Abstract
The cell's endomembranes comprise an intricate, highly dynamic and well-organized system. In plants, the proteins that regulate function of the various endomembrane compartments and their cargo remain largely unknown. Our aim was to dissect subcellular trafficking routes by enriching for partially overlapping subpopulations of endosomal proteomes associated with endomembrane markers. We selected RABD2a/ARA5, RABF2b/ARA7, RABF1/ARA6, and RABG3f as markers for combinations of the Golgi, trans-Golgi network (TGN), early endosomes (EE), secretory vesicles, late endosomes (LE), multivesicular bodies (MVB), and the tonoplast. As comparisons we used Golgi transport 1 (GOT1), which localizes to the Golgi, clathrin light chain 2 (CLC2) labeling clathrin-coated vesicles and pits and the vesicle-associated membrane protein 711 (VAMP711) present at the tonoplast. We developed an easy-to-use method by refining published protocols based on affinity purification of fluorescent fusion constructs to these seven subcellular marker proteins in Arabidopsis thaliana seedlings. We present a total of 433 proteins, only five of which were shared among all enrichments, while many proteins were common between endomembrane compartments of the same trafficking route. Approximately half, 251 proteins, were assigned to one enrichment only. Our dataset contains known regulators of endosome functions including small GTPases, SNAREs, and tethering complexes. We identify known cargo proteins such as PIN3, PEN3, CESA, and the recently defined TPLATE complex. The subcellular localization of two GTPase regulators predicted from our enrichments was validated using live-cell imaging. This is the first proteomic dataset to discriminate between such highly overlapping endomembrane compartments in plants and can be used as a general proteomic resource to predict the localization of proteins and identify the components of regulatory complexes and provides a useful tool for the identification of new protein markers of the endomembrane system.
Membrane compartmentalization is an essential mechanism for eukaryotic life, by which cells separate and control biological processes. Plant growth, development, and adaptation to biotic and abiotic stress all rely on the highly dynamic endomembrane system, yet we know comparatively little about the proteins regulating these dynamic trafficking events. The plasma membrane (PM) provides the main interface between the cell and its environment, mediating the transfer of material to and from the cell and is a primary site for perception of external signals. Transmembrane proteins are synthesized in the endoplasmic reticulum (ER) and trafficked to the PM via the Golgi, although there are other secretory routes for soluble cargo (discussed in (1–4)). Post-Golgi trafficking is the main route by which newly synthesized transmembrane proteins and cell wall glycans are delivered to the PM. In plants, secretory and endocytic traffic converge at the trans-Golgi network (TGN), which also functions as an early endosome (EE). Multivesicular bodies (MVBs) are the other main endosomal compartment in plants and serve as prevacuolar compartments (PVCs) or late endosomes (LE) destined for vacuolar degradation (reviewed (1, 5, 6)).
Recycling and sorting of plasma membrane proteins is essential for generating the polar localization of auxin efflux transporters (discussed in (7)), formation of the cell plate during cell division (8–11), and in defense such as localized deposition of papilla reviewed in (12, 13). Furthermore, the subcellular localization of transporters and receptors is dynamically regulated. For example, the boron transporter (BOR1) exhibits polar localization and is internalized and degraded under conditions of high boron to reduce toxicity (14, 15). Similarly the receptor-like kinases (RLKs) flagellin-sensing 2 (FLS2) and brassinosteroid insensitive 1 (BRI1), important transmembrane receptors in antibacterial immunity and plant development, respectively, are constitutively endocytosed and recycled to the PM (16–18). Both receptors and transporters are also cargoes of the LE/MVB trafficking route (16) and are probably sorted to the vacuole for degradation (19, 20). Importantly, FLS2 trafficking via the recycling endocytic or the late endocytic route depends on its activation status; inactive receptors are recycled while ligand-activated receptors are sorted to the late endosomal pathway (16). Similarly, the polar sorting of auxin efflux transporters depends on their phosphorylation status (21). These observations illustrate that membrane compartmentalization underpins important aspects of plant cell biology and has initiated a quest toward a better understanding of the endomembrane compartments and the routes and mechanisms by which cargo is trafficked and sorted within the cell.
Membrane trafficking within the cell requires complex machinery consisting of a plethora of coat and adaptor proteins, small GTPases, targeting, tethering, and scission factors (reviewed in (22, 23)). Homologues of some animal and yeast and endomembrane regulators have been identified in plants, but the localization and function of many of these remain to be characterized. For example, members of the RAB GTPase family have been shown to have markedly different roles and localizations in plants compared with their animal and yeast homologs (24). Therefore, acquiring localization data for tethering complexes and other regulators in plant systems is essential. In Arabidopsis thaliana, some of these proteins have been developed as useful probes to visualize the different endomembrane compartments by fusion with fluorescent reporters (9, 25–27). These include regulators of trafficking events such as RAB GTPases that are molecular switches responsible for the assembly of tethering and docking complexes and compartment identity. RAB proteins are widely used markers of endomembrane compartments, for example RABD2a/ARA5 labels the Golgi and TGN/EE as well as post-Golgi vesicles (4, 24, 26, 28), RABF2b/ARA7 localizes to TGN/EE and LE (25), RABF1/ARA6 is a marker of the LE/MVB vesicles (25, 29), and RABG3f localizes to MVBs and the tonoplast (26, 30).
Fluorescent-tagged marker lines for the live-cell imaging of plant cells have been invaluable in defining the location of proteins within and between organelles and endomembrane compartments (26). However, microscopic investigation of membrane trafficking is limited by throughput, as only few proteins can be studied simultaneously. A powerful approach to large-scale identification of proteins in endomembrane compartments is through subcellular fractionation based on physical properties to directly isolate or enrich for the subcellular compartment of interest. Subcellular fractionation-based proteomics have been successfully used to decipher the steady state and cargo proteomes of, including but not limited to, the ER, the vacuole, PM, mitochondria and chloroplasts, and smaller vesicle-like compartments such as peroxisomes and Golgi (31–41). However, the smaller, transitory vesicles of the secretory and endocytic pathways have proved challenging to purify for reliable proteomic analysis. To overcome this, affinity purification of vesicles was established in animal cells (42, 43) and recently successfully applied in plants in combination with subcellular fractionation. Affinity purification and mass spectrometry (MS) of syntaxin of plants 61 (SYP61)-positive TGN/EE compartments identified 145 proteins specifically enriched in (44), while affinity isolation of VHA-a1-GFP (vacuolar H+ ATPase A1) identified 105 proteins associated with the TGN/EE (45). The VHA-A1 affinity purification data were then further refined using density gradient centrifugation to differentiate cargo and steady-state proteins (45).
We have further explored affinity purification of fluorescent-tagged markers localizing to defined compartments to identify proteins associated with trafficking. Our motivation was to dissect the trafficking routes by enriching for partially overlapping subpopulations of endosomal proteomes associated with small GTPases in the RAB family. We selected RABD2a/ARA5, RABF2b/ARA7, RABF1/ARA6, and RABG3f as markers for Golgi/TGN/EE/secretory vesicles, LE/MVB compartments, LE/MVB compartments and LE/MVB/tonoplast, respectively. Additionally, we used Golgi transport 1 (GOT1), which localizes to the Golgi, clathrin light chain 2 (CLC2) labeling clathrin-coated vesicles (CCVs) and pits and the vesicle-associated membrane protein 711 (VAMP711) present at the tonoplast (26, 27, 29, 46, 47) as comparisons. Our objective was to identify transient cargo proteins, tethers, and docking factors associated with dynamic subdomains of the endomembrane system, to supplement better-characterized “steady-state” components, and to identify components of recycling and vacuolar trafficking pathways.
EXPERIMENTAL PROCEDURES
Plant Materials
The following A. thaliana lines were used in this study (accession Columbia-0 if not otherwise stated): (Wave lines 1, 5, 29, 9, and 18) pUB:mCherry and YFP, YFP-RABG3f, YFP-RABD2a/ARA5, YFP-VAMP711, YFP-GOT1 (26), RABF1/ARA6-RFP, RABF2b/RFP-ARA7 (provided by K. Schumacher, Heidelberg, Germany), p35S:CLC2-GFP in WS-2 (Wassilewskija) background (provided by S. Bednarek, Madison, WI). For protein extraction and affinity purification, 0.1 g of A. thaliana seed for all constructs were grown in sterile 200 ml of Murashige and Skoog medium at 22 °C, 16 h light, shaken at 120 rpm. For microprojectile bombardment assays, 4–6 weeks old A. thaliana plants were grown on soil under controlled conditions of 22 °C, 12 h light, 60% humidity.
Protein Extraction and Affinity Purification
For all constructs, 30–50 g of 8-day-old A. thaliana seedlings were harvested and frozen in liquid nitrogen and ground with a pestle and mortar. Protein extraction buffer (150 mm Na-HEPES (pH7.5), 10 mm EDTA, 10 mm EGTA, 17.5% (w/v) sucrose, 7.5 mm KCl, 0.01% (v/v) Igepal CA-630, 10 mm dithiothreitol, 1% (v/v) protease inhibitors (Sigma), 0.5% (v/v) polyvinylpolypyrrolidone) at 2 ml to 1 g of fresh weight tissue was added. All subsequent steps were performed at 4 °C. Protein concentration was determined (usually 0.4–0.6 g total protein) with BCA assay using BSA as the standard. Homogenate was filtered through two layers of miracloth and centrifuged at 6,000 g for 20 min. 20 μl of chromotek GFP or Red Fluorescent Protein (RTP) trap Sepharose beads (as appropriate) were added per 50 ml homogenate and incubated for 3 h with gentle shaking. The homogenate was then centrifuged at 500 g for 5 min and the supernatant discarded. The bead slurry was washed five times with fresh prechilled extraction buffer (no polyvinylpolypyrrolidone or protease inhibitors) with 3 min incubation. The slurry was collected after the last wash and protein eluted with 2x SDS-PAGE loading buffer and taken for either LC-MS or Western blotting.
Western Blotting and Immunodetection
10% poly-acrylamide SDS-gels were run at 100/200 V and proteins electroblotted onto PVDF membranes at 250 mA (Bio-Rad). Membranes were rinsed in TBS and blocked in 5% (w/v) nonfat milk powder in TBS 0.1% Tween (TBST) (w/v) for 1 h. Primary antibodies were diluted in 0.5% (w/v) nonfat milk, TBST to the following concentrations and incubated at room temperature for 1 h. Primary antibodies were: anti-AHA1 (H+ATPase 1) 1:2,000 (Agrisera AS07 260), anti-BIP2 (luminal binding protein) 1:2,000 (Agrisera AS09 614), anti-RbcL (Rubisco large subunit) 1:10,000 (Agrisera AS03 037), anti-COX2 (cytochrome oxidase 2) 1:5,000 (AS04 053A Agrisera), anti-RFP 1:10,000 (Abcam ab34771). Membranes were washed three times in TBST before 1 h incubation with secondary antibodies anti-rabbit-HRP (Sigma A3687) 1:10,000 or anti-hen-HRP 1:10,000 (Agrisera AS09 603). Signals were visualized using chemiluminescent substrate (Lumigen ECL, GE Healthcare) and GE healthcare Image Quant LAS 3000.
Tryptic Digestion of Proteins
Affinity purified proteins were separated on 4–20% Tris-glycine nUView precast gradient gels (NuSep). The SDS-PAGE gels were cut into seven slices per affinity purification. Gel slices were washed for 30 min with 50% acetonitrile (ACN)/25 mm ammonium bicarbonate (ABC)1 at 37 °C, twice. Then 100% ACN was added for 10 min and the liquid removed. 10 mm DTT in 50 mm ABC was added to cover the gel pieces for 30 min at 56 °C shaking and the supernatant removed. 55 mm chloroacetamide in ABC (in the dark) was applied for 20 min. The gel pieces were washed twice for 15 min with 50% ACN/25 mm ABC and dehydrated with 100% ACN for 10 min. 1 μg of trypsin, 46 mm ABC, 5% ACN was applied at 37 °C overnight, and the supernatant removed and retained. The gel pieces were washed three times by addition of 50% ACN, 5% formic acid, and sonicated for 10 min, and the wash supernatants were then pooled with previous supernatants. The supernatants containing the peptides were then lyophilized. The control samples, from plant expressing the fluorescent tags mCherry or GFP only, were treated as above expect that the tryptic digestions were performed from the affinity beads directly, and these samples were analyzed on an Orbitrap Fusion, not an Orbitrap XL as detailed below.
Mass Spectrometry
LC-MS/MS analysis was performed using a hybrid mass spectrometer LTQ-Orbitrap XL (ThermoFisher Scientific) and a nanoflow-UHPLC system (nanoAcquity, Waters Corp.) Peptides dissolved in 2% acetonitrile, 0.2% trifluoroacetic acid were applied to a reverse phase trap column (Symmetry C18, 5 μm, 180 μm × 20 mm, Waters Corp.) connected to an analytical column (BEH 130 C18, 1.7 μm, 75 μm × 250 mm, Waters Corp.) in vented configuration using a nano-T coupling union. Peptides were eluted in a gradient of 3–40% acetonitrile in 0.1% formic (solvent B) acid over 50 min followed by gradient of 40–60% B over 3 min at a flow rate of 250 nL min−1 at 40 °C. The mass spectrometer was operated in positive ion mode with nano-electrospray ion source with internal diameter 0.02 mm fused silica emitter (New Objective, Woburn, MA). Voltage +2kV was applied via platinum wire held in a PEEK T-shaped coupling union. Transfer capillary temperature was set to 200 °C, no sheath gas, and the focusing voltages in factory default setting were used. The Orbitrap, MS scan resolution of 60,000 at 400 m/z, range 300 to 2,000 m/z was used, and automatic gain control (AGC) target was set to 1,000,000 counts and maximum inject time to 1,000 ms. In the linear ion trap (LTQ), MS/MS spectra were triggered with data-dependent acquisition method for the five most intense ions. The threshold for collision induced dissociation (CID) was above 1,000 counts, normal scan rate, AGC accumulation target was set to 30,000 counts, and maximum inject time to 150 ms. A data-dependent algorithm was used to collect as many tandem spectra as possible from all masses detected in master scan in the Orbitrap. For the latter, Orbitrap prescan functionality, isolation width 2 m/z, and collision energy set to 35% were used. The selected ions were then fragmented in the ion trap using CID. Dynamic exclusion was enabled allowing for one repeat only, with a 60 s exclusion time, and maximal size of dynamic exclusion list 500 items. Chromatography function to trigger MS/MS event close to the peak summit was used with correlation set to 0.9 and expected peak width 7 s. Charge state screening enabled allowed only 2+ charge states and higher to be selected for MS/MS fragmentation. Control samples were analyzed on an Orbitrap Fusion (ThermoScientific) with a Dionex Ultimate 3000 nanoLC system, with a TRAP 50 cm C18 BEH column form ThermoScientific. Peptides were eluted in a gradient of 3–35% acetonitrile in 0.1% formic (solvent B) acid over 90 min followed by gradient of 90% B over 5 min at a flow rate of 300 nL min−1 at 40 °C. The mass spectrometer was operated in positive ion mode with nanoelectrospray ion source with ID 0.02 mm fussed silica emitter (New Objective). Voltage +2.5 kV was applied to the metal-coated tip. Transfer capillary temperature was set to 265 °C, no sheath gas, and the focusing voltages in factory default setting were used. The Orbitrap, MS scan resolution of 120,000 at 400 m/z, range 300 to 1,500 m/z was used, and AGC target was set to 400,000 counts, and maximum inject time to 50 ms. In the dual pressure ion trap, MS/MS spectra were triggered for charge states 2+ to 7+ using Higher-energy collisional dissociation (HCD) fragmentation at 35% and a maximum speed strategy where variable numbers of MS/MS spectra would be acquired within a 2 s window. The threshold for ion selection was 5,000 counts, and selected parent ions were isolated using a mass window of 1.6 in the quadrupole. Fragment ions were detected using the a rapid ion trap scan rate, AGC accumulation target was set to 10,000 counts, and maximum inject time to 200 ms and range 100–2,000 m/z. Dynamic exclusion was enabled allowing for one repeat only, with a 45 s exclusion time, and maximal size of dynamic exclusion list 500 items.
Software Processing and Peptide Identification
Peak lists in format of Mascot generic files were prepared from raw data using Proteome Discoverer v1.2 (ThermoFisher Scientific). Peak picking settings were as follows: m/z range set to 300–5,000, minimum number of peaks in a spectrum was set to 1, Signal/Noise threshold for Orbitrap spectra set to 1.5, and automatic treatment of unrecognized charge states was used. Peak lists were searched on Mascot server v.2.4.1 (Matrix Science, London, UK) against the The Arabidopsis Information Resource (TAIR) (version 10) database with GFP, RFP. and common contaminants such as keratin added. Only tryptic peptides were permitted with up to two possible miscleavages and charge states +2, +3, +4 were allowed in the search. The following modifications were included in the search: oxidized methionine (variable), carbamidomethylated cysteine (static). Data were searched with a monoisotopic precursor and fragment ions mass tolerance 10 ppm and 0.8 Da, respectively. Mascot results were combined in Scaffold v. 4 (Proteome Software, Portland, OR) and exported in Excel (Microsoft Office). Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm with a minimum of two unique peptides at 99% Peptide Prophet probability (48).
Proteome Analysis
Protein identifications and total spectrum counts were exported from Scaffold and the fold enrichment over control samples (containing fluorescent protein baits) was calculated using SaintExpress (49). Default settings were used, where all three replicates counted equally, and we did not use any known interaction information to weight interaction probabilities. Proteins were considered to be statically enriched if the SaintExpress probability score was greater or equal to 0.8 in keeping with recommendations (49, 50).
The Sungear diagram was generated in virtual plant (51). Venn diagrams were produced in R (File S1). To identify transmembrane domains and putative signal peptides in our proteomic data, we parsed a bulk download of protein data from Swiss-Prot and pTREMBL (http://web.expasy.org/docs/swiss-prot_guideline.html). We also extracted protein name information and number of transmembrane domains from TAIR10 by direct download of proteins with transmembrane domains from (http://www.arabidopsis.org/ and http://www.uniprot.org). We used transmembrane domain information from TAIR10 preferentially to that from Swiss-Prot and protein records from the manually curated Swiss-Prot preferentially to data from electronically annotated pTREMBL. Data were then amalgamated with information of acylaton (52) and comparison to published proteomic data (33, 39, 44, 45, 53, 54) in Excel.
Constructs
N-terminal fusions of PRA1.B1 (prenylated Rab acceptor B1), PRA1.B2, and PRA1.F1 to RFP or YFP were generated from PCR amplification from cDNA. The amplicons were then cloned into the entry vector pENTR/d-TOPO (Invitrogen) and resulting clones confirmed by sequencing and cloned into pUB N RFP/YFP (55).
Transient Expression
For microprojectile bombardment, pUB::YFP or pUB::RFP-PRA1.B1/PRA1.B2/PRA1.F1 were coated onto 1 μm gold particles and bombarded into 4- to 5-week-old leaves of pUB::RFP-RABF2b/ARA7, pUB::YFP-GOT1, and pUB::RABF1/ARA6-RFP using a Bio-Rad Biolistic PDS-1000/He particle delivery system. Bombardment sites were imaged 16 h after bombardment by confocal microscopy. Data were collected from at least two independent bombardment events and several independent plants.
Confocal Microscopy
Confocal laser scanning microscopy was performed using the laser point scanning microscope Leica SP5. YFP was excited using the 514 nm argon laser, and fluorescence emissions were captured between 520 and 550 nm for YFP. RFP was excited at 561 nm, and emission was taken between 580 and 620 nm. The sequential scan mode was used for simultaneously imaging of YFP/RFP. Images were processed using the LeicaLite and Adobe Photoshop CS4 software packages. Images are maximum projections of a consecutive series of multiple Z planes 1 μm apart.
RESULTS AND DISCUSSION
Affinity Enrichment of Subcellular Proteomes
To identify the proteomes of various endomembrane compartments, we simplified published affinity purification methods for the enrichment of vesicle-associated proteins (44, 45) by removing the sucrose density gradient step and extending the number of selected subcellular markers to create a large-scale, comparative proteomics approach. For this, we used sterile grown A. thaliana seedlings stably expressing GOT1, RABD2a/ARA5, RABG3f, and VAMP711 fused to YFP; RABF2b/ARA7 and RABF1/ARA6 fused to the monomeric RFP; CLC2-GFP; and as controls, seedlings expressing the fluorescent proteins only (Fig. 1A, Table I). All these markers were under the control of constitutive promoters ensuring sufficient expression levels and are detected in seedling leaves at their assigned subcellular localizations (Fig. 1B). Total seedlings were harvested after 8 days of hydroponic growth, homogenized, and the proteins extracted in buffer followed by single low speed centrifugation. Using the workflow outlined in Fig. 2A, we affinity purified RFP-RABF2b/ARA7 and YFP-RABG3f from the respective transgenic lines and assessed the enrichment for possible organelle contamination using Western blotting. Our approach greatly enriched the relevant bait proteins with no detectable contamination from the ER, mitochondria, chloroplasts, and PM as revealed by our failure to detect BIP2 (AT5G42020), COXII (ATMG00160), RbcL (ATCG00490), and AHA1 (AT2G18960) with αBIP2, COXII, RbcL, and AHA1 antibodies, respectively (Fig. 2B). No bands to either of RFP, YFP, or the organelle markers were identified in affinity purifications from wild-type seedlings, indicating that abundant cytosolic or membrane-associated proteins did not generally bind to the affinity beads (Fig. 2B). We then proceeded to identify and compare the proteomes of seven different subcellular marker lines. We affinity purified YFP-GOT1, YFP-RABD2a/ARA5, YFP-RABG3f and YFP-VAMP711, RFP-RABF2b/ARA7, RABF1/ARA6-RFP, and CLC2-GFP from the respective transgenic A. thaliana seedlings using GFP and RFP beads. We refer to these affinity purifications as “endosomal-enriched proteomes” and acknowledge that, because we used liquid nitrogen to freeze plant material, it is possible that larger compartments might not remain intact. In this manuscript, we use the term “compartment” as a descriptive term for any membrane-bound structure that is not an organelle, regardless of its size.
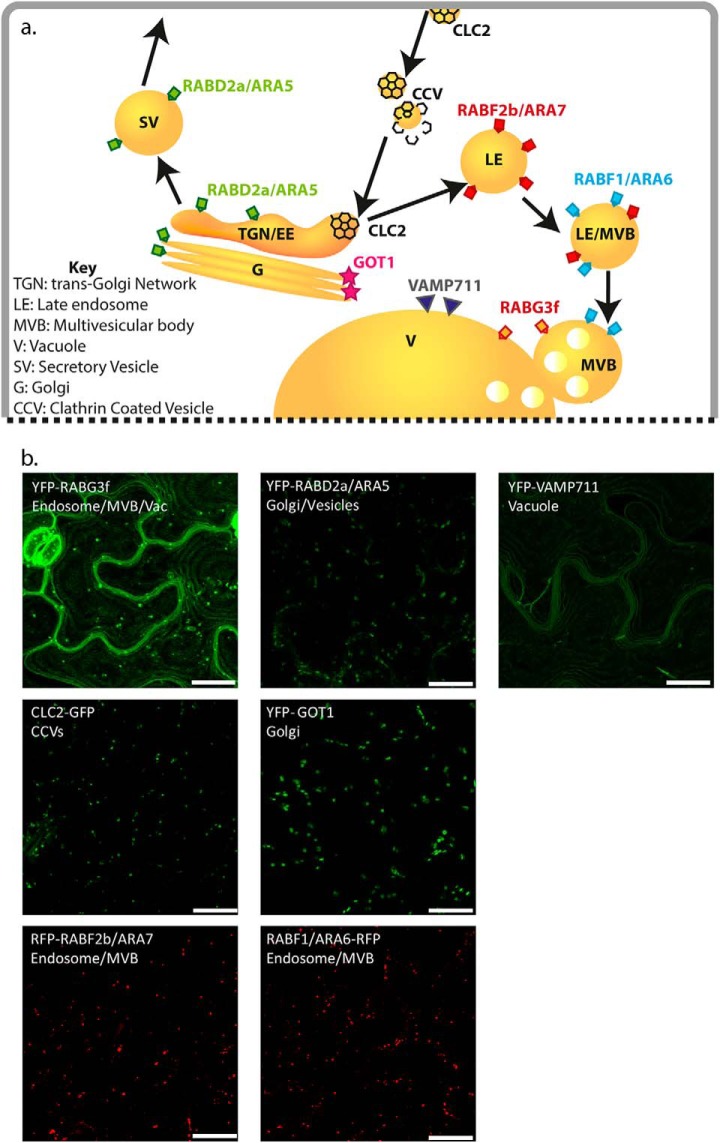
Fig. 1.
Endomembrane targets. (A) Schematic overview of the endomembrane marker proteins used in this study and their localizations. RABD2a/ARA5—post-Golgi/Golgi/TGN/SV, RABF1/ARA6—LE/MVBs, RABF2b/ARA7—LE/MVBs, CLC2—clathrin-coated vesicles, GOT1—Golgi, RABG3f—LE/MVB/vacuole, VAMP711—vacuole. Modified from (16). (B) Localization of fluorescent-tagged marker proteins. Standard confocal micrographs of leaf epidermal cells of A. thaliana transgenic plants stably expressing the indicated recombinant proteins. Scale bars 10 μm. References: pUB:YFP-RabG3f, YFP-RABD2a/ARA5, YFP-VAMP711, YFP-Got1 (26), RABF1/ARA6-RFP, RFP-RABF2b/ARA7 (provided by K. Schumacher, Heidelberg, Germany), p35S:CLC-GFP (provided by S. Bednarek, Madison WI).
Table I. Affinity purification baits and their homologs in mammals and yeasts. ATG numbers and A. thaliana short names are from TAIR (http://www.arabidopsis.org/). Homologs were identified from published works (24, 26, 47, 112). Abbreviations: EE, early endosomes; LE, late endosomes; TGN, trans-Golgi network; SV, secretory vesicles; Ton, tonoplast; MVB, multivesicular body.
| ATG number | A. thaliana | Mammals | Saccharomyces | Saccharomyces | Expected locations | Construct |
|---|---|---|---|---|---|---|
| AT3G18820 | RABG3f | RAB7 | YPT7 | YPT7 | LE/MVB/Ton | YFP-RabG3f |
| AT4G19640 | ARA7/RABF2b | RAB5 | YPT51/52/53 | YPT5 | LE/MVB | RFP-RABF2b/ARA7 |
| AT3G54840 | ARA6/RABF1 | RAB22 | LE/MVB | RABF1/ARA6-RFP | ||
| AT1G02130 | ARA5/RABD2a | RAB1 | YPT1 | YPT1 | Golgi/TGN/EE/SV | YFP-RABD2a/ARA5 |
| AT4G32150 | VAMP711 | VAMP7 | YKT6 | sec22 | Ton | YFP-VAMP711 |
| AT2G40060 | CLC2 | CLTB | CLC1 | CLC | CCV | CLC2-GFP |
| AT3G03180 | GOT1 | GOT1A | GOT1p | GOT1 | Golgi | YFP-GOT1 |
Fig. 2.
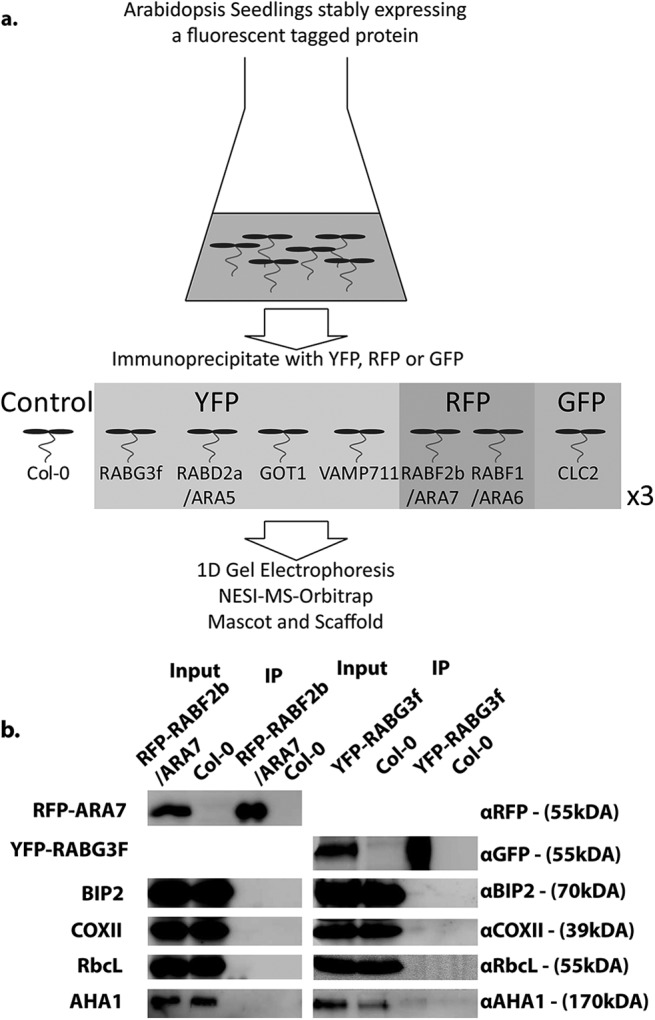
Proteomic workflow and validation. (A) Workflow for the affinity enrichment and analysis of endomembrane proteins from A. thaliana. (B) Immunoblotting of RFP-RABF2b/ARA7 and YFP-RABG3f proteome enrichments to determine organelle contamination. Total protein extracts, from A. thaliana Col-0 or stably expressing RFP-RABF2b/ARA7 or YFP-RABG3f were subjected to immunoaffinity enrichment of RFP or YFP followed by immunoblotting with αRFP, GFP, BIP2, COXII, RbcL, AHA1 as indicated.
Identification and Comparison of Proteins from the Seven Affinity Enrichments
The proteins from each affinity enrichment were separated by one-dimensional gel electrophoresis and identified by mass spectrometry. The full dataset is available from the proteomics database PRIDE (accession numbers PXD001877 to PXD1885 inclusive. Table S1). All affinity enrichments contained peptides from the corresponding bait protein except for YFP-GOT1 that was only identified by peptides from YFP. This is probably because GOT1 is less amenable to tryptic digests, with only two potential peptides between 4 and 24 amino acids in length (Fig. S1). Proteins were accepted as significantly enriched by a specific bait if their SAINTexpress score was 0.8 or higher (49, 50). The SAINT scores and spectrum counts for all proteins assigned to each affinity enrichment, replicate, and associated controls are summarized in Table S2. After SAINT analysis, 433 proteins were accepted being significantly enriched with at least one bait and each affinity enrichment had a number of unique and shared proteins (Table II). Most proteins were identified from the RFP-RABF2b/ARA7 enrichment (279 proteins in total) of which 43% were uniquely assigned to this bait. The CLC2-GFP and YFP-VAMP711 baits had the smallest number of proteins assigned to their enrichments (49 and 51 proteins, respectively) with smaller proportions of uniquely assigned proteins (30 and 23%, respectively). RABF1/ARA6-RFP and YFP-GOT1 both had about 60 proteins assigned to their enrichments but differed in the proportion of unique proteins assigned to each; only nine (14%) were unique to RABF1/ARA6-RFP while 30 (48%) were unique to YFP-GOT1.
Table II. Numbers of proteins identified in affinity purifications. Proteins that were identified in and unique to (according to criteria in materials and methods) each affinity purification. Percentage of proteins identified in selected previous proteomic studies was calculated from whether a protein was identified in Nikolovski et al. (39), 2012, Parsons et al. (37), 2012, Sadowski et al. (53), 2008, Dunkley et al. (33), 2006, Drakakaki et al. (44), 2012. Percentage of proteins with membrane associations was calculated using annotations from Uniprot (http://www.uniprot.org) and TAIR (http://www.arabidopsis.org/) as described in the materials and methods.
| Total number of proteins | Number of unique proteins | % previously identified | % membrane associated | |
|---|---|---|---|---|
| YFP-RABD2a/ARA5 | 120 | 22 | 18% | 34% |
| RABF1/ARA6-RFP | 63 | 9 | 14% | 33% |
| RFP-RABF2b/ARA7 | 279 | 121 | 43% | 33% |
| CLC2-GFP | 49 | 15 | 31% | 24% |
| YFP-GOT1 | 62 | 30 | 48% | 77% |
| YFP-RABG3f | 136 | 42 | 31% | 22% |
| YFP-VAMP711 | 51 | 12 | 24% | 41% |
As several of our bait proteins have related biological functions, it is not surprising that we assigned many proteins to more than one affinity enrichment. In addition to excluding proteins that were enriched with the fluorescent protein baits only, our comparison of seven different baits provides a further measure of likely common contaminant proteins. Only five proteins were significantly enriched in all affinity purifications, and four proteins were enriched with six of the seven baits. These include chaperonin 60A and 60B (CPN60; AT1G55490, AT2G28000), the ABC-type transporter PEN3/ABCG36 (AT1G59870), and two ARM repeat family proteins (AT5G53480, AT5G19820). Several chaperones and ribosomal components are considered to be contaminants in affinity purifications, and we identified five heat shock proteins, six additional CPN60/TCP-1 proteins, and four ribosomal proteins with some, but not all, of our baits. The RFP-RABF2b/ARA7 and YFP-RABG3f enrichments contained several of these proteins, and it is possible that the abundance of proteins associated with protein expression and folding are correlated with the expression level of the bait protein. BIP2 (AT5G42020) and COXII (AT1G02410) RbcL (ATCG00490), which we tested for in our Western blot assessment of contamination from organelles, were not significantly enriched with any bait (Fig. 2B). However, two plasma membrane ATPases were identified with specific baits and might represent contamination with plasma membrane proteins or normal secretion, turnover, and recycling of these abundant proteins. AHA1 (AT4G30190) was identified with RFP-RABF2b/ARA7 and AHA2 (AT2G18960) with three baits.
To determine if proteins with known functions in endomembrane trafficking were identified in our affinity enrichments, we manually curated a set of endomembrane regulators from the literature (Table S3) and present the fold enrichment over controls with each of our seven baits (Table III). This set of established endomembrane proteins was differentially enriched with each of the seven baits and largely corresponds to the known biological functions of our bait proteins.
Table III. Endomembrane regulators identified in affinity purifications. Proteins significantly enriched with each affinity purification are marked with a grey background and the fold enrichment over that of control baits is shown. Significant and fold enrichment was calculated using SAINT Express. NS: proteins that were identified in an enrichment, but not significantly. A: No peptides to this protein were identified. Annotations for inclusion in a complex were manually curated from literature (26, 47, 56, 65, 78, 79, 82, 93, 101, 113–118).
| ATG number | Group | Short name | YFP-RABD2a/ARA5 | RABF1/ARA6-RFP | RFP-RABF2b/ARA7 | CLC2-GFP | YFP-GOT1 | YFP-RABG3f | YFP-VAMP711 |
|---|---|---|---|---|---|---|---|---|---|
| AT5G22780 | CCV | Adaptin α2 | NS | NS | 11 | NS | NS | A | A |
| AT2G25430 | ECA4 | NS | NS | NS | 73 | A | 43 | NS | |
| AT4G32285 | CAP1 | NS | NS | 60 | NS | A | A | A | |
| AT3G11130 | CHC1 | NS | NS | NS | 29 | NS | NS | NS | |
| AT3G08530 | CHC2 | NS | NS | NS | 60 | NS | A | A | |
| AT2G20760 | CLC1 | A | NS | NS | 18 | A | A | A | |
| AT2G40060 | CLC2 | NS | NS | A | 110 | NS | A | A | |
| AT4G33650 | DRP3A | 8 | NS | 10 | 7 | NS | NS | NS | |
| AT1G10290 | DRP2A | NS | NS | NS | NS | NS | NS | 3 | |
| AT4G34660 | SH3PH | A | A | NS | 40 | A | A | A | |
| AT1G52360 | Coatomer | SEC27p | 5 | 4 | 10 | NS | NS | 7 | NS |
| AT4G31480 | SEC26p | NS | A | A | NS | A | 11 | NS | |
| AT4G31490 | SEC26p | NS | NS | 12 | NS | NS | A | A | |
| AT1G62020 | RET1p | 16 | 16 | 21 | NS | NS | 12 | 7 | |
| AT2G21390 | RET1p | 6 | 7 | 9 | NS | NS | 6 | NS | |
| AT5G05010 | RET2p | 10 | NS | 10 | NS | NS | NS | NS | |
| AT3G63460 | SEC31B | 8 | 6 | 10 | NS | NS | NS | NS | |
| AT5G16300 | COG | COG1/VPS51 | 33 | NS | 120 | NS | NS | NS | NS |
| AT4G24840 | COG2 | NS | NS | 37 | NS | NS | NS | A | |
| AT1G67930 | COG5 | NS | NS | 20 | A | NS | A | A | |
| AT5G51430 | COG7 | 27 | NS | 50 | NS | A | NS | NS | |
| AT5G11980 | COG8 | 30 | NS | 47 | NS | NS | NS | A | |
| AT2G27600 | ESCRT | VPS4 | NS | NS | 7 | A | NS | 8 | A |
| AT5G03540 | EXOCYST | EXO70A1 | NS | NS | 60 | NS | A | NS | A |
| AT5G59730 | EXO70H7 | NS | NS | 30 | NS | A | A | A | |
| AT5G49830 | EXO84B/VPS51 | NS | NS | 40 | NS | NS | 23 | NS | |
| AT5G12370 | SEC10 | 9 | NS | 12 | NS | NS | NS | NS | |
| AT1G47550 | SEC3A | NS | NS | NS | NS | NS | 33 | NS | |
| AT1G76850 | SEC5A | NS | NS | 107 | NS | NS | NS | NS | |
| AT3G10380 | SEC8 | 20 | 12 | 29 | 10 | NS | 12 | NS | |
| AT4G02030 | GARP | VPS51 | NS | NS | 8 | NS | A | NS | A |
| AT1G71270 | VPS52 | 33 | 40 | 50 | 40 | NS | 37 | NS | |
| AT1G50500 | VPS53 | 37 | NS | 87 | NS | NS | 60 | NS | |
| AT4G19490 | VPS54 | 15 | NS | 46 | NS | NS | NS | NS | |
| AT3G60860 | GEF | BIG2 | 24 | NS | 60 | NS | A | NS | NS |
| AT1G01960 | BIG3 | 15 | NS | 33 | 8 | NS | 9 | NS | |
| AT3G43300 | BIG5/MIN7 | 16 | NS | 23 | NS | NS | 11 | NS | |
| AT1G13980 | GNOM | 53 | NS | 97 | NS | NS | NS | A | |
| AT2G01470 | SEC12 | NS | NS | NS | A | NS | NS | 33 | |
| AT1G16920 | GTPase | AtRABA1b | 7 | NS | 5 | NS | NS | NS | NS |
| AT4G18800 | AtRABA1d | NS | NS | 33 | NS | NS | NS | 116 | |
| AT3G46830 | AtRABA2c | 14 | NS | NS | A | NS | 38 | NS | |
| AT4G17170 | AtRABB1c | NS | NS | NS | NS | NS | 6 | 4 | |
| AT1G43890 | AtRABC1 | NS | 12 | NS | NS | NS | 6 | A | |
| AT3G11730 | AtRABD1 | A | NS | NS | NS | NS | 30 | NS | |
| AT1G02130 | AtRABD2a | 56 | NS | NS | NS | NS | NS | NS | |
| AT5G47200 | AtRABD2b | NS | NS | 10 | NS | NS | 11 | 7 | |
| AT3G54840 | AtRABF1 | NS | 195 | NS | NS | A | NS | A | |
| AT4G19640 | AtRABF2b | 100 | 272 | 2330 | NS | NS | 76 | NS | |
| AT3G18820 | AtRABG3f | NS | NS | NS | A | NS | 120 | 9 | |
| AT2G44610 | AtRABH1b | NS | NS | NS | NS | NS | 6 | NS | |
| AT3G56110 | RAB regulator | AtPRA1.B1 | NS | A | 117 | A | 53 | NS | A |
| AT2G40380 | AtPRA1.B2 | NS | A | NS | A | 60 | NS | A | |
| AT2G38360 | AtPRA1.B4 | NS | NS | 110 | A | NS | NS | NS | |
| AT2G44100 | GDI1 | NS | A | 57 | A | A | 657 | A | |
| AT3G59920 | GDI2 | NS | A | NS | A | A | 177 | A | |
| AT3G19770 | VPS9a | A | NS | 130 | A | A | A | A | |
| AT4G30260 | YIP4b | 62 | NS | 104 | A | A | NS | NS | |
| AT3G05280 | YIP5b | 80 | A | NS | A | NS | NS | 60 | |
| AT1G75850 | RETROMER | VPS35A | NS | A | NS | A | A | 40 | A |
| AT2G17790 | VPS35B | NS | NS | NS | A | A | 43 | A | |
| AT5G06140 | SNX1 | NS | NS | 53 | NS | A | NS | NS | |
| AT2G45200 | SNARE | GOS12 | NS | A | A | NS | 40 | A | A |
| AT4G04910 | NSF | 70 | NS | 113 | NS | 70 | NS | 93 | |
| AT3G05710 | SYP43 | NS | A | 23 | A | A | A | NS | |
| AT1G16240 | SYP51 | NS | NS | NS | A | A | NS | 53 | |
| AT4G32150 | VAMP711 | A | NS | NS | A | NS | A | 1713 | |
| AT1G04750 | VAMP721 | A | A | A | A | A | A | 167 | |
| AT2G33120 | VAMP722 | 33 | 9 | 16 | NS | 6 | 18 | A | |
| AT3G54300 | VAMP727 | NS | NS | 40 | A | A | A | A | |
| AT3G27530 | Tether | GC6 | NS | NS | 17 | NS | NS | NS | A |
| AT1G21630 | TPLATE complex | EH2 | NS | NS | NS | 20 | A | A | A |
| AT2G07360 | TASH3 | 8 | NS | 6 | 5 | A | A | NS | |
| AT5G57460 | TML | 23 | NS | NS | NS | A | A | A | |
| AT3G01780 | TPLATE | NS | 7 | 10 | NS | NS | NS | NS | |
| AT3G50590 | TWD40–1 | 6 | NS | 8 | NS | NS | A | A | |
| AT5G24710 | TWD40–2 | NS | NS | NS | 10 | A | NS | A | |
| AT5G11040 | TRAPP | TRS120 | NS | 23 | NS | A | A | NS | NS |
| AT5G16280 | TRS85 | 417 | NS | NS | A | NS | 53 | NS | |
| AT3G52850 | VSR | VSR1 | NS | NS | 52 | A | NS | A | NS |
| AT2G14740 | VSR3 | A | A | A | A | A | A | 47 | |
| AT2G14720 | VSR4 | NS | NS | 147 | A | A | NS | A | |
| AT2G20990 | SYPA | 48 | NS | 26 | 46 | NS | NS | NS | |
| AT5G14950 | Golgi marker | GMII | NS | NS | NS | NS | 94 | NS | A |
| AT2G28520 | TGN marker | VHA-A1 | NS | NS | 50 | NS | NS | A | NS |
To visualize the differences and commonalities in the seven affinity enrichments, we used the Sungear tool from Virtual plant (51) (Fig. 3). On the Sungear plot, the CLC2-GFP and YFP-VAMP711 enrichments appear to be distinct from the other enrichments, but, as discussed above, ∼70% of their assigned proteomes are shared with other baits. However, there are few binary overlaps between CLC2-GFP and YFP-VAMP711, rather the shared proteins are common to many baits. The RFP-RABF2b/ARA7 and YFP-RABG3f proteomes have high numbers of uniquely assigned proteins and share a large binary overlap, possibly representing the LE/MVB compartments. There is also substantial pairwise and collective overlaps between the YFP-RABD2a/ARA5, RFP-RABF2b/ARA7, and YFP-RABG3f. The RABF1/ARA6-RFP enrichment has only nine unique proteins and is nearly a subset of RFP-RABF2b/ARA7.
Fig. 3.
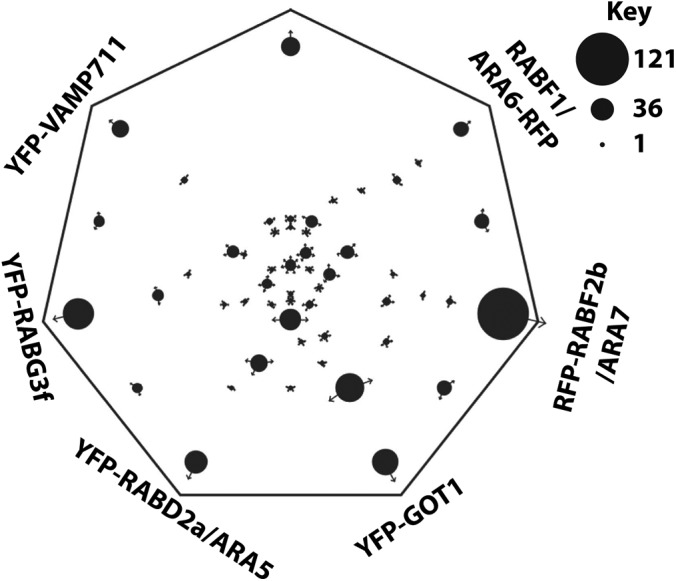
Sungear diagram of proteins assigned to the different proteomes. Sungear diagrams generated in virtual plant (http://virtualplant-prod.bio.nyu.edu/cgi-bin/sungear/index.cgi (51)). Groups of proteins are indicated by the black dots, with a size proportional to the number of proteins in the group. The arrows on each dot indicate the proteome assignment of a group of proteins. Enrichments were performed three times for each bait, proteins were accepted with SAINT scores >0.8.
Our Method Preferentially Enriches Proteins Transiently Associating with Compartments
To compare the proteins we identified with published proteomic studies, we complied a comprehensive dataset of proteins identified in the TGN/EE (44), TGN (45), ER, Golgi, mitochondria/plastid, PM, vacuole (39), Golgi (37), ER, Golgi, mitochondria/plastid/PM/vacuole (combined from (33, 53)) (Table II, Table S4). The YFP-GOT1 enrichment contained the highest proportion of previously identified proteins, likely due to the greater focus on Golgi-derived proteomes in these studies. To assist with comparisons between the sets of proteins associated with each of our baits and previously published proteomes we provide Table S4.
To identify possible cargo proteins from transient interactions of coat proteins and trafficking regulators, we extended our bioinformatic analysis to include predicted transmembrane domains and membrane associated modifications (Table S4). In the 433 proteins enriched by at least one of our affinity baits, we identified 64 proteins with one transmembrane domain and 58 with multiple transmembrane domains, 12 RAB GTPases with membrane association lipid modifications, and a diverse set of 63 potentially S-acylated proteins (52) (Table S4). The remaining 66% of our dataset (285 proteins) did not have any membrane association motifs and are putative cargo proteins. The proportions of proteins in each proteome with membrane association motifs varied across the enrichments; YFP-GOT1 had the greatest at 77%, while CLC2-GFP and YFP-RABG3f the smallest proportion of membrane-associated proteins, 24 and 22%, respectively (Table II, Fig. S2). The proportion of YFP-GOT1 membrane-associated proteins is remarkably similar to Golgi or TGN proteomic studies such as Drakakaki et al. (44) (77%) or Parsons et al. (37) (75%). That our other affinity baits, such as RFP-RABF2b/ARA7, show lower proportions of membrane-associated proteins is likely to be a consequence of our choice of bait protein for affinity purification and the biological functions of these small GTPases. By targeting RAB regulators of vesicle trafficking, we identify more proteins with transient membrane interactions, such as vesicle coats (e.g. COPI and COPII), motor proteins (e.g. myosins), and endomembrane tethers than previous proteomic studies (Table III, Table S4).
Affinity Enrichment of CLC2-GFP Identifies Clathrin Mediated Endocytosis Components
We included CLC2-GFP in this comparative proteomics study because we expected that the proteins associated with CLC2 would be substantially different to those enriched with RAB GTPase baits. Our CLC-GFP enrichment recovered established clathrin-interacting proteins and provides the first independent supportive evidence for the recently defined association of TPLATE complex components with clathrin (Table III). As expected, some of the most abundant proteins in the CLC2-GFP enrichments were; CHC1 (clathrin heavy chain 1, AT3G11130), CHC2 (AT3G08530), CLC1 (AT2G20760), and, the bait, CLC2 (AT2G40060). Several accessory proteins involved in CME were also assigned to the CLC2-GFP enrichment, including components from the TPLATE complex (47, 56) (Table III), specifically: two SH3 (SRC homology) domain-containing proteins (SH3PH, AT4G34660 and TASH3, AT2G07360), an epsin N terminal homology domain-containing protein ECA4 (AT2G25430), a WD40 protein (TWD-40–2, AT5G24710), and a calcium-binding EF hand protein EH2 (AT1G21630).
The TPLATE complex appears to persist at the plasma membrane after removal of AP2 and possibly through scission (56). We identified TPLATE components and dynamin (DRP3a, AT4G33650) in our CLC2-GFP enrichments, (but not AP2 or TPLATE itself), possibly capturing several different stages of interactions. Interestingly, we identified AP2, TPLATE, CAP1, and other members of this complex with RFP-RABF2b/ARA7 and YFP-RABD2a/ARA5 (Table III). Scission of vesicles from membranes is mediated by large GTPases such as dynamins, and we note that several dynamins were abundantly represented in many of our enrichments, including controls, and therefore did not pass our significance threshold. Only DRP3a (AT4G33650) passed significance thresholds with three baits, DRP3b (AT2G14120) with RFP-RABF2b/ARA7 and DRP2a (AT1G10290) with YFP-VAMP711.
One of the less-well-characterized proteins assigned to our CLC2-GFP enrichments was synaptotagmin A (SYTA, AT2G20990) that regulates endocytosis and plasmodesmatal viral movement (57). Lewis et al. (57) demonstrated the colocalization of SYTA with the endocytic tracer dye FM4–64 and with RABF1/ARA6-GFP. We identified SYTA with CLC2-GFP, YFP-RABD2b/ARA5, and RFP-RABF2b/ARA7 baits. A. thaliana has five homologues of synaptotagmin. and little is known about the functions of this family. STY1 is the best characterized and functions in resealing plasma membrane after stress (58, 59), and SYT2 is required for unconventional secretion, namely ER/Golgi-independent pathways and does not colocalize with FM4–64, RABF1/ARA6, or other endosomal markers but only with the Golgi apparatus (60).
The YFP-GOT1 Enrichment Is Consistent with Previously Published Golgi Proteomes
In all eukaryotes, the Golgi is an essential organelle for protein secretion, N-linked protein glycosylation and, in plants, is essential for the biogenesis and secretion of cell wall polysaccharides. YFP-GOT1 is a marker for the Golgi and is homolog of yeast GOT1p that localized to cis-Golgi (26, 61). Little is known about the precise location and function of GOT1 within the Golgi of A. thaliana. However, as several Golgi proteomic studies have been performed, we included YFP-GOT1 as an additional comparison for the RAB GTPase baits and expected it to be an outlier. We compared our YFP-GOT1 enrichment with published proteomic analyses of Golgi compartments (Fig. 4A, Table S4) (33, 37, 39, 53). Two thirds (41/62) of proteins identified with our YFP-GOT1 bait were previously identified in Golgi proteomic analyses (Fig. 4A).
Fig. 4.
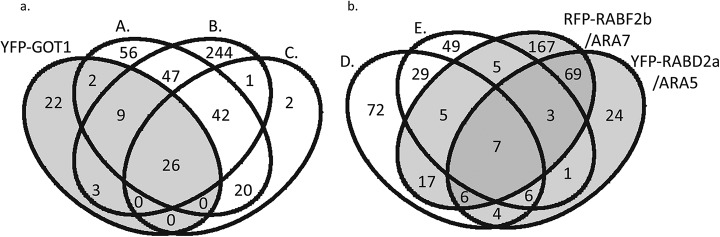
Venn diagrams comparing the proteins assigned to YFP-GOT1 (Golgi) YFP-RABD2a/ARA5 (Golgi/TGN/EE), RFP-RABF2b/ARA7 (LE/MVB) with published endomembrane proteomes. The number in each area of the Venn diagram indicate number of proteins assigned to the proteome or proteomes indicated. Venn diagram comparisons of YFP-GOT1 (Golgi), YFP-RABD2a/ARA5 (Golgi/TGN/EE) and RFP-RABF2b/ARA7 (LE/MVB) proteomes. Proteomes are (A) Nikolovski et al. (39), (B) Parsons et al. (37), (C) Sadowski et al./Dunkley et al. (33, 53). (D) Drakakaki et al. (44). (E) Groen et al. (45). (A) Comparison of our YFP-GOT1 proteome (Golgi). (B) Comparison with our RFP-RABF2b/ARA7 (LE/MVB) and YFP-RABD2a/ARA5 (Golgi/TGN/EE enrichments.
In common with previously published Golgi proteomes, Golgi-localized enzymes were well represented in our YFP-GOT1 enrichment including: galacturonosyltransferases (GAUT, AT2G38650, AT3G02350, AT3G25140, AT3G61130), xylose synthase (AT2G47650) and xylose transferase (KKT5, AT1G74380), and a UDP-d-glucuronate 4-epimerase 6 (GAE1, AT3G23820). The GAUT (galacturonosyltransferase) proteins all have one TM domain are believed to be localized to discrete subcompartments within the Golgi; additionally, GAUT1 is known to interact with GAUT7 (62). We identified nine S-adenosyl-l-methionine-dependent methyltransferases (SAM; AT1G29790, AT1G78240, AT4G00750, AT4G14360, AT5G64030, AT4G18030, AT1G26850, AT1G29470, AT3G23300). Nikolovski et al. (39) suggest that the SAM enzymes are involved in methylation of glycans. Four endomembrane family protein 70 (EMP70; AT4G12650, AT5G35160, AT5G25100, AT1G14670) were also identified in our YFP-GOT1 dataset. EMP70 proteins contain nine transmembrane domains, and their function is not known. Our YFP-GOT1 enrichment also contained the cis-Golgi-localized glycosyltransferase, alpha-mannosidase II (GMII, AT5G14950) that was not identified in previous Golgi proteomes. This protein is frequently used as a marker of the Golgi by Western blotting (e.g. (63)).
Although most of the proteins assigned to our YFP-GOT1 enrichment have been identified in published Golgi studies, our YFP-GOT1 enrichment contained fewer proteins (62 proteins) than were obtained by Parson et al. (37) (371 proteins), Nikoloviski et al. (39) (197 proteins), or Sadowski et al. (53) (91 proteins). Our YFP-GOT1 enrichment shows an overlap of only seven proteins with the 146 proteins identified in the CFP-SYP61 TGN/EE proteome (44) and just four proteins with the TGN proteome (45) (Fig. 4A, Table S4). These differences suggest that YFP-GOT1 labels and enriches for a distinct area of the Golgi, possibly on the cis-Golgi side. To further test the specificity of our method, we compared the YFP-GOT1 enrichment to the YFP-VAMP711 enrichments (Fig. S3). These two proteomes should be distinct due to their spatial and functional differentiation to the Golgi and vacuole, respectively, and the data support this statement (as the H0 of random assignment of proteins can be rejected with a X2 - 43.3, p < .001).
As expected, the YFP-GOT1 enrichment differed from the proteins identified with our RAB baits and most obviously in that few of the known endomembrane regulatory proteins were identified with this bait (Table III). However, two RAB regulatory proteins (PRA1.B2, PRA1.B4) and a Qb-SNARE and a dissociation factor, (GOS12 and NSF) were identified with YFP-GOT1 and were also seen in previous studies. Additionally, the R-SNARE (VAMP722/SAR1) was identified with YFP-GOT1 (and four other of our baits) but was not identified in previous proteomic studies of the Golgi and TGN. Both of these SNARE proteins are known to localize to the Golgi, and in yeast, GOT1 facilitates SNARE fusion (64, 65) (Table III, Table S4).
Plants differ from animals in that they possess numerous highly mobile Golgi bodies that are tightly associated with the ER (66–69). COP-coated vesicles mediate traffic between the ER and the cis-Golgi and might be expected to be enriched with our GOT1 bait. However, the COP proteins we identified assigned to the small GTPase baits (RABD2a/ARA5 and RABF2b/ARA7). The structural roles of the proteins we selected as baits might explain this apparent discrepancy; GOT1 has four TM domains while the RAB GTPases associate transiently with membranes. In yeast, GOT1 is packaged into COPII vesicles and cycles between ER and Golgi (70). It is possible that the YFP-tag interfered with the GOT1 role in COPII vesicles or that the tag was masked from the affinity matrix by coatmers. In yeast, GOT1 has a role in membrane fusion, and we identified a novel fusigenic protein with YFP-GOT1, the large, dynamin-like, GTPase RHD3 (AT3G13870). RHD3 has homology to atlastins and is proposed to mediate ER tubule formation and fusion (60, 68, 71).
ARA5 and ARA7 RAB GTPase Mediate Trafficking Associated with the TGN
RAB GTPases cycle on and off membranes and recruit proteins (known as RAB effectors) such as coat proteins and tethers, to establish a unique, but transient, membrane identity. Our objective in selecting several RAB GTPases as baits for affinity purification was to capture some of the transient trafficking events associated with the TGN and not to characterize a steady-state composition, which has been established by Groen et al. (45). Of all our baits, the YFP-RABD2a/ARA5 and RFP-RABF2b/ARA7 enrichments showed the greatest similarity to the TGN proteomes identified using CFP-SYP61 and VHA-a1-GFP affinity purifications by (44, 45) (Fig. 4B, Table S4).
YFP-RABD2a/ARA5 is localized throughout the TGN and EE in punctate structures that are sensitive to BFA and is (partially) labeled with FM4–64 and colocalizes with VHA-a1 (26, 28). Of the 120 proteins we assigned to the YFP-RABD2a/ARA5 enrichments, 23 proteins were common to CFP-SYP61 and 17 to VHA-a1-GFP enrichments (Fig. 4B). We did not identify VHA-a1 in the YFP-RABD2a/ARA5 enrichment but did identify another VHA-A ATPase (AT1G78900); this protein was also identified in CFP-SYP61 affinity purifications (44). Surprisingly, there was substantial overlap between our YFP-RABD2a/ARA5 and RFP-RABF2b/ARA7 enrichments (85 proteins). The RFP-RABF2b/ARA7 enrichment did not contain RABD2a but did contain RABD2b possibly supporting separate roles for these two RABD2 proteins as reported by (28).
Our RFP-RABF2b/ARA7 enrichments showed an overlap of 20/105 proteins that were identified in VHA-a1-GFP affinity enrichments by Groen et al. (45), including VHA-a1 (AT2G28520). VHA-a-1 was also identified in CFP-SYP61 enrichments by Drakakaki et al. (44), and our RFP-RABF2b/ARA7 enrichment had an overlap of 35/146 with the CFP-SYP61 proteome (44). That we did not identify SYP61 in any of our seven affinity enrichments could suggest distinctness of the proteomes associated with each bait. The 279 RFP-RABF2b/ARA7-associated proteins were the most numerous and diverse of any of our enrichments, suggesting that RABF2b/ARA7 might have a general role in EE/TGN and LE/MVB trafficking.
RAB GTPase Regulator Proteins Exhibit Varied Associations
We identified many proteins that regulate RAB GTPase function in YFP-RABD2a/ARA5 and RFP-RABF2b/ARA7 enrichments (Table III). All RAB GTPases have a hydrophobic membrane association modification, typically prenylation, and as membrane association is a prerequisite for RAB GTPase activation by exchange of GDP for GTP, proteins that bind to this motif prevent the RAB from associating with the membrane. Therefore, these regulatory proteins are known as GDP dissociation inhibitors (GDIs; reviewed in (72–74)). We identified GDI1 with RFP-RABF2b/ARA7 enrichments. For a RAB GTPase to associate with a membrane, the GDI must be removed by the action of a GDI dissociation factor (GDF) (70, 75–77). The homologs of yeast GDFs in A. thaliana are the Ypt-interacting protein (YIP) and prenylated Rab GTPase acceptor 1 (PRA1) family (78, 79). We identified YIP4b with both RFP-RABF2b/ARA7 and YFP-RABD2a/ARA5 baits and PRA1.B4 and PRA1.B1 with RFP-RABF2b/ARA7 and YIP5b and YFP-RABD2a/ARA5. GDI1 was not identified in either CFP-SYP61 or VHA-a1-GFP proteomes, as expected for a protein that associated with the cytoplasmic complex of a RAB. The PRA1 and YIP proteins were identified in previously characterized TGN proteomes.
RABs are poor GTPases, and their activity at membrane surfaces is controlled by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) that facilitate nucleotide exchange and hydrolysis. We identified VPS9a, the activating GEF for RABF GTPases (80) in RFP-RABF2b/ARA7 (and YFP-RABG3f) enrichments. The interaction of RABF2a/ARA7 with VPS9a is well characterized: In yeast-two hybrid assays and by coimmunoprecipitation, VPS9a physically interacts with RABF proteins (80) and the structure of the RABF2b/ARA7–VPS9a complex has been solved (81). In the YFP-RABD2a/ARA5 enriched proteins, we identified one core TRAnsport Protein Particle subunit I (TRAPPI) component, TRS85 (AT5G16280) (82). There are two distinct TRAPP complexes in animal cells: TRAPPI and TRAPPII, and in yeast, the TRAPPI complex is GEF to the RAB1/RABD homologue, Ypt1p (83, 84). Due to their coenrichment, we suggest that the TRAPPI may also be a GEF for RABD2a/ARA5. TRAPP components were identified in CFP-SYP61 and VHA-a1-GFP proteomes, but VPS9a was not.
We identified four other GEFs, of the ADP-ribosylation factor (ARF) family, in both YFP-RABD1a/ARA5 and RFP-RABF2b/ARA7 enrichments, specifically GNOM (AT1G13980), BIG5/MIN7 (AT3G43300), BIG2 (AT3G60860), and BIG3 (AT1G01960). These four GEFs can be all be inhibited by BFA and are associated with coated vesicle formation (1). The best-known plant ARF-GEF is GNOM due to its regulation of auxin transporter (PIN) recycling and partial colocalization with FM4–64 and (85). GNOM has recently been shown to be primarily localized to the Golgi and to regulate TGN structure (86). BFA treatment seems to pull the Golgi into a body that is surrounded by TGN, and this is consistent with recent work disambiguating TGN structure (87) and suggests that GNOM influence on ARA7 localization is indirect (85, 86). Less is known about the functions of BIG1 to BIG4, and BIG5/MIN7 has a role in plant immunity and PIN trafficking at the TGN/EE (88, 89). Of these four ARF-GEFs, only BIG5/MIN7 was previously identified in the VHA-a1-GFP affinity purification.
Identification of Candidate Novel GARP Components
RAB GTPases regulate compartment identity by recruiting intramembrane tethers and SNARE proteins, which promote the direct fusion of membranes once tethering factors have brought them into close contact, reviewed in (87). The combinations of RAB, tethers, and SNARE proteins dictate the interactions a compartment can make and are further regulated by SNARE attachment proteins (SNAP) and Sec1/Munc18 (SM) proteins.
We identified tether proteins and complexes with RFP-RABF2b/ARA7 and YFP-RABD2a/ARA5 baits, including: conserved oligomeric Golgi (COG), Golgi-associated retrograde protein (GARP), and the golgin GC6 (with RFP-RABF2b/ARA7 only). The COG and GARP are both multisubunit complexes of the Complex Associated with Tethering Containing Helical Rods (CATCHR) group of long tethers and are known to be effectors of RAB GTPases (90). These complexes were not identified in previous proteomic analyses of the TGN.
In yeast, the COG complex coordinates retrograde Golgi traffic and interacts with RAB, SNARE, and golgin proteins (reviewed in (88, 91, 92)). In plants, homologues of the COG complex have been predicted on the basis of sequence similarly (93), but there is little experimental evidence for the function and location of this complex in A. thaliana. We identified five of the eight putative COG subunits with RFP-RABF2b/ARA7 (COG1, COG2, COG5, COG7, COG8) and two with YFP-RABD2a/ARA5 enrichments (COG 7, COG8; Table III). The A. thaliana COG2 (AT4G24840) subunit has recently been shown to recruit the exocyst subunit EXO70A1 to punctate structures at cortical microtubules during xylem formation (94). The putative COG1 subunit (AT5G16300) has also been annotated as a possible GARP complex subunit (VPS51) (95). We have placed AT5G16300 in the COG group of Table III, but it could equally belong to the GARP complex.
The primary role of the tetrameric GARP complex is in the reception of endosome-derived vesicles at the TGN (recently reviewed in (90)). GARP is a highly conserved complex consisting of VPS51, VPS52, VPS53, VPS54 subunits (96)). In yeast, the VPS52 subunit interacts with GTP bound RAB, Ypt6 (97, 98); RABH is the homologous group in A. thaliana). We identify the VPS52 subunit with five of our affinity enrichments (RABF1/ARA6-RFP, YFP-RABG3f and CLC2-GFP baits in addition to YFP-RABD2a/ARA5 and RFP-RABF2b/ARA7), whereas the other three GARP components appear to have a more limited distribution (Table III). We suggest that VPS52 might be the GTPase-interacting component of the GARP complex in plants. VPS51 (Unhinged; At4g02030) has recently been shown to interact directly with VPS52 and to colocalize with RABF2a and SYP61 (95). A. thaliana contains three putative VPS51 homologues, and the identification of VPS51 with the other three GARP subunits in our RFP-RABF2a/ARA7 enrichments further supports Pahari et al. Notably VPS51 is not significantly enriched with YFP-RABD2a/ARA5, but another putative VPS51/COG1 homologue (AT5G16300) is.
The Transition from the TGN/EE to LE/MVB
To the best of our knowledge, there are no comparable published proteomes to our YFP-RABG3f, YFP-VAMP711, and RABF1/ARA6-RFP enrichments. Analysis of these proteomes should help elucidate the biological mechanisms of transition from RAB5-labeled LE/MVBs to RAB7-labeled LE/MVBs. Furthermore, 167 proteins of our RFP-RABF2b/ARA7 enrichments did not overlap with TGN-associated proteomes (Fig. 4B). These four baits have many proteins in common and represent the late endosomes to the LE/MVB and vacuole (Fig. 5B).
Fig. 5.
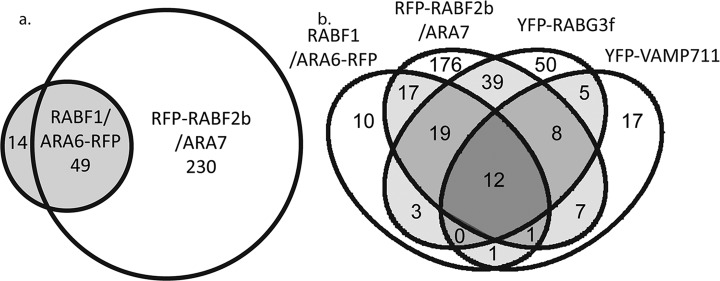
Venn diagrams comparing the proteins assigned to different endomembrane proteomes. The number in each area of the Venn diagram indicate number of proteins assigned to the proteome or proteomes indicated. (A) Comparison of YFP-GOT1 (Golgi) and RFP-RABF2b/ARA7 (LE/MVB) X2 - 78.4, reject Ho (of independent assignment of proteins) p < .001. (B) Comparison of RABF1/ARA6-RFP (LE/MVB, YFP-RABG3f (LE/MVB/vacuole), YFP-VAMP711 (vacuole).
One striking observation is that the proteomes of RFP-RABF2b/ARA7 and RABF1/ARA6-RFP are very, but not significantly (X2 - 0.6, p > .1), similar with an overlap of 49 proteins (Fig. 5A). The RABF1/ARA6-RFP proteome is much smaller than RFP-RABF2b/ARA7, with just 14 proteins not seen in RFP-RABF2b/ARA7, and only eight proteins are unique to RABF1/ARA6-RFP (compared with all our baits). These eight proteins are; TRS120, two ARM repeat family proteins (AT1G65220, AT3G05040), two chloroplast-associated proteins (LHC, AT3G08940; TOC, AT3G46740), and three enzymes (PFK, AT4G29220; KCS19, AT5G04530; GPTA4, AT1G01610). The RABF regulator VPS9a (AT3G19770) was identified with both baits, but only passed our significance test with RFP -RABF2b/ARA7 (80). An important difference between the RFP-RABF2b/ARA7 and RABF1/ARA6-RFP endosomes is the presence of numerous TGN/EE-localized proteins (RABA1b, SYP43, VHA-a1, SCAMP1) and many long tether complexes in the RFP-RABF2b/ARA7 but not RABF1/ARA6-RFP enrichments (8, 72, 99).
That the proteins identified with RABF1/ARA6-RFP are nearly a subset of RFP-RABF2b/ARA7 is in accordance with observations that RABF2b endosome populations mature into the RABF1/ARA6 endosomes (25, 29, 46, 100). Despite the similarities in their associated proteins, RABF1/ARA6- and RABF2b/ARA7-labeled LE/MVBs are distinct. RABF2b/ARA7 LE/MVBs are more susceptible to application of the drug BFA than RABF1/ARA6 LE/MVBs (25). Furthermore, RABF2b/ARA7 colocalizes to a greater extent with the SNARE VAMP727 while RABF1/ARA6 colocalizes to a greater extent with the SNAREs SYP21 and SYP22 (25). The BFA-sensitive ARF-GEFs and VAMP727 were significantly enriched with RFP-RABF2b/ARA7 but not with RABF1/ARA6-RFP. An alternative explanation for some of the differences between our RFP-RABFb/ARA7 and RABF1/ARA6-RFP enrichments might be due to the expression level of these constructs or in their relative efficiency in binding to the affinity matrix.
The YFP-RABG3F enrichment contained 136 proteins and has an overlap of 78 proteins with RFP- RABF2b/ARA7-identified proteins and the RABF1/ARA6-RFP and YFP-VAMP711 baits share 34 and 25 proteins, respectively, with this enrichment (Fig. 5B). The greatest diversity of RAB proteins was identified with RABG3f enrichments, but only two of the five ARF GEFs were identified in total (Table III and Table S4). Other RAB proteins identified with YFP-RABG3f included members of the RABA, RABC, RABD, RABF, and RABH families. The RAB dissociation inhibitors GDI1 and GDI2 were abundantly represented with the YFP-RABG3f bait (assessed by fold enrichment over control samples), and YFP-RABG3f was the only bait to enrich for GDI2. Forty-one other proteins were specific to the YFP-RABG3f proteome (compared with all seven baits).
The LE/MVB is characterized by luminal vesicles that are formed by the endosomal sorting complex required for transport (ESCRT) complex. We did not identify any core ESCRT components with YFP-RABG3f enrichments, but the ATPase that promotes the disassembly of the ESCRT complex, VPS4, was identified with RFP-RABF2b/ARA7 and YFP-RABG3f baits. Two components of the retromer complex were identified only with the YFP-RABG3f enrichments (VPS35A, VPS35B). The retromer recycles vacuolar sorting receptors back to the Golgi/TGN. Zelaznya et al. demonstrated that VPS35 interacts with RABG3f. The retromer consist of two subcomplexes; VPS35-VPS26-VPS29 and a sortin nexin (SNX) (reviewed (101)). We did not identify VPS26 or VPS29 in any enrichment, but we did identify SNX1 with RFP-RABF2b/ARA7, which is known to colocalize with them (102). In addition, two cargo recognition vacuolar sorting receptors, (VSR1 and VSR4) were identified with RFP-RABF2b/ARA7 enrichment only, and VSR3 was identified with YFP-VAMP711 baits. These VSRs are involved with sorting cargo from the TGN to the lytic vacuole and to the seed protein storage vacuole (103).
SNAREs at the Vacuole
Our YFP-VAMP711 enrichment contains 51 proteins, of which 12 are unique to VAMP711. Interestingly, we identified only two SNARE proteins in the YFP-VAMP711 enrichment, VAMP721 and SYP51. These three proteins cannot form a partial SNARE complex, as both VAMP711 and VAMP721 are both R-SNAREs (72). SYP51 is, however, a known interaction partner of VAMP711 and may function as an “interfering” SNARE (104). Of particular interest is the identification of SEC12 in the YFP-VAMP711 proteome; this GEF has been implicated in regulating the exit of vacuolar proteins from the ER. It may be that the vacuole represents another localization for SEC12 (105).
Microscopic Analysis of Golgi and TGN/EE Copurifying Proteins
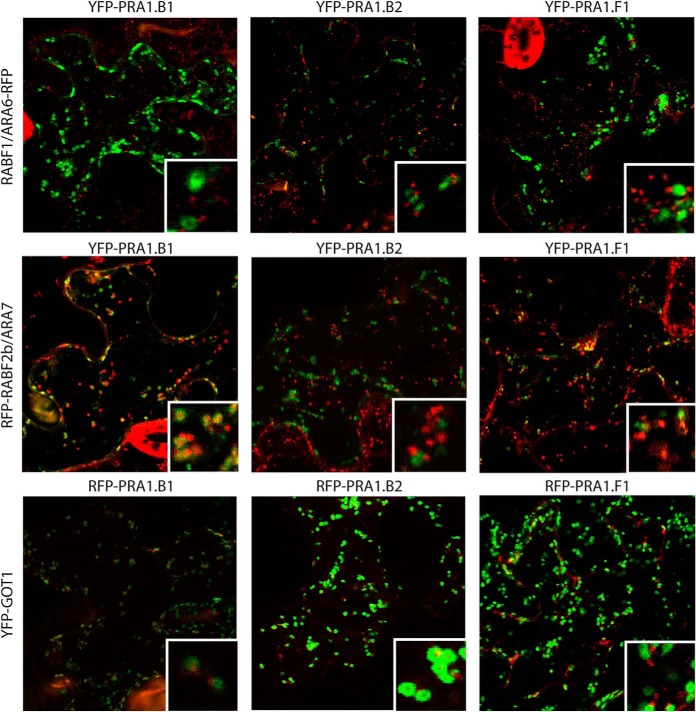
To verify that copurifying proteins are present at the compartments affinity purified with our baits, we monitored the subcellular localization of some of the identified proteins using confocal microscopy. We focused on the YFP-GOT1 and RFP-RABF2b/ARA7 proteomes and selected PRA1 proteins for further study as they are believed to regulate RAB GTPase function through removing the inhibitory GDI (78), although direct evidence is lacking in plants (106). In A. thaliana, the PRA1 family has expanded to include 19 members and their subcellular localizations are thought be varied (78, 107). We identified PRA1.B1 (AT3G56110) in both the YFP-GOT1 Golgi and RFP-RABF2b/ARA7 proteomes, whereas PRA1.B2 (AT2G40380) was found in the YFP-GOT1 proteome only (Table III). By contrast, no spectrum counts matching peptides for PRA1.F1 (AT1G17700) were identified in any of our proteomes (Table S4), although it belongs to the same protein family as PRA1.B1 and PRA1.B2 (78). These observations suggest distinct localization patterns among the PRA1 proteins of the same family.
Using live-cell imaging, we observed partial colocalization of YFP-/RFP-tagged PRA1.B1 with both RFP-RABF2b/ARA7 and YFP-GOT1, respectively (Fig. 6, Fig. S4). Additionally, YFP/RFP-PRA1.B1 was present at some compartments that were not positive for YFP-GOT1 or RFP-RABF2b/ARA7, showing PRA1.B1 has a wider subcellular localization to both Golgi and endosomes. RFP-PRA1.B2 was predominantly colocalized with YFP-GOT1 (Fig. 6, Fig. S4), in agreement with predictions from our proteomic analysis. YFP/RFP-PRA1.F1 was found to be closely associated with YFP-GOT1, RFP-RABF2b/ARA7, and RABF1/ARA6-RFP-positive compartments while not exhibiting strong colocalization (Fig. 6, Fig. S4), which is consistent that this PRA1 protein was not identified in our endomembrane proteomes. All these observations corroborate the findings from our proteome analysis and demonstrate that our data can be used to predict the localization of a protein within the cell. In contradiction to our results that RABF2b/ARA7 and PRA1.F1 do not colocalize, a previous study reports colocalization between mRFP-ARA7 and PRA1.F1-GFP. This is likely to be due to the differences in the systems used for expression (78).
Fig. 6.
Co-localizations of PRA1 family members and RABF1/ARA6 (LE/MVB), RABF2b/ARA7 (LE/MVB) and GOT1 (Golgi). Standard confocal micrographs of leaf epidermis of the indicated A. thaliana transgenic plants stably expressing pUB::YFP-GOT1, RFP-RABF2b/ARA7, or RABF1/ARA6-RFP, transiently transformed using particle bombardment, expressing fluorescent tagged PRA1 family members. Insets show an enlarged section of each image.
CONCLUSIONS
Recent reviews and discussions (13, 54, 108) have highlighted the need to identify additional regulatory proteins and cargo of endosomal and secretory pathways. To meet this need, Table III and Table S4 provide a potential resource for the community to explore the proteomes we present here in the context of published datasets. Our results demonstrate the utility of affinity purification for the exploration of endomembrane proteomes, and our method is comparable in efficacy to other endomembrane enrichment protocols. Furthermore, our proteomic data yield unprecedented biologically significant resolution between separate endocytic compartments (for example, the common and specific proteins in Fig. 5B) and can be used to predict the localization of proteins as evidenced by the PRA1 family localizations (Fig. 6).
It is important to note that endosomes and membrane fractions associated with a specific marker protein exist as heterogeneous populations within the cell and that affinity purification will pool these populations. For example, we identify GDI1 and GDI2 that only associate with the nonmembrane fraction of prenylated RABs, and we identify PRA1 family proteins with YFP-RABG3f and RFP-RABF2b/ARA7. PRA1 proteins are GDFs involved in the positive regulation of RAB GTPases by removing the inhibitory GDIs. We demonstrate differential localization of three related PRA1 GDFs and find there were no recognizable GDFs in the RABF1/ARA6-RFP proteome and that neither PRA1.B1, B2or F1 colocalize with ARA6 (Fig. 6) when analyzed with confocal microscopy. As ARA6 has a different lipid association modification (n-myristolation) (29) to all other RABs (prenylation) (72) and the known GDF machinery is believed to regulate RABs through interaction with the prenylation modification (78), we hypothesize that ARA6 has a set of as yet unidentified noncanonical GDF machinery. Furthermore, we identify known cargoes of endosomes such as PIN3 (109), PEN3 (110), and CESA (111) (Table S4). We emphasize that our data are a starting point for further work and not an end point in itself; additional validation from the community is required to properly establish the proteomes we have pioneered.
Supplementary Material
Acknowledgments
We would like to thank members of the Robatzek and Jones laboratories for fruitful discussions, particularly Martina Beck, Malick Mbengue, Heidrun Häweker, and Dan MacLean for help with scripting and particular thanks to Simon MacMullen for his help with the SAINT analysis. Thank you also to Hannah Kuhn for critical reading and feedback on the manuscript. We acknowledge the help of the WPH Proteomics RTP at Warwick University. We thank N. Geldner, K. Schumacher and S. Bednarek for their contribution of plant materials: pUB:YFP-RABG3f, YFP-RABD2a/ARA5, YFP-VAMP711, YFP-GOT1 (provided by N. Geldner, Université de Lausanne, Switzerland), RABF1/ARA6-RFP, RABF2b/RFP-ARA7 (provided by K. Schumacher, Heidelberg, Germany), p35S:CLC2-GFP (provided by S. Bednarek, Madison, WI).
Footnotes
Author contributions: W.H., S.R., and A.M.J. designed the research; W.H., J.S., and D.F.T. performed the research; W.H., J.S., and A.M.J. analyzed the data; and W.H., S.R., and A.M.J. wrote the paper.
* This research was supported by the Gatsby Charitable Foundation.
 This article contains supplemental material Tables S1 to S4 and Figs. S1 to S4.
This article contains supplemental material Tables S1 to S4 and Figs. S1 to S4.
1 The abbreviations used are:
- ABC
- ammonium bicarbonate
- AGC
- automatic gain control
- ARF-GEFs
- ADP ribosylation factor–GTP exchange factor
- CID
- collision-induced dissociation
- CME
- clathrin-mediated endocytosis
- COG
- conserved oligomeric Golgi
- COP
- coat protein
- EE
- early endosome
- ENTH
- epsin N terminal homology
- ER
- endoplasmic reticulum
- GARP
- Golgi-associated retrograde protein
- GDF
- GDI dissociation factor
- GDI
- GDP dissociation inhibitor
- LE
- late endosomes
- LTQ
- linear ion trap
- MS
- mass spectrometry
- MVB
- multivesicular body
- PM
- plasma membrane
- PVC
- prevacuolar compartments
- TBST
- Tris buffered saline 0.1% (w/v) Tween 20
- TGN
- trans-Golgi network
- Y2H
- yeast two hybrid.
REFERENCES
- 1. Park M., Jurgens G., (2012) Membrane traffic and fusion at post-Golgi compartments. Front Plant Sci. 2, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Drakakaki G., Dandekar A., (2013) Protein secretion: How many secretory routes does a plant cell have? Plant Sci. 203–204, 74–78 [DOI] [PubMed] [Google Scholar]
- 3. Denecke J., Goldman M. H., Demolder J., Seurinck J., Botterman J., (1991) The tobacco luminal binding protein is encoded by a multigene family. Plant Cell 3, 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Batoko H., Zheng H.-Q., Hawes C., Moore I., (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. The Plant Cell 12, 2201–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reyes F. C., Buono R., Otegui M. S., (2011) Plant endosomal trafficking pathways. Curr. Opin. Plant Biol. 14, 666–673 [DOI] [PubMed] [Google Scholar]
- 6. Fujimoto M., Ueda T., (2012) Conserved and plant-unique mechanisms regulating plant post-Golgi traffic. Frontiers Plant Sci. 33 (197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Löfke C., Zwiewka M., Heilmann I., Van Montagu M. C. E., Teichmann T., Friml J., (2013) Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc. Natl. Acad. Sci. U.S.A. 110, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chow C.-M., Neto H., Foucart C., Moore I., (2008) Rab-A2 and Rab-A3 GTPases Define a trans-Golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. The Plant Cell 20, 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dettmer J., Hong-Hermesdorf A., Stierhof Y.-D., Schumacher K., (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18, 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Damme D., Gadeyne A., Vanstraelen M., Inzé D., Van Montagu M. C., De Jaeger G., Russinova E., Geelen D., (2011) Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park E., Diaz-Moreno S. M., Davis D. J., Wilkop T. E., Bulone V., Drakakaki G., (2014) Endosidin 7 specifically arrests late cytokinesis and inhibits callose biosynthesis revealing distinct trafficking events during cell plate maturation. Plant Physiol 165(3), 1019–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kwon C., Bednarek P., Schulze-Lefert P., (2008) Secretory pathways in plant immune responses. Plant Physiol. 147, 1575–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Inada N., Ueda T., (2014) Membrane trafficking pathways and their roles in plant-microbe interactions. Plant Cell Physiol. 55, 672–686 [DOI] [PubMed] [Google Scholar]
- 14. Takano J., Tanaka M., Toyoda A., Miwa K., Kasai K., Fuji K., Onouchi H., Naito S., Fujiwara T., (2010) Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. U.S.A. 107, 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T., (2005) Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. U.S.A. 102, 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beck M., Zhou J., Faulkner C., MacLean D., Robatzek S., (2012) Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24, 4205–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Russinova E., Borst J. W., Kwaaitaal M., Caño-Delgado A., Yin Y. H., Chory J., de Vries S. C., (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16, 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geldner N., Hyman D. L., Wang X., Schumacher K., Chory J., (2007) Endosomal signaling of plant steroid receptor kinase BRI1. Genes Development 21, 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spallek T., Beck M., Ben Khaled S., Salomon S., Bourdais G., Schellmann S., Robatzek S., (2013) ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet 9, e1004035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Viotti C., Bubeck J., Stierhof Y. D., Krebs M., Langhans M., van den Berg W., van Dongen W., Richter S., Geldner N., Takano J., Jürgens G., de Vries S. C., Robinson D. G., Schumacher K., (2010) Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22, 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kleine-Vehn J., Huang F., Naramoto S., Zhang J., Michniewicz M., Offringa R., Friml J., (2009) PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21, 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bassham D. C., Brandizzi F., Otegui M. S., Sanderfoot A. A., (2008) The secretory system of Arabidopsis. Arabidopsis Book 6, e0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Contento A. L., Bassham D. C., (2012) Structure and function of endosomes in plant cells. J. Cell Sci. 125, 3511–3518 [DOI] [PubMed] [Google Scholar]
- 24. Rutherford S., Moore I., (2002) The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 5, 518–528 [DOI] [PubMed] [Google Scholar]
- 25. Ueda T., Uemura T., Sato M. H., Nakano A., (2004) Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40, 783–789 [DOI] [PubMed] [Google Scholar]
- 26. Geldner N., Dénervaud-Tendon V., Hyman D. L., Mayer U., Stierhof Y.-D., Chory J., (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu Y. J., Schornack S., Spallek T., Geldner N., Chory J., Schellmann S., Schumacher K., Kamoun S., Robatzek S., (2012) Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell Microbiol. 14, 682–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pinheiro H., Samalova M., Geldner N., Chory J., Martinez A., Moore I., (2009) Genetic evidence that the higher plant Rab-D1 and Rab-D2 GTPases exhibit distinct but overlapping interactions in the early secretory pathway. J. Cell Sci. 122, 3749–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ueda T., Yamaguchi M., Uchimiya H., Nakano A., (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bottanelli F., Gershlick D. C., Denecke J., (2012) Evidence for sequential action of Rab5 and Rab7 GTPases in prevacuolar organelle partitioning. Traffic 13, 338–354 [DOI] [PubMed] [Google Scholar]
- 31. Carter C., Pan S., Zouhar J., Avila E. L., Girke T., Raikhel N. V., (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. The Plant Cell 16, 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kleffmann T., Russenberger D., von Zychlinski A., Christopher W., Sjölander K., Gruissem W., Baginsky S., (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Current Biology 14, 354–362 [DOI] [PubMed] [Google Scholar]
- 33. Dunkley T. P., Hester S., Shadforth I. P., Runions J., Weimar T., Hanton S. L., Griffin J. L., Bessant C., Brandizzi F., Hawes C., Watson R. B., Dupree P., Lilley K. S., (2006) Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eubel H., Meyer E. H., Taylor N. L., Bussell J. D., O'Toole N., Heazlewood J. L., Castleden I., Small I. D., Smith S. M., Millar A. H., (2008) Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 148, 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jaquinod M., Villiers F., Kieffer-Jaquinod S., Hugouvieux V., Bruley C., Garin J., Bourguignon J., (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteomics 6, 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schmidt U. G., Endler A., Schelbert S., Brunner A., Schnell M., Neuhaus H. E., Marty-Mazars D., Marty F., Baginsky S., Martinoia E., (2007) Novel tonoplast transporters identified using a proteomic approach with vacuoles isolated from cauliflower buds. Plant Physiol. 145, 216–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parsons H. T., Christiansen K., Knierim B., Carroll A., Ito J., Batth T. S., Smith-Moritz A. M., Morrison S., McInerney P., Hadi M. Z., Auer M., Mukhopadhyay A., Petzold C. J., Scheller H. V., Loqué D., Heazlewood J. L., (2012) Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 159, 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elmore J. M., Liu J., Smith B., Phinney B., Coaker G., (2012) Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol. Cell. Proteomics 11, M111.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nikolovski N., Rubtsov D., Segura M. P., Miles G. P., Stevens T. J., Dunkley T. P., Munro S., Lilley K. S., Dupree P., (2012) Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 160, 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eubel H., Heazlewood J., Millar A. H. (2007) Isolation and subfractionation of plant mitochondria for proteomic analysis, in Plant Proteomics, Thiellement H., et al., eds. Humana Press; p. 49–62 [DOI] [PubMed] [Google Scholar]
- 41. Ito J., Batth T. S., Petzold C. J., Redding-Johanson A. M., Mukhopadhyay A., Verboom R., Meyer E. H., Millar A. H., Heazlewood J. L., (2011) Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J. Proteome Res. 10, 1571–1582 [DOI] [PubMed] [Google Scholar]
- 42. Morciano M., Burré J., Corvey C., Karas M., Zimmermann H., Volknandt W., (2005) Immunoisolation of two synaptic vesicle pools from synaptosomes: A proteomics analysis. J. Neurochem. 95, 1732–1745 [DOI] [PubMed] [Google Scholar]
- 43. Steuble M., Gerrits B., Ludwig A., Mateos J. M., Diep T.-M., Tagaya M., Stephan A., Schätzle P., Kunz B., Streit P., Sonderegger P., (2010) Molecular characterization of a trafficking organelle: Dissecting the axonal paths of calsyntenin-1 transport vesicles. Proteomics 10, 3775–3788 [DOI] [PubMed] [Google Scholar]
- 44. Drakakaki G., van de Ven W., Pan S., Miao Y., Wang J., Keinath N. F., Weatherly B., Jiang L., Schumacher K., Hicks G., Raikhel N., (2012) Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 22, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Groen A. J., Sancho-Andrés G., Breckels L. M., Gatto L., Aniento F., Lilley K. S., (2014) Identification of trans-Golgi network proteins in Arabidopsis thaliana root tissue. J Proteome Res. 13, 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee G.-J., Sohn E. J., Lee M. H., Hwang I., (2004) The Arabidopsis Rab5 homologs Rha1 and Ara7 localize to the prevacuolar compartment. Plant Cell Physiol. 45, 1211–1220 [DOI] [PubMed] [Google Scholar]
- 47. Chen X., Irani N. G., Friml J., (2011) Clathrin-mediated endocytosis: The gateway into plant cells. Curr. Opin. Plant Biol. 14, 674–682 [DOI] [PubMed] [Google Scholar]
- 48. Searle B. C., (2010) Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 49. Teo G., Liu G., Zhang J., Nesvizhskii A. I., Gingras A. C., Choi H., (2014) SAINTexpress: Improvements and additional features in significance analysis of interactome software. J. Proteomics 100 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., Nesvizhskii A. I., (2011) SAINT: Probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Poultney C. S., Gutiérrez R. A., Katari M. S., Gifford M. L., Paley W. B., Coruzzi G. M., Shasha D. E., (2007) Sungear: interactive visualization and functional analysis of genomic datasets. Bioinformatics 23, 259–261 [DOI] [PubMed] [Google Scholar]
- 52. Hemsley P. A., Weimar T., Lilley K. S., Dupree P., Grierson C. S., (2013) A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytologist 197, 805–814 [DOI] [PubMed] [Google Scholar]
- 53. Sadowski P. G., Groen A. J., Dupree P., Lilley K. S., (2008) Sub-cellular localization of membrane proteins. Proteomics 8, 3991–4011 [DOI] [PubMed] [Google Scholar]
- 54. Parsons H. T., Drakakaki G., Heazlewood J. L., (2012) Proteomic dissection of the Arabidopsis Golgi and trans-Golgi network. Front Plant Sci. 3, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M. R., (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64, 355–365 [DOI] [PubMed] [Google Scholar]
- 56. Gadeyne A., Sánchez-Rodríguez C., Vanneste S., Di Rubbo S., Zauber H., Vanneste K., Van Leene J., De Winne N., Eeckhout D., Persiau G., Van De Slijke E., Cannoot B., Vercruysse L., Mayers, Jonathan R., Adamowski M., Kania U., Ehrlich M., Schweighofer A., Ketelaar T., Maere S., Bednarek, Sebastian Y., Friml J., Gevaert K., Witters E., Russinova E., Persson S., De Jaeger G., Van Damme D., (2014) The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156, 691–704 [DOI] [PubMed] [Google Scholar]
- 57. Lewis J. D., Lazarowitz S. G., (2010) Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. U.S.A. 107, 2491–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamazaki T., Kawamura Y., Minami A., Uemura M., (2008) Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell 20, 3389–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schapire A. L., Voigt B., Jasik J., Rosado A., Lopez-Cobollo R., Menzel D., Salinas J., Mancuso S., Valpuesta V., Baluska F., Botella M. A., (2008) Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20, 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang H., Zhang L., Gao B., Fan H., Jin J., Botella M. A., Jiang L., Lin J., (2011) Golgi apparatus-localized synaptotagmin 2 is required for unconventional secretion in Arabidopsis. PLoS One 6, e26477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Conchon S., Cao X., Barlowe C., Pelham H. R., (1999) Got1p and Sft2p: Membrane proteins involved in traffic to the Golgi complex. EMBO J. 18, 3934–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oikawa A., Lund C. H., Sakuragi Y., Scheller H. V., (2013) Golgi-localized enzyme complexes for plant cell wall biosynthesis. Trends Plant Sci. 18, 49–58 [DOI] [PubMed] [Google Scholar]
- 63. Waugh M. G., Chu K. M., Clayton E. L., Minogue S., Hsuan J. J., (2011) Detergent-free isolation and characterization of cholesterol-rich membrane domains from trans-Golgi network vesicles. J. Lipid Res. 52, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weinberger A., (2005) Control of Golgi morphology and function by Sed5 t-SNARE phosphorylation. 16(10), 4918–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Uemura T., Ueda T., Ohniwa R. L., Nakano A., Takeyasu K., Sato M. H., (2004) Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Structure Function 29, 49–65 [DOI] [PubMed] [Google Scholar]
- 66. Brandizzi F., Barlowe C., (2013) Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14, 382–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C., (1998) Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447 [DOI] [PubMed] [Google Scholar]
- 68. Stefano G., Hawes C., Brandizzi F., (2014) ER—The key to the highway. Curr. Opin. Plant Biol. 22, 30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sparkes I. A., Frigerio L., Tolley N., Hawes C., (2009) The plant endoplasmic reticulum: A cell-wide web. Biochem. J. 423, 145–155 [DOI] [PubMed] [Google Scholar]
- 70. Lorente-Rodríguez A., Heidtman M., Barlowe C., (2009) Multicopy suppressor analysis of thermosensitive YIP1 alleles implicates GOT1 in transport from the ER. J. Cell Sci. 122, 1540–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen J., Stefano G., Brandizzi F., Zheng H., (2011) Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J. Cell Sci. 124, 2241–2252 [DOI] [PubMed] [Google Scholar]
- 72. Saito C., Ueda T., (2009) Chapter 4 Functions of RAB and SNARE Proteins in Plant Life, in International Review of Cell and Molecular Biology, Kwang W. J., ed. Academic Press; p. 183–233 [DOI] [PubMed] [Google Scholar]
- 73. Ueda T., Matsuda N., Anai T., Tsukaya H., Uchimiya H., Nakano A., (1996) An Arabidopsis gene isolated by a novel method for detecting genetic interaction in yeast encodes the GDP dissociation inhibitor of Ara4 GTPase. Plant Cell 8, 2079–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zarský V., Cvrcková F., Bischoff F., Palme K., (1997) At-GDI1 from Arabidopsis thaliana encodes a rab-specific GDP dissociation inhibitor that complements the sec19 mutation of Saccharomyces cerevisiae. FEBS Lett. 403, 303–308 [DOI] [PubMed] [Google Scholar]
- 75. Sivars U., Aivazian D., Pfeffer S. R., (2003) Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 425, 856–859 [DOI] [PubMed] [Google Scholar]
- 76. Chen C. Z., Collins R. N., (2005) Insights into biological functions across species: examining the role of Rab proteins in YIP1 family function. Biochem. Soc. Trans. 33, 614–618 [DOI] [PubMed] [Google Scholar]
- 77. Kano F., Yamauchi S., Yoshida Y., Watanabe-Takahashi M., Nishikawa K., Nakamura N., Murata M., (2009) Yip1A regulates the COPI-independent retrograde transport from the Golgi complex to the ER. J. Cell Sci. 122, 2218–2227 [DOI] [PubMed] [Google Scholar]
- 78. Alvim Kamei C. L., Boruc J., Vandepoele K., Van den Daele H., Maes S., Russinova E., Inzé D., De Veylder L., (2008) The PRA1 gene family in Arabidopsis. Plant Physiol. 147, 1735–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gendre D., McFarlane H. E., Johnson E., Mouille G., Sjödin A., Oh J., Levesque-Tremblay G., Watanabe Y., Samuels L., Bhalerao R. P., (2013) Trans-Golgi network localized ECHIDNA/Ypt interacting protein complex is required for the secretion of cell wall polysaccharides in Arabidopsis. Plant Cell 25, 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goh T., Uchida W., Arakawa S., Ito E., Dainobu T., Ebine K., Takeuchi M., Sato K., Ueda T., Nakano A., (2007) VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. The Plant Cell 19, 3504–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Uejima T., Ihara K., Goh T., Ito E., Sunada M., Ueda T., Nakano A., Wakatsuki S., (2010) GDP-bound and nucleotide-free intermediates of the guanine nucleotide exchange in the Rab5.Vps9 system. J. Biol. Chem. 285, 36689–36697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Thellmann M., Rybak K., Thiele K., Wanner G., Assaad F. F., (2010) Tethering factors required for cytokinesis in Arabidopsis. Plant Physiol. 154, 720–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wang W., Sacher M., Ferro-Novick S., (2000) TRAPP stimulates guanine nucleotide exchange on Ypt1p. J. Cell Biol. 151, 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sacher M., Kim Y. G., Lavie A., Oh B. H., Segev N., (2008) The TRAPP complex: Insights into its architecture and function. Traffic 9, 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G., (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230 [DOI] [PubMed] [Google Scholar]
- 86. Naramoto S., Otegui M. S., Kutsuna N., de Rycke R., Dainobu T., Karampelias M., Fujimoto M., Feraru E., Miki D., Fukuda H., Nakano A., Friml J., (2014) Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26, 3062–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Uemura T., Ueda T., (2014) Plant vacuolar trafficking driven by RAB and SNARE proteins. Curr. Opin. Plant Biol. 22, 116–121 [DOI] [PubMed] [Google Scholar]
- 88. Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. July 14, 313, (5784), 220–223 [DOI] [PubMed] [Google Scholar]
- 89. Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J., (2009) Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19, 391–397 [DOI] [PubMed] [Google Scholar]
- 90. Bonifacino J. S., Hierro A., (2011) Transport according to GARP: Receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol. 21, 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Miller V. J., Sharma P., Kudlyk T. A., Frost L., Rofe A. P., Watson I. J., Duden R., Lowe M., Lupashin V. V., Ungar D., (2013) Molecular insights into vesicle tethering at the Golgi by the conserved oligomeric Golgi (COG) complex and the golgin TATA element modulatory factor (TMF). J. Biol. Chem. 288, 4229–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hong W., Lev S., (2014) Tethering the assembly of SNARE complexes. Trends Cell Biol. 24, 35–43 [DOI] [PubMed] [Google Scholar]
- 93. Latijnhouwers M., Hawes C., Carvalho C., (2005) Holding it all together? Candidate proteins for the plant Golgi matrix. Curr. Opin. Plant Biol. 8, 632–639 [DOI] [PubMed] [Google Scholar]
- 94. Oda Y., Iida Y., Nagashima Y., Sugiyama Y., Fukuda H., (2015) Novel coiled-coil proteins regulate exocyst association with cortical microtubules in xylem cells via the conserved oligomeric Golgi-complex 2 protein. Plant Cell Physiol. 56, 277–286 [DOI] [PubMed] [Google Scholar]
- 95. Pahari S., Cormark R. D., Blackshaw M. T., Liu C., Erickson J. L., Schultz E. A., (2014) Arabidopsis UNHINGED encodes a VPS51 homolog and reveals a role for the GARP complex in leaf shape and vein patterning. Development 141, 1894–1905 [DOI] [PubMed] [Google Scholar]
- 96. Koumandou V. L., Dacks J. B., Coulson R. M., Field M. C., (2007) Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol. Biol. 7, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Conibear E., Cleck J. N., Stevens T. H., (2003) Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell 14, 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Conibear E., Stevens T. H., (2000) Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell 11, 305–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Feraru E., Feraru M. I., Asaoka R., Paciorek T., De Rycke R., Tanaka H., Nakano A., Friml J., (2012) BEX5/RabA1b Regulates trans-Golgi network-to-plasma membrane protein trafficking in Arabidopsis. The Plant Cell 24, 3074–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ebine K., Fujimoto M., Okatani Y., Nishiyama T., Goh T., Ito E., Dainobu T., Nishitani A., Uemura T., Sato M. H., Thordal-Christensen H., Tsutsumi N., Nakano A., Ueda T., (2011) A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13, 853–859 [DOI] [PubMed] [Google Scholar]
- 101. Schellmann S., Pimpl P., (2009) Coats of endosomal protein sorting: Retromer and ESCRT. Curr. Opin. Plant Biol. 12, 670–676 [DOI] [PubMed] [Google Scholar]
- 102. Jaillais Y., Fobis-Loisy I., Miège C., Rollin C., Gaude T., (2006) AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443, 106–109 [DOI] [PubMed] [Google Scholar]
- 103. Lee Y., Jang M., Song K., Kang H., Lee M. H., Lee D. W., Zouhar J., Rojo E., Sohn E. J., Hwang I., (2013) Functional identification of sorting receptors involved in trafficking of soluble lytic vacuolar proteins in vegetative cells of Arabidopsis. Plant Physiol. 161, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Di Sansebastiano G. P., (2013) Defining new SNARE functions: the i-SNARE. Front Plant Sci. 4, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. González E., Solano R., Rubio V., Leyva A., Paz-Ares J., (2005) Phosphate transporter traffic facilitator1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17, 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bahk J. D., Bang W. Y., Heo J. B., (2009) Plant PRA plays an important role in intracellular vesicular trafficking between compartments as GDF. Plant Signal. Behavior 4, 1094–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lee M. H., Jung C., Lee J., Kim S. Y., Lee Y., Hwang I., (2011) An Arabidopsis prenylated Rab acceptor 1 isoform, AtPRA1.B6, displays differential inhibitory effects on anterograde trafficking of proteins at the endoplasmic reticulum. Plant Physiol. 157, 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Schumacher K., Krebs M., (2010) The V-ATPase: small cargo, large effects. Curr Opin Plant Biol 13, 724–730 [DOI] [PubMed] [Google Scholar]
- 109. Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K., (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 [DOI] [PubMed] [Google Scholar]
- 110. Meyer D., Pajonk S., Micali C., O'Connell R., Schulze-Lefert P., (2009) Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57, 986–999 [DOI] [PubMed] [Google Scholar]
- 111. Paredez A. R., Somerville C. R., Ehrhardt D. W., (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495 [DOI] [PubMed] [Google Scholar]
- 112. Sanderfoot A. A., Assaad F. F., Raikhel N. V., (2000) The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124, 1558–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Robinson D. G., Herranz M.-C., Bubeck J., Pepperkok R., Ritzenthaler C., (2007) Membrane dynamics in the early secretory pathway. Critical Reviews in Plant Sciences 26, 199–225 [Google Scholar]
- 114. Shahriari M., Richter K., Keshavaiah C., Sabovljevic A., Huelskamp M., Schellmann S., (2011) The Arabidopsis ESCRT protein-protein interaction network. Plant Molecular Biol. 76, 85–96 [DOI] [PubMed] [Google Scholar]
- 115. Zhang Y., Liu C.-M., Emons A.-M. C., Ketelaar T., (2010) The plant exocyst. J. Integrative Plant Biol. 52, 138–146 [DOI] [PubMed] [Google Scholar]
- 116. Vernoud V., Horton A. C., Yang Z., Nielsen E., (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Latijnhouwers M., Gillespie T., Boevink P., Kriechbaumer V., Hawes C., Carvalho C. M., (2007) Localization and domain characterization of Arabidopsis golgin candidates. J. Experiment. Botany 58, 4373–4386 [DOI] [PubMed] [Google Scholar]
- 118. Masclaux F. G., Galaud J. P., Pont-Lezica R., (2005) The riddle of the plant vacuolar sorting receptors. Protoplasma 226, 103–108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.