Abstract
Birdshot chorioretinopathy is a rare ocular inflammation whose genetic association with HLA-A*29:02 is the highest between a disease and a major histocompatibility complex (MHC) molecule. It belongs to a group of MHC-I-associated inflammatory disorders, also including ankylosing spondylitis, psoriasis, and Behçet's disease, for which endoplasmic reticulum aminopeptidases (ERAP) 1 and/or 2 have been identified as genetic risk factors. Since both enzymes are involved in the processing of MHC-I ligands, it seems reasonable that common peptide-mediated mechanisms may underlie the pathogenesis of these diseases. In this study, comparative immunopeptidomics was used to characterize >5000 A*29:02 ligands and quantify the effects of ERAP1 polymorphism and expression on the A*29:02 peptidome in human cells. The peptides predominant in an active ERAP1 context showed a higher frequency of nonamers and bulkier amino acid side chains at multiple positions, compared with the peptides predominant in a less active ERAP1 background. Thus, ERAP1 polymorphism has a large influence, shaping the A*29:02 peptidome through length-dependent and length-independent effects. These changes resulted in increased affinity and hydrophobicity of A*29:02 ligands in an active ERAP1 context. The results reveal the nature of the functional interaction between A*29:02 and ERAP1 and suggest that this enzyme may affect the susceptibility to birdshot chorioretinopathy by altering the A*29:02 peptidome. The complexity of these alterations is such that not only peptide presentation but also other potentially pathogenic features could be affected.
Several major histocompatibility complex class I (MHC-I)1 alleles are strongly associated with polygenic inflammatory diseases, including birdshot chorioretinopathy (BSCR: A*29:02), ankylosing spondylitis (AS: HLA-B*27), psoriasis (C*06:02), and Behçet's disease (HLA-B*51). In the three latter disorders, ERAP1, an aminopeptidase of the endoplasmic reticulum performing the final trimming of MHC-I ligands (1, 2), is also a risk factor and is in epistasis with the predisposing MHC-I allele (3–5). These studies suggest common pathogenetic mechanisms involving the MHC-I bound peptidome. ERAP2, a related enzyme that acts in concert with ERAP1 (6, 7), influences the susceptibility to BSCR (8), AS (although not necessarily in epistasis with HLA-B*27) (9), Crohn′s disease (10), and preeclampsia (11–13).
BSCR is a rare and severe form of bilateral posterior uveitis, showing a progressive inflammation of the choroid and retina, whose association with HLA-A*29 is the strongest for any disease and MHC. The frequency of this allele is about 7% in healthy individuals but >95% in BSCR patients (14, 15). This association specifically concerns A*29:02 and not the closely related allotype A*29:01 (8).
Genetic studies on BSCR also showed a highly significant association within the LNPEP gene (rs7705093) in the 5q15 region, which includes the ERAP1 and ERAP2 genes. One single nucleotide polymorphism (SNP) in this region (rs10044354) correlated with ERAP2 expression. This was confirmed at the protein level, leading to the conclusion that ERAP2 expression predisposes to BSCR. Yet, an involvement of functional ERAP1 polymorphisms, not determining protein expression, was not excluded. These polymorphisms have a large influence on the HLA-B*27 peptidome (16, 17). In contrast, the effects of ERAP2 on MHC-I peptidomes are poorly understood and are probably dependent on the particular ERAP1 context since ERAP2 cooperates with ERAP1 in peptide processing. Thus, the present study was conducted to characterize A*29:02-bound peptidomes in various ERAP1 backgrounds and to determine the influence of ERAP1 polymorphism on the amounts and features of A*29:02 ligands in human cells.
EXPERIMENTAL PROCEDURES
Cell Lines
PF97387 (HLA-A*29:02, B*44:03, C*16:01, DRB1*04), MOU (HLA-A*29:02, B*44:03, C*16:01, DRB1*07:01, DRB4*01:01), and SWEIG (HLA-A*29:02, B*40:02, C*02:02, DRB1*11:01, DRB3*02:02) are human lymphoblastoid cell lines (LCL) homozygous for A*29:02. They all were included in the reference panel of the 10th International Histocompatibility Workshop (18). The three cell lines were of Caucasian origin and, to the best of our knowledge, from healthy individuals. These LCL and the human lymphoid cell line C1R (19) were cultured in RPMI 1640 medium with 10% fetal bovine serum (Biowest, Nuaillé, France), 25 mm HEPES buffer, 20 mm L-glutamine, penicillin and streptomycin.
ERAP Genotyping
The exons encompassing eight nonsynonymous SNPs in ERAP1 and 1 in ERAP2, as well as the noncoding sequences including two SNPs associated with loss of ERAP2 expression (Table I) were sequenced as previously described (16).
Table I. ERAP1 and ERAP2 variants expressed in A*29:02- positive cell lines.
| ERAP1 | |||||||
|---|---|---|---|---|---|---|---|
| SNP | Polymorphism | PF97387 |
MOU |
SWEIG |
|||
| Base | AAa | Base | AAa | Base | AAa | ||
| rs26653 | P127R | G | R | C | P | C | P |
| rs26618 | I276M | A | I | A/G | I/M | A | I |
| rs27895 | G346D | G | G | G | G | G | G |
| rs2287987 | M349V | A | M | A | M | A | M |
| rs30187 | K528R | A | K | G | R | A/G | K/R |
| rs10050860 | D575N | G | D | G | D | G | D |
| rs17482078 | R725Q | G | R | G | R | G | R |
| rs27044 | Q730E | C/G | Q/E | G | E | C/G | Q/E |
| ERAP2 | |||||||
|---|---|---|---|---|---|---|---|
| rs2549782 | K392N | G/T | K/N | T | N | G/T | K/N |
| rs2248374 | noncoding | A/G | – | G | – | A/G | – |
| rs10044354 | noncoding | C | – | C | – | C | – |
a AA: amino acid residue.
Quantitative RT-PCR
This was done as in (16) except that standard curves using two ERAP1 variants differing at codon 730 and cloned in pFastBac plasmids were used to estimate the amount of each allele present in our samples.
Western Blotting
This was performed as described elsewhere (20). ERAP1, ERAP2, and γ-tubulin were detected with the 6H9, 3F5 (both from, R&D Systems, Minneapolis, MN), or GTU88 (Sigma-Aldrich, St. Louis, MO) mAbs, respectively.
Flow Cytometry
About 3 × 105 cells were washed twice with 200 μl of PBS, centrifuged, and incubated with the 108–2C5 (IgG1; specific for folded HLA-A molecules) (21) or HCA2 mAb (IgG1; specific for unfolded HLA-A heavy chain) (22) for 30 min at 4 °C at a final concentration of 30 and 10 μg/ml of purified mAb, respectively. The cells were washed three times with 200 μl of PBS and incubated with an FITC-conjugated anti-mouse IgG secondary antibody (eBioscience, San Diego, CA), at 0.5 μg/ml for 30 min and 4 °C. Finally, the cells were washed three times with PBS and fixed with 1% of paraformaldehyde. The detection was carried out in a FACSCalibur instrument (BD Biosciences, San José, CA) using CellQuestTM Pro version 4.0.2. All data were analyzed using FlowJo version 7.5 software (Tree Star, Inc., Ashland, OR).
Isolation of HLA-A*29:02-Bound Peptidomes
This was carried out as previously described (23) with some modifications. Briefly, about 2 × 109 cells were lysed in 1% Igepal CA-630 (Sigma) in the presence of a mixture of protease inhibitors (Roche, Mannheim, Germany). A*29:02/peptide complexes were specifically purified by affinity chromatography using the HLA-A-specific 108–2C5 mAb (21) and eluted with 0.1% TFA. A*29:02 ligands were isolated by filtering with Vivaspin 2, cutoff 5,000 Da, (Sartorious Stedim Biotech, Gottingen, Germany) according to the manufacturer's instructions, concentrated and subjected to reverse phase purification using OMIX tips (Varian Inc. Palo Alto, CA) by elution with 50% acetonitrile in water and 0.1% TFA. The eluted peptides were concentrated to about 100 μl and stored at −20 °C. For each cell line, three peptide preparations were independently obtained from the same cell amounts.
Mass Spectrometry
The samples were analyzed in a Q-Exactive-Plus mass spectrometer fitted with Ultimate 3000 RSLC nano-capillary UHPLC (both from Thermo Fisher). The peptides were resolved on a capillary column (75-μm ID) pressure-packed as in (24) with Reprosil C18-Aqua (Dr. Maisch, GmbH, Germany) resolved using 7–40% acetonitrile gradients in the presence of 0.1% formic acid for 180 min and 0.15 μl/min. The dynamic exclusion was set to 20 s. The top ten most intense masses were selected for higher-energy collision dissociation fragmentations from the survey scan of m/z 300–1,800 AMU at resolution of 70,000. MS/MS spectra were acquired starting at m/z 200 with a resolution of 17,500. The target value was set to 1 × 105 and the isolation window to 1.8 m/z. The maximum injection time was set to 100 ms and normalized collision energy to 25 eV.
MS Data Analyses
The peptides were identified and quantified using the MaxQuant software (version 1.4.0.8) (25) with the Andromeda search engine (26) using the human section of the UniProt/Swiss-Prot database (release 2013 05, 20,255 entries) under the following parameters: both ion precursor mass and fragment mass tolerance 20 ppm, false discovery rate (FDR) 0.01, and peptide-spectrum matching (PSM) FDR 0.05, oxidation (M) as variable modification. The resulting peptides were filtered to eliminate the identifications derived from the reverse database, as well as from common contaminants. To avoid any differences in the peptide intensity attributable to HLA-A29 expression, the intensity assigned to each peptide sequence in a given cell line was normalized by dividing that intensity with the added intensities of all sequenced peptides in each independent experiment.
MHC Binding Affinity
This was estimated as previously described (27) using the NetMHCcons 1.0 Server at http://www.cbs.dtu.dk/services/NetMHCcons/. This server uses three different methods to more accurately estimate the theoretical MHC binding affinity of peptides with 8–15 residues. Statistical analyses were performed with the Mann–Whitney test.
Hydropathy Analysis
This was estimated as the GRAVY score, using the hydropathy index of Kyte and Doolittle (28). A higher GRAVY score indicates higher hydrophobicity. Statistical analyses were performed with the Mann–Whitney test.
RESULTS
The HLA-A*29:02 Peptidome
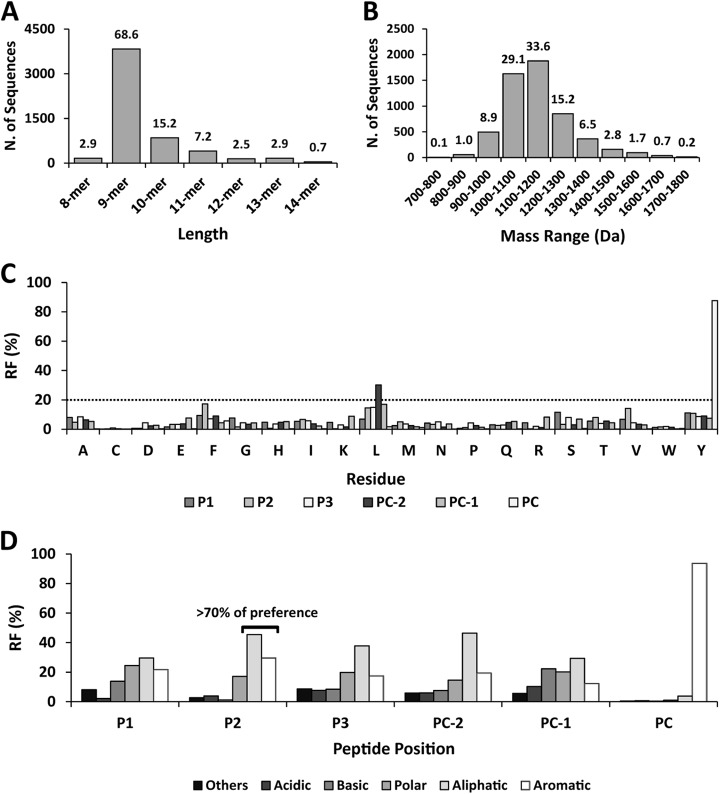
This was characterized by mass spectrometry (MS) of the natural ligands isolated from three A*29:02 homozygous LCL with distinct ERAP1 backgrounds and no expression of ERAP2 (Table I). A total of 5,584 peptides were identified (Table S1), well beyond the ≈300 previously reported human A*29:02 ligands (29, 30). Their length and molecular weight (MW) distributions were similar to other human MHC-I molecules (Figs. 1A and 1B). The main anchor motif was C-terminal Y (87.7%). A secondary motif consisted of aliphatic/aromatic P2 residues (>70%) and L at position PC-2 (30.2%) (Figs. 1C and 1D). With the exceptions mentioned, the frequency of any single residue at any peptide position was <20%, indicating a high allowance throughout (Fig. 1C). This pattern was globally maintained in all peptide lengths, although small length-dependent changes in residue frequencies (i.e.: F2, I3) were occasionally observed (Fig. S1).
Fig. 1.
General features of the A*29:02 peptidome. A total of 5,584 A*29:02 ligands from three A*29:02 homozygous cell lines were identified and analyzed as follows. (A) Length distribution. (B) MW distribution. The percentage of peptides is indicated on top of each bar. (C) Residue frequencies (RF) at the three N-terminal (P1-P3) and three C-terminal (PC-2-PC) positions of the identified peptides. The dotted line indicates a RF of 20%. (D) Individual residues were grouped according to their chemical features: basic (R, K, H), acidic (D, E), aliphatic (A, C, L, I, V, M), polar (N, Q, S, T), aromatic (F, Y, W), and others (G, P). The joint RF of each group is indicated.
ERAP1 and ERAP2 Polymorphism and Expression in A*29:02-Positive Cell Lines
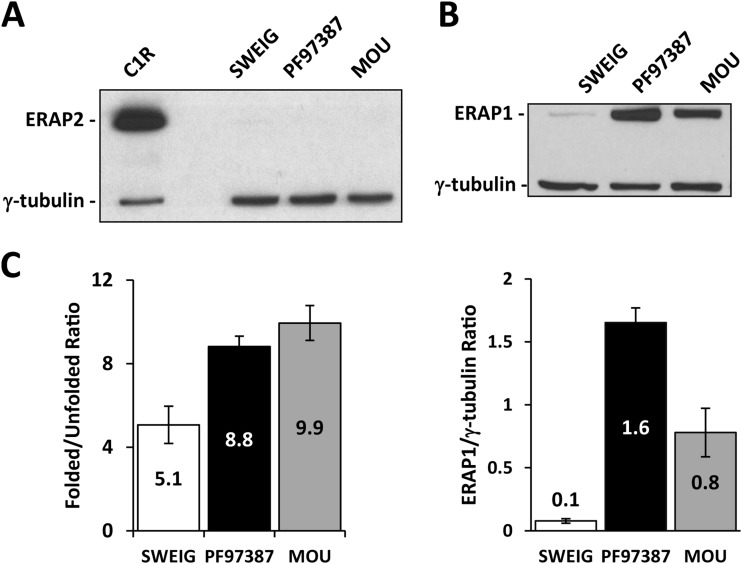
None of the three cell lines used in this study expressed ERAP2 (Fig. 2A). The ERAP1 protein was present in PF97387, MOU and SWEIG in the ratios 1:0.5:0.06 (Fig. 2B). The higher expression level of ERAP1 variants with K528 (PF97387) relative to those with R528 (MOU) has been previously reported (31, 32) and is generally observed. The amount of free HLA-A heavy chain at the cell surface of SWEIG was about 23% of the total HLA-A*29. This was approximately double the amount in the two other LCL (Fig. 2C). Increased cell surface dissociation of A*29:02 in SWEIG is consistent with the known effect of the absence of ERAAP, the mouse ortholog of ERAP1, on decreasing the affinity of MHC-I/peptide complexes (33).
Fig. 2.
ERAP1 and ERAP2 expression among cell lines. ERAP2 (A) and ERAP1 (B) expression was assessed by Western blotting. Gamma-tubulin- was used as internal control. Representative experiments are shown. ERAP1 was normalized as the ERAP1/γ-tubulin ratio to compare the expression levels among cell lines (lower panel). These data are means of three experiments. (C) Surface expression of folded and unfolded A*29:02 was assessed using the 108–2C5 and HCA2 mAbs, respectively, to establish the folded/unfolded HLA-A ratio in each cell line. The data are the mean ± S.D. of at least three experiments.
Sequencing was used to genotype eight nonsynonymous ERAP1 polymorphisms, including those relevant in other MHC-I-associated diseases (Table I). The K528R mutation presumably accounts for the lower expression of ERAP1 in MOU, relative to PF97387, as K528 correlates with higher ERAP1 expression. The Q730 transcript in PF97387 was 2.8-fold more abundant than E730, as determined by quantitative RT-PCR. This demonstrates the codominant expression of both alleles and is compatible with a higher expression of the Q730 variant at the protein level. The noncoding polymorphisms rs2248374 (34) and rs10044354 (8) are both associated with lack of ERAP2 expression. As all three cell lines were homozygous for the loss-of-expression allele of rs10044354 (C) (Table I), this explains the absence of ERAP2.
High sharing of A*29:02 Ligands Among Cell Lines
The distinct contexts of the three cell lines allowed us to address the effect of ERAP1 polymorphism and expression on the A*29:02 peptidome through quantitative pairwise comparisons. With PF97387/MOU, the combined effect of residues R127P, K528R, and QE730E was examined. With the comparisons involving SWEIG, we assessed the influence of low ERAP1 expression, relative to variants with distinct activity. Heterozygosis at residue 276 in MOU must be noted, although there is no evidence that this polymorphism affects ERAP1 function (35).
The A*29:02 peptidome of each cell line was characterized from three independent preparations using the same number of cells in identical conditions. The amount of any given peptide was estimated as the intensity of the ion peaks assigned to that peptide, relative to the total intensity of the identified peptides in that experiment, designated as normalized intensity. The amounts of individual peptides in each cell line were the mean values from all three experiments. The number of peptides identified from each cell line and the shared peptides among cell line pairs are shown in Table II. Pearson correlation of the normalized intensity of shared peptides among cell line pairs indicated a high degree of linearity in the relative peptide amounts (r: 0.8710–0.9266) in all cases (Fig. S2). The fact that these values were somewhat lower in the comparisons involving SWEIG is compatible with the possibility that the presence of ERAP1 (PF97387/MOU) increases the linearity of peptide levels.
Table II. Shared A*29:02 ligands among cell lines in pairwise comparisons.
| Comparison | Ligands | Shareda | % Shareda | PCb |
|---|---|---|---|---|
| PF97387/MOU | 4,931/5,075 | 4,583 | 92.9/90.3 | 0.9266 |
| PF97387/SWEIG | 4,931/3,999 | 3,701 | 75.1/92.5 | 0.8710 |
| MOU/SWEIG | 5,075/3,999 | 3,747 | 73.8/93.7 | 0.8759 |
a Number and percentage of shared A*29:02 ligands between the cell lines compared. The % sharing corresponding to the cell line with less identified ligands (in boldface) was considered as the most reliable value.
b PC: Pearson correlation of the normalized intensities of the shared peptides (Fig. S2): all values were statistically significant: p < .0001.
Influence of ERAP1 Polymorphism and Expression on the Length of A*29:02 Ligands
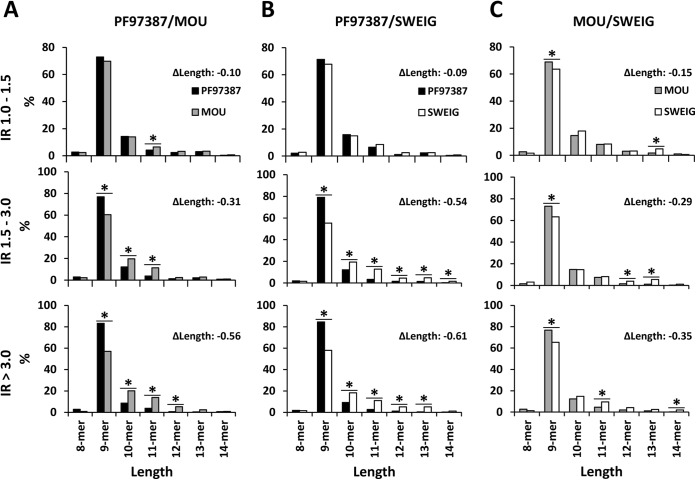
The length and MW distribution of the identified A*29:02 ligands was very similar in all three cell lines (Fig. S3), suggesting that the global peptidome comparisons lacked resolution to detect an influence of ERAP1 on particular peptide subsets. Thus, we performed three pairwise comparisons, PF97387/MOU, PF97387/SWEIG, and MOU/SWEIG, based on the relative intensity of the ion peaks corresponding to any given peptide in each of the two cell lines. Shared peptides were classified in three subsets, according to the normalized intensity ratio (IR) of the corresponding ion peaks: IR>1–1.5, IR>1.5–3, and IR>3. This subdivision was arbitrarily set based on the assumption that quantitative effects of ERAP1 on the expression levels of MHC-I ligands should be best observed among peptides showing the maximal intensity differences among cell lines. It was used in previous studies from our laboratory to reveal the effects of ERAP1 polymorphism on the HLA-B*27 peptidome (16, 17). The relationship of the IR subsets to the statistical significance of the intensity differences observed in the pairwise peptidome comparisons among cell lines is shown in Fig. 3. The length of the peptides from each subset of one cell line was compared with that of the same subset in the other cell line (Fig. 4). The IR>3 subset from PF97387 showed a higher percentage of 9-mers and lower percentage of longer peptides than the corresponding subset from MOU (Fig. 4A). In the comparisons involving SWEIG (Figs. 4B and 4C), the IR>3 peptides from this cell line tended to be longer than the corresponding peptides in the other cell lines in the following order: SWEIG(ERAP1low)>MOU(P127/R528)>PF97387(R127/K528). Since 9-mers are the peptides with optimal length for MHC-I binding, these results indicate that ERAP1 optimizes the length of A*29:02 ligands as a function of its enzymatic activity and expression level, both of which are known to be higher with K528.
Fig. 3.

Volcano plots showing the statistical significance of the of the IR values of the shared A*29:02 ligands in the indicated comparisons among cell lines. The horizontal line indicates the threshold of statistical significance (p: 0.05), which was assessed by the t test. The vertical lines indicate the IR values of 1.5 (discontinuous) and 3.0 (continuous), respectively. Only the shared peptides for which more than one intensity value was obtained in the two cell lines compared are included: 3,822, 2,816, and 2,865 peptides in PF97387/MOU, in PF97387/SWEIG, and MOU/SWEIG, respectively.
Fig. 4.
Length distribution of A*29:02 ligands as a function of their relative abundance in distinct ERAP1 contexts. (A) A total of 4,583 shared peptides were identified in the PF97387/MOU comparison. They were classified in the indicated subsets on the basis of the intensity ratio (IR) of the corresponding ion peaks. The peptides of a given length in each subset from one cell line were compared with those of the corresponding subset from the other cell line. From top to bottom, peptides showing IR>1.0 to 1.5, IR>1.5 to 3.0 and IR>3.0, respectively, were compared. (B) A total of 3,701 shared peptides identified in PF97387/SWEIG were compared as in panel A. (C) The 3,747 shared peptides identified in MOU/SWEIG were analyzed as in panels A and B. In each panel, the differential mean length between the two peptide subsets compared (ΔLength) is indicated. Statistically significant differences (p < .05) were determined by the χ2 test and labeled with asterisks (*).
Length-Independent Effects of ERAP1 on A*29:02 Ligands
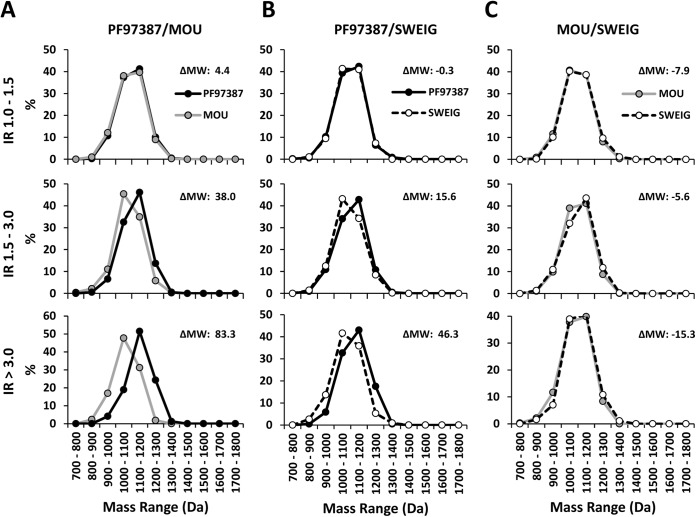
To investigate possible length-independent differences in the MW of A*29:02 ligands among cell lines, further analyses were carried out with peptides of the same length. First, the 9-mers found only in one cell line were considered (Fig. S4). The mean MW of these peptides was: PF97387 (1,125.4 Da) > SWEIG (1,096.0 Da) > MOU (1,073.9 Da). Second, the MW distribution of the shared 9-mers and 10-mers in the various IR subsets (>1–1.5, >1.5–3, and >3) from the pairwise comparisons among cell lines was examined. In PF97387/MOU a shift of the predominant 9-mers from PF97387 (IR>1.5 and IR>3 subsets) toward higher MW, relative to MOU, was observed (Fig. 5A). In the comparisons involving SWEIG, the predominant 9-mers from this cell line (IR>1.5 and IR>3 subsets) showed MW distributions shifted toward lower values relative to PF97387 (Fig. 5B) and little difference relative to MOU (Fig. 5C). Similar results were obtained with 10-mers (Fig. S5).
Fig. 5.
Length-independent differences among A*29:02 ligands expressed on distinct ERAP1 contexts. (A) A total of 3,157 shared nonamers in PF97387/MOU were classified and compared on the basis of their MW distribution as in Fig. 3. (B) The 2,559 shared nonamers in PF97387/SWEIG were comparatively analyzed as in panel A. (C) The 2,577 shared nonamers in MOU/SWEIG were compared as panels A and B. In each panel, the differential mean MW between the peptide subsets compared (ΔMW) is indicated.
These results indicate that, in addition to altering the length of A*29:02 ligands, ERAP1 polymorphism and expression influence their MW in a length-independent way so that peptides with bulkier residues are predominant in the context of the most active variant.
Position-Dependent Differences among A*29:02 Ligands from Distinct Cell Lines
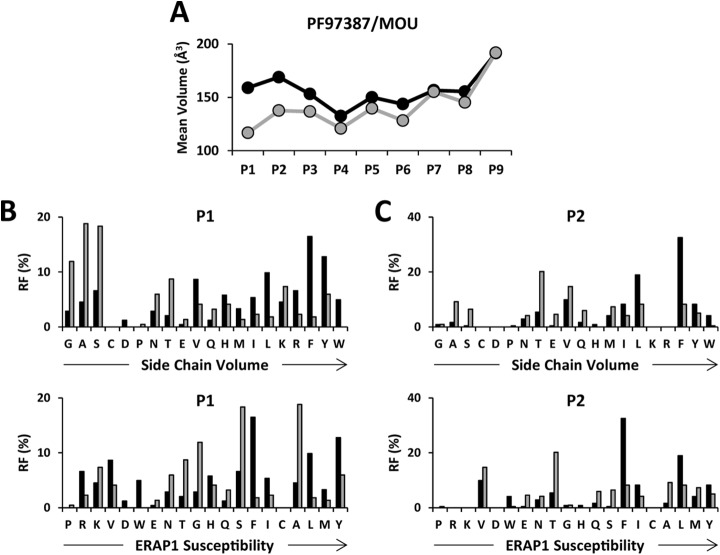
The basis of the length-independent differences in MW among A*29:02 ligands was analyzed by comparing the mean side-chain volume of the most predominant 9-mers (IR>3) in the different pairwise comparisons at each peptide position (Fig. 6). The IR>3 9-mers from PF97387 had bulkier P1-P6 and P8 residues than the corresponding peptides from MOU (Fig. 6A). The largest differences were at P1 and P2. They correlated with the volume of individual residues and not with their susceptibility to ERAP1 trimming (Figs. 6B and 6C).
Fig. 6.
Position-dependent effects of ERAP1 on A*29:02 ligands. The IR>3 nonamers from each cell line in PF97387/MOU (black and gray, respectively) were compared. (A) Mean side-chain volumes at the indicated peptide positions were calculated and compared using the residue frequency (RF) and the experimental side-chain volume of each amino acid. (B) RF values at P1 were ordered as a function of the side-chain volume (upper panel) or ERAP1 susceptibility (52) (lower panel). (C) The RF values at P2 were ordered and compared as above.
A similar analysis carried out with the IR>3 9-mers from PF97387 or MOU, relative to those from SWEIG (Fig. S6), showed that the mean side-chain volume of residues P1-P8 was higher in PF97387, whereas the differences were essentially leveled out in most positions, except P1, P2, and P7, in MOU/SWEIG. For P1 and P2, the relative residue frequencies again correlated better with side-chain volume than with susceptibility to ERAP1.
Highly Active ERAP1 Optimizes the Affinity of A*29:02 Ligands through Length-Dependent and Length-Independent Effects
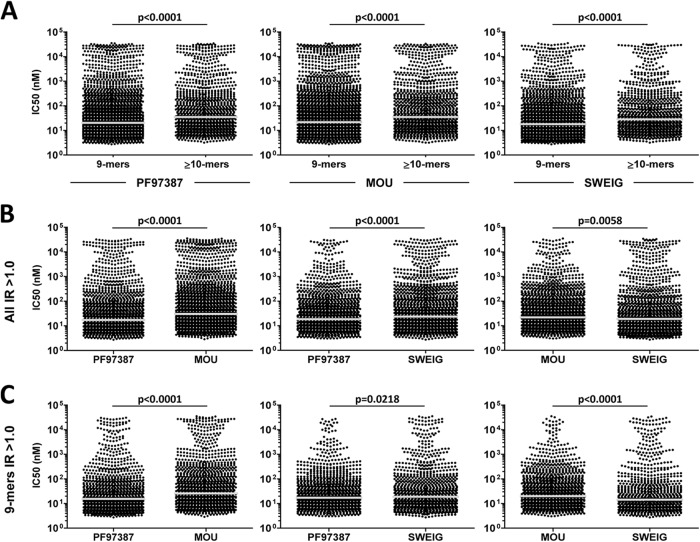
We examined whether the effect of ERAP1 on both peptide length and side chain volume altered the theoretical binding affinity of A*29:02 ligands. The 9-mers showed higher affinity than longer peptides in all 3 cell lines (Fig. 7A). Since a highly active ERAP1 variant increased the frequency of the most abundant 9-mers, relative to longer peptides, in pairwise comparisons (Fig. 4), this implies a length-dependent optimization of affinity in the A*29:02 peptidome. In a similar analysis performed on published peptide data bases, higher theoretical affinity of 9-mers relative to longer peptides was also observed among HLA-A*02 ligands (36) but not in HLA-B*07 (36) or B*27:05 (37).
Fig. 7.
Length-dependent and length-independent influence of ERAP1 on the affinity of A*29:02 ligands. (A) Predicted binding affinity of the nonamers and longer peptides (≥10-mers) from the indicated cell lines. The bars indicate the median of the corresponding values. The average IC50 values for 9-mers/≥10-mers were 888/1619 nm, 1014/1677 nm and 675/1199 nm for PF97387, MOU and SWEIG, respectively (B) Predicted binding affinities of the predominant (IR>1) A*29:02 ligands in the pairwise comparisons between the indicated cell lines. Average IC50 values were 946/1265 nm (PF97387/MOU), 485/1026 nm (PF97387/SWEIG) and 520/1135 nm (MOU/SWEIG) (C) Length-independent effects of ERAP1 on the affinity of A*29:02 ligands were assessed by comparing the IR>1 nonamers. p values, as determined with the Mann–Whitney test, are indicated. Average IC50 values were 659/1053 nm (PF97387/MOU), 325/849 nm (PF97387/SWEIG), and 372/910 nm (MOU/SWEIG). In panels B and C note that in the MOU/SWEIG comparison, the affinity distribution, as indicated by the median, is shifted toward lower values (higher IC50) in MOU. However, the average IC50 values indicate a higher average affinity of the peptides from MOU due to a higher percentage of A*29:02 ligands with very low affinity in SWEIG.
We next compared the affinity of A*29:02 ligands showing differential expression in PF97387/MOU, PF97387/SWEIG, and MOU/SWEIG. The peptides with IR>1 from PF97387 showed higher affinity than those from MOU or SWEIG (Fig. 7B). This was also true when only 9-mers were considered to assess length-independent effects (Fig. 7C). In MOU/SWEIG, the predominant peptides from MOU showed an affinity distribution displaced toward lower affinity values, relative to those from SWEIG, particularly among 9-mers. However, the average affinity of the corresponding peptides from MOU was higher than in SWEIG (mean IC50: 520 versus1,135 nm and 372 versus 910 nm, for all peptide sizes and 9-mers, respectively). This was due to the higher percentage of very low affinity A*29:02 ligands among the predominant peptides from the ERAP1low cell line (Fig. 7B and 7C).
We reasoned that the effect of ERAP1 on the affinity of A*29:02 ligands might be best revealed with the peptides showing the largest expression differences among cell lines. Thus, the same analyses were carried out on the IR>1–1.5, IR>1.5–3, and IR>3 subsets (Fig. S7). As expected, in PF97387/MOU, the peptides from PF97387 showed higher affinity in all three IR subsets but especially in IR>3 and IR>1.5–3, both for all lengths and for 9-mers. A similar trend was observed in PF97387/SWEIG. In MOU/SWEIG, an affinity distribution of the peptides from MOU shifted toward lower affinity values was observed, particularly among 9-mers, in the IR>3 and IR>1.5–3 subsets but with a higher percentage of very low affinity ligands in SWEIG.
The predominant 9-mers from PF97387 were more hydrophobic than those from MOU or SWEIG in the respective comparisons, whereas marginal or no differences were observed between MOU and SWEIG (Fig. S8). This result parallels the length-independent affinity differences (Fig. S7B) and suggests that the basis for the length-independent influence of ERAP1 polymorphism on the affinity of A*29:02 ligands is the increased hydrophobicity of the peptides resulting from selection of bulkier residues by the more active variant.
DISCUSSION
The joint association of A*29:02 and ERAP2 with BSCR strongly suggests a peptide-dependent pathogenetic mechanism. Thus, the characterization of the A*29:02-bound peptidome and its shaping by the aminopeptidases involved in the processing of MHC-I ligands seems critical to our understanding of BSCR pathogenesis.
ERAP2 might influence BSCR in various ways. It may generate a specific autoantigen. Alternatively, ERAP2 may be required for specific epitope production only in certain ERAP1 contexts, being dispensable otherwise. ERAP2 facilitates antigen processing by ERAP1 through its complementary trimming specificity (6). Thus, ERAP2 might be particularly relevant in the context of low activity ERAP1 variants. A third possibility is that, since ERAP2 efficiently degrades 8-mers and shorter peptides, it might influence in this way the substrate-inhibition kinetics of ERAP1 trimming (20, 38) or the affinity-dependent peptide exchange on the MHC-I molecule mediated by very small peptides (39, 40).
We analyzed the effect of functional ERAP1variants, and of decreased enzyme expression, on the A*29:02 peptidome. The focus was on the amounts of shared ligands presented in distinct contexts since it is formally difficult to establish absence of individual epitopes when they are not detected by MS. Substantial alterations affecting many peptides occurred at two different levels: modulation of peptide length and modulation of sequence features based on differential selection of amino acid side chain volumes.
The effect of ERAP1 polymorphism on the length of A*29:02 ligands was analogous as in B*27:04 (16), consisting of a higher percentage of 9-mers, relative to longer peptides, among those predominant in the more active context. Absence of ERAP1 also results in longer MHC-I ligands (41, 42). This was also true for A*29:02, particularly when compared with a highly active ERAP1 background.
The length-independent influence on amino acid side chain volumes demonstrates a further level of ERAP1-induced alterations in the A*29:02 peptidome distinct from substrate length and susceptibility of the P1 residue to trimming. It also confirms that the peptide sequence downstream the N terminus modulates ERAP1 processing (43). The basis for this effect is to be found in the role of substrate affinity on promoting the conformational transition in ERAP1 required for efficient peptide trimming (44, 45). The specific reasons for the predominance of A*29:02 ligands with bulkier side chains in the more active ERAP1 context are less obvious. One possibility is a size effect of residue 730, located in the substrate binding site of ERAP1. Q730 is 5.4 Å3 bigger than E730. Presumably, substrates with smaller side chains are better accommodated in the ERAP1 variant of PF97387 with Q730. This variant also has K528, which confers higher activity than R528, present in MOU (3, 17, 20, 35, 38, 44). Both effects may lead to increased destruction of substrates with smaller side chains by the ERAP1 variant in PF97387, resulting in higher availability of peptides with bulkier side chains for binding to A*29:02. The larger binding site of the ERAP1 variant in MOU, with only E730, and its lower activity, due to R528, would explain the low selection of A*29:02 ligands relative to SWEIG, on the basis of side chain volumes.
Increased expression of free MHC-I heavy chain at the cell surface in the ERAP1low background of SWEIG was also observed in HLA-B*27 (46). The higher expression of free HLA-A heavy chain in SWEIG relative to MOU can be explained by the lower average affinity of the peptides from SWEIG and the higher percentage of very low affinity ligands among those predominant in this cell line. In addition, it is conceivable that, in the absence of ERAP1, a specific subpopulation of peptides binds A*29:02 with low affinity and dissociates at the cell surface. These peptides would hardly be detected in our analyses since this was performed after immunopurification of properly folded peptide/MHC complexes.
There are no studies quantifying the effects of ERAP2 in the context of distinct ERAP1 variants. We propose that ERAP1 influences the susceptibility to BSCR and that the role of ERAP2 in this disease may be particularly prominent in the context of ERAP1 alleles of low activity. Without ruling out a pathogenetic ERAP2-dependent epitope, the influence of ERAP1 on the A*29:02 peptidome may be more complex. For instance, killer cell immunoglobulin-like receptors are risk factors for BSCR in A*29:02+ individuals (47, 48). ERAP1 was recently suggested to modify, killer cell immunoglobulin-like receptor engagement to the MHC-I molecule by altering the affinity of MHC-I-bound peptides (49). Thus, the influence of ERAP1 polymorphism on the affinity of A*29:02 ligands may alter natural killer (NK)-related functions in these individuals, depending on their, killer cell immunoglobulin-like receptor phenotypes, influencing in this way their susceptibility to BSCR. Substantial peptidome alterations may also influence general and potentially pathogenic features of the molecule, such as folding and stability (50).
Future research on BSCR pathogenesis might benefit from a wider standpoint where a comprehensive view of the molecular biology of HLA-A*29 is taken into account. This concept, which is being increasingly substantiated by research in HLA-B*27 (51), might reflect common pathogenetic mechanisms underlying the joint association of MHC-I and ERAP1/2 in inflammatory diseases.
Supplementary Material
Acknowledgments
CAN is a fellow from the Government of Chile. We thank Dr. James Archer for helpful insight.
Footnotes
Author contributions: C.A. and J.A.L. designed the research; C.A., A.M., E.B., A.A., and J.A.L. performed research; C.A., A.M., E.B., A.A., and J.A.L. analyzed data; C.A., A.A., and J.A.L. wrote the paper.
* This work was supported by grants SAF2011/25681 (Plan Nacional de I+D+i) to JALC, Binational Science Foundation Grant 2009393 to AA, and an institutional grant of the Fundación Ramón Areces to the CBMSO.
 This article contains supplemental material Table S1 and Figs. S1-S8.
This article contains supplemental material Table S1 and Figs. S1-S8.
1 The abbreviations used are:
- AS
- ankylosing spondylitis
- BSCR
- birdshot chorioretinopathy
- ERAP
- endoplasmic reticulum aminopeptidase
- FDR
- false discovery rate
- MHC-I
- major histocompatibility complex class I
- RF
- residue frequency
- SNP
- single nucleotide polymorphism.
REFERENCES
- 1. Saric T., Chang S. C., Hattori A., York I. A., Markant S., Rock K. L., Tsujimoto M., Goldberg A. L. (2002) An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 2. Serwold T., Gonzalez F., Kim J., Jacob R., Shastri N. (2002) ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419, 480–483 [DOI] [PubMed] [Google Scholar]
- 3. Evans D. M., Spencer C. C., Pointon J. J., Su Z., Harvey D., Kochan G., Oppermann U., Opperman U., Dilthey A., Pirinen M., Stone M. A, Appleton L., Moutsianas L., Moutsianis L., Leslie S., Wordsworth T., Kenna T. J., Karaderi T., Thomas G. P., Ward M. M., Weisman M. H., Farrar C., Bradbury L. A., Danoy P., Inman R. D., Maksymowych W., Gladman D., Rahman P., Morgan A., Marzo-Ortega H., Bowness P., Gaffney K., Gaston J. S., Smith M., Bruges-Armas J., Couto A. R., Sorrentino R., Paladini F., Ferreira M. A., Xu H., Liu Y., Jiang L., Lopez-Larrea C., Díaz-Peña R., López-Vázquez A., Zayats T., Band G., Bellenguez C., Blackburn H., Blackwell J. M., Bramon E., Bumpstead S. J., Casas J. P., Corvin A., Craddock N., Deloukas P., Dronov S., Duncanson A., Edkins S., Freeman C., Gillman M., Gray E., Gwilliam R., Hammond N., Hunt S. E., Jankowski J., Jayakumar A., Langford C., Liddle J., Markus H. S., Mathew C. G., McCann O. T., McCarthy M. I., Palmer C. N., Peltonen L., Plomin R., Potter S. C., Rautanen A., Ravindrarajah R., Ricketts M., Samani N., Sawcer S. J., Strange A., Trembath R. C., Viswanathan A. C., Waller M., Weston P., Whittaker P., Widaa S., Wood N. W., McVean G., Reveille J. D., Wordsworth B. P., Brown M. A., Donnelly P. (2011) Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strange A., Capon F., Spencer C. C., Knight J., Weale M. E., Allen M. H., Barton A., Band G., Bellenguez C., Bergboer J. G., Blackwell J. M., Bramon E., Bumpstead S. J., Casas J. P., Cork M. J., Corvin A., Deloukas P., Dilthey A., Duncanson A., Edkins S., Estivill X., Fitzgerald O., Freeman C., Giardina E., Gray E., Hofer A., Hüffmeier U., Hunt S. E., Irvine A. D., Jankowski J., Kirby B., Langford C., Lascorz J., Leman J., Leslie S., Mallbris L., Markus H. S., Mathew C. G., McLean W. H., McManus R., Mössner R., Moutsianas L., Naluai A. T., Nestle F. O., Novelli G., Onoufriadis A., Palmer C. N., Perricone C., Pirinen M., Plomin R., Potter S. C., Pujol R. M., Rautanen A., Riveira-Munoz E., Ryan A. W., Salmhofer W., Samuelsson L., Sawcer S. J., Schalkwijk J., Smith C. H., Ståhle M., Su Z., Tazi-Ahnini R., Traupe H., Viswanathan A. C., Warren R. B., Weger W., Wolk K., Wood N., Worthington J., Young H. S., Zeeuwen P. L., Hayday A., Burden A. D., Griffiths C. E., Kere J., Reis A., McVean G., Evans D. M., Brown M. A., Barker J. N., Peltonen L., Donnelly P., Trembath R. C. (2010) A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42, 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kirino Y., Bertsias G., Ishigatsubo Y., Mizuki N., Tugal-Tutkun I., Seyahi E., Ozyazgan Y., Sacli F. S., Erer B., Inoko H., Emrence Z., Cakar A., Abaci N., Ustek D., Satorius C., Ueda A., Takeno M., Kim Y., Wood G. M., Ombrello M. J., Meguro A., Gül A., Remmers E. F., Kastner D. L. (2013) Genome-wide association analysis identifies new susceptibility loci for Behcet's disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 45, 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saveanu L., Carroll O., Lindo V., del Val M., Lopez D., Lepelletier Y., Greer F., Schomburg L., Fruci D., Niedermann G., Van Endert P. M. (2005) Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 6, 689–697 [DOI] [PubMed] [Google Scholar]
- 7. Evnouchidou I., Weimershaus M., Saveanu L., van Endert P. (2014) ERAP1-ERAP2 dimerization increases peptide-trimming efficiency. J. Immunol. 193, 901–908 [DOI] [PubMed] [Google Scholar]
- 8. Kuiper J. J., Van, Setten J., Ripke S, Van T., Slot R., Mulder F., Missotten T., Baarsma G. S., Francioli L. C., Pulit S. L., de Kovel C. G., Ten Dam-Van Loon N., Den Hollander A. I., Huis in het Veld P., Hoyng C. B., Cordero-Coma M., Martín J., Llorenç V., Arya B., Thomas D., Bakker S. C., Ophoff R. A., Rothova A., De, Bakker P. I., Mutis T., Koeleman B. P. (2014) A genome-wide association study identifies a functional ERAP2 haplotype associated with birdshot chorioretinopathy. Hum. Mol .Genet. 23, 6081–6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cortes A., Hadler J., Pointon J. P., Robinson P. C., Karaderi T., Leo P., Cremin K., Pryce K., Harris J., Lee S., Joo K. B., Shim S. C., Weisman M., Ward M., Zhou X., Garchon H. J., Chiocchia G., Nossent J., Lie B. A., Førre Ø., Tuomilehto J., Laiho K., Jiang L., Liu Y., Wu X., Bradbury L. A., Elewaut D., Burgos-Vargas R., Stebbings S., Appleton L., Farrah C., Lau J., Kenna T. J., Haroon N., Ferreira M. A., Yang J., Mulero J., Fernandez-Sueiro J. L., Gonzalez-Gay M. A., Lopez-Larrea C., Deloukas P., Donnelly P., Bowness P., Gafney K., Gaston H., Gladman D. D., Rahman P., Maksymowych W. P., Xu H., Crusius J. B., van, der Horst-Bruinsma I. E., Chou C. T., Valle-Oñate R., Romero-Sánchez C., Hansen I. M., Pimentel-Santos F. M., Inman R. D., Videm V., Martin J., Breban M., Reveille J. D., Evans D. M., Kim T. H., Wordsworth B. P., Brown M. A. (2013) Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45, 730–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Franke A., McGovern D. P., Barrett J. C., Wang K., Radford-Smith G. L., Ahmad T., Lees C. W., Balschun T., Lee J., Roberts R., Anderson C. A., Bis J. C., Bumpstead S., Ellinghaus D., Festen E. M., Georges M., Green T., Haritunians T., Jostins L., Latiano A., Mathew C. G., Montgomery G. W., Prescott N. J., Raychaudhuri S., Rotter J. I., Schumm P., Sharma Y., Simms L. A., Taylor K. D., Whiteman D., Wijmenga C., Baldassano R. N., Barclay M., Bayless T. M., Brand S., Büning C., Cohen A., Colombel J. F., Cottone M., Stronati L., Denson T., de Vos M., D'Inca R., Dubinsky M., Edwards C., Florin T., Franchimont D., Gearry R., Glas J., van Gossum A., Guthery S. L., Halfvarson J., Verspaget H. W., Hugot J. P., Karban A., Laukens D., Lawrance I., Lemann M., Levine A., Libioulle C., Louis E., Mowat C., Newman W., Panés J., Phillips A., Proctor D. D., Regueiro M., Russell R., Rutgeerts P., Sanderson J., Sans M., Seibold F., Steinhart A. H., Stokkers P. C., Torkvist L., Kullak-Ublick G., Wilson D., Walters T., Targan S. R., Brant S. R., Rioux J. D., D'Amato M., Weersma R. K., Kugathasan S., Griffiths A. M., Mansfield J. C., Vermeire S., Duerr R. H., Silverberg M. S., Satsangi J., Schreiber S., Cho J. H., Annese V., Hakonarson H., Daly M. J., Parkes M. (2010) Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson M. P., Roten L. T., Dyer T. D., East C. E., Forsmo S., Blangero J., Brennecke S. P., Austgulen R., Moses E. K. (2009) The ERAP2 gene is associated with preeclampsia in Australian and Norwegian populations. Hum. Genet. 126, 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hill L. D., Hilliard D. D., York T. P., Srinivas S., Kusanovic J. P., Gomez R., Elovitz M. A., Romero R., Strauss J. F., III (2011) Fetal ERAP2 variation is associated with preeclampsia in African Americans in a case-control study. BMC Med. Genet. 12, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vanhille D. L., Hill L. D., Hilliard D. D., Lee E. D., Teves M. E., Srinivas S., Kusanovic J. P., Gomez R., Stratikos E., Elovitz M. A., Romero R., Strauss J. F., III (2013) A novel haplotype structure in a Chilean population: Implications for ERAP2 protein expression and preeclampsia risk. Mol. Genet. Genomic. Med. 1, 98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shah K. H., Levinson R. D., Yu F., Goldhardt R., Gordon L. K., Gonzales C. R., Heckenlively J. R., Kappel P. J., Holland G. N. (2005) Birdshot chorioretinopathy. Surv. Ophthalmol. 50, 519–541 [DOI] [PubMed] [Google Scholar]
- 15. Brézin A. P., Monnet D., Cohen J. H., Levinson R. D. (2011) HLA-A29 and birdshot chorioretinopathy. Ocul. Immunol. Inflamm. 19, 397–400 [DOI] [PubMed] [Google Scholar]
- 16. Garcia-Medel N., Sanz-Bravo A., Van Nguyen D., Galocha B., Gomez-Molina P., Martin-Esteban A., Alvarez-Navarro C., López de Castro. J. A. 2012. Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Mol. Cell. Proteomics. 11, 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanz-Bravo A., Campos J., Mazariegos M. S., Lopez de Castro J. A. (2015). Dominant role of the ERAP1 polymorphism R528K in shaping the HLA-B27 peptidome through differential processing determined by multiple peptide residues. Arthritis Rheumatol. 67, 692–701 [DOI] [PubMed] [Google Scholar]
- 18. Yang S. Y., Milford E. L., Hämmerling U., Dupont B. (1989). Description of the reference panel of B-lymphoblastoid cell lines for factors of the HLA system: The B-cell line panel designed for the Tenth International Histocompatibility Workshop. In: Immunobiology of HLA. Vol. I. Histocompatibility Testting 1987, p. 11–19. In Dupont B. (ed.). Springer-Verlag, New York. [Google Scholar]
- 19. Zemmour J., Little A. M., Schendel D. J., Parham P. (1992) The HLA-A,B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J. Immunol. 148, 1941–1948 [PubMed] [Google Scholar]
- 20. Martín-Esteban A., Gómez-Molina P., Sanz-Bravo A., Lopez de Castro J. A. (2014) Combined effects of ankylosing spondylitis-associated ERAP1 polymorphisms outside the catalytic and peptide-binding sites on the processing of natural HLA-B27 ligands. J. Biol. Chem. 289, 3978–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lozano F., Santos-Aguado J., Borche L., Places L., Doménech N., Gayá A., Vilella R, Vives J. (1989) Identification of the amino acid residues defining an intralocus determinant in the alpha 1 domain of HLA-A molecules. Immunogenetics 30, 50–53 [DOI] [PubMed] [Google Scholar]
- 22. Stam N. J., Vroom T. M., Peters P. J., Pastoors E. B., Ploegh H. L. (1990) HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in Western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. Int. Immunol. 2, 113–125 [DOI] [PubMed] [Google Scholar]
- 23. Paradela A., Garcia-Peydro M., Vazquez J., Rognan D., Lopez de Castro J. A. 1998. The same natural ligand is involved in allorecognition of multiple HLA-B27 subtypes by a single T cell clone: Role of peptide and the MHC molecule in alloreactivity. J. Immunol. 161, 5481–5490 [PubMed] [Google Scholar]
- 24. Ishihama Y., Rappsilber J., Andersen J. S., Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 25. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 26. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome. Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 27. Karosiene E., Lundegaard C., Lund O., Nielsen M. (2012) NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 64, 177–186 [DOI] [PubMed] [Google Scholar]
- 28. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 29. Granados D. P., Yahyaoui W., Laumont C. M., Daouda T., Muratore-Schroeder T. L., Côté C., Laverdure J. P., Lemieux S., Thibault P., Perreault C. (2012) MHC I-associated peptides preferentially derive from transcripts bearing miRNA response elements. Blood 119, e181-e191 [DOI] [PubMed] [Google Scholar]
- 30. Granados D. P., Sriranganadane D., Daouda T., Zieger A., Laumont C. M., Caron-Lizotte O., Boucher G., Hardy M. P., Gendron P., Côté C., Lemieux S., Thibault P., Perreault C. (2014) Impact of genomic polymorphisms on the repertoire of human MHC class I-associated peptides. Nat. Commun. 5, 3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harvey D., Pointon J. J., Evans D. M., Karaderi T., Farrar C., Appleton L. H., Sturrock R. D., Stone M. A., Oppermann U., Brown M. A., Wordsworth B. P. (2009) Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum. Mol. Genet. 18, 4204–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Costantino F., Talpin A., Evnouchidou I., Kadi A., Leboime A., Said-Nahal R., Bonilla N., Letourneur F., Leturcq T., Ka Z., van Endert P., Garchon H. J., Chiocchia G., Breban M. 2015. ERAP1 gene expression is influenced by non-synonymous polymorphisms associated with predisposition to spondyloarthritis. Arthritis Rheumatol. doi: 10.1002/art.39072. [DOI] [PubMed] [Google Scholar]
- 33. Hammer G. E., Gonzalez F., James E., Nolla H., Shastri N. (2007) In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat. Immunol. 8, 101–108 [DOI] [PubMed] [Google Scholar]
- 34. Andres A. M., Dennis M. Y., Kretzschmar W. W., Cannons J. L., Lee-Lin S. Q., Hurle B., Schwartzberg P. L., Williamson S. H., Bustamante C. D., Nielsen R., Clark A. G., Green E. D. (2010) Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS. Genet. 6, e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goto Y., Hattori A., Ishii Y., Tsujimoto M. (2006) Reduced activity of the hypertension-associated Lys528Arg mutant of human adipocyte-derived leucine aminopeptidase (A-LAP)/ER-aminopeptidase-1. FEBS Lett. 580, 1833–1838 [DOI] [PubMed] [Google Scholar]
- 36. Hassan C., Kester M. G., de Ru A. H., Hombrink P., Drijfhout J. W., Nijveen H., Leunissen J. A., Heemskerk M. H., Falkenburg J. H., van Veelen P. A. (2013) The human leukocyte antigen-presented ligandome of B lymphocytes. Mol. Cell. Proteomics 12, 1829–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garcia-Medel N., Sanz-Bravo A., Alvarez-Navarro C., Gómez-Molina P., Barnea E., Marcilla M., Admon A., Lopez de Castro J. A. (2014) Peptide handling by HLA-B27 subtypes influences their biological behavior, association with ankylosing spondylitis and susceptibility to ERAP1. Mol. Cell. Proteomics 13, 3367–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evnouchidou I., Kamal R. P., Seregin S. S., Goto Y., Tsujimoto M., Hattori A., Voulgari P. V., Drosos A. A., Amalfitano A., York I. A., Stratikos E. (2011) Cutting edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J. Immunol. 186, 1909–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saini S. K., Ostermeir K., Ramnarayan V. R., Schuster H., Zacharias M., Springer S. (2013) Dipeptides promote folding and peptide binding of MHC class I molecules. Proc. Natl. Acad. Sci. U.S.A. 110, 15383–15388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saini S. K., Schuster H., Ramnarayan V. R., Rammensee H. G., Stevanoviæ S., Springer S. (2015) Dipeptides catalyze rapid peptide exchange on MHC class I molecules. Proc. Natl. Acad. Sci. U.S.A. 112, 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blanchard N., Kanaseki T., Escobar H., Delebecque F., Nagarajan N. A., Reyes-Vargas E., Crockett D. K., Raulet D. H., Delgado J. C., Shastri N. (2010) Endoplasmic reticulum aminopeptidase associated with antigen processing defines the composition and structure of MHC class I peptide repertoire in normal and virus-infected cells. J. Immunol. 184, 3033–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen L., Fischer R., Peng Y., Reeves E., McHugh K., Ternette N., Hanke T., Dong T., Elliott T., Shastri N., Kollnberger S., James E., Kessler B., Bowness P. (2014) Critical role of endoplasmic reticulum aminopeptidase 1 in determining the length and sequence of peptides bound and presented by HLA-B27. Arthritis Rheumatol. 66, 284–294 [DOI] [PubMed] [Google Scholar]
- 43. Evnouchidou I., Momburg F., Papakyriakou A., Chroni A., Leondiadis L., Chang S. C., Goldberg A. L., Stratikos E. (2008) The internal sequence of the peptide-substrate determines its N-terminus trimming by ERAP1. PLoS ONE. 3, e3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kochan G., Krojer T., Harvey D., Fischer R., Chen L., Vollmar M., von, Delft F., Kavanagh K. L., Brown M. A., Bowness P., Wordsworth P., Kessler B. M., Oppermann U. (2011) Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. U.S.A. 108, 7745–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen T. T., Chang S. C., Evnouchidou I., York I. A., Zikos C., Rock K. L., Goldberg A. L., Stratikos E., Stern L. J. (2011) Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat. Struct. Mol. Biol. 18, 604–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haroon N., Tsui F. W., Uchanska-Ziegler B., Ziegler A., Inman R. D. (2012) Endoplasmic reticulum aminopeptidase 1 (ERAP1) exhibits functionally significant interaction with HLA-B27 and relates to subtype specificity in ankylosing spondylitis. Ann. Rheum. Dis. 71, 589–595 [DOI] [PubMed] [Google Scholar]
- 47. Levinson R. D., Du Z., Luo L., Monnet D., Tabary T., Brezin A. P., Zhao L., Gjertson D. W., Holland G. N., Reed E. F., Cohen J. H., Rajalingam R. (2008) Combination of KIR and HLA gene variants augments the risk of developing birdshot chorioretinopathy in HLA-A*29-positive individuals. Genes Immun. 9, 249–258 [DOI] [PubMed] [Google Scholar]
- 48. Levinson R. D. (2011) Killer immunoglobulin-like receptor genes in uveitis. Ocul .Immunol. Inflamm. 19, 192–201 [DOI] [PubMed] [Google Scholar]
- 49. Cifaldi L., Romania P., Falco M., Lorenzi S., Meazza R., Petrini S., Andreani M., Pende D., Locatelli F., Fruci D. (2015) ERAP1 regulates natural killer cell function by controlling the engagement of inhibitory receptors. Cancer Res. [DOI] [PubMed] [Google Scholar]
- 50. Marcilla M., López de Castro J. A. (2008) Peptides: The cornerstone of HLA-B27 biology and pathogenetic role in spondyloarthritis. Tissue Antigens 71, 495–506 [DOI] [PubMed] [Google Scholar]
- 51. Alvarez-Navarro C., Lopez de Castro J. A. (2014) ERAP1 structure, function and pathogenetic role in ankylosing spondylitis and other MHC-associated diseases. Mol. Immunol. 57, 12–21 [DOI] [PubMed] [Google Scholar]
- 52. Hearn A., York I. A., Rock K. L. (2009) The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J. Immunol. 183, 5526–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.