Abstract
Reliable antibody based-assays are needed to evaluate the immunogenicity of current vaccines, impact of altered dosing schemes or of new vaccine formulations. An ideal assay platform would allow multiplex type-specific detection with minimal sample requirement.
We used the Meso Scale Discovery (MSD) electrochemiluminescence based detection platform to develop a multiplex direct virus-like particle (VLP) ELISA to detect antibodies to HPV 6, 11, 16, and 18 with a protocol developed for detection using the SI 6000 imager (M4ELISA). MSD prepared the plates in the 7-spot/well format, using the purified VLPs (4 spots) and PBS + BSA pH 7.4 (3 blank spots). Three-point titrations and the parallel line method were used to calculate antibody levels. Dynamic range, precision, and stability of pre-printed plates were determined using a panel of previously characterized sera. Cut-off values using children’s sera were established using 99% RLU limits based on the 4-parameter Johnson Su best fit curve. Results of the M4ELISA were compared to competitive Luminex Immunoassay (cLIA) on n = 4454 sera from a predominantly unvaccinated cohort.
Using a VLP coating concentration of 80 μg/ml with BSA provided the most robust RLU signal for all types. The dynamic range of the assay was about 1000 fold, with assay variability under 25% for each of the four vaccine types. Long-term stability of the plates extended to about 7 months from the time plates was received in the laboratory after printing. There was moderate agreement (κ = 0.38–0.54) between M4ELISA and cLIA, with antibody detection for each of the 4 types more frequent with M4ELISA. Quantitative analysis however showed a good correlation between concordant samples by both assays (ρ ≥ 0.6). The MSD platform shows promise for simultaneous quantitation of the antibody responses to four HPV vaccine types in a high-throughput manner.
Keywords: HPV antibodies, Serology, ELISA, Multiplex, Meso Scale Discovery
1. Introduction
Human papillomavirus (HPV) serology has been used as a measure of lifetime exposure to HPV. In HPV vaccine trials, serology was used to identify naïve individuals, to monitor the response to vaccination and for immunobridging (Schiller et al., 2012). In the absence of immune correlates of protection, non-inferiority of the immune response has been used in evaluating altered dosing schedules and new vaccine formulations.
Neutralizing assays, such as the pseudovirion based neutralizing assays (PBNA), are the gold standard for HPV serology as protection is believed to be due to neutralizing antibodies. These neutralizing assays are labor and time intensive, so most large scale studies use other antibody detection assays. As the major immune response is to neutralizing epitopes, these assays generally correlate with PBNA. Both direct ELISA [with conformationally intact L1 viral-like particles (VLP) as antigen] and competitive Luminex Immunoassay (cLIA) [a bead-based liquid array (Luminex Corporation, Austin, TX) with labeled type-specific neutralizing monoclonal antibody that competes with test serum for binding] have been used in HPV vaccine trials (Schiller and Lowy, 2009). Inherent differences in the type of antibody response measured, lack of standardized reagents as well as lack of uniform methods to establish cut-off values have plagued comparisons between these assays.
Multiplexed assays are increasingly needed as current vaccines target 4 types, and a candidate 9-valent vaccine is under review. Multiplexing allows for use of reduced sample volume and increased throughput for large studies. The two most widely used platforms for multiplex assays are the bead-based liquid arrays (Luminex) and electrochemiluminescent multi-spot assays. Meso Scale Discovery (MSD, Rockville, MD) uses electrochemiluminesence technology which lends itself to a large dynamic range for the assay. The spots are printed in an array within each well of the carbon electrode multi-spot plates. The small area of each spot requires less L1-VLP capture reagent. Conformationally intact L1-VLPs are the key to the assay, and conserving this key reagent reduces cost of production and the time for the extensive quality control (QC) that is required. Because the capture reagent binds directly to the plate, the assay is similar to a traditional single-plex ELISA and competition between analytes is reduced. The current plate formats offered by MSD allow for multiplexing 4, 7 or 10 analytes in a single well. The equipment does not require any fluidics as it is based on images of the electrochemical signal captured by the CCD camera with fast read times (about 70 s/plate). We developed a direct L1-VLP ELISA using the MSD platform to simultaneously detect antibodies to HPV 6,11,16 and 18 and performed studies to validate the performance in terms of reproducibility, linear range, lower limits of detection, stability of antigen and agreement with cLIA on a cohort of samples from the general population of the US prior to vaccine introduction.
2. Materials and methods
2.1. VLP production and purification
HPV L1/L2 VLPs were produced in Human Embryonic Kidney cells 293TT, and purified using Optiprep™ density gradient (Sigma-Aldrich, St. Louis, MO) as described (Pastrana et al., 2004). Plasmid constructs for HPV 6, 11, 16 and 18 were obtained as a gift from J. Schiller, National Cancer Institute, Bethesda, MD. Optiprep™ was removed from the VLP preparations using manual gravity-flow based agarose gel filtration. Each VLP fraction was stored at −80 °C with a small aliquot saved for downstream quality analysis. Protein determinations were done using Coomasie Plus Protein Assay (Thermo Scientific, Waltham, MA). The integrity and quality of the VLPs in each fraction were determined using type-specific monoclonal antibodies (Gift of Dr. Neil Christensen, Pennsylvania State University, USA) that recognize a conformational epitope on intact VLPs (WHO HPV Labnet, 2009). VLP fractions with reactivity equal or higher to the laboratory reference VLP stock were pooled to make a new lot of VLP stock. New VLP stock was aliquoted and stored at −80 °C. One aliquot of pooled VLP was again verified for uniformity of shape and size using transmission electron microscopy (TEM) as well as by conformation specific ELISA for reactivity equal or higher to the laboratory reference VLP stock. The fully verified pooled VLP stock was considered qualified for plate coating.
2.2. Selection of assay parameters
HPV 16 single-plex assays were used to evaluate the type of MSD plate, blocking buffer, antigen coating concentration, secondary antibody concentration and incubation times. Single-plex plate printing was performed in-house. A 30 μl aliquot of HPV 16 VLPs diluted in phosphate buffered saline (PBS) (pH7.4) was added to each well of single-spot plate being evaluated and incubated at 4 °C overnight prior to use for testing. ELISA was performed using known control sera with protocol shown in Section 2.4. Evaluation criteria were comparison of the relative light units (RLU) of positive, negative and blank wells.
Additional parameters, including buffer for VLP antigens (Dulbecco’s-Phosphate Buffered Saline (D-PBS) (pH7.4) + 0.5MNaCl with or without Optiprep™), VLP coating concentrations of 20–80 μg/ml, and agents for stabilizing VLP antigen (with or without bovine serum albumin [BSA] or proprietary stabilizer agent) were evaluated on multiplex plates printed by MSD in 7-spot format. HPV 6, 11, 16, and 18 VLPs occupied 4 spots and PBS-BSA alone was used in the remaining 3 spots. Frozen VLP aliquots were sent to MSD on dry-ice where they were stored at −80 °C and thawed prior to printing. The proprietary MSD printing protocol includes QC for spotting accuracy. Plates were shipped to CDC on ice packs within 3–5 days of printing and stored at 4 °C until used for testing. A panel of 16 to 19 residual individual sera that were high, low, and negative in reactivity to the four VLP types based on cLIA (Gift from Merck, Inc.) was used to evaluate the assay parameters using washing and detection protocol as given in Section 2.4.
2.3. Serum samples and controls
For validation experiments, a heat-inactivated pooled serum sample positive for HPV 6, 11, 16 and 18 (Gift from Merck, Inc.) was used as the reference. This sample was calibrated to the HPV16 International Standard (05–134, National Institute for Biological Standards and Controls [NIBSC], Potters Bar, UK) to determine international units (IU)/ml, whereas arbitrary units/ml (AU/ml) were used for the other HPV types. Pooled serum that was high, low and/or negative in reactivity to the different HPV types were used as controls for assay stability. Residual sera from U.S. population-based samples collected between 2005 and 2006, prior to widespread vaccine introduction (n = 4454) that had been previously tested by cLIA (Pharmaceutical Product Development LLC (PPD), Wilmington, NC) were used to compare the two assays. A subset of these samples (n = 100) was used for reproducibility testing across different plate lots and to determine the appropriate dilution series for sample testing. Serum from children (n = 49, gift of Dr. Joakim Dillner, Lund University, Sweden) were used to determine cut-off values (COV) for RLUs for each type.
2.4. MSD 4-plex L1 VLP ELISA (M4ELISA) protocol
Serial 3.16 fold dilutions of serum were prepared with assay diluent [1% ECL™ Blocking Agent (GE Healthcare Biosciences, Piscataway, NJ) in 1X PBST (PBS—0.1% Tween 20)], starting with 1:10 dilution or 1:100 dilution and higher. For each sample, a minimum of 3 dilutions were tested. Serial dilution was performed by Janus® automated liquid handling workstation (PerkinElmer, Waltham, MA). All other steps were performed manually. Plates were blocked for 1 h with 5% ECL™ Blocking Agent in 1X PBST at room temperature (24 °C ± 2), 150 μl per well, on a lab rotator set at 650 rpm. All incubations for subsequent steps were at 37 °C for 1 h with shaking at 650 rpm. After removal of the blocking agent, 25 μl of sample per well was added and the plate incubated. After each incubation plates were washed 4 times with 150 μl per well of 1X PBST using an automated plate washer (ELx405VRS, Biotek, Winooski, VT). 25 μl of biotin-labeled mouse anti-human IgG (Fc specific) (Biotrend Chemicals LLC, Destin, FL) at 1 μg/ml in assay diluent, followed by 25 μl of Streptavidin-Sulfo Tag™(MSD) (1:500 dilution in assay diluent) was added to each well in subsequent steps. 150 μl of 1X Read Buffer T (MSD) was added to each well and the plate was immediately read on the Sector Imager 6000 (MSD) (read time: 70 s/plate). RLU for each spot was exported to Microsoft Excel.
2.4.1. Calculations
Net RLU was calculated by subtracting RLU blank from RLU of each VLP spot in the same well. Net RLUs were used in determination of dynamic range and determination of antibody concentrations with the parallel line method (PLL) as described in the WHO HPV Labnet Manual (Grabowska et al., 2002; WHO HPV Labnet, 2009). Samples with RLU below COV that failed PLL conditions were assigned a zero titer. The RLU COV for each HPV type was determined for each plate lot based on RLUs of 1:100 dilution of children’s sera. The RLU distribution was not normal and best fit a 4-parameter Johnson Su distribution (JOHNSON, 1949). The improved fit to the Johnson Su distribution more than compensated for the two extra parameters needed beyond the normal distribution, as determined by parsimony metrics such as the Akaike Information Criterion (AIC) (Akaike, 1974) and the Bayes Information Criterion (BIC) (Schwarz, 1978). The Lognormal distribution was not a candidate for this data since, for the unexposed sera, a significant portion of the values were <0 after background subtraction. The RLU at the 99th percentile probability distribution (approximately equal to average RLU + 2.33 Standard Deviations (SD) if the data were normally distributed) was used as the COV unless otherwise specified. When noted, the RLU at the 99.87 percentile (approximately equal to average RLU + 3 SD) was used as an alternative COV.
2.5. Establishing assay performance characteristics
2.5.1. Dynamic range
Serial dilutions of high titer sera and plasma from vaccinated individuals, diluted from 1:10 to 1:3.14 × 106, were used to determine the linear dynamic range of the net RLU for three lots of plates. The minimum detection signal was based on average + 3SD over the background signal using only sample diluent for a given VLP type. The maximum signal was set at 1 × 106 RLU, the manufacturer’s designated upper limit for reproducible signal.
2.5.2. Assay variability
Inter-assay coefficient of variation (CV) for quantifiable antibody concentrations within plate lot was determined for two plate lots printed with different VLP preparations using 17–20 serum samples tested in three runs per plate lot, calculated as median CV for both lots. Lot-to-lot inter-assay coefficient of variation was also determined using 11–22 serum samples tested in one run per lot for three plate lots printed with the same VLP preparation and 3 plate lots printed with different VLP preparations. All assays were conducted by the same operator. Inter-operator coefficient of variation was calculated between two operators based on one run on the same plate lot with 23 samples. Intra-assay coefficient of variation was calculated for two operators based on one sample tested on 10–12 plates of the same lot over 4–6 days.
2.5.3. Assay reproducibility
Samples from a mostly unvaccinated cohort were selected based on results of cLIA to include high, low and negative reactivity to the HPV 6, 11, 16 and 18 (n = 100). Samples were tested on two plate lots using a different starting dilution (1:10 or 1:100). Kappa scores were used to evaluate agreement of positive and negative results when tested at different dilutions and different plate lots.
2.5.4. Assay stability
A set of 19 serum samples was assayed once a month on a plate from same lot stored at 4 °C. One lot was tested over a 12 month period and a second lot was tested for 5 months of storage. Time in months was based on the date of receipt in the laboratory not the date of printing. Average fold difference in antibody levels was calculated at each month compared to the first run.
2.6. Assay comparison with cLIA
Final assay parameters established for the M4ELISA were validated by testing 4454 residual samples previously tested with cLIA (Dias, 2005). M4ELISA assay antibody levels (expressed as IU/ml or AU/ml) were compared with cLIA results (mMU/ml or IU/ml). For HPV16, 1 IU/ml = 11.8 mMU/ml (Personal Communication, PPD). Qualitative concordance between the assays was evaluated using the Cohen’s kappa score as well as positive and negative percent agreements. McNemar’s test was performed to identify the test most likely to be positive for antibody detection. Geometric mean concentrations (GMCs) were calculated by type and assay restricted to type-specific positive samples. Except for HPV16, GMCs between assays cannot be compared due to arbitrary units. For HPV16, GMC comparisons were restricted to samples positive in both cLIA and M4ELISA. Spearman’s correlation was calculated for all samples as well as for concordant quantitatifiable concentrations between assays. All statistical analyses were conducted on SPSS ver 21.0 software (IBM, NY) and GraphPad Prism ver 5.0 (La Jolla, CA). Correlation graphs were created using the ggplot2 package in the R program (http://www.R-project.org) (R Core Team, 2014; Wickham, 2009). For graphing purposes only, all values below the detection limit for cLIA were assigned a value of 5 for HPV types 6, 11, 18 and 0.5 for HPV16.
3. Results
3.1. Assay performance characteristics
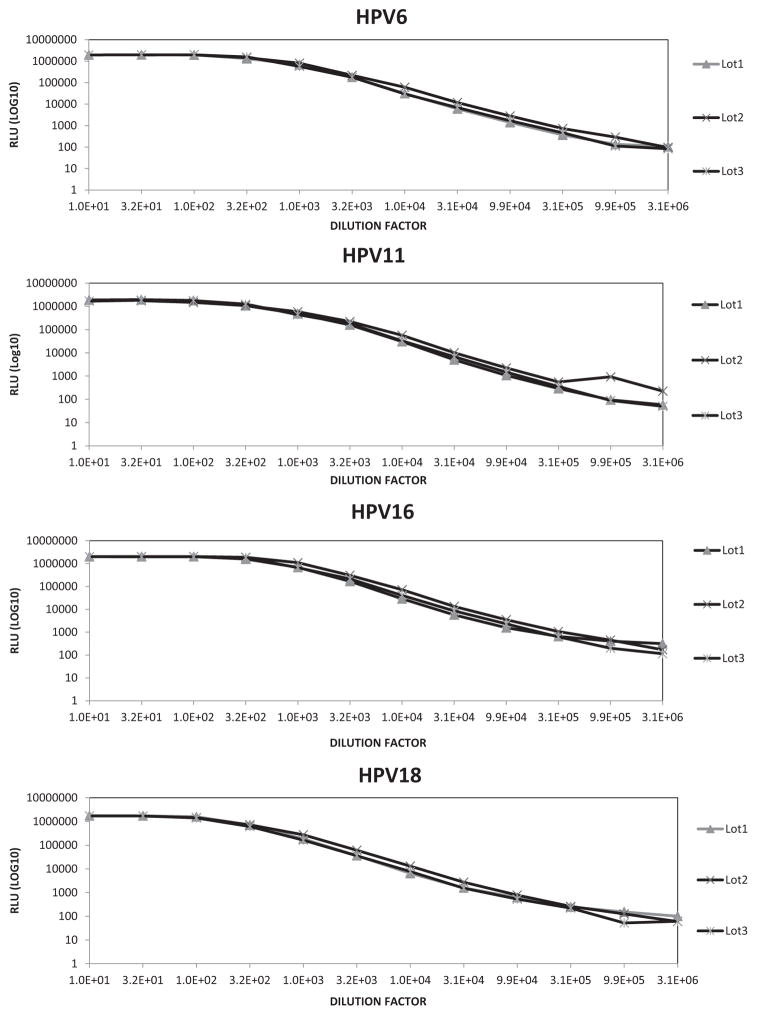
Table 1 summarizes the set of conditions evaluated and selected as the final assay conditions. The dynamic range of the assay was about 1000 fold as evidenced by the linear response seen across 7 dilution steps of a high titer vaccinated serum sample for each HPV type (Fig. 1). The range was consistent over three printed plate lots, with the lower limit of detection based on wells containing only sample diluent ranging from 350 to 650 RLU depending on plate lot.
Table 1.
Assay conditions evaluated for optimization of M4ELISA.
| Parameter | Variations tested | Selected condition |
|---|---|---|
| Evaluated in single-plex assay | ||
| MSD plate | Standard, high-bind | Standard |
| Blocking buffer/assay diluent | Superblock (ThermoFisher, Rockford, IL) ECL blocking agent (GE Healthcare Biosciences, Pittsburgh, PA) |
ECL blocking agent |
| Secondary antibody concentration | 0.5, 1 μg/ml | 1 μg/ml |
| Secondary antibody incubation time | 1 h, 2 h | 1 h |
| Evaluated in multiplex assay | ||
| VLP storage solution | Optiprep Dulbecco’s-PBS + 0.5 M NaCl |
Dulbecco’s-PBS + 0.5 M NaCl |
| VLP coating concentration | 20, 40, 80 μg/ml | 80 μg/ml |
| BSA co-spotting | With BSA Without BSA |
With BSA |
| Stabilizera co-spotting | With stabilizer Without stabilizer |
Without stabilizer |
Proprietary to MSD.
Fig. 1.
Dynamic range of M4ELISA for HPV6, 11, 16 and 18.
The median inter-assay variability was less than 20% for all VLP types irrespective of the plate and reagent lots. Fig. 2 illustrates the inter-assay CVs observed within a plate lot across a range of antibody concentrations measured. Inter-assay variability within the same plate lot was—HPV6 (8.7%), HPV11 (8.9%), HPV16 (7.5%) and HPV 18 (15.8%) (Fig. 2A). The variability between two plate lots with the same VLP preparation was 12.8% for HPV 6, 10.3% for HPV11, 8.3% for HPV16 to 7.9% for HPV 18 (Fig. 2B). Variability among three plate lots with different VLP preparations was slightly higher; 15.8% for HPV6, 14.5% for HPV11, 16.9% for HPV16 and 17.4% for HPV18 (Fig. 2C). Intra-assay variability ranged from 2.9 to 4.8% for operator 1 and 3.5 to 6.7% for operator 2. Inter-operator variability was not extensively evaluated, but median CVs ranged from 5.5 to 13.8% for the four types.
Fig. 2.
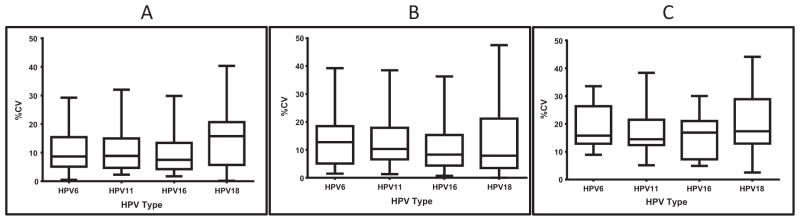
Coefficient of variation. A—Within plate lot (17–20 serum samples tested on two plate lots with three runs per plate lot); B—Lot-to-lot CV (11–22 serum samples tested on three plate lots printed with the same VLP preparation with one run per plate lot); and C—Lot-to-lot CV (11–22 serum samples tested on three plate lots printed with different VLP preparations with one run per plate lot).
Antibody levels for HPV types did not vary more than 2-fold over 5 months for both plate lots evaluated during storage at 4 °C. For the lot evaluated through 12 months of storage, HPV6, 11 and 16 levels remained stable, whereas results for HPV18 varied after month 5 (Fig. 3).
Fig. 3.
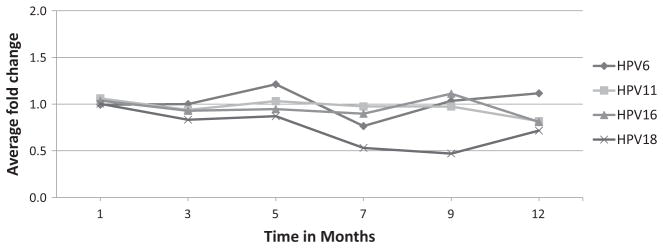
Stability of printed assay plates stored at 4 °C for 12 months.
The quantitative correlation of antibody levels between samples tested at 1:10 and 1:100 starting dilutions on two plate lots was r2 > 0.90 for HPV6, 11, 16 and r2 > 0.87 for HPV18. The kappa score for qualitative agreement between the two lots was 0.8–0.9 for each of the types, with neither lot significantly differing in detection even though different starting dilutions were used (McNemar’s p-value >0.6 for all types). Type-specificity of the M4ELISA was confirmed by testing HPV16 international standard and HPV18 candidate international standard, which were positive only for the respective type and negative for the other three types.
3.2. Assay comparison with cLIA
The overall agreement between the assays ranged from 86.3 to 91.5% for the four types. Positive percent agreement between the two assays ranged from 41.1 to 61.7%. The kappa statistic showed poor to fair agreement between the assays, with the lowest score for HPV18 (κ = 0.38) (Table 2).
Table 2.
Concordance statistics comparing the M4ELISA to the competitive Luminex Assay (cLIA).
| HPV Type | #Positive (%)
|
Kappa score (95%CI) | Positive percent agreement | Negative percent agreement | |
|---|---|---|---|---|---|
| cLIA | M4-ELISA | ||||
| HPV6 | 571 (12.8) | 1020 (22.9) | 0.54 (0.51–0.57) | 61.7 | 91.7 |
| HPV11 | 202 (4.5) | 520 (11.6) | 0.44 (0.39–0.48) | 47.4 | 95.4 |
| HPV16 | 401 (9.0) | 796 (17.8) | 0.53 (0.49–0.56) | 58.3 | 93.5 |
| HPV18 | 147 (3.3) | 496 (11.1) | 0.38 (0.33–0.43) | 41.1 | 95.4 |
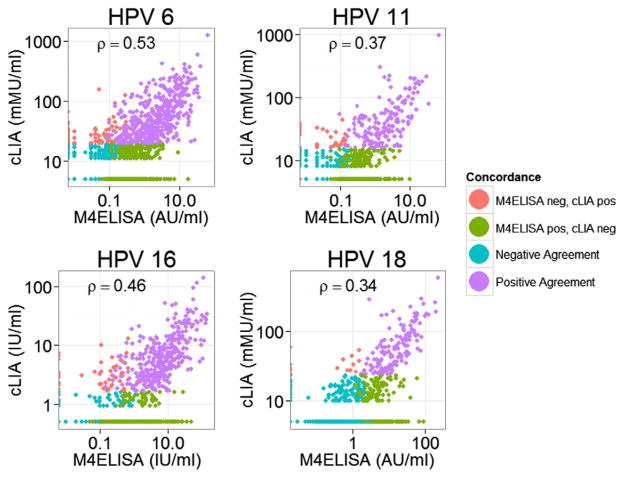
The maximum HPV16 antibody concentration was 141.5 and 142.8 IU/ml for cLIA and M4ELISA respectively. There was no significant difference for HPV16 GMCs calculated for concordant samples (6.2 IU/ml cLIA [95% CI 5.7–6.8] vs 6.7 IU/ml MSD [95% CI 5.8–7.7] (p = 0.23) and HPV16 GMCs were within two-fold if all positive samples by each assay were included: 5.7 IU/ml [95% CI 5.2–6.2 IU/ml] for cLIA positive (n = 401) and 2.6 IU/ml [95% CI 2.4–2.9 IU/ml] for M4ELISA positive (n = 796). Correlations of antibody levels for all samples were ρ = 0.53 for HPV6, 0.37 for HPV11, 0.46 for HPV16 and 0.34 for HPV18 assays (Fig. 4). For concordant samples between each assay, the correlations were ρ = 0.59 HPV6, 0.56 for HPV11, 0.62 for HPV16 and 0.69 for HPV18.
Fig. 4.
Correlation of antibody levels between M4ELISA and cLIA for all samples.
As shown in Table 2, more samples were seropositive by M4ELISA than cLIA (HPV6—22.9% vs 12.8%; HPV11—11.6% vs 4.5%; HPV16—17.8% vs 9.0%; HPV18—11.1% vs 3.3%; p-value <0.001 for all types). The proportions of cLIA negative M4ELISA positive were 11.8% HPV 6, 7.8% HPV11, 10.0% HPV16 and 8.2% HPV18, and the proportions of cLIA positive M4ELISA negative were 1.8% HPV6, 0.7% HPV11, 1.2% HPV16, and 0.3% HPV18 (McNemar’s p < 0.000001 for all types).
Raising the M4ELISA RLU COV to the 99.87-percentile raised the agreement to κ = 0.6–0.7 for all types. The positive percent agreement was 72.1% for HPV6, 64.4% HPV11, 68.3%HPV16, and 65.2% HPV18. McNemar’s differed by type indicating that M4ELISA was more likely to be positive than cLIA for HPV11 and 18 p < 0.002; but not for HPV6 p = 0.34 and HPV 16 p = 0.07. The proportion of cLIA negative M4ELISA positive samples decreased, 3.3% HPV6, 2.2% HPV11, 3.3% HPV16 and 1.6% HPV18.
4. Discussion
This report describes a multiplex ELISA based on the traditional direct L1-VLP ELISA plate format using MSD electrochemiluminescence platform with multi-spot printing to simultaneously detect antibody responses to HPV 6, 11, 16, and 18, the 4 types in the quadrivalent HPV vaccine. The M4ELISA was characterized by large dynamic range for each type (~1000 fold), low background, and plate read times of 70 s. Inter-assay variability as measured by median lot-to-lot CV with same or different VLP preparations was less than 20% for all four types, a value within acceptable limits for multiplexed assays (Marchese et al., 2009; Opalka et al., 2003).
We used results of cLIA in an unvaccinated cohort to verify type-specificity and provide overall comparison to the M4ELISA. It is recognized that direct ELISA and cLIA will not be completely concordant due to differences in their design. ELISA detects IgG binding to all epitopes on VLPs which could include both neutralizing and non-neutralizing epitopes and cLIA detects antibodies of any Ig class that bind to one neutralizing epitope (Schiller and Lowy, 2009). The M4ELISA showed moderate agreement with cLIA for HPV6 and 16 (κ = 0.54 and 0.53 respectively) and fair for HPV 11 and 18 (κ = 0.44 and 0.38 respectively). For all types M4ELISA detected ~2–3 fold more positive samples than cLIA, and this affected both concordance and correlation between the assays. These findings are similar to results observed by other studies comparing VLP-ELISA to cLIA in unvaccinated cohorts with κ ranging from 0.29 to 0.66 (Lin et al., 2013; Scherpenisse et al., 2013; Wentzensen et al., 2011). Most of the discrepant results are near the lower limit of the M4ELISA. Raising the COV for RLUs in M4ELISA from the 99-percentile to the 99.87-percentile (equivalent to + 3SD) improved overall positive agreement and resulted in similar seropositivity rates between the two assays for all four types. Similar results were observed by Safaeian et al. (2012), where positive percent agreement between HPV16 VLP-ELISA and cLIA increased from 49% to 78% with higher COV for ELISA to allow for comparison of the two assay formats.
Determination of cut-off value for HPV serology is difficult as true negatives are hard to determine, leading to varied ways of calculating assay cut-offs. cLIA cut-offs were determined using a clinical sensitivity/specificity algorithm on adult sera, which ensured high specificity of detection at the cost of sensitivity (Dias et al., 2005; Opalka et al., 2003). However, we believe that the 99% RLU limits on children’s sera better reflects the capability of the M4ELISA for detection of antibodies to natural exposure.
The design of the MSD multi-spot wells reduces background noise as each spot forms its own circuit measuring electrochemiluminescent signal bound only to the spot and not to the walls of the microwell. The separate binding surface also reduces competition between antibodies and detection reagent. The low background could contribute to improved detection of antibody response in naturally infected populations, where the antibody levels are low. The Spearman correlation of antibody levels between cLIA and M4ELISA was moderate for each type and ranged between 0.34 and 0.53. For HPV16 and HPV18, correlation was similar to those reported between cLIA and another ELISA platform in unvaccinated population (Wentzensen et al., 2011). Differences between the two assays in high antibody level situations, such as post vaccination, can be anticipated to be smaller. With the exception of HPV16, a direct comparison of GMTs between the assays was not possible. The maximum antibody concentration for HPV16 was in agreement between cLIA and M4ELISA (141.5 and 142.8 IU/ml respectively) as was the GMT for HPV 16 concordant samples (6.2 and 6.7 IU/ml). As the cohort was mostly unvaccinated, only the response to natural exposure was measured. In other reports, GMTs are significantly higher after vaccination than those after natural infection (Romanowski et al., 2009; Schiller et al., 2012). The ability to make this direct comparison between assays for HPV16 highlights the importance of incorporating International Standards into HPV serology assays.
It is recognized that QC of the VLP production is essential for maintaining type-specificity of the reaction (Ferguson et al., 2009). Our results (data not shown) indicate that VLPs in Optiprep™ Gradient Media could be used to coat single-plex MSD plates, but lead to inconsistent antigen printing in 7-spot MSD plates. Removing Optiprep™ resulted in acceptable consistency of antigen printing. We hypothesize that the viscous Optiprep™ media could be interfering with the automated delivery lines designed to deliver very low volumes (nanoliter range) of liquid on each spot. Infrequent random irregularities in spotting, independent of HPV type, were detected in each plate lot as evidenced by 1 out of 3 dilutions of the sample showing low RLU for only one spot within the well. When this occurred, sample was re-tested.
Most published L1-VLP ELISAs specify preparation of plates just prior to use. Multi-spot printing requires sending antigen to the manufacturer for printing. Stability for at least a week was required in order for the assay to be used in any study, but for large cohort testing use of a single lot of reagents would be ideal. Therefore, validating stability of the MSD-printed plates was important. The combination of BSA with VLP concentration of 80 μg/ml allowed for robust detection of antibody for up to twelve months using the same preparation of VLP on printed plates. The effect of BSA on stabilization of protein and enzymes has been well-documented, but the mechanism of action is not clear. It is thought to increase hydrophobic interactions with the protein, thus preventing denaturation (Chang and Mahoney, 1995; Finn et al., 2012).
This is the first study to compare ELISA and cLIA for HPV6 and 11, providing an estimation of antibody response to natural exposure by both assay platforms. We have shown that the multiplexed M4ELISA is reproducible, sensitive, and amenable to high-throughput testing. The assay advantages include the low antigen requirement, rapid reading times, and simplicity of scanner maintenance compared with liquid array platform. The multi-well options for MSD plates include a 10-spot format, suggesting that the current assay could be expanded to include additional HPV types required for testing antibody response to the candidate 9-valent HPV vaccine. The assay also has limitations. The antigens have to be printed by the manufacturer and the plates stored until use. The spot drop-out, noted above, required more repeat testing than would normally occur. Several factors need to be evaluated further such as inter-operator variability and intra-assay variation within a range of antibody concentrations.
“The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the funding agency.”
References
- Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19:716. [Google Scholar]
- Chang BS, Mahoney RR. Enzyme thermostabilization by bovine serum albumin and other proteins: evidence for hydrophobic interactions. Biotechnol Appl Biochem. 1995;22(Pt 2):203. [PubMed] [Google Scholar]
- Dias D, Van DJ, Schlottmann S, Kelly S, Puchalski D, Ruiz W, Boerckel P, Kessler J, Antonello JM, Green T, Brown M, Smith J, Chirmule N, Barr E, Jansen KU, Esser MT. Optimization and validation of a multiplexed luminex assay to quantify antibodies to neutralizing epitopes on human papillomaviruses 6, 11, 16, and 18. Clin Diagn Lab Immunol. 2005;12:959. doi: 10.1128/CDLI.12.8.959-969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, Wilkinson DE, Zhou T. WHO meeting on the standardization of HPV assays and the role of the WHO HPV Laboratory Network in supporting vaccine introduction held on 24–25 January 2008, Geneva, Switzerland. Vaccine. 2009;27:337. doi: 10.1016/j.vaccine.2008.10.062. [DOI] [PubMed] [Google Scholar]
- Finn TE, Nunez AC, Sunde M, Easterbrook-Smith SB. Serum albumin prevents protein aggregation and amyloid formation and retains chaperone-like activity in the presence of physiological ligands. J Biol Chem. 2012;287:21530. doi: 10.1074/jbc.M112.372961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska K, Wang X, Jacobsson A, Dillner J. Evaluation of cost-precision ratios of different strategies for ELISA measurement of serum antibody levels. J Immunol Methods. 2002;271:1. doi: 10.1016/s0022-1759(02)00334-4. [DOI] [PubMed] [Google Scholar]
- Johnson NL. Systems of frequency curves generated by methods of translation. Biometrika. 1949;36:149. [PubMed] [Google Scholar]
- Lin SW, Ghosh A, Porras C, Markt SC, Rodriguez AC, Schiffman M, Wacholder S, Kemp TJ, Pinto LA, Gonzalez P, Wentzensen N, Esser MT, Matys K, Meuree A, Quint W, van Doorn LJ, Herrero R, Hildesheim A, Safaeian M. HPV16 seropositivity and subsequent HPV16 infection risk in a naturally infected population: comparison of serological assays. PLoS ONE. 2013;8:e53067. doi: 10.1371/journal.pone.0053067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese RD, Puchalski D, Miller P, Antonello J, Hammond O, Green T, Rubinstein LJ, Caulfield MJ, Sikkema D. Optimization and validation of a multiplex, electrochemiluminescence-based detection assay for the quantitation of immunoglobulin G serotype-specific antipneumococcal antibodies in human serum. Clin Vaccine Immunol. 2009;16:387. doi: 10.1128/CVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka D, Lachman CE, MacMullen SA, Jansen KU, Smith JF, Chirmule N, Esser MT. Simultaneous quantitation of antibodies to neutralizing epitopes on virus-like particles for human papillomavirus types 6, 11, 16, and 18 by a multiplexed luminex assay. Clin Diagn Lab Immunol. 2003;10:108. doi: 10.1128/CDLI.10.1.108-115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana DV, Buck CB, Pang YY, Thompson CD, Castle PE, FitzGerald PC, Kruger KS, Lowy DR, Schiller JT. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology. 2004;321:205. doi: 10.1016/j.virol.2003.12.027. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. ( http://www.R-project.org/) [Google Scholar]
- Romanowski B, de Borba PC, Naud PS, Roteli-Martins CM, De Carvalho NS, Teixeira JC, Aoki F, Ramjattan B, Shier RM, Somani R, Barbier S, Blatter MM, Chambers C, Ferris D, Gall SA, Guerra FA, Harper DM, Hedrick JA, Henry DC, Korn AP, Kroll R, Moscicki AB, Rosenfeld WD, Sullivan BJ, Thoming CS, Tyring SK, Wheeler CM, Dubin G, Schuind A, Zahaf T, Greenacre M, Sgriobhadair A. Sustained efficacy and immunogenicity of the human papillomavirus (HPV)-16/18 AS04-adjuvanted vaccine: analysis of a randomised placebo-controlled trial up to 6.4 years. Lancet. 2009;374:1975. doi: 10.1016/S0140-6736(09)61567-1. [DOI] [PubMed] [Google Scholar]
- Safaeian M, Ghosh A, Porras C, Lin SW, Rodriguez AC, Schiffman M, Wacholder S, Kemp T, Gonzalez P, Wentzensen N, Esser M, Meuree A, Matys K, Quint W, van Doorn LJ, Sherman ME, Herrero R, Pinto LA, Hildesheim A. Direct comparison of HPV16 serological assays used to define HPV-naive women in HPV vaccine trials. Cancer Epidemiol Biomarkers Prev. 2012;21:1547. doi: 10.1158/1055-9965.EPI-12-0558. [DOI] [PubMed] [Google Scholar]
- Scherpenisse M, Schepp RM, Mollers M, Mooij SH, Meijer CJ, Berbers GA, van der Klis FR. Comparison of different assays to assess human papillomavirus (HPV) type 16- and 18-specific antibodies after HPV infection and vaccination. Clin Vaccine Immunol. 2013;20:1329. doi: 10.1128/CVI.00153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller JT, Lowy DR. Immunogenicity testing in human papillomavirus virus-like-particle vaccine trials. J Infect Dis. 2009;200:166. doi: 10.1086/599988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller JT, Castellsague X, Garland SM. A review of clinical trials of human papillomavirus prophylactic vaccines. Vaccine. 2012;30(Suppl 5):F123. doi: 10.1016/j.vaccine.2012.04.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461. [Google Scholar]
- Wentzensen N, Rodriguez AC, Viscidi R, Herrero R, Hildesheim A, Ghosh A, Morales J, Wacholder S, Guillen D, Alfaro M, Safaeian M, Burk RD, Schiffman M. A competitive serological assay shows naturally acquired immunity to human papillomavirus infections in the Guanacaste Natural History Study. J Infect Dis. 2011;204:94. doi: 10.1093/infdis/jir209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO HPV Labnet. Human Papillomavirus Laboratory Manual (WHO/IVB/ 10.12) 2009 http://whqlibdoc.who.int/hq/2010/WHO_IVB_10.12_eng.pdf?ua=1.
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]