Abstract
Packaging of viral genomes inside empty procapsids is driven by a powerful ATP-hydrolyzing motor, formed in many double strand DNA (dsDNA)-viruses by a complex of a small (S-terminase) and a large terminase (L-terminase) subunit, transiently docked at the portal vertex during genome-packaging. Despite recent progress in elucidating the structure of individual terminase subunits and their domains, little is known about the architecture of an assembled terminase complex. Here, we describe a bacterial co-expression system that yields milligram quantities of the S:L-terminase complex of the Salmonella-phage P22. In vivo assembled terminase complex was affinity-purified and stabilized by addition of non-hydrolysable ATP, which binds specifically to the ATPase domain of L-terminase. Mapping studies revealed the N-terminus of L-terminase ATPase domain (residues 1-58) contains a minimal S-terminase Binding Domain (SBD) sufficient for stoichiometric association with residues 140-162 of S-terminase, the L-terminase Binding Domain (LBD). Hydrodynamic analysis by analytical ultracentrifugation sedimentation velocity and native mass spectrometry revealed the purified terminase complex consists predominantly of one copy of the nonameric S-terminase bound to two equivalents of L-terminase (1S-terminase:2L-terminase). Direct visualization of this molecular assembly in negative stained micrographs yielded a three-dimensional asymmetric reconstruction that resembles a ‘nutcracker’ with two L-terminase protomers projecting from the C-termini of a S-terminase ring. This is the first direct visualization of a purified viral terminase complex analyzed in the absence of DNA and procapsid.
Keywords: viral genome-packaging motor, Large terminase, Small terminase Bacteriophage P22, Salmonella virus, electron microscopy
Graphical Abstract
INTRODUCTION
Viral genome-packaging is a complex, non-spontaneous reaction, catalyzed in many dsDNA viruses by a powerful genome-packaging motor [1-3]. This motor consists of a portal protein, which occupies one of the vertices of the icosahedral procapsid, and a terminase complex that converts ATP hydrolysis into linear translation of dsDNA. In most tailed phages, the terminase complex is formed by two subunits known as small and large terminase (referred to as S- and L-terminase), whereas Herpesviruses also have a third subunit, of unknown function and structure [4]. The ATPase activity necessary to power genome-packaging resides in the L-terminase subunit, which binds directly to the portal protein [5]. In contrast, the S-terminase subunit binds packaging initiation sites on the double stranded DNA (dsDNA) (referred to as pac in P22 [6]) to prepare for genome packaging [7] and regulates the ATPase activity of L-terminase [8, 9]. These functions are likely to be very important in vivo to sustain the enormous rate of genome-packaging, which can be as high as ~2,000 bp/sec [10].
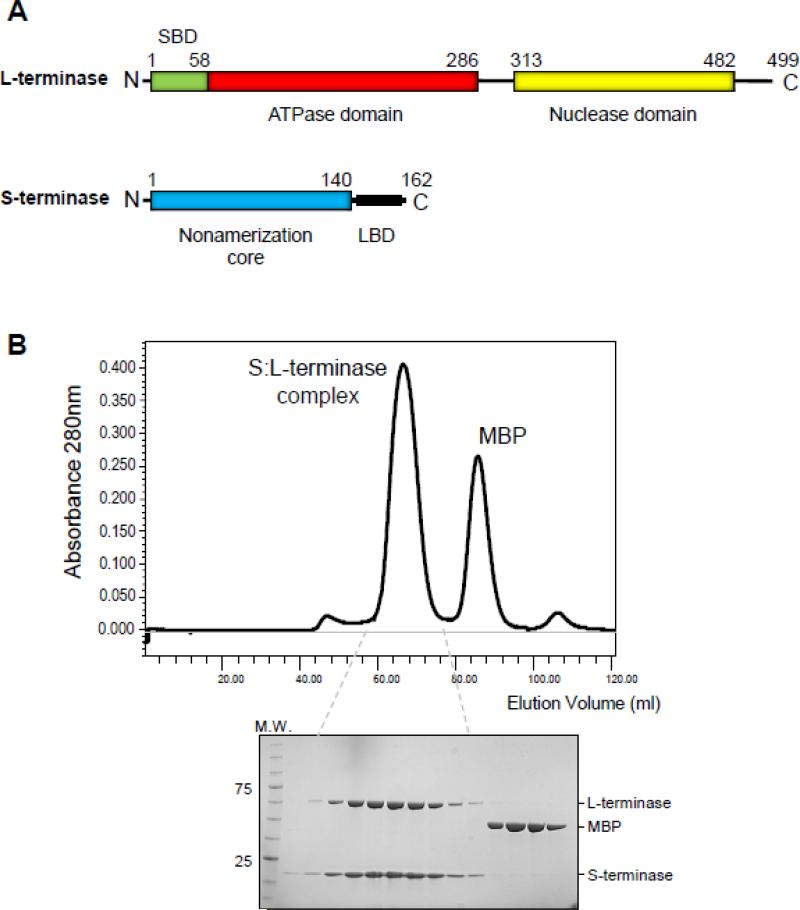
We previously characterized the L-terminase (499 amino acids, 57.6 kDa [11]) and S-terminase (162 amino acids, 18.6 kDa [7]) subunits of the bacteriophage P22 [12] (Fig. 1A), a prototypical member of the Podoviridae family of short tailed bacteriophages. In this phage, S-terminase assembles in solution and in crystals as a hollow nonamer [13-15], similar to the S-terminase of the Siphoviridae SPP1-like phage SF6 [16]. This oligomer is surprisingly different from the octameric S-terminase of phage Sf6 [17], also a Podoviridae, and the distant Myoviridae T4-like 44RR, which was determined crystallographically as a mixture of undecamer and dodecamers [18]. The way S-terminases bind to DNA varies in different viruses. In P22, all DNA-binding determinants are confined in a C-terminal basic moiety comprising residues 140-162, which also overlaps with the L-terminase Binding domain (LBD) [13] (Fig. 1A). In contrast, in phage λ S-terminase (gpNu1) [19] and possibly in Sf6 [20] and T4 [18], DNA-binding is thought to occur via an N-terminal winged helix-turn-helix motif. Unlike S-terminases that are highly divergent in sequence, structure and possibly mechanisms of DNA-binding [21], all known L-terminase subunits have an N-terminal ATPase domain [22, 23] that contains ATP-binding Walker A and B motifs, flexibly linked to a C-terminal RNAse H-fold nuclease [24, 25] also conserved in Herpesviridae [26, 27].
Figure 1. Co-expression and purification of P22 S:L-terminase complex.
(A) Domain organization of P22 S- and L-terminase subunits. (B) Chromatogram of the S:L-terminase complex purified in the presence of mild detergent, magnesium chloride and glycerol. The S:L-terminase complex was separated on a Superdex 200 gel filtration column after o/n digestion with PreScission Protease. Fractions corresponding to the eluted peaks were analyzed by SDS-PAGE (bottom gel) revealing pure S- and L-terminase subunits and free MBP.
DNA-packaging in P22 proceeds by a ‘headful packing’ mechanism, a packaging strategy where the length of the DNA encapsulated inside the procapsid is determined by the interior volume of the mature phage particle [28, 29]. The exact molecular mechanisms by which S- and L-terminase subunits orchestrate headful packaging are poorly understood. It was reported that P22 S- and L-terminase form a complex that can be purified from infected cells [30], but the stoichiometry of this terminase complex is unknown. Genetic evidence has shown that the DNA-recognition subunit, S-terminase, binds to packaging initiation sites (pac) [31] in the P22 genome and positions viral dsDNA for the packaging L-terminase subunit, which uses ATP hydrolysis to translocate a single genome copy into an empty procapsid. The substrate for DNA-packaging in P22 is a repeating polymer containing up to 10 copies of phage genome, known as concatemer [32]. To facilitate packaging into empty P22 procapsid, the L-terminase is thought to oligomerize at the portal vertex, as observed in T4 [22] and T7 [33]. The nuclease domain of L-terminase cleaves concatemeric dsDNA at two stages of the packaging reaction. At the beginning of packaging, the terminase makes sequence-specific cleavages in the pac region (referred to as “series initiation cleavage”) to generate a DNA end and initiate a packaging series. The DNA end is then inserted into the procapsid unidirectionally from the initiation cleavage point in an ATP-dependent process catalyzed by the ATPase domain of L-terminase. Upon insertion typically between 102% and 110% of the genome length [34], the headful nuclease of L-terminase cleaves the DNA, releasing the concatemer from the newly filled particle and resulting in dissociation of the terminase complex from the capsid. This enables binding of the tail proteins gp4 [35-39], gp10 [40] and gp26 [40-42] that seal the portal protein and stabilize the genome inside the capsid, followed by the attachment of six copies of the tailspike gp9 [43]. Subsequent packaging events follow sequentially in a processive fashion, and each round of infection results in about 2% of newly replicated particles that carry host DNA instead of the viral chromosome [44].
Despite a growing number of structures of isolated S- and L-terminase subunits, a complete view of a terminase complex is lacking. In this work, we purified the S:L-terminase complex of bacteriophage P22 and provide a structural characterization of its architecture by identifying biochemical interactions and employing hybrid structural methods.
RESULTS
Purification of a homogeneous complex of P22 S- and L-terminase subunits
Large terminases are intrinsically unstable enzymes, notoriously difficult to purify and prone to aggregation [22, 30, 33, 45, 46]. Previous attempts to reconstitute the S:L-terminase complex of bacteriophage P22 from purified nonameric S-terminase and monomeric L-terminase yielded a heterogeneous mixture [13]. As an alternative approach, we formed the terminase complex in vivo by co-expressing a plasmid encoding Maltose Binding Protein (MBP)-tagged S-terminase and untagged L-terminase in bacteria (Fig. 1B), followed by purification of the S:L-terminase complex on amylose beads. The bead-immobilized complex was then incubated with 1 mM AMP-PNP to stabilize the ATPase domain of L-terminase [30] and MBP cleaved off using PreScission Protease. We found that including traces of the non-ionic detergent n-Dodecyl-β-D-maltoside (DDM) during cell lysis, in addition to magnesium chloride and 5% glycerol during purification greatly reduced the tendency of terminase subunits to aggregate. The purified S:L-terminase complex migrated on a size exclusion chromatography (SEC) column as a monodisperse species with estimated molecular mass of ~300 kDa and was >90% pure by SDS-PAGE (Fig. 1B). Unlike individual terminase subunits that are highly susceptible to proteolysis [25], the complex remained stably assembled for days to weeks.
Solution biophysical analysis
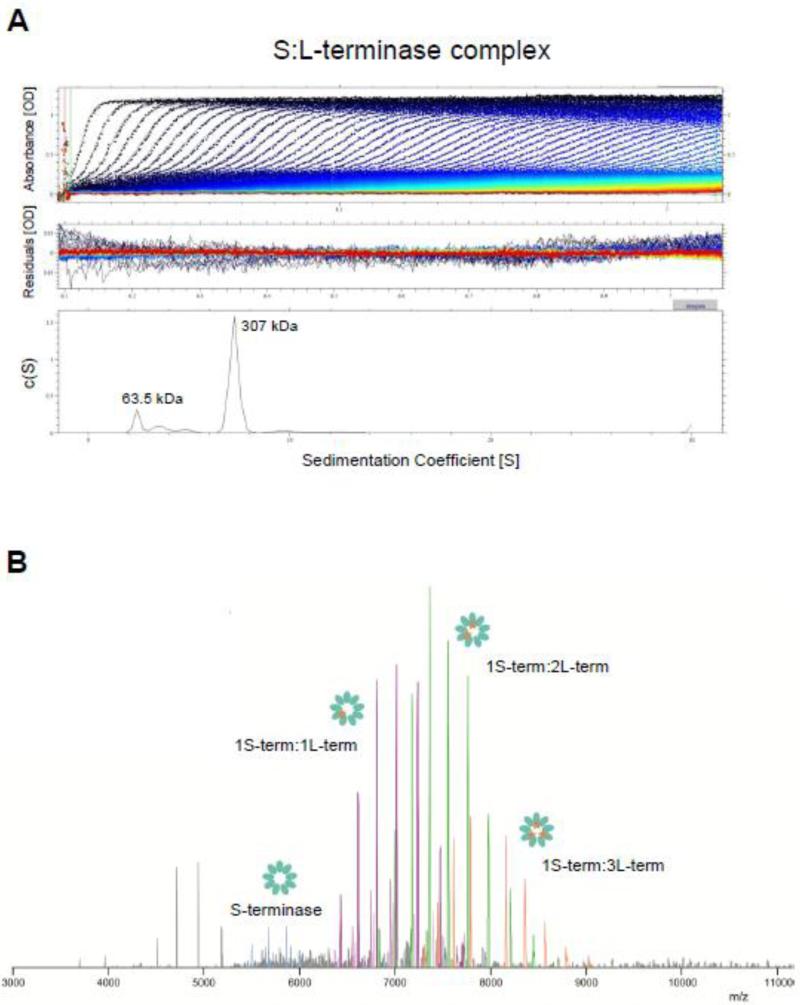
To investigate the oligomeric state of the purified S:L-terminase complex, we subjected this species to analytical ultracentrifugation (AUC) sedimentation velocity analysis. Fig. 2A shows a typical sedimentation profile of P22 terminase complex obtained in 150 mM sodium chloride, at 10°C. In a range of concentration between 1-10 μM, the complex migrated as a largely homogeneous species characterized by one major component with an apparent sedimentation coefficient (s*) of 7.2S (absolute sedimentation coefficient, S20,w = 9.6S). Conversion of this parameter to molecular mass revealed a M.W. ~307 ± 0.5 kDa possibly corresponding to a nonamer of S-terminase (~176 kDa) bound to 2 copies of L-terminase:AMP-PNP (~176 kDa + 2 × (55.9 + 0.5) = 288.8 kDa) or 3 copies of L-terminase:AMP-PNP (~176 kDa + 3 × (55.9 + 0.5) = 344.3 kDa). Furthermore, the frictional ratio estimated based on sedimentation data was f/fo ~1.7, suggestive of an elongated molecular assembly. A smaller species (< 15% of the total sample) was also observed with a sedimentation coefficient consistent with free L-terminase, possibly resulting from complex dissociation during centrifugation.
Figure 2. Biophysical analysis of the purified S:L-terminase complex.
(A) Sedimentation velocity profiles of the S:L-terminase complex measured in 20 mM Tris-Cl pH 8.0, 150 mM NaCl, 3 mm DTT, 5% glycerol, 1 mM MgCl2 at 10 °C. Top panel: raw absorbance at 280 nm plotted as a function of the radial position. Data at intervals of 20 min are shown as dots for sedimentation at 35,000 rpm. Middle panel: the residuals between fitted curve and raw data. Bottom panel: the fitted distribution of the apparent sedimentation coefficient (s*) calculated for S:L-terminase is 7.2S (~85% sample) and 2.7S (<15% sample) corresponding to an estimated molecular mass of ~307 kDa and ~63.5 kDa, respectively. (B) Native MS analysis of the S:L-terminase complex. The following masses are observed: 1S-terminase:1L-terminase: 231.465 ± 0.002 kDa (purple peaks); 1S-terminase:2L-terminase: 286.989 ± 0.012 kDa (green peaks); 1S-terminase:3L-terminase: 342.514 ± 0.008 kDa (orange peaks); nonamer of S-terminase: 175.955 ± 0.002 kDa (magenta peaks).
The same purified S:L-terminase complex was also subjected to native mass spectrometry analysis. In agreement with AUC, we observed charge distributions corresponding to the mass of a single S:L-terminase complex. No larger aggregated species were observed. The spectrum at m/z 6500-9000 in figure 2B originates from one nonamer of the S-terminase bound to different numbers of copies of L-terminase. Up to 3 copies of L-terminase were confidently assigned binding to one S-terminase nonamer, based on accurate mass measurement made possibly by using the Orbitrap EMR platform [47]. Nonameric S-terminase and free L-terminase were also observed in the MS, at low abundance (< 20%), possibly due to in-source dissociation during electrospray ionization. Thus, the predominant S:L-terminase complex assembled in vivo and purified in vitro consists of a nonameric S-terminase ring bound to 2 to 3 copies of L-terminase.
Stoichiometric binding of L-terminase to S-terminase LBD
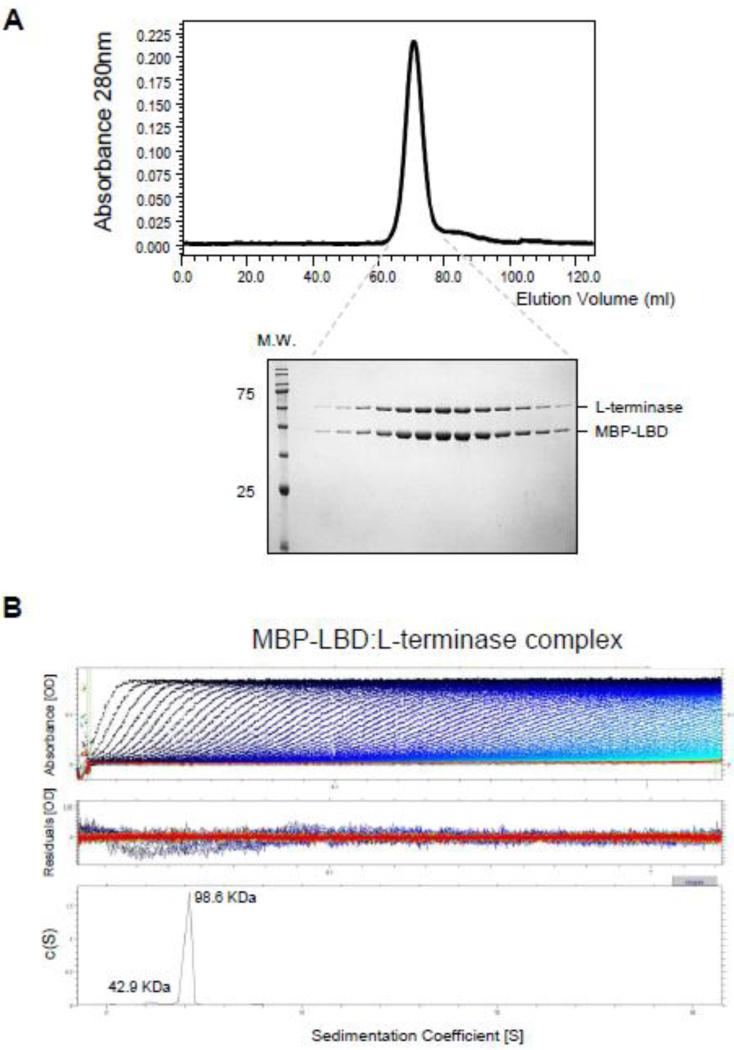
The substoichiometric presence of L-terminase in the terminase complex, and the small size of S-terminase LBD (~22 residues) [13], prompted us to investigate if each L-terminase associates with multiple LBDs in the context of an assembled terminase complex. To address this question, we co-expressed in bacteria untagged L-terminase with MBP-tagged LBD (MBP-LBD) and purified milligram quantity of the complex using amylose beads. By SEC, the MBP-LBD:L-terminase complex migrated as a homogeneous species of ~100 kDa (Fig. 3A), consistent with a heterodimer of the two proteins in 1:1 stoichiometry. A more accurate quantification was obtained by AUC-SV, which gave an apparent sedimentation coefficient (s*) for the complex of 4.2S (absolute sedimentation coefficient, S20,w = 4.4S) (Fig. 3B), corresponding to a mass of 98.6 kDa, unambiguously consistent with one copy of L-terminase:AMP-PNP bound to one copy of MBP-LBD (expected M.W. ~ 98.4 kDa). AUC studies also revealed the MBPLBD remains monomeric in solution at all concentrations tested (data not shown). Thus each copy of L-terminase in the S:L-terminase complex is likely to associate with only one LBD.
Figure 3. Stoichiometric binding of L-terminase to the LBD.
(A) Chromatogram of the purified MBD-LBD:L-terminase complex eluted with 10 mM maltose from amylose beads and separated on a Superdex 200 gel filtration column. Fractions corresponding to the eluted peak were analyzed by SDS-PAGE (bottom gel) revealing a 1:1 stoichiometry of association between L-terminase and MBP-LBD. (B) Sedimentation velocity profiles of the MBP-LBD:L-terminase complex measured in 20 mM Tris-Cl pH 8.0, 150 mM NaCl, 3 mm DTT, 5% glycerol, 1 mM MgCl2 at 10 °C. Top panel: raw absorbance at 280 nm plotted as a function of the radial position. Data at intervals of 20 min are shown as dots for sedimentation at 40,000 rpm. Middle panel: the residuals between fitted curve and raw data. Bottom panel: the fitted distribution of the apparent sedimentation coefficient (s*) calculated for S:L-terminase is 4.2S (~90% sample) and 2.7S (<10% sample) corresponding to an estimated molecular mass of ~98.6 kDa and ~42.9 kDa, respectively.
Biochemical mapping of domains involved in terminase subunits association
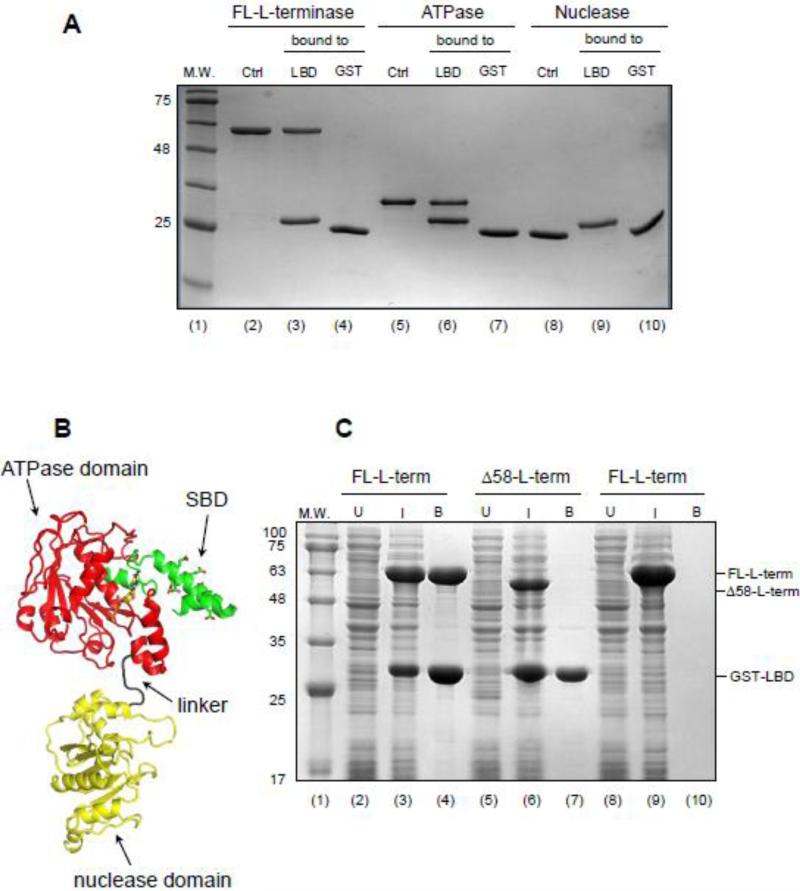
We used an on-bead assay to determine the domain of L-terminase that associates with S-terminase. GST-tagged LBD (GST-LBD) was immobilized on glutathione beads and used to pull down either the full length L-terminase (FL-L-terminase) or individually purified ATPase or nuclease domains. The ATPase domain alone associated specifically with GST-LBD with comparable avidity as the FL-L-terminase (Fig. 4A, lane 6 and 3), but failed to bind to glutathione beads not coupled to LBD (Fig. 4A, lane 7), suggesting that all binding determinants in L-terminase necessary for S-terminase assembly are confined in the N-terminal, ATPase domain. In contrast, no binding was observed between L-terminase nuclease domain and GST-LBD (Fig. 4A, lane 9), ruling out the involvement of this domain in terminase subunit assembly.
Figure 4. The N-terminus of L-terminase ATPase domain binds specifically to the LBD.
(A) Pull-down assay on glutathione beads coupled to GST-LBD reveals L-terminase ATPase domain is sufficient for binding to S-terminase. (B) Ribbon diagram of a homology model of P22 L-terminase (generated using i-TASSER [67]) with S-terminase Binding Domain (SBD), ATPase and nuclease domains colored in green, red and yellow, respectively. (C) Co-expression of FL- and Δ58-L-terminase with GST-LBD in bacteria followed by pull-down on glutathione beads and SDS-PAGE analysis reveals the SBD is essential for association with the LBD.
To further map the region of L-terminase ATPase domain involved in LBD, we made the striking observation that P22 L-terminase contains an N-terminal extension of ~58 aminoacids not found in closely related P22-like phages like Sf6 [17], but conserved in phages whose S-terminase also bears a C-terminal LBD [48]. A homology-model of P22 L-terminase revealed this N-terminal extension folds into a helix-loop-helix, positioned near the ATP binding pocket, whereby the central helix is highly acidic and has propensity to form coiled-coil structures (Fig. 4B). Intrigued by the idea that this moiety in L-terminase represents a dedicated platform for binding to the highly basic S-terminase LBD, we generated a deletion construct of L-terminase lacking residues 1-58 (Δ58-L-terminase) and co-expressed it in bacteria with GST-LBD (Fig. 4C, lane 7). Unlike a positive control of FL-L-terminase (Fig. 4C, lane 4), we found no specific interaction of Δ58-L-terminase with LBD, comparable to a negative control where a lysate expressing FL-L-terminase was passed on uncoupled GST-beads (Fig. 4C, lane 10). Thus, S-terminase LBD associates with an N-terminal acidic extension of L-terminase ATPase domain that we will refer to as SBD (S-terminase Binding Domain) (Fig. 4B).
Visualization of the S:L-terminase complex by TEM
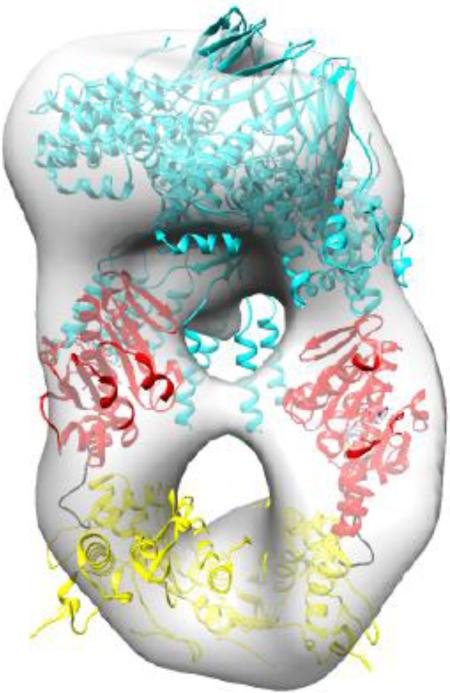
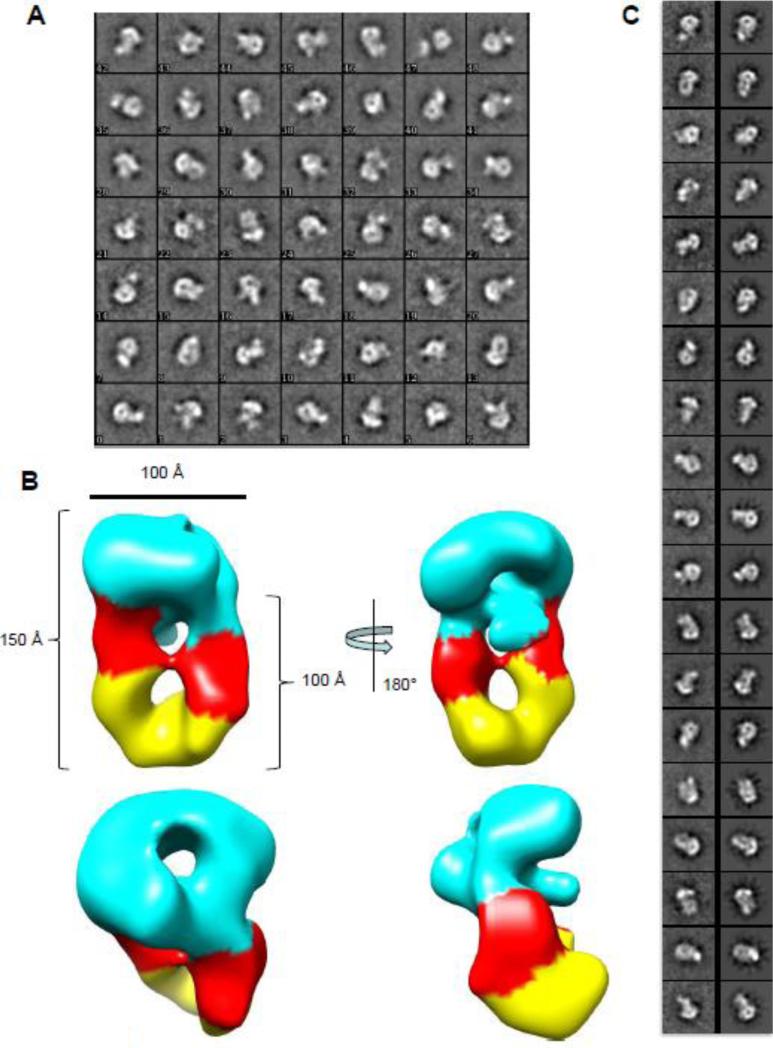
We used single particle transmission electron microscopy to analyze the S:L-terminase complex in negative stain (Fig. S1). From a data set consisting of 44 images containing 6,562 particles, analysis of the 2-D class averages via Iterative Stable Alignment and Clustering (ISAC) revealed several orientations of the S:L-terminase complex (Fig. 5A). Although some heterogeneity was observed, many class averages resemble different views of one S-terminase nonamer bound to two L-terminase molecules. A total of 2,062 particles were identified as belonging to the 1S-terminase:2L-terminase species using 3-D classification analysis with RELION. To obtain an asymmetric 3-D reconstruction, we used the crystal structure of the S-terminase nonamer low-pass filtered to 60 Å as an initial model (Fig. S2A). In addition to low-pass filtering, this model was unbiased in that it lacked density corresponding to L-terminase and comprised less than a third of the mass of the complex. On the other hand the S-terminase nanomer was clearly visible by eye in many of the individual complexes making it an excellent initial model. Because of heterogeneity seen in 2-D class averages, three classes were used during RELION 3-D classification and refinement to separate poor and broken complexes from well-aligned particles. Two classes produced non-interpretable density, while a third showed two “new” elongated densities approximately 100 Å long, not included in the starting model, and presumed to correspond to two L-terminase molecules, beneath the S-terminase ring (Fig. 5B). The L-terminase molecules were oriented with their ATPase domains adjacent to the S-terminase since biochemical analysis described above confirmed that mode of interaction. Overall, the S:L-terminase complex resembles a ‘nutcracker’, with two parallel L-terminase molecules positioned just below the mushroom shaped S-terminase. The complex is approximately 100 Å wide and 150 Å long and presents a central lumen emanating from the S-terminase nonamer, which is readily visible in the reconstruction. The resolution of this reconstruction was estimated to be 30 Å using the FSC=0.143 criterion (Fig. S2B) [49]. To validate this reconstruction, the 2D class averages produced by ISAC analysis were matched to corresponding 2D projections of the model obtained by RELION 3D classification and refinement and are shown at corresponding angles in figure 5C. This analysis revealed a striking similarity between the terminase complex visualized on grid and the model generated by RELION.
Figure 5. Details of the S:L-terminase model.
(A) Iterative Stable Alignment and Clustering (ISAC) 2D class averages reveal numerous orientations of S:L-terminase. The class identifier number appears in the lower left for each class average. Approximately 20-40 particles establish each class average. The box size for each class average is 295 Å. (B) Various views of the 3-D reconstruction of 2,062 particles obtained by RELION. Two molecules of L-terminase (with the ATPase in red and nuclease domain in yellow), are located just below the S-terminase nonamer (cyan). The dimensions of the particle components are shown. (C) ISAC 2D class averages (left) and matching 2D projections of 3D model from RELION (right). Comparisons were generated based using a correlation coefficient with e2classvsproj.py in EMAN2 [68].
A pseudo-atomic model of P22 S:L-terminase complex
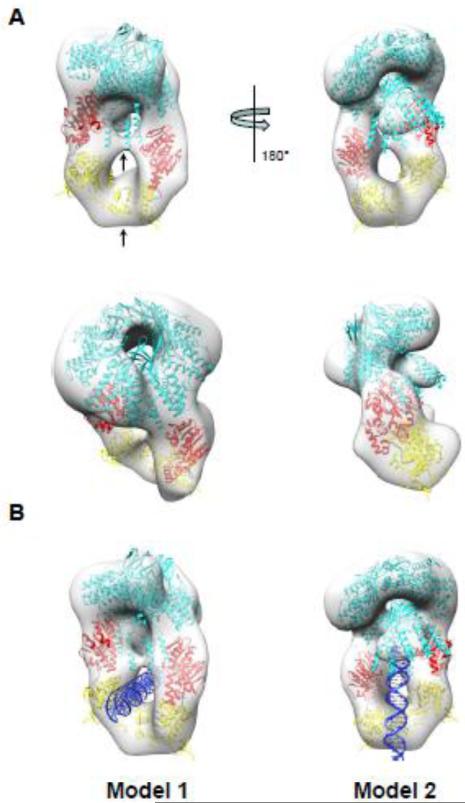
Having established the shape of P22 S:L-terminase complex (Fig. 5B) and knowing the exact domains involved in assembly (Fig. 3-4), we next generated a pseudo-atomic model of the terminase complex. A crystal structure of P22 S-terminase lacking the LBD is available [13]. We then modeled the LBD domain (res 140-162) based on secondary structure prediction as a helix. Similarly, the L-terminase subunit of bacteriophage P22 was modeled based on the ATPase domain of the close relative Sf6 [23] and the crystal structure of P22 nuclease [25] (Fig. 4B). At first, the X-ray model of nanomeric S-terminase was docked into the hollow density orienting the LBDs toward the new density features corresponding to L-terminase (Fig. 6A). In turn, two L-terminases were fit into the ellipsoid density by positioning the ATPase domain proximal to S-terminase and by slightly rotating the ATPase and nuclease domain with respect to each other. These atomic models were then refined as rigid bodies against the EM-density using Chimera [50]. In the final pseudo-atomic model, the two L-terminase subunits are not parallel but slightly twisted and contact each other at two distinct points, corresponding to ATPase domains and far C-termini of the nuclease domain (indicated by arrows in Fig. 6A).
Figure 6. Pseudo-atomic model of S:L-terminase holoenzyme.
(A) Various views of the 3-D reconstruction in Fig. 5B colored as a transparent gray surface with docked models of one nonameric S-terminase (cyan) and two L-terminase subunits with ATPase and nuclease domains colored in red and yellow, respectively. Each L-terminase has an ATPase domain (red) and a nuclease domain (yellow) separated by a short flexible linker (black). Models were rigid-body refined into density using the “Fitmap” function in Chimera [50]. (B) Models of S:L-terminase binding to dsDNA. In model 1, dsDNA passes through the lower hole between the L-terminase domains. In model 2, the dsDNA is vertically positioned between the two L-terminase domains and abuts the S-terminase C-terminal helices. In this conformation, dsDNA could fit in the S-terminase central channel.
Attempts to dock dsDNA inside the pseudo-atomic model of P22 terminase complex suggest two possible modes of binding. DNA could fit through the S-terminase hole (Fig. 6B, model 1) and still interact with both domains of the L-terminase, as previously proposed for the L-terminase of phage Sf6 [23]. Alternatively DNA could be orthogonal to the S-terminase ring going through the ‘nutcracker’ (Fig. 6B, model 2), similar to the ‘inchworm’ mechanism proposed by Sun et al. for T4 L-terminase [22] which, however, forms pentamers upon binding to procapsid. In both models, the topology of the terminase complex is such that the LBD and L-terminase can simultaneously make contacts with dsDNA.
DISCUSSION
Viral packaging motors are fundamental molecular machines that power the delivery of viral genomes into preformed procapsid shells. Despite growing interest in this field of biology [51], a plethora of individual S- and L-terminase subunit crystal structures [13, 16, 18, 20, 22, 23, 25] and recent advances in single-molecule biophysical analysis of packaging motors [52], not even moderate resolution information exists for a terminase complex. In this study, we purified an in vivo assembled complex of P22 terminase subunits and characterized its architecture using biochemical and hybrid structural methods. We found that P22 terminase subunits assemble preferentially into a stable complex containing one nonameric S-terminase bound to two L-terminase subunits (1S-terminase:2L-terminase). While an assembly containing three copies of L-terminase bound to S-terminase (1S-terminase:3L-terminase) was also observed in gas phase, this oligomer was not significantly populated in solution and on grid, suggesting a transient or unstable complex. Unexpectedly, each copy of L-terminase bound to S-terminase associates with only one LBD, suggesting the entire nonameric S-terminase is not required to bind L-terminase and LBDs helices not participating in L-terminase binding are possibly available to interact with DNA. Only two L-terminase subunits assemble stably onto S-terminase possibly due to steric hindrance between ATPase domains, although we cannot rule out that transient terminase complexes containing more than two copies of L-terminase also form in solution.
What is the functional role of the 1S-terminase:2L-terminase complex in DNA-packaging? A satisfactory answer to this question requires further analysis and will be determined by the ability of the terminase complex to bind to and package dsDNA inside empty procapsids. In analogy to phage λ, P22 1S-terminase:2L-terminase complex could represent just a protomer, able to further assemble into larger molecular complexes upon binding to DNA or upon docking to portal protein. Similarly, in λ[45, 46], the terminase subunits can be isolated as a ~114.2 kDa ‘protomer’, consisting of one L-terminase (gpA) associated with two S-terminase subunits (gpNu1), which is in slow equilibrium with a heterogeneous 13.3 S species of ~530 kDa (the ‘mix’), consisting of four protomers. In the absence of procapsid and viral DNA, polymorphic assemblies of terminase subunits are able to form because of the lack of assembly restraints dictated by other viral proteins and DNA. We observed these aggregates when P22 terminases were co-expressed and purified in the absence of DDM in lysis buffer, and glycerol and magnesium chloride were not maintained throughout the purification (data not shown). A similar polymorphic distribution of recombinant protein is observed for viral portal proteins that assemble with variable stoichiometry in vitro, but are always dodecameric in the context of the virion [53-55].
DNA-dependent stimulation of ATPase activity associated to genome-packaging
In P22 and other phages, the ATPase activity associated with genome packaging is stimulated by the S-terminase subunit, by a mechanism that remains unknown [5, 13]. We previously made the unexpected discovery that the S-terminase-dependent stimulation of ATPase activity in P22 is specifically enhanced by the DNA encoding small terminase [13], suggesting that the terminase and viral DNA assemble into a functional complex in preparation to docking to procapsid. The stoichiometry of the S:L-terminase complex elucidated in this paper requires only two of the nine LBDs of S-terminase contact a pair of L-terminase subunits, leaving ‘unoccupied’ seven LBDs. Because LBD is highly basic and recruits both the L-terminase and pac DNA [13], the proposed 9:2 stoichiometry is expected to expose a large number of basic residues at the base of the terminase complex. We propose that P22 S-terminase functions as an ‘assembly scaffold’ to initiate DNA-packaging. Using its oligomeric architecture, S-terminase recruits both P22-DNA and L-terminase to form a stable ‘pre-packaging’ complex. In this complex, the S-terminase subunit can simultaneously ‘recognize’ a specific pac site in P22 genome and ‘present’ it to the nuclease domain of L-terminase that introduces nicks. The proposed role of S-terminase as an ‘assembly scaffold’ for DNA-packaging may help reconcile the different oligomeric state of S-terminase observed in different viruses [13]. Oligomeric rings of different stoichiometry would be sufficient to bring viral DNA and L-terminase in close proximity regardless of the exact number of subunits, possibly explaining why S-terminases have evolved as oligomers of different stoichiometry even in similar viruses like P22 and Sf6. We postulate that a structural rearrangement must occur in the terminase complex to switch from a ‘pre-packaging’ conformation (possibly bound to DNA) to an active ‘packaging’ state, where L-terminase is oligomerized at the portal vertex [56]. The latter state has been studied in bacteriophages T4 and T7, where a pentameric stoichiometry of L-terminase was reported ([22] [33]). These studies, however, were carried out in the absence of S-terminase, whose actual involvement in DNA-packaging remains controversial.
In conclusion, this paper presents the first structural characterization of the terminase complex of bacteriophage P22 and sets up the ground for future higher resolution structural studies by cryo-EM.
MATERIALS AND METHODS
Molecular biology techniques
Cloning of the full length S-terminase (plasmid pMAL-S-terminase) and L-terminase (plasmid pET30b-L-terminase) and L-terminase nuclease domain (residues 289-499) (plasmid pET30b-nuclease) of bacteriophage P22 was previously described [13, 14, 25]. L-terminase ATPase domain (plasmid pET30b-ATPase) (residues 1-287) was generated by introducing a stop codon at residue 288 of plasmid pET30b-L-terminase. The LBD (residues 140-162 of S-terminase) was amplified by PCR and inserted into a pGEX-6P vector (GE Healthcare Life Sciences) between BamHI and XhoI sites (plasmid pGEX-S-LBD). MBP-LBD was generated by splicing off residues 1-139 from plasmid pMAL-S-terminase. All constructs of L-terminase were also generated by PCR using as template plasmid pET28-L-terminase.
Biochemical techniques
The S:L-terminase and MBP-LBD:L-terminase complexes were expressed in E.coli strain BL21-AI (Life Technologies) by inducing at 18 °C for 12-16 hours with a final concentration 0.2% L-arabinose and 0.1 mM IPTG. Cell pellets were dissolved in Lysis buffer containing 20 mM Tris-Cl pH 8.0, 300 mM NaCl, 1 mM MgCl2, 5 % glycerol, 0.1 % DDM , 3 mM β-mercaptoethanol, 1.0 mM phenylmethylsulfonyl fluoride and cells were disrupted by sonication. Terminase complexes were purified on amylose beads (New England Biolabs), and after washing with 500 ml of Lysis buffer, the complexes was incubated with 1 mM AMP-PNP (Sigma) and PreScission Protease to cleave off MBP. The day after, cleaved species coming off beads were further purified on a Superdex 200 16/60 gel filtration column (GE Healthcare) in GF-buffer (20 mM Tris-Cl pH 8.0, 150 mM NaCl, 1 mM MgCl2, 5% glycerol, 3 mM β-mercaptoethanol). The gel filtration column was calibrated with MW markers as previously described [57]. Isolated S:L-terminase complex were concentrated to ~10 mg/ml using a 30 MWCO ultrafiltration spin column (Vivaspin 20, Sartorius Stedim Biotech GmbH). Pull-downs assay were carried out on glutathione beads (GenScript) as previously described [58, 59].
Sedimentation velocity Analytical Ultracentrifugation
AUC analysis was carried out in a Beckman XL-A Analytical Ultracentrifuge operating under velocity sedimentation mode available at the Sydney Kimmel Cancer Center X-ray Crystallography and Molecular Interaction Facility. Purified S:L-terminase and MBP-LBD:L-terminase complexes dissolved at 0.25 mg ml-1 in 20 mM Tris-Cl pH 8.0, 150 mM NaCl, 3 mm DTT, 5% glycerol, 1 mM MgCl2 were spun at 35,000 rpm and 40,000 rpm, respectively at 10° C. Absorbance values at 280 nm were fit to a continuous sedimentation coefficient (c(s)) distribution model in SEDFIT [60].
Native Mass Spectrometry
Prior to native mass spec measurement, the purified S:L-terminase complex was buffer exchanged into 150 mM aqueous ammonium acetate (AmAc) (pH 8.0), by ultrafiltration (Vivaspin 500, Sartorius Stedim Biotech, Germany) with a 10 kDa cut off. 1-2 μL of sample, at a final concentration of 2 μM, was loaded into a nanoflow gold-plated borosilicate electrospray capillary (made in house). The higher order oligomers of S:L-terminase complex were analyzed on a modified QTOF-2 (Waters/MS Visions) operated on positive ion mode. Xenon was used as collision gas. MS parameters: backing pressure 10 mbar; capillary 1300 V; cone 60 V; extracted cone 0 V; pressure in the collision cell 2×10-2 mbar; collision energy 30 V. The sample was analyzed on a modified Exactive Plus EMR Orbitrap instrument (Thermo Fisher Scientific, Bremen) over m/z range 500-20000 [61, 62]. Manual tuning of the voltage offset on the flatapole, transport multipole, ion lenses was used for mass filtering of the incoming protein ions, as previously described [47]. Nitrogen was used for the HCD cell at a gas pressure of 6-8 × 10 -10 bar. Mass spec parameters: spray voltage 1.3-1.4 V, source fragmentation 30 V, source temperature 250 °C, collision energy 40-50 V, 10000 resolution at m/z 200.
EM specimen prep
To determine the structure and organization of the S:L-terminase complex (Fig. 1B) was analyzed with negative stain EM. Continuous carbon grids were glow discharged, and 3 μl of sample at a concentration of 0.014 mg/ml was applied to the grid for 1 minute. The grid was gently blotted and passed through four 50 μl volumes of 2% uranyl formate. Subsequently, the grid was blotted, air-dried, and stored under desiccation.
EM Data collection and processing
Images were acquired using a Tecnai 12 electron microscope operating at 120 KeV, with a dose near 20 e-/Å2 and a nominal range of 1.0-2.0 μm underfocus. The continuous carbon grid areas were targeted using Leginon [63] software at a nominal magnification of 52,000× (pixel size of 0.205 nm). Images were recorded using a 4k × 4k Tietz F416 CMOS detector (Fig. S1). Approximately 8,623 particles were picked from 44 micrographs using Difference of Gaussians (Dog) Picker [64]. These particles were subjected to reference-free XMIPP Clustering 2D [65], and well-ordered particles extracted, resulting in 6,562 particles. To obtain the best 2D class averages in the presence of any heterogeneity, the sharpest, and best aligned, class averages were produced with the Interactive Stable Alignment Clustering (ISAC) program [66] (Fig. 5A). Using only the S-terminase nonamer low-pass filtered to 60 Å as an initial model (Fig. S2A), 3-D classification and subsequent gold standard refinement was used to obtain an asymmetric reconstruction using RELION (Fig. S2). The resolution of the reconstruction was estimated to be approximately 30 Å using the gold standard FSC=0.143 criterion (Fig. S2B-C) [49]. The 3D EM map has been deposited to the EMDataBank with accession code EMD-XXXX.
Placement of 3D models in EM density
The crystal structures of S-terminase with modeled extended helices (PDB id 3P9A) and L-terminase from Sf6 [23] (PDB id 4IEE) were manually oriented in the EM density based on analysis of 2D class averages and biochemical data shown in figure 3 and 4. Subsequently, the models were rigid body refined in the density using the “Fitmap” feature in Chimera (Fig. 6).
Supplementary Material
Highlights.
We assembled the terminase holoenzyme of the bacteriophage P22
The ATPase domain of P22 L-terminase associates with S-terminase C-terminus
P22 terminase complex consists of 1S-terminase:2L-terminase
Acknowledgements
This work was supported by NIH grants R01GM100888 to G.C., R01GM54076 to J.J. and F32GM108310 to R.M. Native MS measurement was performed by YY and AJRH, who have been supported by the ManiFold project, grant number 317371, and in part by the PRIME-XS project, grant number 262067, both funded by the European Union 7th Framework Programme. Research in this publication includes work carried out at the Sidney Kimmel Cancer Center X-ray Crystallography and Molecular Interaction Facility, which is supported in part by NCI Grant P30 CA56036. Electron microscopic imaging and reconstruction were conducted at the National Resource for Automated Molecular Microscopy which is supported by the NIH through a P41 program grant (RR17573) from the National Center for Research Resources.
Abbreviations used
- S-terminase
small terminase
- SBD
S-terminase Binding Domain
- L-terminase
large terminase
- LBD
L-terminase Binding Domain
- dsDNA
double stranded DNA
- SEC
size exclusion chromatography
- MBP
Maltose Binding Protein
- MS
mass spectrometry
- DDM
n-Dodecyl-β-D-maltoside
- AMP-PNP
5′-adenylyl-β,γ-imidodiphosphate
- AUC
analytical ultracentrifugation
- SDS-PAGE
Sodium dodecyl sulfate - polyacrylamide gel electrophoresis
- EM
electron microscopy
- TEM
transmission electron microscopy
- cryo-EM
cryo-electron microscopy
- RELION
Regularized Likelihood Optimization
- ISAC
Iterative Stable Alignment and Clustering
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sun S, Rao VB, Rossmann MG. Genome packaging in viruses. Curr Opin Struct Biol. 2010;20:114–20. doi: 10.1016/j.sbi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–57. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 3.Bhardwaj A, Olia AS, Cingolani G. Architecture of viral genome-delivery molecular machines. Curr Opin Struct Biol. 2014;25:1–8. doi: 10.1016/j.sbi.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heming JD, Huffman JB, Jones LM, Homa FL. Isolation and characterization of the herpes simplex virus 1 terminase complex. Journal of virology. 2014;88:225–36. doi: 10.1128/JVI.02632-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–81. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 6.Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J Mol Biol. 1978;118:365–88. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- 7.Casjens S, Sampson L, Randall S, Eppler K, Wu H, Petri JB, et al. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J Mol Biol. 1992;227:1086–99. doi: 10.1016/0022-2836(92)90523-m. [DOI] [PubMed] [Google Scholar]
- 8.Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J Biol Chem. 2003;278:4618–27. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- 9.Leffers G, Rao VB. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J Biol Chem. 2000;275:37127–36. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- 10.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16868–73. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eppler K, Wyckoff E, Goates J, Parr R, Casjens S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–38. doi: 10.1016/0042-6822(91)90981-g. [DOI] [PubMed] [Google Scholar]
- 12.Teschke CM, Parent KN. 'Let the phage do the work': using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology. 2010;401:119–30. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy A, Bhardwaj A, Datta P, Lander GC, Cingolani G. Small terminase couples viral DNA binding to genome-packaging ATPase activity. Structure. 2012;20:1403–13. doi: 10.1016/j.str.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy A, Bhardwaj A, Cingolani G. Crystallization of the Nonameric Small Terminase Subunit of Bacteriophage P22. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2011;F67:104–10. doi: 10.1107/S174430911004697X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemecek D, Gilcrease EB, Kang S, Prevelige PE, Jr., Casjens S, Thomas GJ., Jr Subunit conformations and assembly states of a DNA-translocating motor: the terminase of bacteriophage P22. J Mol Biol. 2007;374:817–36. doi: 10.1016/j.jmb.2007.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buttner CR, Chechik M, Ortiz-Lombardia M, Smits C, Ebong IO, Chechik V, et al. Structural basis for DNA recognition and loading into a viral packaging motor. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:811–6. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, et al. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1971–6. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:817–22. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, et al. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol Cell. 2002;9:981–91. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H, Kamau YN, Christensen TE, Tang L. Structural and functional studies of the phage Sf6 terminase small subunit reveal a DNA-spooling device facilitated by structural plasticity. J Mol Biol. 2012;423:413–26. doi: 10.1016/j.jmb.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- 22.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–62. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Christensen TE, Kamau YN, Tang L. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8075–80. doi: 10.1073/pnas.1301133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smits C, Chechik M, Kovalevskiy OV, Shevtsov MB, Foster AW, Alonso JC, et al. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 2009;10:592–8. doi: 10.1038/embor.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy A, Cingolani G. Structure of p22 headful packaging nuclease. J Biol Chem. 2012;287:28196–205. doi: 10.1074/jbc.M112.349894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadal M, Mas PJ, Blanco AG, Arnan C, Sola M, Hart DJ, et al. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16078–83. doi: 10.1073/pnas.1007144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selvarajan Sigamani S, Zhao H, Kamau YN, Baines JD, Tang L. The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. Journal of virology. 2013;87:7140–8. doi: 10.1128/JVI.00311-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casjens SWP. Viral genome packaging machines : genetics, structure, and mechanism. Landes Bioscience/Eurekah.com ; Kluwer Academic/Plenum Publishers; Georgetown, Tex. New York, N.Y.: 2005. [Google Scholar]
- 29.Catalano CE. Viral genome packaging machines : genetics, structure, and mechanism. Landes Bioscience/Eurekah.com ; Kluwer Academic/Plenum Publishers; Georgetown, Tex. New York, N.Y.: 2005. [Google Scholar]
- 30.Poteete AR, Botstein D. Purification and properties of proteins essential to DNA encapsulation by phage P22. Virology. 1979;95:565–73. doi: 10.1016/0042-6822(79)90509-9. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Sampson L, Parr R, Casjens S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Molecular microbiology. 2002;45:1631–46. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- 32.Botstein D, Levine M. Intermediates in the synthesis of phage P22 DNA. Cold Spring Harbor symposia on quantitative biology. 1968;33:659–67. doi: 10.1101/sqb.1968.033.01.075. [DOI] [PubMed] [Google Scholar]
- 33.Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL. Large terminase conformational change induced by connector binding in bacteriophage T7. J Biol Chem. 2013;288:16998–7007. doi: 10.1074/jbc.M112.448951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casjens SR, Gilcrease EB. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol. 2009;502:91–111. doi: 10.1007/978-1-60327-565-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olia AS, Al-Bassam J, Winn-Stapley DA, Joss L, Casjens SR, Cingolani G. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J Mol Biol. 2006;363:558–76. doi: 10.1016/j.jmb.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Lorenzen K, Olia AS, Uetrecht C, Cingolani G, Heck AJ. Determination of stoichiometry and conformational changes in the first step of the P22 tail assembly. J Mol Biol. 2008;379:385–96. doi: 10.1016/j.jmb.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olia AS, Prevelige PE, Jr., Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng H, Olia AS, Gonen M, Andrews S, Cingolani G, Gonen T. A conformational switch in bacteriophage p22 portal protein primes genome injection. Mol Cell. 2008;29:376–83. doi: 10.1016/j.molcel.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang J, Lander GC, Olia A, Li R, Casjens S, Prevelige P, Jr., et al. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in p22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olia AS, Bhardwaj A, Joss L, Casjens S, Cingolani G. Role of gene 10 protein in the hierarchical assembly of the bacteriophage P22 portal vertex structure. Biochemistry. 2007;46:8776–84. doi: 10.1021/bi700186e. [DOI] [PubMed] [Google Scholar]
- 41.Bhardwaj A, Olia AS, Walker-Kopp N, Cingolani G. Domain organization and polarity of tail needle GP26 in the portal vertex structure of bacteriophage P22. J Mol Biol. 2007;371:374–87. doi: 10.1016/j.jmb.2007.05.051. [DOI] [PubMed] [Google Scholar]
- 42.Olia AS, Casjens S, Cingolani G. Structural plasticity of the phage P22 tail needle gp26 probed with xenon gas. Protein Sci. 2009;18:537–48. doi: 10.1002/pro.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King J, Lenk EV, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J Mol Biol. 1973;80:697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- 44.Ebel-Tsipis J, Botstein D, Fox MS. Generalized transduction by phage P22 in Salmonella typhimurium. I. Molecular origin of transducing DNA. J Mol Biol. 1972;71:433–48. doi: 10.1016/0022-2836(72)90361-0. [DOI] [PubMed] [Google Scholar]
- 45.Maluf NK, Yang Q, Catalano CE. Self-association properties of the bacteriophage lambda terminase holoenzyme: implications for the DNA packaging motor. J Mol Biol. 2005;347:523–42. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–68. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 47.Snijder J, van de Waterbeemd M, Damoc E, Denisov E, Grinfeld D, Bennett A, et al. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by Orbitrap mass spectrometry. Journal of the American Chemical Society. 2014;136:7295–9. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leavitt JC, Gilcrease EB, Wilson K, Casjens SR. Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor. Virology. 2013;440:117–33. doi: 10.1016/j.virol.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Guo P, Zhao Z, Haak J, Wang S, Wu D, Meng B, et al. Common mechanisms of DNA translocation motors in bacteria and viruses using one-way revolution mechanism without rotation. Biotechnology advances. 2014;32:853–72. doi: 10.1016/j.biotechadv.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S, Chistol G, Hetherington CL, Tafoya S, Aathavan K, Schnitzbauer J, et al. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell. 2014;157:702–13. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cingolani G, Moore SD, Prevelige PE, Jr., Johnson JE. Preliminary crystallographic analysis of the bacteriophage P22 portal protein. J Struct Biol. 2002;139:46–54. doi: 10.1016/s1047-8477(02)00512-9. [DOI] [PubMed] [Google Scholar]
- 54.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, et al. The Structure of an Infectious p22 Virion Shows the Signal for Headful DNA Packaging. Science. 2006;312:1791–5. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 55.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. Journal of virology. 2004;78:12668–71. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrews BT, Catalano CE. Strong subunit coordination drives a powerful viral DNA packaging motor. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5909–14. doi: 10.1073/pnas.1222820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lokareddy RK, Bhardwaj A, Cingolani G. Atomic structure of dual-specificity phosphatase 26, a novel p53 phosphatase. Biochemistry. 2013;52:938–48. doi: 10.1021/bi301476m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J Biol Chem. 2008;283:7877–84. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 59.Lott K, Bhardwaj A, Mitrousis G, Pante N, Cingolani G. The importin beta binding domain modulates the avidity of importin beta for the nuclear pore complex. J Biol Chem. 2010;285:13769–80. doi: 10.1074/jbc.M109.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJ. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nature methods. 2012;9:1084–6. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 62.Rosati S, Rose RJ, Thompson NJ, van Duijn E, Damoc E, Denisov E, et al. Exploring an orbitrap analyzer for the characterization of intact antibodies by native mass spectrometry. Angewandte Chemie. 2012;51:12992–6. doi: 10.1002/anie.201206745. [DOI] [PubMed] [Google Scholar]
- 63.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 64.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–13. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sorzano CO, Bilbao-Castro JR, Shkolnisky Y, Alcorlo M, Melero R, Caffarena-Fernandez G, et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol. 2010;171:197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–47. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.