Abstract
Cells of the immune system communicate with their environment through immunoreceptors. These receptors often harbor intracellular tyrosine residues, which, when phosphorylated upon receptor activation, serve as docking sites to recruit downstream signaling proteins containing the Src Homology 2 (SH2) domain. A systematic investigation of interactions between the SH2 domain and the immunoreceptor tyrosine-based regulatory motifs (ITRM), including inhibitory (ITIM), activating (ITAM), or switching (ITSM) motifs, is critical for understanding cellular signal transduction and immune function. Using the B cell inhibitory receptor CD22 as an example, we developed an approach that combines reciprocal or bidirectional phosphopeptide and SH2 domain array screens with in-solution binding assays to identify a comprehensive SH2-CD22 interaction network. Extending this approach to 194 human ITRM sequences and 78 SH2 domains led to the identification of a high-confidence immunoreceptor interactome containing 1137 binary interactions. Besides recapitulating many previously reported interactions, our study uncovered numerous novel interactions. The resulting ITRM-SH2 interactome not only helped to fill many gaps in the immune signaling network, it also allowed us to associate different SH2 domains to distinct immune functions. Detailed analysis of the NK cell ITRM-mediated interactions led to the identification of a network nucleated by the Vav3 and Fyn SH2 domains. We showed further that these SH2 domains have distinct functions in cytotoxicity. The bidirectional protein-peptide array approach described herein may be applied to the numerous other peptide-binding modules to identify potential protein–protein interactions in a systematic and reliable manner.
Immune receptor signaling, critical for proper immune response, involves signaling pathways mediated by specific tyrosine residues that are phosphorylated upon receptor activation (1). These tyrosine phosphorylation sites are frequently found in one of the three types of immunoreceptor tyrosine-based regulatory motifs (ITRMs)1. The immunoreceptor tyrosine-based activation motifs (ITAMs) with the consensus sequence YxxI/Lx(6–12)YxxI/L, where x represents any amino acid, are typically associated with positive or activating immune response (2). In contrast, the immune system elicits negative or inhibitory response through receptors bearing the immunoreceptor tyrosine-based inhibition motifs (ITIMs) with the degenerated sequence S/I/V/LxYxxI/V/L (3). Interestingly, SLAM/CD150 and related receptors of the CD2 subfamily contain a third type of signaling motif called Immunoreceptor Tyrosine-based Switch Motif (ITSM) with the consensus sequence TxYxx(V/I) (4). An ITSM can convey either an activating or inhibitory signal, depending on the type of immune cell, the receptor and the bound protein (5).
In general, phosphorylated ITIMs recruit the SH2 domain-containing tyrosine phosphatase SHP-1 or SHP-2 (6), and phosphorylated ITAMs are recognized by protein tyrosine kinases such as ZAP70 in T cells and SYK in B cells (7). Nevertheless, these distinctions are only relative and both motifs may be involved in either positive or negative immune functions. For example, ITIM-containing inhibitory receptors found on phagocytes can suppress or enhance inflammatory cytokine production depending on the downstream proteins that they recruit (6). Similarly, ITAM sequences have been found to mediate inhibitory signaling (8). Indeed, even within the same cell type, an ITAM-containing receptor may mediate functions as diverse as microbial killing, antigen presentation, cytokine production, T-Cell instruction, and tissue repair (2). Moreover, exquisite binding specificities have been observed for different ITRMs. For example, the CD31 ITIMs recruit SHIP-1, SHP-1, and SHP-2 (9). In contrast, the archetypal ITIM identified in the cytoplasmic domain of the inhibitory IgG Fc receptor FcγRIIB (10) recruits SHIP-1 and -2, but not SHP-1 or -2. Although the FcγRIIB ITIM has an affinity for the SH2 domain of either SHP-1 or -2, it does not recruit these tyrosine phosphatases as do other ITIM-bearing receptors (11). Besides SHIP-1/2 and SHP-1/2, other SH2-containing proteins have also been found associated with certain ITIMs. For example, through their respective ITIM sequences, the LAIR-1 receptor recruits tyrosine kinase CSK (12), Lax interacts with GRB2, PIK3R1 and GRAP2 (13), and CD72 forms a complex with SHP-1 and GRB2 (14). These findings indicate that immunoreceptor signaling is context-dependent and that the precise pairing of an ITRM sequence with an SH2-containing protein plays a critical role in dictating the signaling and biological outcome.
Given the critical importance of SH2 domain-phosphotyrosine (pY) interaction in immune signaling, we set to systematically identify SH2 domain-ITRM interactions. A variety of different high-throughput methods have been developed for the identification of protein–protein or protein-peptide interactions to date. These include yeast two-hybrid (15), affinity purification coupled to mass spectrometry (AP-MS) (16), phage display (17), and protein (18) and peptide arrays (19). We employed protein and peptide arrays in our study because of their simplicity and amenability to post-translational modifications. Specifically, we combined SH2 domain array with phosphotyrosine peptide array to decipher an SH2-ITRM interactome for immunoreceptors. We showed that the peptide and SH2 domain arrays were not simple mirror images of each other, but rather, they exhibited distinct, but complementary binding patterns. Although neither array platform alone was sufficient to detect all authentic interactions, integration of data from reciprocal peptide and protein array screens allowed us to identify a high-confidence SH2-ITRM interactome. The interactome has not only provided a systematic view of immunoreceptor signaling network, but also generated unique insights into the signaling specificity for the different immune signaling motifs.
MATERIALS AND METHODS
Peptide Synthesis
Peptides were synthesized on Tentagel resin on an Intavis-AG MultiPep peptide synthesizer using N-(9-fluorenyl) methoxycarbonyl (Fmoc) chemistry. For a typical pY-containing peptide, the pY site was flanked by three residues at the N terminus and seven at the C terminus (19). All peptides were synthesized with biotin at the N terminus followed by a Gly-Gly spacer. Identities of the peptides were verified by mass spectrometry.
Peptide Array Preparation and Probing
Peptides were printed on glass slides as neutravidin complexes (20). Specifically, the biotin-peptide was incubated with neutravidin in 3:1 ratio in PBS, pH 7.5, to form a complex. Unbound peptides were removed by ultrafiltration through a 3 kDa-cutoff membrane (Millipore, Billerica, MA). SuperAB glass slides (Fisher, Waltham, MA) were pre-activated in a buffer containing 50 mm NaIO4, 0.1 m sodium acetate, pH 5.5, for 0.5h at RT and used for printing immediately. The peptide-neutravidin conjugates were printed onto the slide using a Bio-Rad VersArray Chipwriter-Pro system. After printing, the peptide array chips were washed in 3% bovine serum albumin (BSA)/TBST (containing 0.1 m Tris-HCl, pH 7.4, 150 mm NaCl, and 0.1% Tween 20) for three times. GST-SH2 domains (1.0 μm) were added directly to the above buffer and incubated with the peptide chips for 1h at RT. Following three TBST washes, the peptide array chips were incubated with a rabbit anti-GST antibody (Abcam #ab3416, Toronto, Canada) for 1h at RT. DyLight 649-labeled goat anti-rabbit IgG antibody (Pierce #35565, Rockford, IL) was used to visualize the bound SH2 protein. The peptide chips were scanned on a microarray laser scanner (Tecan Co., Morrisville, NC) and the binding signals quantified using the embedded software of the scanner.
SH2 Domain Array Preparation and Probing
SH2 domains were expressed as GST fusion proteins in E. coli as previously described (19). Each SH2 domain was purified using glutathione (GSH) agarose beads followed by FPLC on a Sephadex-75 column (AKTA, GE, Mississauga, Canada). Purified proteins were buffer-exchanged into the printing buffer (37 mm Bicine, pH 8.25, 150 mm NaCl, 0.37 mm EDTA, 0.5 m sucrose) at a final protein concentration of 40 μm.
GST-SH2 proteins were printed at three concentrations (40, 10 and 2.5 μm) on Whatman Fast nitrocellulose slides using a VersArray Chipwriter Pro (Bio-Rad, Mississauga, Canada) equipped with four SMP15Xb quill pins (Telechem, Sunnyvale, CA). Spot-to-spot distance was 950 μm, with three reprints of the same protein and each concentration printed in duplicate in the x dimension. A dwell time of 0.1 s was used for each spot with an approach speed of 12.5 mm/sec. Samples were printed at RT with a controlled humidity of 65%. Following printing, all slides were stored in sealed containers at 4 °C.
To screen the SH2 microarray for peptide binding, the microarray slide was incubated for 1 h at 4 °C in a blocking buffer (3% BSA in TBST) to prevent nonspecific binding. The slide was then washed in TBST three times and incubated with 5 μm biotin-labeled peptide (1h, 4 °C). The bound biotin-peptide was detected using 2.5 μg/ml streptavidin-Alexa Fluor 647 (Anaspec, Fremont, CA) in 3% BSA/TBST (1h, 4 °C). The slide were washed another three times in TBST, removed from the incubation tray, spun at 800g for 5 min, and air-dried for 30 min in the dark. The slide was then imaged on a microarray laser scanner (Tecan Co., Morrisville, NC). To validate array quality, a slide was incubated with an anti-GST antibody (Abcam #ab3416, Toronto, Canada; 1:5000 dilution in 3% BSA/TBST) in a similar manner as described above and detected using DyLight 649 donkey anti-rabbit-IgG (Anaspec, Fremont, CA).
Fluorescence Polarization Measurements
Each SH2 protein was serially diluted in a 384-well plate, followed by the addition of fluorescein-labeled peptide in PBS buffer. The mixtures were incubated in the dark for 30 min prior to fluorescent polarization measurements at RT on an EnVision Multilabel Plate Reader (PerkinElmer, Waltham, MA) with excitation set at 480 nm and emission at 535 nm. Binding curves were generated by fitting the isothermal titration data to a hyperbolic nonlinear regression model using Prism 3.0 (GraphPad software, Inc., San Diego, CA), which also produced the corresponding dissociation constants (Kd).
Cell Culture, Immunoprecipitation, GST Pull Down, Western Blot and ELISA
U937 and YT cells (kindly provided by Dr. Christopher Mody, University of Calgary) were cultured in RPMI 1640 medium (ATCC) supplemented with 2 mm l-glutamine, 10 mm HEPES, 1 mm sodium pyruvate, 4.5 g/L glucose, 1.5 g/L sodium bicarbonate, 10% FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin. Cells were washed in PBS and lysed in lysis buffer (1% Nonidet P-40, 50 mm sodium fluoride, 150 mm sodium chloride, 2 mm EDTA and 50 mm Tris-HCl, pH = 7.4) containing protease and phosphatase inhibitors (i.e. 1 μm phenylmethylsulfoyl fluoride, 2 μm E64, 1 μm bestatin, 1 μm pepstatin A, 2 μg/ml aprotinin, 10 μm leupeptin and 1 mm sodium orthovanadate) (Sigma-Aldrich, St. Louis, MO). After centrifugation to remove cell debris, the lysate was incubated with a desired antibody immobilized on protein G or a peptide immobilized on streptavidin beads for 2 h at 4 °C with rocking. The precipitated proteins were separated on 10–15% SDS-PAGE and transferred under semidry conditions to PVDF membrane (Bio-Rad, Mississauga, Canada). The membranes were blocked in 5% BSA (Roche, Mississauga, Canada) for 1 h at RT. This was followed by incubation with a primary antibody for 1 h at RT. After 3 washes in TBST buffer, a goat anti-mouse or anti-rabbit IgG-HRP conjugate (Bio-Rad, Mississauga, Canada) was added to the membrane and incubated for 40 min at RT. The membrane was developed by enhanced chemiluminescence (ECL) (PerkinElmer, Waltham, MA).
The IFN-γ ELISA (Enzyme-Linked ImmunoSorbent Assay) was carried out following the method described by Chuang et al. (38). For delivery of SH2 domains into cells by nanoparticles, FPLC purified proteins were used to saturate 10 nm gold nanoparticles (AuNP) in 1%PBS. Cells (5 × 105) were incubated with 10 nm AuNP-SH2 for 4h at 37 °C, followed by stimulation with C1.7mAb (400 ng/ml) for 1h at 37 °C. Target K562 cells (5 × 104) were then added. After incubation for 16 h at 37 °C, 300 μl of cell-free supernatant were collected. IFN-γ concentration was then quantitated with mouse anti IFN-γ (Santa Cruz sc-8423, Dallas, TX) or rabbit anti IFN-γ (Santa Cruz sc-8308, Dallas, TX).
Array Data Analysis
The binding signal for each peptide or SH2 probe was normalized across the entire domain or peptide array. In the case that multiple spots on an array represented a single peptide or protein, the average binding signal was used in normalization. For example, the binding signal for an SH2 domain on the peptide array in Fig. 1A was calculated as the average of the binding signals of the quadruplets. For the binding of a pTyr peptide to the SH2 domain array in Fig. 1B, we used binding data for the first two rows (corresponding to 40 μm and 10 μm SH2 domains) to calculate the final binding signal. Specifically, for each concentration, the binding signal is an average of the doublet. This signal was then normalized across the entire array for each concentration. The average of the normalized signals (from 0 to 1) for the two concentrations was used as the final binding signal for that pTyr peptide.
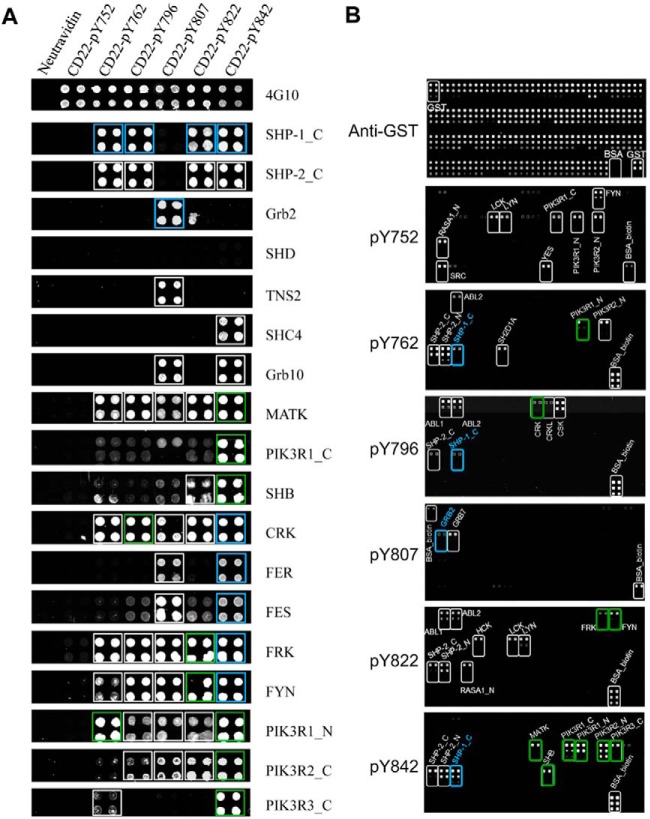
Fig. 1.
Using the bidirectional SH2 domain-pY peptide array screening approach to identify interactions mediated by CD22. A, A pY-containing peptide array representing the cytoplasmic Tyr phosphorylation sites in CD22 was probed for binding, respectively, to human SH2 domains (in GST fusion). Shown are anti-GST Western blots for representative SH2 domains with the names of the corresponding proteins given on the right. Spots framed by blue squares denote known interactions, the remaining novel candidates. Green squares denote interaction candidates detected on both the peptide and domain arrays (B). B, Probing of an SH2 domain array by individual pY peptides. A panel of 78 purified human SH2 domains (in GST fusion), GST, and BSA were printed at three different concentrations (40, 10, 2.5 μm from top to bottom) with duplication for each concentration. The quality of the protein array was verified by an anti-GST Western blot (top panel). Biotinylated pY-peptides were used to probe the array and the bound peptides detected using fluorescein-labeled streptavidin. Spots framed by blue rectangles denote known interactions, white novel interaction candidates, green candidates identified in both peptide and domain arrays.
The following equation was used for normalizing binding signals across a peptide array
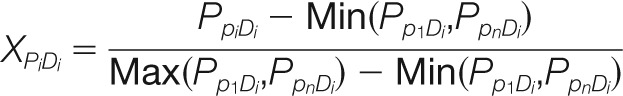 |
Where XpiDi represents the normalized peptide array binding score for the interaction between SH2 domain i (Di) and peptide i (Pi) on the peptide array, and PpiDi represents the strength of the original fluorescent signal for SH2 domain Di to peptide Pi on the peptide array. Max denotes the strongest binding signal produced by domain Di whereas Min the weakest signal for the domain on the peptide array that contains “n” number of peptides. The value of XPiDi ranges from 0 (weakest binding) to 1 (strongest binding).
Similarly, the fluorescent intensity of each spot on the SH2 domain array was normalized from 0 to 1 according the equation:
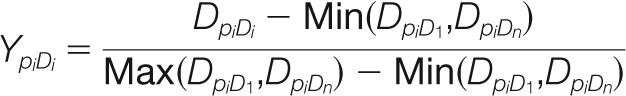 |
Where YpiDi represents the normalized domain array score for the interaction between Pi and Di from the domain array, and DpiDi represents the original fluorescent intensity corresponding to the interaction between Pi and Di on the domain array.
The bidirectional array score, BPiDi, for a given peptide-domain pair is defined by the average of the corresponding peptide array score XpiDi and domain array score YpiDi as in:
The consistence score between the peptide and protein array binding signals for each SH2 domain-pY peptide interaction is defined as in:
Where CpiDi represents the array consistence value of the interaction between peptide Pi and domain Di.
The selectivity score ZpiDi of an SH2 domain for an ITRM peptide is defined according to the formula (22):
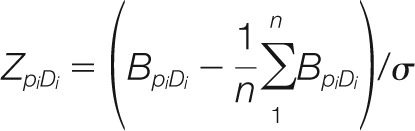 |
Where σ is the standard deviation of BpiDi
The binding free energy between peptide Pi and domain Di was calculated using the equation:
Where ΔGpiDi represents the free energy of binding between Pi and Di, KdpiDi the in-solution binding dissociation constant between Pi and Di in μm.
RESULTS
Combining Peptide and Protein Arrays to Probe SH2-pTyr Interactions
To establish a reliable method to map modular domain-peptide interactions, we used both peptide and protein arrays to identify binding partners for CD22, a coreceptor for the B-cell receptor (BCR) (23). The intracellular portion of human CD22 contains six potential tyrosine phosphorylation (pTyr or pY) sites (supplemental Table S1), four of which conform to the ITIM consensus (24). Although CD22 has been shown to bind a number of SH2-containing proteins, including SHP-1, LYN, SYK, GRB2, SHC1, SHIP, PI3K, and PLCγ1 (25), a comprehensive SH2-CD22_pY interaction network is not yet available.
We employed 78 purified human SH2 domains to probe, respectively, an array containing peptides representing the CD22 pY sites (Fig. 1). Biotin-tagged peptides were printed on glass slides in quadruplets as neutravidin conjugates (20). The peptide array was subsequently probed for binding to SH2 domains (supplemental Fig. S1). The bound SH2 domains were detected by a rabbit anti-GST antibody and visualized by DyLight 649-labeled goat anti-rabbit IgG (Fig. 1A). This analysis not only recapitulated many known interactions (25), but also uncovered numerous novel candidate interactions. Of note, the SHC4 SH2 domain was found to bind exclusively to the peptide pY842 whereas the TNS2_SH2 domain interacted only with the peptide pY807, a site recognized also by the GRB2 SH2 domain (26) (Fig. 1A).
To confirm interactions identified from the peptide arrays, we carried out reciprocal screens of protein arrays using the pY peptides to probe an array of 78 SH2 domains (as GST fusion, supplemental Table S2). To obtain semiquantitative information on binding, we printed each SH2 domain in three concentrations and in doublets for each concentration. An anti-GST Western blot showed that the spots containing 40 or 10, but not 2.5 μm proteins, produced strong and uniform signals (Fig. 1B, top panel). Thus, data points corresponding to 2.5 μm SH2 domains were excluded from subsequent analysis. Next, the SH2 domain arrays were incubated, respectively, with biotin-labeled CD22_pY peptides. Alexa Fluor 647-labeled streptavidin was used to visualize the bound peptides. As shown in Fig. 1B and supplemental Fig. S2, each peptide produced a unique binding profile on the SH2 domain array, indicating distinct functions for these pY sites. For example, although the SHP-1 or -2 SH2 domain bound to the CD22 ITIM peptides, the PIK3R family of SH2 domains recognized only the pY762 and pY842 peptides and the ABL1/2 SH2 domains interacted specifically with the pY796 and pY822 peptides (Fig. 1B; supplemental Fig. S2).
Identification of Authentic Interactions by Integrating Peptide and Protein Arrays
Although the peptide and protein array data agreed with each other in some cases, they differed significantly in others. For example, the SHP-1/2, MATK, PIK3R1_C, PIK3R1_N, PIK3R2_N, PIK3R3_C and SHB_SH2 domains displayed robust binding to the pY842 peptide on the peptide array (Fig. 1A); Conversely, the pY842 peptide probe recognized the same set of SH2 domains on the protein array (Fig. 1B, bottom panel; supplemental Fig. S2). In contrast, although numerous SH2 domains bound to the pY807 peptide on the peptide array (Fig. 1A), the same peptide recognized only the GRB2_SH2 and GRB7_SH2 domains on the protein array (Fig. 1B). Moreover, the strength of binding signal for an SH2-pY pair varied markedly between the two array formats. Whereas the pY752 peptide exhibited the strongest binding to the FYN_SH2 domain on the protein array (Fig. 1B), the latter only showed weak binding to the former on the peptide array (Fig. 1A).
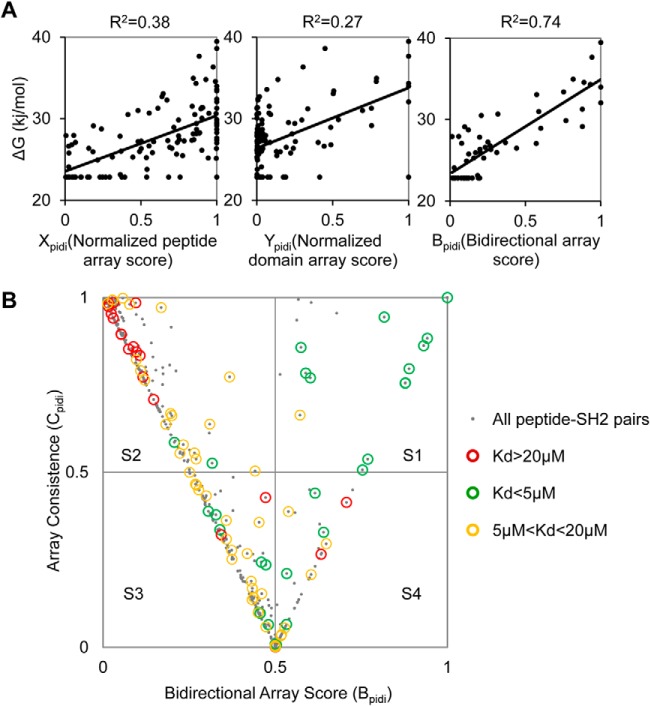
To find out if the array screening could identify authentic SH2-pY interactions, we determined the in-solution affinities (Kd) of the six pY peptides for a panel of 22 SH2 domains that showed binding to the peptides on either the peptide or the protein array (supplemental Table S3). The free energy (ΔG) for an SH2-pY peptide interaction was then calculated based on the corresponding Kd value. The ΔG value was then plotted against the corresponding peptide or protein array signal normalized to a scale of [0, 1] (22). As shown in Fig. 2A, the binding free energy showed a poor correlation with either the peptide (Xpidi) or the protein (Ypidi) array score. However, for the interactions that showed consistent binding on the two array formats (i.e. Xpidi-Ypidi < 0.5), a significantly improved correlation (R2 = 0.74) was obtained when the ΔG was plotted against the bidirectional array score (Bpidi) defined as the average of the peptide and protein array scores, i.e. Bpidi = (Xpidi +Ypidi)/2 (Fig. 2A).
Fig. 2.
Integration of binding signals from reciprocal peptide–protein array screens produced a set of high-confidence interactions for CD22. A, Correlation of pY peptide-SH2 in-solution binding free energy (ΔG) with the corresponding, normalized score on the peptide array (Xpidi, left) or SH2 domain array (Ypidi, middle), or with the bidirectional array score (Bpidi, right). B, A plot of the corresponding bidirectional array (Bpidi) and array consistence (Cpidi) scores for the SH2-CD22_pY peptide pairs. The graph is divided into four squares, namely S1-S4, when intercepted by perpendicular lines drawn at Bpidi = 0.5and Cpidi = 0.5. The SH2-pY pairs are represented in gray dots. Pairs for which in-solution binding data are available are identified by open circles: Kd<5 μm (green circle); 5 μm<Kd<20 μm (yellow circle) and Kd>20 μm (red circle).
We sought next to use the integrated bidirectional array data to distinguish true positive from negative interactions. To this end, we introduced the term, array consistency or “Cpidi”, as defined in Cpidi = 1 − (Xpidi − Ypidi), which measures the degree of agreement between the peptide and protein array binding scores (with Cpidi = 1 indicating 100% agreement) (supplemental Table S4). We then plotted Bpidi against Cpidi for all SH2 domain-pY peptide pairs for which the Kd values had been determined. The graph was sectioned into four squares, namely S1, S2, S3, and S4 (Fig. 2B), by perpendicular lines drawn at Bpidi/Cpidi = 0.5. The SH2-pY peptide pairs were divided into three groups according to the corresponding Kd values. The pairs with Kd <5 μm were considered true positive interactions whereas those with Kd>20 μm true negatives. The pairs with 5 μm<Kd<20 μm, which could fall in either category, would be assigned later after taking the array binding signals into account. We found that square S1 contained 13 interactions with Kd<5 μm (true positive), 1 with 5 μm<Kd<20 μm (potential binders), and none with Kd >20 μm (true negative). In contrast, S2, which is characterized with Bpidi and Cpidi<0.5, is populated mainly with negative interactions (Fig. 3A). Thus, the SH2-pY pairs with Bpidi and Cpidi>0.5 represented true positive interactions with high confidence.
Fig. 3.
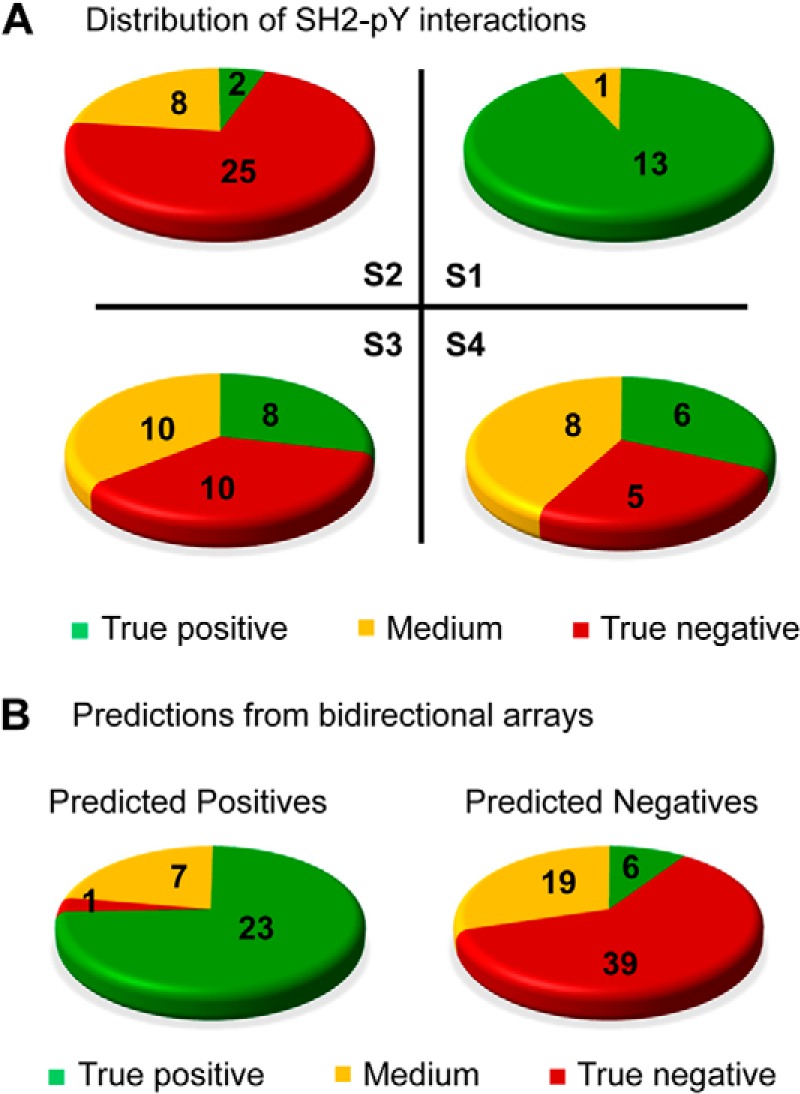
Bidirectional array screens accurately differentiated positive from negative interactions. A, Distribution of the SH2-CD22_pY pairs in squares S1 to S4 based on in-solution Kd values. Green indicates high affinity (Kd<5 μm, true positive); red low affinity (Kd>20 μm, true negative), and yellow medium affinity (5 μm< Kd<20 μm). B, The bidirectional arrays reliably differentiated positive SH2-pY interactions from negative ones when the Bpidi and Cpidi values were combined with a “rescue” scheme described in the main text.
The predictive accuracy fell drastically for SH2-pY pairs with Cpidi <0.5. For example, square S3 contains eight pairs with Kd<5 μm, 10 with 5 μm<Kd<20 μm, 10 with Kd>20 μm. To rescue the true positive interactions from this latter pool, interactions occupying square S1 were taken as the “gold standards” for positive interactions. Moreover, if an SH2 domain or pY peptide produced an array score (Xpidi or Ypidi) greater than the average value for the pool of gold-standard interactions, the corresponding SH2-pY interaction was considered positive and thus “rescued”. Using this strategy, we were able to rescue 10 out of the 16 SH2-pY pairs with Kd<5 μm and 6 out of the 26 pairs with 5 μm <Kd<20 μm from squares S2, S3 and S4. Importantly, only 1 of the 40 pairs with Kd>20 μm was falsely identified as a positive (Fig. 3B, left panel). When the interactions with B and C values above the cut-off (i.e. B>0.5; C>0.5) are combined with the “rescued” ones, the bidirectional array screening predicted 31 positive interactions of which 23 are true positives (Kd<5 μm), 7 with 5 μm <Kd<20 μm, and 1 with Kd>20 μm (corresponding to a false identification rate of 3%). It also identified 39 true negatives (Kd>20 μm) with a 10% (6/64) false identification rate (Fig. 3B).
A High-confidence SH2-CD22_pY Interaction Network Obtained from Bidirectional Peptide–protein Array Screening
Combining the bidirectional array with in-solution binding assay, we were able to identify a high-confidence interaction network for CD22. This network not only recapitulated a number of interactions reported in the literature, but also uncovered many novel ones (Fig. 4A; supplemental Table S4). Notably, all of the ITIM peptides, except for pY796, interacted with the tyrosine phosphatases SHP-1 (PTPN6) and SHP-2 (PTPN11), suggesting that the corresponding pTyr sites mediate inhibitory signaling (6). The pY796 peptide, in contrast, bound to the tyrosine kinases ABL2 and SYK, which are associated with activating signaling in B-cells (27), and to CSK, which inactivates SRC by phosphorylating a C-terminal Tyr residue in the kinase (28). The variety of different binding partners engaged by the ITIM peptides, including kinases (e.g. FYN, SYK, LCK, SRC, and ABL1/2), lipid kinases and phospholipases (e.g. PI3K and PLCG1), and adaptor proteins (e.g. SAP/SH2D1A and SHF), suggests that the function of the ITIM goes well beyond inhibitory signaling.
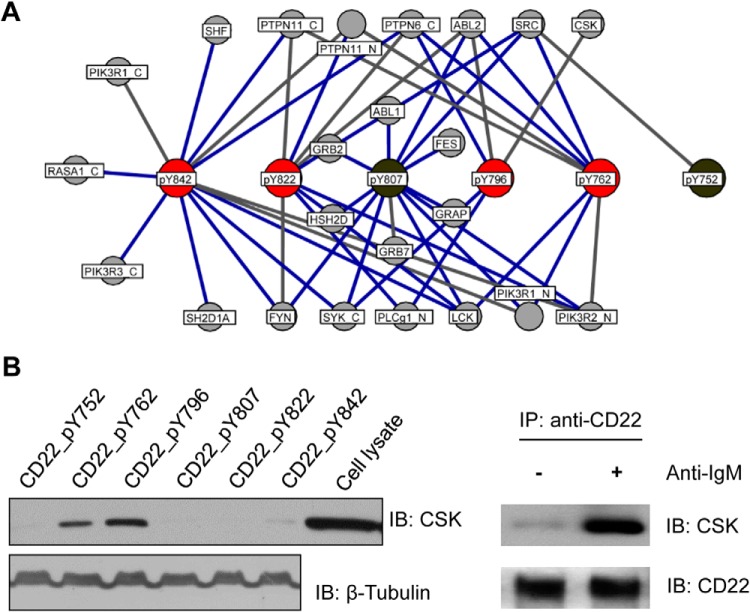
Fig. 4.
Bidirectional array screen produced a high-confidence CD22 interactome. A, A comprehensive SH2 domain-CD22_pY interactome obtained by combining the bidirectional array data with predictions based on in-solution binding Kds. ITIM sequences in CD22 are shown in red, whereas the remaining pY sites in gray. SH2 domains were identified by the corresponding protein names. Known interactions are represented by blue edges, novel ones gray edges. B, Left panel: biotinylated CD22_pY peptides were used to pull down CSK from U937 cells. Right panel: a Western blot to show that CSK and CD22 co-immunoprecipitated from BJAB cells.
It is intriguing that CD22 was capable of binding to both SRC and its negative regulator CSK (Fig. 4A). To find out whether the CSK-CD22 interaction occur in vivo, we employed biotinylated CD22_pY peptides to pull down CSK from the U937 macrophages. Indeed, CSK appeared to bind more strongly to the pY796 peptide than to the pY762 peptide (Fig. 4B). This is in agreement with their corresponding Kd values measured by fluorescence polarization (2.4 μm for CSK_SH2-pY796 and 24 μm for CSK_SH2-pY762, supplemental Table S3). Moreover, endogenous CSK co-immunoprecipitated with CD22 from BJAB cells upon B cell receptor activation following anti-IgM stimulation (Fig. 4B). Collectively, these data indicate that pY796 is the major binding site in CD22 for CSK binding. It is interesting to note that the ITIM-containing receptor PECAM-1 signals sequentially through both the SRC family kinase and CSK (29). It is likely that a similar mechanism regulates signal transduction by CD22.
Systematic Identification of SH2-ITRM Interactions
We next applied the bidirectional array approach to identify a comprehensive interaction network mediated by ITRMs. To this end, we searched the UniProt database (30) and identified 194 ITIM, ITAM, or ITSM peptides from 129 immunoreceptors (supplemental Table S5). Each peptide was synthesized as an 11-mer with 3 residues N-terminal and 7 C-terminal to the pY site. The peptides also contained an N-terminal biotin to facilitate array printing and subsequent probing of the SH2 domain array. The raw data of the reciprocal peptide-domain array screens were shown in supplemental Figs. S3 and S4, with normalized array scores reported in supplemental Table S6.
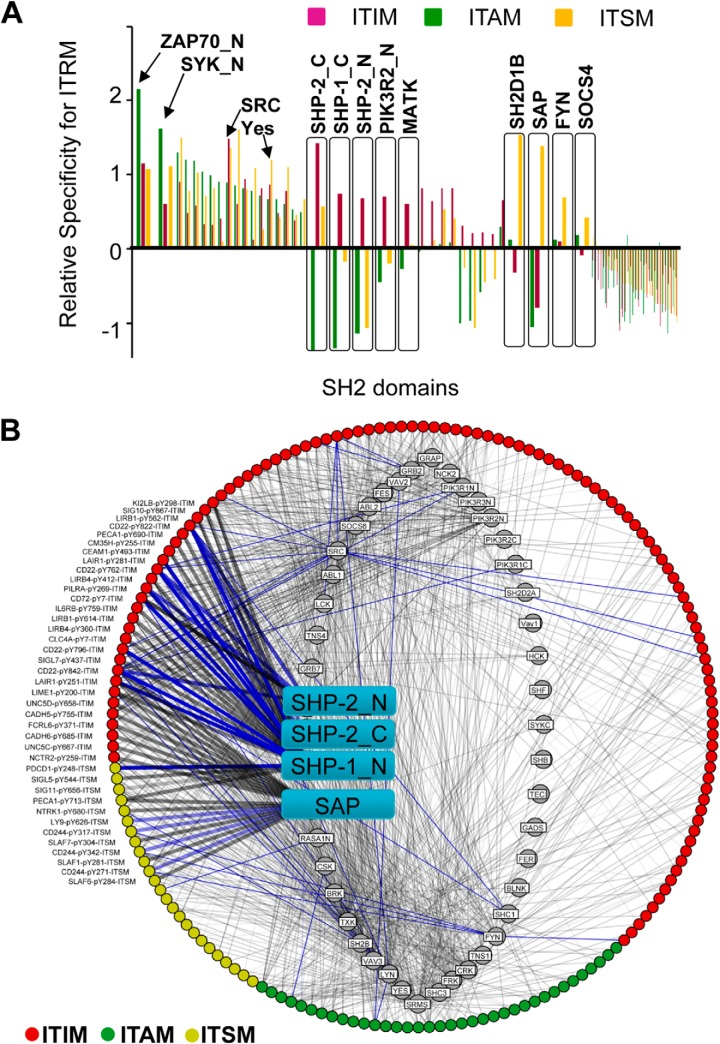
To gauge the preference of different SH2 domains for a given motif quantitatively, we calculated the corresponding Z scores (22). The relative specificity of an SH2 domain for a motif type was defined as the average Z score of that SH2 domain for the entire collection of peptides within that motif category. For example, the relative specificity of the SAP/SH2D1A SH2 domain for ITIM is calculated as its average Z score for all ITIM peptides examined. The same approach was taken to derive the relative specificity for ITAM or ITSM (supplemental Table S7). This analysis led to a comprehensive specificity map for the human SH2 domains in immunoreceptor signaling mediated by tyrosine phosphorylation (Fig. 5A).
Fig. 5.
A comprehensive SH2-ITRM interactome generated from bidirectional peptide–protein array screens. A, The specificity profiles of individual SH2 domains for different motifs. The average Z scores of an SH2 domain for ITIM, ITAM or ITSM peptides obtained from bidirectional array screens were used to measure the relative specificity of the SH2 domain. Green bars represent the relative specificity for ITAM, red for ITIM, and yellow for ITSM. A larger relative specificity value corresponds to a greater preference for that ITRM. B, A predicted human ITRM-SH2 domain interactome based on bidirectional array screens. ITIMs are shown in red, ITAMs in green and ITSMs in yellow. The interactome contains 1137 binary interactions mediated by 46 SH2 domains and 173 pY peptides. Known interactions extracted from STRING database (30) are identified by blue lines. Gray lines denote novel candidate interactions. Strength of binding was drawn proportionate to the line thickness. The SAP and SHP-1/2 SH2 domains are identified in cyan squares.
We found that different SH2 domains have distinct specificity profiles. As expected, the SH2 domains of ZAP70 and SYK, tyrosine kinases mediating positive signaling in T and B cells respectively (7), exhibited the greatest relative specificity for ITAM. In contrast, the SHP-1 and -2 SH2 domains exhibited the strongest preference for ITIM-binding. Interestingly, ITAM and ITSM sequences were excluded from binding the latter SH2 domains. These results are in excellent agreement with the literature demonstrating that SHP-1 and SHP-2 are involved in inhibitory immune response (6). Intriguingly, the MATK and PI3K SH2 domains showed the same preference for ITIM as the SHP-1/2 SH2 domains, suggesting that these proteins are associated with inhibitory signaling. Strong selectivity for ITSM was observed for the SAP/SH2D1A and its homolog SH2D1B, and to a lesser degree, for the Fyn and SOCS4 SH2 domains. Although the former three have been shown to play critical roles in signaling by the SLAM and related receptors through ITSM (4), the function of SOCS4 in immune signaling awaits further investigation.
It should be noted that the selectivity of an SH2 domain for a given motif type is not absolute. But rather, most SH2 domains are capable of binding to more than one type of motif. For example, although the ZAP70 and SYK SH2 domains have a strong preference for ITAM, they can also bind to ITIM and ITSM sequences, an observation consistent with our data on CD22 (Fig. 1). Some SH2 domains, including those from SRC and YES, are capable of binding to all three types of ITRMs without a clear proclivity.
An SH2-ITRM Interaction Network Uncovered by Bidirectional Array Screens
In order to predict authentic interactions from the reciprocal peptide–protein array screens, we calculated the bidirectional scores (B) for all SH2-ITRM peptide pairs and plotted them against the corresponding array consistency score (C). Although the majority of SH2-ITRM pairs (80%) fell in square S2 (i.e. low probability binding events), 4% fell in square S1 as high-confidence binding events (supplemental Fig. S5A, supplemental Table S6). We used the corresponding 116 pairs of interactions within S1 (supplemental Fig. S5B) as gold-standards to rescue potential interactions that fell within squares S3, S4, and S2 using the same approach as described for CD22. Combining the interactions represented by square S1 with the rescue, the bidirectional array screens identified 1137 interactions between 46 SH2 domains and 173 ITRM peptides (supplemental Fig. S6). This interactome recapitulated 130 interactions that are included in the STRING database for protein–protein interaction (31) and uncovered 1007 novel interactions. As shown in Fig. 5B, except for a few SH2 domains that have a clear preference for a given motif type, such as the selective binding of the SHP1/2 SH2 domains to ITIM and SAP to ITSM sequences, most SH2 domains are promiscuous in that they were capable of binding to two or three types of motifs (Fig. 5B, supplemental Table S8).
Regulation of Immune Signaling by SH2-ITRM Interactions
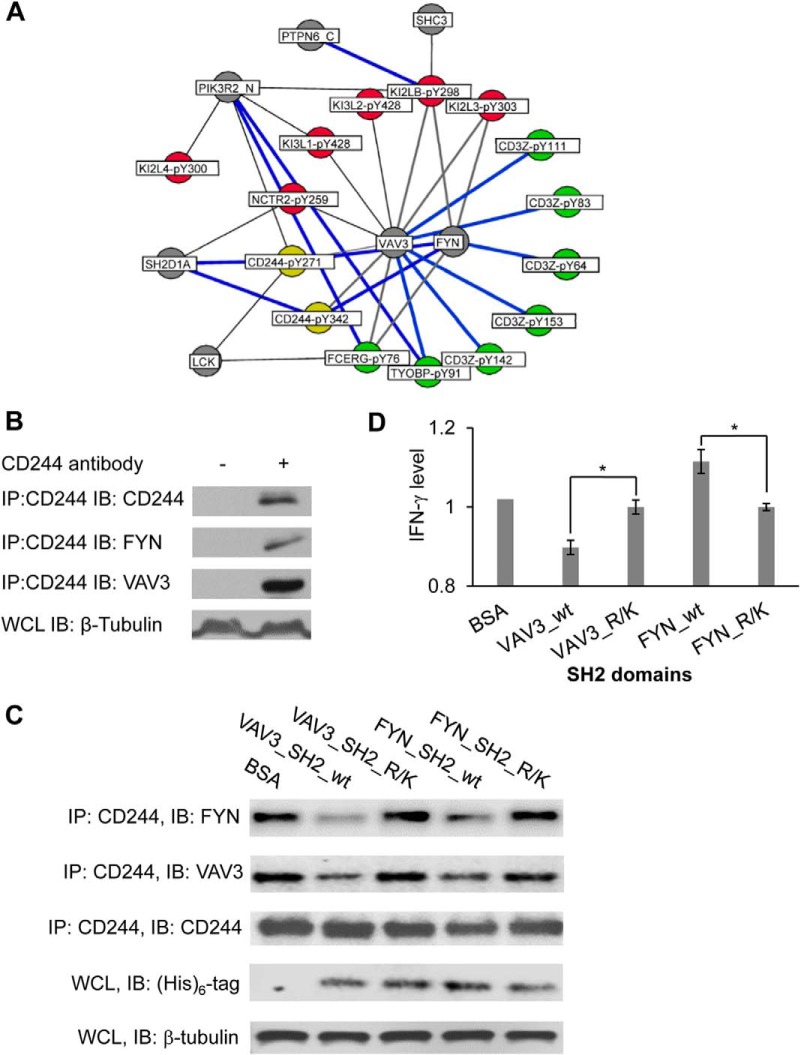
Inherent for any in vitro analysis, the interactions identified from the bidirectional array do not necessarily occur in vivo. The physiological relevance of the resulting SH2-pY interactome was analyzed in the context of known signaling databases such as the KEGG pathway database (32) that lists 34 immune signaling pathways, 15 of which involve ITRMs. The analysis associated 70 SH2-ITRM pairs to 9 immune signaling pathways (supplemental Table S9). In addition to providing site-specific information for these previously reported binding events, our systematic analysis identified numerous novel interactions between a number of receptors involved in NK cell signaling and SH2 domain-containing proteins (Fig. 6A). Our pathway analysis placed FYN and VAV3 at the center of a regulatory network in NK cell cytotoxicity (33) (Fig. 6A). The inhibitory receptor CD244 (34) is capable of binding to both the VAV3 and FYN SH2 domains, in addition to SAP/SH2D1A (4) and LCK (35). Because VAV3 and FYN shared the same binding partners, both proteins might play an important role in NK cell-mediated cytotoxicity (36).
Fig. 6.
An SH2-ITRM signaling network associated with cytotoxicity. A, The SH2-ITRM interactome was mapped onto known NK cell-mediated cytotoxicity pathways (KEGG: hsa04650) (31). ITIMs are shown in red dots, ITAMs in green, and ITSMs in yellow. Gray dots denote SH2 domains. Blue lines indicate known interactions, gray novel ones. B, CD244 bound to both VAV3 and FYN. CD244 was immunoprecipitated from the lysate of YT cells and Western blotted for VAV3 and FYN, respectively. An anti-β-tubulin blot was included as a loading control. WCL, whole cell lysate; IP, immunoprecipitation; IB, immunoblot. C, The SH2 domains disrupted endogenous interactions of VAV3 and FYN with CD244. YT cells were treated with 10 μm AuNP loaded with the wild-type FYN or VAV3 SH2 domain or a binding-deficient (R/K) mutant. The cells were subjected to anti-CD244 immunoprecipitation followed by Western blotting for associated VAV3 or FYN. An anti-(His)6 IB was used to show the amount of transduced SH2 domains. D, The VAV3 SH2 domain significantly inhibited, whereas the FYN SH2 domain promoted, the secretion of interferon gamma (IFN-γ). The INF-γ levels shown are normalized values against that in AuNP-BSA-treated YT cells, set as 1. *, p < 0.01, n = 3.
To test this assumption, we first confirmed that both VAV3 and FYN were able to bind to CD244 by co-immunoprecipitation from YT, a natural killer cell line (Fig. 6B). Because the SH2 domain mediated the binding of VAV3 or FYN to CD244, we predicted that an isolated SH2 domain would disengage endogenous VAV3 or FYN with CD244, and thereby disabling their normal functions in cytotoxicity. To test this, recombinant VAV3 or FYN SH2 domain or the corresponding binding-deficient mutant (i.e. Fyn_SH2 R176K or VAV3_SH2 R697K) (37) was transduced into YT cells using gold nanoparticles (AuNP) (38). The transduction efficiency of AuNP was verified with fluorescein-labeled BSA (supplemental Fig. S7). The wild-type (WT) SH2 domains, but not the binding-deficient mutants, were capable of blocking interactions of CD244 with VAV3 or FYN (Fig. 6C). The cytotoxicity of the YT cells transduced with a WT or mutant SH2 domain was measured in the presence of the K562 target cells following stimulation by an anti-CD244 antibody (c1.7) (21). Compared with the corresponding mutant, the VAV3_SH2 domain significantly inhibited, whereas the FYN_SH2 domain significantly stimulated IFN-γ secretion (Fig. 6D). The stimulatory function of the VAV3_SH2 domain may be attributed to the observation that it blocked CD244 interaction not only with VAV3, but also with FYN, thereby potentially disrupting the CD244-SAP-FYN complex (39). However, the stimulatory effect of the FYN_SH2 domain on IFN-γ secretion was intriguing as it also partially blocked the endogenous VAV3-CD244 interaction (Fig. 6C). Because numerous other proteins are capable of binding to VAV3 and/or FYN (Fig. 6A), the outcome of NK cell signaling is likely dependent on the particular SH2-ITRM sub-network formed under a given condition.
DISCUSSION
A Bidirectional Array Strategy for Systematic Identification of Protein–Protein Interactions Mediated by Peptide-binding Domains
Protein–protein interaction (PPI) plays a central role in normal cell biology (40). However, interactions driven by post-translational modifications such as phosphorylation, which comprises a significant part of the PPI network, have proven difficult to decipher systematically by high throughput methods such as yeast two-hybrid (15) and AP-MS (16). Although systematic yeast two-hybrid screen has been applied to human open-reading frames (ORFs) (15), it is not suitable for deciphering interactions mediated by phosphorylation. AP-MS, a widely used method for PPI mapping involving phosphorylation, is limited by the transient nature of most phosphorylation events occurring in the cell. Moreover, AP-MS analysis, in most cases, neither provides information on direct physical interactions between the probe and the bait proteins, nor does it identify the binding sites. In contrast, peptide and protein arrays, which are not only amenable to PTM-mediated interactions (41, 42), but also semiquantitative and high throughput. Although either peptide or protein arrays may be used in systematic PPI identification in vitro (42, 43), the combined use has rarely been attempted. We found that peptide and protein array screens produced inconsistent data for the SH2 domain-ITRM peptide interactions. Although it is difficult to pinpoint the exact cause of this intriguing observation, the immobilization of a short phosphopeptide onto a solid support may restrict its freedom of motion or accessibility to an SH2 domain. By the same token, the immobilization of an SH2 domain by simple surface absorption may alter its conformation and thereby binding activity. Because of the sensitivity of the detection method (which was based on fluorescence), subtle changes in the physiochemical properties of the phosphopeptide and/or SH2 domain could be amplified as large differences in apparent binding signals.
To overcome this limitation, we combined the binding signals from the peptide and SH2 domain array screens and used the average signal as a measurement for the corresponding SH2-ITRM interaction. This simple manipulation produced significantly improved correlation between the peptide–protein bidirectional array data and in-solution binding affinity. Moreover, we developed an approach to “rescue” interactions that are likely authentic but exhibited large variance between the peptide and the protein array screens. This approach, when applied to the 120 human SH2 domains (of which 78 were purified to homogeneity suitable for the array analysis) and the majority of ITRM sequences contained in immunoreceptors, yielded a comprehensive, high-confidence network of immune signaling mediated by tyrosine phosphorylation. It should be cautioned, however, that interactions identified by our in vitro approach may or may not occur in vivo. Although the physiological relevance of the majority of interactions within this network awaits validation, our data provides a framework on which to systematically explore the functions of Tyr phosphorylation in immune signaling. It can be envisioned that the same approach may be used to identify protein–protein interactions mediated other peptide-binding modules in a systematic and unbiased manner (44).
Specificity and Promiscuity of Immune Signaling via the SH2 Domain
Tyr-based regulatory motifs and the associated SH2 domains play a critical role in immune response. Of the three types of ITRMs, ITIM is generally linked to inhibitory whereas ITAM to stimulatory immune signaling. However, this distinction is far from being absolute. For instance, some ITAMs recruit SHP-1, a phosphatase found associated usually with ITIMs in inhibitory immune response (45). Conversely, certain ITIMs have been shown to propagate activation signals (46). Our work provides support to the notion that a given ITRM can assume multiple roles during immunoreceptor signal transduction depending on the specific SH2 domain proteins that it recruits. Apart from the strong motif-selectivity exhibited by a limited number of SH2 domains, most SH2 domains bind to ITRM sequences with a high degree of degeneracy. This latter group includes SH2 domains from TXK, SRC, and ABL1/2 (supplemental Fig. S6).
Our interactome data suggests that it is likely that ITIMs may mediate positive and ITAM negative immune signaling under certain conditions. For instance, in addition to binding SHP-1 (24), the CD22_pY762 ITIM peptide was found to bind SH2 domains of the tyrosine kinases SRC, LCK, and ABL2. In contrast, although most ITAMs were found to associate with tyrosine kinases, some, including those from CLC7A and FLT3, were capable of binding to tyrosine phosphatases SHP-1 or SHP-2 (supplemental Fig. S6, supplemental Table S8).
Because how cells respond to an immune stimulus is dependent on the activation of specific receptors, which may contain multiple ITRMs (47), the SH2-ITRM interactome identified herein provides a valuable resource for understanding signal integration in immune regulation. Our data suggests that the whether a specific ITRM convey an activating or inhibitory signal is not only dependent on the residues flanking the pY site, but more importantly, it is dictated by the downstream SH2 domain-containing effectors recruited by the motif. Although the high degree of degeneracy in the SH2-ITRM interaction network casts a rather promiscuous view, specificity of immune signaling may be enhanced by the activation of specific receptors in different cells, the formation of specific protein complexes facilitated by scaffolding proteins (48), or by spatial restrains such as those found in immunological synapses or membrane microdomains (49).
In summary, we have shown that integration of signals from peptide and protein arrays greatly improve the quality of the identified PPI network. Using the bidirectional array approach, we identified the largest immunoreceptor signaling network mediated by phosphotyrosine to date. Numerous novel interactions have been identified, which would provide a valuable resource for the cell signaling community. Moreover, our interactome data could guide the design of experiments to interrogate the mechanism of immune regulation and the development of inhibitors for disease intervention. We expect that the bidirectional array strategy to play an important part in the identification of the functional human proteome, particularly protein–protein interactions mediated by peptide-binding domains.
Supplementary Material
Acknowledgments
We thank Dr. David Schibli for technical assistance.
Footnotes
Author contributions: H.L. and S.S.L. designed research; H.L. and C.V. performed research; F.W. and J.L. contributed new reagents or analytic tools; H.L., L.L., F.W., J.L., and S.S.L. analyzed data; H.L. and S.S.L. wrote the paper.
* This work was supported, in part, by grants from the Canadian Cancer Society Research Institute and from the Ontario Research Fund. SSCL holds a Canada Research Chair in Functional Genomics and Cellular Proteomics.
 This article contains supplemental Figs. S1 to S7 and Tables S1 to S9.
This article contains supplemental Figs. S1 to S7 and Tables S1 to S9.
1 The abbreviations used are:
- ITRM
- immunoreceptor tyrosine-based regulatory motifs
- ITAM
- immunoreceptor tyrosine-based activation motifs
- ITIM
- immunoreceptor tyrosine-based inhibition motifs
- ITSM
- immunoreceptor tyrosine-based switch motifs.
REFERENCES
- 1. Guy C. S., Vignali K. M., Temirov J., Bettini M. L., Overacre A. E., Smeltzer M., Zhang H., Huppa J. B., Tsai Y. H., Lobry C., Xie J., Dempsey P. J., Crawford H. C., Aifantis I., Davis M. M., Vignali D. A. (2013) Distinct TCR signaling pathways drive proliferation and cytokine production in T cells. Nat. Immunol. 14, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Leeuwen J. E., Samelson L. E. (1999) T cell antigen-receptor signal transduction. Current Opinion Immunol. 11, 242–248 [DOI] [PubMed] [Google Scholar]
- 3. Ravetch J. V., Lanier L. L. (2000) Immune inhibitory receptors. Science 290, 84–89 [DOI] [PubMed] [Google Scholar]
- 4. Dong Z., Veillette A. (2010) How do SAP family deficiencies compromise immunity? Trends Immunol. 31, 295–302 [DOI] [PubMed] [Google Scholar]
- 5. Ostrakhovitch E. A., Li S. S. (2006) The role of SLAM family receptors in immune cell signaling. Biochem. Cell Biol. 84, 832–843 [DOI] [PubMed] [Google Scholar]
- 6. Steevels T. A., Meyaard L. (2011) Immune inhibitory receptors: essential regulators of phagocyte function. Eur. J. Immunol. 41, 575–587 [DOI] [PubMed] [Google Scholar]
- 7. Bezbradica J. S., Medzhitov R. (2012) Role of ITAM signaling module in signal integration. Current Opinion Immunol. 24, 58–66 [DOI] [PubMed] [Google Scholar]
- 8. Hamerman J. A., Lanier L. L. (2006) Inhibition of immune responses by ITAM-bearing receptors. Science's STKE 2006, re1. [DOI] [PubMed] [Google Scholar]
- 9. Pumphrey N. J., Taylor V., Freeman S., Douglas M. R., Bradfield P. F., Young S. P., Lord J. M., Wakelam M. J., Bird I. N., Salmon M., Buckley C. D. (1999) Differential association of cytoplasmic signalling molecules SHP-1, SHP-2, SHIP and phospholipase C-gamma1 with PECAM-1/CD31. FEBS Lett. 450, 77–83 [DOI] [PubMed] [Google Scholar]
- 10. Bonnerot C., Briken V., Amigorena S. (1997) Intracellular signaling and endosomal trafficking of immunoreceptors. Shared effectors underlying MHC class II-restricted antigen presentation. Immunology Lett. 57, 1–4 [DOI] [PubMed] [Google Scholar]
- 11. Daeron M., Jaeger S., Du Pasquier L., Vivier E. (2008) Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol. Rev. 224, 11–43 [DOI] [PubMed] [Google Scholar]
- 12. Verbrugge A., Rijkers E. S., de Ruiter T., Meyaard L. (2006) Leukocyte-associated Ig-like receptor-1 has SH2 domain-containing phosphatase-independent function and recruits C-terminal Src kinase. Eur. J. Immunol. 36, 190–198 [DOI] [PubMed] [Google Scholar]
- 13. Zhu M., Janssen E., Leung K., Zhang W. (2002) Molecular cloning of a novel gene encoding a membrane-associated adaptor protein (LAX) in lymphocyte signaling. The J. Biol. Chem. 277, 46151–46158 [DOI] [PubMed] [Google Scholar]
- 14. Fusaki N., Tomita S., Wu Y., Okamoto N., Goitsuka R., Kitamura D., Hozumi N. (2000) BLNK is associated with the CD72/SHP-1/Grb2 complex in the WEHI231 cell line after membrane IgM cross-linking. Eur. J. Immunol. 30, 1326–1330 [DOI] [PubMed] [Google Scholar]
- 15. Lamesch P., Li N., Milstein S., Fan C., Hao T., Szabo G., Hu Z., Venkatesan K., Bethel G., Martin P., Rogers J., Lawlor S., McLaren S., Dricot A., Borick H., Cusick M. E., Vandenhaute J., Dunham I., Hill D. E., Vidal M. (2007) hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics 89, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brettner L. M., Masel J. (2012) Protein stickiness, rather than number of functional protein-protein interactions, predicts expression noise and plasticity in yeast. BMC Syst. Biol. 6, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim T., Tyndel M. S., Huang H., Sidhu S. S., Bader G. D., Gfeller D., Kim P. M. (2012) MUSI: an integrated system for identifying multiple specificity from very large peptide or nucleic acid data sets. Nucleic Acids Res. 40, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Melton L. (2004) Protein arrays: proteomics in multiplex. Nature 429, 101–107 [DOI] [PubMed] [Google Scholar]
- 19. Huang H., Li L., Wu C., Schibli D., Colwill K., Ma S., Li C., Roy P., Ho K., Songyang Z., Pawson T., Gao Y., Li S. S. (2008) Defining the specificity space of the human SRC homology 2 domain. Mol. Cell. Proteomics 7, 768–784 [DOI] [PubMed] [Google Scholar]
- 20. Liu H., Galka M., Iberg A., Wang Z., Li L., Voss C., Jiang X., Lajoie G., Huang Z., Bedford M. T., Li S. S. (2010) Systematic identification of methyllysine-driven interactions for histone and nonhistone targets. J. Proteome Res. 9, 5827–5836 [DOI] [PubMed] [Google Scholar]
- 21. Chuang S. S., Kumaresan P. R., Mathew P. A. (2001) 2B4 (CD244)-mediated activation of cytotoxicity and IFN-gamma release in human NK cells involves distinct pathways. J. Immunol. 167, 6210–6216 [DOI] [PubMed] [Google Scholar]
- 22. Aksoy S., Haralick R. M. (2001) Feature normalization and likelihood-based similarity measures for image retrieval. Pattern Recogn. Lett. 22, 563–582 [Google Scholar]
- 23. Haso W., Lee D. W., Shah N. N., Stetler-Stevenson M., Yuan C. M., Pastan I. H., Dimitrov D. S., Morgan R. A., Fitzgerald D. J., Barrett D. M., Wayne A. S., Mackall C. L., Orentas R. J. (2012) Anti-CD22-chimeric antigen receptors targeting B cell precursor acute lymphoblastic leukemia. Blood. 121, 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nitschke L. (2009) CD22 and Siglec-G: B-cell inhibitory receptors with distinct functions. Immunol. Rev. 230, 128–143 [DOI] [PubMed] [Google Scholar]
- 25. Fujimoto M., Kuwano Y., Watanabe R., Asashima N., Nakashima H., Yoshitake S., Okochi H., Tamaki K., Poe J. C., Tedder T. F., Sato S. (2006) B cell antigen receptor and CD40 differentially regulate CD22 tyrosine phosphorylation. J. Immunol. 176, 873–879 [DOI] [PubMed] [Google Scholar]
- 26. Poe J. C., Fujimoto M., Jansen P. J., Miller A. S., Tedder T. F. (2000) CD22 forms a quaternary complex with SHIP, Grb2, and Shc. A pathway for regulation of B lymphocyte antigen receptor-induced calcium flux. J. Biol. Chem. 275, 17420–17427 [DOI] [PubMed] [Google Scholar]
- 27. Tolar P., Hanna J., Krueger P. D., Pierce S. K. (2009) The Constant Region of the Membrane Immunoglobulin Mediates B Cell-Receptor Clustering and Signaling in Response to Membrane Antigens. Immunity 30, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bjorge J. D., Jakymiw A., Fujita D. J. (2000) Selected glimpses into the activation and function of Src kinase. Oncogene 19, 5620–5635 [DOI] [PubMed] [Google Scholar]
- 29. Tourdot B. E., Brenner M. K., Keough K. C., Holyst T., Newman P. J., Newman D. K. (2013) Immunoreceptor tyrosine-based inhibitory motif (ITIM)-mediated inhibitory signaling is regulated by sequential phosphorylation mediated by distinct nonreceptor tyrosine kinases: a case study involving PECAM-1. Biochemistry 52, 2597–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Apweiler R., Martin M. J., O'Donovan C., Magrane M., Alam-Faruque Y., Antunes R., Barrell D., Bely B., Bingley M., Binns D., Bower L., Browne P., Chan W. M., Dimmer E., Eberhardt R., Fazzini F., Fedotov A., Foulger R., Garavelli J., Castro L. G., Huntley R., Jacobsen J., Kleen M., Laiho K., Legge D., Lin Q. A., Liu W. D., Luo J., Orchard S., Patient S., Pichler K., Poggioli D., Pontikos N., Pruess M., Rosanoff S., Sawford T., Sehra H., Turner E., Corbett M., Donnelly M., van Rensburg P., Xenarios I., Bougueleret L., Auchincloss A., Argoud-Puy G., Axelsen K., Bairoch A., Baratin D., Blatter M. C., Boeckmann B., Bolleman J., Bollondi L., Boutet E., Quintaje S. B., Breuza L., Bridge A., deCastro E., Coudert E., Cusin I., Doche M., Dornevil D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gehant S., Ferro S., Gasteiger E., Gateau A., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Hulo N., James J., Jimenez S., Jungo F., Kappler T., Keller G., Lara V., Lemereier P., Lieberherr D., Martin X., Masson P., Moinat M., Morgat A., Paesano S., Pedruzzi I., Pilbout S., Poux S., Pozzato M., Redaschi N., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stanley E., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A. L., Wu C. H., Arighi C. N., Arminski L., Barker W. C., Chen C. M., Chen Y. X., Dubey P., Huang H. Z., Mazumder R., McGarvey P., Natale D. A., Natarajan T. G., Nchoutmboube J., Roberts N. V., Suzek B. E., Ugochukwu U., Vinayaka C. R., Wang Q. H., Wang Y. Q., Yeh L. S., Zhang J. A., Consortium U. (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39, D214-D219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Du J., Yuan Z., Ma Z., Song J., Xie X., Chen Y. (2014) KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol. Biosyst. 10, 2441–2447 [DOI] [PubMed] [Google Scholar]
- 33. Bloch-Queyrat C., Fondaneche M. C., Chen R., Yin L., Relouzat F., Veillette A., Fischer A., Latour S. (2005) Regulation of natural cytotoxicity by the adaptor SAP and the Src-related kinase Fyn. J. Exp. Med. 202, 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clarkson N. G., Simmonds S. J., Puklavec M. J., Brown M. H. (2007) Direct and indirect interactions of the cytoplasmic region of CD244 (2B4) in mice and humans with FYN kinase. J. Biol. Chem. 282, 25385–25394 [DOI] [PubMed] [Google Scholar]
- 35. Clarkson N. G., Brown M. H. (2009) Inhibition and activation by CD244 depends on CD2 and phospholipase C-gamma1. J. Biol. Chem. 284, 24725–24734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lowin-Kropf B., Kunz B., Schneider P., Held W. (2002) A role for the src family kinase Fyn in NK cell activation and the formation of the repertoire of Ly49 receptors. Eur. J. Immunol. 32, 773–782 [DOI] [PubMed] [Google Scholar]
- 37. Charvet C., Canonigo A. J., Billadeau D. D., Altman A. (2005) Membrane localization and function of Vav3 in T cells depend on its association with the adapter SLP-76. J. Biol. Chem. 280, 15289–15299 [DOI] [PubMed] [Google Scholar]
- 38. Tsai D. H., DelRio F. W., Keene A. M., Tyner K. M., MacCuspie R. I., Cho T. J., Zachariah M. R., Hackley V. A. (2011) Adsorption and conformation of serum albumin protein on gold nanoparticles investigated using dimensional measurements and in situ spectroscopic methods. Langmuir 27, 2464–2477 [DOI] [PubMed] [Google Scholar]
- 39. Lanier L. L. (2008) Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9, 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kathiriya J. J., Pathak R. R., Clayman E., Xue B., Uversky V. N., Dave V. (2014) Presence and utility of intrinsically disordered regions in kinases. Mol. Biosyst. 10, 2876–2888 [DOI] [PubMed] [Google Scholar]
- 41. Deng Y., Alicea-Velazquez N. L., Bannwarth L., Lehtonen S. I., Boggon T. J., Cheng H. C., Hytonen V. P., Turk B. E. (2014) Global analysis of human nonreceptor tyrosine kinase specificity using high-density peptide microarrays. J. Proteome Res. 13, 4339–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones R. B., Gordus A., Krall J. A., MacBeath G. (2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439, 168–174 [DOI] [PubMed] [Google Scholar]
- 43. Tong A. H. Y., Drees B., Nardelli G., Bader G. D., Brannetti B., Castagnoli L., Evangelista M., Ferracuti S., Nelson B., Paoluzi S., Quondam M., Zucconi A., Hogue C. W. V., Fields S., Boone C., Cesareni G. (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295, 321–324 [DOI] [PubMed] [Google Scholar]
- 44. Kyle K., Biggar S. S.-C. L. (2014) Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. 1, 5–17 [DOI] [PubMed] [Google Scholar]
- 45. Pfirsch-Maisonnas S., Aloulou M., Xu T., Claver J., Kanamaru Y., Tiwari M., Launay P., Monteiro R. C., Blank U. (2011) Inhibitory ITAM signaling traps activating receptors with the phosphatase SHP-1 to form polarized “inhibisome” clusters. Sci. Signaling 4, ra24. [DOI] [PubMed] [Google Scholar]
- 46. Barrow A. D., Trowsdale J. (2006) You say ITAM and I say ITIM, let's call the whole thing off: the ambiguity of immunoreceptor signalling. Eur. J. Immunol. 36, 1646–1653 [DOI] [PubMed] [Google Scholar]
- 47. Van Roey K., Gibson T. J., Davey N. E. (2012) Motif switches: decision-making in cell regulation. Current Opin. Structural Biol. 22, 378–385 [DOI] [PubMed] [Google Scholar]
- 48. Pawson T., Scott J. D. (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science 278, 2075–2080 [DOI] [PubMed] [Google Scholar]
- 49. Horejsi V., Hrdinka M. (2014) Membrane microdomains in immunoreceptor signaling. FEBS Lett. 588, 2392–2397 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.