The functional characterization of two Arabidopsis floral repressors unravels their role and regulation at low ambient temperatures.
Abstract
Plants integrate day length and ambient temperature to determine the optimal timing for developmental transitions. In Arabidopsis (Arabidopsis thaliana), the floral integrator FLOWERING LOCUS T (FT) and its closest homolog TWIN SISTER OF FT promote flowering in response to their activator CONSTANS under long-day inductive conditions. Low ambient temperature (16°C) delays flowering, even under inductive photoperiods, through repression of FT, revealing the importance of floral repressors acting at low temperatures. Previously, we have reported that the floral repressors TEMPRANILLO (TEM; TEM1 and TEM2) control flowering time through direct regulation of FT at 22°C. Here, we show that tem mutants are less sensitive than the wild type to changes in ambient growth temperature, indicating that TEM genes may play a role in floral repression at 16°C. Moreover, we have found that TEM2 directly represses the expression of FT and TWIN SISTER OF FT at 16°C. In addition, the floral repressor SHORT VEGETATIVE PHASE (SVP) directly regulates TEM2 but not TEM1 expression at 16°C. Flowering time analyses of svp tem mutants indicate that TEM may act in the same genetic pathway as SVP to repress flowering at 22°C but that SVP and TEM are partially independent at 16°C. Thus, TEM2 partially mediates the temperature-dependent function of SVP at low temperatures. Taken together, our results indicate that TEM genes are also able to repress flowering at low ambient temperatures under inductive long-day conditions.
Plants constantly monitor environmental and endogenous signals to control their growth and adjust developmental responses to daily and seasonal cues (Penfield, 2008). During the juvenile phase, plants are not competent to flower; they are insensitive to inductive environmental factors, such as favorable conditions of day length or temperature. The transition to the adult phase permits reaching the competence to respond to those signals, which is essential to trigger flowering during the reproductive phase (Bergonzi and Albani, 2011; Huijser and Schmid, 2011). Consequently, the control of flowering time is a key determinant of reproductive success and plays an essential role in plant adaptation to seasons and geography.
Flowering time is controlled by an intricate network of interdependent genetic pathways that monitor and respond to both endogenous and environmental signals. These pathways include age, photoperiod and light quality, GA, thermosensory (ambient temperature), vernalization, and autonomous pathways (Fornara et al., 2010; Srikanth and Schmid, 2011). In Arabidopsis (Arabidopsis thaliana), it is well documented the noteworthy regulation of the timing of flowering by day length or photoperiod and temperature (for review, see Andrés and Coupland, 2012; Song et al., 2013; Chew et al., 2014; Romera-Branchat et al., 2014). However, in contrast to the finely described photoperiod and light quality pathways, the nature of the primary perception of temperature and the molecular characterization of its signaling remain limited (McClung and Davis, 2010).
Lately, several studies have reported how changes in ambient temperature, defined as the physiological nonstressful temperature range of a given species, modulate many processes in plant development and in particular, how they affect flowering time (for review, see Wigge, 2013; Capovilla et al., 2015). Genetic analyses unraveled the existence of the ambient temperature pathway that mediates temperature responses in Arabidopsis (Blázquez et al., 2003; Balasubramanian et al., 2006; Lee et al., 2007; Kumar et al., 2012). It has been described that a slight decrease from 23°C to 16°C is sufficient to cause a remarkable delay in flowering, even under an inductive long-day (LD) photoperiod (Blázquez et al., 2003). Temperature-dependent differences in flowering time are controlled by multiple factors that mainly affect the expression levels of one of the key floral activators, the FLOWERING LOCUS T (FT) gene (Kardailsky et al., 1999; Kobayashi et al., 1999). Low ambient temperatures reduce the expression of FT, although this decrease is not caused by changes in its transcriptional activator CONSTANS (CO), which reveals the importance of floral repressors controlling flowering time under low ambient temperatures (16°C; Blázquez et al., 2003; Lee et al., 2013). However, the levels of TWIN SISTER OF FLOWERING LOCUS T (TSF), the closest homolog of FT, are similar at both temperatures, resulting in a higher expression of TSF than FT at 16°C (Blázquez et al., 2003; Lee et al., 2012, 2013). Although TSF plays a secondary but redundant role, FT and TSF act as floral pathway integrators (Kardailsky et al., 1999; Kobayashi et al., 1999; Yamaguchi et al., 2005; Jang et al., 2009), and their main function is the promotion of photoperiodic flowering (Kardailsky et al., 1999; Kobayashi et al., 1999), with TSF playing a secondary but redundant role. A similar relation is also observed at 16°C (Kim et al., 2013; Lee et al., 2013), and flowering of the double mutant ft tsf is insensitive to ambient temperature changes (Kim et al., 2013), which indicates that FT and TSF play an important role in the regulation of ambient temperature-responsive flowering (Kim et al., 2013).
Previously, the TEMPRANILLO (TEM) genes were identified as main players in the control of flowering time at 22°C, and they were shown to directly repress FT (Castillejo and Pelaz, 2008) and the GA biosynthetic genes GA 3-OXIDASE1 (GA3OX1) and GA3OX2 (Osnato et al., 2012).
Recent genome-wide analysis has identified TEM1 and TEM2 as direct targets of the MCM1-AGAMOUS-DEFICIENS-SRF (MADS)-box transcription factor SHORT VEGETATIVE PHASE (SVP) under LD at 22°C (Tao et al., 2012). TEM1 and TEM2 are expressed at low levels in svp-41 plants and high levels in SVP-overexpressing plants compared with wild-type plants, indicating a positive regulation of TEMs by SVP, which is more evident on TEM2 than on TEM1 (Tao et al., 2012). Interestingly, previous genetic studies identified svp mutants as insensitive to a wide range of ambient temperature changes (5°C–27°C; Lee et al., 2007, 2013). Thus, svp mutants show an early flowering phenotype, producing almost the same number of leaves at flowering at all temperatures (Lee et al., 2013). Similar to TEM, SVP delays flowering by direct repression of FT (Lee et al., 2007; Li et al., 2008). Moreover, SVP represses TSF in the vascular tissue of leaves and plays an antagonistic role with another MADS-box transcription factor, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), in the meristem (Li et al., 2008; Jang et al., 2009).
Here, we characterized the role of TEM genes as repressors of flowering at moderately low ambient temperature of 16°C under LD conditions. We show that TEM genes act as floral repressors at 16°C under LD conditions by regulating both FT and TSF expression. Furthermore, we show that SVP specifically regulates TEM2 at 16°C to repress flowering under LD conditions. Therefore, our results provide additional information regarding the genetic relation between SVP and TEM at low temperatures.
RESULTS
tem Mutants Are Early Flowering at 16°C But Still Sensitive to Low Temperature
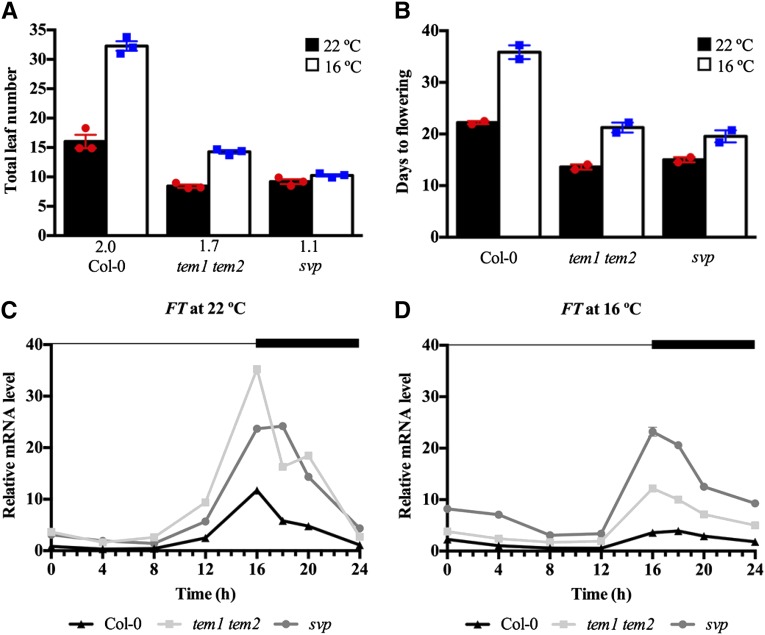
To study the function of TEM1 and TEM2 as floral repressors at low ambient temperature, we first analyzed the flowering phenotype of the loss-of-function double mutant tem1 tem2 at 16°C compared with 22°C under LD conditions. tem1 tem2 showed early flowering at 22°C, in agreement with previous results (Castillejo and Pelaz, 2008; Osnato et al., 2012), but also, at 16°C and showed a bigger difference with wild-type plants at 16°C than at 22°C. However, tem1 tem2 plants grown at 16°C produced more leaves than those grown at 22°C (Fig. 1; Supplemental Table S1), indicating that this double mutant is still thermosensitive but significantly less sensitive than wild-type plants, which produced double numbers of leaves at 16°C than at 22°C (the wild type, 16°C/22°C = 2.0; tem1 tem2, 16°C/22°C = 1.7; P = 0.0191). By contrast, svp mutants, described as insensitive to temperature (Lee et al., 2007), flowered with a similar number of leaves at both temperatures. We found that tem1 tem2 plants flowered with slightly fewer leaves than svp plants at 22°C, although this difference was not statistically significant, whereas svp plants were clearly earlier than tem1 tem2 at 16°C (Fig. 1A; Supplemental Table S1). tem1 tem2 plants were also earlier than wild-type plants in terms of the number of days to flowering at both temperatures (Fig. 1B). Interestingly, tem1 tem2 plants grown at 16°C flowered with a similar number of leaves and days to wild-type plants grown at 22°C. These flowering time data are directly correlated with the FT expression levels observed in wild-type, tem1 tem2, and svp mutant plants at different temperatures (Fig. 1, C and D; Supplemental Fig. S1). In tem1 tem2 plants, FT expression was slightly higher than in svp mutants and clearly up-regulated compared with that in wild-type plants at 22°C (Fig. 1C). At 16°C, FT levels of tem1 tem2 plants exhibited a clear increase compared with wild-type levels, but this increase was lower than in svp plants (Fig. 1D). Both results clearly correlated with the flowering time of those plants at both temperatures. Interestingly, the similar flowering time phenotype observed in wild-type plants grown at 22°C and tem1 tem2 plants grown at 16°C was associated with similar FT levels.
Figure 1.
tem1 tem2 mutant plants are early flowering at 16°C but still sensitive to changes in ambient growth temperature. Flowering time was measured as the number of total leaves produced at flowering (A) and the number of days to flowering (B) for wild-type (Col-0) plants and tem1 tem2 and svp mutants grown under LD conditions at 22°C (black) or 16°C (white). Data are reported as mean ± sem of three independent experiments (each dot plot represents an independent experiment; red circles indicate experiments performed at 22°C, and blue squares indicate experiments performed at 16°C). A minimum of 12 plants per genotype and experimental condition was analyzed in each independent experiment. The numbers below the bars denote the leaf number ratio (16°C/22°C). For more details, see Supplemental Table S1. C and D, Reverse transcription followed by quantitative real-time PCR (RT-qPCR) analysis of FT expression in wild-type (black triangles), tem1 tem2 (gray squares), and svp (gray circles) plants in 9-d-old seedlings grown under LD conditions at 22°C (C) or 16°C (D). Samples were collected over a 24-h period. The dark period is denoted by the black bar. Two independent experiments gave similar results (Supplemental Fig. S1), and one was chosen as representative. RNA levels were normalized to UBQ10. Error bars show sd of three technical replicates.
Although GAs seem to play a minor role in flowering under LD (Reeves and Coupland, 2001), we tested if GA3OX1 was also repressed at 22°C and 16°C, because TEM1 directly represses GA3OX1 under short day at 22°C (Osnato et al., 2012). We found a similar derepression in tem1 tem2 double mutants at both temperatures (Supplemental Fig. S2).
All of these results indicate that TEM genes have a role in the control of flowering time at low temperature.
Low Temperature Keeps High TEM Levels before Floral Transition
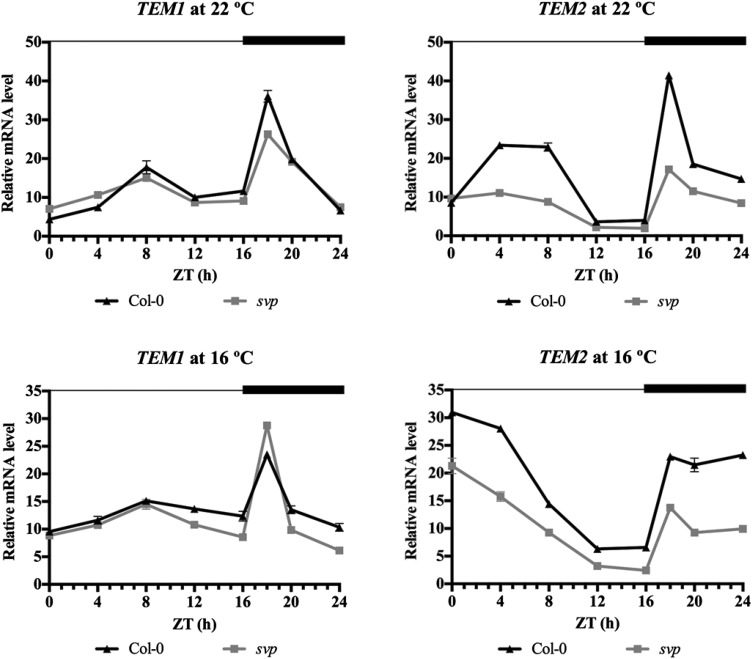
To characterize the response of TEM1 and TEM2 to low temperatures, we first performed diurnal analyses in 9-d-old wild-type plants grown under LD conditions at 16°C and 22°C. We harvested samples every 4 h for 24 h and added an extra point of collection at Zeitgeber time 18 (ZT18) during the peak of TEM1 and TEM2 expression. Our results indicate that TEM mRNA levels were not affected by low temperature at this stage, because TEM daily oscillation showed a similar pattern at both temperatures (Supplemental Fig. S3). In particular, TEM1 exhibited a clear peak of expression at ZT18 as previously described (Osnato et al., 2012) and a small peak during the day between ZT8 and ZT12 at 22°C and also, 16°C; TEM2 showed high levels during the night until the beginning of the day, when it was gradually reduced. Because of the importance of TEMs as repressors of flowering time genes along development (Castillejo and Pelaz, 2008; Osnato et al., 2012), we then analyzed TEM expression pattern later in development under LD conditions. As we already knew, under LD at 22°C, TEM genes showed high expression levels during early stages of development, which prevents a precocious activation of FT and a consequent early flowering (Castillejo and Pelaz, 2008). After that, there was a gradual decline of their levels until TEMs reached their minimum expression around day 12, when FT activation takes place (Supplemental Fig. S4).
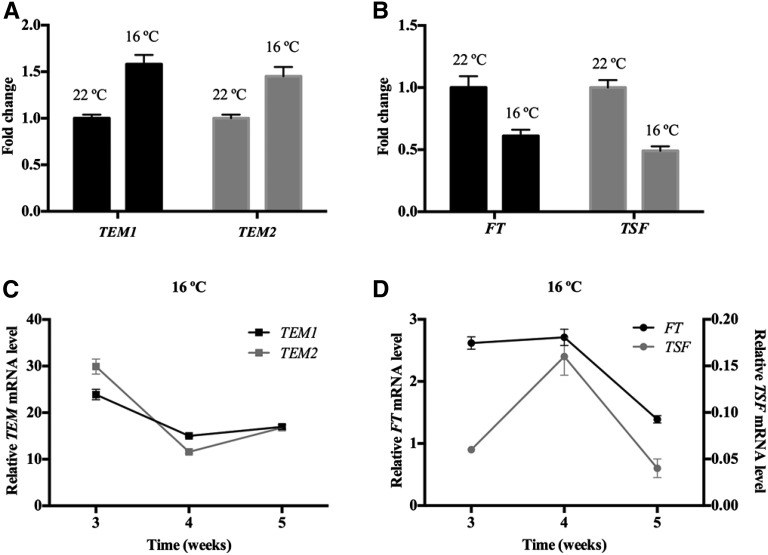
To test our hypothesis of a possible thermal regulation, we compared TEM expression in 12-d-old wild-type plants grown at 16°C and 22°C. Our results show that plants grown at 16°C keep high TEM1 and TEM2 levels longer than those grown at 22°C, maintaining high levels at 16°C at a developmental phase in which they normally reached their minimum at 22°C (Fig. 2; Supplemental Fig. S4A). This indicates that a thermal regulation of TEM exists under LD conditions. Moreover, the increased TEM levels at 16°C (Fig. 2A) were correlated with a reduction of FT expression at low temperature (Fig. 2B). In addition to FT, we decided to include the analysis of TSF in our experiments, because it is known to play a role in the regulation of ambient temperature-responsive flowering (Kim et al., 2013). We found that TSF and FT display a similar expression pattern throughout development in wild-type plants grown at 22°C under LD conditions (Supplemental Fig. S4B), a pattern opposite to TEM abundance (Supplemental Fig. S4A).
Figure 2.
Opposite expression pattern of TEM and FT/TSF genes at 16°C. Expression analysis of TEM1, TEM2, FT, and TSF in 12-d-old wild-type (Col-0) plants grown at 22°C or 16°C (A and B) and wild-type plants grown at 16°C (C and D) for 5 weeks. Fold change in transcript levels at 16°C is depicted compared with 22°C. All samples were collected at ZT18. Three independent experiments gave similar results (Supplemental Fig. S5), and one was chosen as representative. Error bars show sd of three technical replicates. RNA levels were determined by RT-qPCR and normalized to UBQ10.
Next, to better characterize the TEM thermal regulation along development and determine when TEM abundance reaches the minimum at low temperatures, we performed time course analyses of wild-type plants grown at 16°C during 5 weeks. The relative mRNA levels of TEM1 and TEM2 showed the expected gradual decrease along development. In contrast to what happens at 22°C (Supplemental Fig. S4A), at low temperature, their levels dropped later (around the third to fourth week; Fig. 2C; Supplemental Fig. S5). In accordance with that, plants grown at 16°C showed a later rise of FT and TSF levels than at 22°C, and this rise occurred almost simultaneously with the descent of TEM expression (around the third to fourth week; Fig. 2D; Supplemental Fig. S5B). These results indicate that there is a correlation between the decrease of TEM abundance and the increase of FT and TSF levels at 16°C and that this happens later than at 22°C, which is in agreement with the delayed flowering at low temperatures.
TEMs Directly Repress FT and TSF at 16°C
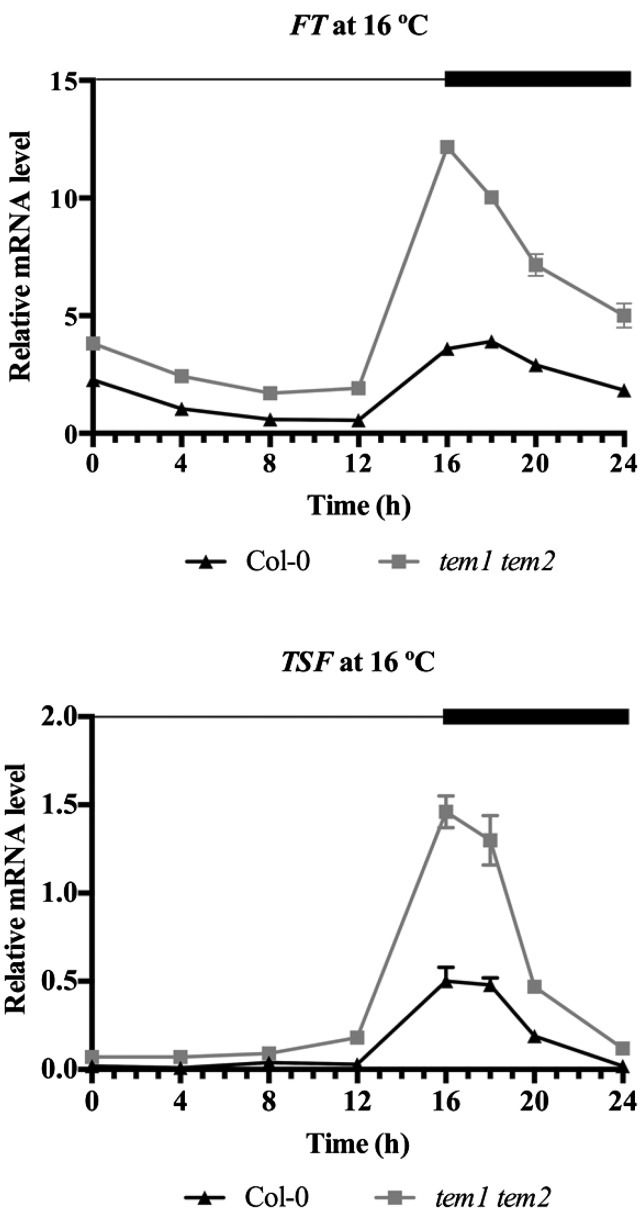
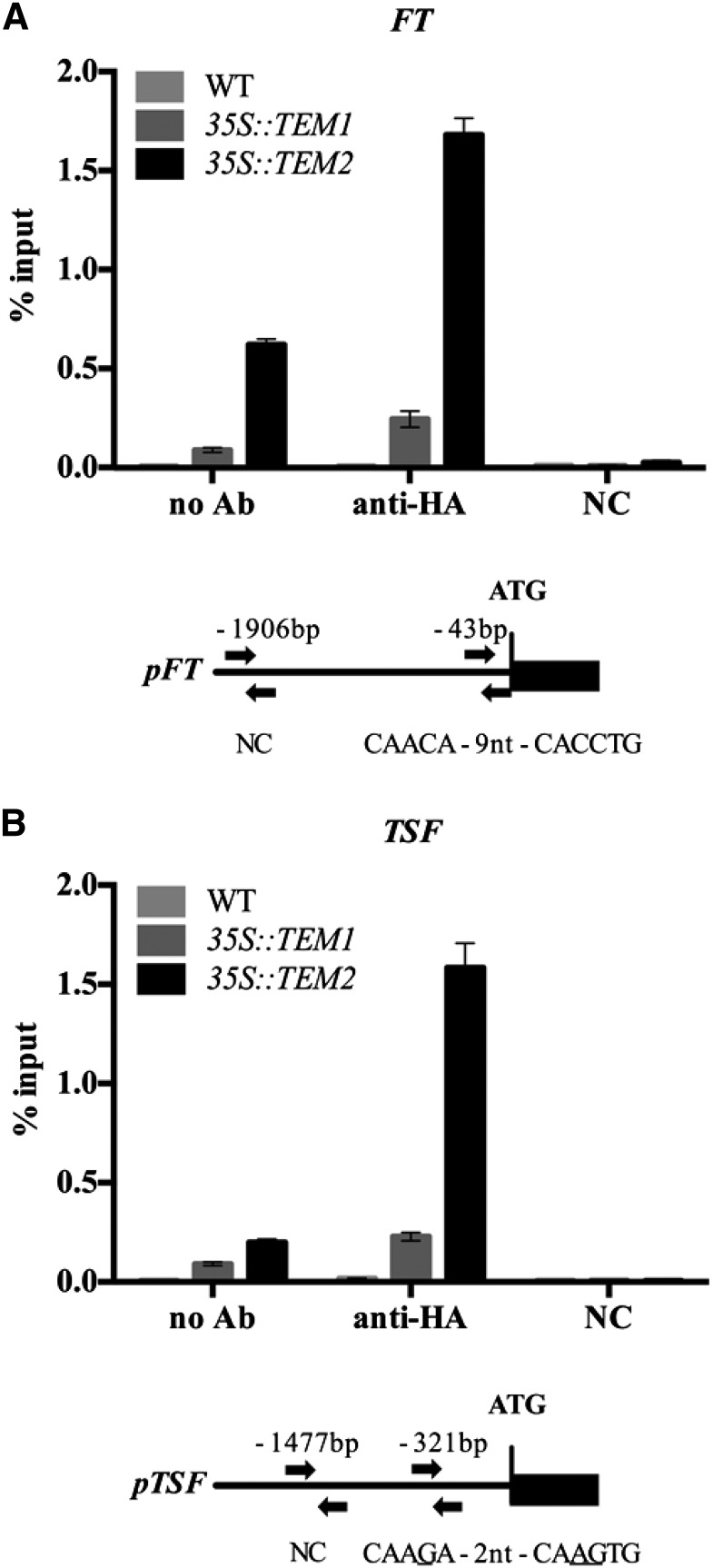
To further confirm whether TEM genes regulate FT and/or TSF, we carried out expression analyses in wild-type and tem1 tem2 mutant plants grown at 16°C. As we expected, FT and TSF were clearly up-regulated in tem1 tem2 (Fig. 3; Supplemental Fig. S6). To understand whether the repression of FT and TSF was direct or indirect, we performed chromatin immunoprecipitation (ChIP) experiments at low-temperature conditions. TEM proteins, like other Related to ABI3/VP1 (RAV) members, recognize and bind a canonical sequence known as RAV binding site (Kagaya et al., 1999). Direct binding of TEM1 to the RAV binding site of the 5′-untranslated region of FT was previously reported under LD at 22°C (Castillejo and Pelaz, 2008). Here, we show that TEM2 binds in vivo specifically to both FT and TSF chromatin at 16°C. We found a significant enrichment of the 5′-untranslated region of FT containing the canonical RAV binding site (5′-CAACAN9CACCTG-3′; Fig. 4; Supplemental Fig. S7) 43 nucleotides upstream of the ATG start codon in 35S::TEM2 plants, whereas only a slight enrichment was found in 35S::TEM1 plants. In addition, a clear significant enrichment of the TSF promoter was detected in 35S::TEM2 but not 35S::TEM1 plants in a region 321 nucleotides upstream of the ATG, which contains a noncanonical RAV binding site (5′-CAAGAN2CAAGTG-3′; underlined nucleotides indicate those that are different from the consensus RAV binding site; Fig. 4B; Supplemental Fig. S7B). Taken together, these data show that TEM2 specifically binds to both FT and TSF and directly regulates their expression at 16°C.
Figure 3.
TEMs regulate FT and TSF levels at 16°C. Relative FT and TSF mRNA levels in tem1 tem2 mutant compared with wild-type (Col-0) plants. Nine-day-old seedlings were sampled at 4-h intervals, except from ZT16 to ZT20, when samples were collected every 2 h. Two independent experiments gave similar results (Supplemental Fig. S6), and one was chosen as representative. Error bars show sd of three technical replicates. RNA levels were determined by RT-qPCR and normalized to UBQ10.
Figure 4.
Binding of TEM2 protein to the FT and TSF promoters at 16°C. ChIP assay of binding of TEM1-HA and TEM2-HA proteins to the RAV motifs in the FT (A) and TSF (B) promoters. Fragments containing the canonical RAV binding site for FT, a putative RAV binding site for TSF, and noncontaining RAV binding sequences (used as negative controls [NCs]) were analyzed by ChIP using 9-d-old 35S::TEM1 and 35S::TEM2 plants carrying an HA tag. Precipitated chromatin was used as a template in qPCR. Immunoprecipitated DNA enrichment is presented as a percentage of input DNA. Two (FT) or three (TSF) independent experiments gave similar results (Supplemental Fig. S7), and one was chosen as representative. Error bars show sd of three technical replicates. Schematic diagrams of the FT and TSF promoters are shown below graphs. Arrows indicate fragments amplified by qPCR after ChIP. WT, Wild type.
SVP Positively Regulates TEM2 at 16°C
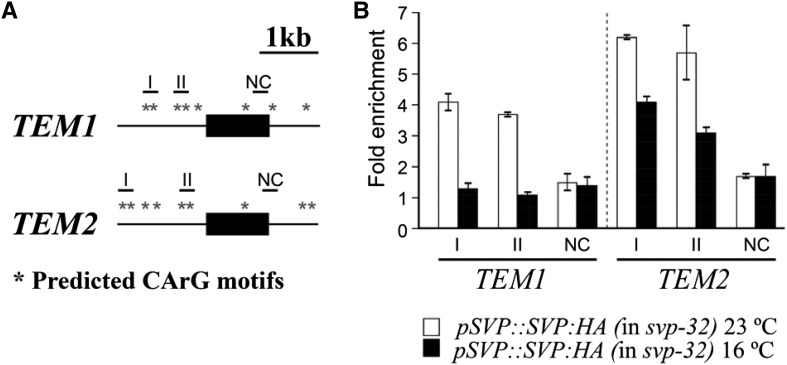
As a result of genome-wide analyses, TEM genes were identified as targets of SVP under LD conditions at 23°C (Tao et al., 2012). Given that SVP is involved in the thermosensory pathway, our next question was whether SVP regulates TEM genes at low temperatures. To test the possibility that SVP protein regulates TEM1 and TEM2 expression through direct binding to the CArG motifs, where the MADS domain proteins are known to bind (West et al., 1997), present in the TEM1 and TEM2 genomic loci, we performed ChIP assays using pSVP::SVP:hemagglutinin tag (HA) svp-32 plants (Lee et al., 2013; Supplemental Fig. S8) under the two temperature conditions (23°C and 16°C). We chose two regions containing CArG motifs (Tao et al., 2012) in the TEM1 and TEM2 promoter sequences (Fig. 5; Supplemental Fig. S9). A region lacking a CArG motif was used as a negative control. Strong binding of SVP protein was observed in regions I and II of TEM1 and TEM2 at 22°C, whereas interestingly, at 16°C, we only observed a clear binding to TEM2 but not TEM1 regulatory regions (Fig. 5B).
Figure 5.
Binding of SVP protein to the TEM1 and TEM2 genomic loci. A, Schematic diagram of the TEM1 and TEM2 genomic regions. Black boxes and thin lines represent exons and introns, respectively. Asterisks indicate the predicted CArG and variant CArG motifs. Short horizontal lines indicate amplicons in ChIP-qPCR assays. Regions I and II, carrying CArG motifs, were selected to amplify; negative control (NC) was the amplicon used as a negative control. B, ChIP analysis of binding of SVP protein to the TEM1 and TEM2 genomic regions at 23°C and 16°C in 9-d-old pSVP::SVP:HA svp-32 plants. An anti-HA antibody was used for immunoprecipitation. Black bars denote the amplified fragments in qPCR: region I (−1,005 to −920; relative to ATG), region II (−350 to −271), and NC (+1,011 to +1,085) for TEM1 and region I (−1,429 to −1,385; relative to ATG), region II (−434 to −345), and NC (+1,005 to +1,065) for TEM2. Two independent experiments gave similar results (Supplemental Fig. S9), and one was chosen as representative. Error bars show sd of three technical replicates.
To determine if the binding of SVP to TEM1 and TEM2 genomic loci affects TEM expression, we carried out expression analysis in wild-type and svp mutant plants at 22°C and 16°C. A strong down-regulation of TEM2 was observed in svp plants at both 22°C and 16°C, whereas a slight or no TEM1 reduction was found in svp at 22°C and 16°C, respectively (Fig. 6; Supplemental Fig. S10). Taken together, these data show that SVP regulates the expression levels of TEM genes by direct binding to the CArG motifs in TEM1 and TEM2 genomic loci at 22°C but only to TEM2 at 16°C for the regulation of ambient temperature-responsive flowering.
Figure 6.
SVP positively regulates TEM2 expression at 22°C and 16°C. Relative mRNA levels of TEM1 and TEM2 at 22°C (upper) and 16°C (lower) in svp mutant compared with wild-type (Col-0) plants. Nine-day-old seedlings were sampled at 4-h intervals, except from ZT16 to ZT20, when samples were collected every 2 h. Two independent experiments gave similar results (Supplemental Fig. S10), and one was chosen as representative. Error bars show sd of three technical replicates. RNA levels were determined by RT-qPCR and normalized to UBQ10.
To test whether there could be reciprocal regulation between TEM and SVP, we examined the expression levels of SVP in tem1 tem2 plants. However, we did not find changes in the expression levels of SVP in tem1 tem2 compared with those in wild-type plants at either 22°C or 16°C (Supplemental Fig. S11), suggesting that TEMs do not regulate SVP levels.
Genetic Interactions between svp and tem Mutations at Different Temperatures
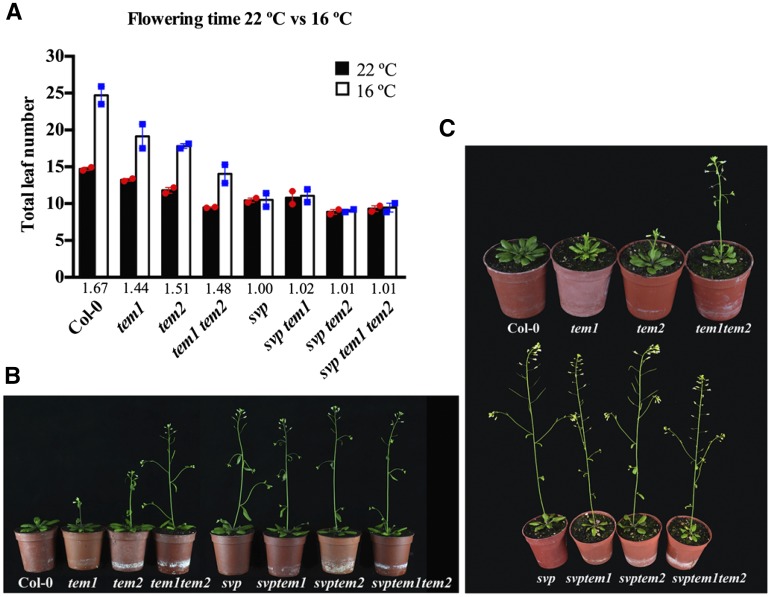
Finally, to examine the genetic relationship between SVP and TEMs in the thermosensory pathway, we measured the flowering times of tem single mutants, tem1 tem2 double mutant, svp mutant, and the double and triple combinations of svp and tem mutations under LD conditions at 22°C and 16°C (Fig. 7; Supplemental Table S2). As shown above, at 16°C, tem1 tem2 flowered earlier than wild-type plants, and tem single mutants also showed an early flowering phenotype. Despite their early flowering, we observed that tem double mutants flowered later than any combination with svp mutation. svp, svp tem1, svp tem2, and the triple svp tem1 tem2 all flowered earlier at 16°C than tem1 tem2 plants (Fig. 7). Accordingly, FT levels are higher in triple mutants at 16°C than in tem1 tem2 double mutants (Supplemental Fig. S12). By contrast, at 22°C, tem1 tem2 flowered with a similar number of leaves as mutants including tem and svp mutations, and the small differences observed were not statistically significant (Fig. 7). We found that these differences were correlated with FT expression levels (Supplemental Fig. S12); tem1 tem2 double mutants showed similar FT levels as triple mutants. Interestingly, tem2 single mutants flowered with a similar number of leaves to svp at 22°C but flowered later than svp at 16°C, whereas tem2 showed a slight delay compared with svp tem2 at 22°C, which was more evident at 16°C (Fig. 7). Taken together, these results indicate that TEMs regulate flowering at 22°C in an SVP-dependent manner and 16°C in a partially SVP-independent manner.
Figure 7.
Genetic interaction between tem and svp mutants. A, Flowering time measured as the number of total leaves produced at flowering for the wild type (Col-0), tem mutants, and svp tem double and triple mutants grown under LD conditions at 22°C or 16°C. Data are reported as mean ± sem of two independent experiments (each dot plot represents an independent experiment; red circles indicate experiments performed at 22°C, and blue squares indicate experiments performed at 16°C). A minimum of 10 plants per genotype and experimental condition was analyzed in each independent experiment. The numbers below the bars denote the leaf number ratio (16°C/22°C). For more details, see Supplemental Table S2. B and C, Photographs of plants used in flowering time analysis grown for 24 d at 22°C (B) and 32 d at 16°C (C).
DISCUSSION
Plants have developed mechanisms to perceive and respond to environmental fluctuations by adjusting their growth as well as predict upcoming daily and seasonal cues, which result in massive developmental plasticity (Franklin, 2009). Photoperiod and ambient temperature provide relevant information for the adaptation to seasonal changes, which would allow plants to respond to a cold snap or a sudden warmup and optimize flowering time. Hence, light and temperature cues have a key role in flowering time regulation (Andrés and Coupland, 2012).
TEM Genes Repress Flowering at Low Ambient Temperatures
It has been shown that a slight decrease from 23°C to 16°C down-regulates FT expression, even under a favorable photoperiod (Blázquez et al., 2003; Fig. 1). This suggests that, under low ambient temperature and LD conditions, floral repressors gain a relevant role in maintaining low FT expression levels, despite the presence of its activator CO. One of the most studied repressors acting in these conditions is SVP, which interacts with other floral repressors, such as FLOWERING LOCUS M (FLM; Lee et al., 2013), FLOWERING LOCUS C (FLC; Lee et al., 2007; Li et al., 2008), and possibly, other members of the FLC clade (MADS AFFECTING FLOWERING genes; Gu et al., 2013) to repress FT, TSF, and SOC1.
The role of TEM genes as flowering repressors under LD at 22°C has been previously reported (Castillejo and Pelaz, 2008); however, their function at low ambient temperatures was unknown. The early flowering observed in the tem1 tem2 mutant at both 22°C and 16°C compared with wild-type plants indicates that TEMs act as floral repressors as well at low ambient temperature. However, this double mutant flowered later at 16°C than at 22°C (Figs. 1A and 7). Therefore, in contrast to other genes, which have loss of function that causes insensitivity to ambient temperature, such as SVP or FLM (Balasubramanian et al., 2006; Lee et al., 2007, 2013; Posé et al., 2013), tem mutants are still sensitive to low ambient temperature, although their response is reduced compared with that of wild-type plants. Thus, TEM genes do not seem to control the low temperature-responsive pathway but somehow, act downstream of SVP in transmitting the response signal. Indeed, there is a clear correlation between the flowering time of tem1 tem2 and svp mutants and the FT relative expression at both temperatures (Fig. 1, C and D; Supplemental Fig. S1). At 22°C, the FT up-regulation displayed in tem1 tem2 and svp plants gave rise to the same earlier flowering, probably because in both mutant plants, FT exceeded the expression threshold required to induce the floral transition (Fig. 1, A and C). By contrast, at 16°C, the higher level of FT in svp mutants leads to an earlier flowering than in tem1 tem2 (Fig. 1, A and D). Therefore, the later flowering of tem mutants relative to svp at 16°C seems to be caused by the presence of active SVP, which keeps some repression on FT expression in tem mutants. In that sense, SVP directly represses FT expression through direct binding to the FT promoter (Lee et al., 2007, 2013).
Late Decay in TEM Gene Expressions at 16°C Delays Flowering
Like SVP, which shows practically the same mRNA levels at 16°C and 22°C to 23°C (Lee et al., 2007; Supplemental Fig. S11), TEM expression is not increased at 16°C at early stages of development, because we did not detect changes in TEM1 or TEM2 mRNA levels in 9-d-old wild-type seedlings at 16°C relative to 22°C (Supplemental Fig. S3). However, our results showed that, in 12-d-old seedlings, TEM genes are expressed at higher levels at 16°C than at 22°C, which correlated with a decrease in FT and TSF expression at 16°C (Fig. 2, A and B). These data indicate that TEM gene expression decays later at low ambient temperatures, a conclusion supported by the analysis of TEM expression throughout development at 16°C and 22°C (Fig. 2, C and D; Supplemental Fig. S4).
FT and TSF act as floral promoters at 22°C to 23°C and 16°C (Michaels et al., 2005; Yamaguchi et al., 2005; Jang et al., 2009; Kim et al., 2013; Lee et al., 2013), with FT being the main player. We previously reported that TEM genes are floral repressors under LD and short-day conditions at 22°C by controlling FT (Castillejo and Pelaz, 2008) and GA biosynthetic genes GA3ox1/2 (Osnato et al., 2012). Here, we report that TEM genes also delay flowering and repress FT as well as TSF and GA3OX1 at 16°C under LD conditions. The FT and TSF daily patterns of expression were mostly unchanged in tem1 tem2 mutants, but their abundance was increased at both 22°C and 16°C (Figs. 1 and 3; Supplemental Figs. S1 and S6). This up-regulation of FT and TSF in tem1 tem2 plants (Fig. 3) together with the binding of TEM to FT and TSF regulatory regions (Fig. 4) indicate that TEM and more specifically, TEM2 directly repress FT and TSF at low ambient temperatures. Thus, the later drop of TEM gene expression at 16°C results in a longer FT and TSF repression and therefore, a later flowering compared with plants growing at 22°C.
SVP Up-Regulates TEM2 through Direct Binding in Response to Low Temperatures
Previous high-throughput experiments indicated that SVP positively regulates TEM genes at 22°C under LD conditions (Tao et al., 2012). Here, we show that the effect of low temperature on TEM2 can be explained by the positive and direct regulation that SVP exerts over it as well at 16°C (Figs. 5 and 6). Bioinformatic analyses detected several MADS binding sites in the promoters of TEM1 and TEM2 where SVP could putatively bind, and these sites were experimentally tested by ChIP assays at 23°C and 16°C. Indeed, our results confirmed the binding of SVP to TEM1 and TEM2 at 23°C described by Tao et al. (2012) through ChIP-chip analysis and also, provide unique data on the specific regulation of TEM2 but not TEM1 by SVP at 16°C (Figs. 5 and 6). At both temperatures, SVP binding on TEM2 seemed to be stronger than that on TEM1, which correlated with the strong down-regulation of TEM2 observed in svp mutants at 22°C and 16°C (Fig. 6). Furthermore, in svp mutants, TEM2 presented practically the same relative mRNA levels at 22°C and 16°C, which were reduced in both cases compared with wild-type plants. This indicates that, although TEM genes are redundant in function as repressors of FT and GA3ox1/2 (Castillejo and Pelaz, 2008; Osnato et al., 2012), they are differentially regulated by SVP in the ambient temperature pathway. The different binding of SVP to TEMs at different temperatures might be because of the interaction of SVP with FLM-β at 16°C; TEM2 but not TEM1 has been identified as a target of FLM-β (Posé et al., 2013). In addition, TEM chromatin modifications might occur at low temperatures, making them differentially accessible.
TEM Acts at Least Partially Independently of SVP at Low Ambient Temperatures
The analyses of tem1, tem2, and svp mutants in multiple combinations at 22°C indicate that TEM1 and TEM2 act basically on the same genetic pathway as SVP. However, at 16°C, the global analysis of our flowering time data indicates that a slight additive effect of TEM and SVP exists, which suggests that they may act in a partially independent manner at low ambient temperature (Fig. 7). Analyzed in detail, at 22°C, the flowering time of tem2 and svp was almost the same, because the difference in the total leaves produced was not statistically significant, and we did not find a significant difference between the double tem1 tem2 mutant and svp plants. However, when we compared tem2 and svp tem2, we found that svp tem2 is slightly earlier, which indicates that not all of the effect of SVP is through TEM2. These flowering time data correlate with the strong binding of SVP to TEM2 obtained by ChIP-quantitative PCR (qPCR; Fig. 5) and the down-regulation of TEM2 observed in svp mutant plants (Fig. 6). At 16°C, svp tem1 tem2 is not much earlier than the single svp mutant, and the difference obtained was not statistically significant, which is in agreement with the similar FT levels observed (Supplemental Fig. S12). This suggests that loss of SVP activity masks the effect of TEM on flowering time at lower temperatures. Therefore, at 16°C, SVP represses flowering partly through TEM, specifically TEM2, and partly through direct binding to FT (Lee et al., 2007, 2013).
CONCLUSION
In conclusion, we have identified new players, TEM genes, in the ambient temperature pathway as well as their regulation by SVP. SVP and TEM can reinforce the temperature responses by signaling partially through distinct pathways to control common outputs, such as FT and TSF. Moreover, SVP protein accumulation is higher during the day than during the night under LD conditions (Yoshida et al., 2009), in agreement with its mRNA expression (Supplemental Fig. S11), whereas TEM1 protein, and most probably, TEM2 have the opposite pattern (Osnato et al., 2012). This could suggest that SVP and TEM could regulate FT and TSF in different moments of the day. SVP represses FT during the morning (for review, see Song et al., 2013), and TEM would repress FT, TSF, and GA3ox1/2 during the night. This work and previous reports indicate that TEM genes are involved in several genetic pathways that regulate flowering.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) ecotype was used as the wild-type control in all of the experiments.
All mutants and transgenic lines are in the Col-0 background. tem1-1, 35S::TEM1, 35S::TEM2 (Castillejo and Pelaz, 2008), tem2-2, tem1-1 tem2-2 (Osnato et al., 2012), and svp-32 (Lee et al., 2007) have been described previously. The svp-41 mutant (Hartmann et al., 2000) was donated by Martin Kater. The tem1-1 svp-41, tem2-2 svp-41, and tem1-1 tem2-2 svp-41 combinations were generated by crosses. Genotypes were confirmed by PCR using published oligonucleotides (Supplemental Table S3). Seeds were stratified in darkness at 4°C for 3 d and sown on soil. All plants were grown in chambers under a controlled LD photoperiod (16-h-light/8-h-dark cycle) at 22°C to 23°C or 16°C under a mixture of cool white (TL5 54 W; 965) and warm white (TL5 54 W; 840) fluorescent lights, with a fluence rate of 80 to 90 μmol m−2 s−1.
Generation of Transgenic Plants
To generate the pSVP::SVP:HA construct, the open reading frame of SVP was amplified by reverse transcription (RT)-PCR using RNA isolated from 8-d-old seedlings. The resulting amplicons were cloned into the pCHF3 vector harboring the approximately 2.5-kb promoter fragment of SVP. This construct was introduced into svp-32 plants (Lee et al., 2007) using the floral dip method with minor modifications (Weigel and Glazebrook, 2002). Subsequently, transformants were selected for kanamycin resistance, and about 30 to 40 T1 seedlings were analyzed (Lee et al., 2013). Oligonucleotide primers used for cloning are listed in Supplemental Table S3. In pSVP::SVP:HA svp-32 plants, the production of the SVP-HA protein was confirmed (Supplemental Fig. S8A), and the early flowering and ambient temperature-insensitive flowering phenotypes of svp-32 mutants were rescued by pSVP::SVP:HA (Supplemental Fig. S8B), indicating that HA-tagged SVP protein is functional.
Phenotypic and Statistical Analyses
For flowering time measurements, plants were randomized with the respective controls and grown on soil in controlled environment growth chambers. Flowering time was determined by counting the number of cauline and rosette leaves of at least 12 individual plants. The number of days to flowering was determined when the floral bud was visible to the naked eye. Data are reported as a mean value of the total leaf number ± sd for each genotype and experimental condition used; we use the mean value of the total leaf number ± sem to compare the mean of independent experiments. All flowering time assays were performed at least two times. Flowering time data were subjected to ANOVAs. Post hoc tests were performed using Tukey’s multiple comparisons test after two-way ANOVA. Statistical analyses were performed with Prism 6 software (GraphPad Software, Inc.).
RNA Isolation and Expression Analysis
Samples consisted of pools of seedlings (12–59 individuals for each time point depending on the time of collection) sown on soil, which were quickly frozen in liquid nitrogen and powdered before RNA extraction. RNA was extracted using the PureLink Micro-to-Midi Total RNA Purification Kit (Invitrogen-Ambion) and DNAse treated using the DNA-Free Kit (Ambion). RNA integrity was checked on agarose gels, and concentration was measured using an ND-1000 Spectrophotometer (Thermo Scientific). Between 1 and 1.5 μg of DNAse-treated RNA was used for cDNA synthesis by using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. The resulting cDNA was diluted before subsequent expression analyses. qPCR was performed on a LightCycler 480 (Roche) using SYBR Premix ExTaq (Takara). Three technical replicates were made per sample. The relative expression was calculated using the 2−ΔΔCt (Livak and Schmittgen, 2001). Ubiquitin10 (UBQ10) was used as a reference gene. Results from biological duplicates are shown. Oligonucleotide primers used for qPCR are listed in Supplemental Table S3.
ChIP Assays
Two grams of pSVP::SVP:HA seedlings grown on soil under LD conditions at 23°C or 16°C were cross linked in 1% (v/v) formaldehyde on ice using vacuum infiltration. Nuclear extracts were isolated, and the immunoprecipitation assays were conducted as described previously (Kim et al., 2012). After shearing chromatin by sonication, rabbit anti-HA polyclonal antibody (about 5 µg; Santa Cruz Biotechnology) was used to immunoprecipitate genomic DNA fragments. qPCR was performed using DNA recovered from immunoprecipitation or 10% input DNA with a number of primer sets spanning the regulatory regions of TEM1 and TEM2 (Supplemental Table S3). The relative enrichment of each fragment was calculated by comparing samples immunoprecipitated with HA and cMyc (negative control) antibodies (Livak and Schmittgen, 2001). ChIP experiments were performed in two biological replicates (samples independently harvested on different days) with three technical replicates each with similar results.
From 1 to 1.5 g of 35S::TEM1:HA and 35S::TEM2:HA seedlings were grown on soil under LD conditions at 22°C and 16°C to test direct binding of TEM to FT and TSF loci. Cross linked DNA was immunoprecipitated with an anti-HA antibody (Sigma), purified using Protein A-Agarose Resin (Millipore), and tested by qPCR using specific primer sets (Supplemental Table S3) on regulatory regions of FT and TSF. ChIP experiments were performed in at least two biological replicates (samples independently harvested on different days) with three technical replicates each with similar results.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Upregulation of FT in tem1 tem2 and svp mutants.
Supplemental Figure S2. Upregulation of GA3ox1 in tem1 tem2.
Supplemental Figure S3. Low temperature does not increase TEM levels early in development.
Supplemental Figure S4. Opposite expression pattern of TEM and FT/TSF at 22°C.
Supplemental Figure S5. Opposite expression pattern of TEM and FT/TSF at 16°C.
Supplemental Figure S6. TEMs regulate FT and TSF levels at 16°C.
Supplemental Figure S7. TEM2 protein binds to the FT and TSF promotors at 16°C.
Supplemental Figure S8. Characterization of pSVP::SVP:HA svp-32 transgenic plants.
Supplemental Figure S9. Binding of SVP protein to the TEM1 and TEM2 genomic loci.
Supplemental Figure S10. SVP positively regulates TEM2 expression at 22°C and 16°C.
Supplemental Figure S11. TEMs do not regulate SVP levels.
Supplemental Figure S12. Upregulation of FT in tem1 tem2, svp, and svp tem1 tem2.
Supplemental Table S1. Flowering time and statistical analysis of data in Figure 1.
Supplemental Table S2. Flowering time and statistical analysis of data in Figure 7.
Supplemental Table S3. Oligonucleotide sequences.
Supplementary Material
Acknowledgments
We thank Martin Kater for svp-41 seeds. A.E.A.-J. performed this work within the framework of a PhD program of the Universitat Autònoma de Barcelona.
Glossary
- cDNA
complementary DNA
- ChIP
chromatin immunoprecipitation
- Col-0
Columbia-0
- LD
long day
- qPCR
quantitative PCR
- RT
reverse transcription
- ZT
Zeitgeber time
Footnotes
This work was supported by the Ministerio de Economía y Competitividad/European Regional Development Fund (grant no. BFU2012–33746), the Spanish Government (Formación de Personal Investigador fellowship to E.M.-G.), the Investigator Training Program of the Catalonian Government (predoctoral fellowship to A.E.A.-J.), and the Catalonian Government (Consolidated Research Group no. 2014 SGR 1406 to the research group of S.P.). J.H.A. and J.H.L. were supported by a National Research Foundation of Korea grant funded by the South Korean Government (Ministry of Science, ICT, and Future Planning; 2008-0061988) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1D1A4A0101941), respectively.
Articles can be viewed without a subscription.
References
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC (2011) Reproductive competence from an annual and a perennial perspective. J Exp Bot 62: 4415–4422 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33: 168–171 [DOI] [PubMed] [Google Scholar]
- Capovilla G, Schmid M, Posé D (2015) Control of flowering by ambient temperature. J Exp Bot 66: 59–69 [DOI] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chew YH, Smith RW, Jones HJ, Seaton DD, Grima R, Halliday KJ (2014) Mathematical models light up plant signaling. Plant Cell 26: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141: 550e1–550.e2 [DOI] [PubMed] [Google Scholar]
- Franklin KA. (2009) Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol 12: 63–68 [DOI] [PubMed] [Google Scholar]
- Gu X, Le C, Wang Y, Li Z, Jiang D, Wang Y, He Y (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun 4: 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G (2009) Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J 60: 614–625 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH (2012) The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol 159: 461–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Park TI, Yoo SJ, Jun AR, Ahn JH (2013) Generation and analysis of a complete mutant set for the Arabidopsis FT/TFL1 family shows specific effects on thermo-sensitive flowering regulation. J Exp Bot 64: 1715–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim JJ, Kim SH, Cho HJ, Kim J, Ahn JH (2012) The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana. Plant Cell Physiol 53: 1802–1814 [DOI] [PubMed] [Google Scholar]
- Lee JH, Ryu HS, Chung KS, Posé D, Kim S, Schmid M, Ahn JH (2013) Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342: 628–632 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- McClung CR, Davis SJ (2010) Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Curr Biol 20: R1086–R1092 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S (2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- Penfield S. (2008) Temperature perception and signal transduction in plants. New Phytol 179: 615–628 [DOI] [PubMed] [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M (2013) Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417 [DOI] [PubMed] [Google Scholar]
- Reeves PH, Coupland G (2001) Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol 126: 1085–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera-Branchat M, Andrés F, Coupland G (2014) Flowering responses to seasonal cues: what’s new? Curr Opin Plant Biol 21: 120–127 [DOI] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H (2012) Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J 70: 549–561 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA [Google Scholar]
- West AG, Shore P, Sharrocks AD (1997) DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol Cell Biol 17: 2876–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA. (2013) Ambient temperature signalling in plants. Curr Opin Plant Biol 16: 661–666 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yoshida RO, Fekih R, Fujiwara S, Oda A, Miyata K, Tomozoe Y, Nakagawa M, Niinuma K, Hayashi K, Ezura H, et al. (2009) Possible role of early flowering 3 (ELF3) in clock-dependent floral regulation by short vegetative phase (SVP) in Arabidopsis thaliana. New Phytol 182: 838–850 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.