivity within breast cancer subtypes was enabled by analysis of grade 1 and grade 3 tumors in the same study design and was based on agreements between proteomics, transcriptomics and IHC data. Table III shows clinicopathological selectivity of proteins/mRNAs that were up-regulated in lymph node positive versus lymph node negative grade 1 tissues: CPB1, RNF25, PDLIM2, STMN1, TMSB10, RELA, YWHAH, and TRAF3IP2. Up-regulation of these proteins and corresponding transcripts in lymph node positive tissues was observed in grade 1 tumors (G1:N1–2/N0 in Table III) but not in grade 3 tumors (G3:N1–2/N0), with the exception of TRAF3IP2 that was up-regulated in lymph node positive tissues regardless of grade (Table III). Further stratification of these potential biomarkers was based on their differential expression between grade 3 and grade 1 tumors (G3/G1). Multiple comparisons in Table III clearly show that higher level of CPB1, RNF25 and PDLIM2 proteins and transcripts were typical of highly differentiated grade 1 tumors. On the other hand, STMN1 and TMSB10 that were up-regulated in grade 1 lymph node positive versus negative tumors, however, were up-regulated also in grade 3 versus grade 1 tumors. The above protein stratification into groups according to the clinicopathological selectivity was fully reflected in hierarchical clustering (Fig. 2).
Expression of Metastasis-correlating Genes and Patient Survival Data in Independent Public Data Sets
To further investigate correlations with metastatic behavior of luminal A tumors in independent sample sets, we analyzed a published (11) data set SUPERTAM_HGU133A of gene expression data on 856 patients, including 348 luminal A-like breast tumors. The analysis confirmed up-regulation of CPB1 (p = 0.00155), PDLIM2 (p = 0.02027) and RELA (p = 0.00015) in lymph node positive luminal A tumors, see Fig. 4.
Fig. 4.

Validation of expression of selected genes on SUPERTAM_HGU133A independent sample set (n = 343). Log2 intensity of gene expression of CPB1, PDLIM2, and RELA genes is plotted for lymph node negative and positive luminal A samples. Names of specific probes are shown in brackets.
Moreover, we tested the relationship with patient survival in publically searchable microarray database of 4142 breast cancer tissues (13). The analysis showed statistically significant connection with relapse free survival in luminal A group (n = 1678) for CPB1 (p = 0.0073), PDLIM2 (p = 0.00013), RNF25 (p = 0.016), STMN1 (p = 0.0038), and TMSB10 (p = 0.0011) (Fig. 5A–5E). In addition, we obtained the highest significance of CPB1 connection with relapse free survival (p = 0.00091) and distant metastasis free survival (p = 0.00062) for luminal A grade 1 tumors, Fig. 5F–5G. To investigate prognostic value of the putative biomarkers, we further restricted these groups to lymph node negative (N0) patients and received statistically significant connection of CPB1 (p = 0.0248), PDLIM2 (p = 0.0034) and TMSB10 (p = 0.018) with relapse free survival, Fig. 5H–5J. Control analysis of relapse-free survival in high grade luminal B tumors showed similar trends as in low grade luminal A tumors for STMN1 and opposite trends for CPB1, in agreement with their clinicopathological selectivity observed in our discovery set.
Fig. 5.
Kaplan-Meier plots of CPB1, PDLIM2, RNF25, STMN1 and TMSB10 expression and patient survival derived from http://kmplot.com. Relapse free survival of CPB1 (A), PDLIM2 (B), RNF25 (C), STMN1 (D), and TMSB10 (E) in luminal A (LA) tumors. Relapse free survival (F) and distant metastasis free survival (G) of CPB1 in luminal A grade 1 tumors. Relapse free survival of CPB1 (H) in luminal A grade 1 N0 tumors. Relapse free survival of PDLIM2 (I) and TMSB10 (J) in luminal A N0 tumors. Relapse free survival of CPB1 (K) and STMN1 (L) in luminal B (LB) tumors. The associations not shown here were not statistically significant.
DISCUSSION
Methodological Strategy
The main aim of this study was to identify novel proteins correlating with early lymph node metastasis in low grade breast cancer that could be easily incorporated into clinical use. We used quantitative proteomics approach to identify such proteins by comparing a group of 24 lymph node positive luminal A grade 1 tumors with a corresponding set of 24 lymph node negative tumors. We selected small (pT1c, 11–20 mm) tumors to keep potential sampling error as low as possible, to capture metastasis-responsible cell population. Although this approach is sufficient for this purpose, we also wished to study whether low grade metastasis-associated proteins differ from those connected to metastasis in high grade tumors. The distinctive identification of proteins associated specifically with metastatic spread of tumors of different grades would imply different mechanisms employed during their progression, requiring different therapeutic approaches for each. Identifying markers of potential metastasis in low grade tumors specifically is also important clinically because these are less sensitive to chemotherapy than high grade tumors and may therefore require different targeted therapeutics. However, markers that indicate metastasis in all breast cancer patients regardless of grade are also very important clinically and are discovered as of potential general applicability through this methodological strategy.
We also analyzed individual patients at the transcript level for a large panel of putative protein biomarkers and using IHC for a more limited set for which antibodies are available. In addition to providing independent verification of the shotgun proteomic data, these approaches are imperative for the adoption of biomarkers into clinical practice, either through incorporation into multiplex expression assays (e.g. Oncotype DX and MammaPrint) or for use in current routine diagnostic histopathology laboratories. For mRNA analysis, discrepancies between mRNA and protein levels are not uncommon, but selecting targets correlating at two distinct biological levels reduces false positive findings inherent to a single screening approach and allowed us to interrogate independent large patient data sets for further validation and investigation of clinical correlation (Fig. 4) and impact on survival (Fig. 5), which is not otherwise possible from our limited number of samples.
Proteins Correlating With Lymph Node Positivity in Grade 1 Tumors and Their Potential Prometastatic Functions
As shown in Table I–III, CPB1 exhibited the largest increase in lymph node positive versus lymph node negative tumors grade 1 (G1:N1/N0 FCH = 4.34, p = 0.002); transcript level (FCH = 6.737, p = 0.025); supported by IHC staining in discovery set (p = 0.03809). IHC in an independent sample set (n = 64) of luminal A grade 1 tumors showed similar trend (Fig. 3) but was not statistically significant (p = 0.13055); this may also reflect the semiquantitative nature of the method as well as the increased proportion of N0 to N1–2 samples in the validation set. Moreover, high protein levels and expression were specific for grade 1 and not grade 3 tumors. Kaplan-Meier plots on independent samples support both the clinical impact of CPB1 (Fig. 5) and its selectivity for luminal A tumors, because different association with patient survival for luminal B tumors of higher grade was found (Fig. 5K). CPB1 is a secreted tissue protease and its potential pro-metastatic role in lymph node positive tumors might be related to matrix metalloprotease activity (http://www.uniprot.org/uniprot/P15086). In this regard, its homolog, plasma carboxypeptidase B2, has been implicated in the pro-metastatic urokinase plasminogen-activator/inhibitor system (17).
Another target correlating with lymph node metastasis of grade 1 tumors identified here was PDLIM2, a protein with pro-metastatic, pro-survival, pro-angiogenic and pro-transformation functional properties through the NF-κB pathway (18). PDLIM2 inhibits the central transcription regulator of the NF-κB pathway, p65/RELA, via its nuclear ubiquitin ligase activity. Both PDLIM2 and RELA were up-regulated in discovery and validation (Fig. 4) sample sets, indicating activation of NF-κB pathway (Fig. 4). In addition, the inhibition effect of PDLIM2 may be reflected in Kaplan-Meier plots, where association of higher PDLIM2 expression with better survival was found (Figs. 5B and 5I). PDLIM2 was recently proposed as a potential therapeutic target in breast cancer (19). A recent study also revealed that PDLIM2 regulates transcription factor activity in epithelial-to-mesenchymal transition via COP9 signalosome (20). The third target identified in the study as up-regulated in grade 1 tumors with lymph node involvement was ring finger protein 25 (RNF25, or AO7), which binds to the transactivation domain of p65/RELA and enhances its transcriptional activity (21). In parallel, RNF25 has a pro-metastatic role as an E3-ubiquitin ligase of Naked2, an antagonist of the pro-metastatic Wnt pathway (22). Moreover, levels of these NF-κB associated proteins were down-regulated in grade 3 tumors compared with grade 1 tumors, indicating the components of NF-κB pathway being more typical of grade 1 tumors (Table III and supplemental Files 9, 10).
Other proteins correlating with lymph node metastasis are STMN1 and TMSB10 which were linked more generally with an aggressive phenotype because of their overexpression in grade 3 and triple negative tumors (Table I). STMN1 (also known as oncoprotein 18, OP-18) is a microtubule destabilizing phosphoprotein with a key role in the control of mitosis. Recent evidence supports a role for STMN1 in advanced invasive and metastatic cancer because of its pro-survival role (23) and is associated with high risk and lymph node metastasis (24, 25). High STMN1 expression also negatively influences tamoxifen response in estrogen positive breast cancer (26). These observations support the role of STMN1 as a marker of poor prognosis and a target for antitumoral and anti-metastatic therapies (23, 27). On the other hand, it was reported that STMN1 plays a protumorigenic role in early stages of carcinogenesis but may act as a tumor suppressor and inhibit metastasis formation in later stages (28). The role of TMSB10 in cancer is mainly related to cytoskeletal alterations. TMSB10 is part of a gene expression signature for predicting lymph node metastasis of early stage cervical carcinomas (29) and is up-regulated in metastatic papillary thyroid carcinomas (30). The roles of STMN1 and TMSB10 in breast cancer metastasis are supported by their correlation and clustering with other metastasis-associated gene products within G1 metastasis related cluster 2, e.g. epithelial cell adhesion molecule (EPCAM), metastasis-suppressor KiSS-1 (KISS1) and metastasis-associated protein 1 (MTA1). Furthermore, their neighboring cluster in Fig. 2 contains two markers recommended for prediction of breast cancer metastasis (4), PLAU and SERPINE1. The last marker identified here is 14–3-3η (YWHAH). Bergamaschi and Katzenellenbogen recently reported high YWHAH protein levels in correlation with early breast cancer recurrence and regulation by tamoxifen via down-regulation of microRNA-451 (31).
Different Prometastatic Proteins and Mechanisms Observed in Grade 3 Tumors
In contrast to low grade tumors, fewer proteins correlated with lymph node status in grade 3 cancers. The protein and corresponding transcript most significantly up-regulated in lymph node positive versus negative grade 3 tumors was integrin B1 (ITGB1) which functions in cell-to-cell and cell-to-extracellular matrix (ECM) adhesion, transducing signals from the ECM to the cell and vice versa to influence cell migration and invasion (32). Overexpression of plasminogen activator inhibitor 1 (SERPINE1) in lymph node positive versus negative tumors supported the validity of the experiment and was specific for grade 3 tumors (p = 0.024 for grade 3 versus p = 0.241 for grade 1, Table II). The same applied for osteopontin (SPP1), a known pro-metastatic and pro-survival protein from previous breast cancer studies (33, 34).
To determine prognosis in early stage breast cancer, two gene expression-based tests are available (MammaPrint and Oncotype DX). In our study, we analyzed several genes included in these tests (MMP11, ESR1, PGR, ERBB2, ACTB, TGFB1, STMN1, and MMP9) and observed a significant relation to metastasis only in the case of STMN1. Changes in MMP11, MMP9, ESR, and PGR were more related to differences between breast cancer subtypes with different tumor grade (Table II). Our data also indicated that generally accepted prometastatic markers in breast cancer (urokinase plasminogen activator/inhibitor system, osteopontin and most MammaPrint and OncotypeDX genes tested here, with the exception of STMN1) are effective mainly in high grade tumors and may not be useful for predicting metastatic potential of low grade carcinomas.
In addition to the proteins and corresponding transcripts discussed above, the design of our study enabled the identification of proteins and transcripts correlating with tumor grade, ER and HER2 receptors status. As identification of such targets was not aim of the study, we discuss the most interesting observations in supplemental File 11 and the data sets are publically available for future inspection and analysis.
CONCLUSIONS
A combination of state of the art proteomics, transcriptomics and IHC, together with validation in independent database sets led to identification of CPB1, PDLIM2, RNF25, RELA, STMN1, TMSB10, TRAF3IP2, and YWHAH (listed according to tumor grade specificity and verification) as proteins correlating with lymph node positivity of low grade breast cancer. Our findings indicate that pro-metastatic mechanisms in low grade breast tumors may involve overexpression of CPB1, activation of NF-κB pathway, pro-survival mechanisms and changes in cytoskeleton, and are different from those in high grade tumors. These data provide candidates for further characterization and validation toward clinically usable diagnostic and therapeutic targets in low grade breast cancer patients and may be useful to predict those rare low grade luminal A breast cancer patients that should receive more regular follow-up and intensive therapy.
Supplementary Material
Acknowledgments
We thank the women who provided their tissue for this research and all of the clinically related staff involved in their treatment. We thank Dana Knoflíčková for RIN determination, Alice Hlobilková, M.D. for her help with TMA preparation, Zina Hanzelková for her excellent technical assistance with TMA analysis and Andrea Janotová for the statistical analysis of TMA data. We also thank Dr. Michaela Ščigelová and Dr. Martin Hornshaw for their kind cooperation with mass spectrometry measurement of peptide fractions, Dr. Peter Baker (University of California, San Francisco) for displaying MS/MS data in MS-Viewer and PRIDE Team for public accessibility of raw MS data. We are very grateful to Dr. Philip J. Coates for critical reading of the manuscript and correction of language.
Footnotes
Author contributions: P.B., E.B., R.V., S.D.G., B.V., and R.N. designed research; P.B., M.D., T.R., I.P., J.M., S.D.G., and R.N. performed research; J.T.H. and S.D.G. contributed new reagents or analytic tools; P.B., Z.B., I.I., H.I., E.B. and S.D.G. analyzed data; P.B., B.V., and R.N. wrote the paper.
* This work was supported by Czech Science Foundation (project No. 14-19250S), European Regional Development Fund and the State Budget of the Czech Republic (RECAMO; CZ.1.05/2.1.00/03.0101), Czech Ministry of Education, Youth and Sports (BBMRI:LM2010004), by the project MEYS - NPS I - LO1413, MH CZ - DRO (MMCI, 00209805), Amgen, the Wessex Cancer Trust and Medical Research, UK, Hope for Guernsey and the University of Southampton “Annual Adventures in Research.”
 This article contains supplemental Files 1 to 11.
This article contains supplemental Files 1 to 11.
1 The abbreviations used are:
- ER
- estrogen receptor
- ASCO
- American Society for Clinical Oncology
- CPB1
- carboxypeptidase B1 protein encoded by CPB1 gene
- ESR1
- estrogen receptor alpha protein encoded by ESR1 gene
- FCH
- fold change
- G1
- grade 1
- G3
- grade 3
- HER2
- Her2/neu receptor encoded by ERBB2 gene
- iTRAQ
- isobaric tags for relative and absolute quantification
- LA
- luminal A subtype breast tumors
- MMCI
- Masaryk Memorial Cancer Institute
- N0
- lymph node negative tumors
- N1–2
- lymph node positive tumors
- PDLIM2
- PDZ and LIM domain protein 2 encoded by PDLIM2 gene
- PLAU
- urokinase plasminogen activator encoded by PLAU gene
- qRT-PCR
- quantitative real-time polymerase chain reaction
- PR
- progesterone receptor protein encoded by PGR gene
- RELA
- NF-κB transcription factor p65 protein encoded by RELA gene
- RNF25
- ring finger protein 25 protein encoded by RNF25 gene
- SERPINE1
- plasminogen activator inhibitor 1 (also known as PAI-1) encoded by SERPINE1 gene
- STMN1
- stathmin 1 protein encoded by STMN1 gene
- TN
- triple negative subtype breast tumors
- TRAF3IP2
- TRAF3-interacting protein 2 encoded by TRAF3IP2 gene
- TMSB10
- thymosin beta 10 protein encoded by TMSB10 gene
- YWHAH
- 14–3-3η protein encoded by YWHAH gene.
REFERENCES
- 1. Ross J. S., Hortobagyi G.N. (Eds) (2005) Molecular Oncology of Breast Cancer, Jones and Barlett Publishers, Sadbury, MA, U.S.A. [Google Scholar]
- 2. Cress A. E., Nagle R.B., (Eds.) (2006) Cell Adhesion and Cytoskeletal Molecules in Metastasis, Springer, Dodrecht, The Netherlands [Google Scholar]
- 3. Mansel R. E., Fodstad O., Jiang W.G., (Eds.) (2007) Metastasis of Breast Cancer, Springer, Dodrecht, The Netherlands [Google Scholar]
- 4. Harris L., Fritsche H., Mennel R., Norton L., Ravdin P., Taube S., Somerfield M. R., Hayes D. F., Bast R. C., Jr. (2007) American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. 25, 5287–5312 [DOI] [PubMed] [Google Scholar]
- 5. Stephens R. W., Brunner N., Janicke F., Schmitt M. (1998) The urokinase plasminogen activator system as a target for prognostic studies in breast cancer. Breast Cancer Res. Treat. 52, 99–111 [DOI] [PubMed] [Google Scholar]
- 6. Maryas J., Faktor J., Dvorakova M., Struharova I., Grell P., Bouchal P. (2014) Proteomics in investigation of cancer metastasis: functional and clinical consequences and methodological challenges. Proteomics 14, 426–440 [DOI] [PubMed] [Google Scholar]
- 7. Bouchal P., Roumeliotis T., Hrstka R., Nenutil R., Vojtesek B., Garbis S. D. (2009) Biomarker discovery in low-grade breast cancer using isobaric stable isotope tags and two-dimensional liquid chromatography-tandem mass spectrometry (iTRAQ-2DLC-MS/MS) based quantitative proteomic analysis. J. Proteome Res. 8, 362–373 [DOI] [PubMed] [Google Scholar]
- 8. Scigelova M., Hornshaw M., Giannakopulos A., Makarov A. (2011) Fourier Transform Mass Spectrometry. Mol. Cell. Proteomics 10, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. R_Development_Core_Team (2008) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 10. Bookout A. L., Mangelsdorf D. J. (2003) Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Receptor Signal. 1, e012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haibe-Kains B., Desmedt C., Loi S., Culhane A. C., Bontempi G., Quackenbush J., Sotiriou C. (2012) A three-gene model to robustly identify breast cancer molecular subtypes. J. Natl. Cancer Inst. 104, 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wirapati P., Sotiriou C., Kunkel S., Farmer P., Pradervand S., Haibe-Kains B., Desmedt C., Ignatiadis M., Sengstag T., Schutz F., Goldstein D. R., Piccart M., Delorenzi M. (2008) Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 10, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gyorffy B., Lanczky A., Eklund A. C., Denkert C., Budczies J., Li Q., Szallasi Z. (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 [DOI] [PubMed] [Google Scholar]
- 14. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paik S., Tang G., Shak S., Kim C., Baker J., Kim W., Cronin M., Baehner F. L., Watson D., Bryant J., Costantino J. P., Geyer C. E., Jr., Wickerham D. L., Wolmark N. (2006) Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 24, 3726–3734 [DOI] [PubMed] [Google Scholar]
- 16. van 't Veer L. J., Dai H., van de Vijver M. J., He Y. D., Hart A. A., Mao M., Peterse H. L., van der Kooy K., Marton M. J., Witteveen A. T., Schreiber G. J., Kerkhoven R. M., Roberts C., Linsley P. S., Bernards R., Friend S. H. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 [DOI] [PubMed] [Google Scholar]
- 17. Swaisgood C. M., Schmitt D., Eaton D., Plow E. F. (2002) In vivo regulation of plasminogen function by plasma carboxypeptidase B. J. Clin. Invest. 110, 1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prasad S., Ravindran J., Aggarwal B. B. (2010) NF-kappaB and cancer: how intimate is this relationship. Mol. Cell. Biochem. 336, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qu Z., Fu J., Yan P., Hu J., Cheng S. Y., Xiao G. (2010) Epigenetic repression of PDZ-LIM domain-containing protein 2: implications for the biology and treatment of breast cancer. J. Biol. Chem. 285, 11786–11792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bowe R. A., Cox O. T., Ayllon V., Tresse E., Healy N. C., Edmunds S. J., Huigsloot M., O'Connor R. (2014) PDLIM2 regulates transcription factor activity in epithelial-to-mesenchymal transition via the COP9 signalosome. Mol. Biol. Cell 25, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Asamitsu K., Tetsuka T., Kanazawa S., Okamoto T. (2003) RING finger protein AO7 supports NF-kappaB-mediated transcription by interacting with the transactivation domain of the p65 subunit. J. Biol. Chem. 278, 26879–26887 [DOI] [PubMed] [Google Scholar]
- 22. Ding W., Li C., Hu T., Graves-Deal R., Fotia A. B., Weissman A. M., Coffey R. J. (2008) EGF receptor-independent action of TGF-alpha protects Naked2 from AO7-mediated ubiquitylation and proteasomal degradation. Proc. Natl. Acad. Sci. U.S.A. 105, 13433–13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Belletti B., Baldassarre G. (2011) Stathmin: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin. Therapeutic Targets 15, 1249–1266 [DOI] [PubMed] [Google Scholar]
- 24. Trovik J., Wik E., Stefansson I. M., Marcickiewicz J., Tingulstad S., Staff A. C., Njolstad T. S., MoMaTec Study, G., Vandenput I., Amant F., Akslen L. A., Salvesen H. B. (2011) Stathmin overexpression identifies high-risk patients and lymph node metastasis in endometrial cancer. Clin. Cancer Res. 17, 3368–3377 [DOI] [PubMed] [Google Scholar]
- 25. Jeon T. Y., Han M. E., Lee Y. W., Lee Y. S., Kim G. H., Song G. A., Hur G. Y., Kim J. Y., Kim H. J., Yoon S., Baek S. Y., Kim B. S., Kim J. B., Oh S. O. (2010) Overexpression of stathmin1 in the diffuse type of gastric cancer and its roles in proliferation and migration of gastric cancer cells. Br. J. Cancer 102, 710–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Golouh R., Cufer T., Sadikov A., Nussdorfer P., Usher P. A., Brunner N., Schmitt M., Lesche R., Maier S., Timmermans M., Foekens J. A., Martens J. W. (2008) The prognostic value of Stathmin-1, S100A2, and SYK proteins in ER-positive primary breast cancer patients treated with adjuvant tamoxifen monotherapy: an immunohistochemical study. Breast Cancer Res. Treat. 110, 317–326 [DOI] [PubMed] [Google Scholar]
- 27. Rana S., Maples P. B., Senzer N., Nemunaitis J. (2008) Stathmin 1: a novel therapeutic target for anticancer activity. Expert Rev. Anticancer Therapy 8, 1461–1470 [DOI] [PubMed] [Google Scholar]
- 28. Williams K., Ghosh R., Giridhar P. V., Gu G., Case T., Belcher S. M., Kasper S. (2012) Inhibition of stathmin1 accelerates the metastatic process. Cancer Res. 72, 5407–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang L., Zheng M., Zhou Q. M., Zhang M. Y., Jia W. H., Yun J. P., Wang H. Y. (2011) Identification of a gene-expression signature for predicting lymph node metastasis in patients with early stage cervical carcinoma. Cancer 117, 3363–3373 [DOI] [PubMed] [Google Scholar]
- 30. Feher L. Z., Pocsay G., Krenacs L., Zvara A., Bagdi E., Pocsay R., Lukacs G., Gyory F., Gazdag A., Tarko E., Puskas L. G. (2012) Amplification of thymosin beta 10 and AKAP13 genes in metastatic and aggressive papillary thyroid carcinomas. Pathol. Oncol. Res. 18, 449–458 [DOI] [PubMed] [Google Scholar]
- 31. Bergamaschi A., Katzenellenbogen B. S. (2012) Tamoxifen downregulation of miR-451 increases 14–3-3zeta and promotes breast cancer cell survival and endocrine resistance. Oncogene 31, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mizejewski G. J. (1999) Role of integrins in cancer: survey of expression patterns. Proc. Soc. Exp. Biol. Med. 222, 124–138 [DOI] [PubMed] [Google Scholar]
- 33. Tuck A. B., O'Malley F. P., Singhal H., Harris J. F., Tonkin K. S., Kerkvliet N., Saad Z., Doig G. S., Chambers A. F. (1998) Osteopontin expression in a group of lymph node negative breast cancer patients. Int. J. Cancer 79, 502–508 [DOI] [PubMed] [Google Scholar]
- 34. Tuck A. B., Chambers A. F. (2001) The role of osteopontin in breast cancer: clinical and experimental studies. J. Mammary Gland Biol. Neoplasia 6, 419–429 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.