Abstract
Bone samples from several vertebrates were collected from the Ziegler Reservoir fossil site, in Snowmass Village, Colorado, and processed for proteomics analysis. The specimens come from Pleistocene megafauna Bison latifrons, dating back ∼120,000 years. Proteomics analysis using a simplified sample preparation procedure and tandem mass spectrometry (MS/MS) was applied to obtain protein identifications. Several bioinformatics resources were used to obtain peptide identifications based on sequence homology to extant species with annotated genomes. With the exception of soil sample controls, all samples resulted in confident peptide identifications that mapped to type I collagen. In addition, we analyzed a specimen from the extinct B. latifrons that yielded peptide identifications mapping to over 33 bovine proteins. Our analysis resulted in extensive fibrillar collagen sequence coverage, including the identification of posttranslational modifications. Hydroxylysine glucosylgalactosylation, a modification thought to be involved in collagen fiber formation and bone mineralization, was identified for the first time in an ancient protein dataset. Meta-analysis of data from other studies indicates that this modification may be common in well-preserved prehistoric samples. Additional peptide sequences from extracellular matrix (ECM) and non-ECM proteins have also been identified for the first time in ancient tissue samples. These data provide a framework for analyzing ancient protein signatures in well-preserved fossil specimens, while also contributing novel insights into the molecular basis of organic matter preservation. As such, this analysis has unearthed common posttranslational modifications of collagen that may assist in its preservation over time. The data are available via ProteomeXchange with identifier PXD001827.
During the last decade, paleontology and taphonomy (the study of decaying organisms over time and the fossilization processes) have begun to overlap with the field of proteomics to shed new light on preserved organic matter in fossilized bones (1–4). These bones represent a time capsule of ancient biomolecules, owing to their natural resistance to post mortem decay arising from a unique combination of mechanical, structural, and chemical properties (4–7).
Although bones can be cursorily described as a composite of collagen (protein) and hydroxyapatite (mineral), fossilized bones undergo three distinct diagenesis pathways: (i) chemical deterioration of the organic phase; (ii) chemical deterioration of the mineral phase; and (iii) (micro)biological attack of the composite (6). In addition, the rate of these degradation pathways are affected by temperature, as higher burial temperatures have been shown to accelerate these processes (6, 8). Though relatively unusual, the first of these three pathways results in a slower deterioration process, which is more generally mitigated under (6) specific environmental constraints, such as geochemical stability (stable temperature and acidity) that promote bone mineral preservation. Importantly, slower deterioration results in more preserved biological materials that are more amenable to downstream analytical assays. One example of this is the controversial case of bone and soft-tissue preservation from the Cretaceous/Tertiary boundary (9–22). In light of these and other studies of ancient biomolecules, paleontological models have proposed that organic biomolecules in ancient samples, such as collagen sequences from the 80 million-year-(my)-old Campanian hadrosaur, Brachylophosaurus canadensis (16) or 68-my-old Tyrannosaurus rex, might be protected by the microenvironment within bones. Such spaces are believed to form a protective shelter that is able to reduce the effects of diagenetic events. In addition to collagen, preserved biomolecules include blood proteins, cellular lipids, and DNA (4, 5). While the maximum estimated lifespan of DNA in bones is ∼20,000 years (ky) at 10 °C, bone proteins have an even longer lifespan, making them an exceptional target for analysis to gain relevant insights into fossilized samples (6). Indeed, the survival of collagen, which is considered to be the most abundant bone protein, is estimated to be in the range 340 ky at 20 °C. Similarly, osteocalcin, the second-most abundant bone protein, can persist for ≈45 ky at 20 °C, thus opening an unprecedented analytical window to study extremely old samples (2, 4, 23).
Although ancient DNA amplification and sequencing can yield interesting clues and potential artifacts from contaminating agents (7, 24–28), the improved preservation of ancient proteins provides access to a reservoir of otherwise unavailable genetic information for phylogenetic inference (25, 29, 30). In particular, mass spectrometry (MS)-based screening of species-specific collagen peptides has recently been used as a low-cost, rapid alternative to DNA sequencing for taxonomic attribution of morphologically unidentifiable small bone fragments and teeth stemming from diverse archeological contexts (25, 31–33).
For over five decades, researchers have presented biochemical evidence for the existence of preserved protein material from ancient bone samples (34–36). One of the first direct measurements was by amino acid analysis, which showed that the compositional profile of ancient samples was consistent with collagens in modern bone samples (37–39). Preservation of organic biomolecules, either from bone, dentin, antlers, or ivory, has been investigated by radiolabeled 14C fossil dating (40) to provide an avenue of delineating evolutionary divergence from extant species (3, 41, 42). It is also important to note that these parameters primarily depend on ancient bone collagen as the levels remain largely unchanged (a high percentage of collagen is retained, as gleaned by laboratory experiments on bone taphonomy (6)). Additionally, antibody-based immunostaining methods have given indirect evidence of intact peptide amide bonds (43–45) to aid some of the first evidence of protein other than collagen and osteocalcin in ancient mammoth (43) and human specimens (46).
In the past, mass spectrometry has been used to obtain MS signals consistent with modern osteocalcin samples (2, 47), and eventually postsource decay peptide fragmentation was used to confirm the identification of osteocalcin in fossil hominids dating back ∼75 ky (48). More recently, modern “bottom-up” proteomic methods were applied to mastodon and T. rex samples (10), complementing immunohistochemistry evidence (13, 17). The results hinted at the potential of identifying peptides from proteolytic digest of well-preserved bone samples. This work also highlighted the importance of minimizing sources of protein contamination and adhering to data publication guidelines (20, 21). In the past few years, a very well-preserved juvenile mammoth referred to as Lyuba was discovered in the Siberian permafrost and analyzed using high-resolution tandem mass spectrometry (29). This study was followed with a report by Wadsworth and Buckley (30) describing the analysis of proteins from 19 bovine bone samples spanning 4 ky to 1.5 my. Both of these groups reported the identification of additional collagen and noncollagen proteins.
Recently, a series of large extinct mammal bones were unearthed at a reservoir near Snowmass Village, Colorado, USA (49, 50). The finding was made during a construction project at the Ziegler Reservoir, a fossil site that was originally a lake formed at an elevation of ∼2,705 m during the Bull Lake glaciations ∼140 ky ago (49, 51). The original lake area was ∼5 hectares in size with a total catchment of ∼14 hectares and lacked a direct water flow inlet or outlet. This closed drainage basin established a relatively unique environment that resulted in the exceptional preservation of plant material, insects (52), and vertebrate bones (49). In particular, a cranial specimen from extinct Bison latifrons was unearthed from the Biostratigraphic Zone/Marine Oxygen Isotope Stage (MIS) 5d, which dates back to ∼120 ky (53, 54).
Here, we describe the use of paleoproteomics, for the identification of protein remnants with a focus on a particularly unique B. latifrons cranial specimen found at the Ziegler site. We developed a simplified sample processing approach that allows for analysis of low milligram quantities of ancient samples for peptide identification. Our method avoids the extensive demineralization steps of traditional protocols and utilizes an acid labile detergent to allow for efficient extraction and digestion without the need for additional sample cleanup steps. This approach was applied to a specimen from B. latifrons that displayed visual and mechanical properties consistent with the meninges, a fibrous tissue that lines the cranial cavity. Bioinformatics analysis revealed the presence of a recurring glycosylation signature in well-preserved collagens. In particular, the presence of glycosylated hydroxylysine residues was identified as a unique feature of bone fossil collagen, as gleaned through meta-analyses of raw data from previous reports on woolly mammoth (Mammuthus primigenius) and bovine samples (29, 30). The results from these meta-analyses indicate a common, unique feature of collagen that coincides with, and possibly contributes to its preservation.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Ammonium bicarbonate, ammonium acetate, Tris(2-carboxyethyl)phosphine hydrochloride, acetonitrile, and iodoacetamide were purchased from Sigma-Aldrich (St. Louis, MO). Formic acid (FA) and trifluoroacetic acid were purchased from Fluka (Buchs, Switzerland). Trypsin (sequencing grade, TPCK treated) and sodium 3-((1-(furan-2-yl)undecyloxy)carbonylamino)propane-1-sulfonate (ProteaseMAX) were purchased from Promega (Madison, WI). Steps taken to minimize protein contamination and carry over included 1) the use of new labware for each sample; 2) bone samples were fractured in new, single-use foil sleeves; and 3) only fresh chromatography columns (trap- and nano-columns) were used for analysis. In addition, blank runs were scheduled before every sample to ensure identifications were unique to the sample being run. Keratins, the ubiquitous protein contaminant, were identified in all samples along with porcine trypsin, the protease used for digestion. As expected, no protein hits were reported when assaying soil samples (negative controls) other than those just listed.
Protein Sample Preparation
Bone samples were collected from the Ziegler reservoir in Snowmass Village, Colorado, and processed directly. The skull sample (B. latifrons, identifier 13.009a) was removed from the underlying bone with a razor blade using mild force. The sample was brought up in 0.1% ProteaseMax in 25 mm ammonium bicarbonate (100 μl/mg bone) and vortexed at room temperature for 1 h. Tris(2-carboxyethyl)phosphine hydrochloride was added to 10 mm and incubated for 30 min at 70 °C to reduce disulfide bonds. After cooling to room temperature, iodoacetamide was added to 15 mm, and the samples were incubated in the dark at room temperature for 45 min. Supernatants from centrifuged samples were subjected to standard overnight digestion. Digestion was carried out by adding 5 μg trypsin and incubating at 37 °C overnight. An aliquot representing 5% of the B. latifrons sample was removed, acidified using formic acid, and spun for 1 h at 5,000 × g to remove insoluble material. This sample was run in duplicate by LC-MS/MS to generate the preliminary unfractionated dataset (BL_UF). The remainder of the sample was spun in a similar fashion before high pH reversed phase chromatographic separation on an AKTA micro HPLC system (GE Healthcare Life Sciences). The sample was run on a Gemini-NX C18 column (250 mm x 4.60 mm) (Phenomenex; Torrance, CA) over a gradient of 5–40% acetonitrile with 20 mm ammonium acetate (pH 10.0). 20 fractions were pooled (fraction 1 + 11, 2 + 12, etc.), concentrated on a speed-vac, and acidified using FA to generate 10 samples for LC-MS/MS analysis (Bison_HCD_Run1_Frac01–10).
Liquid Chromatography - Tandem Mass Spectrometry Analysis
Samples were analyzed on an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) coupled to an Eksigent nanoLC-2D system through a nano-electrospray LC-MS interface. Eight μl of sample was injected and desalted using a trapping column (ZORBAX 300SB-C18, dimensions 5 × 0.3 mm 5 μm). Here, the sample was washed with 5% acetonitrile, 0.1% FA at a flow rate of 10 μl/min for 3 min. Afterward, the trapping column was put online with the nano-pump at a flow rate of 350 nl/min. Separations were performed using 15 cm long Kasil fritted, fused silica capillary columns (100 μm inner diameter, 360 μm outer diameter) (Polymicro Technologies, Phoenix, AZ) slurry packed in house with 4 μm, Synergi C18 resin (Phenomenex; Torrance, CA). The mobile phase consisted of water with 0.1% FA (solvent A) and acetonitrile with 0.1% FA (solvent B). A 90 min gradient from 6% acetonitrile to 35% acetonitrile was used to separate peptides. Data acquisition was performed using the instrument supplied Xcalibur (version 2.0.6) software. The LC runs were monitored in positive ion mode by sequentially recording survey MS scans (m/z 400–2,000) in the Orbitrap mass analyzer followed by MS/MS scans in the high-energy collision dissociation cell for the 10 most-intense ions detected in the full MS survey scan. automatic gain control was set to 1 × 106, trigger intensity was set to 40,000, and the normalized collision energy was set to 35. Dynamic exclusion was set to 90 s with a repeat count of 2 for 30 s.
Data Analysis
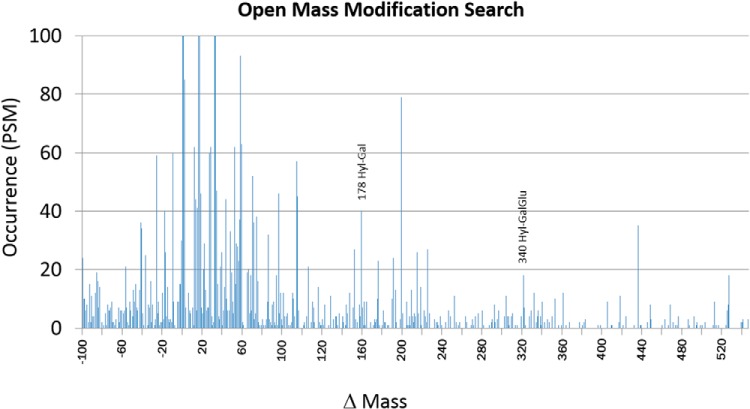
Raw files were converted to peak lists using a script; PAVA (UCSF). Exploratory searches were performed on the 47,291 spectra from the fractionated samples to determine the most appropriate search conditions. Database searches were performed using Protein Prospector, Mascot, and Byonic (55). As expected, most peptide matches were to bovine proteins (Bos taurus or Bos indicus extant species) with the exception of trypsin (porcine) and human keratins, both common processing contaminants. B. taurus sequences were isolated from UniprotKB, and concatenated with decoy-randomized sequences, human keratins, latex contaminants, and porcine trypsin. This database was used for all searches (release date 2013.6.17 for Protein Prospector and 2014.9.18 for Mascot and Byonic). Protein annotations were determined when necessary using the Uniref90 or Uniref50 databases. Initial searches revealed that deamidation of Asn and Gln, carbamidomethylation of peptide N termini (due to unquenched alkylating reagent during sample prep), and oxidation of Met and Pro (Hyp) were all common modifications. These modifications were used in Protein Prospector's Batch-Tag utility (v5.12.1) as variable modifications, with a maximum of three per peptide, to generate initial protein identifications. Additional parameters included semi-trypsin specificity, precursor ion tolerance of 8 ppm, and fragment ion tolerance of 0.05 Da (used for consistency with our version of Mascot, which does not permit relative fragment ion tolerance). Protein identifications were filtered at a max E-value of 1.0E-5, and the resulting accession numbers were searched with an “open mass” modification search with limits of -100 to +400 Da mass addition on any residue to identify additional modifications (Fig. 3). 178 Da and 340 Da mass additions, indicative of lysine galactosylation and glucosylgalactosylation, were identified. A final Batch-Tag search, “BT_Final,” added oxidation, galactosylation, and glucosylgalactosylation of lysine to the “base” search variable modifications and allowed for a max of four modifications per peptide and up to one missed cleavage. 3,060 unique peptides were identified from bovine proteins with an E-value ≤ 0.005. With the high abundance of modifications in our dataset, assigning a modification site localization score to each peptide allowed us to filter out peptides with a high probability of a modification false localization (Ex. deamidation of Q adjacent to N; hydroxylation of P adjacent to P). Thus, we filtered identifications to require a minimum Site Localization in Peptide (SLIP)1 score of 6, outputting only peptides with high probability modification site assignments (supplemental Table 1) (56).
Fig. 3.
Occurrence plot from open mass modification search using Protein Prospector Batch-Tag. The Y-axis is limited to <100 peptide spectra matches to improve visualization of lower abundant mass modifications. Mass additions consistent with galactosylation and glucosylgalactosylation of hydroxylysine are highlighted.
Byonic searches were performed using aforementioned “BT_Final” parameters, yielding 4,003 unique peptide identifications from bovine proteins at an estimated 1.3% spectrum-level FDR. Mascot searches were performed on all data files, including those from Cappellini et al. (29) and Wadsworth and Buckley (30). Datasets were either publicly available or kindly provided by the authors. “BT_Final” search parameters were used with a significance threshold of p < .05. 4,124 unique peptides were identified from bovine proteins at an estimated 4% spectrum-level FDR.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD001827 and 10.6019/PXD001827. Peptide Spectral Matches (PSMs) for our Mascot and Protein Prospector results can be viewed using Protein Prospector's public MS-Viewer utility (57). The search keys are: czdpqztspo (Protein Prospector) and md0te2ewgb (Mascot). We have included the Byonic results separately, as they can be viewed using Byonic Viewer.
RESULTS
During sample preparation, it was observed that the surface of a bone sample from B. latifrons 13.009a was pliable with mild force applied through the tip of surgical forceps. The image of this surface is shown in Fig. 1. This material was excised and prepared for LC-MS/MS analysis as described above.
Fig. 1.

Cranial bone sample from B. latifrons (13. 009a) with inside surface shown. Presence of material that is consistent with meninges connective tissue is shown. Structure that may be the site of a former blood vessel is found in the upper right of the bottom image (boxed area).
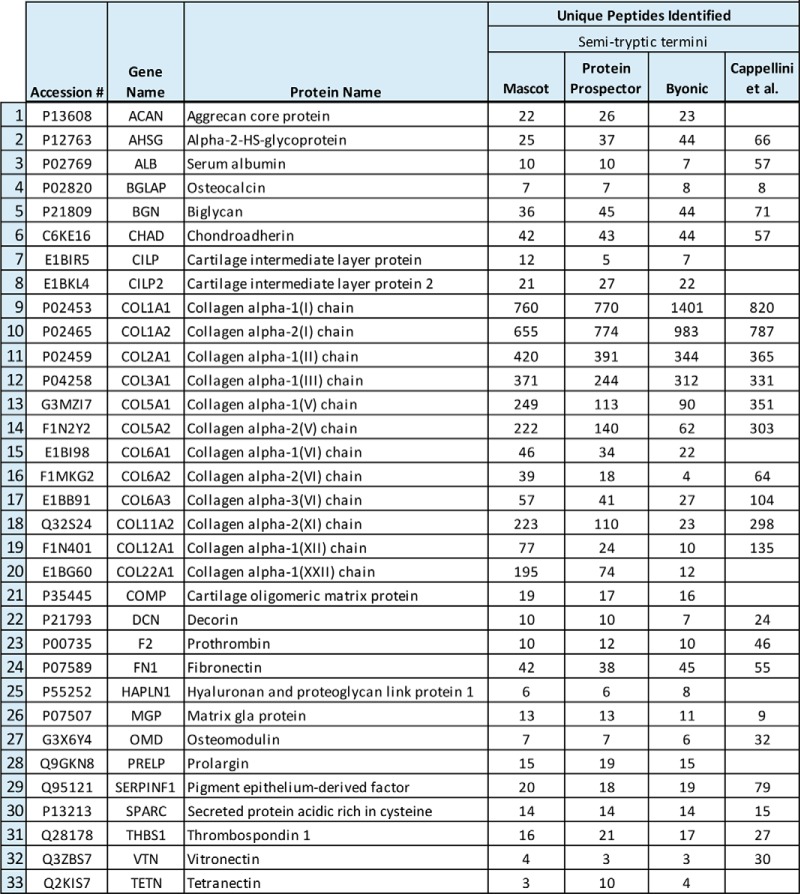
The unfractionated digest was run in duplicate, yielding 12,668 spectra, while the fractionated samples cumulatively resulted in 47,291 spectra. Bioinformatic analyses were performed on the spectra in order to identify peptide sequences that match to extant bovine protein sequence databases. The high number of variable modifications prompted the use of three search engines to increase confidence in identifications as our searches became more computationally demanding. Semi-trypsin specificity was required due to a high occurrence within our sample, as can be seen from Supplemental Table 2, likely due to protein degradation. Although the bulky glycan modifications hinder tryptic cleavage, we observe several glycosylated lysine residues at peptide C termini, which we infer is a result of non-enzymatic cleavage. As expected, the fractionated dataset from B. latifrons resulted in higher sequence coverage and a larger number of peptides that mapped to at least 33 proteins identified from all three search engines as seen in Table I. A comprehensive summary of protein and peptide identifications from the analysis can be found in Supplemental Table 3.
Table I. Proteins identified in ice age bison skull (identified in all three search engines and requiring at least three unique peptides). Comparison to Cappellini et al.'s mammoth bone data (29) is shown to highlight similarities and differences in protein identifications (data were processed using Mascot; see methods section for details).
Of these identifications, peptides that map to collagen are very abundant in the B. latifrons datasets. Spectral matches to the α1 and α2 chains of type 1 collagen account for ∼60% of all PSMs. Another 27% match to other fibrillar collagens (COL2A1, COL3A1, COL5A1/2, COL11A1/2) and ∼1% account for additional collagen families. A total of 83% of peptides were found to have at least one of five modifications that were variables in the base searches, and 53% contained two or more of the five modifications.
Glutamine and Asparagine Deamidation
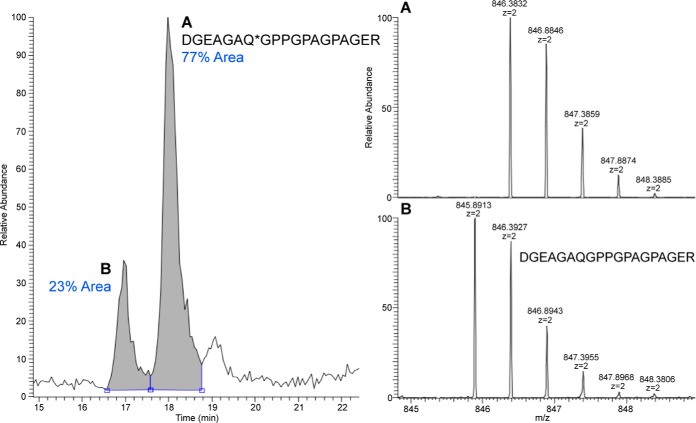
Deamidation of glutamine and asparagine residues was one of the most commonly observed peptide modifications in our datasets. Deamidation is widely observed in both new and ancient protein samples and minor deamidation can occur during sample processing. On the other hand, extensive Q and N deamidation (Δ +0.984 Da) has been proposed to be a potential marker of bone collagen deterioration levels (58). We examined the level of deamidation for several of the collagen peptides reported by Van Doorn et al. by integrating the area of extracted ion chromatograms that cover the entire ion envelopes for both native and deamidated-containing peptides. Fig. 2 shows that the deamidated peptide DGEAGAQ*GPPGPAGPAGER (COL1A1–0441-0 Van Doorn et al. nomenclature (58)) can be distinguished by the MS ion envelope and elutes after the non-deamidated form (59). Interestingly, peptides derived from noncollagen proteins showed a very high to complete level of deamidation. For example, all significant peptides that mapped to matrix gla protein, serum albumin, biglycan, and thrombospondin 1 and contained N and Q residues showed complete deamidation.
Fig. 2.
Relative quantification of COL1A1 peptide deamidation using extracted ion chromatogram of the ion envelopes. Area under the curve was used to calculate percentage of peptide with glutamine deamidation. Insets: MS signal for the right and left peak showing the COL1A1 deamidated form of DGEAGAQ*GPPGPAGPAGER (top) and the parent COL1A1 peptide DGEAGAQGPPGPAGPAGER (bottom).
Glycosylation of Hydroxylysine
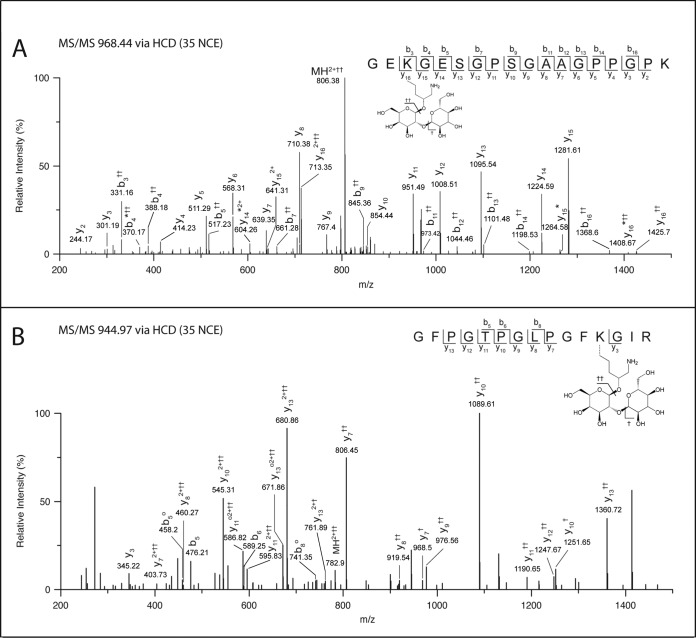
Open mass modification searches were utilized to detect unexpected posttranslational modifications in the samples. Though plagued by high false discovery rates and thus requiring manual validation, the method has the ability to identify unexpected modifications. A plot of mass addition or subtraction from the unmodified peptide mass (Δ Da) versus occurrence is shown in Fig. 3. A fairly surprising modification was +178 Da, consistent with galactosyl-hydroxylysine (†) and +340 Da, consistent with glucosylgalactosyl-hydroxylysine (††). Figure 4 shows the spectra used to identify the COL5A1 peptide GEK††GESGPSGAAGPPGPK (K753) and the COL1A2 peptide GFPGTPGLPGFK††GIR (K175). To the best of our knowledge, this is the first identification of a glycan moiety in an ancient sample. It is important to note that, while our MS data can verify the mass additions of galactose and glucosylgalactose, assigning these hexose groups as Hyl O-linked is based on the well-characterized modification of fibrillar collagens (60–63). high-energy collision dissociation fragmentation often removes the O-linked glycan while maintaining the integrity of the formerly modified residue, thus providing limited fragment ions containing a mass shift due to the modification. Additionally, beam-type fragmentation typically results in poor fragmentation of O-linked glycopeptides, as is the case with many of our spectra (63).
Fig. 4.
Spectra assignments for glucosylgalactosyl-hydroxylysine containing peptides are reported in (A) COL5A1 peptide GEK††GESGPSGAAGPPGPK (K753) with oxidation of P15 and (B) COL1A2 peptide GFPGTPGLPGFK††GIR (K175) with oxidation of P3, P6, and P9. †† denotes site of modification (Δ 340.10 Da). For y and b ion identifications; † denotes the loss of 162.05 Da (Hex), †† denotes the loss of 324.11 Da (2 Hex), * denotes the loss of NH3, and o denotes the loss of H2O. Assigning these hexose groups as Hyl O-linked galactose and glucosylgalactose is consistent with the well-characterized modification of fibrillar collagens (75–79). Consistent with this assignment, all modifications are found in the triple helix regions.
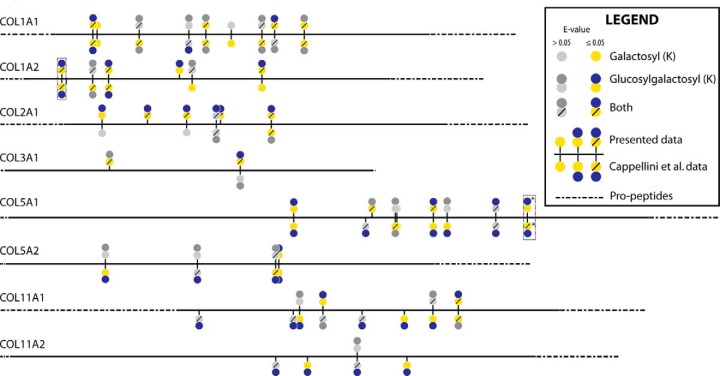
In comparison to type I collagen of modern bone, five specific Hyl-galactose/glucosylgalactose sites have recently been characterized in a young bovine femur (60), to which our B. latifrons specimen shows the same glycosylation pattern. We identified all five sites containing both glycan modifications, with three of the five being identified with high probability (E-value < 0.05). Of note, the previous studies label each residue relative to the helical region of the protein, whereas we label each residue relative to the full protein sequence. Additionally, all reported B. latifrons COL2A1 glycosylated Hyl sites in Fig. 5 (K287, K419, K527, K608, K620, K764) were recently identified by LC-MS/MS in a modern bovine sample with the same glycosylation pattern (61).
Fig. 5.
Glycosylated hydroxylysine residues identified within several collagen proteins. Line lengths mirror the primary structure organization of the represented proteins. Colored circles represent significant modifications (expectation value ≤ 0.05), while grayscale circles represent modifications with an e-value > 0.05 and are included only for sites where a significant glycan modification was identified in at least one of the two datasets. PTMs are either oriented upwards or downwards, indicating whether a specific glycosylation pattern has been identified in the present B. latifrons dataset (up) or in a recent proteomics report on woolly mammoth (M. primigenius) femur proteomics by Cappellini et al. (29) (down). The boxed COL1A2 and COL5A1 glycosylation sites represent the peptides shown in Fig. 4(A) and 4(B). * denotes that this specific site, K753, is found in a different isoform of the α1-chain of type V collagen (Accession # K7QCV6) than the one used for the figure (Accession # G3MZI7). The placement of this site at residue #1491 was determined by homology searching.
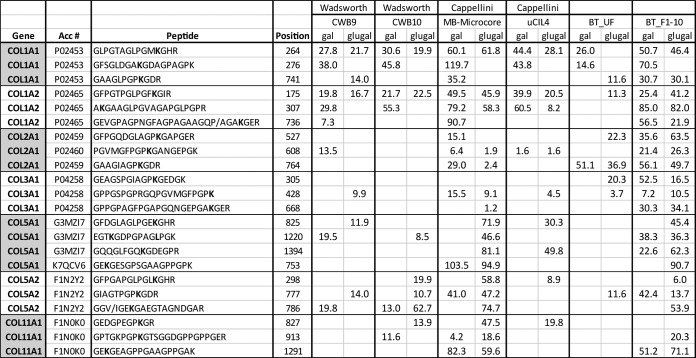
To explore the possibility that these unexpected modifications are not unique to our B. latifrons sample and rather represented a common feature in bone fossil samples, a meta-analysis of raw MS files from previously published proteomics work on fossil bones from M. primigenius and bovine (29, 30) was performed. A comparison of collagen glycosylation patterns to the dataset from Cappellini et al. (29), is reported in Fig. 5, Table I, and Table II. Additionally, Table II compares glycosylated collagen peptides from search results of data from Cappellini et al. (29) and Wadsworth and Buckley (30). Percentage protein coverage for several collagens, specifically collagen type II and type III, are much higher in our dataset compared with being very low or not identified in the other datasets. Not surprisingly, fractionation allows for increased coverage (and score of identifications) of modified peptides as can be seen from the comparison of our unfractionated (BL_UF) and fractionated datasets (BL_F1–10).
Table II. Potential fibrillar collagen glycoside modifications in ancient protein datasets. Mascot scores are reported for the lysine modifications indicated. Differences in the elephant and bovine sequences used to identify mammoth or bison proteins, respectively, are separated by a forward slash (/). The probable site of modification is bolded.
The dataset from our fractionated sample is very similar to the two datasets from Cappellini et al. for collagen 1 and 5 chains. In general, the profile of galactosyl, glucosylgalactosyl, or both modifications observed at a given site are consistent between the datasets. For example, COL1A2 position K175 was identified in both forms, whereas COL1A1 K276 was only found in the galactosyl form across all datasets. Several peptides matched with low scores to the same site as observed with high confidence in one or more datasets. For example, COL1A2 K175 and COL2A1 K527 in our BL_UF and BL_F1–10 datasets (Table II). The reliable detection of these preserved post-translational modifications (PTMs) from high-quality samples/spectra might enable the inference of these peptides in lower-quality samples as well (Supplemental Fig. 1). Lower-quality spectra might be obtained through the analysis of unfractionated samples, by means of less sensitive analytical platforms, or from samples that are less well preserved. Identification based on spectral matching offers a means to further explore common features of ancient samples such as those described here.
DISCUSSION
Although historical and fossil records show that the bison arose in Eurasia, evolutionary divergence and migration have led to extensive bison colonies inhabiting North America (54). The divergence of bison to North America has given rise to species not found elsewhere, such as the giant-horned B. latifrons. Speciation of B. latifrons dates back to the Middle and Late Pleistocene periods, extending back at least to late Illinoian (Riss) time (191–130 ky ago). The B. latifrons samples found at the Ziegler site date back ∼120 ky, thus opening a previously unexplored window to the biology of extinct ice age North American fauna (51).
Multiple bone samples from the Ziegler Reservoir site were analyzed using a bottom-up mass-spectrometry-based proteomic analysis. A rapid sample preparation method was developed that yielded confident peptide identifications to bovine proteins from low milligram quantities of archeological samples. Analysis of the inner surface from the skull of B. latifrons resulted in several thousand peptide identifications that map to several dozen proteins, including COL1A1 and COL1A2. Notably, our method lent itself to the extraction an identification of several noncollagenous proteins, in like fashion to recent reports in the literature about ad hoc sample processing strategies for fossil bones (29, 30).
Despite differences in species and sample location, the protein profile of B. latifrons was similar to that reported for a bone sample from Lyuba, a 43-ky-old woolly mammoth (M. primigenius) found preserved in the Siberian permafrost (29). Furthermore, the overall number of proteins identified, 33, is close (both quantitatively and qualitatively) to the number that was recently reported by Wadsworth and Buckley for bovine fossilized samples treated in an attempt to degrade collagen (via digestion with bacterial collagenase) and enable/enrich for detection of noncollagenous species (30). Similarities include a subset of serum proteins, some abundant in plasma such as alpha-2-HS-glycoprotein and serum albumin, and some found at lower levels, such as thrombospondin 1. There is a high correlation of ECM proteins identified between these samples, and ours that include abundant collagens, major ECM glycoproteins, and plasma proteins. Peptide identifications not represented in Table I include additional ECM, plasma, and intracellular proteins that show a high degree of similarity with peptide identifications from Cappellini et al. (24) Notably, both datasets include identifications to fibromodulin, lumican, and olfactomedin-like protein 3.
Differences in identified proteins between the aforementioned datasets include several that are specific to connective tissue, namely type II collagen, aggrecan, hyaluronan and proteoglycan link protein 1, and cartilage intermediate layer proteins (CILP and CILP2). Identification of these proteins is consistent with our hypothesis that this sample was derived, at least in part, from the connective tissue meninges. The second most abundant bone protein (2, 6, 65) osteocalcin (BGLAP), was identified here by all of the software packages used for searching (Table I). We identified 25 of 49 residues from the carboxy-terminus, with the region containing the namesake gamma-carboxyglutamic acid (gla) residues missing. The identified peptides start with a non-tryptic termini after Cys at position 29 in the mature protein (C)DELADHIGFQEAY(R)RFYGPV(-). Interestingly, no significant matches were made to peptides containing cysteine residues in our dataset; however, a cysteine residue was identified from osteocalcin in Cappellini et al.'s dataset (29), possibly suggesting differing degrees of preservation between samples. It is likely that these, and possibly other reactive residues, progressed down degradative pathways that resulted in additional modifications yielding mass defects and/or possible backbone cleavage. Consistent with this finding, infrared mapping of proteins from 50-my-old soft tissues suggested that diagenesis might affect thiol stability, thereby favoring -SH group modifications hindering MS-detection (66).
Owing to its small size, diagenetic stability, and residence in the sheltered microenvironment of bone, osteocalcin is ideal for exploiting the technology of proteomics to extend sequencing of biomolecules to ancient samples (6, 23, 67). Osteocalcin was even thought to survive in Mesozoic dinosaurs until the challenges and possibility of false positives using immunological methods for fossil protein detection were realized (68). However, despite being very abundant, identification of osteocalcin from bone samples might be dependent upon specific bone protein extraction protocols and bioinformatic elaboration of mass spectra results. Identification of osteocalcin and hyaluronan and proteoglycan link protein 1 in our dataset required semi-tryptic enzyme specificity as a search parameter (Supplemental Table 2), which is indicative of partial degradation of osteocalcin as a recurring taphonomic event in post mortem bones. We matched six peptides to osteocalcin from our B. latifrons dataset yielding 50% sequence coverage of the 49-residue mature protein. This identification is strongly suggestive that protein was derived, in part, from bone of this inner skull specimen. Although osteocalcin was not reported by Cappellini et al. (29), our search conditions were used to match eight unique peptides yielding 78% sequence coverage. While osteocalcin has historically been one of the most studied proteins from ancient bone samples, it is worth noting that previous reports describe the use of matrix-assisted laser desorption/ionization (67) and often times ion exchange chromatographic enrichment, as opposed to electrospray ionization and no protein-level enrichment used in the recent report by Cappellini et al. (29) and here.
Advanced analytical platforms that extract difficult protein samples for enzymatic digestion, offer high mass accuracy with more sensitive detection of the resulting peptide species is allowing for acquisition of more useful and complete ancient protein datasets. Efficient analysis of these datasets is challenging with current bioinformatics workflows due to the large number of modifications that accumulate both in vivo and during diagenesis. Open mass modification searches offer the potential to identify numerous modifications but currently require extensive manual validation. Further development of these methods and the characterization of additional ancient samples will be required to refine the methods of analysis and ultimately draw conclusions regarding the role of molecular features and environmental exposure on protein survival.
Lack of cellular proteins, abundance of non-tryptic termini, and specific peptide modifications are all markers of taphonomy and diagenesis of organic matter, resulting from extensive post mortem protein decomposition and chemical/enzymatic degradation. High levels of Gln and Asn deamidation were observed in these samples with variable levels of modification at the intra- and interprotein levels. Some of the lowest levels of deamidations were found in the fibrillar collagens, suggesting the highest level of protection from diagenetic events. A high number of hydroxyproline (Hyp) modifications were observed in fibrillar collagens on the G-X-Y repeat motif where X is often Pro and Y is often Hyp. Hyp formation (69) is an ascorbic-acid-dependent reaction catalyzed by the prolyl hydroxylase enzyme family and is part of the in vivo maturation of collagen. The 4-hydroxyproline modification in the Y position stabilizes the triple-helix bundle and is largely responsible for the unique properties that infer long-term stability on the fibrillar collagen molecules (70). These modifications give rise to the high number of unique peptides identified that map to the collagen proteins. Several of the ECM proteins identified are known to be involved in collagen fiber organization and development. Whether these identified ECM and secreted proteins are part of a specific matrix network encapsulated in higher-order collagen structures or are more randomly trapped and protected in mineralized material is unknown.
Together, our data suggest that under unique environmental circumstances the mineralized matrix of bone favors protein preservation and may be dispensable in the case of well-preserved specimens. ECM fibrillar structures, most likely due to stable covalent protein–protein bonds, such as those found in collagen fibers, may be sufficient to preserve protein material and allow for peptide identification using modern proteomic techniques. Such interactions might be further promoted by the presence of unique glycosylation profiles found in modern (60–62) and ancient collagen. Meta-analyses of key datasets in the field revealed recurring collagen hydroxylysine glycosides in ancient collagen molecules, suggesting the universality of this feature in Middle and Late Pleistocene fossils. Similarities in glycosylation patterns between our dataset and modern datasets suggest that these modifications are bona fide PTMs and not post mortem artifacts. For example, the hydroxylysine glycosylation of COL1A2 site K175 has recently been identified in collagen isolated from mouse osteoblast cells at 80% stoichiometry (62) and from a modern bovine femur at ∼60% stoichiometry (60). In our B. latifrons dataset, nearly all tryptic peptides containing K175 from COL1A2 are glycosylated, hinting at the possible correlation between glycosylation and protection from diagensis.
Both of the aforementioned O-linked glycosylations are unique to fibrillar collagens (63) and have been previously reported in other proteomics datasets (60, 62, 71, 72). Although their biological function still remains somewhat unclear (73), they are believed to play a role in modulating the structural stability of collagen (74, 75). Indeed, collagen glycosylation plays a role in bone mineralization; however, overglycosylation has been shown to be associated with disorders such as osteogenesis imperfecta (76), chondrodysplasias (77), rheumatoid arthritis (78), postmenopausal osteoporosis, osteosarcoma, osteofibrous dysplasia, and Kashin–Beck disease (73)—further suggesting a role for these particular modifications in modulating collagen architecture. From a structural standpoint, the bulky nature of the sugars attached to the side chain of hydroxylysine residues suggests specific functions for these PTMs, including their involvement in the control of collagen fibrillogenesis, crosslinking, remodeling, collagen-cell/collagen extracellular matrix interactions, and induction of vessel-like structures (79–81) (reviewed in Yamauchi and Sricholpech (73)).
Notably, the COL1A1 glycopeptide fragment consisting of residues 256–270 and including the orthologue to K264 (glycosylated in our study and in mammoth and bovine bone samples assayed by Cappellini and Buckley, respectively—Table II), stimulates the majority of the autoimmune helper T cells obtained in a murine model of rheumatoid arthritis (82). Human K265 is a well-established target of O-glycosylation events involving collagen-induced autoimmune responses, such as in rheumatoid arthritis, or bone mineralization. Furthermore, hydroxylysine galactosylation at position 264 (mouse orthologue) was revealed to form critical contacts with the T cell receptor, while Ile260 and Phe263 anchored the glycopeptide in the P1 and P4 pockets of the disease associated H-2Aq MHC molecule, thereby promoting autoimmune reactions underlying rheumatoid arthritis (83). These modifications are found in modern samples, play multiple roles in normal mammalian physiology and pathologies, and are consistent with the O-glycosylation of key lysine residues from B. latifrons collagen reported here and in other ancient protein specimens.
Similarities between ancient and modern datasets can be interpreted in several ways: 1) These PTMs are part of specific collagen structures that promote, or are markers of, collagen survival over time; 2) these modifications stem from a selective negative enrichment process through diagenesis of un-modified sequences; 3) albeit unlikely, despite being analytically consistent with hydroxylysine-galactosyl/glucosylgalactosyl modifications (accurate mass, retention time shift, modification site consistency with modern samples, and signature tandem MS features) the modification assignments are false positives; 4) these modifications are products of a yet uncharacterized diagenetic pathway; or 5) these modifications, although prevalent in several ancient and modern specimens, are not related to or effect preservation of ancient protein. At this stage, we hypothesize that interpretation 1 or 2 is correct: that these findings are suggestive of a specific biochemical mechanism that occurs during in vivo maturation of the extracellular matrix (72) to increase the robustness of macromolecular structures that contain hydroxylysine glycosides at key locations in fibrillar collagens. While further studies will be required to determine the validity of this hypothesis, the evidence presented here proposes novel information on the potential role of collagen PTMs in the suppression of post mortem taphonomy and diagenesis, which might foster future developments in the field of paleoproteomics, molecular taxonomy, and biomaterials research.
Supplementary Material
Acknowledgments
We would like to thank Mike Buckley for sharing raw data. Sharon Rose for assisting with open mass searches and deamidation analysis, Chelsey Zaharris for assisting with preliminary data analysis, and Ryan Walsh for assisting with sample preparation. We would like to thank Richard Stucky, Carol Lucking, and Kirk Johnson for sample procurement and discussions. Specimens referenced in this paper were collected at the Ziegler Reservoir locality, 5PT1264, under State of Colorado Paleontological Permits 2010-106 and 2011-17. We thank History Colorado/The Colorado Historical Society, the Snowmass Water and Sanitation District, and the Town of Snowmass Village for their assistance with this excavation. Contents are the authors' sole responsibility and do not necessarily represent official NIH views.
Footnotes
Author contributions: R.C.H., M.D., and K.C.H. designed the research; R.C.H., M.D., and K.C.H. performed the research; R.C.H., M.J.W., T.N., A.B., A.D., and K.C.H. analyzed the data; and R.C.H., M.J.W., T.N., A.B., A.D., and K.C.H. wrote the paper.
* This work was supported in part by NIH/IMAT R21 CA132741, NIH/NCI P30 CA046934, and NIH/NCATS Colorado CTSI Grant Number UL1 TR000154.
 This article contains supplemental material Supplemental Tables 1 to 3 and Supplemental Fig. 1.
This article contains supplemental material Supplemental Tables 1 to 3 and Supplemental Fig. 1.
1 The abbreviations used are:
- ID
- Identification
- SLIP
- Site Localization in Peptide
- FDR
- False Discovery Rate
- PRIDE
- Proteomics Identifications Database
- PSM
- Peptide Spectral Match
- PTM
- Post-translational Modification
- ECM
- Extracellular Matrix
- LC-MS/MS
- Liquid Chromatography-Tandem Mass Spectrometry
- ACN
- Acetonitrile
- IAM
- Iodoacetamide
- TPCK
- Tosyl phenylalanyl chloromethyl ketone
- TCEP
- Tris(2-carboxyethyl)phosphine
- HCD
- High-energy collision dissociation.
REFERENCES
- 1. Smejkal G. B., Schweitzer M. H. (2007) Will current technologies enable dinosaur proteomics? Expert Rev. Proteomics 4, 695–699 [DOI] [PubMed] [Google Scholar]
- 2. Ostrom P. H., Schall M., Gandhi H., Shen T.-L., Hauschka P. V., Strahler J. R., Gage D. A. (2000) New strategies for characterizing ancient proteins using matrix-assisted laser desorption ionization mass spectrometry. Geochim. Cosmochim. Acta 64, 1043–1050 [Google Scholar]
- 3. Marota I., Rollo F. (2002) Molecular paleontology. Cell. Mol. Life Sci. 59, 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hofreiter M., Collins M., Stewart J. R. (2012) Ancient biomolecules in Quaternary palaeoecology. Quat. Sci. Rev. 33, 1–13 [Google Scholar]
- 5. Bada J. L., Wang X. S., Hamilton H. (1999) Preservation of key biomolecules in the fossil record: current knowledge and future challenges. Philos. Trans. R. Soc. London B 354, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collins M. J., Nielsen–Marsh C. M., Hiller J., Smith C. I., Roberts J. P., Prigodich R. V., Wess T. J., Csapò J., Millard A. R., Turner–Walker G. (2002) The survival of organic matter in bone: a review. Archaeometry 44, 383–394 [Google Scholar]
- 7. Schwarz C., Debruyne R., Kuch M., McNally E., Schwarcz H., Aubrey A. D., Bada J., Poinar H. (2009) New insights from old bones: DNA preservation and degradation in permafrost preserved mammoth remains. Nucleic Acids Res. 37, 3215–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dobberstein R. C., Collins M. J., Craig O. E., Taylor G., Penkman K. E. H., Ritz-Timme S. (2009) Archaeological collagen: Why worry about collagen diagenesis? Archaeol. Anthropol. Sci. 1, 31–42 [Google Scholar]
- 9. Asara J. M., Garavelli J. S., Slatter D. A., Schweitzer M. H., Freimark L. M., Phillips M., Cantley L. C. (2007) Interpreting sequences from mastodon and T. rex. Science 317, 1324–1325 [DOI] [PubMed] [Google Scholar]
- 10. Asara J. M., Schweitzer M. H., Freimark L. M., Phillips M., Cantley L. C. (2007) Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry. Science 316, 280–285 [DOI] [PubMed] [Google Scholar]
- 11. Avci R., Schweitzer M. H., Boyd R. D., Wittmeyer J. L., Terán Arce F., Calvo J. O. (2005) Preservation of bone collagen from the late Cretaceous period studied by immunological techniques and atomic force microscopy. Langmuir ACS J. Surf. Colloids 21, 3584–3590 [DOI] [PubMed] [Google Scholar]
- 12. Schweitzer M. H., Chiappe L., Garrido A. C., Lowenstein J. M., Pincus S. H. (2005) Molecular preservation in Late Cretaceous sauropod dinosaur eggshells. Proc. Biol. Sci. 272, 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schweitzer M. H., Wittmeyer J. L., Horner J. R., Toporski J. K. (2005) Soft-tissue vessels and cellular preservation in Tyrannosaurus rex. Science 307, 1952–1955 [DOI] [PubMed] [Google Scholar]
- 14. Schweitzer M. H., Wittmeyer J. L., Horner J. R. (2005) Gender-specific reproductive tissue in ratites and Tyrannosaurus rex. Science 308, 1456–1460 [DOI] [PubMed] [Google Scholar]
- 15. Schweitzer M. H. (2010) Blood from stone. Sci. Am. 303, 62–69 [DOI] [PubMed] [Google Scholar]
- 16. Schweitzer M. H., Zheng W., Organ C. L., Avci R., Suo Z., Freimark L. M., Lebleu V. S., Duncan M. B., Vander Heiden M. G., Neveu J. M., Lane W. S., Cottrell J. S., Horner J. R., Cantley L. C., Kalluri R., Asara J. M. (2009) Biomolecular characterization and protein sequences of the Campanian hadrosaur B. canadensis. Science 324, 626–631 [DOI] [PubMed] [Google Scholar]
- 17. Schweitzer M. H., Suo Z., Avci R., Asara J. M., Allen M. A., Arce F. T., Horner J. R. (2007) Analyses of soft tissue from Tyrannosaurus rex suggest the presence of protein. Science 316, 277–280 [DOI] [PubMed] [Google Scholar]
- 18. Schweitzer M. H., Zheng W., Cleland T. P., Bern M. (2013) Molecular analyses of dinosaur osteocytes support the presence of endogenous molecules. Bone 52, 414–423 [DOI] [PubMed] [Google Scholar]
- 19. Dalton R. (2008) Fresh doubts over T. rex chicken link. Nat. News 454, 1035–1035 [DOI] [PubMed] [Google Scholar]
- 20. Pevzner P. A., Kim S., Ng J. (2008) Comment on “Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry.” Science 321, 1040–1040 [DOI] [PubMed] [Google Scholar]
- 21. Buckley M., Walker A., Ho S. Y. W., Yang Y., Smith C., Ashton P., Oates J. T., Cappellini E., Koon H., Penkman K., Elsworth B., Ashford D., Solazzo C., Andrews P., Strahler J., Shapiro B., Ostrom P., Gandhi H., Miller W., Raney B., Zylber M. I., Gilbert M. T. P., Prigodich R. V., Ryan M., Rijsdijk K. F., Janoo A., Collins M. J. (2008) Comment on “Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry.” Science 319, 33; author reply 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bern M., Phinney B. S., Goldberg D. (2009) Reanalysis of Tyrannosaurus rex mass spectra. J. Proteome Res. 8, 4328–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collins M. J., Gernaey A. M., Nielsen-Marsh C. M., Vermeer C., Westbroek P. (2000) Slow rates of degradation of osteocalcin: Green light for fossil bone protein? Geology 28, 1139–1142 [Google Scholar]
- 24. Binladen J., Gilbert M. T., Willerslev E. (2007) 800,000 year old mammoth DNA, modern elephant DNA or PCR artifact? Biol. Lett. 3, 55—56; discussion 60–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buckley M. (2013) A molecular phylogeny of Plesiorycteropus reassigns the extinct mammalian order “Bibymalagasia.” PloS One 8, e59614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hagelberg E., Thomas M. G., Cook C. E., Jr., Sher A. V., Baryshnikov G. F., Lister A. M. (1994) DNA from ancient mammoth bones. Nature 370, 333–334 [DOI] [PubMed] [Google Scholar]
- 27. Hedges S. B., Schweitzer M. H. (1995) Detecting dinosaur DNA. Science 268, 1191–1192; author reply 1194 [DOI] [PubMed] [Google Scholar]
- 28. Rohland N., Reich D., Mallick S., Meyer M., Green R. E., Georgiadis N. J., Roca A. L., Hofreiter M. (2010) Genomic DNA sequences from mastodon and woolly mammoth reveal deep speciation of forest and savanna elephants. PLoS Biol 8, e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cappellini E., Jensen L. J., Szklarczyk D., Ginolhac A., da Fonseca R. A., Stafford T. W., Holen S. R., Collins M. J., Orlando L., Willerslev E., Gilbert M. T., Olsen J. V. (2012) Proteomic analysis of a pleistocene mammoth femur reveals more than one hundred ancient bone proteins. J. Proteome Res. 11, 917–926 [DOI] [PubMed] [Google Scholar]
- 30. Wadsworth C., Buckley M. (2014) Proteome degradation in fossils: investigating the longevity of protein survival in ancient bone. Rapid Commun. Mass Spectrom. RCM 28, 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buckley M., Collins M., Thomas-Oates J. (2008) A method of isolating the collagen (I) α2 chain carboxytelopeptide for species identification in bone fragments. Anal. Biochem. 374, 325–334 [DOI] [PubMed] [Google Scholar]
- 32. Buckley M., Collins M., Thomas-Oates J., Wilson J. C. (2009) Species identification by analysis of bone collagen using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. RCM 23, 3843–3854 [DOI] [PubMed] [Google Scholar]
- 33. Buckley M., Collins M. J. (2011) Collagen survival and its use for species identification in Holocene-lower Pleistocene bone fragments from British archaeological and paleontological sites. Antiqua 1, 1 [Google Scholar]
- 34. Eastoe J. E., Eastoe B. (1954) The organic constituents of mammalian compact bone. Biochem. J. 57, 453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Isaacs W. A., Little K., Currey J. D., Tarlo L. B. H. (1963) Collagen and a cellulose-like substance in fossil dentine and bone. Nature 197, 192. [DOI] [PubMed] [Google Scholar]
- 36. Wyckoff R. W., Wagner E., Matter P., 3rd, Doberenz A. R. (1963) Collagen in fossil bone. Proc. Natl. Acad. Sci. U.S.A. 50, 215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wyckoff R. W., McCaughey W. F., Doberenz A. R. (1964) the amino acid composition of proteins from Pleistocene bones. Biochim. Biophys. Acta 93, 374–377 [DOI] [PubMed] [Google Scholar]
- 38. Ho T. Y. (1965) The amino acid composition of bone and tooth proteins in late Pleistocene mammals. Proc. Natl. Acad. Sci. U.S.A. 54, 26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goodman M., Birk D. E., Romero-Herrera A. E., Lande M. A., Dene H., Barnhart M. I. (1980) Collagen preservation in soft tissue from the Magadan mammoth. FEBS Lett. 114, 30–34 [Google Scholar]
- 40. Pessenda L. R., Gouveia S. M., Aravena R. (2006) Radiocarbon dating of total soil organic matter and humin fraction and its comparison with (super 14) C ages of fossil charcoal. Radiocarbon 43, 595–601 [Google Scholar]
- 41. Lynch M. (2010) Evolution of the mutation rate. Trends Genet. 26, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pál C., Papp B., Lercher M. J. (2006) An integrated view of protein evolution. Nat. Rev. Genet. 7, 337–348 [DOI] [PubMed] [Google Scholar]
- 43. Prager E. M., Wilson A. C., Lowenstein J. M., Sarich V. M. (1980) Mammoth albumin. Science 209, 287–289 [DOI] [PubMed] [Google Scholar]
- 44. Ulrich M. M., Perizonius W. R., Spoor C. F., Sandberg P., Vermeer C. (1987) Extraction of osteocalcin from fossil bones and teeth. Biochem. Biophys. Res. Commun. 149, 712–719 [DOI] [PubMed] [Google Scholar]
- 45. Schmidt-Schultz T. H., Schultz M. (2004) Bone protects proteins over thousands of years: extraction, analysis, and interpretation of extracellular matrix proteins in archeological skeletal remains. Am. J. Phys. Anthropol. 123, 30–39 [DOI] [PubMed] [Google Scholar]
- 46. Cattaneo C., Gelsthorpe K., Phillips P., Sokol R. J. (1990) Blood in ancient human bone. Nature 347, 339. [DOI] [PubMed] [Google Scholar]
- 47. Nielsen-Marsh C. M., Ostrom P. H., Gandhi H., Shapiro B., Cooper A., Hauschka P. V., Collins M. J. (2002) Sequence preservation of osteocalcin protein and mitochondrial DNA in bison bones older than 55 ka. Geology 30, 1099–1102 [Google Scholar]
- 48. Nielsen-Marsh C. M., Richards M. P., Hauschka P. V., Thomas-Oates J. E., Trinkaus E., Pettitt P. B., Karavanić I., Poinar H., Collins M. J. (2005) Osteocalcin protein sequences of Neanderthals and modern primates. Proc. Natl. Acad. Sci. U.S.A. 102, 4409–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pigati J. S., Miller I. M., Johnson K. R., Honke J. S., Carrara P. E., Muhs D. R., Skipp G., Bryant B. (2014) Geologic setting and stratigraphy of the Ziegler Reservoir fossil site, Snowmass Village, Colorado. Quat. Res. 82, 3, 477–489 [Google Scholar]
- 50. Johnson K. R., Miller I. M., Pigati J. S. (2014) The Snowmastodon Project. Quat. Res. 82, 3, 473–476 [Google Scholar]
- 51. Sertich J. J. W., Stucky R. K., McDonald H. G., Newton C., Fisher D. C., Scott E., Demboski J. R., Lucking C., McHorse B. K., Davis E. B. (2014) High-elevation late Pleistocene (MIS 6–5) vertebrate faunas from the Ziegler Reservoir fossil site, Snowmass Village, Colorado. Quat. Res. 82, 3, 504–517 [Google Scholar]
- 52. Elias S. A. (2014) Environmental interpretation of fossil insect assemblages from MIS 5 at Ziegler Reservoir, Snowmass Village, Colorado. Quat. Res. 82, 3, 592–603 [Google Scholar]
- 53. Miller I. M., Pigati J. S., Scott Anderson R., Johnson K. R., Mahan S. A., Ager T. A., Baker R. G., Blaauw M., Bright J., Brown P. M., Bryant B., Calamari Z. T., Carrara P. E., Cherney M. D., Demboski J. R., Elias S. A., Fisher D. C., Gray H. J., Haskett D. R., Honke J. S., Jackson S. T., Jiménez-Moreno G., Kline D., Leonard E. M., Lifton N. A., Lucking C., Gregory McDonald H., Miller D. M., Muhs D. R., Nash S. E., Newton C., Paces J. B., Petrie L., Plummer M. A., Porinchu D. F., Rountrey A. N., Scott E., Sertich J. J. W., Sharpe S. E., Skipp G. L., Strickland L. E., Stucky R. K., Thompson R. S., Wilson J. (2014) A high-elevation, multi-proxy biotic and environmental record of MIS 6–4 from the Ziegler Reservoir fossil site, Snowmass Village, Colorado, U.S.A. Quat. Res. 82, 3, 618–634 [Google Scholar]
- 54. Mahan S. A., Gray H. J., Pigati J. S., Wilson J., Lifton N. A., Paces J. B., Blaauw M. (2014) A geochronologic framework for the Ziegler Reservoir fossil site, Snowmass Village, Colorado. Quat. Res. 82, 3, 490–503 [Google Scholar]
- 55. Bern M., Kil Y. J., Becker C. (2012) Byonic: Advanced peptide and protein identification software. Curr. Protoc. Bioinforma./editoral board, Andreas D Baxevanis. [et al], Chapter 13, Unit13.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baker P. R., Trinidad J. C., Chalkley R. J. (2011) Modification site localization scoring integrated into a search engine. Mol. Cell. Proteomics 10, M111.008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baker P. R., Chalkley R. J. (2014) MS-viewer: A web-based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Van Doorn N. L., Wilson J., Hollund H., Soressi M., Collins M. J. (2012) Site-specific deamidation of glutamine: a new marker of bone collagen deterioration. Rapid Commun. Mass Spectrom. 26, 2319–2327 [DOI] [PubMed] [Google Scholar]
- 59. Dasari S., Wilmarth P. A., Rustvold D. L., Riviere M. A., Nagalla S. R., David L. L. (2007) Reliable detection of deamidated peptides from lens crystallin proteins using changes in reversed-phase elution times and parent ion masses. J. Proteome Res. 6, 3819–3826 [DOI] [PubMed] [Google Scholar]
- 60. Terajima M., Perdivara I., Sricholpech M., Deguchi Y., Pleshko N., Tomer K. B., Yamauchi M. (2014) Glycosylation and cross-linking in bone type I collagen. J. Biol. Chem. 289, 22636–22647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song E., Mechref Y. (2013) LC-MS/MS identification of the O-glycosylation and hydroxylation of amino acid residues of collagen α-1 (II) chain from bovine cartilage. J. Proteome Res. 12, 3599–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sricholpech M., Perdivara I., Yokoyama M., Nagaoka H., Terajima M., Tomer K. B., Yamauchi M. (2012) Lysyl hydroxylase 3-mediated glucosylation in type I collagen: Molecular loci and biological significance. J. Biol. Chem. 287, 22998–23009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Spiro R. G. (1967) The structure of the disaccharide unit of the renal glomerular basement membrane. J. Biol. Chem. 242, 4813–4823 [PubMed] [Google Scholar]
- 64. Wells L. (2010) Handbook of Glycomics, Pierce R. D. C. M. Academic Press, San Diego, 45–57 [Google Scholar]
- 65. Colombo G., Fanti P., Yao C., Malluche H. H. (1993) Isolation and complete amino acid sequence of osteocalcin from canine bone. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 8, 733–743 [DOI] [PubMed] [Google Scholar]
- 66. Edwards N. P., Barden H. E., van Dongen B. E., Manning P. L., Larson P. L., Bergmann U., Sellers W. I., Wogelius R. A. (2011) Infrared mapping resolves soft tissue preservation in 50 million year-old reptile skin. Proc. R. Soc. B 278, 3209–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Deleted in proof.
- 68. Muyzer G., Sandberg P., Knapen M. H. J., Vermeer C., Collins M., Westbroek P. (1992) Preservation of the bone protein osteocalcin in dinosaurs. Geology 20, 871 [Google Scholar]
- 69. Millard C. J., Campbell I. D., Pickford A. R. (2005) Gelatin binding to the 8F19F1 module pair of human fibronectin requires site-specific N-glycosylation. FEBS Lett. 579, 4529–4534 [DOI] [PubMed] [Google Scholar]
- 70. Jenkins C. L., Raines R. T. (2002) Insights on the conformational stability of collagen. Nat. Prod. Rep. 19, 49–59 [DOI] [PubMed] [Google Scholar]
- 71. Taga Y., Kusubata M., Ogawa-Goto K., Hattori S. (2012) Development of a novel method for analyzing collagen O-glycosylations by hydrazide chemistry. Mol. Cell. Proteomics 11, M111.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hermansson M., Sawaji Y., Bolton M., Alexander S., Wallace A., Begum S., Wait R., Saklatvala J. (2004) Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin βA (activin A), a regulatory molecule for chondrocytes. J. Biol. Chem. 279, 43514–43521 [DOI] [PubMed] [Google Scholar]
- 73. Yamauchi M., Sricholpech M. (2012) Lysine post-translational modifications of collagen. Essays Biochem. 52, 113–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang C., Luosujärvi H., Heikkinen J., Risteli M., Uitto L., Myllylä R. (2002) The third activity for lysyl hydroxylase 3: Galactosylation of hydroxylysyl residues in collagens in vitro. Matrix Biol. 21, 559–566 [DOI] [PubMed] [Google Scholar]
- 75. Ruotsalainen H., Sipilä L., Vapola M., Sormunen R., Salo A. M., Uitto L., Mercer D. K., Robins S. P., Risteli M., Aszodi A., Fässler R., Myllylä R. (2006) Glycosylation catalyzed by lysyl hydroxylase 3 is essential for basement membranes. J. Cell Sci. 119, 625–635 [DOI] [PubMed] [Google Scholar]
- 76. Tenni R., Valli M., Rossi A., Cetta G. (1993) Possible role of overglycosylation in the type I collagen triple helical domain in the molecular pathogenesis of osteogenesis imperfecta. Am. J. Med. Genet. 45, 252–256 [DOI] [PubMed] [Google Scholar]
- 77. Murray L. W., Bautista J., James P. L., Rimoin D. L. (1989) Type II collagen defects in the chondrodysplasias. I. Spondyloepiphyseal dysplasias. Am. J. Hum. Genet. 45, 5–15 [PMC free article] [PubMed] [Google Scholar]
- 78. Holm B., Broddefalk J., Flodell S., Wellner E., Kihlberg J. (2000) An improved synthesis of a galactosylated hydroxylysine building block and its use in solid-phase glycopeptide synthesis. Tetrahedron 56, 1579–1586 [Google Scholar]
- 79. Di Lullo G. A., Sweeney S. M., Körkkö J., Ala-Kokko L., Antonio J. D. (2002) Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J. Biol. Chem. 277, 4223–4231 [DOI] [PubMed] [Google Scholar]
- 80. Orgel J. P., San Antonio J. D., Antipova O. (2010) Molecular and structural mapping of collagen fibril interactions. Connect. Tissue Res. 52, 2–17 [DOI] [PubMed] [Google Scholar]
- 81. Orgel J. P., Persikov A. V., Antipova O. (2014) Variation in the helical structure of native collagen. PloS One 9, e89519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Broddefalk J., Bäcklund J., Almqvist F., Johansson M., Holmdahl R., Kihlberg J. (1998) T cells recognize a glycopeptide derived from type II collagen in a model for rheumatoid arthritis. J. Amer. Chem. Soc. 120, 7676–7683 [Google Scholar]
- 83. Kjellén P., Brunsberg U., Broddefalk J., Hansen B., Vestberg M., Ivarsson I., Engström A., Svejgaard A., Kihlberg J., Fugger L., Holmdahl R. (1998) The structural basis of MHC control of collagen-induced arthritis; binding of the immunodominant type II collagen 256–270 glycopeptide to H-2Aq and H-2Ap molecules. Eur. J. Immunol. 28, 755–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.