The comparative study of the gene targets of the KANADI1 transcription factor indicates that it is part of a basic growth-promoting module.
Abstract
An intricate network of antagonistically acting transcription factors mediates the formation of a flat leaf lamina of Arabidopsis (Arabidopsis thaliana) plants. In this context, members of the class III homeodomain leucine zipper (HD-ZIPIII) transcription factor family specify the adaxial domain (future upper side) of the leaf, while antagonistically acting KANADI transcription factors determine the abaxial domain (future lower side). Here, we used a messenger RNA sequencing approach to identify genes regulated by KANADI1 (KAN1) and subsequently performed a meta-analysis combining our data sets with published genome-wide data sets. Our analysis revealed that KAN1 acts upstream of several genes encoding auxin biosynthetic enzymes. When exposed to shade, we found three YUCCA genes, YUC2, YUC5, and YUC8, to be transcriptionally up-regulated, which correlates with an increase in the levels of free auxin. When ectopically expressed, KAN1 is able to transcriptionally repress these three YUC genes and thereby block shade-induced auxin biosynthesis. Consequently, KAN1 is able to strongly suppress shade-avoidance responses. Taken together, we hypothesize that HD-ZIPIII/KAN form the basis of a basic growth-promoting module. Hypocotyl extension in the shade and outgrowth of new leaves both involve auxin synthesis and signaling, which are under the direct control of HD-ZIPIII/KAN.
A fundamental question in plant developmental biology is how plant organs achieve their final form. Leaves of flowering plants are so-called lateral organs that initiate from small populations of founder cells in the periphery of the shoot apical meristem. The initiation and proper spacing of leaves around the shoot apex are mediated by polar auxin transport (Reinhardt et al., 2000, 2003). Once initiated, polarity axes (proximodistal, dorsoventral, and mediolateral) are established, guiding the fast-dividing primordia cells in order for the leaf to attain its final shape (Hudson, 2000). A complex network of transcription factors and small RNAs acts to divide the leaf primordium along the dorsoventral axis into distinct zones: (1) the adaxial zone producing cells and tissues that will form the upper part of the leaf blade; (2) the abaxial zone that will form the lower side of the leaf blade (Byrne, 2006); and (3) the middle domain required for blade outgrowth (Nakata et al., 2012). It is important to note that, besides the molecular framework that is required for proper leaf initiation and development, the environment strongly influences organ shape and physiology. The latter is exemplified in shade, where the petiole elongates to allow better spacing between the light-capturing leaf blades (Kozuka et al., 2005); increased stomata density in response to elevated CO2 levels (Woodward, 1987); and decreased leaf size in response to cold temperature (Gurevitch, 1992).
Members of the plant-specific class III homeodomain leucine zipper (HD-ZIPIII) transcription factor family act as major regulators of adaxial leaf development (McConnell et al., 2001; Emery et al., 2003). HD-ZIPIII mRNAs are highly expressed in the adaxial domain and absent in the abaxial domain. This expression pattern is achieved by a gradient of microRNAs, miR165/6, functioning in opposite directions (Emery et al., 2003; Juarez et al., 2004; Mallory et al., 2004). KANADI (KAN), transcription factors of the GARP family, literally mirror HD-ZIPIII expression and are most abundant in abaxial tissue (Kerstetter et al., 2001; Emery et al., 2003). HD-ZIPIII and KAN act antagonistically, thus maintaining a stable dorsoventral axis that allows proper outgrowth of the leaf blade. Besides their complementary patterns of expression, HD-ZIPIII and KAN also exhibit opposite biological activities; whereas HD-ZIPIIIs mostly function as transcriptional activators, KAN proteins seem to predominantly act as transcriptional repressors. Recently, direct target genes of HD-ZIPIII protein, REVOLUTA (REV) and KAN1, have been identified (Brandt et al., 2012; Merelo et al., 2013; Reinhart et al., 2013; Huang et al., 2014). These genome-wide screens revealed that, besides their opposite expression patterns and biological activities, the HD-ZIPIII/KAN antagonism is also manifested in the opposite regulation of a set of shared target genes (Brandt et al., 2012; Merelo et al., 2013; Reinhart et al., 2013).
In addition to the determination of polarity in the early leaf, KAN1 plays additional roles in other polarity setup processes in the ovule, vasculature, and root (Hawker and Bowman, 2004; Ilegems et al., 2010; Kelley et al., 2012). We recently discovered that, in addition to the basic patterning function of the HD-ZIPIII/KAN module, both gene families seem to be also required for adaptive developmental processes. Both REV and KAN1 impinge on a set of genes known to be required for shade-dependent growth initiation. These genes comprise components of the auxin biosynthesis machinery and transcription factors of the class II HD-ZIP (HD-ZIPII) family (Bou-Torrent et al., 2012; Brandt et al., 2012). Furthermore, HD-ZIPIIs are also expressed in the adaxial domain and, together with HD-ZIPIIIs, promote adaxial cell fate (Brandt et al., 2012; Turchi et al., 2013).
Auxin is required for both the initiation and polarization of leaf primordia. Our previous studies revealed that two genes encoding auxin biosynthetic enzymes, TRYPTOPHAN AMINO TRANSFERASE OF ARABIDOPSIS1 (TAA1) and YUCCA5 (YUC5), are direct and negative KAN1 targets (Brandt et al., 2012; Merelo et al., 2013). Moreover, in kan mutant plants, members of the PINFORMED (PIN) family of proteins that encode auxin efflux carrier show a disturbed localization pattern, indicating that one of the functions of KAN proteins is to regulate auxin transport (Eshed et al., 2001, 2004). These findings indicate that both auxin synthesis and transport are regulated by KAN1. Recently published genome-wide approaches to isolate KAN1 target genes confirmed the regulation of genes encoding components of auxin biosynthesis and transport and further revealed that a number of factors involved in transducing auxin signals, such as members of the INDOLE-3-ACETIC ACID INDUCIBLE (IAA), AUXIN RESPONSE FACTOR, and NONPHOTOTROPIC HYPOCOTYL (NPH)-like families of proteins, also are potentially under the direct regulation by KAN1 (Merelo et al., 2013; Reinhart et al., 2013; Huang et al., 2014).
Here, we used an additional messenger RNA sequencing (mRNA-Seq) approach to characterize genes regulated by KAN1 in Arabidopsis (Arabidopsis thaliana). Transgenic plants expressing 35S::FLAG-GR-KAN1 were exposed to either mock treatment or dexamethasone (DEX) to induce KAN1 release from its cytoplasmic blockage. Illumina sequencing of mRNAs isolated from these plants in comparison with wild-type plants revealed approximately 1,000 transcripts that change significantly in expression in response to KAN1 induction. We employed a meta-analysis comparing this new data set of KAN1-regulated genes with three recently published data sets that used chromatin immunoprecipitation sequencing (ChIP-Seq), DNA-tiling arrays, or DNA microarrays and identified a set of 72 high-confidence KAN1 targets. Because our previous work suggested that KAN1 has an additional role in the shade-avoidance response, we also performed RNA-Seq in simulated shade conditions. Here, we determined shade-regulated transcripts in Columbia-0 (Col-0) wild-type plants and discovered that KAN1 antagonizes shade growth by repressing a large number of genes encoding auxin biosynthesis and signaling components. Determination of free auxin levels in shade revealed that KAN1 represses auxin production, which strongly inhibits shade-avoidance responses in transgenic plants misexpressing KAN1 at high levels. We conclude that the module of HD-ZIPIII/KAN transcription factors that inter alia patterns young leaf primordia forms the basis of a basic growth-promoting module.
RESULTS
Comparative Analysis of Gene Expression Profiling Data Sets Using an Inducible Version of the KAN1 Protein
Constitutive overexpression of the KAN1 protein causes severe developmental defects and has led to the development of inducible systems of KAN1 induction using the rat glucocorticoid receptor (GR; Brandt et al., 2012; Merelo et al., 2013; Reinhart et al., 2013; Huang et al., 2014). Plants that constitutively overexpress the GR-KAN1 fusion protein accumulate high levels of protein in the cytoplasm, which, upon DEX exposure, translocates to the nucleus and induces KAN1 target genes. Using this inducible system, microarray-based expression profiling experiments have been carried out with the goal to identify genes regulated by KAN1 (Merelo et al., 2013; Reinhart et al., 2013; Huang et al., 2014). Complementary to these approaches, we generated transgenic plants overexpressing GR-KAN1 with an additional N-terminal FLAG epitope (35S::FLAG-GR-KAN1) and performed ChIP-Seq studies to identify genomic regions bound by the KAN1 transcription factor (Merelo et al., 2013). These approaches resulted in the identification of approximately 500 genes that change in expression in response to KAN1 induction and approximately 3,000 genes for which a significant enrichment of KAN1-bound chromatin was identified in the proximal promoter region.
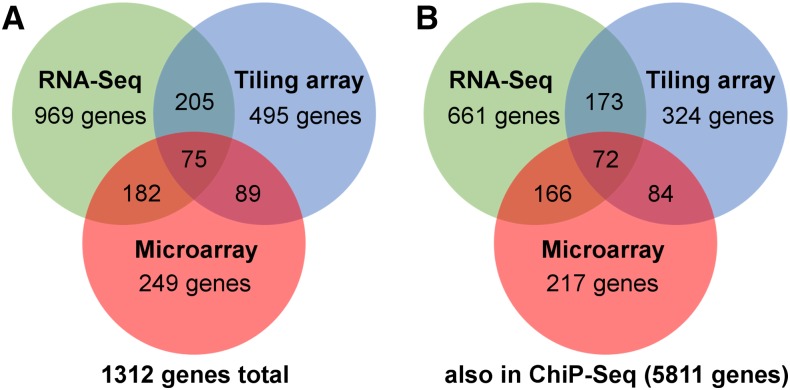
To enhance our understanding of the function of KAN1, it is important to better define its direct targets. Knowing the nature of these direct targets will allow us to predict how KAN1 functions in patterning and adaptive growth processes. So far, all expression profiling approaches relied on microarray-based systems. In order to identify transcription units not represented on these expression arrays or genes expressed at low levels precluding previous identification, we performed KAN1 induction experiments followed by Illumina mRNA sequencing. Wild-type and transgenic 35S::FLAG-GR-KAN1 plants were grown for 10 d in white-light conditions. The two genotypes were then treated with either a mock solution (0.5% [v/v] ethanol) or with a 50 μm DEX solution for 90 min. Altogether, we collected two biological replicates for each genotype and treatment, isolated RNA, constructed sequencing libraries, and sequenced these on the Illumina HiSeq platform. In total, approximately 550 million paired-end reads were produced, and each sample contained above 90% correctly aligned read pairs. Comparative mRNA-Seq analysis of genes whose expression significantly decreases in response to KAN1 induction resulted in the identification of 969 transcripts (Fig. 1A; Supplemental Data Set S1). When compared with the ChIP-Seq data set, we can identify 661 genes (corresponding to 66%) being potentially directly regulated (Fig. 1B; Supplemental Data Set S1). When compared with the tiling array gene expression profiling data (Merelo et al., 2013) and the microarray data (Reinhart et al., 2013), 75 genes can be identified that are down-regulated in all three data sets (Fig. 1A; Supplemental Data Set S1). A further focus on only the genes that are also bound by KAN1 in the ChIP-Seq data set reveals a total of 72 genes that are bound and down-regulated by KAN1 (Fig. 1B; Table I). It is reasonable to assume that many of these 661 genes that were identified in our combinatory analysis can be directly regulated by KAN1 and, thus, may perform functions downstream of KAN1.
Figure 1.
Comparison of available genome-wide data sets aiming at the identification of genes down-regulated by KAN1. A, Venn diagram showing genes regulated by RNA-Seq from this study, DNA tiling array (Merelo et al., 2013), and DNA microarray (Reinhart et al., 2013). B, Venn diagram showing the numbers of genes that are potentially directly regulated by KAN1.
Table I. The 72 high-confidence KAN1 targets identified through meta-analysis.
Microarray data were retrieved from Reinhart et al. (2013).
| Arabidopsis Genome Initiative Code | Annotation | ChIP-Seq Binding Site | RNA-Seq | Microarray |
|---|---|---|---|---|
| kb to gene | fold change | |||
| At1g01120 | KCS1 | −2.3 | −0.1 | 0.14 | 0.36 |
| At1g01140 | CIPK9 | −0.9 | −0.4 | 0.42 | 0.37 |
| At1g01490 | HMT superfamily | −1.0 | −0.3 | 0.0 | 0.5 | 0.24 | 0.26 |
| At1g21590 | Protein kinase | −0.5 | 0.19 | 0.37 |
| At1g23090 | AST91 | −4.6 | −3.1 | 0.15 | 0.33 |
| At1g25550 | Myb-like transcription factor family | −0.3 | 0.5 | 0.25 | 0.19 |
| At1g25560 | TEMPRANILLO1 (TEM1) | −3.1 | −2.1 | −0.9 | 3.0 | 0.32 | 0.23 |
| At1g28660 | GDSL-like superfamily | −1.0 | −0.2 | 0.47 | 0.39 |
| At1g33240 | AT-GTL1 | 0.2 | 0.54 | 0.48 |
| At1g35350 | PHO1-H8 | −1.8 | −0.2 | 0.43 | 0.23 |
| At1g51805 | LRR kinase family | −0.4 | 0.07 | 0.08 |
| At1g52290 | PERK15 | −0.1 | 0.16 | 0.14 |
| At1g61660 | BHLH112 | −10.7 | −3.2 | −1.4 | 0.43 | 0.46 |
| At1g66840 | PMI2 | −0.2 | 0.13 | 0.26 |
| At1g69760 | Unknown protein | −6.1 | −3.0 | −2.4 | 0.13 | 0.29 |
| At1g71880 | SUC1 | −10.1 | −5.9 | −5.5 | −4.6 | 0.09 | 0.46 |
| At1g72450 | JAZ6 | −2.1 | −1.3 | 0.27 | 0.22 |
| At1g76990 | ACR3 | −3.6 | −1.3 | 0.33 | 0.50 |
| At2g01420 | PIN4 | −0.4 | 0.9 | 0.16 | 0.21 |
| At2g02950 | PKS1 | −0.3 | 0.29 | 0.21 |
| At2g16400 | BLH7 | −0.4 | 0.34 | 0.33 |
| At2g17820 | ATHK1 | −0.2 | 0.18 | 0.31 |
| At2g21210 | SAUR6 | −0.2 | 0.58 | 0.45 |
| At2g27050 | EIL1 | 0.4 | 0.50 | 0.61 |
| At2g30520 | RPT2 | −3.2 | −1.0 | 0.61 | 0.44 |
| At2g30990 | Unknown protein | −0.9 | −0.2 | 0.44 | 0.39 |
| At2g31070 | TCP10 | −0.1 | 0.55 | 0.23 |
| At2g38310 | PYL4 | 0.6 | 1.2 | 0.40 | 0.46 |
| At2g39360 | Protein kinase superfamily | −0.3 | 2.2 | 0.44 | 0.29 |
| At2g40270 | Protein kinase superfamily | −0.8 | −0.2 | 0.9 | 0.38 | 0.37 |
| At2g41940 | ZFP8 | −3.9 | −2.9 | 1.6 | 0.16 | 0.23 |
| At2g42690 | α/β-Hydrolase superfamily | −0.2 | 0.29 | 0.51 |
| At2g43820 | GT | −1.0 | 0.56 | 0.34 |
| At3g05120 | ATGID1A | −1.7 | −0.6 | −0.1 | 0.30 | 0.42 |
| At3g06750 | Hyp-rich glycoprotein family | −1.8 | −0.7 | 0.0 | 0.9 | 0.26 | 0.30 |
| At3g13110 | ATSERAT2 | −3.1 | 0.44 | 0.43 |
| At3g15570 | NPH3 family protein | −1.1 | 0.11 | 0.07 |
| At3g19850 | NPH3 family protein | −1.0 | 0.04 | 0.03 |
| At3g19930 | STP4 | −1.7 | −0.9 | 0.40 | 0.50 |
| At3g23820 | GAE6 | −9.4 | −8.0 | −3.9 | −2.5 | −1.3 | 0.38 | 0.48 |
| At3g49220 | PME34 | −0.2 | 0.0 | 0.0 | 0.15 | 0.26 |
| At3g55560 | AGF2 | −1.1 | 0.32 | 0.46 |
| At3g58120 | ATBZIP61 | −1.2 | 0.46 | 0.42 |
| At3g61460 | BRASSINOSTEROID-RESPONSIVE RING-H2 (BRH1) | −2.3 | −0.4 | 0.40 | 0.29 |
| At4g16980 | Arabinogalactan protein family | −0.4 | 0.3 | 0.34 | 0.41 |
| At4g16990 | RLM3 | −2.0 | 0.19 | 0.26 |
| At4g18010 | IP5PII | −9.0 | −7.3 | −6.1 | −3.5 | −2.2 | 0.0 | 0.36 | 0.14 |
| At4g18340 | Glycosyl hydrolase superfamily | −1.5 | 0.30 | 0.27 |
| At4g22190 | Unknown protein | −3.0 | −1.8 | 0.30 | 0.32 |
| At4g24060 | Zinc finger family protein | −0.1 | 1.5 | 0.31 | 0.50 |
| At4g24660 | ATHB22 | −1.5 | −0.9 | 0.22 | 0.20 |
| At4g25620 | Hyp-rich glycoprotein family | −3.5 | −1.7 | 0.0 | 0.25 | 0.24 |
| At4g25990 | CIL | −3.0 | −1.9 | 0.12 | 0.18 |
| At4g27300 | SD11 | −3.4 | −0.3 | 0.20 | 0.19 |
| At4g33050 | EDA39 | 0.0 | 2.8 | 0.08 | 0.22 |
| At4g34220 | LRR protein kinase family | −1.5 | −1.0 | 0.05 | 0.12 |
| At4g37590 | NPY5 | −2.0 | −1.5 | 0.8 | 0.50 | 0.62 |
| At4g38840 | SAUR14 | −0.3 | 0.48 | 0.32 |
| At5g03150 | JACKDAW (JKD) | −3.1 | 0.22 | 0.28 |
| At5g09850 | MED26C | 0.0 | 0.46 | 0.59 |
| At5g11090 | Ser-rich protein-related | 1.6 | 2.3 | 4.3 | 0.41 | 0.35 |
| At5g17860 | CAX7 | −1.1 | −0.2 | 0.0 | 0.7 | 0.17 | 0.08 |
| At5g28300 | GT2L | −0.3 | 0.7 | 0.44 | 0.36 |
| At5g40450 | Unknown protein | −5.1 | −3.6 | −2.7 | −1.5 | −0.2 | 0.24 | 0.41 |
| At5g47370 | HAT2 | −0.7 | −0.1 | 0.35 | 0.44 |
| At5g47560 | ATTDT | −2.0 | −0.9 | 0.0 | 0.64 | 0.29 |
| At5g52900 | MAKR6 | −0.8 | 2.3 | 4.4 | 0.07 | 0.21 |
| At5g54250 | ATCNGC4 | −1.5 | 0.4 | 0.27 | 0.18 |
| At5g59780 | MYB59 | −6.0 | −2.6 | −1.3 | 0.36 | 0.32 |
| At5g61590 | ERF107 | −0.7 | −0.2 | 0.08 | 0.03 |
| At5g63410 | LRR protein kinase family | −1.3 | −0.2 | 0.22 | 0.30 |
| At5g67440 | NPY3 | −0.5 | 0.0 | 0.1 | 0.36 | 0.48 |
We also performed a comparative analysis of our RNA-Seq data and the published microarray and tiling array data with a recently published study where the authors treated wild-type and transgenic 35S::GR-KAN1 plants with cycloheximide (CHX) prior to DEX treatment and RNA isolation (Huang et al., 2014). This latter analysis resulted in the identification of 231 differentially expressed transcripts in response to GR-KAN1 induction. CHX was used to block protein synthesis, with the aim of enriching for transcripts under the direct regulation of GR-KAN1. Comparative analysis of the CHX data set with our RNA-Seq data sets and the recently published tiling array data set eliminated approximately 90% of all regulated genes. Of the 72 potential directly regulated genes identified in this study, only 24 genes are regulated in the presence of CHX (Supplemental Fig. S1). We suspect that the addition of CHX and the combination of CHX and DEX did not aid in the identification of KAN1 direct targets but rather produced artifacts that hampered the identification of real direct targets. This is especially apparent in the case of the best-known direct KAN1 target gene, ASYMMETRIC LEAVES2 (AS2; Wu et al., 2008), which we can identify in our ChIP-Seq and RNA-Seq data sets but is absent from the CHX data set (Huang et al., 2014).
Classification of Genes Underlying Potential Direct KAN1 Regulation
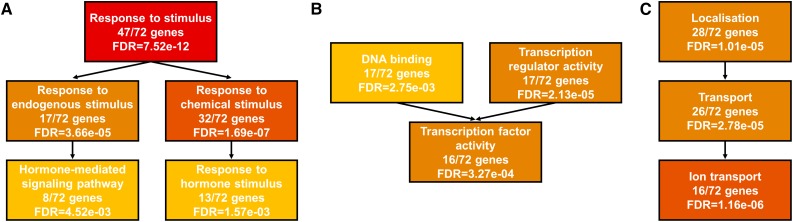
As reported previously, KAN1 induction causes profound changes in genes encoding components of the auxin signaling machinery from synthesis and transport to signal dissipation. We used the agriGO tool (Du et al., 2010) to search for enriched Gene Ontology (GO) terms in the set comprising the 72 potential high-confidence direct targets. This analysis revealed that the response to stimulus term is significantly enriched, and besides the PIN4 auxin transporter, many genes encoding auxin-related SMALL AUXIN UP-REGULATED (SAUR) and NPH3-type proteins are potential direct KAN1 targets (Fig. 2A). In addition, the second-strongest enriched term is DNA-binding/transcriptional regulation, because several of the downstream target genes encode for transcriptional regulators (Fig. 2B). Besides these two obvious GO terms, we also found enrichment for the term transport processes and several genes encoding ion transporter, which seem to underlie direct KAN1 regulation (Fig. 2C; Table I). It might be important to note that sugar transport also seems to be negatively regulated by KAN1. Taken together, KAN1 might act by regulating not only transcription factors and hormone biosynthesis but also ion and sugar homeostasis, thus strongly influencing the cellular environment.
Figure 2.
GO analysis of KAN1 targets. Analysis of the 72 potentially direct KAN1 targets reveals a strong enrichment for genes involved in response to stimuli (A), transcription factors (B), and localization and transport processes (C). FDR, False discovery rate.
Identification of cis-Elements Responsible for KAN1 DNA Binding
The analysis of ChIP-Seq data yielded the identification of a possible KAN1-binding site (Merelo et al., 2013). Using MEME-ChIP (Machanick and Bailey, 2011), high-confidence ChIP-Seq peaks were analyzed and the (A/G/C)GAATA(T/A) motif was found to be enriched in those peaks. Using the purified KAN1 DNA-binding domain as bait in PCR-assisted in vitro DNA-binding site selection experiments (Huang et al., 2014), the slightly different but similar GNATA(A/T) motif was discovered as a potential KAN1 cis-element. However, since only the DNA-binding domain of KAN1 was used in these experiments, it cannot be excluded that the full-length KAN1 protein interacts with motifs that are different from this in vitro-identified element.
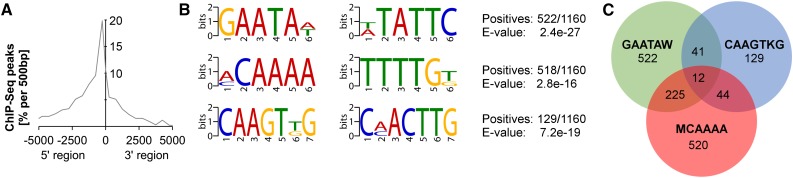
To identify the in vivo DNA-binding motif bound by KAN1, we exploited the power of high-throughput sequencing and compared our KAN1 ChIP-Seq and mRNA-Seq data sets. First, we selected peaks identified by ChIP-Seq that show at least 3-fold enrichment over the control sample. This analysis resulted in the identification of 4,183 peaks corresponding to 5,811 genes potentially regulated by KAN1 (Merelo et al., 2013). Analysis of the peak position relative to the potentially regulated transcription units using the combination of ChIP-Seq and RNA-Seq revealed that the majority of peaks (about 20%) are located in the first 500 bp upstream of the respective transcription starts (Fig. 3A). Our RNA-Seq analysis resulted in the identification of 661 genes that are transcriptionally responsive to KAN1 induction and also showed binding of KAN1 to chromatin regions that are in close proximity to these genes (Fig. 1B). In total, this analysis yielded the identification of 1,160 chromatin regions that are in close proximity to the 661 genes that are regulated by KAN1. We subjected these 1,160 sequences to MEME-ChIP (Machanick and Bailey, 2011) and identified three motifs that are significantly enriched in this data set: E1, GAATA(A/T); E2, (A/C)CAAAA; and E3, CAAGT(T/G)G (Fig. 3B). The finding that KAN1 interacts with chromatin regions containing the GAATA(A/T) motif is consistent with previous findings in which we found the (A/G/C)GAATA(T/A) element to be enriched in the ChIP-Seq data set (Merelo et al., 2013). The second element, E2, (A/C)CAAAA, is currently unknown. The third enriched element, E3, CAAGT(T/G)G, strongly resembles an E-box (CANNTG) and therefore might represent an enhancer element for KAN1 or an element recognized by a KAN1-interacting protein.
Figure 3.
Identification of cis-sequences identified by ChIP-Seq. A, Locations of KAN1-binding peaks identified by ChIP-Seq/RNA-Seq. B, Sequence logos of three potential KAN1-binding sites identified by MEME-ChIP analysis. C, Venn diagram showing the occurrence of three potential cis-elements in the promoters of the 661 RNA-Seq/ChIP-Seq KAN1 target genes.
Having identified these elements, we analyzed how many of the 661 potential direct KAN1 target genes contain one of these elements in the DNA region identified by ChIP-Seq. For several of the 661 target genes, binding to multiple genomic regions was observed, and in total, we could identify 1,160 genomic regions bound by KAN1. We found 522 of the 1,160 genomic regions to contain element E1, GAATA(A/T), in the central position of the peak, 520 peaks have element E2, (A/C)CAAAA, in the central position, and 129 have element E3, CAAGT(T/G)G, in the central position (Fig. 3C). We also realized that several peaks have combinations of the three identified elements in the central position of the peak, and the combination of GAATA(A/T) and (A/C)CAAAA is found in approximately one-half of all peaks having either GAATA(A/T) or (A/C)CAAAA. It is interesting that the KAN1-binding site identified in the AS2 promoter (Wu et al., 2008) is composed of an (A/C)CAAAA motif followed by two GAATA(A/T) motifs. This finding implies that both motifs might have relevance for the association of KAN1 with DNA. Therefore, we decided to experimentally test (1) whether KAN1 interacts with chromatin regions containing these motifs and (2) whether genes harboring such motifs in their respective promoters are transcriptionally regulated by KAN1.
Experimental Validation of Potential KAN1 Target Genes
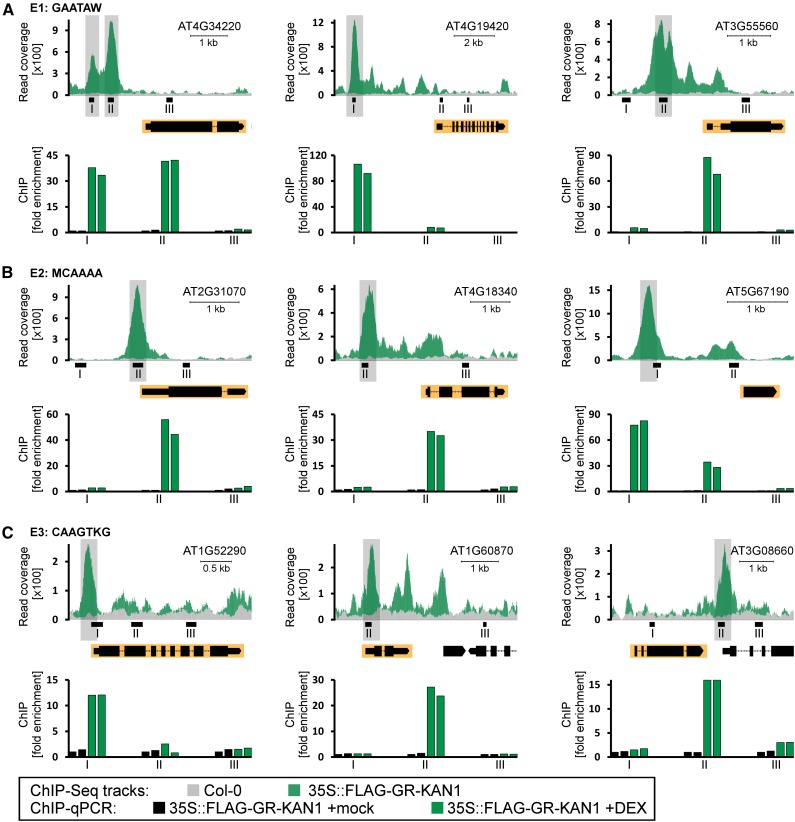
Previous studies have shown that KAN1 interacts with the GAATA(A/T) E1 cis-element, which is the most enriched element in our previous ChIP-Seq study (Merelo et al., 2013). In order to test in vivo that KAN1 associates with the newly identified elements in this study, we filtered the list of potential direct targets to identify those genes having only one of the three elements (E1, E2, or E3) in the central region of the ChIP-Seq peak in proximity to the gene. For each of those cases, we selected three potential target genes and performed individual chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) experiments to detect the binding of KAN1 to regions containing only the respective element. Candidate target genes having an E1, GAATA(A/T), element in their proximal promoter are At4g34220 (Leu-rich repeat protein kinase family protein), At4g19420 (pectinacetylesterase family protein), and At3g55560 (HOOK PROTEIN OF GA FEEDBACK2 and HOOK MOTIF NUCLEAR-LOCALIZED PROTEIN15). Target genes with E2 element, (A/C)CAAAA, are At2g31070 (TCP DOMAIN PROTEIN10), At4g18340 (glycosyl hydrolase superfamily protein), and At5g67190 (DREB AND EAR MOTIF PROTEIN2). E3, CAAGT(T/G)G, elements are found in promoters of the following candidate targets: At1g52290 (protein kinase superfamily protein), At1g60870 (MATERNAL EFFECT EMBRYO ARREST9), and At3g08660 (phototropic-responsive NPH3 family protein). For each of these genes, we selected three positions (I, II, and III) at which either strong or no enrichment was observed and tested by individual ChIP-qPCR whether KAN1 associates with the regions containing the respective elements (Fig. 4). For all the genes we analyzed, we detected significant binding of KAN1 to regions containing E1, E2, or E3 elements in their respective promoters, which supports the validity of our ChIP-Seq data set. These findings suggest that KAN1 associates with the chromatin of these target genes by interacting with the identified elements.
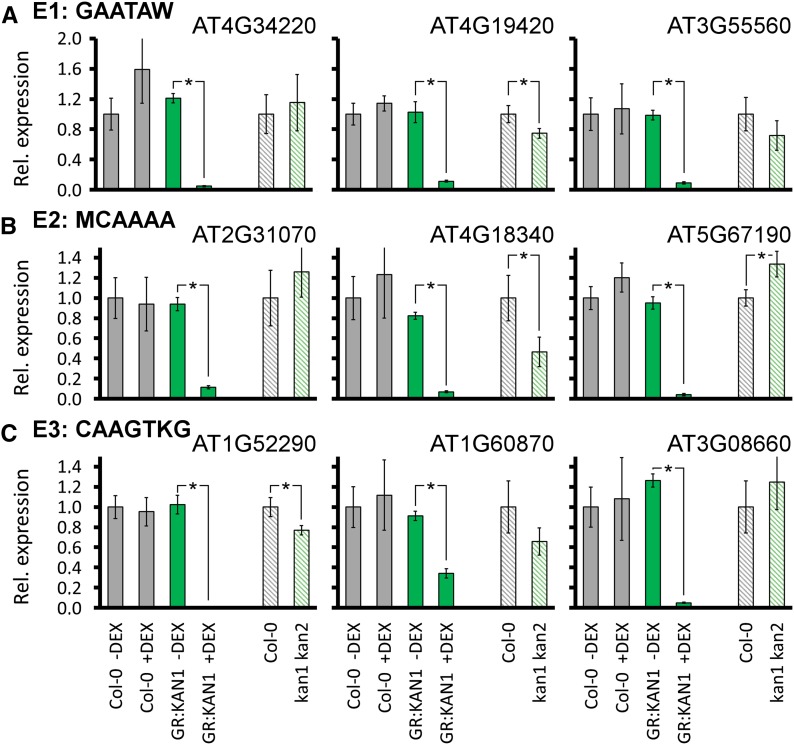
Figure 4.
Validation of KAN1 interacting with the E1, E2, and E3 elements of selected target genes. ChIP-qPCR analysis is shown for three KAN1 targets with the E1 element (GAATAW) in the promoter (A), three KAN1 targets with the E2-binding site (MCAAAA) in the proximal promoter (B), and three KAN1 targets with the E3 element (CAAGTKG) in the proximal promoter (C). Top rows depict the read coverage of the respective transcription units obtained from ChIP-Seq of Col-0 (gray) and 35S::FLAG-GR-KAN1 (green) plants. Gene models are shown under the ChIP-Seq tracks, and respective genes have a yellow background. Shaded peaks harbor the respective elements. Bottom rows show ChIP-qPCR experiments with two biological replicates for 35S::FLAG-GR-KAN1 plants that were mock treated (black bars) and 35S::FLAG-GR-KAN1 plants treated with DEX (green bars). Each genomic region was tested with three primer pairs (I–III). Primer pairs not present in the read coverage plots are located outside the depicted region. The y axis shows the fold enrichment normalized to the mock-treated immunoprecipitates.
We next tested whether these potential KAN1 target genes are also transcriptionally regulated by KAN1, so we performed quantitative reverse transcription (RT)-PCR experiments with Col-0 wild-type and 35S::FLAG-GR-KAN1 transgenic plants treated with either a mock or a DEX solution (Fig. 5). All selected genes harboring E1, E2, or E3 elements in their proximal promoters were strongly down-regulated in 35S::FLAG-GR-KAN1 transgenic plants in response to DEX application. Our results demonstrate that KAN1 is able to actively repress these genes. To understand whether these genes are also controlled by KAN1 in wild-type plants, we compared their levels of expression between the wild type and kan1 kan2 double mutant plants. We found diverging levels of expression in only four (At4g19420, At4g18340, At5g67190, and At1g52290) out of the nine genes that we analyzed. KAN1 acts as a strong transcriptional repressor of these nine candidate genes; therefore, we expected to find elevated levels of expression of these target genes in kan1 kan2 double mutants relative to wild-type plants. This, however, is the case for only At5g67190 encoding for the DEAR2 transcriptional regulator (Fig. 5B). Three more genes have slightly elevated levels of expression in kan1 kan2 double mutants relative to wild-type plants, but these differences are not statistically significant. In summary, we can conclude that KAN1 associates with the chromatin of these target genes via different cis-elements and, when ectopically expressed, is able to strongly repress the expression of these genes.
Figure 5.
Expression of potential KAN1 target genes. Real-time quantitative RT-PCR experiments show expression changes of three KAN1 targets with the E1 element (GAATAW) in the promoter (A), three KAN1 targets with the E2-binding site (MCAAAA) in the proximal promoter (B), and three KAN1 targets with the E3 element (CAAGTKG) in the proximal promoter (C). Plotted are relative expression levels in Col-0 (gray) and 35S::FLAG-GR-KAN1 (green) in response to 90 min of DEX induction or mock treatment. Striped bars depict gene expression levels in Col-0 (gray) and kan1 kan2 (Col-0 background; green). Average expression levels of three biological replicates were normalized to glyceraldehyde-3-phosphate dehydrogenase and plotted with se. *, P < 0.01.
KAN1 Activity Suppresses Auxin Synthesis and Signaling
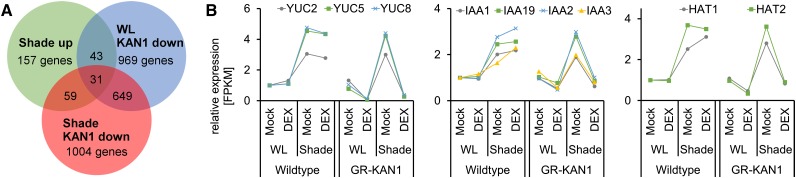
Many of the potential direct target genes we identify here are involved in auxin-mediated signaling processes. Auxin levels have been shown to increase when Arabidopsis plants experience shade (Tao et al., 2008; Won et al., 2011). This increase in auxin and the shuttling via PIN auxin carriers (Keuskamp et al., 2010) are both essential for a full shade-avoidance response. To investigate the effect of KAN1 on the regulation of auxin-related genes in a situation where these genes are actively turned on, we compared the transcriptomes of plants grown in white-light and shade conditions. For this reason, we germinated and cultivated Arabidopsis seedlings for 10 d in white-light conditions and transferred one part to a true shade environment for 45 min. To identify KAN1 targets in shade conditions, we also cultivated transgenic 35S::FLAG-GR-KAN1 plants like the wild-type control plants but induced the translocation of GR-KAN1 by DEX application 45 min prior to transfer to shade. In order to exclude light/DEX effects, all plant material was treated with DEX or a mock solution in white-light and shade conditions employing the same protocol. After these treatments, we isolated mRNA followed by RT and Illumina short-read sequencing. Our analysis revealed that 400 genes change significantly in wild-type plants in response to shade (Supplemental Table S1). Of these 400 genes, 241 genes are transcriptionally down-regulated while 159 genes are transcriptionally up-regulated. Among the shade-induced genes are several known players, such as members of the HD-ZIPII gene family (HOMEOBOX FROM ARABIDOPSIS1 [HAT1], HAT2, HAT3, HAT4/ARABIDOPSIS HOMEOBOX PROTEIN-2 [ATHB-2], and ATHB4), the auxin efflux transporter gene PIN3, and several YUC genes encoding auxin biosynthesis genes (YUC2, YUC5, YUC8, and YUC9). When compared with the set of KAN1-regulated genes in white light and shade, we can identify 43 genes that are shade induced and repressed by ectopic KAN1 induction (Fig. 6A). A slightly larger set of genes (59) contains genes that are only repressed by KAN1 in shade conditions. Because these 59 genes are all shade induced, the role of KAN1 is to keep their expression low, even under inductive light conditions (Fig. 6A). Inspection of these 59 shade-regulated KAN1 targets revealed a strong overrepresentation of genes whose products function in hormone biosynthesis or hormone-mediated signaling (Table II). Four genes are associated with abscisic acid (ABA) signaling processes: PYRABACTIN RESISTANCE1-LIKE5/REGULATORY COMPONENT OF ABSCISIC ACID RECEPTOR8 encoding an ABA receptor; A5g02760, an ABA down-regulated PROTEIN PHOSPHATASE 2C phosphatase; ABSCISIC ACID INSENSITIVE (ABI) FIVE BINDING PROTEIN4, an ABI5-binding protein involved in ABA signal transduction; and POTASSIUM CHANNEL IN ARABIDOPSIS1 encoding an ABA-regulated potassium channel. Another four genes are involved in brassinosteroid (BR) synthesis/signaling: DWARF4 (DWF4) encoding a P450 enzyme involved in BR synthesis; PHYB ACTIVATION TAGGED SUPPRESSOR1 (BAS1) encoding a P450 enzyme involved in BR catabolism; BRASSICOSTEROID-ENHANCED EXPRESSION1 (BEE1) encoding a basic helix-loop-helix transcription factor; and BRH1 encoding a ring-finger protein. The vast majority of genes, however (23 in total), encode auxin synthesis, transport, and signaling components. Among these genes are several SAUR genes, AUXIN/IAA transcriptional regulators, as well as three of the four shade-induced YUC-type auxin biosynthesis enzymes (YUC2, YUC5, and YUC8), all of which are no longer induced by shade when KAN1 is ectopically expressed (Fig. 6B). The 59 genes (Table II) that no longer respond to shade in a situation where KAN1 is ectopically active might represent the core set of genes required for activating growth in response to shade. It also demonstrates that one of the major functions of KAN1 is to block auxin action at all levels.
Figure 6.
Shade RNA-Seq. A, Venn diagram depicting the numbers of genes identified by mRNA-Seq of shade-exposed plants and KAN1 target genes. B, Responses of known shade-regulated genes involved in dissipating the auxin signal. Plotted are relative expression levels of the average of two biological replicates obtained from mRNA-Seq. FPKM, Fragments per kilobase of transcript per million mapped reads (calculated by Cufflinks package); WL, white light.
Table II. Shade-regulated KAN1 target genes.
| Arabidopsis Genome Initiative Code | Annotation | ChIP-Seq Binding Site | Shade Col-0 Mock White Light Col-0 Mock | Shade GR:KAN1 DEX Shade GR:KAN1 Mock |
|---|---|---|---|---|
| kb to gene | fold change | |||
| AT1G02640 | BXL2 | −0.6 | 2.2 | 0.2 |
| AT1G04240 | SHY2 | −6.1 | −5.1 | −3.3 | 1.6 | 0.4 |
| AT1G14920 | GAI | −4.1 | −3.5 | −1.3 | 0.1 | 1.6 | 0.5 |
| AT1G15670 | KMD2 | −3.7 | −3.1 | −1.9 | −1.4 | 1.8 | 0.5 |
| AT1G18400 | BEE1 | 0.0 | 0.9 | 1.7 | 2.2 | 2.0 | 0.1 |
| AT1G21050 | Unknown protein | −1.4 | 1.2 | 3.0 | 4.0 | 1.8 | 0.1 |
| AT1G21830 | Unknown protein | −2.4 | −0.9 | 1.3 | 1.7 | 0.1 |
| AT1G29430 | SAUR-like | 2.0 | 0.4 | |
| AT1G29440 | SAUR63 | 2.4 | 0.5 | |
| AT1G29450 | SAUR64 | 2.8 | 0.5 | |
| AT1G29460 | SAUR65 | 2.2 | 0.4 | |
| AT1G29500 | SAUR66 | 2.1 | 0.4 | |
| AT1G29510 | SAUR68 | 2.2 | 0.5 | |
| AT1G67900 | NPH3 family | 0.0 | 0.0 | 0.0 | 0.1 | 1.8 | 0.5 |
| AT1G70940 | PIN3 | −0.1 | 2.7 | 1.7 | 0.4 |
| AT1G72416 | DnaJ domain superfamily | 0.2 | 1.7 | 0.1 |
| AT2G23170 | GH3.3 | 1.9 | 0.3 | |
| AT2G26710 | BAS1 | −6.0 | −0.1 | 0.0 | 2.3 | 0.4 |
| AT2G42870 | PAR1 | −2.3 | −1.1 | 3.5 | 0.4 |
| AT2G43820 | GT | −1.0 | 1.7 | 0.5 |
| AT3G02140 | TMAC2 | −4.5 | −3.1 | −2.8 | −0.8 | −0.3 | 1.8 | 0.3 |
| AT3G03820 | SAUR29 | −0.4 | 3.0 | 0.2 |
| AT3G03850 | SAUR26 | 2.7 | 0.1 | |
| AT3G12920 | BRG3 | −4.6 | −1.8 | −0.8 | 0.9 | 2.3 | 3.0 | 4.3 | 1.7 | 0.5 |
| AT3G15540 | IAA19 | −2.1 | 2.5 | 0.3 |
| AT3G21330 | BHLH87 | −3.4 | −2.1 | 8.1 | 0.1 |
| AT3G23030 | IAA2 | −5.1 | −3.0 | −2.6 | −1.3 | −0.2 | 2.8 | 0.3 |
| AT3G47570 | LRR protein kinase family | 1.7 | 0.6 | |
| AT3G50340 | Unknown protein | 2.1 | 0.5 | |
| AT3G50660 | DWF4 | −4.9 | −3.8 | −2.4 | 0.0 | 4.2 | 1.8 | 0.4 |
| AT3G55500 | ATEXPA16 | 2.6 | 0.2 | |
| AT3G58120 | ATBZIP61 | −1.2 | 1.9 | 0.4 |
| AT3G61460 | BRH1 | −2.3 | −0.4 | 1.7 | 0.3 |
| AT4G13260 | YUC2 | 4.0 | 3.1 | 0.1 |
| AT4G14560 | IAA1 | 2.0 | 0.3 | |
| AT4G17460 | HAT1 | −1.1 | −0.1 | 2.5 | 0.3 |
| AT4G18170 | WRKY28 | 0.4 | 1.6 | 2.6 | 0.3 |
| AT4G25260 | Plant invertase superfamily | 1.8 | 0.5 | |
| AT4G27450 | Aluminum induced | −0.4 | 2.3 | 0.5 |
| AT4G28720 | YUC8 | −8.8 | 1.6 | 4.2 | 4.8 | 0.1 |
| AT4G31820 | ENP | −0.1 | 1.8 | 0.5 |
| AT4G36850 | PQ loop repeat family | −0.5 | 2.3 | 0.3 |
| AT4G37770 | ACS8 | −2.7 | 5.0 | 0.1 |
| AT5G02760 | APD7 | −0.2 | 2.6 | 0.1 |
| AT5G05440 | PYL5 | −0.9 | 0.6 | 1.6 | 0.4 |
| AT5G12050 | Unknown protein | −0.7 | 4.4 | 0.2 |
| AT5G18010 | SAUR19 | 4.1 | 0.3 | |
| AT5G18030 | SAUR21 | 2.4 | 0.4 | |
| AT5G18050 | SAUR22 | 4.0 | 0.3 | |
| AT5G18060 | SAUR23 | 2.8 | 0.4 | |
| AT5G39860 | PRE1 | 2.5 | 0.5 | |
| AT5G43890 | YUC5 | −17.1 | 4.5 | 0.1 |
| AT5G44260 | TZF5 | −0.1 | 0.4 | 2.1 | 0.4 |
| AT5G46240 | KAT1 | 2.6 | 0.1 | |
| AT5G47370 | HAT2 | −0.7 | −0.1 | 3.7 | 0.2 |
| AT5G48900 | Pectin lyase-like superfamily | 0.7 | 1.9 | 0.3 |
| AT5G52900 | MAKR6 | −0.8 | 2.3 | 4.4 | 1.9 | 0.1 |
| AT5G62280 | Unknown protein | 2.4 | 0.1 | |
| AT5G66580 | Unknown protein | 0.4 | 4.2 | 0.3 |
Shade, Auxin, Action
To further probe the possibility of KAN1 controlling auxin-mediated growth responses, we exposed Col-0 wild-type plants and 35S::FLAG-GR-KAN1 transgenic plants to either white-light or simulated canopy shade conditions in the presence or absence of DEX. When mock treated, both Col-0 and 35S::FLAG-GR-KAN1 transgenic plants develop long hypocotyls upon exposure to shade (Fig. 7A). In response to DEX treatment, however, wild-type plants show normal responses whereas 35S::FLAG-GR-KAN1 transgenic plants are completely shade insensitive (Fig. 7A). These findings support a role for KAN1 in the control of auxin-mediated growth responses. The fact that several of the shade-induced KAN1 target genes have a role in the production of auxin (Table II; Fig. 6) prompted us to assess levels of free auxin in both Col-0 and 35S::FLAG-GR-KAN1 transgenic plants. When exposed to true shade, we can detect elevated levels of free auxin in Col-0 plants, and this increase is unaffected by DEX application (Fig. 7B). In contrast to wild-type plants, the amount of free auxin in shade is strongly reduced in 35S::FLAG-GR-KAN1 transgenic plants only when treated with DEX (Fig. 7B). These findings suggest that the growth-repressing effect of ectopic KAN1 expression in shade is partly due to the inability of these transgenic plants to boost auxin synthesis.
Figure 7.
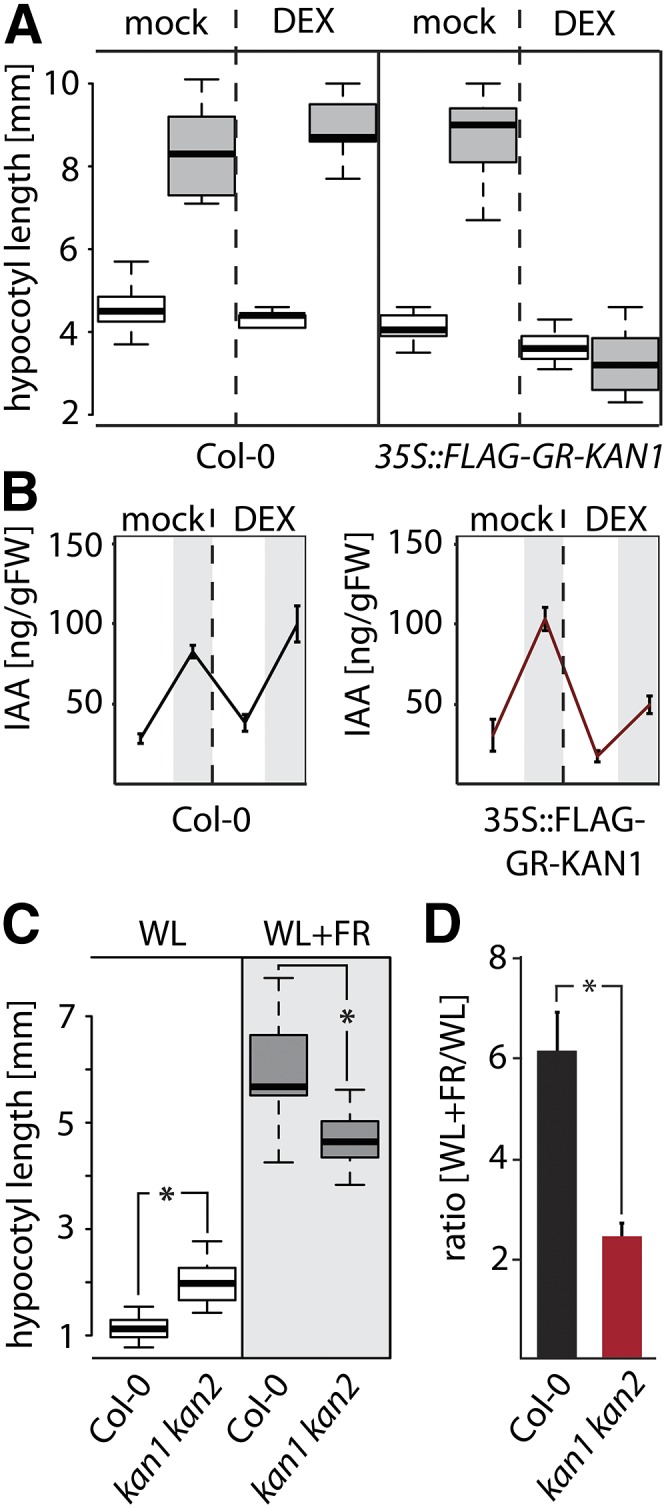
Influence of KAN1 on shade-induced auxin production. A, Hypocotyl lengths of 35S::FLAG::GR-KAN1 and wild-type Col-0 without 50 μm DEX in white light and simulated canopy shade (far-red light-enriched white light) are significantly shorter in the DEX-induced transgenic line. Box plots show the observed experimental data; white boxes represent hypocotyls grown in white light, and gray boxes represent shade-grown hypocotyls. B, Determination of free auxin levels in Col-0 and in 35S::FLAG-GR-KAN1 transgenic plants using gas chromatography-mass spectrometry. Plants were grown on petri dishes for 10 d and treated with either mock solution or a 5 μm DEX solution for 45 min. One-half of the plants were then transferred to simulated canopy shade conditions for another 45 min (gray background). Plotted are levels of free IAA per gram fresh weight (FW). C, Hypocotyl lengths of kan1 kan2 double mutant and wild-type Col-0 seedlings in white light (WL) and far-red light (FR)-supplemented white light. Box plots show the observed experimental data; white boxes represent hypocotyls grown in white light, and gray boxes represent WL+FR hypocotyls. *, P < 0.001. D, Ratio of the lengths of hypocotyls grown in far-red light-enriched white-light conditions compared with white light. Plotted is the average of three biological experiments with sd. *, P < 0.05.
Since ectopic expression of KAN1 is able to suppress both the production of auxin and the elongation of the hypocotyl, we tested the shade-avoidance response of kan1 kan2 loss-of-function double mutant plants. When grown in white light, kan1 kan2 double mutant plants develop significantly longer hypocotyls compared with wild-type plants (Fig. 7C). In far-red light-enriched conditions, hypocotyl elongation is significantly reduced in kan1 kan2 double mutant plants compared with wild-type plants (Fig. 7, C and D). These findings indicate that KAN1 is not only able to suppress elongation growth but that its activity is also required for a full shade-avoidance response in wild-type plants.
DISCUSSION
We have employed a meta-analysis approach to gain a deeper understanding of the function of the KAN1 transcription factor by identifying its direct target genes. Using this approach, we can identify a set of putative KAN1 target genes that act downstream of KAN1 in patterning and growth-promoting pathways (Fig. 1; Table I). Furthermore, by performing gene expression analysis of transgenic 35S::FLAG-GR-KAN1 plants in shade conditions, we found that a large number of auxin-related genes underlie direct KAN1 regulation and that plants ectopically expressing KAN1 cannot induce auxin biosynthesis in shade conditions (Fig. 7).
Identification of a Set of Direct KAN1 Target Genes Using Genome-Wide Comparative Approaches
Genome-wide transcriptional profiling approaches have been employed to study the regulation of KAN1 target genes using 35S::KAN1-GR transgenic plants (Merelo et al., 2013; Reinhart et al., 2013). All approaches resulted in the identification of a relatively large number of potential direct KAN1 target genes (greater than 200). Using a comparative approach, we filtered all available data sets to condense the number of genes regulated by KAN1. Even though all data sets were generated with transgenic plants harboring a similar inducible overexpression construct of KAN1 (either FLAG-GR-KAN1 or KAN1-GR), there are differences in the genes that are regulated. Between the tiling array study and the microarray study, only 35% of the genes (89 out of 249) of the microarray study were identified using tiling arrays and only 41% of the genes identified to be altered on the tiling array were confirmed by RNA-Seq (205 out of 495). Interestingly, 73% (182 out of 249) of the genes identified by microarray could be verified by mRNA-Seq (Fig. 1A). Further comparison of all data sets with our ChIP-Seq data revealed that 68% and 65% of the genes that are regulated using RNA-Seq or tiling arrays, respectively, had KAN1-binding regions close to or in their coding regions (Fig. 1B). Also in this comparison, the microarray data set yielded the largest overlap, with approximately 87% of the identified genes (217 out of 249) being bound by KAN1. The finding that, of the 75 genes that were identified in all data sets, 72 contain KAN1-binding regions (corresponding to 96%) further indicates that our meta-analysis enriched for genes that are likely direct KAN1 targets (Table I).
KAN1 is well known for its role as a patterning factor of leaves, shoots, roots, and ovules. Surprisingly, when analyzing the 72 high-confidence target genes, we found a strong overrepresentation of genes involved in responding to stimulus (47 out of 72, corresponding to approximately 65%), and several of the genes are involved in hormone-mediated signaling processes (Fig. 2A; Table I). A significant number of KAN1 targets are involved in the regulation of ion transport, which is intriguing and suggests that, upon activation, KAN1 could strongly influence the cellular environment (e.g. redox state). HD-ZIPIII transcription factors counteract KAN activity in the patterning process. The finding that REV, a member of the HD-ZIPIII family, is redox sensitive (Xie et al., 2014) would allow the regulation by KAN1 via changing the cellular redox state. Such regulation could potentially add to the HD-ZIPIII/KAN antagonism and thus contribute to patterning and growth processes.
Identification of Novel cis-Elements in Genes Regulated by KAN1
Combinatory analysis of genes regulated by KAN1 (using RNA-Seq) and promoters bound by KAN1 (using ChIP-Seq) revealed three sequence motifs (E1–E3) to be enriched in the respective target gene promoters (Fig. 3). These novel elements could be recognized by KAN1 directly or might represent binding sites for proteins that interact with KAN1 and bind DNA as a heteromeric complex. The selection of genes with a single binding site of E1, E2, or E3 in the proximal promoter revealed that these genes are bound by KAN1 and also underlie negative regulation by KAN1 (Fig. 5). This suggests that KAN1 can recognize these novel cis-elements and is able to transcriptionally control the expression of the respective genes. To confirm that KAN1 is involved in the regulation of these putative target genes, we also examined their levels of expression in wild-type and kan1 kan2 double mutant plants. We found that only one of the examined genes is significantly increased in expression in kan1 kan2 plants relative to the wild type (Fig. 2B). The expression levels of three genes are significantly lower in kan1 kan2 plants relative to the wild type, and all other genes are not significantly changed (Fig. 2). This discrepancy could be explained by the fact that we isolated RNA from whole seedlings, but both KAN1 and KAN2 exhibit a strong cell type-specific pattern of expression. In addition, KAN1 itself is a transcriptional repressor that negatively regulates other transcription factors that also function as repressors (e.g. TEM1, JKD, or HAT2). Removal of the repressive activity of genes encoding repressors (in the kan1 kan2 double mutant background) can potentially result in the up-regulation of shared secondary target genes. Another hypothesis is that the loss of KAN1 and KAN2 changes cell type identity and that the cells in which KAN1 acts to repress these target genes are no longer present, resulting in no overall change in expression of these putative targets.
KAN1 Represses a Large Number of Shade-Induced Genes and Counteracts Auxin-Mediated Shade Growth
Previous work has shown that both HD-ZIPIII and KAN1 oppositely regulate a number of shared targets that are associated with the shade-avoidance response (Brandt et al., 2014). These common targets include genes encoding the auxin biosynthesis enzymes YUC5 and TAA1 as well as the HAT2 transcription factor. To better understand the role of KAN1 in shade, we used mRNA-Seq to identify shade-regulated genes that are also regulated by KAN1. We can identify 59 such genes that include a large number of genes encoding auxin synthesis and signaling components, providing evidence that one of the main functions of KAN1 is to inhibit both the production and dissipation of the auxin signal. This is in line with the finding that, when treated with DEX, 35S::FLAG-GR-KAN1 transgenic plants are unable to elongate their hypocotyls (Fig. 7A). This effect might be related to the fact that KAN1 is a strong repressor of auxin biosynthesis genes and auxin levels in shade are strongly affected in transgenic 35S::FLAG-GR-KAN1 plants treated with DEX (Fig. 7B). Three YUC genes (YUC2, YUC5, and YUC8) show strong transcriptional up-regulation in response to shade treatment in the wild type but not in DEX-induced transgenic 35S::FLAG-GR-KAN1 plants. These findings indicate that YUC2, YUC5, and YUC8 are instrumental for the induction of auxin in response to shade. However, KAN1 seems to affect the shade-induced production of auxin at multiple levels, as it can also repress the expression of TAA1 that converts Trp to indole-3-pyruvic acid (Stepanova et al., 2011), which is subsequently converted to auxin by YUC-type cytochrome P450 monooxygenases. Thus, KAN1 blocks both steps in this two-step biosynthetic process.
In contrast to the findings that KAN1 acts as a repressor of auxin-related gene expression and, thus, as an inhibitor of shade-induced growth responses, we found that kan1 kan2 loss-of-function double mutant plants show elongated hypocotyls in white light conditions but reduced hypocotyl growth in far-red light-enriched white light conditions (Fig. 7, C and D). These findings show that KAN1 activity is essential for a full shade-avoidance response and suggest that the auxin gradient, which is established by the opposing activities of HD-ZIPIII and KAN, generates the driving force for elongation growth. If the gradient is weakened by the loss of HD-ZIPIII activity (less auxin production in adaxial tissue) or the loss of KAN activity (higher auxin in abaxial tissue), this driving force is weakened and hypocotyl growth is reduced.
CONCLUSION
Using a meta-analysis approach, we performed a comparative analysis with genome-wide expression and ChIP-Seq data sets for KAN1. We were able to produce a set of 72 high-confidence KAN1 target genes that are transcriptionally down-regulated by ectopic KAN1 expression and are directly bound by KAN1. By performing RNA-Seq in white light and shade conditions, we identified several genes involved in auxin biology. Concordantly, our data show that ectopic KAN1 suppresses auxin production in shade, which accounts for the non-shade-avoiding phenotype of transgenic plants ectopically expressing KAN1.
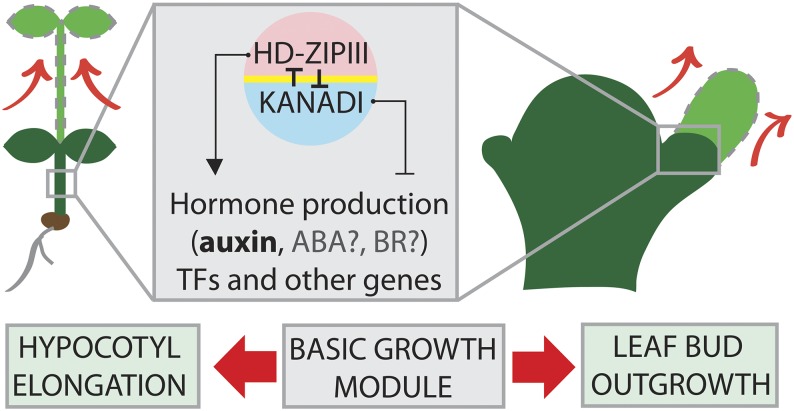
Plants carrying loss-of-function mutant alleles in genes encoding for HD-ZIPIII transcription factors exhibit reduced shade-avoidance responses (Brandt et al., 2012; Baima et al., 2014), while gain-of-function mutants seem to be slightly hypersensitive to shade. In addition, HD-ZIPIII can induce auxin production and also positively regulate several other shade-induced genes. HD-ZIPIII and KAN transcription factors have opposing roles in controlling development, which is evidenced by loss- and gain-of-function phenotypes: loss of HD-ZIPIII activity causes developmental defects similar to KAN gain of function, while HD-ZIPIII gain of function resembles KAN loss of function. In addition, both gene families seem to impinge on a set of shared target genes (Brandt et al., 2012; Merelo et al., 2013; Reinhart et al., 2013). In short, these findings point toward the existence of a basic growth regulatory module driven by the opposite activities of HD-ZIPIII and KAN (Fig. 8). We hypothesize that this module is at the basis of shade-induced auxin production, which is a prerequisite for elongation growth processes. After leaf primordia initiation, HD-ZIPIII and KAN act to establish a pattern that partitions the primordium into an adaxial (future upper leaf side) domain and an abaxial (future lower leaf side) domain. We think that it is possible that the same forces that are being produced in response to shade, which act to elongate the hypocotyl, also take place in the leaf primordium, driving its outgrowth from the shoot apex.
Figure 8.
Speculative model for the action of the HD-ZIPIII/KAN growth-promoting module. HD-ZIPIIIs and KANs show polar expression in both the vasculature of the hypocotyl and the early leaf primordium. Activation of the HD-ZIPIII/KAN module causes the spatial induction/repression of a number of direct target genes that results in the production of auxin and outgrowth of the leaf primordium or elongation of the hypocotyl.
MATERIALS AND METHODS
Plant Material and Growth Conditions
For hypocotyl measurements, Arabidopsis (Arabidopsis thaliana) plants were grown for 2 d in constant white light conditions to induce germination and then kept for another 4 to 5 d either in the same growing conditions or were transferred to simulated canopy shade conditions (Brandt et al., 2012). The kan1 kan2/+ (Landsberg erecta) mutant plants were backcrossed into the Col-0 background three times to obtain kan1 kan2 (Col-0) plants for comparative analysis with Col-0 wild-type plants.
For hypocotyl measurements, gene expression studies, and the determination of free auxin, wild-type and mutant plants were grown in white light and in simulated canopy shade conditions. The following light regime was applied: long-day conditions at 22°C for 2 d in a Fi-totron 600H growth chamber (Fisons) in white light (blue light [460–480 nm] = 2.34 μmol m−2 s−1, red light [650–670 nm] = 1.93 μmol m−2 s−1, far-red light [720–755 nm] = 0.65 μmol m−2 s−1, and photosynthetically active radiation [395–710 nm] = 89.3). For shade avoidance hypocotyl measurements, plants were transferred at day 2 to a shaded compartment (using a combination of LEE filters [LEE] and far-red light bulbs [Narva; http://www.narva-bel.de/]) in the growth chamber and irradiated with far-red light-enriched white light (blue light = 0.88 μmol m−2 s−1, red light = 1.65 μmol m−2 s−1, far-red light = 2.56 μmol m−2 s−1, and photosynthetically active radiation = 39.8). Seedlings were kept under these conditions for 4 d. Seedlings were photographed and hypocotyls were measured using ImageJ. Comparison of the wild type and kan1 kan2 double mutant was performed by growing them for 7 d under white light (20–25 μmol m−2 s−1 photosynthetically active radiation; red:far-red light ratio of 5.6) or for 2 d in white light and another 5 d under far-red light-enriched white light (20–25 μmol m−2 s−1 photosynthetically active radiation; red:far-red light ratio of 0.06).
Chromatin Immunoprecipitation
For the chromatin immunoprecipitation experiments, Col-0 and transgenic 35S::FLAG-GR-KAN1 plants were grown in liquid Murashige and Skoog medium for 10 d and induced with 50 μm DEX for 90 min prior to harvesting. Chromatin immunoprecipitation experiments were carried out as described by Kwon et al. (2005), except that anti-FLAG M2 magnetic beads (Sigma-Aldrich) were used and immunoprecipitation experiments were performed for only 2 h.
RNA Extraction and Quantitative PCR
RNA was isolated from seedlings using the Roboklon GeneMATRIX Universal RNA Purification Kit following the manufacturer’s recommendations. One microgram of total RNA was reverse transcribed using the Fermentas RevertAid Premium reverse transcriptase with oligo(dT) primers. Complementary DNAs were diluted 10-fold, and 3.5 μL was used for RT-PCR. Quantitative measurements were performed on a Bio-Rad CFX384 using the Fermentas SYBR Green qPCR master mix. Relative quantities were calculated using the comparative threshold cycle method by determining the expression of a gene of interest relative to an internal housekeeping gene. Oligonucleotides are listed in Supplementary Table S2.
RNA-Seq Analysis
Sequencing libraries were prepared using the Illumina TruSeq RNA Library Preparation Kit according to the manufacturer’s recommendations. Libraries were sequenced on the Illumina HiSeq2000 platform, and between 14 and 22 million read pairs per sample were obtained. Approximately 550 million paired-end reads were loaded into Galaxy (version 15.05.rc1; Giardine et al., 2005; Blankenberg et al., 2010; Goecks et al., 2010), and quality was assessed using FastQC (version 0.10.1). Tophat2 (version 2.0.9) aligned above 90% of read pairs of each sample correctly to The Arabidopsis Information Resource 10 genome (Kim et al., 2013). Galaxy’s Cufflinks package (version 0.0.7) was employed for differentially expressed gene calling (cutoff q-value of 0.05; Trapnell et al., 2010).
Auxin Measurements
For the determination of free auxin (IAA) levels, approximately 200 mg of Arabidopsis seedlings was harvested and homogenized in liquid nitrogen. Extraction of the free analytes was carried out at 28°C for 90 min with 1.5 mL of ethyl acetate containing 0.1% (v/v) formic acid and the internal standards 3-hydroxybenzoic acid and indole-5-formic acid. After centrifugation at 10,000g at 4°C for 10 min, 1.2-mL supernatants were collected, the ethyl acetate was removed, and the samples were dried overnight in a SpeedVac (100 mbar). Derivatization was performed with 70 μL of N-methyl-N-(trimethylsilyl) trifluoracetamide (Sigma-Aldrich) for 60 min at 40°C; 1 μL was injected onto the gas chromatograph column. Determination of the analytes was done by gas chromatography-mass spectrometry (Agilent 6890 gas chromatograph and Agilent 5973 single-quadrupole mass spectrometer; Agilent Technologies) using split-injection mode and an SPB-50 column (30 m, 0.25 mm internal diameter; Supelco, Sigma-Aldrich). The gas chromatograph oven temperature was held at 70°C for 5 min, then ramped at 5°C min−1 to 265°C followed by 2°C min−1 from 265°C to 280°C, and afterward held for an additional 8 min at 280°C. Helium was used as the carrier gas with a flow rate of 1 mL min−1. Detection of analytes was performed by electron impact ionization single-quadrupole mass spectrometry operated in selected ion monitoring mode.
Sequence data from this article can be found in the Gene Expression Omnibus Database under accession number GSE68684.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of available genome-wide data sets aiming at the identification of genes down-regulated by KAN1.
Supplemental Table S1. Genes whose expression is altered in response to shade in Col-0 plants.
Supplemental Table S2. Oligonucleotide sequences used in this study.
Supplemental Data Set S1. Lists of KAN1-regulated genes.
Supplementary Material
Acknowledgments
We thank Gesine Seibold, Ingrid Blumberg, and Rocio Alonso for excellent technical support and Marcus Heisler for critically reading the article.
Glossary
- RNA-Seq
RNA sequencing
- ChIP-Seq
chromatin immunoprecipitation sequencing
- Col-0
Columbia-0
- DEX
dexamethasone
- mRNA-Seq
messenger RNA sequencing
- CHX
cycloheximide
- GO
Gene Ontology
- ChIP-qPCR
chromatin immunoprecipitation-quantitative PCR
- RT
reverse transcription
- ABA
abscisic acid
- BR
brassinosteroid
Footnotes
This work was supported by the European Union (Marie-Curie International Reintegration grant no. 256502 to S.W.), the Deutsche Forschungsgemeinschaft Collaborative Research Centre (grant no. SFB1101 to S.W.), the European Research Council (grant no. 336295 to S.W.), and the Spanish MINECO (grant no. BIO2011–23489 to J.F.M.-G.).
Articles can be viewed without a subscription.
References
- Baima S, Forte V, Possenti M, Peñalosa A, Leoni G, Salvi S, Felici B, Ruberti I, Morelli G (2014) Negative feedback regulation of auxin signaling by ATHB8/ACL5-BUD2 transcription module. Mol Plant 7: 1006–1025 [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19: Unit 19.10.11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Torrent J, Salla-Martret M, Brandt R, Musielak T, Palauqui JC, Martínez-García JF, Wenkel S (2012) ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal Behav 7: 1382–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R, Cabedo M, Xie Y, Wenkel S (2014) Homeodomain leucine-zipper proteins and their role in synchronizing growth and development with the environment. J Integr Plant Biol 56: 518–526 [DOI] [PubMed] [Google Scholar]
- Brandt R, Salla-Martret M, Bou-Torrent J, Musielak T, Stahl M, Lanz C, Ott F, Schmid M, Greb T, Schwarz M, et al. (2012) Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Byrne ME. (2006) Shoot meristem function and leaf polarity: the role of class III HD-ZIP genes. PLoS Genet 2: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL (2004) Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitch J. (1992) Sources of variation in leaf shape among two populations of Achillea lanulosa. Genetics 130: 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawker NP, Bowman JL (2004) Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol 135: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Harrar Y, Lin C, Reinhart B, Newell NR, Talavera-Rauh F, Hokin SA, Barton MK, Kerstetter RA (2014) Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 26: 246–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. (2000) Development of symmetry in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 349–370 [DOI] [PubMed] [Google Scholar]
- Ilegems M, Douet V, Meylan-Bettex M, Uyttewaal M, Brand L, Bowman JL, Stieger PA (2010) Interplay of auxin, KANADI and class III HD-ZIP transcription factors in vascular tissue formation. Development 137: 975–984 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC (2004) MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS (2012) ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LACJ, Peeters AJM, Pierik R (2010) Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T, Horiguchi G, Kim GT, Ohgishi M, Sakai T, Tsukaya H (2005) The different growth responses of the Arabidopsis thaliana leaf blade and the petiole during shade avoidance are regulated by photoreceptors and sugar. Plant Cell Physiol 46: 213–223 [DOI] [PubMed] [Google Scholar]
- Kwon CS, Chen C, Wagner D (2005) WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev 19: 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Merelo P, Xie Y, Brand L, Ott F, Weigel D, Bowman JL, Heisler MG, Wenkel S (2013) Genome-wide identification of KANADI1 target genes. PLoS ONE 8: e77341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Matsumoto N, Tsugeki R, Rikirsch E, Laux T, Okada K (2012) Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24: 519–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Liu T, Newell NR, Magnani E, Huang T, Kerstetter R, Michaels S, Barton MK (2013) Establishing a framework for the ad/abaxial regulatory network of Arabidopsis: ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell 25: 3228–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchi L, Carabelli M, Ruzza V, Possenti M, Sassi M, Peñalosa A, Sessa G, Salvi S, Forte V, Morelli G, et al. (2013) Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development 140: 2118–2129 [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward FI. (1987) Stomatal numbers are sensitive to increases in CO2 from preindustrial levels. Nature 327: 617–618 [Google Scholar]
- Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA (2008) KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc Natl Acad Sci USA 105: 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Huhn K, Brandt R, Potschin M, Bieker S, Straub D, Doll J, Drechsler T, Zentgraf U, Wenkel S (2014) REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development 141: 4772–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.