A thylakoid membrane protein shares functional similarity and physical interactions with the homologous protein insertase, despite sequence-level divergence in a critical C-terminal domain.
Abstract
ALBINO3 (ALB3) is a well-known component of a thylakoid protein-targeting complex that interacts with the chloroplast signal recognition particle (cpSRP) and the cpSRP receptor, chloroplast filamentous temperature-sensitive Y (cpFtsY). Its protein-inserting function has been established mainly for light-harvesting complex proteins, which first interact with the unique chloroplast cpSRP43 component and then are delivered to the ALB3 integrase by a GTP-dependent cpSRP-cpFtsY interaction. In Arabidopsis (Arabidopsis thaliana), a subsequently discovered ALB3 homolog, ALB4, has been proposed to be involved not in light-harvesting complex protein targeting, but instead in the stabilization of the ATP synthase complex. Here, however, we show that ALB3 and ALB4 share significant functional overlap, and that both proteins are required for the efficient insertion of cytochrome f and potentially other subunits of pigment-bearing protein complexes. Genetic and physical interactions between ALB4 and ALB3, and physical interactions between ALB4 and cpSRP, suggest that the two ALB proteins may engage similar sets of interactors for their specific functions. We propose that ALB4 optimizes the insertion of thylakoid proteins by participating in the ALB3-cpSRP pathway for certain substrates (e.g. cytochrome f and the Rieske protein). Although ALB4 has clearly diverged from ALB3 in relation to the partner-recruiting C-terminal domain, our analysis suggests that one putative cpSRP-binding motif has not been entirely lost.
Although the machinery responsible for the import of proteins into chloroplasts is largely a eukaryotic invention, the internal sorting of chloroplast proteins depends on components derived from the original endosymbiont (Jarvis and López-Juez, 2013). One such component was revealed through analysis of the albino3 (alb3) mutant of Arabidopsis (Arabidopsis thaliana), identified using a Dissociation element insertion screen adapted from maize (Zea mays; Long et al., 1993). The mutant was reported to have white to light-yellow cotyledons and leaves, abnormal chloroplasts, and reduced levels of chlorophyll (Sundberg et al., 1997). The affected protein, ALB3, is homologous to the bacterial membrane protein YidC and to the mitochondrial cytochrome c oxidase complex assembly factor Oxidase assembly1 (Oxa1), and was shown to be localized in chloroplast thylakoids. Thus, it was suggested to play a role similar to that of Oxa1 in the assembly of thylakoid membrane complexes (Sundberg et al., 1997). Since ALB3 antibodies could inhibit the insertion of light-harvesting complex proteins (LHCPs), the affected thylakoid membrane complexes were identified to be the light-harvesting complexes (Moore et al., 2000). The YidC/Oxa1/Alb3 family of proteins are now widely recognized to have conserved functions in most organisms, from bacteria to plants, and are generally involved in the insertion of proteins into membranes, the folding of membrane proteins, and the assembly of membrane protein complexes (Zhang et al., 2009; Saller et al., 2012; Dalbey et al., 2014).
For the in vitro reconstitution of LHCPs into thylakoid membranes, it was previously found that the stroma could be replaced by the chloroplast signal recognition particle (cpSRP), the cpSRP receptor chloroplast filamentous temperature-sensitive Y (cpFtsY), and GTP (Tu et al., 1999). The necessity of ALB3 for LHCP insertion suggested that these stromal factors can deliver LHCPs to the thylakoids via an interaction with ALB3 at the thylakoid membrane (Moore et al., 2000). The chloroplast SRP is a heterodimer of cpSRP54 and cpSRP43 in Arabidopsis (Groves et al., 2001; Stengel et al., 2008). These components interact with each other and with their LHCP cargo in the stroma (Tu et al., 2000; Jonas-Straube et al., 2001), whereas cpFtsY is required for thylakoid docking of the cpSRP-LHCP complex and interacts GTP dependently with cpSRP54 (Moore et al., 2003). A conserved membrane insertase function was confirmed for ALB3 by showing that it could complement a bacterial deletion mutant of YidC and restore the ability to insert membrane proteins (Jiang et al., 2002).
Mutants of Alb3 homologs have been isolated in several distantly related species, and very similar defects in the accumulation of pigments and in thylakoid organization have been reported in every case. In Chlamydomonas reinhardtii, the knockout of Alb3.1, one of two paralogues, causes a drastic reduction in both chlorophyll and LHCPs, as in Arabidopsis (Bellafiore et al., 2002). In the cyanobacterium Synechocystis sp. PCC 6803, the knockout of the Alb3 homolog slr1471 also leads to impaired thylakoid organization, reduced levels of photosynthetic pigments, and consequently reduced photosynthetic performance (Spence et al., 2004).
Although the importance of ALB3 for the insertion and assembly of pigment-bearing LHCPs in an evolutionary conserved way seems well established, the contribution of ALB3 to the insertion or assembly of other photosystem proteins or components of other thylakoid complexes is less well understood. In Arabidopsis, the insertion of PSII subunits PsbS, PsbX, PsbW, and PsbY was unaffected by the blocking of ALB3 with anti-ALB3 antibodies, whereas in Chlamydomonas reinhardtii, the knockout of Alb3.1 led to a general reduction of PSII but left PSI, the cytochrome b6f complex, and the ATP synthase complex unaffected (Woolhead et al., 2001; Bellafiore et al., 2002). Interaction studies showed that ALB3 is able to bind to PSII core components, and that it is likely involved in the assembly of the PSII core (Ossenbühl et al., 2004; Pasch et al., 2005), which would explain the reduced level of PSII in the C. reinhardtii Alb3.1 knockout (Bellafiore et al., 2002). ALB3 was also shown to interact with the chloroplast secretion system homolog Y (cpSecY) translocase, and it was suggested that these components act together as a cpSRP-specific translocase (Klostermann et al., 2002); however, as removal of the main, thylakoidal cpSecY isoform (cpSecY1, also called SCY1) did not inhibit ALB3 function, the relevance of this interaction for protein insertion via ALB3 was disputed (Mori et al., 1999; Moore et al., 2003).
In Arabidopsis, a homolog of ALB3 exists, which is called ALB4 (Gerdes et al., 2006). A knockdown mutant of ALB4 was reported to be visibly normal under standard growth conditions, but to have altered chloroplast ultrastructure; the organelles were more spherical in shape and had a deteriorated thylakoid structure (Gerdes et al., 2006). A later study reported a slightly reduced growth rate of the alb4 knockdown line (Benz et al., 2009). Knockout mutants of alb4 were shown to have reduced amounts of ATP synthase subunits, whereas transcription of those components was not reduced (Benz et al., 2009). Moreover, larger ATP synthase complexes were decreased in favor of smaller intermediate complexes, and the photophosphorylation capacity of the mutant was consequently reduced, pointing to a role for ALB4 in the stabilization of ATP synthase intermediates during complex assembly (Benz et al., 2009). Although both ALB4 and ALB3 were localized to the same subfraction of stromal lamellae, gel filtration and coimmunoprecipitation revealed no interaction between ALB3 and ALB4, whereas ALB4 clearly interacted with ATP synthase subunits, and ALB3 with cpSecY (Benz et al., 2009). Given the strong difference between the phenotypes of the Arabidopsis alb3 and alb4 mutants, preferences of ALB3 for light-harvesting complex and PSII assembly and ALB4 for ATP synthase assembly were proposed. However, the consequences of the loss of both ALB3 and ALB4 had not been investigated. Therefore, to address the possibility that ALB3 and ALB4 share some functions, the alb3 alb4 double mutant was identified and analyzed, and genetic and physical interactions of ALB4 with ALB3 and with other components of the cpSRP-targeting pathway were investigated.
RESULTS
Basic Molecular and Genetic Analyses of Arabidopsis alb Mutants
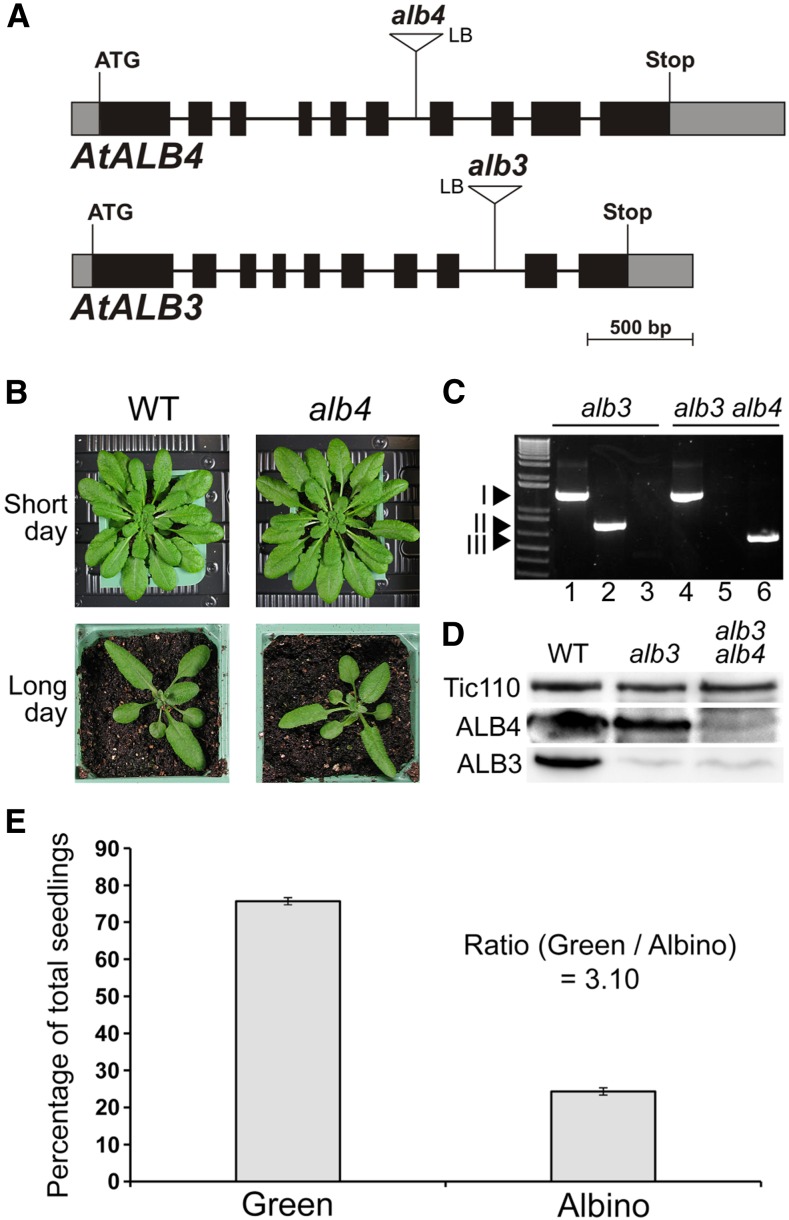
We wished to gain a better understanding of the in vivo role of ALB4, and of its functional relationship with ALB3. For the analysis of alb4 mutants, the line Salk_136199 was chosen, which has a transfer DNA (T-DNA) insertion in the sixth intron (Fig. 1A) and was described previously (Gerdes et al., 2006; Benz et al., 2009). This line has been reported to accumulate less than 10% of wild-type levels of ALB4 (Gerdes et al., 2006) and to have a growth retardation defect (Benz et al., 2009). To further investigate this potential growth retardation defect, homozygous alb4 mutants were grown alongside the wild type directly on soil. However, under neither long- nor short-day conditions could a growth retardation defect of alb4 mutant plants be detected, which is in contrast with the results of Benz et al. (2009, figure 1B). Even under various stress conditions, we did not observe significant differences between alb4 mutant plants and the wild type (Supplemental Fig. S1). This confirms the earlier finding that this alb4 mutant line does not have a visible phenotype under normal growth conditions (Gerdes et al., 2006). The expression level of ALB4 was compared with those of ALB3 and other components of the cpSRP pathway to determine if the large difference between the phenotypes of alb4 and alb3 mutants could be due to differential gene expression (Supplemental Fig. S2). However, although the expression level of ALB4 is lower than that of ALB3, the difference is not large (ratio of expression levels is approximately 0.8), and ALB4 actually has a very similar expression level to cpFtsY. The mutants of cpFtsY, cpSRP54, and cpSRP43 all have pronounced chlorotic phenotypes, but the corresponding gene expression levels are in a similar range to those of ALB3 and ALB4. Thus, the difference in phenotype between the alb4 and alb3 mutants is unlikely to be caused by differential expression alone.
Figure 1.
Basic characteristics and genetic analysis of the Arabidopsis alb4 and alb3 mutants. A, Gene diagrams of Arabidopsis ALB4 and ALB3 with the T-DNA insertions of the alb4 and alb3 mutant lines shown. Protein-coding exons are represented by black boxes, untranslated regions by gray boxes, and introns by thin lines between the boxes. T-DNA insertion sites are indicated precisely, but insertion sizes are not to scale. ATG, Translation initiation codon; Stop, translation termination codon; LB, T-DNA left border. B, Wild-type (WT) and alb4 mutant plants (Salk_136199) were directly grown on soil under short-day (8-h-light/16-h-dark) conditions or long-day (16-h-light/8-h-dark) conditions. Pictures were taken after 7 weeks (short day) and 4 weeks (long day). C, Genotype analysis by genomic PCR. Heterozygous alb3 mutants (GK_293B08) and alb3/+ alb4/alb4 plants were visibly indistinguishable from the wild type, but both contained the alb3 T-DNA insertion (I; lanes 1 and 4); alb3/+ alb4/alb4 plants additionally contained the homozygous alb4 T-DNA insertion (III; lanes 3 and 6) but not the wild-type ALB4 allele (II; lanes 2 and 5). The ladder (lane 0, left side) includes standards of the following sizes, starting at the bottom: 0.3, 0.4, 0.5, 0.65, 0.85, 1.0, 1.65, and 2.0 kb. D, Immunoblot analysis of total protein extracts from 3-week-old albino plants. The alb3 alb4 double-mutant seedlings contained considerably less ALB4 protein than alb3 single mutants. The translocon at the inner chloroplast envelope membrane component of 110 kD (Tic110) has been used as a loading control. E, Segregation analysis of the progeny of three individual alb3/+ alb4/alb4 (heterozygous/homozygous) plants. Values are means of the percentages of green and albino progeny from each of the three parent plants. The error bars denote sd.
To assess the functional relationship between ALB3 and ALB4, the alb4 mutant was crossed to a heterozygous alb3 mutant (as the homozygous alb3 genotype is seedling lethal). An alb3 line containing a T-DNA insertion in the eighth intron was chosen for this analysis (Fig. 1A), which contains a greatly reduced amount of ALB3 protein (Fig. 1D; Supplemental Fig. S3) and is, similar to alb4, in the Columbia-0 background. In the resulting F2 generation, green plants that were homozygous for alb4 and heterozygous for alb3 were selected (Fig. 1C); the F3 progeny of these plants segregated one-quarter of albino alb3 alb4 double mutants (Fig. 1E), indicating that the double-homozygous mutant genotype is viable. This contrasts with the envelope-localized protein import system for which double mutants lacking homologous components cause embryo lethality (Constan et al., 2004; Kubis et al., 2004). The identified double-homozygous mutants have a reduced amount of ALB4 protein, as was described earlier for this alb4 line (Gerdes et al., 2006), and a greatly reduced amount of ALB3 protein (Fig. 1D); the specificity of the ALB3 and ALB4 antibodies used in this and later analyses was tested as shown in Supplemental Figure S3.
Double-Mutant alb3 alb4 Plants Are Visibly Paler than the alb3 Single Mutants with Reduced Pigment Levels
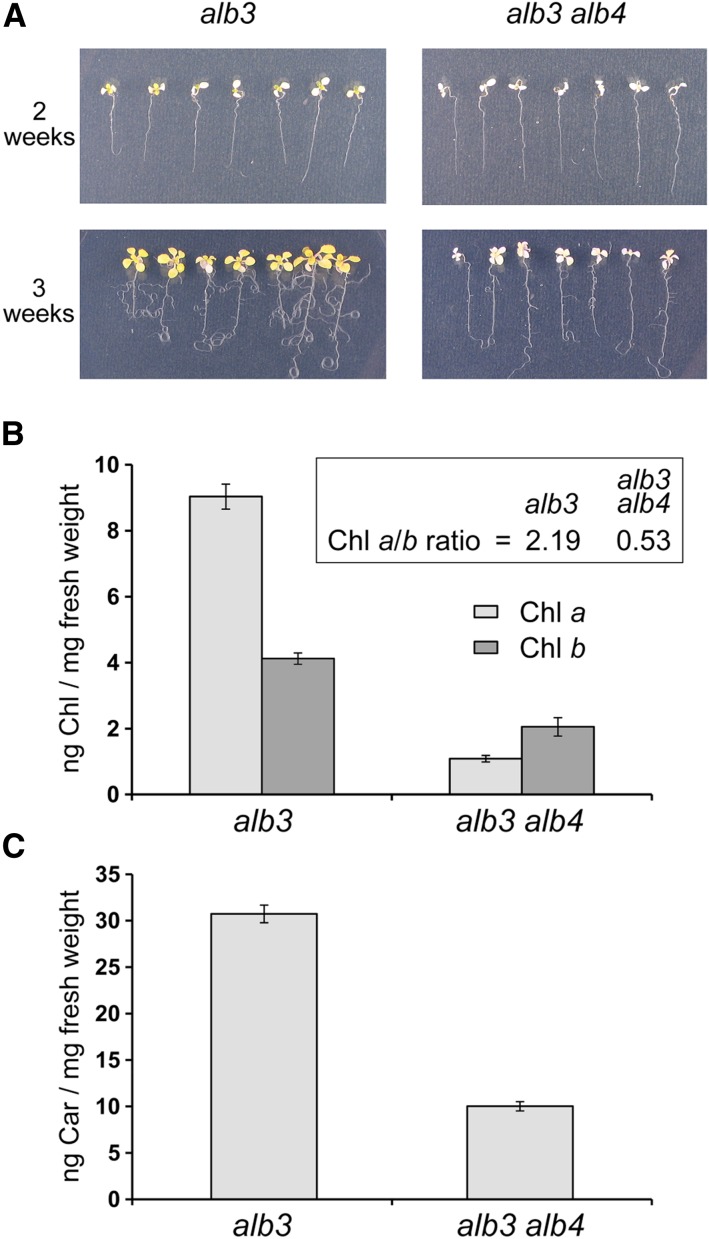
Interestingly, when the double mutants were grown carefully alongside control alb3 single-mutant plants, it became clear that the alb3 alb4 double mutants are visibly paler than the alb3 single mutants (Fig. 2A). Although the cotyledons did not appear to be significantly different between the two genotypes, after 2 weeks of growth on medium supplemented with 0.5% (w/v) Suc, the first true leaves that emerged from the alb3 mutant seedlings were considerably more pigmented (yellowish) than those emerging from the alb3 alb4 double-mutant seedlings. The 2-week-old seedlings were then transferred onto medium supplemented with 3% (w/v) Suc to support further growth for another week, and the resulting 3-week-old alb3 seedlings were considerably larger and more yellowish than the alb3 alb4 double-mutant seedlings (Fig. 2A). This implies that alb3 mutants are able to accumulate photosynthetic pigments more effectively than the alb3 alb4 double mutants.
Figure 2.
Phenotypic analysis of alb3 alb4 double-mutant plants. A, Albino seedlings derived from the segregating progeny of alb3/+ or alb3/+ alb4/alb4 plants grown in vitro on Murashige and Skoog (MS) medium supplemented with 0.5% (w/v) Suc were transferred to MS medium containing 3% (w/v) Suc 2 weeks after germination (top) and allowed to grow for an additional 1 week (bottom). B, Chlorophyll a (Chl a) and Chl b were measured in ng mg−1 fresh weight using 3-week-old seedlings grown in vitro as described above. The ratio of chlorophyll a to chlorophyll b (Chl a/b) is larger than 1 for alb3 but smaller than 1 for alb3 alb4. C, Total carotenoid (Car) content was measured in ng mg−1 fresh weight using similarly grown 3-week-old seedlings. All error bars denote se (n = 3).
To directly assess photosynthetic pigment accumulation in the alb3 alb4 double mutants, chlorophyll and carotenoids from both genotypes were extracted and quantified. Significantly, chlorophyll a was strongly reduced in the double mutant compared with alb3, whereas the reduction in chlorophyll b was relatively minor (Fig. 2B). Thus, the ratio of chlorophyll a to chlorophyll b was larger than 2 for alb3 mutants but smaller than 1 for alb3 alb4 double mutants. This is intriguing since chlorophyll a binds mainly to photosynthetic core proteins, suggesting that the predominant loss of chlorophyll a in the alb3 alb4 double mutants could be caused by a loss of photosynthetic core proteins. In addition to chlorophyll, the amount of total carotenoids (including carotenes and xanthophylls) was measured and found to be reduced to approximately one-third in the alb3 alb4 double mutants compared with alb3 single mutants (Fig. 2C). The reduction in both chlorophylls and carotenoids implies that ALB4 is responsible for the accumulation of some pigment-bearing proteins in the alb3 background. Severe deficiencies in such proteins in the alb3 alb4 double mutants could have a profound effect on the accumulation of thylakoid membranes, which is examined in detail in the next section.
The Chloroplasts of alb3 alb4 Double Mutants Have Fewer Thylakoid Membranes Than alb3 Chloroplasts
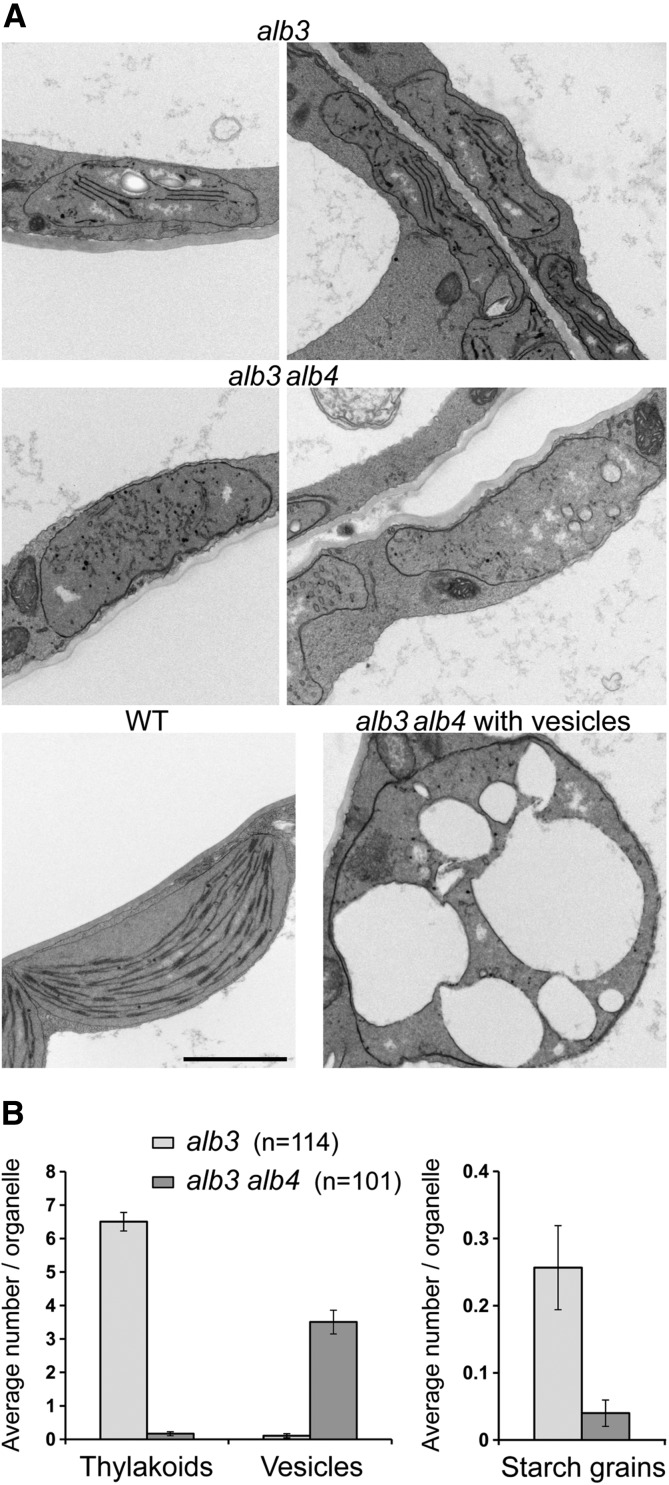
Plastids lacking ALB3 have been shown previously to form significantly fewer thylakoid membranes than the wild type (Sundberg et al., 1997). Here, the mesophyll cell plastids of the first true leaves of 17-d-old seedlings of both alb3 and alb3 alb4 double mutants were analyzed by transmission electron microscopy. Consistent with the previous report (Sundberg et al., 1997), it was found that alb3 plastids contain considerably fewer thylakoid membranes, and no granal stacks could be observed (Fig. 3A). Interestingly, the alb3 alb4 double-mutant plastids were almost completely devoid of thylakoids, with many instead accumulating large vesicles (Fig. 3A), which might be indicative of photooxidative damage (Jakob et al., 1997). In the alb3 alb4 double mutant, large, round plastids containing large vesicles occurred with roughly the same frequency as rather flat plastids without vesicles, although intermediate forms were also present. Generally, the plastids of the alb3 alb4 double mutant were larger than those of the alb3 single mutant. Quantification of thylakoid membranes and vesicles in a large number of plastids from both alb3 single mutants and alb3 alb4 double mutants showed that alb3 mutant plastids contain, on average, six to seven thylakoids per section, whereas thylakoids were virtually absent from alb3 alb4 double mutants (Fig. 3B); on the other hand, alb3 alb4 double mutants accumulated, on average, three to four large vesicles per organelle cross section, whereas vesicles did not accumulate in alb3 single mutants. Although it was shown previously that starch grains are absent from alb3 mutant chloroplasts (Sundberg et al., 1997), some starch grains could be found in a low number of alb3 mutant chloroplasts (Fig. 3, A [top left] and B). However, essentially no starch grains could be observed in the alb3 alb4 double mutants. This further indicates that the alb3 alb4 double mutants have a more severe phenotype than the alb3 single mutants, with reduced photosynthetic capacity, and that therefore there is some functional overlap between ALB3 and ALB4.
Figure 3.
Transmission electron micrographs of alb3 single and alb3 alb4 double mutants. A, Transmission electron micrograph images of mesophyll cell plastids from the first true leaves of 17-d-old seedlings grown in vitro. All images show plastids at the same scale; the black scale bar in the wild-type image corresponds to 2 μm. The left image of alb3 shows a starch grain in the middle of the plastid. The alb3 plastids generally contain a few thylakoid membranes, whereas these are lacking almost completely in the alb3 alb4 double mutants. In the alb3 alb4 double mutants, large round plastids with many vesicles occur roughly at the same frequency as relatively flat plastids without vesicles. B, Average number of thylakoids, vesicles, and starch grains found in plastids from alb3 single and alb3 alb4 double mutants. Numbers in parentheses denote total numbers of plastids used for counting. The plastid images analyzed were derived from three biological replicates. Error bars denote se (n shown in parentheses).
ALB4 Plays a Role in the Accumulation of Some Cytochrome b6f Components in the Thylakoid Membranes
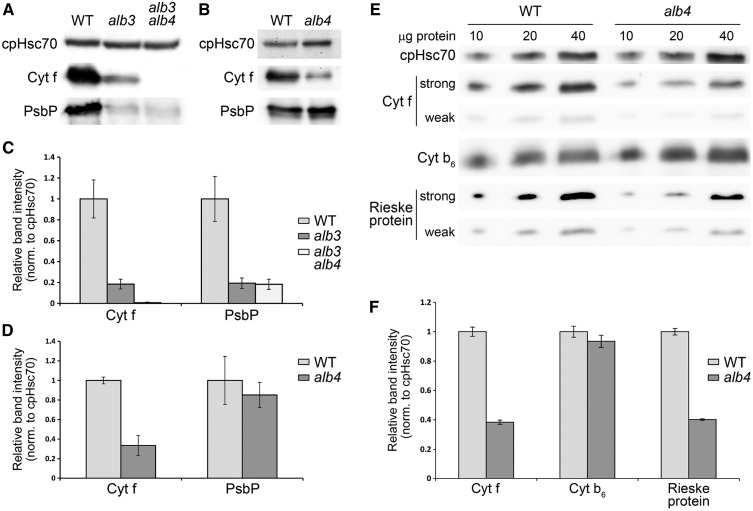
Since alb3 single mutants accumulate some pigments (of which a large part is chlorophyll a) and still contain a few thylakoid membranes, we asked which pigment-binding proteins might be present in the remaining thylakoid membranes in alb3. By using western blotting, we found that many photosynthesis-related proteins were already undetectable in the alb3 single mutant, making it difficult to assess for a role of ALB4 in their assembly by analysis of the double mutant (data not shown). Nonetheless, we found that cytochrome f (Cyt f) accumulates in the alb3 mutant but not in the thylakoid-less alb3 alb4 double mutants (Fig. 4, A and C), suggesting that ALB4 alone can insert some Cyt f into the remaining thylakoids of alb3 mutants. Thus, Cyt f is a potential client of ALB4. It was shown previously that the cytochrome b6f complex contains one molecule of chlorophyll a per molecule of Cyt f (Dashdorj et al., 2005; Baniulis et al., 2011), and so this complex may be (partly) responsible for the residual pigment-binding activity present in alb3. Interestingly, even though the alb4 single mutant has a wild-type visible phenotype, Cyt f is also significantly reduced in alb4 compared with the wild type (Fig. 4, B and D). This suggests that ALB3 alone cannot efficiently insert Cyt f, and that ALB4 plays an important role in the Cyt f insertion process. Nevertheless, alb4 accumulates some Cyt f, suggesting that only the concerted action of ALB3 and ALB4 is able to efficiently insert Cyt f.
Figure 4.
Accumulation of thylakoid proteins in alb3, alb4, and double-mutant plants. A, Western-blot analysis of total protein extracts from 3-week-old seedlings. All seedlings were grown on MS medium containing 0.5% (w/v) Suc. The alb3 and alb3 alb4 seedlings were transferred to medium containing 3% (w/v) Suc at 2 weeks postgermination. Sample loading was normalized according to the intensity of the chloroplast Hsp70 cognate (cpHsc70) band. B, Western-blot analysis of total protein extracts from 2-week-old seedlings grown on MS medium containing 0.5% (w/v) Suc. C, Band intensity quantification of the data shown in A, normalized according to the cpHsc70 data. Error bars denote sd (n = 3). The average Cyt f value for the wild type (WT) has been set to 1. D, Band intensity quantification of the data shown in B, prepared exactly as described for C. E, Western-blot analysis of total protein extracts from 2-week-old seedlings grown on MS medium containing 0.5% (w/v) Suc. A series of 10, 20, and 40 μg of total protein was loaded for each genotype, as indicated. For Cyt f and the Rieske protein, two different exposures are shown (strong and weak) to better illustrate the differences in signal intensity. F, Band intensity quantification of the data shown in E, prepared exactly as described for C.
To assess a potential role of ALB4 in the insertion of other components of the cytochrome b6f complex, we quantified the amounts of cytochrome b6 (Cyt b6) and the Rieske protein in the alb4 mutant relative to the wild type by immunoblotting (Fig. 4, E and F). To enable better quantification, this analysis was conducted using a dilution series of each sample, and so Cyt f was analyzed again to provide further corroboration of its reduced abundance in the alb4 mutant. The data revealed that the Rieske protein is reduced in alb4 mutants to a similar extent as Cyt f, whereas Cyt b6 was not significantly reduced. It should be noted that Cyt f and Cyt b6 are both encoded by the chloroplast genome, but only one of them is depleted. Therefore, a secondary effect because of reduced chloroplast translation seems unlikely. The two depleted components (Cyt f and the nucleus-encoded Rieske protein) both have only one transmembrane domain, and so the insertion mechanism used (possibly involving ALB4) might be different from that used by Cyt b6 with four transmembrane spans. As two components of the cytochrome b6f complex are significantly reduced in the alb4 mutant, we reasoned that photosynthetic capacity might also be reduced in this mutant. To test this, we measured the maximum photochemical efficiency of PSII (the Fv/Fm ratio) in wild-type and alb4 mutants using a chlorophyll fluorescence imaging system. Indeed, there was a small but significant reduction in Fv/Fm in the alb4 mutant plants compared with the wild type (Student’s two-sample t test based on equal variances, P < 0.05; Supplemental Fig. S4).
Overall, it can be concluded that ALB4, together with ALB3, is involved in the insertion of some cytochrome b6f subunits. This suggests close functional and potentially physical interactions between ALB3 and ALB4. However, a physical interaction between ALB3 and ALB4 was not detected in a previous study (Benz et al., 2009). To clarify this discrepancy, we examined whether ALB3 and ALB4 can interact in vivo.
ALB3 and ALB4 Can Interact Physically in Vivo
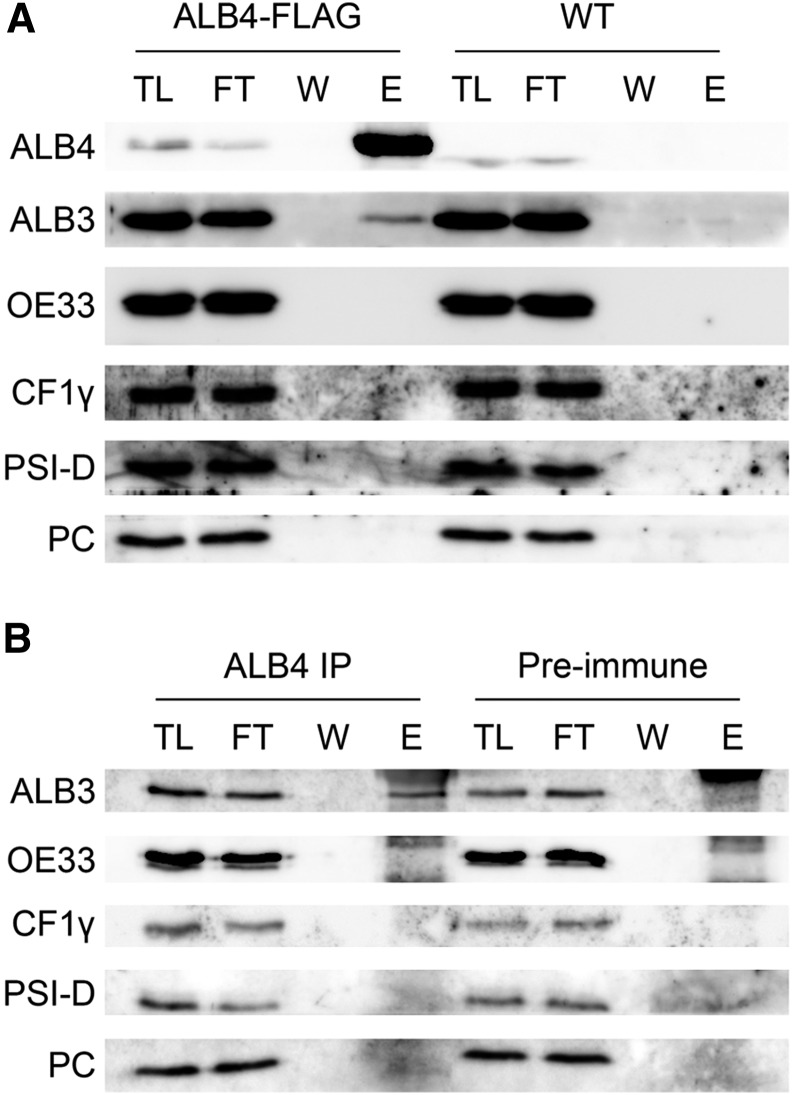
Given that the above analysis revealed significant functional overlap between ALB3 and ALB4, it seemed conceivable that the two proteins interact physically with each other or with the same partners in vivo. To address this possibility, we used immunoprecipitation from isolated chloroplasts following solubilization using the nonionic detergent n-dodecyl-β-d-maltopyranoside (DDM). An interaction between ALB3 and ALB4 could not be found in uncross-linked chloroplasts, neither in a previous study (Benz et al., 2009) nor in the current study (data not shown). However, by using chloroplasts from transgenic plants expressing FLAG-tagged ALB4, a weak interaction of ALB3 with ALB4-FLAG could be detected after cross linking with 0.5 mm dithiobis(succinimidyl propionate) (DSP; Fig. 5A); note that ALB3 and ALB4 migrate at significantly different positions during electrophoresis and so were easily distinguished in this analysis (Supplemental Fig. S3). Several control proteins, all of which are thylakoid membrane-associated proteins, did not interact with ALB4-FLAG, indicating that the detected ALB3-ALB4 interaction is specific. That the ALB3-ALB4 interaction could only be detected following stabilization using a chemical cross linker suggests that the ALB3-ALB4 interaction is not stable, but rather is transient in nature or possibly mediated by other factors that bridge the interaction in vivo.
Figure 5.
Physical interaction between ALB3 and ALB4 in vivo. A, Immunoprecipitation with Anti-FLAG M2 Affinity Gel using 100 million chloroplasts isolated from plants stably overexpressing an ALB4-FLAG fusion under the Cauliflower mosaic virus 35S promoter. The chloroplasts were cross linked with 0.5 mm DSP. TL, Total chloroplast lysate (0.45%); FT, flow through (0.45%); W, final (sixth) wash (approximately 4.5%); E, eluate (36%). ALB4 was detected with anti-ALB4 antibody; the size shift of ALB4 in the ALB4-FLAG sample compared with the wild type corresponds to the size of the FLAG tag. B, Immunoprecipitation (IP) with anti-ALB4 antibody (or with corresponding preimmune serum) using 100 million chloroplasts isolated from wild-type plants. Chloroplasts were cross linked with 0.5 mm DSP. OE33, Oxygen-evolving complex 33 kD subunit; PSI-D, photosystem I subunit D; PC, plastocyanin.
Because the ALB4-FLAG lines contain an overexpressed and tagged form of ALB4, one could argue that the observed interaction is an artifact caused by the artificial nature of the experimental system. To exclude this possibility, further immunoprecipitation experiments were performed using an anti-ALB4 antibody and wild-type chloroplasts (Fig. 5B). In the immunoprecipitation with anti-ALB4, a weak but specific interaction between ALB3 and ALB4 could be observed, confirming the finding described above. It is particularly interesting that the ATP synthase subunit CF1γ could not be cross linked to either native ALB4 (Fig. 5B) or overexpressed ALB4-FLAG (Fig. 5A), even though it was shown previously that ALB4 comigrates with CF1γ in two-dimensional blue native/SDS-PAGE experiments (Benz et al., 2009). This finding argues against a nonspecific interaction caused by over-cross linking of the proteins, and therefore supports the relevance of the ALB3-ALB4 interaction detected in this study.
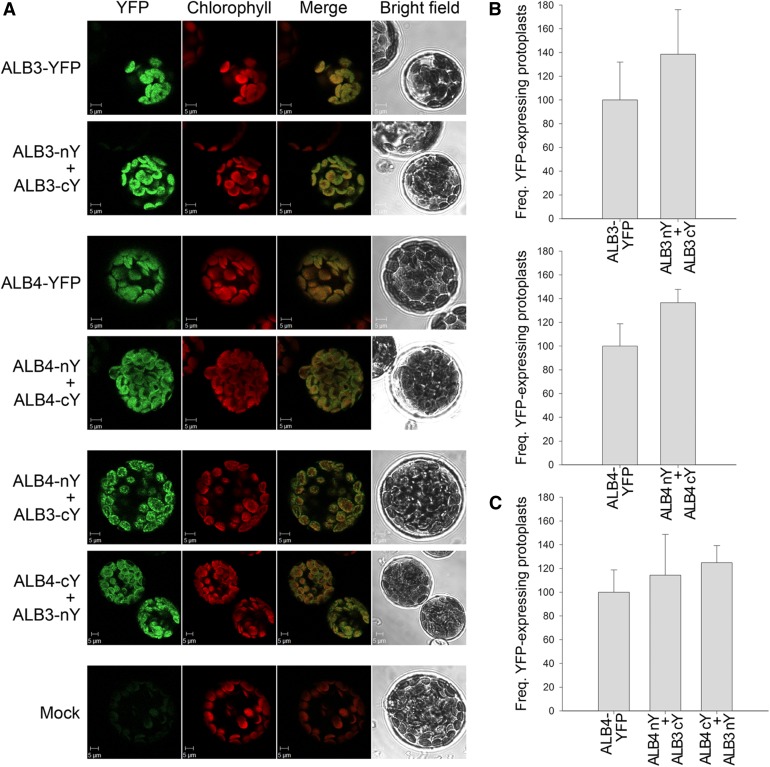
To corroborate the immunoprecipitation data, we performed bimolecular fluorescence complementation (BiFC) experiments using constructs that express ALB3-cYFP, ALB3-nYFP, ALB4-cYFP, and ALB4-nYFP under the 35S promoter (nYFP and cYFP are complementary N- and C-terminal fragments of yellow fluorescent protein [YFP]). The data clearly showed that ALB3 and ALB4 can interact in vivo. Both combinations of cotransfections (i.e. ALB4-nYFP with ALB3-cYFP and ALB4-cYFP with ALB3-nYFP) produced a signal localized to the thylakoid membranes, in a range of patterns from particulate to filamentous (Fig. 6A). Moreover, the previous finding that ALB3 can form dimers at the thylakoid membrane (Dünschede et al., 2011) was confirmed by our analysis and extended to include the ALB4 homolog (Fig. 6A), further emphasizing their functional similarity. The frequency of the transformed, fluorescent protoplasts compared with untransformed, nonfluorescent protoplasts was comparable for ALB3 and ALB4 homodimers and the respective full-length YFP constructs (Fig. 6B) and for ALB3-ALB4 heterodimers compared with the ALB4-YFP full-length construct (Fig. 6C), suggesting that all interactions are equally efficient in vivo (note that the frequencies of positive protoplasts seen for ALB3-YFP and ALB4-YFP were very similar; data not shown). The specificity of the observed interactions was confirmed by the analysis of the ALB fusions in conjunction with fusions to two different control proteins (Supplemental Fig. S5).
Figure 6.
Bimolecular fluorescence complementation analysis of ALB3 and ALB4 interactions. A, Wild-type protoplasts were (co)transfected with the indicated constructs and analyzed by confocal microscopy for YFP fluorescence and chlorophyll autofluorescence, and by bright-field illumination; YFP denotes fusions to full-length YFP, whereas nY and cY denote fusions to complementary N- and C-terminal fragments of YFP. The formation of ALB3 or ALB4 homodimers, and the interaction between ALB3 and ALB4 in heterodimers, is shown by the reconstitution of the YFP protein leading to fluorescence, as shown by the typical images in the respective panels. No YFP signal was detected in untransformed control protoplasts (Mock). B, The frequency (Freq.) of protoplasts that displayed YFP reconstitution via ALB3 or ALB4 homodimerization. Values for formation of the homodimers are normalized relative to the frequency observed for the respective full-length YFP construct. C, The frequency of protoplasts that displayed YFP reconstitution via ALB3-ALB4 heterodimerization (in both directions). Values for formation of heterodimers are normalized relative to the frequency observed for the full-length ALB4-YFP construct. In B and C, all error bars denote sd (n = 3).
ALB4 Can Indirectly Associate with cpSRP Components in Vivo
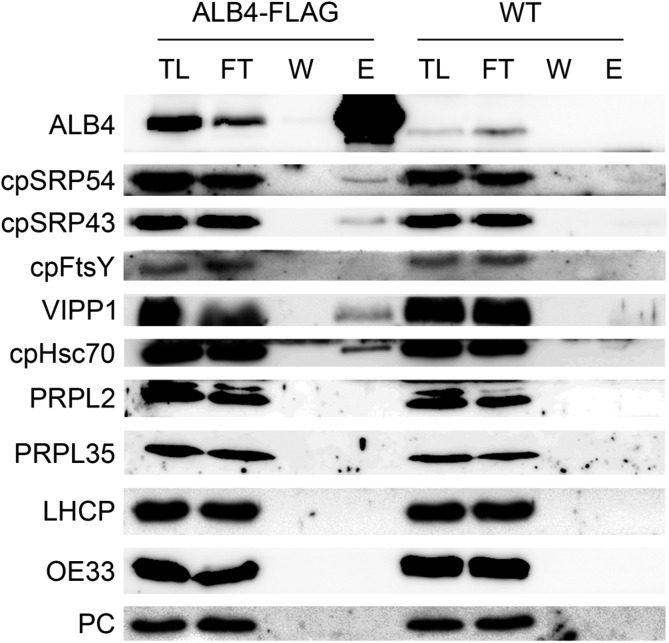
Since ALB4 could be cross linked specifically to ALB3, the question arises as to whether it could also be cross linked to the cpSRP components and cpFtsY (which are well known to act in the same pathway as ALB3). Because anti-FLAG and anti-ALB4 immunoprecipitations gave comparable results for the interactions of ALB3 and ALB4 (Fig. 5), further interaction studies were performed by anti-FLAG immunoprecipitation using solubilized ALB4-FLAG chloroplasts. In this way, an interaction between ALB4 and both cpSRP components, but not cpFtsY, could be observed (Fig. 7), supporting the notion that ALB4, similar to ALB3, acts in the cpSRP pathway. The detected interactions were specific, since ALB4-FLAG could not be cross linked to abundant components of the photosynthetic apparatus, including LHCP, oxygen-evolving complex 33-kD subunit, and plastocyanin, which were used as negative controls in this experiment.
Figure 7.
Physical interaction of ALB4 with chloroplast SRP components in vivo. Immunoprecipitation with Anti-FLAG M2 Affinity Gel using 100 million chloroplasts isolated from plants stably overexpressing an ALB4-FLAG fusion under the Cauliflower mosaic virus 35S promoter. Chloroplasts were cross linked with 0.5 mm DSP. TL, total chloroplast lysate (0.45%); FT, flow through (0.45%); W, final (sixth) wash (approximately 4.5%); E, eluate (36%). ALB4-FLAG interacts with the signal recognition particle components cpSRP43 and cpSRP54, as well as with vesicle-inducing protein of plastids1 (VIPP1) and cpHsc70. However, no interaction between ALB4-FLAG and PRPL2 and PRPL35 could be observed. The abundant photosynthesis-related proteins LHCP, oxygen-evolving complex 33 kD subunit (OE33), and plastocyanin (PC) were used as negative controls. WT, Wild type.
A previous study reported an interaction between C. reinhardtii Alb3.2 and vesicle inducing protein in plastids1 (VIPP1; Göhre et al., 2006), which is involved in thylakoid formation and maintenance (Kroll et al., 2001). In Alb3.2 RNA interference mutants, both VIPP1 and chloroplast Hsp70 are overproduced, and it was suggested that the VIPP1/chloroplast heat shock protein of 70 kD (cpHsp70) system is involved in the targeting of Alb3.2 in C. reinhardtii (Göhre et al., 2006). Therefore, we assessed for possible interactions of ALB4 with VIPP1 and cpHsc70 in Arabidopsis. Indeed, clear interactions of ALB4-FLAG with VIPP1 and cpHsc70 could be observed after cross linking (Fig. 7). This implies functional similarity between ALB4 and ancestral Alb3-type proteins. The VIPP1/cpHsp70 system might be involved in the targeting of Arabidopsis ALB proteins, or alternatively, it might act concomitantly with ALB4 to maintain thylakoid stability and complex assembly, as suggested for C. reinhardtii (Göhre et al., 2006).
Because it has been previously reported that Oxa1, a mitochondrial homolog of ALB3 and ALB4, can interact with components of the large ribosomal subunit in mitochondria (Jia et al., 2003; Szyrach et al., 2003), and because it was speculated that two conserved motifs in the ALB3 and ALB4 C termini could be responsible for ribosome binding (Falk et al., 2010), the interaction of ALB4-FLAG with two plastid ribosomal large-subunit proteins (PRPLs) was also investigated. However, neither PRPL2 nor PRPL35 could be cross linked to ALB4-FLAG, arguing against a role for ALB4 similar to that of Oxa1 (Fig. 7).
Phylogenetic Analysis of ALB3 and ALB4 Supports Functional Divergence
Both ALB3 and ALB4 are multispanning transmembrane proteins with a substantial C-terminal domain (starting after the fifth transmembrane domain) exposed to the stroma. It has been proposed that differences in their functions are caused by differences in their C termini, resulting in differing interactions with partner proteins. By the alignment of sequences from several species, including Arabidopsis, a previous analysis identified four motifs (denoted I, II, III, and IV) in the C-terminal domain of ALB3 (Falk et al., 2010). Motifs II and IV were shown to be essential for the binding of ALB3 to cpSRP43, and found to be absent from ALB4; motifs I and III were present in both ALB proteins and dispensable for the cpSRP43 interaction (Falk et al., 2010). These results are not entirely consistent with our observations that ALB4 associates with cpSRP components in vivo and shares functional redundancy with ALB3. To begin to address the discrepancy, we first conducted a reassessment for the presence of the motifs using a computer program called MEME (Bailey and Elkan, 1994). In accordance with the previous analysis, all four motifs were identified in ALB3, whereas only motifs I and III were clearly present in ALB4 (Fig. 8). Our analysis also identified an ALB4-specific motif running from the C-terminal end of motif I and across the position occupied by motif II in ALB3. This indicates that motifs I and II may be part of one larger motif, with its C-terminal end (identified as motif II) being responsible for substrate binding. We also identified an additional conserved region in ALB3 that overlaps with the C-terminal end of motif II, and is very similar to a corresponding region in ALB4 (at least in the Brassicaceae, Arabidopsis, and Brassica rapa; Fig. 8).
Figure 8.
Motifs identified in the C-terminal domains of ALB3 and ALB4. A multiple sequence alignment of the C-terminal regions of ALB3 and ALB4 homologs from different species is shown. The first residue is position 339 in Arabidopsis ALB3 (and the corresponding position in all other sequences), and represents the start of motif I as defined by Falk et al. (2010). Positions of the motifs are indicated above the alignment. The red box indicates an ALB4-specific motif spanning the positions of motifs I and II, which was identified by the program meme. The green box indicates the position of the extended C-terminal part of motif II, which is present in both ALB3 and ALB4, and is hypothesized to be part of the substrate binding motif. Highly conserved sites are identified by a black background, and similar amino acids by a gray background.
Thus, we hypothesize that the interaction between ALB4 and cpSRP43 observed in our study (Fig. 7) is either indirect (e.g. mediated by a mutual partner protein) or occurs via the aforementioned extended motif II that is present in both ALB3 and ALB4. Interestingly, our analysis also showed that all four motifs are present in the ALB3 homolog of the moss Physcomitrella patens, implying that the ancestor of all land plants contained an Alb3-type protein with all four motifs, and that two of them were subsequently lost, or highly modified, in ALB4 (Fig. 8).
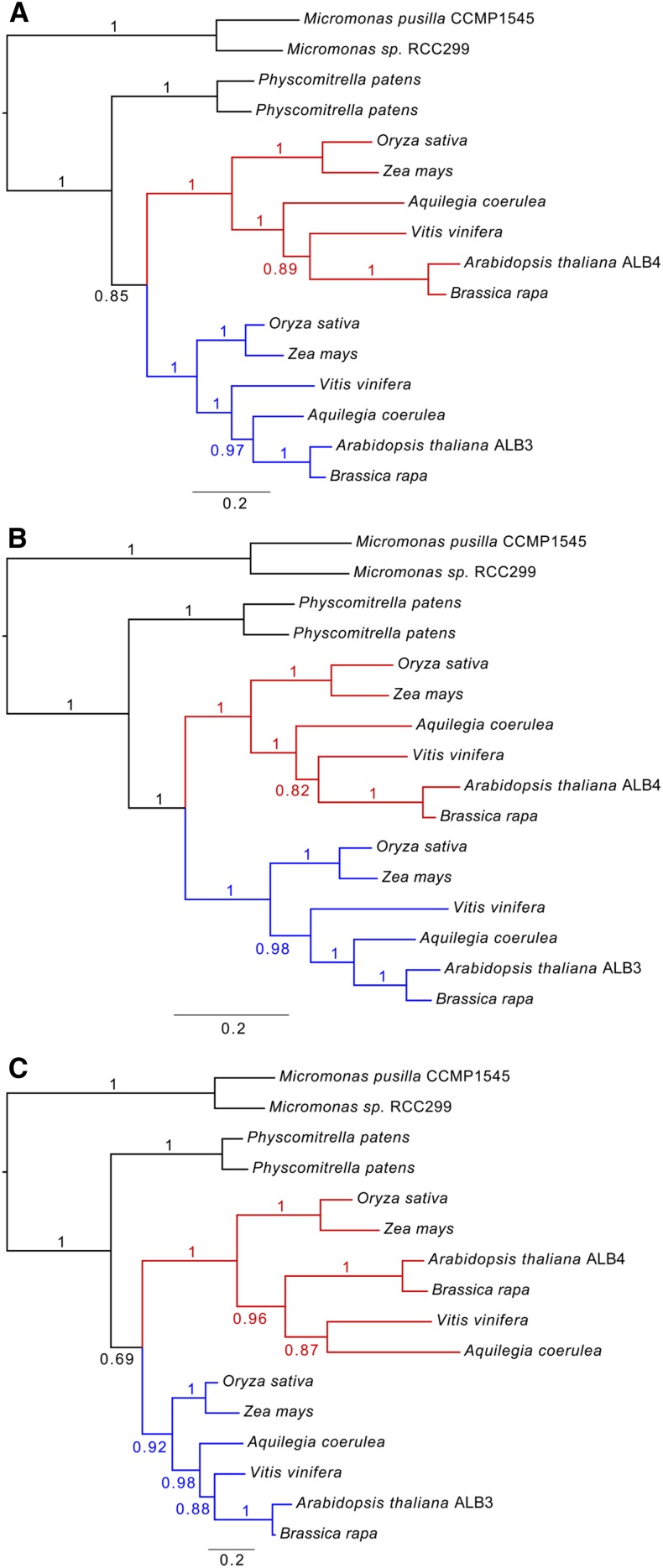
To further assess whether the common ancestor of the ALB proteins was more similar to ALB3 or ALB4, and whether the subsequent evolutionary divergence was greater in the C termini relative to other parts of the protein, a phylogenetic analysis was conducted. In this work, we analyzed the full-length sequences, as well as the N-terminal and C-terminal domains independently. The results indicated that the different parts of the proteins have indeed evolved at different rates. Analysis of the full-length sequences showed that the evolutionary rate in the ALB4 clade has been greater than that in ALB3, as indicated by the differences in branch length between the ALB3 and ALB4 clades (Fig. 9A; see Supplemental Fig. S6 for a more extensive analysis). This is consistent with the notion that ALB4 has undergone greater functional divergence from the ancestral protein. Interestingly, although the N-terminal parts evolved at an equal rate in the two clades (Fig. 9B), accelerated evolutionary rates occurred in the C termini of the ALB4 proteins (Fig. 9C), thus explaining the overall difference seen with the full-length sequences. These results are consistent with the aforementioned motif analyses and support the notion that ALB4 functions have diverged, at least to some extent, from those of ALB3 due to changes in the C-terminal domain.
Figure 9.
Phylogenetic analysis of ALB sequences from different species. The full-length sequences of ALB3 and ALB4 proteins, as well as their N-terminal and C-terminal domains, were analyzed independently. Comparison of the branch lengths indicates that the evolutionary rates have varied in different parts of the proteins. A, Analysis of full-length sequences shows that the overall rate has been greater in the ALB4 clade (red) than in the ALB3 clade (blue). B, The N-terminal domain has evolved at an equal rate in the two main clades. C, An accelerated evolutionary rate occurred in relation to the C-terminal part of the ALB4 proteins. The scale bar below each tree indicates the number of expected changes per site along the branches, and posterior probability values are indicated at each branch.
DISCUSSION
In this study, we showed that Arabidopsis alb3 alb4 double mutants have a lower chlorophyll and carotenoid content than alb3 single mutants, and that the chloroplast ultrastructure of the double mutant is further deteriorated. Therefore, one role of ALB4 in the alb3 mutant background is likely to be the insertion of some pigment-bearing proteins into the thylakoid membrane, which was not predicted in earlier models of ALB4 function (Benz et al., 2009). Indeed, we could show that Cyt f accumulates in alb3 mutants but is almost completely absent in alb3 alb4 double mutants. Moreover, both Cyt f and the Rieske protein are significantly reduced in alb4 single mutants, even though these mutants have no obvious visible defects. Since it is known that the cytochrome b6f complex binds chlorophyll a and β-carotene molecules (Dashdorj et al., 2005; Baniulis et al., 2011), differences in the accumulation of cytochrome complex components between the alb3 single mutant and the alb3 alb4 double mutant could be (partly) responsible for the pigmentation differences. The data suggest that ALB4 is involved in the insertion of Cyt f and the Rieske protein, and thus possibly in the biogenesis of the cytochrome b6f complex. This notion is supported by the observation that photosynthetic performance (as indicated by the Fv/Fm ratio) is slightly but significantly reduced in alb4 mutants compared with the wild type. That Cyt f is still present in the alb4 single mutant but not in the alb3 alb4 double mutant suggests that both ALB3 and ALB4 may act together in the biogenesis of the cytochrome b6f complex.
In a previous study, ALB3 and ALB4 could not be coimmunoprecipitated from isolated thylakoid membranes in the absence of a cross linker (Benz et al., 2009). Likewise, an interaction between ALB3 and ALB4 could not be detected in the current study using uncross-linked isolated chloroplasts. This suggests that any interaction between these components may be transient, indirect, or lost during protein preparation. Nonetheless, by using cross-linked chloroplasts, a specific interaction between ALB3 and ALB4 could be detected. The function of this interaction is unlikely to be reciprocal membrane insertion (i.e. a client-insertase relationship), as ALB4 is still prominently detected in albino alb3 mutants (Fig. 1D), and ALB3 is also detected in alb4 mutants (Supplemental Fig. S3). Our BiFC study not only verified the interaction between ALB3 and ALB4 in vivo, but also suggested that this heterodimerization is as frequent as homodimerization. In C. reinhardtii, the two paralogous Alb3 proteins, Alb3.1 and Alb3.2, were found to interact with each other and with reaction center polypeptides, suggesting that both play a role in the assembly of reaction centers (Göhre et al., 2006). This shows that the interaction between different ALB isoforms for the concerted insertion of a subset of substrate proteins might not be uncommon.
The weak phenotype of alb4 single mutants suggests that most thylakoid proteins can be targeted by ALB3 alone. However, as ALB4 is able to interact with ALB3 in vivo, it may form heterodimers that potentially enable optimized insertion of a subset of thylakoid proteins, relative to ALB3 homodimers, which might be inefficient in these cases. The discovery that both ALB3 and ALB4 are required for efficient insertion of Cyt f suggests that this may be one such substrate protein. It is tempting to speculate that ALB4 evolved to optimize the insertion of specific substrates: that ALB3 alone may insert LHCPs efficiently and other substrates less efficiently, and that ALB4 increases efficiency when acting together with ALB3 on the latter substrates. It should be noted that a previous study associated the release of soluble Cyt f with fewer stacked thylakoid membranes and a more swollen thylakoid lumen in Chlorella saccharophila chloroplasts (Zuppini et al., 2009). Interestingly, a very similar phenotype has been described for Arabidopsis chloroplasts of alb4 mutants (Benz et al., 2009), again supporting the role of ALB4 in the insertion of Cyt f.
As it is well known that proteins from the YidC/Oxa1/Alb3 family can function in both post- and cotranslational modes of membrane protein insertion, a role for ALB3 in cotranslational membrane protein insertion that might involve SecY-related SCY1 and cpSRP54 has been suggested (Zhang and Aro, 2002; Moore et al., 2003). In fact, the ALB3 and ALB4 C termini both contain two conserved motifs, I and III, which have been suggested to be involved in SCY1 binding (Falk et al., 2010). Therefore, the question arises as to whether ALB3 and ALB4 are involved in cotranslational protein targeting. The C terminus of Oxa1, the mitochondrial homolog of ALB3 and ALB4, is known to interact with the ribosomal large subunit during cotranslational protein insertion (albeit independently of SRP and the Sec translocon, both of which are absent in mitochondria; Glick and Von Heijne, 1996; Szyrach et al., 2003). However, here, we show that ALB4 does not interact with two plastid ribosomal proteins of the large subunit, PRPL2 and PRPL35. Also, alb4 mutants are not specifically affected in chloroplast-encoded proteins, as the reduced Cyt f and Rieske components are of different genetic origins, whereas the chloroplast-encoded Cyt b6 component is not reduced. Although this does not exclude the possibility that ALB4 transiently interacts with other ribosomal proteins or translating proteins, it does argue against an active participation of ALB4 in the early steps of cotranslational membrane protein insertion.
It was previously shown in vitro that the C terminus of ALB3 can interact with cpSRP43 based on motifs that are absent in the ALB4 C terminus, which consequently cannot interact with cpSRP43 (Falk et al., 2010). Surprisingly, in the current study, we found that both cpSRP43 and cpSRP54 can be cross linked to ALB4 in vivo, suggesting an interaction involving other ALB4 motifs (such as the motif II extension identified in this study) or an indirect, bridged interaction between ALB4 and cpSRP43. Since ALB4 also interacts with ALB3, it cannot be determined if ALB4 interacts with the cpSRP components directly or via ALB3. Nevertheless, our results provide evidence that ALB4 can participate in processes that involve a close physical proximity to the whole ALB3-cpSRP system. The fact that ALB4 can interact with ALB3, cpSRP43, and cpSRP54 is consistent with a function of ALB4 in the protein insertion process of the ALB3-cpSRP system for a subset of thylakoid proteins, as outlined above. In our experiments, cpFtsY was not cross linked to ALB4, which is in line with the earlier finding that cpFtsY does not form significant interactions with cpSRP components or ALB3 (Tu et al., 2000). Perhaps the GTP-dependent interaction of cpSRP54 with cpFtsY is too transient to be caught with the conditions used here, even following cross linking. The detection of ALB4 interactions with VIPP1 and cpHsc70 in the current study is in line with previous studies showing interactions between Alb3.2 and VIPP1 (Göhre et al., 2006), and between soluble VIPP1 and cpHsc70 (Liu et al., 2007), in C. reinhardtii. Perhaps the VIPP1 system and the ALB3-cpSRP system cooperate in thylakoid biogenesis, coordinating the extension of the lipid layers with protein insertion.
Our phylogenetic analyses showed that the N-terminal parts of the ALB3/4 proteins evolved at similar rates in angiosperms, and that there was a shift in evolutionary rate in relation to the C-terminal domains following a gene duplication in an ancient angiosperm Alb3 protein. The gene duplication would have led to a redundancy of Alb3 genes that enabled one of the copies (the ALB4 progenitor) to mutate and eventually evolve a divergent function, while the other (the ALB3 progenitor) retained the original function performed by the ancestral protein. The accelerated evolutionary rate led to the loss (or extensive modification) of two potential binding motifs (II and IV) in ALB4. Nonetheless, our results show that cpSRP43 can to some extent interact with ALB4 in vivo, suggesting an interaction involving motifs other than II and IV (e.g. the extended motif II described here, or others present elsewhere in the C-terminal domain or even in one of the loops) or an indirect, bridged interaction. Motifs I and III, which are present in both ALB3 and ALB4, have been suggested to be responsible for ribosome binding (Falk et al., 2010). However, we were unable to detect an interaction of ALB4 with two chloroplast ribosome components.
In summary, our results point to a role for ALB4 in inserting some proteins (e.g. Cyt f and potentially other components of pigment-bearing complexes) into thylakoids in the alb3 background, whereas in the wild-type background, ALB4 might act together with ALB3 and thus optimize its function for the assembly of the cytochrome b6f complex and possibly other complexes. Although ALB3 is certainly crucial for the insertion of light-harvesting complex components, and ALB4 might influence the stability of nonphotosynthetic thylakoid complexes (Benz et al., 2009), a sharp separation of the functions of the two paralogues is not supported by the presented data. Both genetic and physical interaction studies suggest some functional overlap between ALB3 and ALB4, but they may have evolved such that ALB3 plays key roles during membrane protein insertion while ALB4 optimizes this process for a subset of substrates. Thus, ALB3 and ALB4 likely contribute differentially but synergistically to the same process of protein insertion into the thylakoids via the ALB3-cpSRP pathway.
MATERIALS AND METHODS
Plant Growth and Genotyping
Mutant seeds were ordered from the Nottingham Arabidopsis Stock Centre (alb3: GABI_293B08/N324478; alb4: Salk_136199/N636199). They were grown directly on soil (alb4, kanamycin resistance marker silenced), or in vitro on one-half-strength MS plates containing 0.5% (w/v) Suc and 11.25 μg mL−1 sulfadiazine (for alb3 selection) after surface sterilization of the seeds and 2-day stratification at 4°C as described previously (Aronsson and Jarvis, 2002). After initial germination and growth for 14 d on the standard medium indicated above, albino seedlings (homozygous alb3 mutants and alb3 alb4 double mutants) were transferred to one-half-strength MS plates containing 3% (w/v) Suc for further growth. Plants were grown in approximately 100 μmol m−2 s−1 white light under long-day cycles (16-h light, 8-h dark) except where short-day conditions (8-h light, 16-h dark) are specifically indicated.
For genotyping, the following primers were used: alb4 forward, 5′-CCTTGCA-GGTACAGTATGTTA-3′; alb4 reverse, 5′-CTGTTGCATAGAAGGATTTCG-3′; alb3 forward, 5′-CGCTTCGTATTGAGAGATATA-3′; alb3 reverse, 5′-GAGAGGA-TACAACTAGAGACA-3′. For T-DNA-specific PCRs, the alb4 forward primer was combined with SALK LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′, and the alb3 reverse primer was combined with GK-LB, 5′-CCCATTTGGACGTGAATGTAGACAC-3′.
Total Chlorophyll and Carotenoid Measurements, Transmission Electron Microscopy, and Fluorescence Measurements
Pigment extraction and measurement was based on a method described previously (Czarnecki et al., 2011). Approximately 50 mg of 3-week-old seedlings were frozen in liquid nitrogen, ground, and then covered with 300 μL of 80% (v/v) acetone containing 10 μm KOH. The samples were vortexed for at least 1 min and then centrifuged at 10,000g for 10 min in a microcentrifuge. Absorbances of the supernatant were measured at 663, 647, and 470 nm, and the amounts of total chlorophyll per milligram fresh weight were calculated as previously described (Czarnecki et al., 2011).
For transmission electron microscopy, the first true leaves from 17-d-old plants were used, and the service of the University of Leicester Electron Microscope Laboratory (Faculty of Medicine and Biological Sciences) was used. Leaves were excised from the plant and fixed overnight in 4% (v/v) glutaraldehyde/2% (v/v) formaldehyde in 0.1 m sodium cacodylate buffer with 2 mm calcium chloride (pH 7.2), and then washed in the same cacodylate/CaCl2 buffer. The tissue was then fixed with 1% (w/v) OsO4/1.5% (w/v) potassium ferricyanide for 3 h, washed with distilled, deionized water, and finally tertiary fixed with 2% (w/v) uranyl acetate for 1 h. The tissue was serially dehydrated through ethanol and propylene oxide and embedded in Spurr’s modified low-viscosity resin. Thin sections of approximately 80-nm thickness were cut using a Reichert Ultracut S ultramicrotome, collected onto copper mesh grids, and stained with Reynolds’ lead citrate. The grids were viewed under a JEOL JEM-1400 electron microscope at 80 kV. Digital images were recorded using a SIS digital camera and iTEM software (Olympus; Glauert, 1998; Hyman and Jarvis, 2011).
The Fv/Fm ratio was measured using a CF Imager chlorophyll fluorescence imaging system (Technologica). Wild-type and alb4 mutants were grown on one-half-strength MS medium containing 0.5% (w/v) Suc as described above. Four plates with 17-d-old seedlings from each genotype were dark adapted for 1 h and subsequently recorded under saturating light pulses of 6,000 μmol photons m−2 s−1. The Fv/Fm ratio values were determined by the CF Imager’s software (Baker, 2008; Gorecka et al., 2014) for four quarters of each plate independently, resulting in 16 individual measurements per genotype.
Antibody Production and Immunoblotting
The anti-ALB4 antibody was generated by cloning the coding sequence of the C-terminal 155 amino acids of ALB4 (soluble part) into pQE-30 (Qiagen) using BamHI and PstI following amplification using primers ALB4-His-forward, 5′-GGGGGATCCCCAGTGGAGAAATTCACTAA-3′, and ALB4-His-reverse, 5′-GGCCTGCAGGTTACCTCTTCTCTGTTTCAT-3′. Transformed XL1-Blue (Agilent Technologies) cells were grown in the presence of 1% (w/v) Glc to repress the lactose operon and avoid leaky expression. Expression of His-tagged ALB4 C terminus was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside, and the cells were lysed by sonication in lysis buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, pH 8.0). The lysate was put onto polypropylene columns containing Ni-NTA resin (Qiagen), washed with wash buffer (50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole, pH 8.0), and the proteins were eluted in elution buffer (50 mm NaH2PO4, 300 mm NaCl, 250 mm imidazole, pH 8.0). Purified protein was sent to Harlan Sera-Lab for antibody production in rabbits, and the antiserum was affinity purified using the original antigen. The anti-ALB3 antibody was a gift from D. Schünemann (AG Molekularbiologie, Ruhr-Universität Bochum; Gerdes et al., 2006; Asakura et al., 2008). The cpSRP and cpFtsY antibodies were a gift from M. Nakai (Institute for Protein Research, Osaka University). Immunoblots were performed using standard methods with 12% (w/v) polyacrylamide gels and nitrocellulose membranes (Kovacheva et al., 2005).
Bimolecular Fluorescence Complementation
For BiFC, the vectors pSAT4(A)-cEYFP-N1 and pSAT4(A)-nEYFP-N1 were used (Lee et al., 2008). For the ALB4 constructs, amplification of the wild-type complementary DNA used the primers ALB4-BiFC-forward, 5′-AAGAGATCTCAAAGCAAGAACACAACAACA-3′ and ALB4-BiFC-reverse, 5′-AAGGTCGACTCCTCCTCTCTGTTTCATGAGA-3′; for the ALB3 constructs, the primers were ALB3-BiFC-forward, 5′-AAAGAATTCTCTCTATCTTCTTCTTCGTCTCTT-3′ and ALB3-BiFC-reverse, 5′-AAAGTCGACATACAGTGCGTTTCCGCTTCGA-3′; for the OEP7 constructs, the primers were OEP7-5 EcoRI, 5′-AAGAATTCATGGGAAAAACTTCGGGA-3′ and OEP7-3 SalI, 5′-TTGTCGACACAAACCCTCTTTGGATGT-3′; for the PSI-D constructs, the primers were PSI-D XhoI forward, 5′-TTCTCGAGATGGCAACTCAAGCCGCC-3′ and PSI-D BamHI reverse, 5′-CCGGATCCCCAAATCATAACTTTGTTTG-3′. Purified PCR products and vectors were digested with BglII and SalI (ALB4), EcoRI and SalI (ALB3 and OEP7), or XhoI and BamHI (PSI-D). The digested PCR products were ligated into the linearized vectors.
Protoplasts from 4- to 5-week-old wild-type plants grown on soil were isolated using the tape-Arabidopsis-sandwich method (Wu et al., 2009). Approximately 1 × 105 protoplasts and 5 μg of plasmid DNA were used per (co)transfection in 40% (w/v) polyethylene glycol 4000 solution (Sigma-Aldrich; Wu et al., 2009). Samples were analyzed 16 to 18 h after (co)transfection using a Zeiss LSM 510 META laser-scanning confocal microscope using a C-Apochromat 40×/1.2 W Korr objective. To detect YFP, a 514-nm excitation from a 5-mW Argon ion laser with an HFT 458/514 primary dichroic mirror (Carl Zeiss) and a 535- to 590-nm emission filter was used. To simultaneously detect chlorophyll autofluorescence, an NFT 635 vis long-pass filter was used. The frequency of protoplasts that successfully expressed YFP fluorescence was estimated by counting the number of positives in the total number of protoplasts observed per microscope field, in 20 to 25 microscope fields per (co)transfection. The frequency of YFP-expressing protoplasts in the cotransfections was normalized to the frequency of the respective full-length YFP fusion used as a control. Images were processed with the Zeiss LSM Image Browser software. Each (co)transfection was conducted three times, with the same result, and typical images are shown.
Creation of ALB4-FLAG Lines
The ALB4 coding sequence without the stop codon was cloned using Gateway technology (Invitrogen) into a C-terminal FLAG tag (Sigma-Aldrich) binary vector derived from pEarleyGate 302 (Earley et al., 2006) containing a 35S promoter and spectinomycin and hygromycin resistance markers. Agrobacterium tumefaciens GV3101 was transformed by the freeze-thaw method (Holsters et al., 1978), and 6-week-old flowering wild-type plants were transformed by the floral dip method (Clough and Bent, 1998). ALB4-FLAG-overexpressing T2 lines were confirmed by immunoblotting using our anti-ALB4 antibody and using an anti-FLAG antibody (Sigma-Aldrich). Finally, an ALB4-FLAG line was selected (line 32) that showed a 1:4 segregation on selective medium, indicating a single T-DNA insertion locus, and that showed a high transgene transcription rate by reverse transcription-PCR.
Chloroplast Isolation and Cross Linking
Chloroplasts of 2-week-old wild-type plants or plants overexpressing the ALB4-FLAG construct were isolated as described previously (Aronsson and Jarvis, 2002). Freshly isolated chloroplasts were counted using a counting chamber (Weber Scientific) and normalized to 100 million chloroplasts in 300 mL of HEPES-MgSO4-sorbitol buffer (Aronsson and Jarvis, 2002). For cross linking, 0.5 mm DSP (in dimethyl sulfoxide) was added to the chloroplasts prior to incubation on ice for 15 min. Reactions were then quenched with 50 mm Gly for 15 min before the chloroplasts were pelleted by centrifugation at maximum speed and 4°C for 30 s using an Eppendorf Centrifuge 5417 R with an EL 082 rotor. The supernatant was removed and the pellet was used for immunoprecipitations.
Immunoprecipitation
Anti-FLAG-immunoprecipitation was performed with 100 million cross-linked chloroplasts from the ALB4-FLAG-expressing line, and with an equal number of wild-type chloroplasts as control. The chloroplasts were solubilized in solubilization buffer (20 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 10% [v/v] glycerol, 1% [v/v] DDM, pH 7.5) containing 1× protease inhibitor cocktail (cOmplete, Mini, EDTA-free, Roche). Following the manufacturer’s guidelines, 60 μL of Anti-FLAG M2 Affinity Gel (Sigma-Aldrich) per sample was preequilibrated in solubilization buffer, and ALB4-FLAG (and control) was incubated for 2 h by rotating at 4°C. The Anti-FLAG M2 Affinity Gel was then washed six times with wash buffer (20 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 10% [v/v] glycerol, 0.3% [v/v] DDM, pH 7.5), and each time, the resin was collected by quick centrifugation before 50 μL of 2× SDS-PAGE sample loading buffer was added. Per experiment, 18 μL of the denatured eluate was loaded.
Anti-ALB4 immunoprecipitation was performed with 100 million cross-linked chloroplasts from wild-type plants. The chloroplasts were solubilized as described above, and equal volumes of anti-ALB4 antibody and preimmune serum were added to the lysate of the sample and the control, respectively. Both were incubated for 3 h by rotating at 4°C. Protein A sepharose CL-4B (GE Healthcare; 50 mg) was preequilibrated in 200 μL of sterile water on ice for 30 min, then washed three times with water and once with solubilization buffer. To the samples, 100 μL of this slurry was added, prior to rotation for 2 h at 4°C and then washing as described above. Again, 50 μL of 2× SDS-PAGE sample loading buffer was added to the slurry, and 18 μL of the denatured eluate was loaded per experiment.
Phylogenetic Analysis
Homologous ALB3 and ALB4 amino acid sequences from land plant species were obtained using BLAST 2.2.26+ (Camacho et al., 2009). In addition, green algal sequences where included in the analysis to root the tree (Supplemental Table S1). Data sets were downloaded from www.phytozome.net, and the BLAST searches were run locally. The sequences were aligned using mafft-linsi (Katoh and Standley, 2013), and the independently analyzed regions of the alignment matrix were obtained by excluding regions (the N terminus and the C terminus, respectively) using the alignment editor SeaView (Gouy et al., 2010). Independent analyses on the full-length sequences, the N termini corresponding to residues 0 to 338 in Arabidopsis ALB3 (Falk et al., 2010), and the C termini (corresponding to residues 339–462, and including all four motifs of Arabidopsis ALB3) were performed using MrBayes v.3.2.2 (Ronquist et al., 2012) and the Beagle library (v1.0) for likelihood calculations (Ayres et al., 2012). A mixed model of amino acid substitutions was used for all three analyses, and the Markov chain Monte Carlo chains were run for 1,000,000 generations, sampling trees every 1,000 generations. An extended set of sequences (see Supplemental Table S1; Supplemental Fig. S2) were also analyzed, and the mcmc sampling was then done for 2,000,000 generations. Convergence and mixing of the mcmc chains was analyzed using Tracer v.1.5 (Rambaut and Drummond, 2007), after which 50% majority-rule consensus phylograms were produced using the default settings.
Identification of Sequence Motifs
The motif search analysis was performed on the MEME web server (http://meme.nbcr.net/meme/; Bailey and Elkan, 1994) by searching for a maximum of 20 motifs, allowing each motif to occur zero or one time and to be 6 to 300 residues wide. The analysis was performed on the two different data sets containing 25 angiosperm ALB3 and ALB4 sequences each and an out-group data set of seven sequences from green algae and the moss P. patens. For further details, see http://dx.doi.org/10.6084/m9.figshare.1061978.
Sequence data from this article can be found in the GenBank/EMBL amino acid database and the Phytozome repository under the accession numbers found in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic assessment of alb4 mutant plants under different stress conditions.
Supplemental Figure S2. Expression levels of genes relevant to this study as determined using public microarray data.
Supplemental Figure S3. Specificity analysis of the anti-ALB3 and anti-ALB4 antibodies.
Supplemental Figure S4. Measurement of photosynthetic performance in the alb4 mutant using a chlorophyll fluorescence imaging system.
Supplemental Figure S5. Control analyses for the BiFC study.
Supplemental Figure S6. Expanded phylogenetic analysis of ALB proteins and domains employing sequences from 39 different species.
Supplemental Table S1. Sequences of the ALB proteins used in the phylogenetic analyses.
Supplementary Material
Acknowledgments
We thank Natalie Allcock and Stefan Hyman for electron microscopy, Paula Töpel for graphical support, Ramesh Patel for technical support, Feijie Wu for the OEP7 BiFC constructs, Qihua Ling for help with the CF Imager chlorophyll fluorescence imaging system, Danja Schünemann and Masato Nakai for providing antibodies, and Stanton Gelvin for providing the BiFC vectors. This article is dedicated to the memory of Stefan Hyman, who passed away on March 26, 2015.
Glossary
- LHCP
light-harvesting complex protein
- T-DNA
transfer DNA
- MS
Murashige and Skoog
- DDM
n-dodecyl-β-d-maltopyranoside
- DSP
dithiobis(succinimidyl propionate)
- BiFC
bimolecular fluorescence complementation
- PSI-D
photosystem I subunit D
Footnotes
This work was supported by the Gatsby Charitable Foundation (Sainsbury PhD studentship to R.T.), Carl Tryggers Stiftelse (grant no. CTS 12:507 to M.T.), and the Biotechnology and Biological Sciences Research Council (grant nos. BB/D016541/1, BB/F020325/1, BB/H008039/1, BB/J009369/1, and BB/J017256/1 to P.J.).
Articles can be viewed without a subscription.
References
- Aronsson H, Jarvis P (2002) A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Asakura Y, Kikuchi S, Nakai M (2008) Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. Plant J 56: 1007–1017 [DOI] [PubMed] [Google Scholar]
- Ayres DL, Darling A, Zwickl DJ, Beerli P, Holder MT, Lewis PO, Huelsenbeck JP, Ronquist F, Swofford DL, Cummings MP, et al. (2012) BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst Biol 61: 170–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Baker NR. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Baniulis D, Zhang H, Zakharova T, Hasan SS, Cramer WA (2011) Purification and crystallization of the cyanobacterial cytochrome b6f complex. Methods Mol Biol 684: 65–77 [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Ferris P, Naver H, Göhre V, Rochaix JD (2002) Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell 14: 2303–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz M, Bals T, Gügel IL, Piotrowski M, Kuhn A, Schünemann D, Soll J, Ankele E (2009) Alb4 of Arabidopsis promotes assembly and stabilization of a non chlorophyll-binding photosynthetic complex, the CF1CF0-ATP synthase. Mol Plant 2: 1410–1424 [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Constan D, Patel R, Keegstra K, Jarvis P (2004) An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J 38: 93–106 [DOI] [PubMed] [Google Scholar]
- Czarnecki O, Peter E, Grimm B (2011) Methods for analysis of photosynthetic pigments and steady-state levels of intermediates of tetrapyrrole biosynthesis. Methods Mol Biol 775: 357–385 [DOI] [PubMed] [Google Scholar]
- Dalbey RE, Kuhn A, Zhu L, Kiefer D (2014) The membrane insertase YidC. Biochim Biophys Acta 1843: 1489–1496 [DOI] [PubMed] [Google Scholar]
- Dashdorj N, Zhang H, Kim H, Yan J, Cramer WA, Savikhin S (2005) The single chlorophyll a molecule in the cytochrome b6f complex: unusual optical properties protect the complex against singlet oxygen. Biophys J 88: 4178–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünschede B, Bals T, Funke S, Schünemann D (2011) Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. J Biol Chem 286: 35187–35195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Falk S, Ravaud S, Koch J, Sinning I (2010) The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem 285: 5954–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes L, Bals T, Klostermann E, Karl M, Philippar K, Hünken M, Soll J, Schünemann D (2006) A second thylakoid membrane-localized Alb3/OxaI/YidC homologue is involved in proper chloroplast biogenesis in Arabidopsis thaliana. J Biol Chem 281: 16632–16642 [DOI] [PubMed] [Google Scholar]
- Glauert AM. (1998) Practical Methods in Electron Microscopy, Vol 17. Portland Press Ltd., London [Google Scholar]
- Glick BS, Von Heijne G (1996) Saccharomyces cerevisiae mitochondria lack a bacterial-type sec machinery. Protein Sci 5: 2651–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V, Ossenbühl F, Crèvecoeur M, Eichacker LA, Rochaix JD (2006) One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18: 1454–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecka M, Alvarez-Fernandez R, Slattery K, McAusland L, Davey PA, Karpinski S, Lawson T, Mullineaux PM (2014) Abscisic acid signalling determines susceptibility of bundle sheath cells to photoinhibition in high light-exposed Arabidopsis leaves. Philos Trans R Soc Lond B Biol Sci 369: 20130234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Groves MR, Mant A, Kuhn A, Koch J, Dübel S, Robinson C, Sinning I (2001) Functional characterization of recombinant chloroplast signal recognition particle. J Biol Chem 276: 27778–27786 [DOI] [PubMed] [Google Scholar]
- Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163: 181–187 [DOI] [PubMed] [Google Scholar]
- Hyman S, Jarvis RP (2011) Studying Arabidopsis chloroplast structural organisation using transmission electron microscopy. Methods Mol Biol 774: 113–132 [DOI] [PubMed] [Google Scholar]
- Jakob B, Gamalei Y, Wolf R, Heber U, Gross HJ (1997) Photooxidative damage in young leaves of declining grapevine: does it result from a new and possibly viroid-related disease? Plant Cell Physiol 38: 1–9 [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Jia L, Dienhart M, Schramp M, McCauley M, Hell K, Stuart RA (2003) Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J 22: 6438–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Yi L, Moore M, Chen M, Rohl T, Van Wijk KJ, De Gier JW, Henry R, Dalbey RE (2002) Chloroplast YidC homolog Albino3 can functionally complement the bacterial YidC depletion strain and promote membrane insertion of both bacterial and chloroplast thylakoid proteins. J Biol Chem 277: 19281–19288 [DOI] [PubMed] [Google Scholar]
- Jonas-Straube E, Hutin C, Hoffman NE, Schünemann D (2001) Functional analysis of the protein-interacting domains of chloroplast SRP43. J Biol Chem 276: 24654–24660 [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klostermann E, Droste Gen Helling I, Carde JP, Schünemann D (2002) The thylakoid membrane protein ALB3 associates with the cpSecY-translocase in Arabidopsis thaliana. Biochem J 368: 777–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, Ríos G, Koncz C, Jarvis P (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P (2001) VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA 98: 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S, Patel R, Combe J, Bédard J, Kovacheva S, Lilley K, Biehl A, Leister D, Ríos G, Koncz C, et al. (2004) Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB (2008) Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Willmund F, Golecki JR, Cacace S, Hess B, Markert C, Schroda M (2007) The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J 50: 265–277 [DOI] [PubMed] [Google Scholar]
- Long D, Martin M, Sundberg E, Swinburne J, Puangsomlee P, Coupland G (1993) The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: identification of an albino mutation induced by Ds insertion. Proc Natl Acad Sci USA 90: 10370–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Goforth RL, Mori H, Henry R (2003) Functional interaction of chloroplast SRP/FtsY with the ALB3 translocase in thylakoids: substrate not required. J Cell Biol 162: 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Harrison MS, Peterson EC, Henry R (2000) Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J Biol Chem 275: 1529–1532 [DOI] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Ma X, Cline K (1999) Component specificity for the thylakoidal Sec and Delta pH-dependent protein transport pathways. J Cell Biol 146: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbühl F, Göhre V, Meurer J, Krieger-Liszkay A, Rochaix JD, Eichacker LA (2004) Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16: 1790–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasch JC, Nickelsen J, Schünemann D (2005) The yeast split-ubiquitin system to study chloroplast membrane protein interactions. Appl Microbiol Biotechnol 69: 440–447 [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ (2014) Tracer v1.5. http://tree.bio.ed.ac.uk/software/tracer/ (December 4, 2012)
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saller MJ, Wu ZC, de Keyzer J, Driessen AJ (2012) The YidC/Oxa1/Alb3 protein family: common principles and distinct features. Biol Chem 393: 1279–1290 [DOI] [PubMed] [Google Scholar]
- Spence E, Bailey S, Nenninger A, Møller SG, Robinson C (2004) A homolog of Albino3/OxaI is essential for thylakoid biogenesis in the cyanobacterium Synechocystis sp. PCC6803. J Biol Chem 279: 55792–55800 [DOI] [PubMed] [Google Scholar]
- Stengel KF, Holdermann I, Cain P, Robinson C, Wild K, Sinning I (2008) Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science 321: 253–256 [DOI] [PubMed] [Google Scholar]
- Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G (1997) ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM (2003) Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J 22: 6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu CJ, Peterson EC, Henry R, Hoffman NE (2000) The L18 domain of light-harvesting chlorophyll proteins binds to chloroplast signal recognition particle 43. J Biol Chem 275: 13187–13190 [DOI] [PubMed] [Google Scholar]
- Tu CJ, Schuenemann D, Hoffman NE (1999) Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J Biol Chem 274: 27219–27224 [DOI] [PubMed] [Google Scholar]
- Woolhead CA, Thompson SJ, Moore M, Tissier C, Mant A, Rodger A, Henry R, Robinson C (2001) Distinct Albino3-dependent and -independent pathways for thylakoid membrane protein insertion. J Biol Chem 276: 40841–40846 [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Aro EM (2002) Synthesis, membrane insertion and assembly of the chloroplast-encoded D1 protein into photosystem II. FEBS Lett 512: 13–18 [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Tian HF, Wen JF (2009) The evolution of YidC/Oxa/Alb3 family in the three domains of life: a phylogenomic analysis. BMC Evol Biol 9: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuppini A, Gerotto C, Moscatiello R, Bergantino E, Baldan B (2009) Chlorella saccharophila cytochrome f and its involvement in the heat shock response. J Exp Bot 60: 4189–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.