Abstract
Background
The Clinical and Functional Translation of CFTR project (CFTR2) classified some cystic fibrosis transmembrane conductance regulator (CFTR) gene variants as non cystic fibrosis (CF)-causing. To evaluate this, the clinical status of children carrying these mutations was examined.
Methods
We analyzed CF disease-defining variables over 2–6 years in two groups of California CF screen- positive neonates born from 2007–2011: (1) children with two CF-causing variants and (2) children with one CF-causing and one non-CF causing variant, as defined by CFTR2.
Results
Children carrying non CF-causing variants had significantly higher birth weight, lower immunoreactive trypsinogen and sweat chloride values, higher first year growth curves, and a lower rate of persistent Pseudomonas aeruginosa colonization compared to children with two CF-causing variants.
Conclusions
The outcomes in children 2–6 years of age with the L997F, G576A, R1162L, V754M, R668C, R31C, and S1235R variants are consistent with the CFTR2 non CF-causing classification.
Keywords: Cystic Fibrosis, newborn screening, CFTR2, non CF-causing variants, CF-causing variants, genotype-phenotype associations
Introduction
Soon after the discovery of the cystic fibrosis transmembrane conductance regulator (CFTR) gene, it was recognized that in addition to known deleterious variants, there were neutral variants that differed from wild-type but did not cause cystic fibrosis (CF).1 So far, nearly 2,000 CFTR variants have been described in the CF Mutation Database (http://www.genet.sickkids.on.ca/cftr/Home.html). A mechanism-based classification system (classes I–VI)2 has existed for over a decade and remains relevant for therapy development but does not differentiate disease liability.3,4 More recently, the Clinical and Functional Translation of CFTR project (CFTR2) began to address this gap by using worldwide clinical data from CF patient registries, functional assays using cell cultures, and population data to further characterize CFTR variants into four categories. To date, a total of 206 CFTR variants have been characterized by CFTR2: 176 are CF-causing, 12 are non CF-causing, 11 have varying clinical consequences, and 7 are still unknown due to inadequate information (http://www.CFTR2.org). For variants that had evidence of residual function in experimental models, CFTR2 used penetrance analysis among families with children with CF to distinguish non CF-causing variants from those categorized as varying clinical consequence. Variants seen on the opposite chromosome of the transmitted CF-causing allele in fathers of CF offspring were classified as non CF-causing on the basis that those variants did not result in infertility.5 The conclusions from the penetrance analysis are somewhat tentative because the analyses were based on small sample sizes for each non CF-causing variant, and it could not be confirmed that the fathers studied did not have CF or did not use assisted reproductive technology.
In this paper, we seek to evaluate variants in the CFTR2 non CF-causing classification through analysis of various clinical outcomes in a comprehensively genotyped and diverse newborn screening population followed from birth to age 2–6 years.
Methods
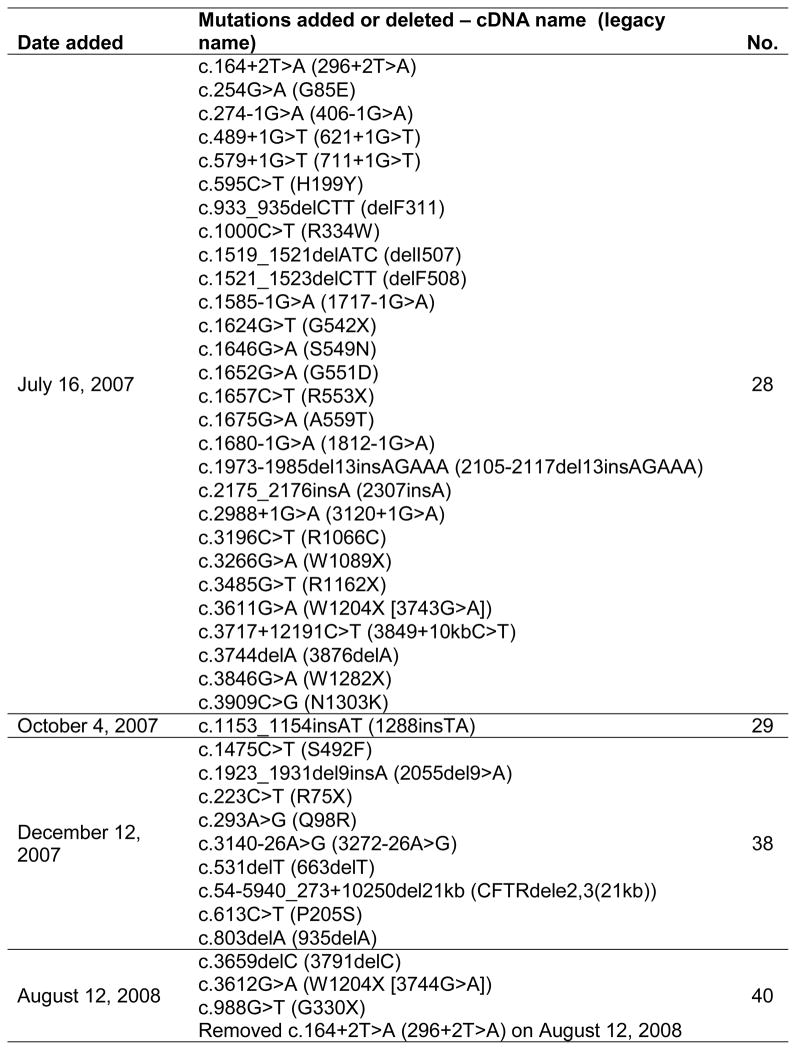
This is a retrospective cohort study of cystic fibrosis screening test positive newborns born in California from July 2007 to July 2011 and followed until August 2013. Newborns were screened for CF using a unique 3-step model: (1) measuring immunoreactive trypsinogen (IRT) levels in all newborn blood spot specimens (median age at collection: 30 hours), (2) testing for 28–40 selected CFTR variants (Figure 1) in specimens with high IRT values (≥ 62 ng/mL, top 1.6%), and (3) testing by DNA sequencing in specimens found to have only one variant in Step 2. Infants with 2 or more variants (including any DNA sequence listed in the CF Mutation Database or published in the literature [except those documented to be benign polymorphisms], Poly 5T of any TG tract length in intron 9, or any novel variant) were referred to 17 CF Centers for sweat chloride testing and follow up. Each CF Center entered clinical and laboratory data into a state database for all referrals in the initial assessment visits (quarters 1 to 3) until the final diagnosis was established and yearly thereafter. For this study, the data on study subjects were de-identified, and sent to the Children’s Hospital Los Angeles (CHLA) research team for analysis. The study was approved by the CHLA Institutional Review Board and the California Health and Human Services Agency Committee for the Protection of Human Subjects.
Figure 1.
CFTR mutation panel used by the California Newborn Screening Program.
Two genotype groups were created from screen-positive children: 1) those identified with two CF-causing variants (CF-C) from the California 40 panel or from the panel plus CFTR sequencing known to be CF-C and 2) those with one CF-causing from the California 40 panel and one or more non CF-causing variants (N-CF) from CFTR sequencing. All but three of the variants on the panel (F311del, 935delA and Q98R) have been evaluated by CFTR2 and determined to be CF-causing. Outcome variables included IRT, highest sweat chloride concentration, pancreatic insufficiency status, growth parameters, and rate of first acquisition and persistent colonization with Pseudomonas aeruginosa.6–9 If sweat chloride values were <60 mmol/L at the first visit, repeat tests were done at around 6, 12, and 24 months.7,8 Pancreatic insufficiency was defined as fecal elastase <200 mcg/g6, or use of pancreatic enzymes if fecal elastase values were not available. P. aeruginosa infection was assessed by throat cultures done at every visit. The rate of first acquisition in the first year of life and persistent colonization (defined as two positive cultures less than 12 months apart) were recorded.9,10 Lost to follow up was defined as ≥18 months from the last clinic visit.
Comparisons between the two genotype groups were made with t-tests or Wilcoxon rank sum tests for continuous variables and Chi-square or Fisher’s exact test for categorical variables. Longitudinal weight-for-height Z scores (based on CDC growth curves from year 2000) were analyzed using quadratic repeated measures mixed models, with CF-C further divided into pancreatic sufficient (PS) and pancreatic insufficient (PI) and only including subjects with at least 3 data points. Outlier growth data points were removed if not biologically plausible as part of the data cleaning process. Dunnett’s post hoc comparisons of least squares means were made with CF-C-PS children as the reference group. Time to first positive culture for P. aeruginosa during the first year of life was analyzed with the Kaplan-Meier method and compared between groups with the log-rank test. Comparisons between groups were also made adjusting for ethnicity (Hispanic vs. all others) with Cochran- Mantel–Haenszel test for categorical variables, 2-way ANOVA for continuous variables and Cox regression for time to fist PSA. Statistical analysis was performed with SAS/STAT® v9.2 software. Statistical tests were 2-sided with a Type I error of p<0.05.
Results
Among 2,178,829 births from 2007 to 2011, 848 had a positive CF screen (≥2 CFTR variants identified), and 283 met inclusion criteria for the study (CF-C: 226 and N-CF: 57). There were seven non CF-causing variants identified by the newborn screening program. Functional data from CFTR2 for these seven variants showed residual function compared to wild type (Table 1). Altogether these seven variants comprised 6.7% of all CF screening test positive newborns. Of the seven variants, L997F was found most frequently among screening test positive newborns (34/848, 4%), with each of the other variants being <1%.
Table 1.
Non CF-causing variants identified among positive screen children in California and their reference functional data from CFTR2. Functional experiments were performed previously. 5,27
| Non CF-causing variants from CF NBS in California | c. DNA name | Number of patients identified from the CA CF NBS | Mean [Cl-] conductance (as % WT-CFTR)b | C/(B+C) (as % of WT-CFTR) in HeLa cellsc | C/(B+C) (as % of WT-CFTR) in FRT cellsd | CFTR Protein Quantity (as % WT-CFTR)e |
|---|---|---|---|---|---|---|
| L997F | c.1408A>G | 34 | 22 | 97 | 104 | 100 |
| G576Aa | c.1727G>C | 7 | 147 | 98 | 110 | 104 |
| R1162L | c.3485G>T | 6 | 130 | 93 | 94 | 94 |
| V754M | c.2260G>A | 4 | 140 | 98 | 107 | 102 |
| R668Ca | c.2002C>T | 2 | 58 | 97 | 106 | 102 |
| R31C | c.91C>T | 2 | 105 | 92 | 86 | 89 |
| S1235R | c.3705T>G | 2 | 79 | 96 | 106 | 101 |
All subjects who had G576A from this study also had R668C in cis. Two other subjects had R668C without G576A. G576A and R668C were analyzed as single alleles in CFTR2.
CFTR function assessed by short circuit current in Fisher Rat Thyroid (FRT) cells transfected with either mutant or wild type CFTR. Results are expressed as percentage of wild type (WT) CFTR.
CFTR protein processing assessed by the ratio of fully glycosylated (C-band) to partially glycosylated B-band + C-band in Western blots of transfected HeLa (c) and FRT (d) cells. The ratio is expressed as a percentage of wild type CFTR C/(B+C) ratio.
CFTR protein quantity tested in transfected HeLA cells and expressed as a percentage of wild type transfected cells.
CA CF NBS = California cystic fibrosis newborn screening program.
The N-CF group, as a whole, had a higher percentage of Hispanics compared to the CF-C group, as well as a higher lost to follow up rate. The N-CF group had a more benign phenotype, including a lower IRT, higher mean birth weight, no cases of meconium ileus, and no deaths (Table 2).
Table 2.
Selected characteristics of two study groups
| Population Characteristics | CF-Ca(n=226) | N-CFb(n=57) | p valueg |
|---|---|---|---|
|
| |||
| Age at last follow up, mean (SD), months | 49 (14) | 52 (12) | 0.18 |
|
| |||
| Female gender, n (%) | 97 (43) | 31 (54) | 0.14 |
|
| |||
| Race/ethnicity, n (%) | 0.036c | ||
| Whites | 108 (47.8) | 22 (38.6) | |
| Hispanics | 90 (39.8) | 32 (56.1) | |
| Non-Hispanics Blacks | 12 (5.3) | 0 (0.0) | |
| Multiple and others | 15 (6.6) | 3 (5.3) | |
| Missing | 1(0.4) | 0 (0.0) | |
|
| |||
| Birth weight, mean (SD), kg | 3.11 (0.59) n=225 |
3.39 (0.53) n=56 |
0.004 |
|
| |||
| IRT median (IQR)d, ng/mL | 162 (110-241) | 75 (70-96) | <0.0001 |
|
| |||
| Meconium Ileus, n (%) | 36 (16) | 0 (0.0) | 0.0002 |
| Highest Sweat Chloride, median (range), mmol/L | 95 (25-135) | 18 (7-52) | <0.0001 |
| Sweat Chloride ≥ 60 mmol/L, n (%) | 184 (89%) n=207 |
0 (0.0) n=56 |
|
|
| |||
| Pancreatic Insufficiency, n (%) | 180/220 (81.8) | 1/57 (1.8) | <0.0001 |
|
| |||
| P. aeruginosa acquisition in the first year of life, probability ± s.e. | 0.18 ± 0.03 | 0.16 ± 0.06 | 0.54 |
|
| |||
| P. aeruginosa Colonization, n (%) | n/ae | ||
| Unknown | 39 (17.3) | 40 (70.2) | |
| Never | 84 (37.2) | 8 (4.0) | |
| Yes-but not persistent | 52 (23.0) | 6 (0.5) | |
| Persistent | 51 (22.6) | 3 (5.3) | |
|
| |||
| Lost to follow up, n (%) | 41 (18.1) | 31 (54.4) | <0.0001 |
|
| |||
| Deaths, n (%) | 6 (2.6)f | 0 (0.0) | 0.21 |
Two CF-causing variants (Two from the 40-panel or one from the panel and one sequenced)
- a,bDenominators are the total of subjects per group unless otherwise specified
Hispanics compared to all other race/ethnicities.
IRT=Immunoreactive Trypsinogen, IQR=Interquartile Range.
Statistical comparison is inappropriate due to a large percentage of missing data in the N-CF group.
The causes of death were: complications from prematurity including one confirmed case of acute respiratory distress syndrome (n=3), complications from Biotinidase deficiency (n=1), sepsis post-intestinal perforation repair (n=1), and acute respiratory failure secondary to CF-related pneumonia and liver disease (n=1).
p values after adjusting for Hispanic versus all others are as follows: birth weight 0.001, IRT <0.0001, meconium ileus 0.0017, sweat chloride <0.0001, pancreatic insufficiency <0.0001, P. aeruginosa acquisition 0.56, lost to follow up <0.0001, death 0.23.
There were no sweat chloride results meeting CF diagnostic criteria of ≥60 mmol/L in the N-CF group, and only one subject with three variants (G542X, G576A, R668C) had values in the “possible CF” category (≥40 mmol/L) beyond 6 months. Median sweat chloride values were significantly lower in the N-CF versus the CF-C group (Table 2).
Fecal elastase levels were available and used to determine the pancreatic status in 81% of N-CF and in 79% of CF-C group. Pancreatic status was inferred for the remaining subjects from use of pancreatic enzymes. Those from CF-C group missing fecal elastase values and labeled as PI based on enzyme use had genotypes frequently associated with pancreatic dysfunction (56% were homozygous F508del). Compared to the CF-C group, pancreatic insufficiency was significantly lower in the N-CF group (Table 2, p<0.0001) as there was only one subject in this group classified as PI. This individual had fecal elastase of 196 mcg/g and never received pancreatic enzymes.
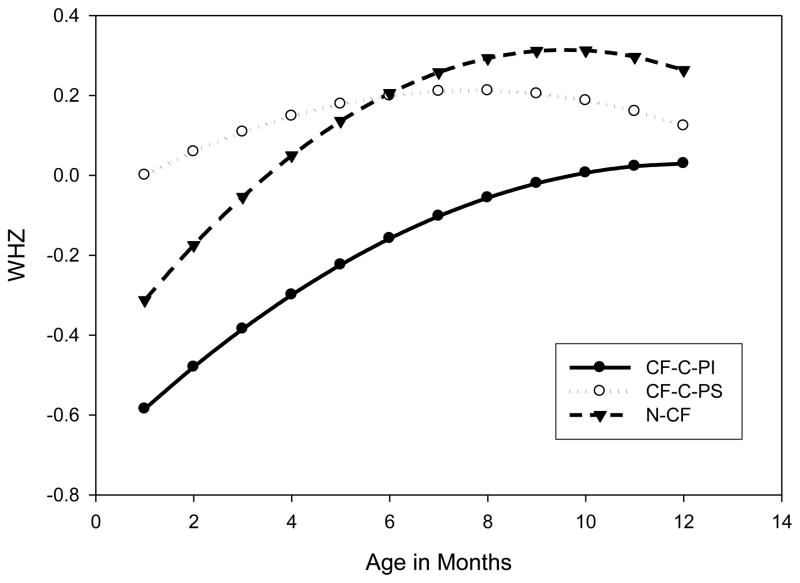
Longitudinal analysis of growth data did not include birth measurements, as birth length was not consistently available. This analysis showed that the N-CF group had normal growth: weight for height Z scores (WHZ) were similar comparing N-CF to CF-C-PS children (p=0.851), while CF-C-PI scores were significantly lower than in the CF-C-PS group (p=0.023; Figure 2).
Figure 2. Predicted Weight for Height Z score (WHZ).
by age in months plotted for 3 groups of patients using coefficients derived from a quadratic mixed model. WHZ least squares estimated means±s.e. over 12 months: CF-C-PS 0.11±0.14, CF-C-PI -0.28±0.07, N-CF 0.03±0.13. Analysis by genotype group and pancreatic status; comparison to CF-C-PS (n=37): CF-C-PI (n=138; p=0.023) and N-CF (n=46; p=0.851).
CF-C-PS: CF causing variant group and pancreatic sufficiency; PI=pancreatic insufficiency; and N-CF = Non CF-causing variant group.
P. aeruginosa infection rate was assessed by a range of 2–6 throat cultures per subject per year during 2–6 years of follow up. While the probability of first year acquisition did not differ between the CF-C and N-CF groups (p=0.54), persistent colonization was observed in only 5% of N-CF vs. 23% of CF-C children (Table 2).
Secondary analyses - comparing the groups after adjusting for ethnicity (Hispanic vs. others) - showed that the higher prevalence of Hispanics in N-CF compared to CF-C children did not explain differences between the groups in CF-related phenotype variables (birth weight, IRT, meconium ileus, sweat chloride, pancreatic insufficiency, and P. aeruginosa acquisition; Table 2, footnote).
Discussion
In this 2–6 year follow up study of CF screening positive newborns, those with non CF-causing variants had a more benign phenotype compared to those with two CF-causing variants. None of the N-CF subjects met criteria for CF diagnosis based on sweat chloride, and only one was classified as pancreatic insufficient (although never clinically treated as such). Additionally, children with non CF-causing genotypes had normal birth weights, growth, and a lower rate of persistent P. aeruginosa colonization.
Although inconsistent with CF, it is unclear if and how frequently non CF-causing variants would cause atypical forms of disease or CFTR-related disorders (CFTR-RD).11 This disorder, associated with CFTR dysfunction causing symptoms that do not fulfill diagnostic criteria for CF, is manifested by single organ disease such as congenital absence of vas deferens, recurrent/chronic pancreatitis, rhino-sinusitis, or bronchiectasis.11 All non CF-causing variants have been previously associated with CFTR-RD,11,12–17 but have also been identified in unaffected persons.18–20 As the subjects from our study are still young, it is difficult to predict which, if any, individuals carrying non CF-causing variants will be at risk for CFTR-RD. Some variants may alter function in a manner that produces mild or incomplete forms of disease, while others may cause no discernable change in phenotype.21
One implication of these findings has been to change the manner that newborns with one CF-causing and one or more non CF-causing variants are followed up after screening. Because none to date have had meconium ileus, a positive sweat chloride value or other frank CF signs or symptoms, the California newborn screening program in May 2014 instituted a genetic-counseling-only follow up protocol for these newborns and their parents. Unless requested specifically by the family or physician, a physical exam and sweat chloride test are not routinely conducted. Instead, parents are educated about the CFTR variants of the child and given a list of common symptoms associated with CFTR dysfunction to look out for. Should the child exhibit any of these symptoms in the future, they are instructed to return immediately to a CF Center for evaluation. The objective is to identify individuals at risk for CFTR-RD at a younger age so that they may have the benefit of closer clinical monitoring, more rapid diagnosis, and prompt clinical interventions.
Complex alleles (multiple variants on the same chromosome) may complicate interpretation of disease liability. The notion of assigning a phenotype to an individual variant needs to consider other genetic changes that may affect CFTR function. An example of this is the poly-T repeat within intron 9 that influences the penetrance of R117H.22 In another example, the combination of R117L and L997F on the same allele causes a more severe phenotype than L997F alone, though this combination was not observed in this study.23 There could be other reasons that CF cases with non CF-causing variants exist, including incomplete genotyping of the CFTR gene and gene-gene interactions.
There was a high lost to follow up rate in the N-CF group (54%), and the period of this study (2–6 years) provided short follow up. It is unclear why families were choosing not to continue follow up at the CF centers. One can speculate that the children were clinically well and only followed by their pediatricians for routine care, which could have potentially introduced a selection bias towards sicker children in our study. However, eliminating this bias would only make the N-CF group appear healthier. Longer periods of follow up and more extensive clinical information such as description of symptoms and chest radiographs or chest computed tomography are needed for a better understanding of the penetrance of these variants.
One individual in the N-CF group was labeled as PI based on a borderline fecal elastase. However, this individual was never prescribed pancreatic enzymes. The use of enzymes alone as criteria to define pancreatic status can lead to misclassification bias 24, which is minimized in this study considering that ~80% of children had fecal elastase values available. Likewise, there may be overestimation of the rate of PI in the CF-C group. We can speculate that though fecal elastase measurement is encouraged by guidelines, some clinicians may choose to start pancreatic enzymes supplementation based on genotype and/or clinical suspicion of malabsorption.8
Furthermore, the observed rate of persistent P. aeruginosa colonization in N-CF subjects could be related to frequent surveillance, increased exposure to CF patients, and/or some degree of CFTR dysfunction. Alternatively, this observation may be a normal phenomenon as transient colonization has been described in healthy children.25,26 Considering the high rate of “unknown colonization status” among N-CF children (70%), which is explained by a high rate of lost to follow up and missing culture results, it would be inappropriate to make a statistical comparison of persistent colonization between groups.
The CFTR2 classification of non CF-causing variants has great relevance in the design of CFTR mutation panels for carrier and newborn screening programs, making accurate diagnoses, and prioritizing new therapies. We found that nearly 7% of the newborns with a positive CF screening test in California had one CF-causing variant and one or more non CF-causing variants. The majority of these newborns carried the L997F variant. As more CFTR variants are evaluated by CFTR2, an even higher percentage of newborns will likely have non CF-causing variants. We conclude that the outcomes seen to date in comprehensively genotyped hypertrypsinogenemic children 2–6 years of age were consistent with the CFTR2 non CF-causing classification.
However, it is not known how many of these individuals will later develop respiratory, gastrointestinal, or infertility manifestations. Improved CFTR function assays that more accurately predict outcomes pre-clinically could be invaluable in advancing our understanding of these non CF-causing variants.
Highlights.
CFTR2 non CF-causing variant classification applied to a NBS population
Six years of clinical data from 17 California CF centers were analyzed
Children carrying non CF-causing variants did not meet diagnostic criteria for CF
The outcomes seen are consistent with the CFTR2 non CF-causing classification.
Acknowledgments
Funding: this study received support from the National Institute of Health: USC-CTSI (NIH/NCRR/NCATS; Grant # KL2TR000131) and the Division of Pediatric Pulmonology at the Children’s Hospital Los Angeles.
Footnotes
Additional contributions: all members of the California Cystic Fibrosis Newborn Screening Consortium comprised of providers from 17 CF care centers in the state contributed with patient data.
Author Contributions: Dr. Danieli Salinas, Dr. Martin Kharrazi and Colleen Azen had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Salinas, Azen, Keens, Sosnay and Kharrazi. Data acquisition and analysis: Salinas, Azen, Young and Kharrazi. Interpretation: Salinas, Keens, Azen, Young, Raraigh, Sosnay and Kharrazi. Drafting of the manuscript: Salinas, Keens, Raraigh, Sosnay and Kharrazi. Critical revision of the manuscript for important intellectual content: Salinas, Raraigh, Sosnay and Kharrazi. Obtained funding: Salinas and Keens. Statistical analysis: Azen.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Danieli Barino Salinas, Email: dsalinas@chla.usc.edu, Children’s Hospital Los Angeles – University of Southern California, Department: Pediatrics-Pediatric Pulmonology, Institution: Keck School of Medicine-University of Southern California (USC), Mailing Address: 4650 Sunset Boulevard, MS 83., Los Angeles, CA 90027, Phone# 323-361 2101.
Patrick R. Sosnay, Email: psosnay@jhmi.edu, McKusick-Nathans Institute of Medical Genetics - Johns Hopkins University, Department: Medicine-Pulmonary and Critical Care Medicine, Institution: Johns Hopkins University School of Medicine, Mailing address: 1830 E. Monument St. 5th Floor, Baltimore, MD, 21287, Phone: 410 502-7044.
Colleen Azen, Email: cazen@chla.usc.edu, Children’s Hospital Los Angeles – University of Southern California, Department: Southern California Clinical and Translational Science Institute, Institution: Children’s Hospital Los Angeles, Mailing address: 4650 Sunset Boulevard, MS 142., Los Angeles, CA 90027.
Suzanne Young, Email: young.suzy@gmail.com, The Sequoia Foundation, Institution: The Sequoia Foundation, Mailing address: 2166 Avenida De La Playa, La Jolla, CA 92037.
Karen S. Raraigh, Email: kraraigh@jhmi.edu, McKusick-Nathans Institute of Medical Genetics - Johns Hopkins University, Institution: Johns Hopkins University School of Medicine, Mailing address: 733 North Broadway, MRB 553, Baltimore, MD 21287.
Thomas G. Keens, Email: tkeens@chla.usc.edu, Children’s Hospital Los Angeles – University of Southern California, Department: Pediatrics-Pediatric Pulmonology, Institution: Keck School of Medicine-University of Southern California (USC), Mailing address: 4650 Sunset Boulevard, MS 83., Los Angeles, CA 90027.
Martin Kharrazi, Email: marty.kharrazi@cdph.ca.gov, California Department of Public Health, Department: Environmental Health Investigations Branch, Institution: California Department of Public Health, Mailing address: 850 Marina Bay Parkway, P-3, Richmond, CA, 94804, Phone: 510 412-1480.
References
- 1.Quere I, Guillermit H, Mercier B, Audrezet MP, Ferec C. A polymorphism in intron 20 of the CFTR gene. Nucleic acids research. 1991;19:5453. doi: 10.1093/nar/19.19.5453-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welsh MJ, Smith AE. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–4. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 3.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. The New England journal of medicine. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 4.Moskowitz SM, et al. Clinical practice and genetic counseling for cystic fibrosis and CFTR-related disorders. Genetics in medicine: official journal of the American College of Medical Genetics. 2008;10:851–68. doi: 10.1097/GIM.0b013e31818e55a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sosnay PR, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nature genetics. 2013;45:1160–7. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell PM, et al. Guidelines for diagnosis of cystic fibrosis in newborns through older adults: Cystic Fibrosis Foundation consensus report. The Journal of pediatrics. 2008;153:S4–S14. doi: 10.1016/j.jpeds.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borowitz D, et al. Cystic Fibrosis Foundation practice guidelines for the management of infants with cystic fibrosis transmembrane conductance regulator-related metabolic syndrome during the first two years of life and beyond. The Journal of pediatrics. 2009;155:S106–16. doi: 10.1016/j.jpeds.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borowitz D, et al. Cystic Fibrosis Foundation evidence-based guidelines for management of infants with cystic fibrosis. The Journal of pediatrics. 2009;155:S73–93. doi: 10.1016/j.jpeds.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pressler T, et al. Chronic Pseudomonas aeruginosa infection definition: EuroCareCF Working Group report. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2011;10 (Suppl 2):S75–8. doi: 10.1016/S1569-1993(11)60011-8. [DOI] [PubMed] [Google Scholar]
- 10.Lee TW, Brownlee KG, Conway SP, Denton M, Littlewood JM. Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2003;2:29–34. doi: 10.1016/S1569-1993(02)00141-8. [DOI] [PubMed] [Google Scholar]
- 11.Bombieri C, et al. Recommendations for the classification of diseases as CFTR-related disorders. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2011;10 (Suppl 2):S86–102. doi: 10.1016/S1569-1993(11)60014-3. [DOI] [PubMed] [Google Scholar]
- 12.Bishop MD, et al. The cystic fibrosis transmembrane conductance regulator gene and ion channel function in patients with idiopathic pancreatitis. Human genetics. 2005;118:372–81. doi: 10.1007/s00439-005-0059-z. [DOI] [PubMed] [Google Scholar]
- 13.LaRusch J, et al. Mechanisms of CFTR functional variants that impair regulated bicarbonate permeation and increase risk for pancreatitis but not for cystic fibrosis. PLoS genetics. 2014;10:e1004376. doi: 10.1371/journal.pgen.1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monaghan KG, et al. Frequency and clinical significance of the S1235R mutation in the cystic fibrosis transmembrane conductance regulator gene: results from a collaborative study. American journal of medical genetics. 2000;95:361–5. [PubMed] [Google Scholar]
- 15.Girodon E, et al. CFTR gene mutations in adults with disseminated bronchiectasis. European journal of human genetics: EJHG. 1997;5:149–55. [PubMed] [Google Scholar]
- 16.Pignatti PF, Bombieri C, Marigo C, Benetazzo M, Luisetti M. Increased incidence of cystic fibrosis gene mutations in adults with disseminated bronchiectasis. Human molecular genetics. 1995;4:635–9. doi: 10.1093/hmg/4.4.635. [DOI] [PubMed] [Google Scholar]
- 17.Ziedalski TM, Kao PN, Henig NR, Jacobs SS, Ruoss SJ. Prospective analysis of cystic fibrosis transmembrane regulator mutations in adults with bronchiectasis or pulmonary nontuberculous mycobacterial infection. Chest. 2006;130:995–1002. doi: 10.1378/chest.130.4.995. [DOI] [PubMed] [Google Scholar]
- 18.Derichs N, et al. Homozygosity for L997F in a child with normal clinical and chloride secretory phenotype provides evidence that this cystic fibrosis transmembrane conductance regulator mutation does not cause cystic fibrosis. Clinical genetics. 2005;67:529–31. doi: 10.1111/j.1399-0004.2005.00437.x. [DOI] [PubMed] [Google Scholar]
- 19.Ghanem N, et al. Identification of eight mutations and three sequence variations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics. 1994;21:434–6. doi: 10.1006/geno.1994.1290. [DOI] [PubMed] [Google Scholar]
- 20.Stanke F, et al. Diversity of the basic defect of homozygous CFTR mutation genotypes in humans. Journal of medical genetics. 2008;45:47–54. doi: 10.1136/jmg.2007.053561. [DOI] [PubMed] [Google Scholar]
- 21.Sosnay PR, Cutting GR. Interpretation of genetic variants. Thorax. 2014;69:295–7. doi: 10.1136/thoraxjnl-2013-204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiesewetter S, et al. A mutation in CFTR produces different phenotypes depending on chromosomal background. Nature genetics. 1993;5:274–8. doi: 10.1038/ng1193-274. [DOI] [PubMed] [Google Scholar]
- 23.Lucarelli M, et al. A new complex allele of the CFTR gene partially explains the variable phenotype of the L997F mutation. Genetics in medicine: official journal of the American College of Medical Genetics. 2010;12:548–55. doi: 10.1097/GIM.0b013e3181ead634. [DOI] [PubMed] [Google Scholar]
- 24.Borowitz D, et al. Use of fecal elastase-1 to classify pancreatic status in patients with cystic fibrosis. The Journal of pediatrics. 2004;145:322–6. doi: 10.1016/j.jpeds.2004.04.049. [DOI] [PubMed] [Google Scholar]
- 25.Carlson D, et al. Oropharyngeal flora in healthy infants: observations and implications for cystic fibrosis care. Pediatric pulmonology. 2009;44:497–502. doi: 10.1002/ppul.21029. [DOI] [PubMed] [Google Scholar]
- 26.Rosenfeld M, et al. Prevalence of cystic fibrosis pathogens in the oropharynx of healthy children and implications for cystic fibrosis care. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2012;11:456–7. doi: 10.1016/j.jcf.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Van Goor F, Yu H, Burton B, Hoffman BJ. Effect of ivacaftor on CFTR forms with missense mutations associated with defects in protein processing or function. Journal of cystic fibrosis: official journal of the European Cystic Fibrosis Society. 2014;13:29–36. doi: 10.1016/j.jcf.2013.06.008. [DOI] [PubMed] [Google Scholar]