An actin nucleation protein affects rice panicle development as well as root growth.
Abstract
Panicle development, a key event in rice (Oryza sativa) reproduction and a critical determinant of grain yield, forms a branched structure containing multiple spikelets. Genetic and environmental factors can perturb panicle development, causing panicles to degenerate and producing characteristic whitish, small spikelets with severely reduced fertility and yield; however, little is known about the molecular basis of the formation of degenerating panicles in rice. Here, we report the identification and characterization of the rice panicle degenerative mutant tutou1 (tut1), which shows severe defects in panicle development. The tut1 also shows a pleiotropic phenotype, characterized by short roots, reduced plant height, and abnormal development of anthers and pollen grains. Molecular genetic studies revealed that TUT1 encodes a suppressor of cAMP receptor/Wiskott-Aldrich syndrome protein family verprolin-homologous (SCAR/WAVE)-like protein. We found that TUT1 contains conserved functional domains found in eukaryotic SCAR/WAVE proteins, and was able to activate Actin-related protein2/3 to promote actin nucleation and polymerization in vitro. Consistently, tut1 mutants show defects in the arrangement of actin filaments in trichome. These results indicate that TUT1 is a functional SCAR/WAVE protein and plays an important role in panicle development.
Rice (Oryza sativa) yield is closely associated with panicle development, including panicle structure and panicle number. The basic unit of the rice panicle is the spikelet, which consists of multiple florets (each having a lodicule, carpel, and six stamens) enclosed by glumes (Yoshida and Nagato, 2011). During rice growth and development, panicle degeneration may occur under various conditions (Komatsu et al., 2001; Ashikari et al., 2005; Ikeda et al., 2005). The abortion of spikelets usually causes degeneration of floral organs, leading to reduction of rice yield. Spikelet degeneration can occur from the basal or apical end of the panicle and lead to defective, whitish spikelets, and eventually fewer spikelets per panicle (Li et al., 2009; Cheng et al., 2011).
Various genetic and environmental factors cause panicle degeneration, but the underlying mechanisms remain poorly understood (Xing and Zhang, 2010). Several quantitative trait loci related to panicle apical spikelet abortion have been reported. One such quantitative trait locus, panicle apical abortion8, affects the formation of anther and pollen, and level of reactive oxygen species (ROS) in spikelets (Cheng et al., 2011). Several reports also indicate that cuticle deficiency can cause degenerated anthers and abnormal pollen grains (Jung et al., 2006; Li et al., 2010). Cuticles, the hydrophobic, waxy structures produced by the epidermal cells in most land plants, play an important role in plant development and in the tolerance to environmental stresses. The constituents of the cuticle include fatty acids, esters, alcohols, aldehydes, ketones, and alkanes (Kunst and Samuels, 2003; Panikashvili et al., 2007). Defects in fatty acid metabolism or transport can also cause anther degeneration. Mutations in rice cytochrome P450 family gene, CYP704B2, encoding an enzyme catalyzing the production of Ω-hydroxylated C16 and C18 fatty acids, cause a degenerative phenotype in anther development, associated with defective anther cutin biosynthesis (Li et al., 2010). OsC6 is a fatty acid transporter that affects cutin formation in rice anther, and mutations in the OsC6 gene also produce a degenerative anther phenotype (Zhang et al., 2010).
In plants, a complicated genetic regulatory network controls cytoskeletal organization (Hussey et al., 2006). Actin filaments, one of the major components of the cytoskeleton, are composed of linear polymers that polymerize from G-actin subunits under the control of regulatory proteins (Elzinga et al., 1973). The actin-related protein 2/3 (Arp2/3) complex forms a nucleation site to initiate new F-actin branches and is essential for actin filament polymerization in animal, yeast (Saccharomyces cerevisiae), and plant cells (Vartiainen and Machesky, 2004; Yanagisawa et al., 2013). During actin polymerization, suppressor of cAMP receptor/Wiskott-Aldrich syndrome protein family verprolin-homologous (SCAR/WAVE) proteins activate the Arp2/3 complex. The carboxyl regions of SCAR/WAVE proteins contain a conserved verprolin, cofilin, and acidic (VCA) domain that has three subdomains: the verprolin-homology domain, the cofilin-homology domain, and the acidic domain (Takenawa and Miki, 2001). SCAR/WAVE proteins bind to both actin monomer and the Arp2/3 complex through the VCA domain to promote actin polymerization by activating the Arp2/3 complex (Miki and Takenawa, 2003; Frank et al., 2004).
Work in plant systems has identified and characterized a number of the components of the highly conserved SCAR-Arp2/3 pathway (Le et al., 2003; Mathur et al., 2003; El-Din El-Assal et al., 2004a; Deeks and Hussey, 2005; Harries et al., 2005). SCAR proteins share limited amino acid sequence similarity among various species, but the VCA domain shows high evolutionary conservation in animals and plants (Frank et al., 2004; Basu et al., 2005; Zhang et al., 2005, 2008). Studies in Arabidopsis (Arabidopsis thaliana) and maize (Zea mays) showed that the VCA domain of SCARs can activate the Arp2/3 complex (Frank et al., 2004; Basu et al., 2005; Uhrig et al., 2007; Zhang et al., 2008). Mutations in the genes encoding SCAR/WAVE complex proteins show various defects in actin arrangement and trichome development (Frank and Smith, 2002; Frank et al., 2003; Mathur et al., 2003; Basu et al., 2005; Szymanski, 2005; Zhang et al., 2005, 2008; Uhrig et al., 2007; Yanagisawa et al., 2015). Moreover, mutations in all of the identified Arabidopsis Arp2/3 complex subunit genes also display a similar phenotype, with defects in actin organization and aberrant leaf trichome morphology (Le et al., 2003; Mathur et al., 2003; El-Din El-Assal et al., 2004a; Saedler et al., 2004; Kotchoni et al., 2009; Yanagisawa et al., 2015), suggesting that the Arp2/3 and SCAR/WAVE complexes genetically act in a linear pathway. In addition to SCAR proteins, several proteins have been identified in the Arabidopsis SCAR/WAVE complex (Deeks et al., 2004; El-Din El-Assal et al., 2004b; Le et al., 2006; Jörgens et al., 2010). Among those, BRICK1 (BRK1) is involved in the stabilization of SCAR protein in vivo (Le et al., 2006), and mutations in the BRK genes result in defects of epidermal cell morphogenesis and cell division in maize (Frank et al., 2003, 2004). However, given the critical role of the Arp2/3 complex and SCAR/WAVE in the organization of the cytoskeleton, it is somewhat unexpected that none of these mutations substantially affects plant growth and development in Arabidopsis and maize.
In this study, we report the characterization of TUTOU1 (TUT1), which encodes a functional SCAR/WAVE protein and plays an important role in rice growth and development. The tut1 shows a pleiotropic phenotype with severe defects in panicle development. We show that TUT1 can activate the Arp2/3 complex to promote actin polymerization in vitro, and is involved in modulating actin organization in vivo. We demonstrate that TUT1 plays important roles in growth and development of rice plants.
RESULTS
Identification and Characterization of the Rice tut1 Mutant
To understand the cellular and molecular mechanisms that regulate panicle development, we performed a genetic screen for tut (which means bald head in Chinese) mutants that have a panicle apical degeneration phenotype from a gamma ray-mutagenized M2 population in the japonica variety ‘Zhonghua 11’ background. From this screen, we identified several tut mutants, and we report a detailed study of tut1. tut1 was backcrossed to wild-type plants (cv Zhonghua 11) three times, and progeny showing the tut1 phenotypes were collected for all studies described below, unless otherwise indicated.
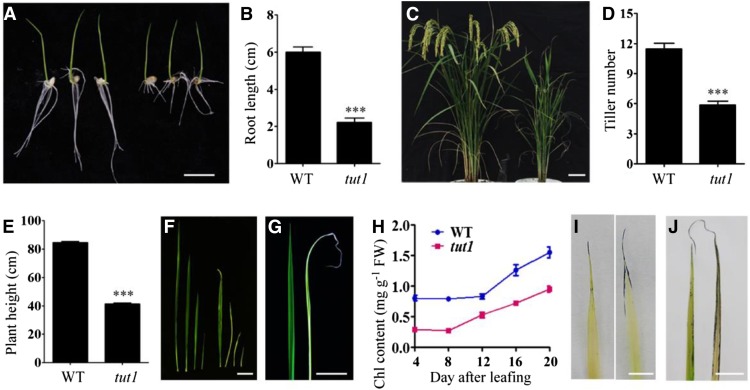
During early seedling growth, the aerial part of tut1 seedlings showed a wild-type-like phenotype (Fig. 1A). However, the tut1 roots were shorter than wild-type roots (Fig. 1, A and B). Compared with wild-type plants, the mature tut1 plants were shorter and had fewer tillers (Fig. 1, C–E). During all growth stages, the tut1 leaves were smaller than wild-type leaves and showed degeneration that initiated from the leaf tips, accompanied by a reduction of the chlorophyll levels (Fig. 1, F–H). Young tut1 leaves were usually pale white and then turned green upon maturation (Fig. 1F). Consistent with the observation of degenerated leaf tips in tut1, massive cell death was detected by staining with Evans blue along the leaf tips of tut1, but was rarely detectable in wild-type leaves (Fig. 1I). Moreover, tut1 leaves had higher levels of superoxide than did the wild type (Fig. 1J), suggesting that degenerative cell death in tut1 leaves was related to the production of ROS.
Figure 1.
Phenotypes of tut1. A, Seedling phenotype of the wild type (cv Zhonghua 11, left three seedlings) and tut1 (right three seedlings). Seeds were germinated at 30°C for 3 d and then transferred to a culture chamber with a 16-h/8-h photoperiod at 30°C. Bar = 1 cm. B, Statistical analysis of root length in the wild type (WT) and tut1 as shown in A. The data presented are mean values of two independent experiments (n > 45). Error bars indicate sd; ***, P < 0.01 (Student’s t test). C, Phenotypes of wild-type (left) and tut1 (right) mutants at the grain-filling stage. Bar = 5 cm. D and E, Statistical analysis of tiller number and plant height at maturity of the wild type and tut1. The data presented are mean values (n > 50). Error bars indicate sd; ***, P < 0.01 (Student’s t test). F, Leaves of the wild type (left four leaves) and tut1 (right four leaves) at different growth stages. Bar = 5 cm. G, Leaves of the wild type (left) and tut1 (right) at 10 d after leafing. Bar = 2 cm. H, Analysis of chlorophyll (Chl) content of wild-type and tut1 leaves at different times after leafing. Blue line indicates the wild type, red line indicates tut1. Leaves at 4, 8, 12, 16, and 20 d after leafing were used in the analysis. Fresh leaves were used for measurement of chlorophyll contents. Two technical repeats were made in the test. Error bars indicate sd. I, Cell death analysis of wild-type (left) and tut1 (right) seedling leaves using Evans blue staining. Bar = 0.5 cm. J, ROS observation of wild-type (left) and tut1 (right) mature leaves using nitroblue tetrazolium (NBT) staining. FW, Fresh weight. Bar = 2 cm.
Degenerated Panicles in tut1
tut1 showed a delayed-heading phenotype, approximately 15 d later than the wild-type plants (Fig. 1C). The tut1 panicles were significantly shorter than wild-type panicles and showed severe degeneration of spikelets in the apical portion of the panicle (Fig. 2A). In the tut1 panicles, the degenerating spikelets occurred in the apical parts of all branches, including the primary and secondary branches (Fig. 2B; Supplemental Fig. S1). Similar to that in leaves, we also observed significantly increased ROS levels in the degenerated tut1 spikelets compared with that in the wild type (Fig. 2, C and D). tut1 had smaller flower filaments than the wild type, and the tut1 anthers were usually pale white and showed various degrees of degeneration (Fig. 2E; Supplemental Fig. S2).
Figure 2.
Morphology of panicle and spikelet in tut1. A, Panicle rachis of wild-type (WT; left) and tut1 (right) plants at the grain-filling stage. Bar = 2 cm. B, The expanding panicle branch of wild-type (left) and tut1 (right) plants at the grain-filling stage. The part of the tut1 panicle shown in the white box was enlarged. The arrows show the degenerated branches and spikelets in the tut1. Bars = 2 cm. C, ROS analysis of the wild type (bottom) and tut1 (top) by NBT staining. Bar = 2 mm. D, ROS analysis of spikelets showing different degrees of degeneration in tut1 at the heading stage by NBT staining. Bar = 2 mm. E, Comparison of wild-type (left one) and tut1 spikelets (right three) after removing the lemma and palea at the heading stage. Right three spikelets, named Type 1, Type 2 and Type 3, show varying degrees of developmental defects in tut1. Bar = 1 mm. F to K, Scanning electron microscopy (SEM) observation of spikelet at the tip of the panicle at stage In7 (F), stage In8 (arrows indicate the apical spikelets in the wild type and the tut1 [G]), stages In8 and In9 (H; the white square frame region shown in H is enlarged in I), and stage In9 (J; the white square frame region shown in J is enlarged in K). Bars = 100 μm (F and K), 400 μm (G), 200 μm (H), 10 μm (I), and 500 μm (J).
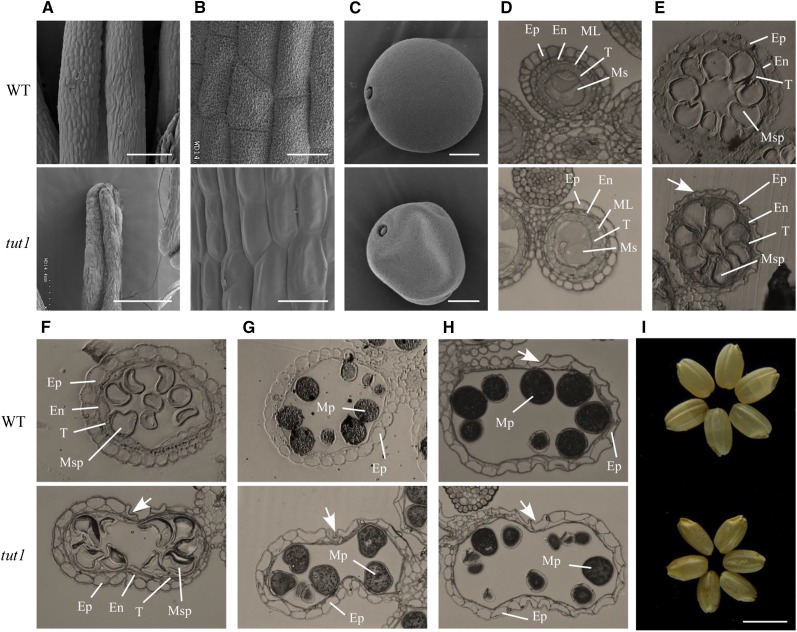
To analyze the cellular basis of panicle degeneration in tut1, we examined panicle development of the wild type and tut1 by SEM over the nine stages (Inflorescence development stage1–9 [In1–In9]) of rice panicle development (the panicle development stages were followed as defined by Ikeda et al., 2004). In the early stages, the spikelets at the tip of wild-type and tut1 panicles showed no significant difference, and spikelets at the In7 stage are shown in Figure 2F. At stage In8, the difference between the wild type and tut1 became apparent, as the panicle apical spikelet of tut1 was smaller than the wild type (Fig. 2G). At stages In8 and In9, the tut1 spikelets were misshapen and disorganized compared with the wild type (Fig. 2H). A closer examination revealed that the surface of wild-type spikelets was well organized and smooth, whereas the surface of the tut1 spikelet contained shrunken and disorganized cell files (Fig. 2I). During later developmental stages, the degenerative phenotype of tut1 panicles became more severe, resulting in the formation of small, aborted spikelets that lacked recognizable cell files on the surface of the tut1 spikelets (Fig. 2, J and K). These results suggested that TUT1 plays an important role in panicle development and is likely involved in cell expansion.
Defective Anther and Pollen Development in tut1
Anthers in tut1 also showed a degenerative phenotype, as mutant anthers lacked organized cell files on the epidermis and abortive or shrunk pollen grains (Figs. 2E and 3, A–C). We analyzed pollen development in tut1 in comparison with that of the wild type by light microscopy. The developing tut1 pollen showed no abnormalities at the microspore mother cell stage (Fig. 3D). However, after entering the vacuolated pollen stage, the tut1 microspores became misshapen and disorganized (Fig. 3E), and eventually aborted (Fig. 3, F–H). Notably, the degeneration of the epidermal cell layer usually occurred after the mature pollen stage in the wild type, whereas the premature degeneration of the epidermal cells in tut1 anthers was observed at the vacuolated pollen stage (Fig. 3E). Moreover, the tut1 pollen sac also had an irregular shape compared with the wild type at the first pollen mitosis stage (Fig. 3, F and G). At the mature pollen stage, tut1 had significantly fewer mature pollen grains compared with the wild type (Fig. 3H). Due to these defects, tut1 showed poor fertility and produced smaller seeds (Fig. 3I; Supplemental Fig. S1).
Figure 3.
SEM and semithin section analysis of wild-type and tut1 anther morphology. A to C, SEM observation of mature anthers of the wild type (WT) and tut1 (A), enlarged view of the anther surface of the wild type and tut1 (B), and mature pollen grain of the wild type and the tut1 (C). Bars = 200 μm (A), 20 μm (B), and 10 μm (C). D to H, The cross sections were stained with toluidine blue. Ep, Epidermis; En, endothecium; ML, middle layer; Ms, microsporocyte; Msp, microspore; Mp, mature pollen; T, tapetum. Cross section of single locule at the microspore mother cell stage; Ep, En, ML, T, and Ms are indicated (D); vacuolated pollen stage; Ep, En, T, and Msp are indicated (E); pollen first mitosis stage; Ep, En, T, and Msp are indicated (F); pollen second mitosis stage; Ep and Mp are indicated (G); and mature pollen stage; Ep and Mp are indicated (H). The arrows show the degenerated epidermal cell. I, Phenotype of mature seeds without glumes of the wild type (top) and tut1 (bottom). Bar = 5 mm.
Defective Cuticle Formation in tut1
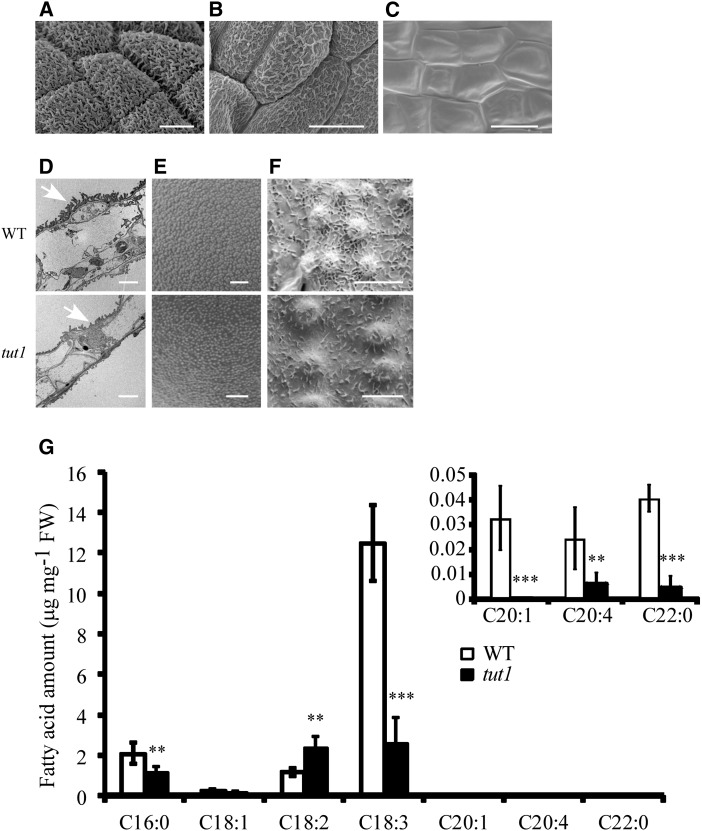
The epidermis of wild-type anthers was coated with epicuticular wax crystals, but these wax crystals were substantially fewer and disorganized on the surface of the tut1 anthers (Fig. 3, A and B). On the tut1 anthers, the formation of a wax cuticle was reduced in the basal panicle and completely absent in the apical panicle (Fig. 4, A–C). Consistent with this, transmission electron microscopy revealed that the tut1 cuticle had significantly fewer wax structures compared with the wild type (Fig. 4D). Similarly, the tut1 mutation also caused abnormalities in pollen exine development (Fig. 4E) and formation of a cuticle on the surface of leaves and leaf stomata (Fig. 4F; Supplemental Fig. S3). These results suggested that TUT1 is involved in cuticle formation, which may explain the defective development of anthers and pollen in tut1.
Figure 4.
Cuticle formation and fatty acid metabolism in the wild type (WT) and tut1. A to C, SEM observation of mature anther cutin of the wild type (A) and tut1 at the middle panicle (B) and apical panicle (C). tut1 anthers showing different degrees of degeneration are shown in B and C. Bars = 10 μm. D, Transmission electron microscopy observation of wild-type and tut1 mature anthers. Arrowhead indicates cuticle. Bar = 5 μm. E and F, SEM observation of the surface of mature pollen grains (E) and the cuticle of mature leaves (F) of the wild type and tut1. Bars = 2 μm (E) and 5 μm (F). G, Analysis of fatty acid contents in the wild type and tut1 using gas chromatography. Fresh panicles at the heading stage of the wild type and tut1 were used in the measurement. C17 was added as an internal standard. Seven types of fatty acid, C16:0, C18:1, C18:2, C18:3, C20:1, C20:4, and C22:0, were measured, and three biological repeats were conducted. FW, Fresh weight. Error bars indicate sd. **, P < 0.05; ***, P < 0.01 (Student’s t test).
We reasoned that the defective development of wax cuticles might be related to abnormal metabolism of fatty acids. To test this possibility, we analyzed the fatty acid levels in heading panicles by gas chromatography. We found that the levels of palmitic acid (C16:0), eicosenoic acid (C20:1), arachidonic acid (C20:4), and docosanoic acid (C22:0) were reduced in tut1 panicles compared with the wild type (Fig. 4G), suggesting that altered fatty acid metabolism may be related to the tut1 phenotype.
Molecular Cloning of TUT1
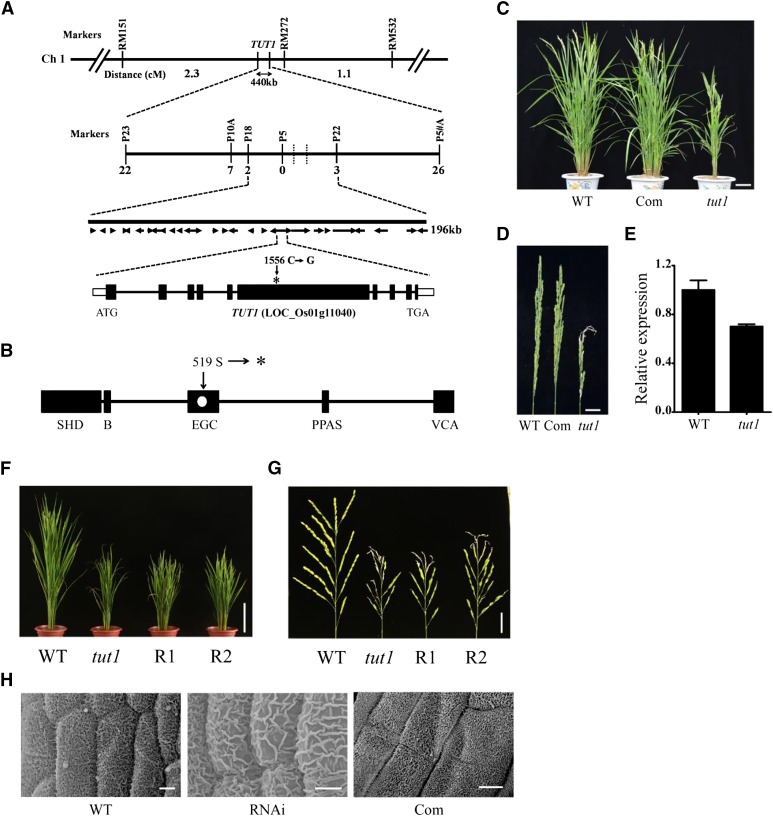
In a cross between the wild type (cv Zhonghua 11) and tut1, all F1 progenies displayed a wild-type phenotype. In the self-pollinated F2 population, the wild-type and tut1 mutant plants segregated in a ratio of approximately 3:1 (wild type:tut1 = 203:54; χ2 = 2.17, P < 0.05), indicating that tut1 is a recessive mutation in a single nuclear gene. To genetically map TUT1, we generated an F2 population (approximately 10,000 plants) derived from the cross between tut1 and cv Zhenshan 97 (an indica cultivar). TUT1 was roughly mapped on the short arm of chromosome 1, between the simple sequence repeat (SSR) markers RM151 and RM272 (Fig. 5A). Using 2,600 F2- and F3-derived tut1 segregants, we mapped TUT1 to an interval of 196 kb between the SNP markers P18 and P22 (Fig. 5A). This region contained 28 annotated genes, and DNA sequencing analysis of tut1 revealed a C-to-G transition at nucleotide 1,556 of the coding sequence of LOC_Os01g11040. The mutation occurred in exon 6, which converted a Ser into a premature stop codon (TCA to TGA), resulting in the formation of a truncated polypeptide lacking 815 amino acid residues of the carboxyl region (Fig. 5, A and B). qRT-PCR revealed that the transcript levels of the putative TUT1 gene were slightly lower in tut1 than in the wild type (Fig. 5E). Also, the tut1 mutation occurred in the highly conserved EGC domain and caused the loss of the VCA domain at the carboxyl region (Fig. 5B), which is functionally important for the SCAR/WAVE proteins (Machesky et al., 1999; Basu et al., 2005; Zhang et al., 2008). Therefore, tut1 is likely a null mutant allele.
Figure 5.
Map-based cloning of TUT1. A, TUT1 maps on chromosome 1. TUT1 was roughly mapped to a 440-kb interval between the SSR markers RM151 and RM272. The number of recombinants between the molecular markers and TUT1 are indicated. TUT1 was fine mapped to an interval of about 196 kb between the single nucleotide polymorphism (SNP) markers P18 and P22. This region contained 28 predicted genes. A one-nucleotide transversion mutation was found in a gene annotated as a SCAR-like protein2 (LOC_Os01g11040). The mutation site at nucleotide 1,556 (C–G) in the sixth exon is indicated by the asterisk. Ch 1, Chromosome 1. B, Conserved domain organization of TUT1, showing the SCAR/WAVE homolog domain (SHD), basic domain (B), envelope glycoprotein C (EGC) domain, pantetheine attachment site (PPAS), and VCA domain as black rectangles. The mutation alters the 519th codon (Ser) to a termination codon (shown as a white dot in the EGC domain). The asterisk indicates the termination codon. C, Phenotypes of the wild type (WT), complemented transgenic line (Com), and tut1 at the heading stage. Bar = 10 cm. D, Mature panicle phenotypes of the wild type, complemented line, and tut1. Bar = 2 cm. E, TUT1 expression analysis of the wild type and tut1 by quantitative reverse transcription (qRT)-PCR. F, Morphology of wild type, tut1, and RNA interference (RNAi) transgenic lines (R1 and R2) at the heading stage. Bar = 20 cm. G, Panicles of wild type, tut1, and RNAi transgenic lines (R1 and R2) at the flowering stage. Bar = 3 cm. H, Anther cuticle of wild type, tut1, and representative complemented line. Bars = 20 cm (F), 3 cm (G), and 5 μm (H).
To verify the identity of the putative TUT1 gene, we conducted a complementation assay. To this end, we transformed a full-length wild-type complementary DNA (cDNA) fragment of LOC_Os01g11040 under the control of the maize ubiquitin promoter into the TUT1/tut1 heterozygous plants. The transgene fully rescued the tut1 phenotype, including the degenerated panicle phenotype (Fig. 5, C and D). Moreover, the transgenic plants carrying an RNAi transgene had a reduced expression level of LOC_Os01g11040 (Supplemental Fig. S4) and showed tut1-like phenotypes, including reduced plant height, degenerated panicles, cuticle defects, blunt trichomes, and increased ROS levels (Fig. 5, F–H; Supplemental Fig. S4). These results demonstrate that LOC_Os01g11040 represents the TUT1 gene.
Analysis of TUT1 Expression
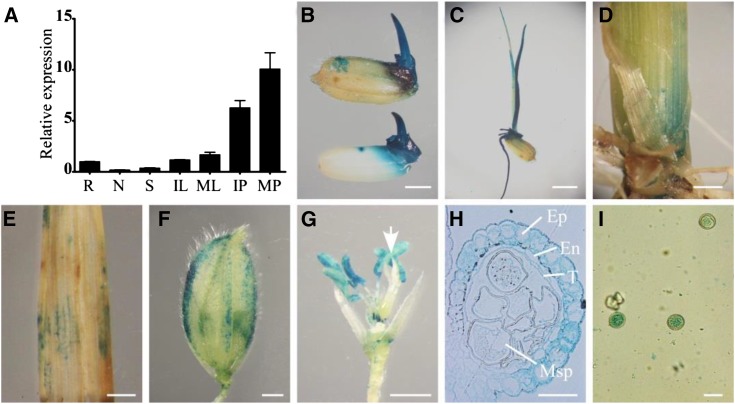
To investigate the expression pattern of TUT1 in rice, we used qRT-PCR to measure the TUT1 mRNA levels in various organs or tissues of wild-type plants, including root, node, stem, immature leaf, mature leaf, immature panicle, and mature panicle (Fig. 6A). Consistent with the tut1 phenotype, the level of TUT1 mRNA was highest in the panicles, followed by leaves. We also detected TUT1 expression in roots, stems, and nodes, but at lower levels compared with that in panicles and leaves (Fig. 6A).
Figure 6.
Spatial expression pattern of TUT1. A, TUT1 mRNA levels were measured by qRT-PCR. Total RNA was extracted from the root (R), node (N), stem (S), immature leaf (IL), mature leaf (ML), immature panicle (IP), and mature panicle (MP) of the wild type. B to I, Expression of TUT1, as revealed by promoter-GUS fusion analysis in transgenic plants. GUS activity was observed in the mature glume, seed, and seed germ (B), root, coleoptile, and seedling (C); basal node of the shoot (D), leaf (E), and glume at the heading stage (F); and anther filaments and anthers. The arrow indicates the pistil (G). GUS staining of a cross section of the anther, with epidermis (Ep), endothecium (En), tapetum (T), and microspore (Msp) indicated (H) and mature pollen (I). Bars = 2 mm (B, D, and E), 4 mm (C), 1 mm (F and G), and 30 μm (H and I).
To analyze the spatial expression pattern of TUT1, we generated transgenic plants harboring a TUT1:GUS reporter gene. Five independent TUT1:GUS transgenic lines were analyzed and gave similar results. We found GUS activity in mature glume, seed germ, root, coleoptile, node, and leaf (Fig. 6, B–E), consistent with the tut1 phenotype of short roots, small seeds, dwarf plants, and degenerating leaves. Close examination of TUT1:GUS expression in the panicle revealed TUT1 expression in glumes, anthers, and mature pollen grains (Fig. 6, F–I). In the anther, TUT1:GUS expression mainly occurred in the epidermis and endothecium, with a lower level in the tapetum and microspore (Fig. 6H). TUT1:GUS was also highly expressed in mature pollen grains (Fig. 6I). These results suggest that the expression pattern of TUT1 is consistent with its important role in panicle development, especially in anther development.
TUT1 Encodes a Functional SCAR/WAVE Protein
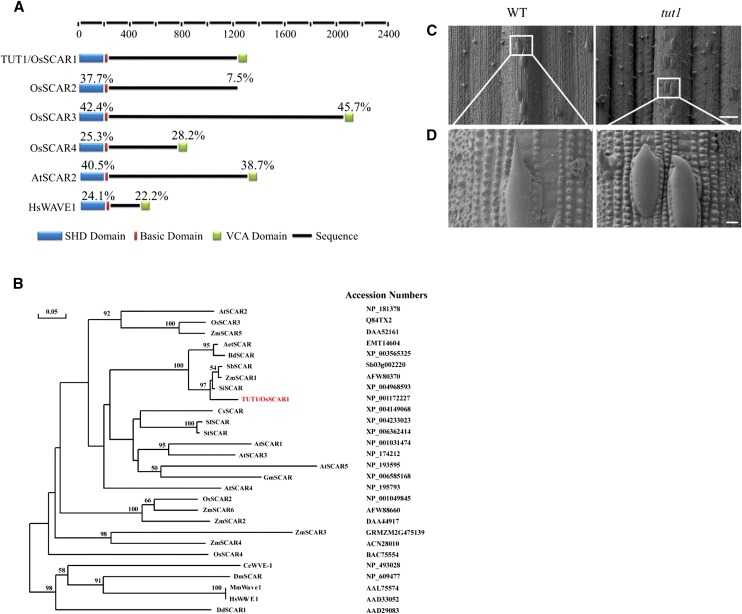
Sequence alignment suggested that TUT1 encodes a SCAR/WAVE-like protein (Supplemental Fig. S5; Supplemental Methods S1). The classic SCAR/WAVE family proteins contain a SHD, a short stretch of basic amino acids in the N-terminal region, and a VCA domain in the C-terminal region (Fig. 7A). A sequence comparison indicated that TUT1 contains all functionally important domains found in eukaryotic SCAR/WAVE proteins (Fig. 7A). TUT1 exists as a single-copy gene in the rice genome. In a previous study, TUT1 was also named OsSCAR1 (Zhang et al., 2008). Three additional rice genes encode proteins containing the SCAR homolog domain, annotated as OsSCAR2, OsSCAR3, and OsSCAR4, of which OsSCAR2 lacks the putative VCA domain (Fig. 7A; Supplemental Fig. S5). TUT1/OsSCAR1 shared the highest sequence similarity with SCAR-like proteins from Brachypodium distachyon (68% identity), Sorghum bicolor (63% identity), Setaria italica (62% identity), and Aegilops tauschii (62% identity), followed by those from S. bicolor and maize, Arabidopsis, and other plant species (Fig. 7B; Supplemental Fig. S5). Arabidopsis has one SCAR-like protein (AtSCAR5/SCARL) and four SCAR proteins (AtSCAR1-4; Zhang et al., 2008), of which AtSCAR1 and AtSCAR3 show higher sequence similarity to TUT1 than do AtSCAR2 and AtSCAR4 (Fig. 7B; Supplemental Fig. S6).
Figure 7.
TUT1 belongs to the SCAR/WAVE family. A, Domain structure of SCAR/WAVE proteins. The rice genome encodes four SCAR proteins: TUT1/OsSCAR1, OsSCAR2, OsSCAR3, and OsSCAR4. All four have SHD and basic domains, but OsSCAR2 lacks the VCA domain. Percentages above each domain indicate the amino acid sequence identities of the SHD and VCA domains in each protein compared with TUT1. The domains are color coded: SHD (blue), basic domain (red), VCA domain (green), and the black line represents the amino acid sequence. B, A Phylogenetic tree of TUT1 and its homologous proteins in different species. The phylogenetic tree was constructed using DNAMAN software with 1,000 bootstrapping trials. Bootstrapping values over 50% and the scale bar are shown in the phylogenetic tree. The tree was constructed using the distance method with maximum likelihood. TUT1 is shown in red. Accession numbers for each sequence are shown. Species are as indicated: A. tauschii (Aet), Arabidopsis (At), B. distachyon (Bd), Caenorhabditis elegans (Ce), Cucumis sativus (Cs), Dictyostelium discoideum (Dd), Drosophila melanogaster (Dm), Glycine max (Gm), Homo sapiens (Hs), Mus musculus (Mm), rice (Os), S. italica (Si), Solanum lycopersicum (Sl), Solanum tuberosum (St), S. bicolor (Sb), and maize (Zm). C and D, Trichome phenotype of the wild type (WT) and tut1 shown by SEM. Mature trichomes of the wild type and tut1 are shown in C. Enlarged view of wild-type trichome and tut1 trichome (D) from regions indicated by white squares in C. Bars = 100 μm (C) and 10 μm (D).
Mutations in the SCAR/WAVE complex genes of Arabidopsis (atscar2) and maize (brk1) cause defective trichome development (Frank and Smith, 2002; Frank et al., 2003; Basu et al., 2005; Zhang et al., 2005). In the tut1 mutant, development of trichomes was also impaired, characterized by blunted tips of the trichomes (Fig. 7, C and D), suggesting that TUT1 plays an evolutionarily conserved role in trichome development. Moreover, similar to that in the maize brk mutants (Frank et al., 2003), the cell wall at the junction between two epidermal cell layers was affected in tut1. The tut1 cells were more rounded compared with the wild type (Supplemental Fig. S7).
The VCA domain of SCAR/WAVE proteins acts to activate the Arp2/3 complex, and consequently, increases the actin filament nucleation and branching activities of the complex (Stradal et al., 2004). Sequence alignment analysis showed that the carboxyl-terminal region of the TUT1 protein contains a conserved Wiskott-Aldrich Homology2 region and acidic region (Supplemental Fig. S5). In an actin polymerization assay, the VCA domain of TUT1 significantly increased actin filament assembly in the presence of bovine Arp2/3 (Supplemental Fig. S8), indicating that the TUT1-VCA domain is capable of activating Arp2/3. Taken together, these results indicate that TUT1 is a functional SCAR/WAVE protein.
Impaired Actin Filament Organization in tut1
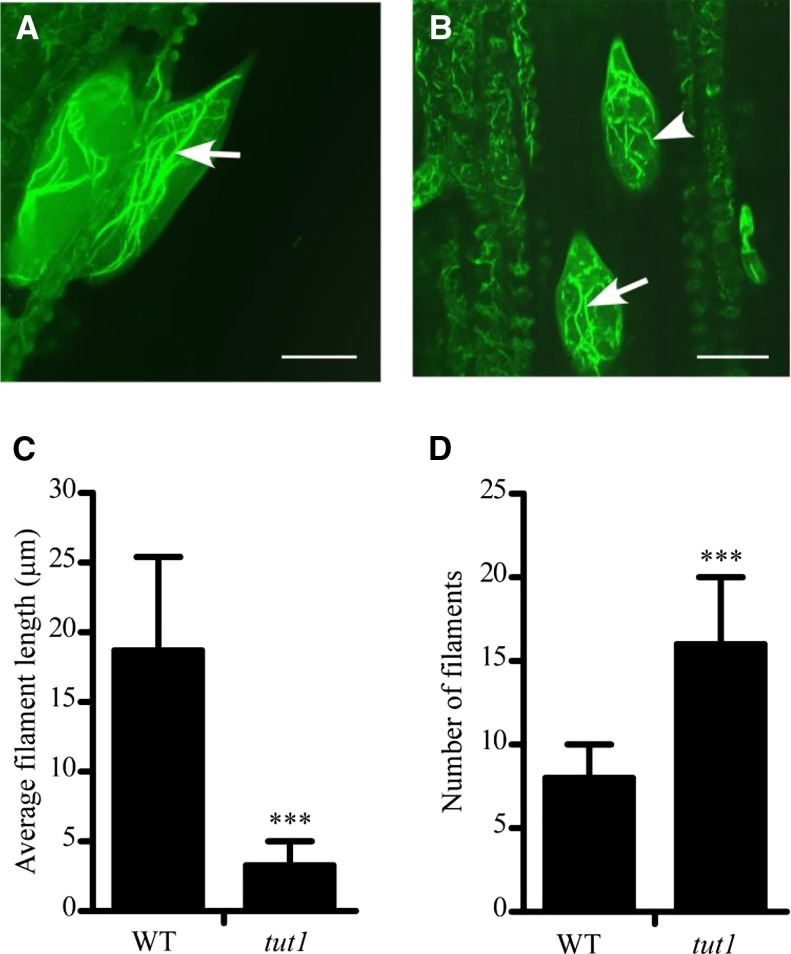
The data presented above indicate that TUT1 can activate Arp2/3. The SCAR/WAVE-activated Arp2/3 complex is involved in the regulation of the formation of loosely aligned actin bundles at the earliest stages of trichome branch elongation (El-Din El-Assal et al., 2004b). To assess the possible regulatory role of TUT1, we examined the organization of F-actin in the trichomes of wild-type and tut1 leaves by Alexa Fluor 488-phalloidin staining (Life Technologies). In the trichomes of wild-type leaves, the F-actin filaments were well organized along the longitudinal axis (Fig. 8A). However, in tut1 leaves, we observed significantly fragmented F-actin filaments (Fig. 8B), consistent with the altered shape of these cells in tut1 (Fig. 7, C and D). Statistical analysis showed that the mean length of actin filaments in tut1 trichomes was significantly shorter compared with the wild type (Fig. 8C). The number of actin filaments increased in tut1 trichomes (Fig. 8D). Our results suggest that TUT1 is a SCAR/WAVE protein and is involved in F-actin organization in rice.
Figure 8.
F-actin organization in the wild type (WT) and tut1. A and B, F-actin organization in wild-type (A) and tut1 (B) mutant leaf trichomes visualized by staining with Alexa Fluor 488-phalloidin. Arrow indicates long actin filaments and arrowhead indicates fragmented F-actin. Bars = 20 μm. C, Analysis of average filament length in the wild type and tut1. ***, P < 0.01 (Student’s t test). D, Analysis of number of filaments in trichomes of the wild type and tut1. ***, P < 0.01 (Student’s t test). Over 20 trichomes were used in these analyses.
DISCUSSION
In this study, we functionally characterized rice TUT1 and presented multiple lines of evidence showing that TUT1 encodes a functional SCAR/WAVE protein that plays an important role in regulating F-actin organization. We also demonstrate TUT1 functioning in multiple aspects of rice growth and development, especially in panicle development.
Our present work shows that TUT1, a SCAR/WAVE-like protein in rice, can activate the Arp2/3 complex to modulate actin polymerization. The SCAR/WAVE complex plays an important role in regulating actin filament arrangement by activating the Arp2/3 complex to modify actin filament organization (Blanchoin et al., 2000; Frank et al., 2004; Beltzner and Pollard, 2008). Mutations in key components of the SCAR/WAVE-Arp2/3 complex result in various defects in yeast and animals (Winter et al., 1999; Di Nardo et al., 2005). In many cases, these mutations cause embryonic arrest and zygotic lethality by impairing various cellular activities (Severson et al., 2002; Sawa et al., 2003; Vartiainen and Machesky, 2004). However, in Arabidopsis and maize, mutations in the SCAR/WAVE complex genes cause relatively weak phenotypes, affecting cell morphogenesis and development of the trichome and the hypocotyl (Frank and Smith, 2002; Uhrig et al., 2007). Notably, the rice tut1 mutant shows a pleiotropic phenotype during its entire life cycle, including shorter roots, reduced plant height, degenerating leaves, and degenerating panicles, suggesting that TUT1 functions as a housekeeping gene during rice growth and development. A recent study revealed that EARLY SENESCENCE1 (ES1), identical to TUT1, plays an important role in regulating water loss (Rao et al., 2015), suggesting that TUT1/ES1 is involved in various developmental and physiological processes.
The most severe defects in tut1 occur in panicles, consistent with the observation that the panicle has the highest level of TUT1 transcripts among the examined organs and tissues. In the panicle, we detected strong TUT1-GUS expression in glumes and anthers but not in the pistil. Consistent with the expression pattern of TUT1, whereas development of glumes, anthers, and pollen grains is severely affected in tut1, resulting in partial male sterility, the pistils and carpels remain relatively normal. Since TUT1 encodes a SCAR/WAVE-like protein, we presume that TUT1 is involved in panicle development by regulating F-actin organization. The biochemical analysis shows that activation of the TUT1-VCA domain is capable of activating the Arp2/3 complex in vitro. We noticed that the TUT1-VCA polypeptide shows a lower activity compared with SCAR/WAVE proteins of Arabidopsis and maize. We speculate that the reduced activity might be caused by the assay condition, which may not be optimized for the rice TUT1-VCA polypeptide, or by improper folding of TUT1-VCA recombinant polypeptide.
Panicle and spikelet development is controlled by multiple genes that function in different cellular processes, such as cuticle formation and cell differentiation (Komatsu et al., 2001; Ikeda et al., 2004; Zheng et al., 2005; Panikashvili et al., 2009). The cuticle plays an important role in rice flower morphogenesis, anther and pollen development, pathogen resistance, and salt tolerance (Bird et al., 2007; Qin et al., 2011, 2013). However, our understanding of cuticle function in panicle development remains limited. Considering that the panicle apical degeneration phenotype of tut1 is closely associated with cuticle disorganization and deficiency, it is reasonable to presume that the formation of the cuticle affects panicle development in rice. The complex regulation of cuticle formation includes vesicle trafficking, fatty acid metabolism, and transport processes (Yeats and Rose, 2013). Vesicle trafficking that can be regulated by F-actin is important for wax secretion from epidermal cells in Arabidopsis (McFarlane et al., 2014). Consistently, we found that fatty acid metabolism is impaired in tut1, suggesting that abnormal fatty acid metabolism may cause deficient cuticle formation and affect F-actin-regulated vesicle trafficking in tut1. A recent study revealed that Arp2/3 is involved in modulating the cell wall thickness gradient from tip to base of the trichome branch by regulating F-actin meshwork formation (Yanagisawa et al., 2015). Consistently, we also observed the abnormal arrangement of F-actin in the tut1 mutant, suggesting that the Arp2/3-SCAR/WAVE complex plays a conserved role in leaf trichome development in plants. Compared with mutants in Arabidopsis SCAR/WAVE proteins, the rice tut1 mutants show multiple phenotypes, especially in panicle development. Therefore, the diverse functions of SCAR/WAVE family proteins need to be studied in different varieties of plants to provide a comprehensive understanding of SCAR/WAVE functions.
Additionally, an important cause of rice yield reduction is defective panicle development (Xing and Zhang, 2010). Understanding the mechanism of panicle development should be helpful to mitigate the crop yield reduction caused by defective panicle development. However, agricultural applications to avoid panicle degeneration remain relatively limited, and additional research will be required to identify the molecular mechanisms that can be modified to confer fully developed panicles in rice and to understand the complicated regulation of panicle apical spikelet development, including the effects of temperature, nutrition, and genetic background (Cheng et al., 2011). The identification and characterization of genes that function in rice panicle development and production, such as TUT1, provide the first steps toward genetic manipulation to engineer or breed high-production rice.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
tut1 was screened from a gamma ray-induced M2 population in the japonica cultivar (rice [Oryza sativa] ‘Zhonghua 11’). Plants were grown in an experimental field in Beijing (39° 54′ 20″ N, 116° 23′ 29″ E) from May to January under long-day conditions and in Lingshui (18° 30′ 00″ N, 110° 01′ 20″ E) from January to May under short-day conditions. When grown under the laboratory condition, seeds of the wild type and tut1 were germinated in darkness at 30°C for 3 d and then transferred to a SANYO culture chamber (MLR-351H) with a 16-h/8-h photoperiod at 30°C.
Histochemical Analysis
For light microscopy, samples were fixed at 4°C for 12 h in 3.7% (v/v) glutaraldehyde in 0.025 m phosphate buffer (pH 6.8) and dehydrated with ethanol. The samples were embedded overnight at room temperature in embedding medium (Leica Historesin) and then sectioned at 4-μm thickness using a Leica RM2265 rotary microtome. The sections were then stained with 0.05% (w/v) toluidine blue for 1 min and observed with a light microscope (Olympus BX51). To measure chlorophyll contents, chlorophyll was extracted from fresh leaves of wild-type and tut1 plants and analyzed using a spectrophotometer (Beckman Coulter DU800) as described (Lichtenthaler, 1987). For cell death staining, samples were treated with 1% (w/v) Evans blue for 6 h and then treated with ethanol for observation. For ROS staining, samples were treated with 0.05% (w/v) NBT at 28°C overnight and then treated with ethanol for observation.
Gene Expression Analysis
For qRT-PCR analysis, total RNA was extracted from root, node, stem, immature leaves, mature leaves, immature panicle, and mature panicle using the RNAprep Pure Plant RNA Purification Kit (Tiangen Biotech). Total RNA (2 μg) was reverse transcribed into cDNA and then used as a template for qRT-PCR analysis with specific primers (TUT1_ RT-qPCR _F and TUT1_ RT-qPCR _R; Supplemental Table S1). qRT-PCR was performed using the UltraSYBR mixture (CWBIO) according to the manufacturer’s instructions, and the reactions were run in a CFX96TM real-time PCR system (Bio-Rad). The rice glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as an internal control with 40 PCR cycles using primers GAPDH_F and GAPDH_R (Supplemental Table S1).
TUT1:GUS Construct and Histological GUS Assay
The 785-bp fragment immediately upstream of the start codon of TUT1 was amplified using the gene-specific primers GUS_F and GUS_R and inserted in front of the GUS gene in pCAMBIA1300 with PstI and BamHI (Supplemental Table S1). The final construct containing the TUT1:GUS reporter gene was introduced into the wild-type background plant by Agrobacterium tumefaciens-mediated transformation (Hiei et al., 1994). Transgenic plants were selected on hygromycin medium, and T1 transgenic plants were used for analysis of GUS activity.
GUS activity was detected using a standard protocol (Jefferson et al., 1987). Transgenic plant tissues were incubated in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid buffer overnight at 37°C. The samples were cleared of chlorophyll by dehydration with ethanol. The stained samples were photographed using a Cannon digital camera and stereoscope (Olympus SZX12). The stained stamen was sliced using resin sections (Leica Historesin) and analyzed by light microscopy (Olympus BX51).
SEM and Transmission Electron Microscopy
For SEM, samples were fixed overnight at 4°C with 2.5% (v/v) glutaraldehyde in 0.1 m phosphate buffer (pH 7.4). The samples were dehydrated in a graded ethanol series and substituted with a graded isoamyl acetate series. The samples were critical-point dried, sputter coated with gold, and observed using a Hitachi S-3000N scanning electron microscope at 10 kV. For transmission electron microscopy, samples were fixed overnight at 4°C with 2.5% (v/v) glutaraldehyde in 0.1 m phosphate buffer (pH 7.4). After three washes with phosphate buffer, samples were fixed overnight in 2% (v/v) OsO4 in phosphate buffer. After being stained and dehydrated in a graded ethanol series, the samples were submerged in LR White resin (Sigma-Aldrich) and polymerized for 2 d. Ultrathin cross sections were prepared using a Leica EM UC6 ultramicrotome. Ultrathin sections were observed under a FEI Tecnai Spirit (120 kV) transmission electron microscope.
Map-Based Cloning of TUT1
tut1 was crossed with cv Zhenshan 97 (indica) to generate the F2 mapping population. TUT1 was mapped to a 196-kb region on chromosome 1 using SSR markers and SNP markers (Supplemental Table S1). Exons of candidate genes in this region were amplified from tut1 and wild-type plants. The sequences were compared using DNASTAR software and the National Center for Biotechnology Information Web site. The full-length open reading frame of TUT1 was amplified using reverse-transcribed DNA as a template. The primers used were TUT1_OE_F and TUT1_OE_R (Supplemental Table S1). The full-length open reading frame was then cloned into the SK vector using the SalI and SmaI restriction sites and then cloned into the binary vector pTCK303 using the KpnI and SpeI restriction sites (Supplemental Table S1; Wang et al., 2004). A. tumefaciens-mediated transformation was used to transform the resulting construct, pTCK303-TUT1, into a compound callus pool containing wild-type, heterozygous, and homozygous callus generated from TUT1/tut1 heterozygous seeds. More than two independent transgenic lines were produced that complemented the tut1 phenotypes. A specific DNA fragment of TUT1 was amplified from the cv Zhonghua 11 cDNA using the PCR primers RNAi_F (KpnI and SpeI) and RNAi_R (BamHI and SacI; see Supplemental Table S1). The PCR products were then digested with SpeI/SacI and KpnI/BamHI, respectively. The two digested fragments were sequentially cloned into the vector pTCK303, which contains a maize (Zea mays) ubiquitin promoter, to make the final pTCK303:TUT1 RNAi plasmid as previously described (Wang et al., 2004). This final construct was introduced into the wild-type background plant by A. tumefaciens-mediated transformation (Hiei et al., 1994).
Fatty Acid Content Analysis
For fatty acid analysis, 50 mg of tissue was ground in liquid nitrogen and then added to 1.5 mL of extraction buffer (2.5% [v/v] sulfuric acid in methanol) and incubated at 80°C for 2 h. After the sample cooled to room temperature, 2 mL of 0.9% (w/v) NaCl and 1 mL of hexyl hydride were added to the extraction buffer, and the mixture was vortexed. The mixture was centrifuged at 4,000 rpm for 10 min at room temperature, and the supernatant was transferred to a new microfuge tube, then dried to powder with a vacuum pump. The powder was dissolved in 50 μL of isoamyl acetate for analysis.
Fluorescence Microscopy of Actin Filaments
F-actin was stained using the previously described method (Olyslaegers and Verbelen, 1998; Yang et al., 2011; Zhang et al., 2011). Mature rice leaves were incubated in PEM buffer (100 mm PIPES, 10 mm EGTA, 0.3 m mannitol, and 5 mm MgSO4, pH 6.9) and 0.66 μm Alexa Fluor 488-phalloidin (Life Technologies). After overnight incubation at 4°C, samples were observed in 50% (v/v) glycerol with a confocal microscope (Olympus FLUOVIEW FV1000).
Sequence data from this article can be found in the GenBank/EMBL databases and the Gramene Web site under the following accession numbers: TUT1/OsSCAR1 (NP_001172227.1), OsSCAR2 (NP_001049845), OsSCAR3 (Q84TX2), OsSCAR4 (BAC75554), AtSCAR1 (NP_001031474), AtSCAR2 (NP_181378), AtSCAR3 (NP_174212), AtSCAR4 (NP_195793), AtSCAR5 (NP_193595), ZmSCAR1 (AFW80370), ZmSCAR2 (DAA44917), ZmSCAR3 (GRMZM2G475139), ZmSCAR4 (ACN28010), ZmSCAR5 (DAA52161), ZmSCAR6 (AFW88660), AetSCAR (EMT14604), BdSCAR (XP_003565325), SiSCAR (XP_004968593), SbSCAR (Sb03g002220), CsSCAR (XP_004149068), GmSCAR (XP_006585168), SlSCAR (XP_004233023), StSCAR (XP_006362414), HsWAVE1 (AAD33052), MmWAVE1 (AAL75574), CeWVE-1 (NP_493028), DmSCAR (NP_609477), and DdSCAR1 (AAD29083).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Quantitation of agronomic traits in the wild type and tut1.
Supplemental Figure S2. Quantitation of anther degeneration in the wild type and tut1.
Supplemental Figure S3. SEM and transmission electron microscopy examination of cuticle in wild-type and tut1 leaves.
Supplemental Figure S4. The phenotypes of TUT1 RNAi transgenic lines.
Supplemental Figure S5. Amino acid sequence alignment of TUT1 and other SCAR/WAVE family proteins.
Supplemental Figure S6. Phylogenetic tree of SHD of SCAR/WAVE proteins from Arabidopsis and rice.
Supplemental Figure S7. Analysis of mature leaves of the wild type and tut1 by light microscopy.
Supplemental Figure S8. Activation analysis of the TUT1-VCA domain in vitro.
Supplemental Table S1. Primers used in this study.
Supplemental Methods S1. Literature cited.
Supplementary Material
Acknowledgments
We thank Dr. Kang Chong (Institute of Botany, Chinese Academy of Sciences) for providing the pTCK303 vector, and Shufeng Sun (Institute of Biophysics, Chinese Academy of Sciences) for technical assistance in transmission electron microscopy.
Glossary
- ROS
reactive oxygen species
- VCA
verprolin, cofilin, and acidic
- NBT
nitroblue tetrazolium
- SEM
scanning electron microscopy
- SSR
simple sequence repeat
- SNP
single nucleotide polymorphism
- SHD
SCAR/WAVE homolog domain
- EGC
envelope glycoprotein C
- qRT
quantitative reverse transcription
- RNAi
RNA interference
- cDNA
complementary DNA
Footnotes
This work was supported by the Ministry of Science and Technology of China (grant no. 2014CB943402), State Key Laboratory of Plant Genomics (grant no. SKLPG2011A0210), the Ministry of Agriculture of China (grant nos. 2013ZX08009–003 and 2014ZX08009–003), and National Natural Science Foundation of China (grant no. 30700500).
References
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Basu D, Le J, El-Essal SD, Huang S, Zhang C, Mallery EL, Koliantz G, Staiger CJ, Szymanski DB (2005) DISTORTED3/SCAR2 is a putative Arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17: 502–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD (2008) Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem 283: 7135–7144 [DOI] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD (2000) Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404: 1007–1011 [DOI] [PubMed] [Google Scholar]
- Cheng ZJ, Mao BG, Gao SW, Zhang L, Wang JL, Lei CL, Zhang X, Wu FQ, Guo XP, Wan J (2011) Fine mapping of qPAA8, a gene controlling panicle apical development in rice. J Integr Plant Biol 53: 710–718 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Hussey PJ (2005) Arp2/3 and SCAR: plants move to the fore. Nat Rev Mol Cell Biol 6: 954–964 [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Kaloriti D, Davies B, Malhó R, Hussey PJ (2004) Arabidopsis NAP1 is essential for Arp2/3-dependent trichome morphogenesis. Curr Biol 14: 1410–1414 [DOI] [PubMed] [Google Scholar]
- Di Nardo A, Cicchetti G, Falet H, Hartwig JH, Stossel TP, Kwiatkowski DJ (2005) Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc Natl Acad Sci USA 102: 16263–16268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Din El-Assal S, Le J, Basu D, Mallery EL, Szymanski DB (2004a) Arabidopsis GNARLED encodes a NAP125 homolog that positively regulates ARP2/3. Curr Biol 14: 1405–1409 [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Le J, Basu D, Mallery EL, Szymanski DB (2004b) DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J 38: 526–538 [DOI] [PubMed] [Google Scholar]
- Elzinga M, Collins JH, Kuehl WM, Adelstein RS (1973) Complete amino-acid sequence of actin of rabbit skeletal muscle. Proc Natl Acad Sci USA 70: 2687–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Egile C, Dyachok J, Djakovic S, Nolasco M, Li R, Smith LG (2004) Activation of Arp2/3 complex-dependent actin polymerization by plant proteins distantly related to Scar/WAVE. Proc Natl Acad Sci USA 101: 16379–16384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, Cartwright HN, Smith LG (2003) Three brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development 130: 753–762 [DOI] [PubMed] [Google Scholar]
- Frank MJ, Smith LG (2002) A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr Biol 12: 849–853 [DOI] [PubMed] [Google Scholar]
- Harries PA, Pan A, Quatrano RS (2005) Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell 17: 2327–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ (2006) Control of the actin cytoskeleton in plant cell growth. Annu Rev Plant Biol 57: 109–125 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa N, Nagato Y (2005) ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev Biol 282: 349–360 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Sunohara H, Nagato Y (2004) Developmental course of inflorescence and spikelet in rice. Breed Sci 54: 147–156 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörgens CI, Grünewald N, Hülskamp M, Uhrig JF (2010) A role for ABIL3 in plant cell morphogenesis. Plant J 62: 925–935 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee DY, Lee YS, Schreiber L, Franke R, Faust A, Yephremov A, Saedler H, Kim YW, et al. (2006) Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Maekawa M, Shimamoto K, Kyozuka J (2001) The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev Biol 231: 364–373 [DOI] [PubMed] [Google Scholar]
- Kotchoni SO, Zakharova T, Mallery EL, Le J, El-Assal SD, Szymanski DB (2009) The association of the Arabidopsis actin-related protein2/3 complex with cell membranes is linked to its assembly status but not its activation. Plant Physiol 151: 2095–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Samuels AL (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Le J, El-Assal SD, Basu D, Saad ME, Szymanski DB (2003) Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr Biol 13: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Le J, Mallery EL, Zhang C, Brankle S, Szymanski DB (2006) Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr Biol 16: 895–901 [DOI] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, et al. (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Qian Q, Fu Z, Zeng D, Meng X, Kyozuka J, Maekawa M, Zhu X, Zhang J, Li J, et al. (2009) Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J 58: 592–605 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD (1999) Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA 96: 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hülskamp M (2003) Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15: 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane HE, Watanabe Y, Yang W, Huang Y, Ohlrogge J, Samuels AL (2014) Golgi- and trans-Golgi network-mediated vesicle trafficking is required for wax secretion from epidermal cells. Plant Physiol 164: 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Takenawa T (2003) Regulation of actin dynamics by WASP family proteins. J Biochem 134: 309–313 [DOI] [PubMed] [Google Scholar]
- Olyslaegers G, Verbelen JP (1998) Improved staining of F-actin and co-localization of mitochondria in plant cells. J Microsc 192: 73–77 [Google Scholar]
- Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Höfer R, Schreiber L, Chory J, Aharoni A (2007) The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol 145: 1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Shi JX, Schreiber L, Aharoni A (2009) The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiol 151: 1773–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin BX, Tang D, Huang J, Li M, Wu XR, Lu LL, Wang KJ, Yu HX, Chen JM, Gu MH, et al. (2011) Rice OsGL1-1 is involved in leaf cuticular wax and cuticle membrane. Mol Plant 4: 985–995 [DOI] [PubMed] [Google Scholar]
- Qin P, Tu B, Wang Y, Deng L, Quilichini TD, Li T, Wang H, Ma B, Li S (2013) ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol 54: 138–154 [DOI] [PubMed] [Google Scholar]
- Rao Y, Yang Y, Xu J, Li X, Leng Y, Dai L, Huang L, Shao G, Ren D, Hu J, et al. (2015) EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol 169: 1225–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler R, Mathur N, Srinivas BP, Kernebeck B, Hülskamp M, Mathur J (2004) Actin control over microtubules suggested by DISTORTED2 encoding the Arabidopsis ARPC2 subunit homolog. Plant Cell Physiol 45: 813–822 [DOI] [PubMed] [Google Scholar]
- Sawa M, Suetsugu S, Sugimoto A, Miki H, Yamamoto M, Takenawa T (2003) Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J Cell Sci 116: 1505–1518 [DOI] [PubMed] [Google Scholar]
- Severson AF, Baillie DL, Bowerman B (2002) A formin homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol 12: 2066–2075 [DOI] [PubMed] [Google Scholar]
- Stradal TE, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G (2004) Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol 14: 303–311 [DOI] [PubMed] [Google Scholar]
- Szymanski DB. (2005) Breaking the WAVE complex: the point of Arabidopsis trichomes. Curr Opin Plant Biol 8: 103–112 [DOI] [PubMed] [Google Scholar]
- Takenawa T, Miki H (2001) WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J Cell Sci 114: 1801–1809 [DOI] [PubMed] [Google Scholar]
- Uhrig JF, Mutondo M, Zimmermann I, Deeks MJ, Machesky LM, Thomas P, Uhrig S, Rambke C, Hussey PJ, Hülskamp M (2007) The role of Arabidopsis SCAR genes in ARP2-ARP3-dependent cell morphogenesis. Development 134: 967–977 [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Machesky LM (2004) The WASP-Arp2/3 pathway: genetic insights. Curr Opin Cell Biol 16: 174–181 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu YY, Jiang RX, Han Y, Xu ZH, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22: 409–417 [Google Scholar]
- Winter DC, Choe EY, Li R (1999) Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc Natl Acad Sci USA 96: 7288–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61: 421–442 [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Desyatova AS, Belteton SA, Mallery EL, Turner JA, Szymanski DB (2015) Patterning mechanisms of cytoskeletal and cell wall systems during leaf trichome morphogenesis. Nat Plants 1: 15014. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Zhang C, Szymanski DB (2013) ARP2/3-dependent growth in the plant kingdom: SCARs for life. Front Plant Sci 4: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Ren S, Zhang X, Gao M, Ye S, Qi Y, Zheng Y, Wang J, Zeng L, Li Q et al. (2011) BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23: 661–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JK (2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nagato Y (2011) Flower development in rice. J Exp Bot 62: 4719–4730 [DOI] [PubMed] [Google Scholar]
- Zhang C, Mallery EL, Schlueter J, Huang S, Fan Y, Brankle S, Staiger CJ, Szymanski DB (2008) Arabidopsis SCARs function interchangeably to meet actin-related protein 2/3 activation thresholds during morphogenesis. Plant Cell 20: 995–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 154: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dyachok J, Krishnakumar S, Smith LG, Oppenheimer DG (2005) IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17: 2314–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang Y, Tan H, Wang Y, Li G, Liang W, Yuan Z, Hu J, Ren H, Zhang D (2011) RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23: 681–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L (2005) Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.