Several species from an ancient legume lineage independently evolved a novel class of cysteine-rich peptides to impose a differentiation process on their endosymbionts.
Abstract
Nutritional symbiotic interactions require the housing of large numbers of microbial symbionts, which produce essential compounds for the growth of the host. In the legume-rhizobium nitrogen-fixing symbiosis, thousands of rhizobium microsymbionts, called bacteroids, are confined intracellularly within highly specialized symbiotic host cells. In Inverted Repeat-Lacking Clade (IRLC) legumes such as Medicago spp., the bacteroids are kept under control by an arsenal of nodule-specific cysteine-rich (NCR) peptides, which induce the bacteria in an irreversible, strongly elongated, and polyploid state. Here, we show that in Aeschynomene spp. legumes belonging to the more ancient Dalbergioid lineage, bacteroids are elongated or spherical depending on the Aeschynomene spp. and that these bacteroids are terminally differentiated and polyploid, similar to bacteroids in IRLC legumes. Transcriptome, in situ hybridization, and proteome analyses demonstrated that the symbiotic cells in the Aeschynomene spp. nodules produce a large diversity of NCR-like peptides, which are transported to the bacteroids. Blocking NCR transport by RNA interference-mediated inactivation of the secretory pathway inhibits bacteroid differentiation. Together, our results support the view that bacteroid differentiation in the Dalbergioid clade, which likely evolved independently from the bacteroid differentiation in the IRLC clade, is based on very similar mechanisms used by IRLC legumes.
Legumes, thanks to their ability to develop a symbiotic interaction with nitrogen-fixing bacteria, collectively called rhizobia, are among the agronomically and ecologically most important plants. This symbiosis results in the formation of new organs, the nodules, inside which rhizobia differentiate into an endosymbiotic form, the bacteroids, able to fix atmospheric nitrogen to the benefit of the plant. During this differentiation step, profound modifications of the metabolism of the rhizobia are observed, and this can be accompanied by a marked change in the bacterial cell shape and size (Haag et al., 2013). Three distinct bacteroid morphotypes have been observed in different legume species (Oono et al., 2010; Bonaldi et al., 2011; Kondorosi et al., 2013; Supplemental Fig. S1A): (1) elongated or E-morphotype bacteroids described in legumes of the Inverted Repeat-Lacking Clade (IRLC; Medicago, Pisum, and Vicia spp.) and some Aeschynomene spp. such as Aeschynomene afraspera; (2) enlarged, spherical bacteroids (S-morphotype) encountered in some species of the Dalbergioid clade (such as Aeschynomene indica, Aeschynomene evenia, and Arachis hypogaea); and (3) unmodified bacteroids (U-morphotype), which display a rod-shape morphology similar to free-living bacteria found in phaseoloid or robinoid legumes (i.e. Phaseolus, Vigna, Lotus, Glycine, and Sesbania spp.). The fact that the same rhizobium strain nodulating legumes of different clades can display different morphotypes is strong evidence supporting the conclusion that the host plant governs the bacteroid morphotype (Sen and Weaver, 1984; Mergaert et al., 2006; Bonaldi et al., 2011). The change of shape is probably the tip of the iceberg of the control exerted by the plant on the bacteria physiology during symbiosis. Indeed, besides their E-morphotype, the Sinorhizobium meliloti bacteroids in Medicago truncatula differ from their free-living state in several respects: they become polyploid, their membrane permeability increases dramatically, and they lose their reproductive capacity. Bacteroids that display such extreme changes are considered as terminally differentiated because they are unable to revert to their bacterial form (Mergaert et al., 2006).
In M. truncatula, a class of peptides, named nodule-specific cysteine-rich (NCR) peptides, plays a key role in this terminal bacteroid differentiation (Van de Velde et al., 2010). The M. truncatula NCR family is composed of about 600 highly divergent genes, which are nearly all expressed exclusively in nodules (Mergaert et al., 2003; Alunni et al., 2007; Young et al., 2011). NCR peptides are similar to the defensin type of antimicrobial peptides. The peptides contain an N-terminal secretion signal, and the mature peptides are usually no longer than 60 amino acids and have four to six conserved Cys residues (Mergaert et al., 2003). The cleavage of the signal peptide by a nodule-specific signal peptidase complex (SPC) located in the endoplasmic reticulum is required to permit the trafficking of NCR peptides to the symbiosome compartment. Bacteroid and symbiosome development is blocked in M. truncatula defective in nitrogen fixation1 (dnf1) mutant that is affected in the SPC22 subunit of this nodule-specific SPC (Van de Velde et al., 2010; Wang et al., 2010). Some NCR peptides have been found to have antimicrobial activity, killing rhizobium when applied at high concentrations (Van de Velde et al., 2010). However, these antimicrobial peptide-like NCR peptides can, at lower concentrations, induce typical features of E-morphotype bacteroids in vitro on cultured rhizobia (Van de Velde et al., 2010). The NCR gene family has been identified in all investigated legume species of the IRLC clade but in no other plant species outside of this clade. This raises the question about the nature of the factors used by the other legumes to control bacteroid metamorphosis.
In the Dalbergioid legume clade, bacteroids can be of the E- or S-morphotype. For example, within the Aeschynomene genus, A. afraspera has E-type bacteroids but A. indica or A. evenia have S-type bacteroids (Bonaldi et al., 2011; Arrighi et al., 2012). The bacteroid morphogenesis in the Aeschynomene genus is also under plant control, since the Bradyrhizobium spp. strain ORS285 transforms into E-morphotype bacteroids in A. afraspera nodules and in S-morphotype bacteroids in A. indica and A. evenia nodules (Bonaldi et al., 2011; Arrighi et al., 2012). The Aeschynomene symbiosis with Bradyrhizobium spp. has several other particular features, including the formation of nodules on both roots and stems, photosynthesis by the Bradyrhizobium symbionts that is required for optimal stem nodulation (Giraud et al., 2000), and the use of a Nod factor-independent nodulation pathway in some Aeschynomene spp., including A. indica and A. evenia (Giraud et al., 2007; Arrighi et al., 2012). Furthermore, in contrast to classical determinate and indeterminate nodules, aeschynomenoid nodules originate from the successive divisions of only one or a few root cortical cells initially infected by an infection thread-independent mechanism (Bonaldi et al., 2011). This infection mechanism has two consequences: (1) all the nodule primordium cells are infected, and (2) all the nodule primordium cells and bacteria are synchronized. Thus, during the nodule development of A. indica nodules, the differentiation of bacteroids occurs simultaneously for all nodule cells and takes place between days 4 and 5 after inoculation (Bonaldi et al., 2011). The possibility for obtaining samples of homogenous material at different differentiation stages and the existence of two distinct bacteroid morphotypes in closely related species make the Bradyrhizobium-Aeschynomene symbiotic couples an appealing model system for unraveling the mechanisms of bacteroid morphotype determination in legume species outside the IRLC clade.
In this study, we show that E- and S-morphotype bacteroids from A. afraspera and A. indica nodules display typical features of terminal differentiation. Furthermore, in these two species, we reveal the presence of an NCR-like peptide family that is specifically produced in the nodules and targets the bacteroids. Alteration of the trafficking of these peptides by silencing the dnf1 homolog identified in A. evenia correlated with the absence of bacterial differentiation, indicating that the NCR-like peptides identified in Aeschynomene spp. play a similar role to those described in Medicago spp.. Altogether, these data support the hypothesis of a convergent evolution of the host molecular mechanisms governing bacteroid differentiation in legumes.
RESULTS
E- and S-Bacteroids in Aeschynomene spp. Are Terminally Differentiated
Previous studies revealed that Bradyrhizobium sp. strain ORS285 displayed two distinct bacteroid morphotypes according to the Aeschynomene spp. host: E-morphotype in A. afraspera and S-morphotype in A. indica (Fig. 1A; Bonaldi et al., 2011). As this difference in shape could imply different properties, we analyzed in each bacteroid type, isolated from mature nodules at 14 d post inoculation (dpi), several characteristics that have been shown to change during bacteroid metamorphosis in Medicago spp., such as bacterial DNA content, membrane permeability, and viability. As revealed by flow cytometry analysis (Fig. 1B), the DNA content of E- and S-bacteroids peaked at around 7C and 16C, respectively, in comparison with the 1C/2C DNA content of free-living bacteria. These ploidy levels are less than described for S. meliloti bacteroids (24C; Mergaert et al., 2006), although other studies have shown lower ploidy levels also in Medicago spp. (Sinharoy et al., 2013; Berrabah et al., 2014).
Figure 1.
Properties of free-living cultured Bradyrhizobium sp. ORS285 bacteria and ORS285 bacteroids isolated from A. afraspera or A. indica nodules. A, Normaski bright-field (BF; top row) and fluorescence microcopy of bacteria and bacteroids stained with 4′,6-diamidino-2-phenylindole (DAPI; middle row) and PI (bottom row). Bars = 1 µm. B, DNA content of DAPI-stained bacteria and bacteroids measured by flow cytometry. Colors are as follows: blue, free-living bacteria; green, bacteroids isolated from A. afraspera; and red, bacteroids isolated from A. indica.
Bacteroid membrane integrity was estimated using propidium iodide (PI), a DNA stain that is excluded from living cells but penetrates in cells displaying membrane integrity damage (Mergaert et al., 2006). Fluorescence images of the PI-stained bacteroids showed that the membrane permeability of E- and S-morphotype bacteroids was more prominent than that of the free-living bacteria (Fig. 1A). Indeed, 88% of S-bacteroids and 95% of E-bacteroids were found to be permeable to PI against 7% for free-living bacteria. For comparison, PI stained about 50% of S. meliloti bacteroids (Mergaert et al., 2006). The loss of viability measured for S-morphotype bacteroids from A. indica nodules (98%) was also comparable to the one estimated previously for S. meliloti bacteroids (99%; Mergaert et al., 2006). In contrast, intracellular cells of A. afraspera appeared more viable, as 22% formed colonies on agar plates. However, this higher value could result from the fact that A. afraspera nodules displayed the unusual presence of two infected tissues, a large central tissue in which the bacteria are elongated and a superficial tissue in which the shape of the bacteria remained unmodified (Bonaldi et al., 2011). Therefore, the intracellular cells extracted from nodules consisted of a mixture of differentiated and nondifferentiated bacteria, suggesting that the loss of viability of E-morphotype bacteroids should be far more important. Taken together, we observed that the E- and S-morphotype bacteroids from Aeschynomene spp. nodules share the same features as the S. meliloti bacteroids in Medicago spp. and, therefore, that they can be considered as terminally differentiated.
Distribution of S- and E-Morphotype Bacteroids among Aeschynomene spp.
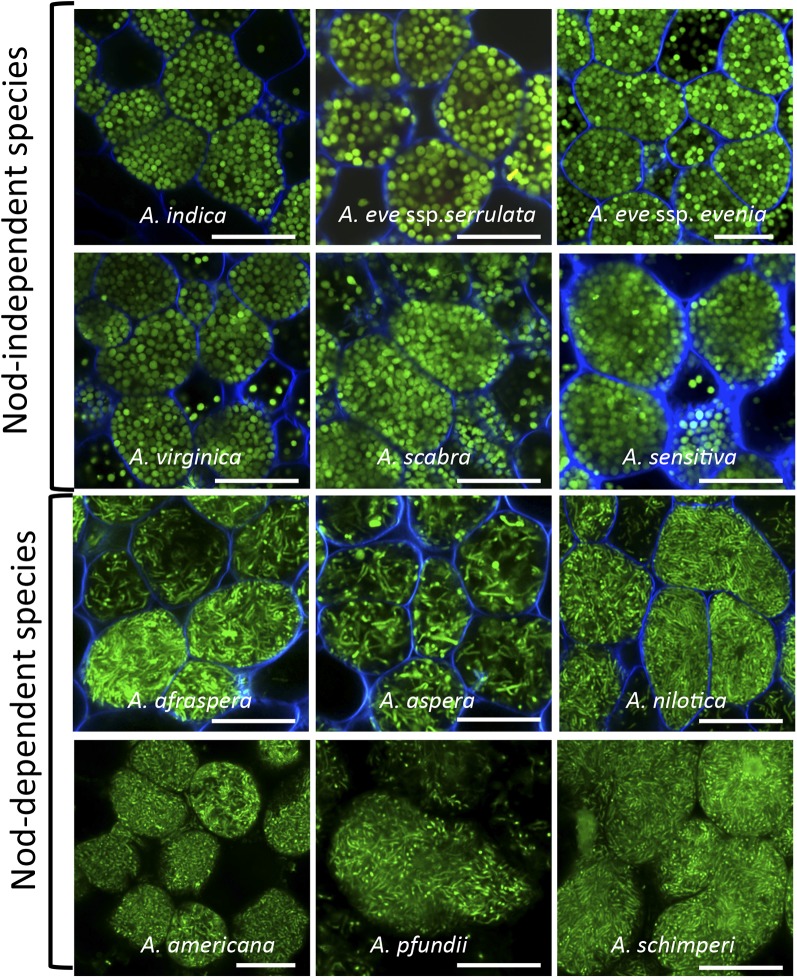
To obtain more insight into the distribution of the E- and S-bacteroid morphotypes among the Aeschynomene spp., we analyzed by confocal microscopy the shape of intracellular bacteria from mature nodules of various Aeschynomene spp. elicited by the ORS285 strain tagged with GFP. Unlike the other photosynthetic Bradyrhizobium spp. strains, such as ORS278 and BTAi1, this bacterium does contain the canonical nodABC genes and displays a broader host range due to its ability to use a Nod factor-dependent and a Nod factor-independent symbiotic process according to the host plant (Bonaldi et al., 2011). We observed in all tested species using a Nod factor-independent process (A. indica, A. evenia ssp. serrulata, A. evenia. ssp. evenia, Aeschynomene virginica, Aeschynomene scabra, and Aeschynomene sensitiva) that the bacteroids displayed an S-morphotype, whereas in the three Nod factor-dependent species tested (A. afraspera, Aeschynomene nilotica, and Aeschynomene aspera), the bacteroids displayed an E-morphotype (Fig. 2; Supplemental Fig. S1B).
Figure 2.
Distribution of S- and E-morphotype bacteroids among Aeschynomene ssp. The species A. indica, A. evenia ssp. serrulata, A. evenia spp. evenia, A. virginica, A. scabra, A. sensitiva, A. afraspera, A. nilotica, and A. aspera were nodulated by the ORS285 GFP-tagged strain; the species A. americana, A. schimperi, and A. pfundii were nodulated by Bradyrhizobium spp. strains ORS301, ORS302, and ORS305, respectively, and bacteroids were stained by the SYTO 9 fluorescent probe of the live/dead stain. Bars = 10 μm.
We also analyzed the bacteroid shape in another group of Aeschynomene spp. not nodulated by the ORS285 strain but by nonphotosynthetic Bradyrhizobium spp. strains containing nod genes such as ORS301, ORS302, and ORS305 (Molouba et al., 1999). We also observed in these species (Aeschynomene americana, Aeschynomene pfundii, and Aeschynomene schimperi) that the bacteroids displayed an E-morphotype. Altogether, these data indicate that S-morphotype bacteroids are specific for the Aeschynomene spp. using a Nod factor-independent symbiotic process, while the E-morphotype is specific to the Aeschynomene spp. using a Nod factor-dependent one.
Aeschynomene spp. Contain a New Class of Nodule-Specific Cys-Rich Peptide-Encoding Genes
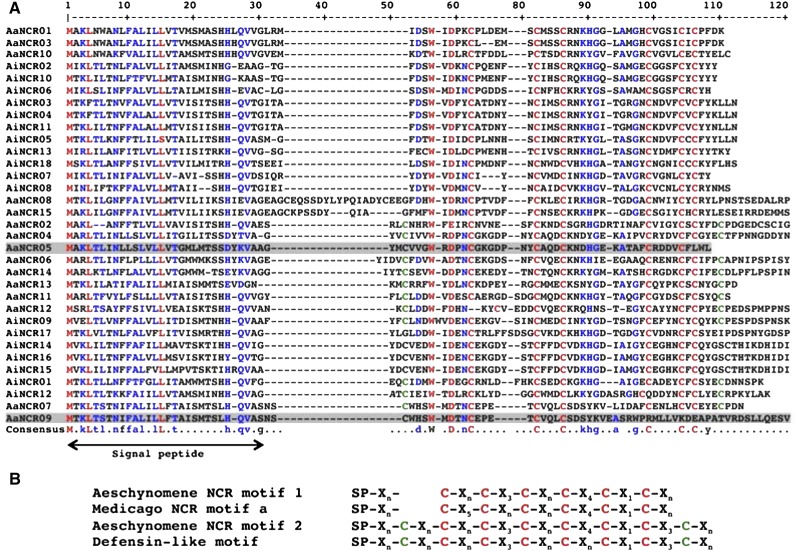
Our results indicate that in Aeschynomene spp. nodules, the endosymbiotic bacteria undergo terminal differentiation similar to that described in nodules of IRLC species. This raises the question of whether NCR peptides are also recruited in Aeschynomene spp. to govern this differentiation step. To check this possibility, we analyzed four EST libraries previously constructed in our laboratory and corresponding to noninoculated roots and mature nodules from A. afraspera and A. indica plants inoculated with the ORS285 strain. Although the total number of complementary DNAs (cDNAs) sequenced for each library was relatively small, around 9,500 ESTs per library (Supplemental Table S1), we postulated that if NCR peptides were also involved in bacteroid differentiation in Aeschynomene spp., they should be specifically and highly expressed in the nodules and, hence, easily detectable. We first performed a BLAST search on the EST libraries using several M. truncatula NCR genes as a query, but no conclusive results were obtained. Considering that NCR genes are rapidly evolving and are highly diverse, even within M. truncatula (Alunni et al., 2007; Branca et al., 2011), we reanalyzed the available EST databases using the following parameters determined from typical features of the NCR family: (1) candidate genes should encode small proteins (less than 100 amino acids) containing a signal peptide; (2) the gene expression should be nodule specific with a significant read count in the EST database (we arbitrarily fixed a lower limit of five reads); and (3) the candidate genes should encode peptides rich in Cys residues (at least four). This second in silico analysis identified 15 genes encoding putative NCR-like peptides from the A. afraspera nodule library and 18 from the library of A. indica nodules (Fig. 3).
Figure 3.
Alignment and Cys signature of Aeschynomene spp. NCR peptides. A, The deduced protein sequences from the up-regulated NCR genes from A. indica (AiNCRs) or A. afraspera (AaNCRs) were aligned using Multalin (http://multalin.toulouse.inra.fr/multalin/), and the alignment obtained was adjusted manually. Highly conserved amino acids are in red, and conservative substitutions are in blue. The putative location of the signal peptide is indicated below the alignment. B, Comparison of the Cys-rich motifs of Aeschynomene spp. NCR peptides with those of Medicago spp. and defensin-like peptides. Red and green Cys residues correspond to the NCR and defensin signatures, respectively. The peptides that do not contain any motif are gray shaded in A.
These genes were named AaNCRs or AiNCRs and numbered according to their rank in read counts (Supplemental Table S2). The deduced mature peptides (i.e. without their signal peptides) are between 37 and 85 residues long and contain four to 10 Cys residues (Fig. 3A). The alignment of these NCR-like peptides highlighted, for 31 out of the 33 sequences, a conserved pattern of six Cys residues within the sequence of the mature peptides (Fig. 3B). This pattern is very close to the one described previously for a group of Medicago spp. NCR peptides but distinct by the spacing between the first three Cys residues. This led us to propose a specific Aeschynomene spp. NCR-like motif. Interestingly, 11 of these NCR-like peptides contained two extra Cys residues, giving rise to a typical defensin signature that was named motif 2 (Fig. 3B).
By analogy with M. truncatula, for which the NCR family is composed of about 600 genes that exhibit distinct temporal and spatial expression patterns in nodules (Mergaert et al., 2006; Alunni et al., 2007; Young et al., 2011; Guefrachi et al., 2014), the involvement of a larger number of NCR genes is also expected in Aeschynomene spp. Therefore, we considered the possibility that our criteria were too selective, leading to a restricted number of candidate genes identified. By performing BLAST searches against the same EST libraries but this time using the first candidate genes that emerged, we identified a total of 38 and 44 NCR-like genes in A. afraspera and A. indica, respectively (Supplemental Table S2). All these additional genes were found to be specifically expressed in the nodules, and a global alignment shows that all of them except three contained the Aeschynomene spp. NCR-like motif 1 or 2.
Furthermore, a BLAST search against transcriptome data obtained from A. evenia ssp. serrulata mature nodules made by the 454 technology, which allows the detection of a larger number of transcripts, revealed an even higher number of NCR-like genes of more than 80 (Supplemental Table S2). Altogether, these data suggest that the NCR-like genes constitute also in Aeschynomene spp. an important family that could count several tens or even hundreds of members.
NCR-Like Genes of Aeschynomene spp. Are Expressed before or Concomitant with Bacteroid Differentiation
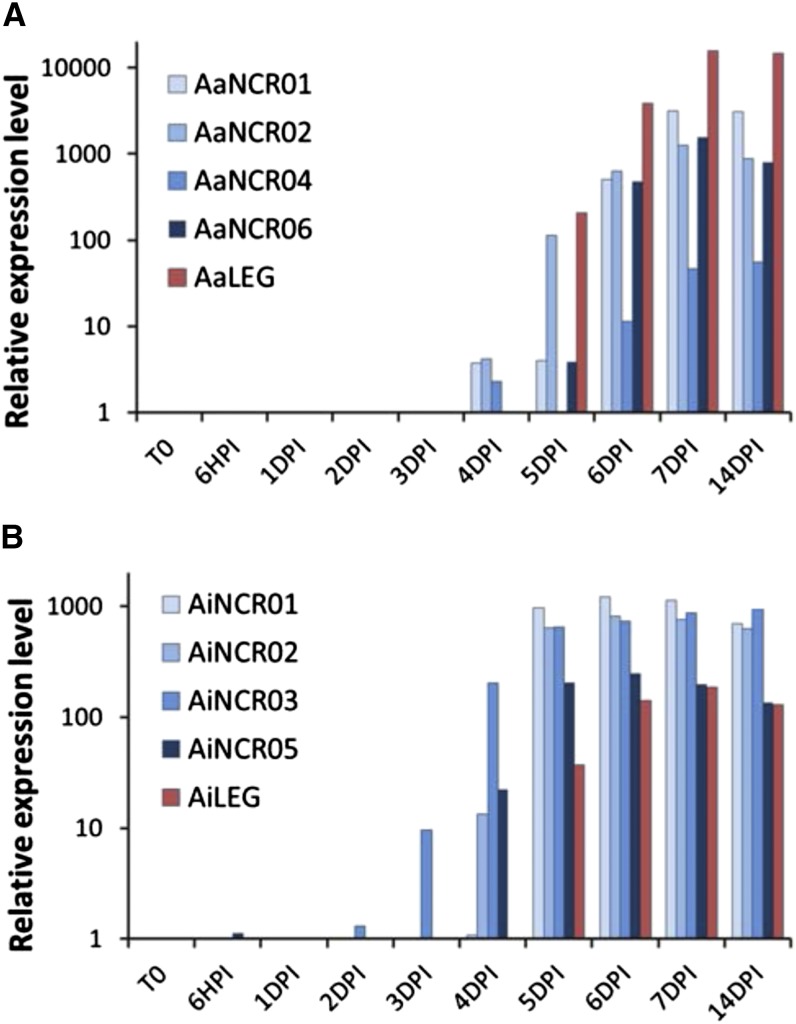
In order to analyze the temporal expression of the AaNCR and AiNCR genes, we performed a reverse transcription-quantitative PCR (RT-qPCR) analysis of four of them for each Aeschynomene spp. Root tissues of A. afraspera and A. indica were sampled at time 0 and at different time points after inoculation with Bradyrhizobium sp. strain ORS285 (6 h and 1, 2, 3, 4, 5, 6, 7, and 14 d). As a symbiotic marker, we also monitored Leghemoglobin (LegHb) mRNA accumulation. As shown in Figure 4, mRNA corresponding to LegHb started to accumulate at 5 dpi for both species (i.e. when the bacteroid differentiation process is completed and a nitrogenase activity is detectable; Bonaldi et al., 2011). Interestingly, while the NCR-like gene transcripts were not detected in control roots or during the early time points of nodule formation, they were all strongly expressed at 5 dpi, and some of them even started to accumulate 1 or 2 d before (Fig. 4), which coincided with the beginning of bacteroid morphogenesis (Bonaldi et al., 2011).
Figure 4.
NCR expression during the symbiotic process. NCR expression is shown during A. afraspera (A) and A. indica (B) nodule development. Plants were inoculated with the Bradyrhizobium sp. ORS285 strain, and the roots were sampled at time 0 (T0) and at different time points as indicated. The relative expression level of the NCR peptides was determined by RT-qPCR and normalized by the expression of Elongation Factor 1α (EF1α). As a control of nodule development, the expression of LegHb (LEG) was also measured.
To complete this expression analysis, we also checked the expression by RT-qPCR, in both roots and nodules, of three candidate NCR genes identified in A. evenia by BLAST. As expected, the three genes appeared specifically and highly expressed in A. evenia nodules (Supplemental Fig. S2).
NCR-Like Genes Are Expressed Specifically in Infected Cells of Aeschynomene spp. Nodules
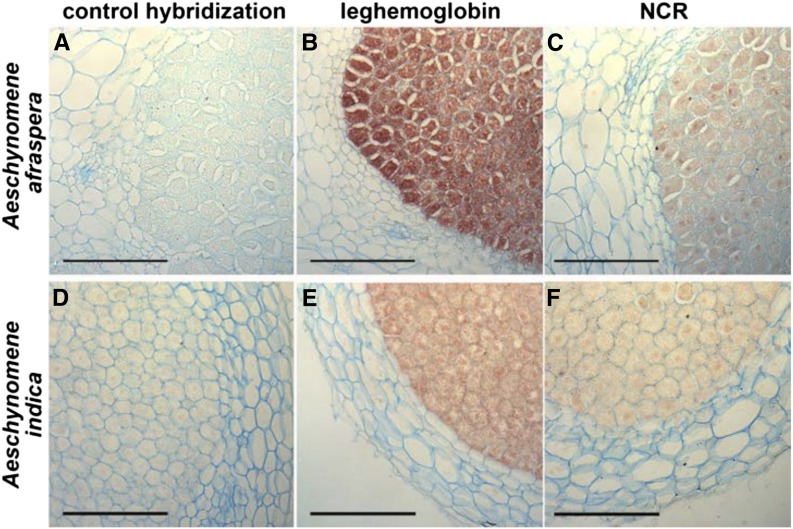
In Medicago spp., the NCR peptides are specifically synthesized in the infected plant cells and are targeted to the bacteria (Van de Velde et al., 2010). To check if the expression of the identified AaNCR and AiNCR genes was also restricted to the infected cells, we performed an in situ hybridization on 14-d-old nodules of A. afraspera and A. indica infected with the ORS285 strain. We used as probes AaNCR01 and AiNCR01, which were found to be the most highly expressed NCR genes in each species. In addition, the LegHb gene, known to be expressed in infected nodule cells, was included as a positive control for the hybridization experiment. As shown in Figure 5, a strong signal was obtained using the LegHb antisense DNA as a probe. The signal was restricted to the infected cells (Fig. 5, B and E), consistent with the known role of this protein in symbiosis. Both A. afraspera and A. indica NCR antisense probes also gave a signal limited to the infected cells (Fig. 5, C and F), similar to what was observed for LegHb. Control hybridizations without probe or with a sense LegHb probe or sense NCR-like probes (Fig. 5, A and D) showed a complete absence of signal.
Figure 5.
NCR genes are specifically expressed in the infected symbiotic cells of Aeschynomene spp. nodules. Sections of 14-dpi A. indica and A. afraspera nodules, infected with the Bradyrhizobium sp. ORS285 strain, were analyzed by in situ hybridization with antisense probes of LegHb (B and E) or NCR (C and F). Control hybridizations were done with the sense NCR probe (A and D). Bars = 100 μm.
NCR-Like Peptides Target Bacteroids in Aeschynomene spp. Nodules
To test the possibility that AaNCR and AiNCR mature peptides are targeted to the endosymbiotic bacteria, we set up a proteome approach. Total protein extracts were prepared from nodules and from purified bacteroids from both A. afraspera and A. indica. In addition, a control extract was prepared from free-living ORS285 bacteria. Proteins were subjected to Tricine-PAGE analysis. Slight differences in the patterns of low-molecular-mass proteins (between 4 and 9 kD), where we expected the AaNCR or AiNCR peptides, could be observed between nodule or bacteroid extracts and extracts from free-living bacteria (Supplemental Fig. S3). We identified the corresponding proteins by mass spectrometry. From the bacteroid extracts isolated from A. afraspera and A. indica nodules, 51 and 42 proteins were identified, respectively (Supplemental Table S3). Among them, in both nodule contexts, four corresponded to plant proteins, of which three were identified as NCR peptides (Table I). The additional plant protein identified in A. indica bacteroids was a putative monosaccharide-proton symporter corresponding probably to a symbiosome-located transporter, whereas there was an extensin-like protein of unknown function in A. afraspera bacteroids. Among the 40 proteins identified in free-living bacterial cells, all corresponded to bacterial proteins and none displayed the characteristics of Cys-rich peptides (Supplemental Table S3).
Table I. Mass spectrometry identification of proteins of plant origin from bacteroids isolated from A. afraspera and A. indica mature nodules.
This table is a compilation of all the plant proteins identified from bacteroid samples and processed by Mascot Distiller (version 2.1.1.0) software (Matrix Science).
| Gene Name | Protein Function | Protein Mass | Mascot Protein Score | Protein Coverage | Peptide Sequences (5′–3′) |
|---|---|---|---|---|---|
| D | % | ||||
| Bacteroid A. afraspera-ORS285 | |||||
| AaNCR15 | NCR like | 4,928 | 74 | 32.5 | GDGECSGIYCHCR |
| AaNCR01 | NCR like | 8,714 | 56 | 16.9 | CPLDEMSCMSSCR |
| AaNCR07 | NCR like | 7,928 | 53 | 31.3 | VLIDAFCENLHCVCEYECPDN |
| AaCL26Contig2 | Extensin-like protein | 3,497 | 56 | 46.9 | NIPISLSLILNVCSR |
| Bacteroid A. indica-ORS285 | |||||
| ADL0ADA10YJ01CM1 | Putative monosaccharide H+ symporter | 20,215 | 52 | 4.3 | LASSILPR |
| AiNCR02 | NCR like | 8,521 | 78 | 25.7 | HGQSAMGECGGSFCYCYYY |
| 52 | 24.3 | HGQSAMGECGGSFCYCYY | |||
| AiNCR18 | NCR like | 9,165 | 64 | 21.8 | HGATANGYCNGNICCCK |
| AiNCR06 | NCR like | 8,471 | 124 | 52.1 | YGSSAWAMCSGSFCR |
| LGSDSWMDINCPGDDSICNFHCK | |||||
The DNF1 SPC Is Recruited for Bacteroid Differentiation in Aeschynomene spp.
To further support the role of the Aeschynomene spp. NCR peptides in bacterial differentiation, we applied a strategy aiming to interfere with their targeting to the symbiosomes, similar to that described before in M. truncatula (Van de Velde et al., 2010; Wang et al., 2010). NCR peptides are secretory proteins with an N-terminal signal peptide. Therefore, their transport requires the removal of this signal peptide by the SPC machinery, which is located in the membrane of the endoplasmic reticulum. Reduced SPC activity is supposed to hamper the transport of the NCR peptides to the symbiosomes, similar to that in the dnf1 mutant of M. truncatula (Van de Velde et al., 2010; Wang et al., 2010).
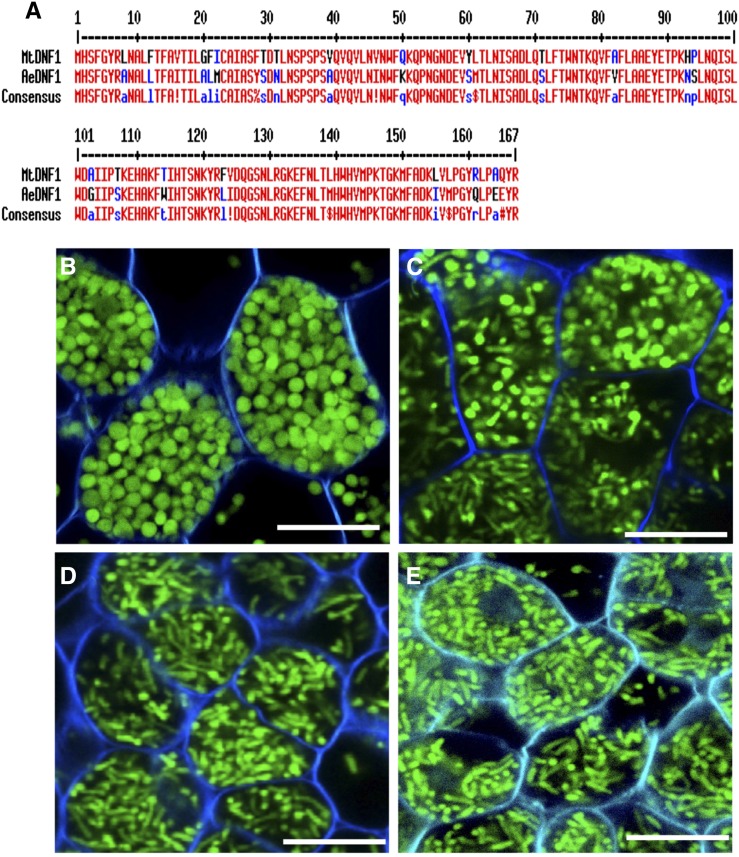
To develop such a loss-of-function strategy in Aeschynomene spp., we chose to conduct this experiment on A. evenia ssp. serrulata, which is diploid, contrary to A. indica and A. afraspera, which are allopolyploids (Renard et al., 1983; Arrighi et al., 2014). A BLAST search using MtDNF1 allowed the identification in the A. evenia ssp. serrulata transcriptome database of a single candidate gene, which we named AeDNF1, encoding a protein of 167 amino acids with 83% identity (Fig. 6A). We used an RNA interference (RNAi) approach to knock down AeDNF1. The morphology of the bacteroids was observed in mature nodules elicited by the GFP-tagged ORS285 strain. In parallel, we monitored the level of AeDNF1 extinction by RT-qPCR. As shown in Figure 6B, in transgenic lines in which the expression level of AeDNF1 was not affected, the bacteroid differentiation occurred normally, giving rise to spherical bacteroids, while in the lines with the highest extinction level (about 65% reduction of AeDNF1 expression), defects in bacteroid differentiation were observed with the presence of various bacteroid morphotypes within the same cell (Fig. 6C) or elongated bacteroids instead of spherical ones (Fig. 6, D and E).
Figure 6.
DNF1 silencing affects bacteroid differentiation in Aeschynomene spp. A, Alignment of the deduced protein sequence of the A. evenia ssp. serrulata DNF1 ortholog with the corresponding Medicago spp. protein (accession no. TC121074). The two protein sequences were aligned using Multalin (http://multalin.toulouse.inra.fr/multalin/). B to E, Roots of different lines of A. evenia ssp. serrulata transformed with an RNAi construct directed against AeDNF1 encoding the SPC22 subunit of the SPC were inoculated with a GFP-tagged Bradyrhizobium sp. ORS285 strain. At 21 dpi, the nodules were harvested, and the shape of the bacteroids was observed by confocal microscopy on nodule sections. As a control, a line with no RNAi effect (1% reduction) was used (B). The lines 3, 7, and 53, with expression levels reduced by 62%, 65%, and 66%, respectively, are presented in C to E. Bars = 10 µm.
DISCUSSION
The use of Cys-rich peptides called NCR peptides to govern bacteroid differentiation was assumed to be specific to legume species belonging to the IRLC clade, no homologs of NCR peptides being found in legume species outside of this clade (Mergaert et al., 2003). However, our data lead us to propose here that Aeschynomene spp. use a similar molecular mechanism to impose a differentiation process on their rhizobium endosymbionts.
First, we demonstrated that the distinguishing characters associated with bacteroid differentiation in nodules of IRLC legumes (i.e. marked cell enlargement, polyploidy, membrane permeability modification, and loss of viability) are conserved in Aeschynomene spp. nodules. Second, we identified in several Aeschynomene spp. a class of Cys-rich peptides that have similar characteristics to the Medicago spp. NCR peptides and that are synthesized specifically in nodule symbiotic cells and targeted to the bacteroids. Finally, silencing of the dnf1 homolog in A. evenia ssp. serrulata impaired bacteroid differentiation. Moreover, it has been shown that Bradyrhizobium sp. strains ORS278 and ORS285 require the Bacteroid development A-like (BclA) peptide transporter for both E- and S-morphotype bacteroid differentiation in Aeschynomene spp. (Guefrachi et al., 2015). BclA is similar to S. meliloti Bacteroid development A (BacA), which is needed for bacteroid differentiation in Medicago spp. and which imports NCR peptides into the bacterial cells. BclA can functionally replace BacA for symbiosis and for peptide transport. Together, these observations suggest that, in Aeschynomene spp., E- or S-morphotype bacteroid differentiation is under the control of the NCR peptide family that we identified here.
These findings might be surprising, considering that, according to the Aeschynomene spp., the bacteroids display two distinct morphotypes (S- and E-morphotypes). This difference of morphological shape suggests distinct mechanisms governing bacterial morphogenesis. The change from a rod to a spherical shape as observed in A. indica and A. evenia during bacteroid differentiation resembles bacterial morphologies resulting from mutations that affect peptidoglycan synthesis and that were described in Escherichia coli (Young, 2003). Similarly, it has been shown in Vibrio spp. that some noncanonical d-amino acids could lead to a rod-to-sphere shape transition both via their incorporation into the peptidoglycan polymer and by regulating enzymes that synthesize and modify it (Lam et al., 2009). Therefore, the bacteroid morphology in A. indica or A. evenia is possibly associated with changes in the peptidoglycan cell wall architecture. This raises the question of whether the morphogenesis to the S-morphotype is governed by a specific class of NCR peptides targeting enzymes or genes involved in peptidoglycan synthesis or if additional plant effectors of a different nature such as particular d-amino acids are involved in the control of bacteroid morphogenesis.
Even if the underlying mechanisms for E- and S-morphotype bacteroid formation are obviously different, some observations strongly suggest that the two processes are closely related. In A. indica and A. evenia, the formation of the spherical bacteroids is taking place synchronously in all nodule cells between 4 and 5 dpi (Bonaldi et al., 2011). In this 1-d time frame, all intracellular rod-shaped bacteria transform into spheres. However, when making observations between 4 and 5 dpi, one can discern intermediate differentiation stages that strongly resemble E-morphotype bacteroids (Supplemental Fig. S4). In addition, we observed in the A. evenia ssp. serrulata knockdown lines, in which the level of expression of AeDNF1 was reduced, bacteroids with an E-morphotype instead of an S-morphotype or a mixture of both shapes within the same cell. Finally, the bacterial gene bclA is similarly required for E-morphotype bacteroid formation in A. afraspera or S-morphotype bacteroid formation in A. indica or A. evenia (Guefrachi et al., 2015).
Interestingly, analysis of the distribution of the E- and S-morphotype bacteroids among Aeschynomene revealed that all the tested species using a Nod factor-independent symbiotic process have S-morphotype bacteroids, whereas those using a Nod factor-dependent process have E-morphotype bacteroids, similar to the IRLC species. Furthermore, a recent phylogenetic analysis revealed that all Aeschynomene spp. using a Nod factor-independent symbiotic process form a monophyletic clade that does not include other species using a Nod factor-dependent process (Supplemental Fig. S1B; Chaintreuil et al., 2013). Therefore, it would be tempting to speculate that the S-morphotype of bacteroids and Nod factor-independent symbiosis are both derived characters, which are correlated. However, in the Dalbergioid clade that contains the Aeschynomene genus, other species, such as A. hypogaea or Stylosanthes hanata, also host spherical bacteroids (Chandler et al., 1982). It has been established that at least A. hypogaea uses a Nod factor-dependent symbiotic process (Ibáñez and Fabra, 2011). This suggests that the S-morphotype character emerged several times during the diversification of the Dalbergioid clade independently from the Nod factor-independent character.
The mode of action of the NCR peptides and their bacterial targets remains unclear in Medicago spp. However, it has been shown that some cationic NCR peptides are able to provoke symptoms of terminal differentiation in vitro (Van de Velde et al., 2010). To examine if the NCR peptides identified in Aeschynomene spp. could induce the same symptoms in vitro, we tested the activity of five synthetic Aeschynomene spp. NCR peptides (AaNCR01, AaNCR02, AiNCR01, AiNCR02, and AiNCR03), which are the most abundant ones identified in the A. afraspera and A. indica nodule EST libraries. No activity could be detected on Bradyrhizobium sp. strain ORS285 (Supplemental Fig. S5A). Nevertheless, one of them (AaNCR01) displayed a weak antimicrobial activity on S. meliloti and could provoke a slight increase in the bacterial ploidy level (Supplemental Fig. S5, B and C). We also tested on the strain ORS285 the Medicago spp. NCR peptides (NCR035, NCR247, and NCR335) known to have an antimicrobial effect on a large panel of bacteria (Tiricz et al., 2013), but surprisingly, no activity was detected for those peptides, whereas on the control S. meliloti strain, strong antimicrobial activity and membrane damage could be observed under the same conditions. The membrane of Bradyrhizobium spp. strains differs from that of rhizobium strains by the presence of hopanoids, which may represent up to 40% of the total lipid extract (Kannenberg et al., 1995). It was recently shown that this class of compounds, which display some structural similarity with eukaryotic sterols, reinforce the stability and rigidity of the outer membrane of photosynthetic Bradyrhizobium spp. (Silipo et al., 2014). It is possible, therefore, that hopanoids reduce the antimicrobial activity of cationic Medicago NCR peptides by reinforcing the outer membrane barrier function. In agreement with this assumption, we observed that a hopanoid mutant of the photosynthetic Bradyrhizobium spp. strain BTAi1 was highly sensitive to the Medicago NCR peptides, in contrast to the wild-type strain (Supplemental Fig. S5A). Nevertheless, this mutant still remained resistant to the Aeschynomene spp. NCR peptides tested (Supplemental Fig. S5A).
The lack of activity of the tested Aeschynomene NCR peptides in vitro on the ORS285 strain or on the BTAi1 hopanoid mutant could result from the fact that they have a net neutral or negative charge. Indeed, in Medicago spp., only cationic NCR peptides, particularly those with a high pI, were reported so far to display in vitro activity. This does not imply that anionic or neutral NCR peptides are not functional in vivo, considering their high level of expression, which is similar to that of the cationic NCR peptides (Mergaert et al., 2003). It was proposed that the cationic NCR peptides interact preferentially with the negatively charged outer membrane of rhizobia and, by compromising its permeability, facilitate the entry of anionic NCR peptides to the bacterial cytosol (Kondorosi et al., 2013). In Aeschynomene spp., except for two A. indica NCR peptides with a pI close to 8, no cationic NCR-like peptides could be identified among the several tens of candidate genes identified in each species (Supplemental Table S2). However, we cannot exclude that such a class of cationic NCR peptides does exist in Aeschynomene spp. Our transcriptomic analyses made on mature nodules at 14 dpi could have missed these NCR peptides that are more likely to act earlier in the differentiation process. Furthermore, the difference in the membrane ultrastructure between bradyrhizobia and rhizobia could also imply a mechanism other than electrostatic binding and a specific class of NCR peptides that we have not yet identified. The understanding of how Aeschynomene spp. NCR peptides can enter in Bradyrhizobium spp. cells and modulate their physiology and morphology will require further studies.
In silico analyses of the available transcriptome databases suggest the NCR-like genes constitute an important family in Aeschynomene spp. that could count several tens or even hundreds of candidates. As observed for Medicago spp., the amino acid sequences of these NCR peptides are highly divergent except for the distribution of six Cys residues, which is conserved in more than 90% of the identified peptides. A phylogenetic analysis based on the sequences of the mature peptides shows that the NCR-like peptides identified in Aeschynomene spp. are distant from the NCR peptides identified in IRLC (Supplemental Fig. S6). A recent study mapping the bacteroid morphology in 40 legume species of the Papilionoideae suggests that legumes inducing bacteroid morphogenesis evolved independently at least five times from an ancestral papilionoid legume hosting U-morphotype bacteroids (Oono et al., 2010). It has been proposed that the NCR family identified in IRLC legumes evolved from defensin ancestors (Mergaert et al., 2003). This large family of antimicrobial peptides is found in all eukaryotes, where they are part of the first line of defense against invading microbes. The fact that the NCR peptides identified in Aeschynomene spp. are phylogenetically distant from the NCR peptides identified in the IRLC legumes but also display a defensin signature suggest that they have similarly evolved from defensins but likely from a distinct ancestral repertoire.
It will be interesting to determine if the use of Cys-rich peptides is a general strategy in legumes to keep their endosymbionts under control and to analyze the transcriptomes of nodules from species belonging to the two other lineages, the genistoids and the mirbelioids, in which bacteroid morphogenesis was reported, using the selective criteria defined in this study. Remarkably, it was shown recently that also actinorhizal plants forming nodules with bacteria from the Frankia genus (Carro et al., 2015), as well as insects, produce symbiosis-specific antimicrobial peptides to control the development of their endosymbiotic bacteria, which display in certain cases a spherical or elongated shape similar to the S- or E-morphotype bacteroids (Login et al., 2011; Shigenobu and Stern, 2013). This suggests that the enrollment of antimicrobial peptides as symbiosis effectors is an optimal modus operandi for a eukaryotic host cell to control large populations of intracellular bacteria.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Bradyrhizobium spp. strain ORS285 tagged or not with GFP (Bonaldi et al., 2011) and Bradyrhizobium spp. strains ORS301, ORS302, and ORS305 (Molouba et al., 1999) were grown at 37°C in yeast mannitol medium (Vincent, 1970). The medium used for ORS285::GFP was supplemented with kanamycin (50 µg mL–1).
Plant Material, Growth Conditions, and Inoculation
All the accessions of Aeschynomene spp. used in this study, their geographic origins, their sources, and the Bradyrhizobium spp. strains used to inoculate them are listed in Supplemental Table S4. Seed germination, plant culture, and bacterial inoculation were performed as before (Bonaldi et al., 2011).
Bacteroid Characterization
Bacteroid isolation was performed as described (Mergaert et al., 2006). Bacteroids and free-living bacteria were stained with DAPI at 50 µg mL–1 and PI at 2 µg mL–1 and observed with a Reichert Polyvar fluorescence microscope equipped with a Nikon dxm 1200 digital camera. DNA measurements were performed with a Beckman-Coulter ELITE ESP flow cytometer. For counting of cultivable bacterial cells, bacteroid suspensions were serially diluted in 10-fold dilutions and plated on yeast mannitol agar plates. At the same time, the cells in the bacteroid suspensions were also directly counted by light microscopy using a Malassez cell-counting chamber.
Analysis of the Bacteroid Morphotype in Various Aeschynomene spp.
Semithin nodule sections (30 to 40 µm thick) were prepared using a Vibratome (VT1000S; Leica). Nodules elicited by the ORS285-tagged GFP strain were visualized as described previously (Bonaldi et al., 2011). For the nodules elicited with the nontagged strains, the nodule sections were incubated for 15 min in live/dead staining solution (5 µm SYTO 9 and 30 µm PI in 50 mm Tris, pH 7 buffer; Live/Dead BacLight; Invitrogen). Sections were then removed and incubated an additional 15 min in 10 mm phosphate saline buffer containing Calcofluor White M2R (Sigma) to a final concentration of 0.01% (w/v) to stain the plant cell wall (Nagata and Takebe, 1970). Analyses were carried out using a confocal laser-scanning microscope (Carl Zeiss LSM 700). Calcofluor White was excited at 405 nm with emission signal collection at 405 to 470 nm. For SYTO 9 and PI, excitation wavelengths of 488 and 555 nm were used with emission signal collection at 490 to 522 nm and 555 to 700 nm, respectively. Images were obtained using the ZEN 2008 software (Zeiss).
EST Libraries
For the Aeschynomene afraspera and Aeschynomene indica EST libraries, total RNA was extracted from uninoculated roots and from mature nodules (14 dpi with Bradyrhizobium spp. strain ORS285) using the RNeasy plant mini kit (Qiagen). The root and nodule EST libraries were constructed using the Creator Smart cDNA Library kit (Clontech). For each library, 15,000 clones were randomly processed through the Genoscope genomic and robotic platform for single-read sequencing from the 5′ end (www.genoscope.cns.fr). For A. afraspera, 9,492 and 9,582 sequences were obtained from roots and nodules, respectively, and 9,621 and 9,847 sequences were obtained for A. indica (http://esttik.cirad.fr).
In Situ Hybridization
A. indica and A. afraspera nodules were harvested at 14 dpi and fixed immediately with 4% (w/v) paraformaldehyde containing 0.1% (w/v) Tween 20 and 0.1% (w/v) Triton X-100 at 4°C overnight. Nodule tissues were dehydrated by incubation in increasing concentrations of ice-cold ethanol series. They were then embedded in paraffin blocks (Javelle et al., 2011). Sections (7 μm thick) were cut with a microtome (RM2155; Leica). Sections were adhered on poly-l-Lys-coated slides, and paraffin was subsequently removed with Histoclear and Histoclear/ethanol series.
To produce the hybridization probes, PCR products of approximately 200 bp of the genes of interest were cloned into the pGEM-T Easy plasmid. The inserts were then amplified by PCR using the vector-specific T7 and SP6 primers flanking the insert. These primers correspond to the bacteriophage SP6 and T7 RNA polymerase promoters. RNA probes, labeled with digoxigenin, were obtained by in vitro transcription using the PCR product as a template and the T7 RNA polymerase of the DIG RNA labeling kit (SP6/T7; Roche). Antisense probes were used to detect the location of transcripts of interest, and sense probes were used as negative controls. A control without probe was also performed. Prehybridization, hybridization, and posthybridization treatments and mounting were done by a method derived from the procedure described by Javelle et al. (2011). To reveal the hybridization signal, an anti-digoxigenin antibody coupled to alkaline phosphatase was used with its substrate, 4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate, which forms a brownish precipitate. The slides were finally observed with a Reichert Polyvar epifluorescence microscope combined with a Retiga 2000 CCD camera (QImaging).
Proteome Analysis
Crude protein extracts of nodules, bacteroids, and cultured Bradyrhizobium sp. ORS285 were half-diluted with Novex Tricine SDS sample buffer (2×). These samples were reduced (5 mm tributylphosphine, 30 min at 60°C) and alkylated (50 mm iodoacetamide, 30 min in the dark at room temperature) before the analyses by Novex 10% to 20% (w/v) Tricine Protein Gels (Life Technologies). Proteins in the range of approximately 4 to 9 kD (indicated with the dashed line in Supplemental Fig. S3) from free-living bacteria and bacteroid extracts were cut out from Tricine SDS-PAGE gels and identified by liquid chromatography-tandem mass spectrometry analysis (for detailed procedure, see Supplemental Text S1).
DNF1 Silencing
The A. evenia ssp. serrulata DNF1 homolog was identified from the transcriptome reference library. A BLAST search using the Medicago spp. signal peptidase protein sequence allowed the identification of a single candidate, CL5310_contig1. This cluster of 1,041 bp encodes a protein of 167 amino acids with 83% identity with MtDNF1 and was named AeDNF1 (Fig. 6A). For the RNAi strategy, a 283-bp internal fragment located between positions 16 to 298 of the AeDNF1 cDNA cluster sequence was PCR amplified from root cDNA using specific primers (Supplemental Table S5). After insertion into pENTR through TOPO cloning (Invitrogen), the fragment was introduced into the binary pK7GWIWG2_II-RedRoot vector (Plant Genetic System) by Gateway technology to generate the hairpin construct. The construct was verified by PCR and DNA sequencing before introduction into the Agrobacterium rhizogenes ARqua1 strain for root transformation as described before (Arrighi et al., 2012). As a control, the same plasmid containing a GUS gene fragment was used. Transformed roots were selected by their red fluorescence under UV light resulting from the constitutive expression of the red fluorescent protein, and one root was conserved per plant. To assess the AeDNF1 extinction level, quantitative reverse transcription-PCR was performed.
Real-Time Quantitative PCR Expression Analyses
Total RNA was extracted from roots or nodules at different time points after inoculation using the SV Total RNA Isolation System (Promega) and quantified using a NanoDrop ND-1000 spectrophotometer. Two hundred nanograms per sample was reverse transcribed using SuperScriptII reverse transcriptase (Invitrogen) and oligo(dT)12–18 primers. Real-time quantitative PCR was performed by using the Brilliant II SYBR Green QPCR Master Mix (Agilent Technologies). The primers used for quantitative PCR are provided in Supplemental Table S5. The real-time SYBR Green cycling PCR program on a Stratagene MX3005P (Agilent Technologies) was as follows: one cycle at 95°C for 10 min; 40 cycles at 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s; followed by one cycle at 95°C for 1 min, 55°C for 30 s, and 95°C for 30s. Reactions were performed in triplicate, and the efficiency-corrected comparative quantification method was used to quantify gene expression (Pfaffl, 2001). The results were standardized with AeEF1α expression levels.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FO981374 to FO99999 and LO000008 to LO016971.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Distribution of bacteroid morphotypes among legumes.
Supplemental Figure S2. Nodule-specific expression of NCR genes in A. evenia.
Supplemental Figure S3. NCR peptides colocalize with bacteroids in A. afraspera and A. indica nodules.
Supplemental Figure S4. Change in the bacterial cell shape during the symbiotic interaction between Bradyrhizobium spp. strain ORS285 and A. indica.
Supplemental Figure S5. In vitro effect of synthetic Aeschynomene spp. and Medicago spp. NCR peptides.
Supplemental Figure S6. Neighbor-joining phylogeny of NCR protein sequences.
Supplemental Table S1. A. afraspera and A. indica root and nodule ESTs and cluster collection statistics.
Supplemental Table S2. Nodule-specific Cys-rich putative peptides from A. indica, A. afraspera, and A. evenia.
Supplemental Table S3. Protein identification by mass spectrometry in ORS285 free-living state and ORS285 bacteroids isolated from A. indica and A. afraspera nodules.
Supplemental Table S4. Accessions and origins of the Aeschynomene spp. used in this study.
Supplemental Table S5. Primer pairs used in this study and their utilization.
Supplemental Text S1. Supplemental materials and methods.
Supplementary Material
Glossary
- IRLC
Inverted Repeat-Lacking Clade
- NCR
nodule-specific cysteine-rich
- SPC
signal peptidase complex
- dpi
days post inoculation
- PI
propidium iodide
- cDNA
complementary DNA
- RT-qPCR
reverse transcription-quantitative PCR
- RNAi
RNA interference
- DAPI
4′,6-diamidino-2-phenylindole
Footnotes
This work was supported by the French National Research Agency (grant nos. ANR–SESAM–2010–BLAN–170801 and ANR–BugsInaCell–13–BSV7–0013).
References
- Alunni B, Kevei Z, Redondo-Nieto M, Kondorosi A, Mergaert P, Kondorosi E (2007) Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol Plant Microbe Interact 20: 1138–1148 [DOI] [PubMed] [Google Scholar]
- Arrighi JF, Cartieaux F, Brown SC, Rodier-Goud M, Boursot M, Fardoux J, Patrel D, Gully D, Fabre S, Chaintreuil C, et al. (2012) Aeschynomene evenia, a model plant for studying the molecular genetics of the nod-independent rhizobium-legume symbiosis. Mol Plant Microbe Interact 25: 851–861 [DOI] [PubMed] [Google Scholar]
- Arrighi JF, Chaintreuil C, Cartieaux F, Cardi C, Rodier-Goud M, Brown SC, Boursot M, D’Hont A, Dreyfus B, Giraud E (2014) Radiation of the Nod-independent Aeschynomene relies on multiple allopolyploid speciation events. New Phytol 201: 1457–1468 [DOI] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Eschstruth A, Cayrel A, Guefrachi I, Mergaert P, Wen J, Jean V, Mysore KS, Gourion B, et al. (2014) A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol 203: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Bonaldi K, Gargani D, Prin Y, Fardoux J, Gully D, Nouwen N, Goormachtig S, Giraud E (2011) Nodulation of Aeschynomene afraspera and A. indica by photosynthetic Bradyrhizobium sp. strain ORS285: the nod-dependent versus the nod-independent symbiotic interaction. Mol Plant Microbe Interact 24: 1359–1371 [DOI] [PubMed] [Google Scholar]
- Branca A, Paape TD, Zhou P, Briskine R, Farmer AD, Mudge J, Bharti AK, Woodward JE, May GD, Gentzbittel L, et al. (2011) Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc Natl Acad Sci USA 108: E864–E870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro L, Pujic P, Alloisio N, Fournier P, Boubakri H, Hay AE, Poly F, François P, Hocher V, Mergaert P, et al. (2015) Alnus peptides modify membrane porosity and induce the release of nitrogen-rich metabolites from nitrogen-fixing Frankia. ISME J 9: 1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaintreuil C, Arrighi JF, Giraud E, Miché L, Moulin L, Dreyfus B, Munive-Hernández JA, Villegas-Hernandez MdelC, Béna G (2013) Evolution of symbiosis in the legume genus Aeschynomene. New Phytol 200: 1247–1259 [DOI] [PubMed] [Google Scholar]
- Chandler MR, Date RA, Roughley RJ (1982) Infection and root-nodule development in Stylosanthes species by Rhizobium. J Exp Bot 33: 47–57 [Google Scholar]
- Giraud E, Hannibal L, Fardoux J, Verméglio A, Dreyfus B (2000) Effect of Bradyrhizobium photosynthesis on stem nodulation of Aeschynomene sensitiva. Proc Natl Acad Sci USA 97: 14795–14800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC, Jaubert M, Simon D, Cartieaux F, Prin Y, et al. (2007) Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316: 1307–1312 [DOI] [PubMed] [Google Scholar]
- Guefrachi I, Nagymihaly M, Pislariu CI, Van de Velde W, Ratet P, Mars M, Udvardi MK, Kondorosi E, Mergaert P, Alunni B (2014) Extreme specificity of NCR gene expression in Medicago truncatula. BMC Genomics 15: 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guefrachi I, Pierre O, Timchenko T, Alunni B, Barriere Q, Czernic P, Villaécija-Aguilar JA, Verly C, Bourge M, Fardoux J, et al. (July 8, 2015) Bradyrhizobium BclA is a peptide transporter required for bacterial differentiation in symbiosis with Aeschynomene legumes. Mol Plant Microbe Interact http://dx.doi.org/10.1094/MPMI-04-15-0094-R [DOI] [PubMed] [Google Scholar]
- Haag AF, Arnold MF, Myka KK, Kerscher B, Dall’Angelo S, Zanda M, Mergaert P, Ferguson GP (2013) Molecular insights into bacteroid development during Rhizobium-legume symbiosis. FEMS Microbiol Rev 37: 364–383 [DOI] [PubMed] [Google Scholar]
- Ibáñez F, Fabra A (2011) Rhizobial Nod factors are required for cortical cell division in the nodule morphogenetic programme of the Aeschynomeneae legume Arachis. Plant Biol (Stuttg) 13: 794–800 [DOI] [PubMed] [Google Scholar]
- Javelle M, Marco CF, Timmermans M (2011) In situ hybridization for the precise localization of transcripts in plants. J Vis Exp 23: e3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannenberg EL, Perzl M, Härtner T (1995) The occurrence of hopanoid lipids in Bradyrhizobium bacteria. FEMS Microbiol Lett 127: 255–262 [DOI] [PubMed] [Google Scholar]
- Kondorosi E, Mergaert P, Kereszt A (2013) A paradigm for endosymbiotic life: cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu Rev Microbiol 67: 611–628 [DOI] [PubMed] [Google Scholar]
- Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK (2009) D-Amino acids govern stationary phase cell wall remodeling in bacteria. Science 325: 1552–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Login FH, Balmand S, Vallier A, Vincent-Monégat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A (2011) Antimicrobial peptides keep insect endosymbionts under control. Science 334: 362–365 [DOI] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, Mausset AE, Barloy-Hubler F, Galibert F, Kondorosi A, et al. (2006) Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA 103: 5230–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, de Lajudie P, Masson-Boivin C (1999) Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol 65: 3084–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Takebe I (1970) Cell wall regeneration and cell division in isolated tobacco mesophyll protoplasts. Planta 92: 301–308 [DOI] [PubMed] [Google Scholar]
- Oono R, Schmitt I, Sprent JI, Denison RF (2010) Multiple evolutionary origins of legume traits leading to extreme rhizobial differentiation. New Phytol 187: 508–520 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard R, Lambinon J, Reekmans M, Van der Veken P, Govaert M (1983) Nombres chromosomiques de quelques Angiospermes du Rwanda, du Burundi et du Kenya. Bull Jard Bot Nat Belg 53: 342–371 [Google Scholar]
- Sen D, Weaver RW (1984) A basis for different rates of N2-fixation by the same strains of Rhizobium in peanut and cowpea root nodules. Plant Sci Lett 34: 239–246 [Google Scholar]
- Shigenobu S, Stern DL (2013) Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc Biol Sci 280: 20121952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A, Vitiello G, Gully D, Sturiale L, Chaintreuil C, Fardoux J, Gargani D, Lee HI, Kulkarni G, Busset N, et al. (2014) Covalently linked hopanoid-lipid A improves outer-membrane resistance of a Bradyrhizobium symbiont of legumes. Nat Commun 5: 5106. [DOI] [PubMed] [Google Scholar]
- Sinharoy S, Torres-Jerez I, Bandyopadhyay K, Kereszt A, Pislariu CI, Nakashima J, Benedito VA, Kondorosi E, Udvardi MK (2013) The C2H2 transcription factor REGULATOR OF SYMBIOSOME DIFFERENTIATION represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula. Plant Cell 25: 3584–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiricz H, Szucs A, Farkas A, Pap B, Lima RM, Maróti G, Kondorosi É, Kereszt A (2013) Antimicrobial nodule-specific cysteine-rich peptides induce membrane depolarization-associated changes in the transcriptome of Sinorhizobium meliloti. Appl Environ Microbiol 79: 6737–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z, Farkas A, Mikulass K, Nagy A, Tiricz H, et al. (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Vincent JM. (1970) A Manual for the Practical Study of Root-Nodule Bacteria. Blackwell Scientific Publications, Oxford [Google Scholar]
- Wang D, Griffitts J, Starker C, Fedorova E, Limpens E, Ivanov S, Bisseling T, Long S (2010) A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 327: 1126–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. (2003) Bacterial shape. Mol Microbiol 49: 571–580 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.