Methylated cytokinins contribute to pathogenesis as hormone-mimics.
Abstract
Cytokinins (CKs), a class of phytohormones that regulate plant growth and development, are also synthesized by some phytopathogens to disrupt the hormonal balance and to facilitate niche establishment in their hosts. Rhodococcus fascians harbors the fasciation (fas) locus, an operon encoding several genes homologous to CK biosynthesis and metabolism. This pathogen causes unique leafy gall symptoms reminiscent of CK overproduction; however, bacterial CKs have not been clearly correlated with the severe symptoms, and no virulence-associated unique CKs or analogs have been identified. Here, we report the identification of monomethylated N6-(∆2-isopentenyl)adenine and dimethylated N6-(∆2-isopentenyl)adenine (collectively, methylated cytokinins [MeCKs]) from R. fascians. MeCKs were recognized by a CK receptor and up-regulated type-A ARABIDOPSIS THALIANA RESPONSE REGULATOR genes. Treatment with MeCKs inhibited root growth, a hallmark of CK action, whereas the receptor mutant was insensitive. MeCKs were retained longer in planta than canonical CKs and were poor substrates for a CK oxidase/dehydrogenase, suggesting enhanced biological stability. MeCKs were synthesized by S-adenosyl methionine-dependent methyltransferases (MT1 and MT2) that are present upstream of the fas genes. The best substrate for methylation was isopentenyl diphosphate. MT1 and MT2 catalyzed distinct methylation reactions; only the MT2 product was used by FAS4 to synthesize monomethylated N6-(∆2-isopentenyl)adenine. The MT1 product was dimethylated by MT2 and used as a substrate by FAS4 to produce dimethylated N6-(∆2-isopentenyl)adenine. Chemically synthesized MeCKs were comparable in activity. Our results strongly suggest that MeCKs function as CK mimics and play a role in this plant-pathogen interaction.
The balance of phytohormones, such as cytokinins (CKs) and auxins, is finely controlled to maintain proper plant growth and development but is often disturbed following pathogen infection (Robert-Seilaniantz et al., 2007; Pieterse et al., 2012). As a virulence strategy, many phytopathogens synthesize phytohormones that cause aberrant organogenesis and modulate primary carbon metabolism that ultimately aids disease establishment (Jameson, 2000; Robert-Seilaniantz et al., 2007). For several pathogens, CK production is essential for virulence, and they carry genes for CK biosynthesis in a harbored plasmid (Jameson, 2000). Fungal pathogens employ CKs to form green islands with delayed senescence, whereas bacterial pathogens develop gall structures (Sakakibara et al., 2005; Walters et al., 2008; Giron et al., 2013). Rhodococcus fascians is a gram-positive actinomycete that causes symptoms ranging from leaf deformation to differentiated shooty outgrowths known as leafy galls in more than 150 different plant species (Goethals et al., 2001; Stes et al., 2011). In ornamental plants, such infections reduce their value and contribute to economic losses worldwide (Putnam and Miller, 2007). Leafy gall symptoms are reminiscent of CK overproduction and can be partially induced by exogenous application of CKs (Thimann and Sachs, 1966; Eason et al., 1996). Although several CKs have been isolated from R. fascians culture filtrates, a clear correlation with pathogenesis is lacking partially owing to the low concentration of bacterial CKs (Eason et al., 1996). A synergistic action by a mixture of bacterially produced CKs has been proposed, leading to persistent accumulation of CKs locally (Pertry et al., 2009). Nevertheless, to date, no virulence-associated CK analogs have been identified that could contribute to the infection symptoms.
Naturally occurring CKs are adenine derivatives with different side chains at the N6 position. Major plant CKs are N6-prenylated adenine derivatives such as N6-(Δ2-isopentenyl)adenine (iP), trans-zeatin (tZ), cis-zeatin (cZ), and dihydrozeatin, collectively known as isoprenoid CKs (Sakakibara, 2006). Among them, iP and tZ are the major CKs in Arabidopsis (Arabidopsis thaliana). iP is synthesized by adenosine phosphate-isopentenyl transferase (IPT) using dimethylallyl diphosphate (DMAPP) and adenosine phosphate as substrates (Sakakibara, 2006). tZ is formed by hydroxylation of the trans-end of the prenyl side chain of the iP nucleotide. CK homeostasis is governed by both biosynthesis and catabolism and has an important regulatory role in plant growth (Sakakibara, 2006; Werner et al., 2006). CK oxidase/dehydrogenase (CKX) is responsible for an irreversible reaction cleaving the unsaturated isoprenoid side chain that results in the formation of adenine and the corresponding aldehyde (Werner et al., 2006). In Arabidopsis, CKs are perceived by a subset of sensory His kinases, ARABIDOPSIS HIS KINASE2 (AHK2) to AHK4, which undergo a His-Asp phosphorelay leading to induction of direct target genes including type-A ARABIDOPSIS RESPONSE REGULATOR (ARR) genes (Kieber and Schaller, 2010). This two-component signaling system has been implicated in mediating basal and pathogen-induced plant immunity (Choi et al., 2010; Argueso et al., 2012). For instance, infection of Arabidopsis plants by R. fascians reportedly activates type-A ARR5 expression with increased expression of AHK3 and AHK4, resulting in mitotic cell divisions that arrest the infected leaves in a meristematic state to establish a nutrient-rich niche (Depuydt et al., 2008, 2009; Pertry et al., 2010; Stes et al., 2011). As the infection progresses, IPT genes are switched off, whereas the expression of all CKX genes are strongly induced in symptomatic tissues (Depuydt et al., 2008).
The virulence determinant of R. fascians is located within the fasciation (fas) locus, an operon encoding several genes involved in CK metabolism, indicating that CKs are essential for this plant-pathogen interaction (Stes et al., 2011). fas4 encodes IPT that catalyzes the rate-limiting step of CK biosynthesis and is vital for virulence (Stes et al., 2013). Interestingly, two methyltransferase-like genes are present upstream of the fas gene, whose functions have been unknown. Despite the presence of the fas genes in R. fascians, fewer known CKs have been detected compared with other gall-causing pathogens such as Pantoea agglomerans, Agrobacterium tumefaciens, and Pseudomonas savastanoi (Goethals et al., 2001). Further, the leafy gall phenotype is unique, not invoked by any of the above-mentioned pathogens, implying that the virulence of R. fascians might not be due to typical CKs alone (Goethals et al., 2001). R. fascians has long been hypothesized to produce CK analogs using similar or modified substrates (Goethals et al., 2001; Galis et al., 2005; Stes et al., 2011), but no such molecules have been discovered so far. Here, we report the identification and mode of biosynthesis for methylated cytokinins (MeCKs) as hormone mimics from R. fascians. These compounds are synthesized by two methyltransferases and FAS4. Their CK-like activity and higher in planta stability suggest a role for the methylated analogs as CK mimics that foster efficient pathogenesis.
RESULTS
Isolation and Structural Determination of MeCKs
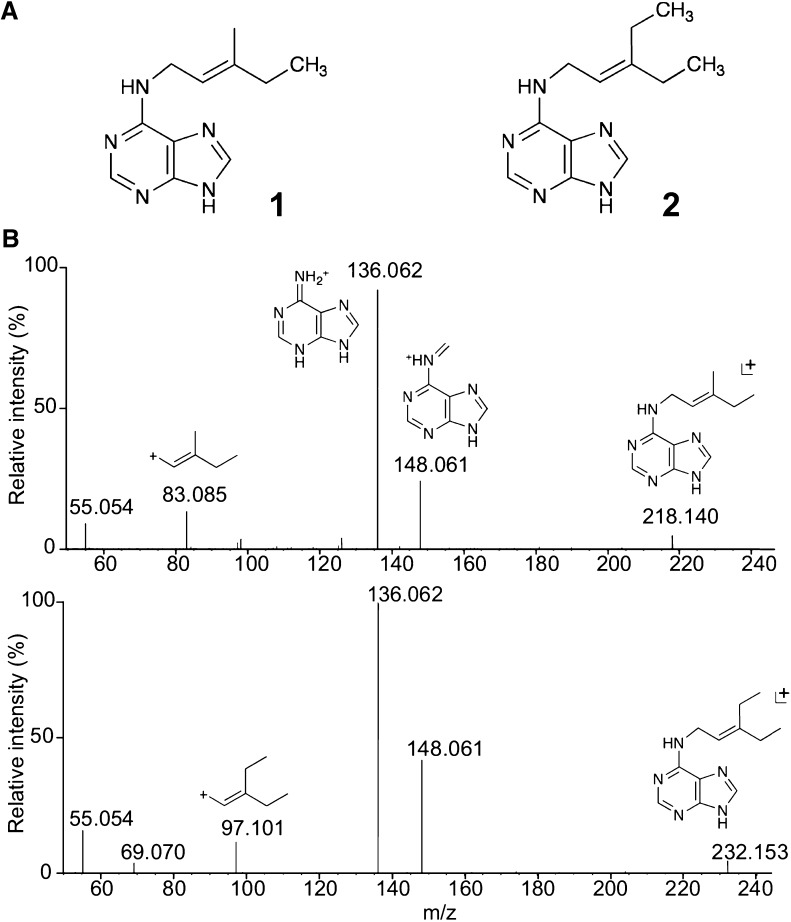
Upstream of the fas genes, there are two putative methyltransferase-like genes, mt1 and mt2, but their function and relation to CK biosynthesis has remained unknown (Stes et al., 2011). To examine the involvement of the mt genes in the synthesis of CKs, fas4mt1mt2, fas4mt1, and fas4mt2 were overexpressed in Escherichia coli, and liquid culture filtrates were analyzed by liquid chromatography (LC)-quadrupole tandem mass spectrometry (MS). In the filtrates of fas4mt1mt2 and fas4mt2, two new CK-like peaks were detected; one peak (peak 2) corresponds to a compound with a mass-to-charge ratio (m/z) of 218 and the other peak (peak 3) has a m/z of 232 (Supplemental Fig. S1, C and E–G). Peak 2 was also detected in small amounts in the filtrate of fas4mt1 (Supplemental Fig. S1D). These compounds were purified from large-scale E. coli cultures. Characterization by Orbitrap MS and NMR analyses revealed that the compounds obtained from the fas4mt2 and fas4mt1mt2 filtrates were monomethylated N6-(Δ2-isopentenyl)adenine (1MeiP) and dimethylated N6-(Δ2-isopentenyl)adenine (2MeiP), respectively (Fig. 1).
Figure 1.
MeCKs produced by R. fascians mts and fas4. A, Chemical structures of 1MeiP and 2MeiP isolated from filtrates of E. coli cultures expressing R. fascians fas4mt2 and fas4mt1mt2, respectively. 1, 1MeiP, 1H NMR (DMSO-d6): δ (ppm) = 12.86 (brs,1H), 8.17 (s,1H), 8.01 (s,1H), 7.61 (s,1H), 5.32 (t, 6.2 Hz,1H), 4.08 (brs, 2H), 1.98 (q, 6.93 Hz, 2H), 1.70 (s, 3H), and 0.95 (t, 7.39 Hz, 3H). 2, 2MeiP, 1H NMR (DMSO-d6): δ (ppm) = 12.91 (brs, 1H), 8.17 (s, 1H), 8.06 (s, 1H), 7.67 (s, 1H), 5.25 (t, 6.2 Hz, 1H), 4.08 (brs, 2H), 2.13 (q, 7.5 Hz, 2H), 1.99 (q, 7.3 Hz, 2H), 0.97 (t, 7.4 Hz, 3H), and 0.94 (t, 7.4 Hz, 3H). B, Fragmentation patterns of MeCKs. MS spectra of 1MeiP (top) and 2MeiP (bottom) were analyzed with a hybrid quadrupole-Orbitrap mass spectrometer in the positive ion mode. Predicted fragmented structures are shown above the peaks.
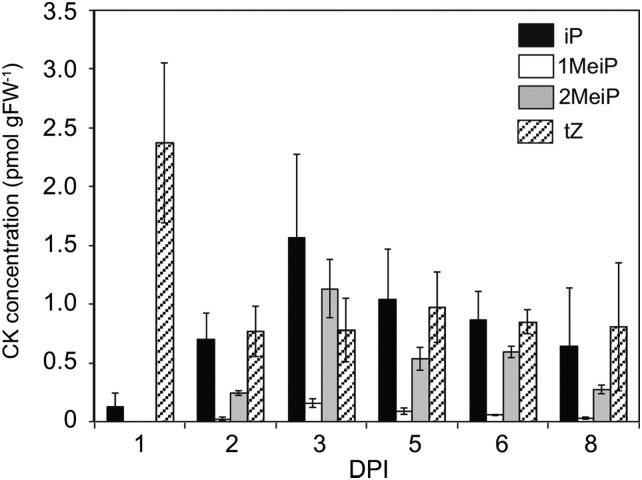
To ascertain whether MeCKs are involved in the infection process, tobacco (Nicotiana tabacum) plants infected with R. fascians were analyzed. In our experimental conditions, leafy galls became visible after 6 to 7 d post infection (dpi). Overall, the levels of all CKs were significantly higher in plants after infection than in the uninfected control plants, in which iP and MeCKs were below the quantification limits (Fig. 2; Supplemental Table S1). Our analyses detected 1MeiP and 2MeiP in the infected leaves of tobacco (Fig. 2). The MeCKs were not detected on 1 dpi, but from 2 dpi, the MeCK levels increased in the infected plants (Fig. 2). The levels of 2MeiP and iP were similar, reaching their highest concentrations at 3 dpi and then gradually decreasing (Fig. 2), whereas 1MeiP was only found at a low concentration. Total MeCKs were 33% of the total active CK concentration at 3 dpi. A steady decrease in the levels of tZ was observed in both infected and uninfected plants (Fig. 2). On the other hand, methylthio derivatives of CK, such as 2-methylthio-N6-(Δ2-isopentenyl)adenosine and 2-methylthio-cis-zeatin, were below the detection limit in this experiment. These results indicate that infection of R. fascians causes the accumulation of MeCKs preceding leafy gall formation.
Figure 2.
CK concentrations in tobacco plants after infection with R. fascians. Error bars represent the sd of three biological replicates. The complete data set, including cZ, is presented in Supplemental Table S1. FW, Fresh weight.
MeCKs Can Mimic CK Action
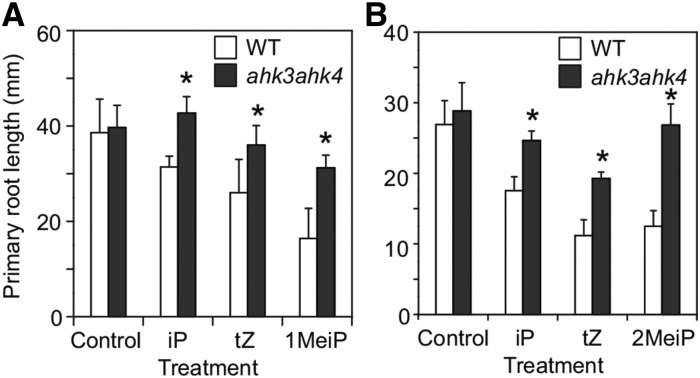
Symptoms of R. fascians infections are characteristic of CK overproduction and are correlated with a strong CK response. To examine whether the MeCKs have CK activity, the Arabidopsis wild type and the ahk3ahk4 double mutant, in which CK perception is deficient, were grown in the presence of 100 nm iP, tZ, 1MeiP, and 2MeiP, and root growth was monitored. Treatment with MeCKs significantly inhibited primary root growth, as did iP and tZ treatments (Fig. 3). The ahk3ahk4 double mutant was insensitive to both iP and MeCK treatments (Fig. 3).
Figure 3.
Effect of MeCKs on primary root growth in Arabidopsis. The Arabidopsis wild type (WT) and the ahk3ahk4 mutant were germinated and grown on Murashige and Skoog agar plates containing 100 nm iP, tZ, or 1MeiP (A) and 100 nm iP, tZ, or 2MeiP (B) for 7 d. The lengths of the primary roots were measured. Asterisks represent statistical significances between the wild type and the mutant (one-way ANOVA, P < 0.05). Error bars represent the sd of 10 biological replicates.
A previous study demonstrated that heterologous expression of Arabidopsis AHK4 in budding yeast (Saccharomyces cerevisiae) complements the yeast synthetic lethal of N-end rule1 (sln1) mutation in a CK-dependent manner (Inoue et al., 2001). Normal yeast growth was observed in the presence of iP and MeCKs, whereas the mutant yeast cells with the empty vector were able to grow only in the presence of Gal (Supplemental Fig. S2).
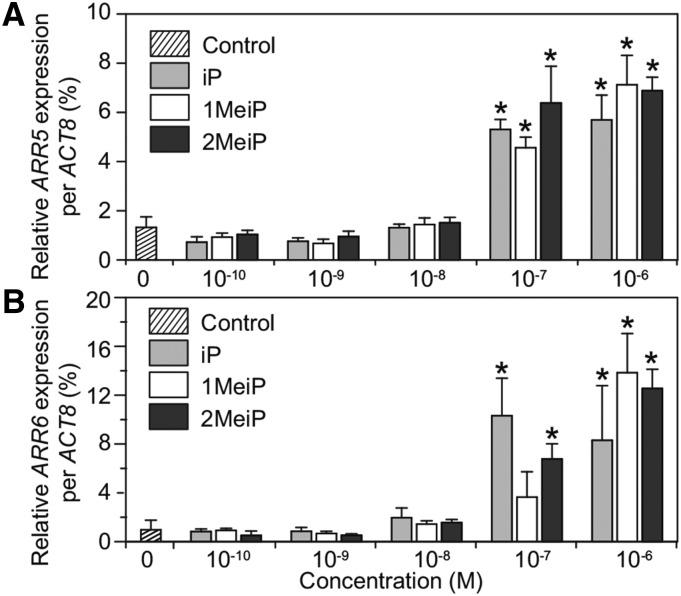
Type-A ARR genes are the primary transcriptional targets of CK signaling and are specifically induced in response to exogenous CKs (To et al., 2004). The effect of MeCKs on inducing the expression of two type-AARR genes, ARR5 and ARR6, was examined (Fig. 4). Quantitative reverse transcription-PCR analysis showed that ARR gene expression was induced by treatment with 0.1 µm MeCKs, and the dose dependency was similar with iP (Fig. 4). Together, these results show that the MeCKs have activity equivalent to canonical CKs.
Figure 4.
Induction of CK-responsive gene expression by MeCKs. Two-week-old Arabidopsis wild-type seedlings were treated with iP, 1MeiP, or 2MeiP at the indicated concentrations by vacuum infiltration for 1 h. Expression levels of ARR5 (A) and ARR6 (B) were analyzed by quantitative reverse transcription-PCR. The expression levels were normalized using ACTIN8 (ACT8) as the internal control. Error bars represent the sd of four or five biological replicates. Asterisks indicate statistically significant differences compared with a mock treatment (one-way ANOVA, P < 0.05).
In Planta Stability of MeCKs
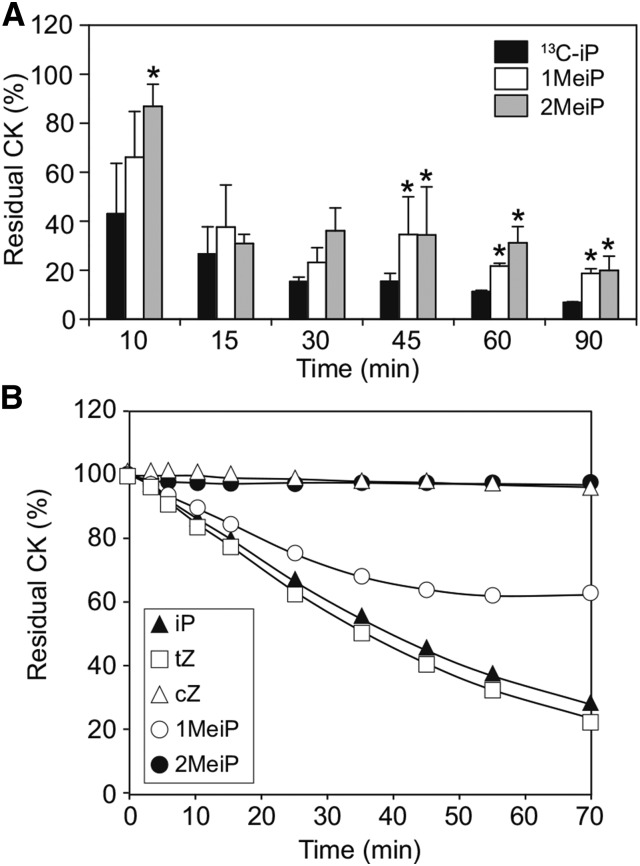
R. fascians infection triggers a homeostasis mechanism in the host plant directed to reduce excess CKs, as shown by the down-regulation of plant IPT genes and activation of CK-degrading CKX enzymes (Depuydt et al., 2008). To monitor the fate of MeCKs in the homeostasis mechanism, we examined their in planta stability (Fig. 5). Wild-type Arabidopsis plants supplied with equal amounts of 13C-labeled iP and MeCKs were found to retain a significantly higher percentage of MeCKs (approximately 20%–30%) after 60 min, whereas levels of [13C]iP were reduced to approximately 10% (Fig. 5A), suggesting that MeCKs are more stable in planta compared with iP. Additionally, an in vitro assay with partially purified recombinant CKX2 showed that MeCKs are poor substrates for this enzyme when compared with iP (Fig. 5B). Similar to cZ, which is barely degraded by CKX, 2MeiP was not degraded by CKX2, and 1MeiP was less suitable as a substrate than iP and tZ (Fig. 5B) as measured by the reduction of the electron acceptor 2,6-dichloroindophenol. These results show that methylation of iP could circumvent or delay the homeostasis mechanisms leading to hyperaccumulation of CKs in host plants.
Figure 5.
Stability of MeCKs. A, Comparison of the metabolism of CKs in planta. Two-week-old Arabidopsis wild-type seedlings were treated with 1 μm 13C-labeled iP, 1MeiP, or 2MeiP by vacuum infiltration for 1 h. The seedlings were then placed in water and harvested at the indicated times. The residual amount of supplied CKs in the seedling tissues was analyzed. Asterisks represent statistically significant differences among the various treatments (one-way ANOVA, Tukey’s honestly significant difference posthoc test, P < 0.05). Error bars represent the sd of three biological replicates. B, Reactivity of MeCKs to CKX in vitro. The indicated CK compounds (50 µm) were incubated with recombinant Arabidopsis CKX2 in the presence of 2,6-dichlorophenolindophenol. Reduction of 2,6-dichlorophenolindophenol was monitored by measuring the A600 at the indicated times.
Biosynthesis of CK Mimics
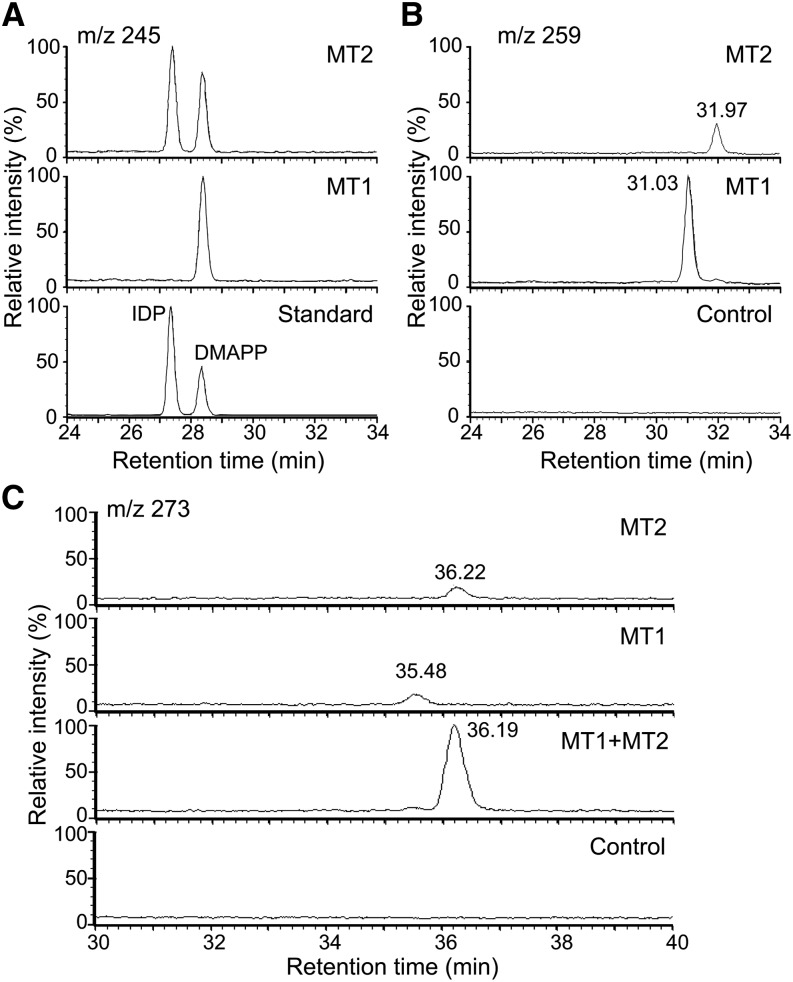
To investigate the biosynthesis of MeCKs, recombinant MTs and FAS4 proteins were purified. The recombinant MTs (MT1 and MT2) were assayed for methyltransferase activity in vitro using S-adenosyl methionine (SAM), various CKs and their conjugates, iP, iP riboside, iP riboside 5′-monophosphate, tZ, tZ riboside, and tZ riboside 5′-monophosphate, and isoprenoid diphosphates such as DMAPP, isopentenyl diphosphate (IDP), and 1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate as substrates. Interestingly, the only substrate that was methylated in our assay was IDP (Fig. 6A). Methylation by MT1 and MT2 led to different reaction products with IDP, as evidenced by their different retention times (Fig. 6B). Both MT1 and MT2 reaction mixtures had very low amounts of the dimethylated IDP (m/z = 273; Fig. 6C). When both MT1 and MT2 enzymes were present together in the reaction, dimethylated IDP was formed in higher amounts (Fig. 6C). These results were further confirmed by gas chromatography (GC)-MS analysis of the corresponding alcohols obtained after dephosphorylation by alkaline phosphatase (Supplemental Fig. S3). The MT2 reaction produced 3-methyl-2-penten-1-ol, as confirmed by comparison with a synthetic standard (Supplemental Fig. S3, C and D). A mixture of MT1 and MT2 enzymes led to 3-ethyl-2-penten-1-ol, also confirmed by comparison with a synthetic standard (Supplemental Fig. S3, E and F). The MT1 product could not be identified due to noncorrespondence with any available candidate standards. Our results clearly showed that MT1 and MT2 transform IDP in different ways, leading to monomethylated products, and when present together, they give rise to dimethylated IDP. Notably, the monomethylated products of MT2 have mass fragments corresponding to allyl alcohol (m/z = 71.1; Supplemental Fig. S3, B–D), suggesting that the MT reaction also isomerizes IDP.
Figure 6.
Characterization of MT1 and MT2 substrates. LC-MS chromatograms of in vitro MT assay reaction products. The reaction mixtures containing SAM, IDP, and DMAPP and MT enzymes in MT buffer were incubated at 30°C for 20 h. The reaction products were analyzed by LC-MS with selected ion modes: the chromatograms of m/z = 245, which corresponds to IDP and DMAPP (A); m/z = 259, which corresponds to a monomethylated prenyl substrate (B); and m/z = 273, which corresponds to a dimethylated prenyl substrate (C). In A, the chromatogram of the standards is shown in the bottom section, and the highest peak in each chromatogram is indicated as 100. In B and C, the chromatograms of the control (–MT enzymes) are shown in the bottom sections, and the highest peak in each section is indicated as 100.
Next, to investigate the order of methylation, we used a sequential enzymatic reaction assay, where one of the MTs was added first, followed by addition of the other MT to the same reaction mixture. When MT2 was added first and allowed to react for a day followed by MT1, both isomers of the methylated IDP were observed, although the amount of the MT2 product was higher (Supplemental Fig. S4A). In this case, a very low level of the dimethylated product was observed (Supplemental Fig. S4B). When the order of addition was reversed, only the MT1 product was observed as a small peak (Supplemental Fig. S4A). Here, an intense peak of the dimethylated product could be seen (Supplemental Fig. S4B). This result shows that the dimethylated product is formed only when MT2 reacts on the MT1 product and not vice versa; in other words, MT2 can utilize IDP as well as the MT1 product as a substrate.
To understand whether the R. fascians FAS4 enzyme can utilize the methylated IDPs as substrates, we linked the FAS4 reaction to follow the MT reaction. The MT2 reaction followed by FAS4 and phosphatase produced 1MeiP riboside (1MeiPR) and small amounts of 2MeiP riboside (2MeiPR; Supplemental Fig. S5). This finding is in line with our earlier results where 1MeiP was purified from fas4mt2-expressing E. coli filtrates. When FAS4 was added to MT1 reaction mixtures, no product was observed, indicating that the MT1 product is not utilized by FAS4. Interestingly, the product of the sequential reaction of MT1 followed by MT2 was readily utilized by FAS4 to form 2MeiPR as a single product (Supplemental Fig. S5). Similarly, in the reaction where MT2 was added first followed by MT1 and later FAS4, 1MeiPR was produced (Supplemental Fig. S5). These results clearly show that FAS4 can utilize the MT2 product directly to synthesize 1MeiPR but cannot use the MT1 product; however, the MT1 product is used as a substrate by MT2 to form dimethylated IDP, which is then utilized by FAS4 to synthesize 2MeiPR (Supplemental Figs. S4 and S5). Thus, methylation occurs prior to prenyl transfer, and only one of the monomethylated IDPs is utilized by FAS4.
Chemical Synthesis of CK Mimics
1MeiP and 2MeiP were chemically synthesized starting from commercially available 2-butanone and 3-pentanone, respectively (Supplemental Fig. S6), and were compared with pathogen-derived MeCKs (Supplemental Fig. S7). The chemically synthesized MeCKs were tested for their inhibition of root elongation compared with the pathogen-derived MeCKs and iP (Supplemental Fig. S8). Both synthesized MeCKs and pathogen-derived MeCKs reduced root elongation in a manner similar to iP. Root elongation in the ahk3ahk4 mutant did not change in any of the treatments. There was no significant difference among various CK treatments. These results demonstrate that synthetic MeCKs are comparable in activity to MeCKs obtained from R. fascians.
DISCUSSION
Many pathogens produce phytohormones such as jasmonic acid (JA), CKs, abscisic acid, and auxins (Crocoll et al., 1991; Costacurta and Vanderleyden, 1995; Mittal and Davis, 1995; MacMillan, 2001). R. fascians is known to secrete CKs and indole-acetic acid, an auxin, but the amounts of these phytohormones are low when compared with other gall-causing pathogens (Galis et al., 2005; Depuydt et al., 2008). The low concentration of bacterial CKs might be one reason that no new CKs or CK analogs have been reported so far. In this study, two MeCKs were identified as new CK analogs from R. fascians using highly sensitive MS analysis and were isolated from E. coli cultures expressing the relevant fas genes. In an older study, 6-(3-methylpent-trans-2-enylamino)purine (Fig. 1A) was chemically synthesized as a zeatin analog and was shown to induce cell expansion in radish (Raphanus sativus) cotyledons (Letham, 1972). In our study, we identified the bacterially produced 1MeiP and 2MeiP and elucidated the biosynthetic pathway.
Methylation plays an important role in plants and is commonly used to modify metabolites. The carboxyl groups of JA, salicylic acid, GAs, and indole-acetic acid are methylated to form the corresponding methyl esters by the SABATH family of methyltransferases (Qu et al., 2010). Methylation facilitates enhanced movement by diffusion through cell membranes, and some methylated phytohormones such as methyl JA and methyl salicylic acid are biologically active as plant defense signals in vivo (Reymond and Farmer, 1998). We hypothesize that by secreting MeCKs, the pathogen can also facilitate movement across cellular membranes without a specific transport system. Nevertheless, whether bacterial MeCKs are transported or metabolized into corresponding conjugates requires further investigation. It is interesting to note that previous studies reported the accumulation of CK-methylthio derivatives in Arabidopsis after R. fascians infection as well as in R. fascians culture (Pertry et al., 2009, 2010; Stes et al., 2013). However, in our study, these CK derivatives were below the detection limit in the recombinant E. coli cultures expressing mts and fas4 as well as in the R. fascians-infected tobacco plants. One possible explanation for this discrepancy is genotype variation of the R. fascians strains used in our respective studies, resulting in a different capacity for CK-methylthio derivative production. However, in all studies, infection of R. fascians induced leafy gall formation. In addition, in vitro studies demonstrated that MT1 and MT2 catalyze the methylation of the prenyl-donor substrate for FAS4. Thus, the two MTs are not involved in the synthesis of CK-methylthio derivatives.
Functional analyses demonstrated that MeCKs mimic CK action. Their recognition by the CK receptor AHK4 and the nonresponsiveness of the ahk3ahk4 double mutant clearly showed that MeCKs are perceived in a manner similar to classical CKs. The Arabidopsis ahk3ahk4 mutants are known to respond poorly toward R. fascians infection (Pertry et al., 2009), an observation that is in line with our findings. MeCKs also induced the expression of downstream ARR genes, corroborating a previous observation that bacterial infection induces expression of the ARR genes (Pertry et al., 2010). Further, MeCKs were retained at higher levels than iP, suggesting a delay in compensatory homeostasis mechanisms. This hypothesis is further supported by our result that MeCKs are not good substrates for CKX2, an enzyme whose transcript strongly accumulates in R. fascians-infected Arabidopsis (Depuydt et al., 2008). On the other hand, in vivo experiments showed a decrease in the MeCK levels with time (Fig. 5) that could be due to either the action of other CKX enzymes or metabolic conversion into the corresponding glucosides, nucleotides, or nucleosides that are common modifications of CKs in planta (Sakakibara, 2006).
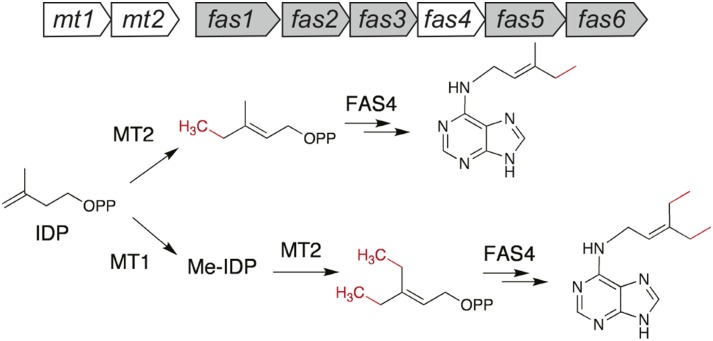
R. fascians virulence is primarily due to the CKs derived from the fas operon (Stes et al., 2011). fas1, encoding a putative cytochrome P450 monooxygenase, and fas4 are absolutely essential for virulence, as evidenced by the fas mutants characterized in the strain D188 (Crespi et al., 1992, 1994). FAS2 and FAS3 are hypothesized accessory proteins that generate the reducing power for FAS1 (Goethals et al., 2001). FAS5 is homologous to CKX, although the actual substrate is still elusive (Pertry et al., 2010; Stes et al., 2011). FAS6, a phosphoribohydrolase, is probably involved in generating CK bases essential for symptom maintenance (Pertry et al., 2010). Our study also suggests a role for mts, which is upstream of the fas genes, in R. fascians pathogenicity. Both mt1 and mt2 mutants of R. fascians are nonpathogenic and incapable of inhibiting seedling growth in tobacco (Pertry, 2009). The proximal location of methyltransferase-like genes is also reported for Streptomyces turgidiscabies, another phytopathogen that carries the fas operon, although function of the genes is not known (Joshi and Loria, 2007). Our results show that MTs are essential for MeCK biosynthesis and that IDP is the most suitable substrate for MT1 and MT2. Interestingly, FAS4 failed to use IDP as a substrate, but methylated IDP was readily utilized. The R. fascians ipt gene (fas4) has low (approximately 30%) overall amino acid identity with ipt sequences of other hyperplasia-inducing pathogens like P. savastanoi and A. tumefaciens. (Crespi et al., 1992; Temmerman et al., 2000). The FAS4 protein is unique in utilizing methylated IDP as a substrate, corroborating its different enzymatic capability. IDP is isomerized to its more active isomer, DMAPP, for canonical CK biosynthesis (Sakakibara, 2006); however, in this study, DMAPP was not a good substrate for R. fascians MTs. MeCKs are thus synthesized via a biochemical pathway similar to that of CKs but with different substrates (Fig. 7).
Figure 7.
A model of reaction schemes for MeCK production by R. fascians. MeCK biosynthesis starts from IDP, which is methylated and then utilized by FAS4. Gene organization of the fas operon and mt1 and mt2 are shown above the schemes. The MT1 and MT2 products are different, and only the MT2 product is directly utilized by FAS4 as shown. The MT1 product (methylated [Me]-IDP) is dimethylated by MT2, and the dimethylated product is used as a substrate by FAS4 to yield 2MeiPR 5′-monophosphate. The nucleotides are hypothesized to be converted to nucleobases by FAS6 or by bacterial metabolism. PP, Diphospholic acid.
We identified 3-methyl-2-pentene diphosphate as the reaction product of MT2 but could not identify the reaction product of MT1. Because the IDP plus MT1 product does not have mass fragments corresponding to allyl alcohol (Supplemental Fig. S3G), MT1 is not expected to have isomerization activity. Based on these observations, 3-methyl-3-pentene diphosphate or 3-methylidene pentane diphosphate are possible candidates.
Upon pathogen attack, a complex phytohormone signaling network is activated in plants causing defensive responses. Pathogens either suppress or hijack these pathways by secreting effectors or phytohormone mimics to promote disease establishment. The best-studied example of such molecules is the JA mimic, coronatine produced by Pseudomonas syringae (Mittal and Davis, 1995; Fonseca et al., 2009), that stimulates opening of the stomata and also suppresses SA-dependent defenses (Brooks et al., 2005). R. fascians is not known to enter through stomata and is usually present on the exterior of the infection sites (Goethals et al., 2001) but secretes highly stable MeCKs. Whether these CK mimics affect the signaling of other phytohormones remains to be elucidated. Synthetic and/or natural phytohormone mimics are vital to the agrochemical industry. Using chemical genomics approaches, many such synthetic compounds have been identified (Fonseca et al., 2014). Natural phytohormone mimics are valuable for elucidating pathogen biochemical pathways and for providing insights into the coevolutionary arms race.
MATERIALS AND METHODS
Bacterial Strains
Rhodococcus fascians 6162 (ATCC 35014, Tilford 1936) was obtained from the RIKEN BioResource Center Microbe Division (Japan Collection of Microorganisms). All experiments using the living strain were conducted in the permitted area of the BioResource Center. Four-week-old tobacco (Nicotiana tabacum) ‘Petit Havana SR1’ was infected by local application of a bacterial culture to the stump of the cut shoot apical meristem (Temmerman et al., 2000).
Overexpression of Rhodococcus spp. Genes in Escherichia coli
For the isolation of MeCKs, the coding regions of fas4, mt1 (National Center for Biotechnology Information accession no. JN093097, Region:76291.0.77142), and mt2 (National Center for Biotechnology Information accession no. JN093097, Region:77182.0.78033) were amplified with extracted DNA from the R. fascians strain using gene-specific primers (Supplemental Table S2). The PCR products were ligated into the pCOLD-IV vector (Takara). The BL21 (DE3) strain harboring pG-Tf2 (Takara) was used as the E. coli host. Expression of these three proteins was induced in modified M9 medium (Takei et al., 2001) for 20 h at 15°C. CKs were purified from the cell-free medium.
Isolation and Purification of MeCKs
The cell-free E. coli supernatants were filtered through 0.45-μm membrane filters before passing through solid phase extraction cartridges (OASIS HLB [6 g] followed by filtration through OASIS MCX [6 g]; Waters). The MCX cartridge was equilibrated with 1 m formic acid. After passing the filtrates, the cartridge was successively washed with methanol followed by 0.35 m NH4OH. The CKs were eluted with 0.35 m NH4OH in 60% (v/v) methanol. The eluent was dried and purified on Sephadex-LH20 (GE Healthcare). The fractions were further purified by semipreparative HPLC (Waters Alliance 2695 HPLC system) with an ODS column (Symmetry C18, 5.0 μm, 19 × 100 mm; Waters).
Mass Fragmentation Analyses
Purified MeCKs from E. coli liquid culture filtrates were analyzed by Q-Exactive (Thermo Scientific), a hybrid quadrupole-Orbitrap mass spectrometer in the positive ion mode, and fragmented structures were predicted with Mass Frontier (Thermo Scientific).
NMR Analyses
Spectra were recorded on an AVANCE ll-700 spectrometer (Bruker) equipped with an inverse triple resonance cryogenic probe with a z-axis gradient operating at 700.154 MHz for 1H (176.061 for 13C). All samples were inserted in 5-mm-diameter tubes and maintained at 298 K.
CK Quantification
Extraction and determination of CKs in plant material were performed as described previously (Kojima et al., 2009).
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used as the wild type. The CK receptor ahk3ahk4 double mutant was characterized previously (Higuchi et al., 2004). Plants were grown on vertical one-half-strength Murashige and Skoog agar plates supplemented with 1% (w/v) Suc at 22°C under fluorescent lights (100 μmol m–2 s–1, 16-h-light/8-h-dark cycle).
CK Activity Assays
Stock solutions of CKs were prepared in dimethyl sulfoxide (DMSO). Root growth assays used Arabidopsis plants germinated on vertical one-half-strength Murashige and Skoog agar plates containing 1% Suc supplemented with the corresponding CK (100 nm) diluted in water. Expression analysis of ARR genes used 2-week-old seedlings that were vacuum infiltrated with diluted CKs for 1 h. Entire seedlings were frozen in liquid N2 immediately and stored at –80°C until further analysis. For measuring CK stability in planta, 2-week-old seedlings were dipped in the corresponding CK solution in a 12-well plate taking care to dip the roots fully. After 1 h of uptake, the seedlings were transferred to water, and samples of whole plants were collected at selected time intervals. The sln1 yeast (Saccharomyces cerevisiae) assay was performed as previously described (Inoue et al., 2001).
Gene Expression Analyses
RNA was extracted with an RNeasy Plant Mini kit (QIAGEN). First-strand complementary DNA was synthesized by a Superscript III First-Strand Synthesis System (Life Technologies) using total RNA with oligo(dT)20 primers. The gene expression analysis was performed by quantitative reverse transcription-PCR with the StepOne Plus Real-Time PCR System (Applied Biosystems) using gene-specific primers (Supplemental Table S2) and the KAPA SYBR FAST qPCR Kit (Kapa Biosystems) according to the manufacturer’s protocol.
MS Analysis of Isoprenoid Phosphates
Analyses were performed by LC-MS equipped with an electrospray interface (Alliance 2695/ZQ2000; Waters) with an AS11-HC column (250 × 2 mm, 13 μm; Dionex) as described (Kollas et al., 2002). The capillary was held at 350°C at a voltage of 3.5 kV, and the peaks were detected by single ion recording in negative mode. For the analyses of MT products by GC-MS, the in vitro assay mixture was treated with alkaline phosphatase (Takara) for 30 min. The reaction was then purified by extraction with diethyl ether, dried over anhydrous MgSO4, evaporated, and suspended in n-hexane for analysis. GC-MS was performed on a 7890A GC system equipped with a 5975C triple-axis mass selective detector (Agilent Technologies).
Recombinant Enzymes and Enzyme Assays
The coding regions of mt1, mt2, and fas4 were ligated into the pCOLD1 (Takara) vector to express the corresponding His-tagged recombinant proteins. BL21 (DE3) harboring pG-Tf2 (Takara) was the host. Detailed procedures for purification of the recombinant proteins were described previously (Takei et al., 2001). The recombinant MT proteins were assayed for methyltransferase activity as follows: MT buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 10 mm MgCl2, 1 mg mL–1 bovine serum albumin, and 0.5 mm dithiothreitol), 2.5 mm SAM (Wako Chemicals), 100 μm IDP, DMAPP, or 1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate, and 40 μg of recombinant enzyme for a 100-μL reaction mixture. Arabidopsis CKX2 recombinant enzyme was prepared as described (Frébortová et al., 2007). The CKX assay was conducted as described (Laskey et al., 2003) with two independent assays.
Chemical Synthesis of 1MeiP and 2MeiP
The procedure was modified from Ogura et al. (1970). NaH (60% [w/v] in mineral oil, 8.6 mmol) was washed with hexane (10 mL) three times. After removing the hexane, the mixture was cooled on ice after the addition of 4 mL of dry tetrahydrofuran. To this, diethyl methoxycarbonylmethyl phosphonate (8.6 mmol) was slowly added in 1-mL aliquots to the dry tetrahydrofuran to avoid a violent reaction. Subsequently, the corresponding methyl alkyl ketone (8.6 mmol) was added and stirred at room temperature for 2 h. After completion, the reaction mixture was extracted into hexane and dried over anhydrous MgSO4 overnight, and the solvents evaporated to yield 1. The products were confirmed by NMR analysis: methyl-3-ethylpent-2-enoate, 1H NMR (CDCl3) δ (ppm) = 5.61 (s, 1H), 3.68 (s, 3H), 2.62 (q, 7.5 Hz, 2H), 2.19 (q, 7.4 Hz, 2H), 1.07 (t, 7.5 Hz, 3H), and 1.07 (t, 7.2 Hz, 3H) and methyl (E)-3-methylpent-2-enoate (purified by distillation), 1H NMR (CDCl3) δ (ppm) = 5.64 (s, 1H), 3.66(s, 3H), 2.14 (q, 7.5 Hz, 2H), 2.14 (s, 3H), and 1.04 (t, 7.5 Hz, 3H).
To prepare the alcohol, the corresponding alkenoate (2.1 mmol) in dry diethyl ether (5 mL) was added drop-wise to a solution of LiAlH4 (3.2 mmol) in dry ether (7–8 mL) cooled on ice and then stirred overnight at room temperature. The next day, ice-cold water was added drop-wise to the cold reaction mixture, followed by 10 mL of 10% (w/v) NaOH, and the precipitate was filtered. The filtrate was extracted in ether and dried to obtain the alcohols: 3-ethylpent-2-en-1-ol, 1H NMR (CDCl3) δ (ppm) = 5.36 (t, 6.9Hz, 1H), 4.17 (d1, 6.4 Hz, d2 6.0 Hz, 2H), 2.10 (q, 7.6 Hz, 2H), 2.06 (q, 7.4 Hz, 2H), 1.02 (t, 7.4 Hz, 3H), and 0.99 (t, 7.66 Hz, 3H) and (E)-3-methylpent-2-en-1-ol, 1H NMR (CDCl3) δ (ppm) = 5.41 (t, 6.9 Hz, 1H), 4.16 (dd, 5.2 Hz, 2H), 2.04 (q, 7.4 Hz, 2H), 1.68 (s, 3H), and 1.02 (t, 2.5 Hz, 3H). The next step in preparing tetracetylated adenosine followed by the Mitsunobu reaction was done according to a published procedure (Taravov et al., 2011) using diisopropyl azodicarboxylate instead of diethyl azodicarboxylate for the synthesis of N6-acetyl-2′,3′5′-tri-O-acetyl-N6(3-ethylpent-2-enyl)-adenosine. This step was followed by an enzymatic reaction with E. coli purine nucleoside phosphorylase and alkaline phosphatase (Takei et al., 2003) to obtain 1MeiP and 2MeiP as the final products, respectively. An outline is shown in Supplemental Figure S6.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JN093097 for mt1 and mt2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Detection of CK-like compounds in liquid culture filtrates of E. coli coexpressing fas4 and mts.
Supplemental Figure S2. CK-dependent growth of yeast sln1Δ expressing Arabidopsis AHK4.
Supplemental Figure S3. Mass spectra of methylated prenyl compounds in GC-MS with electron ionization.
Supplemental Figure S4. Sequential reaction assay.
Supplemental Figure S5. Reaction products generated by the R. fascians FAS4 recombinant protein using the MT products as substrates.
Supplemental Figure S6. Chemical synthesis of 1MeiP and 2MeiP.
Supplemental Figure S7. Fragmentation patterns of purified and synthesized MeCKs.
Supplemental Figure S8. Effect of chemically synthesized MeCKs on primary root growth in Arabidopsis.
Supplemental Table S1. CK concentrations in tobacco plants after infection with R. fascians.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Haruhiko Taneda for tobacco seeds and Dr. Tatsuo Kakimoto for the ahk3ahk4 mutant.
Glossary
- CK
cytokinin
- MeCK
methylated cytokinin
- iP
N6-(∆2-isopentenyl)adenine
- tZ
trans-zeatin
- cZ
cis-zeatin
- DMAPP
dimethylallyl diphosphate
- MS
mass spectrometry
- LC
liquid chromatography
- m/z
mass-to-charge ratio
- 1MeiP
monomethylated N6-(∆2-isopentenyl)adenine
- 2MeiP
dimethylated N6-(∆2-isopentenyl)adenine
- dpi
days post infection
- SAM
S-adenosyl methionine
- IDP
isopentenyl diphosphate
- GC
gas chromatography
- 1MeiPR
monomethylated N6-(∆2-isopentenyl)adenine riboside
- 2MeiPR
dimethylated N6-(∆2-isopentenyl)adenine riboside
- JA
jasmonic acid
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported in part by the Japan Society for the Promotion of Sciences (KAKENHI grant nos. 24370026 and 23–01211 to H.S.), the Japan Society for the Promotion of Sciences (Postdoctoral Fellowship for Overseas Researchers to V.R.), and RIKEN (Foreign Postdoctoral Fellowship to V.R.).
Articles can be viewed without a subscription.
References
- Argueso CT, Ferreira FJ, Epple P, To JP, Hutchison CE, Schaller GE, Dangl JL, Kieber JJ (2012) Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet 8: e1002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DM, Bender CL, Kunkel BN (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol Plant Pathol 6: 629–639 [DOI] [PubMed] [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I (2010) The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell 19: 284–295 [DOI] [PubMed] [Google Scholar]
- Costacurta A, Vanderleyden J (1995) Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol 21: 1–18 [DOI] [PubMed] [Google Scholar]
- Crespi M, Messens E, Caplan AB, van Montagu M, Desomer J (1992) Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J 11: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M, Vereecke D, Temmerman W, Van Montagu M, Desomer J (1994) The fas operon of Rhodococcus fascians encodes new genes required for efficient fasciation of host plants. J Bacteriol 176: 2492–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocoll C, Kettner J, Dörffling K (1991) Abscisic acid in saprophytic and parasitic species of fungi. Phytochemistry 30: 1059–1060 [Google Scholar]
- Depuydt S, Dolezal K, Van Lijsebettens M, Moritz T, Holsters M, Vereecke D (2008) Modulation of the hormone setting by Rhodococcus fascians results in ectopic KNOX activation in Arabidopsis. Plant Physiol 146: 1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Trenkamp S, Fernie AR, Elftieh S, Renou JP, Vuylsteke M, Holsters M, Vereecke D (2009) An integrated genomics approach to define niche establishment by Rhodococcus fascians. Plant Physiol 149: 1366–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eason JR, Morris RO, Jameson PE (1996) The relationship between virulence and cytokinin production by Rhodococcus fascians (Tilford 1936) Goodfellow 1984. Plant Pathol 45: 323–331 [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Rosado A, Vaughan-Hirsch J, Bishopp A, Chini A (2014) Molecular locks and keys: the role of small molecules in phytohormone research. Front Plant Sci 5: 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frébortová J, Galuszka P, Werner T, Schmülling T, Frébort I (2007) Functional expression and purification of cytokinin dehydrogenase from Arabidopsis thaliana (AtCKX2) in Saccharomyces cerevisiae. Biol Plant 51: 673–682 [Google Scholar]
- Galis I, Bilyeu K, Wood G, Jameson PE (2005) Rhodococcus fascians: shoot proliferation without elevated cytokinins? Plant Growth Regul 46: 109–115 [Google Scholar]
- Giron D, Frago E, Glevarec G, Pieterse CMJ, Dicke M (2013) Cytokinins as key regulators in plant-microbe-insect interactions: connecting plant growth and defence. Funct Ecol 27: 599–609 [Google Scholar]
- Goethals K, Vereecke D, Jaziri M, Van Montagu M, Holsters M (2001) Leafy gall formation by Rhodococcus fascians. Annu Rev Phytopathol 39: 27–52 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Jameson PE. (2000) Cytokinins and auxins in plant-pathogen interactions: an overview. Plant Growth Regul 32: 369–380 [Google Scholar]
- Joshi MV, Loria R (2007) Streptomyces turgidiscabies possesses a functional cytokinin biosynthetic pathway and produces leafy galls. Mol Plant Microbe Interact 20: 751–758 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE (2010) The perception of cytokinin: a story 50 years in the making. Plant Physiol 154: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi-Tanaka M, Matsuoka M, Suzuki K, et al. (2009) Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollas AK, Duin EC, Eberl M, Altincicek B, Hintz M, Reichenberg A, Henschker D, Henne A, Steinbrecher I, Ostrovsky DN, et al. (2002) Functional characterization of GcpE, an essential enzyme of the non-mevalonate pathway of isoprenoid biosynthesis. FEBS Lett 532: 432–436 [DOI] [PubMed] [Google Scholar]
- Laskey JG, Patterson P, Bilyeu K, Morris RO (2003) Rate enhancement of cytokinin oxidase/dehydrogenase using 2,6-dichloroindophenol as an electron acceptor. Plant Growth Regul 40: 189–196 [Google Scholar]
- Letham DS. (1972) Cytokinin activities of compounds related to zeatin. Phytochemistry 11: 1023–1025 [Google Scholar]
- MacMillan J. (2001) Occurrence of gibberellins in vascular plants, fungi and bacteria. J Plant Growth Regul 20: 387–442 [DOI] [PubMed] [Google Scholar]
- Mittal S, Davis KR (1995) Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv tomato. Mol Plant Microbe Interact 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Ogura K, Nishino T, Koyama T, Seto S (1970) Enzymic condensation of 3-methyl-2-alkenyl pyrophosphates with isopentenyl pyrophosphate. J Am Chem Soc 92: 6036–6041 [DOI] [PubMed] [Google Scholar]
- Pertry I (2009) How the fas locus contributes to R. fascians cytokinin production: as in-depth molecular and biochemical analysis. PhD thesis. Ghent University, Ghent, Belgium [Google Scholar]
- Pertry I, Václavíková K, Depuydt S, Galuszka P, Spíchal L, Temmerman W, Stes E, Schmülling T, Kakimoto T, Van Montagu MC, et al. (2009) Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc Natl Acad Sci USA 106: 929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertry I, Václavíková K, Gemrotová M, Spíchal L, Galuszka P, Depuydt S, Temmerman W, Stes E, De Keyser A, Riefler M, et al. (2010) Rhodococcus fascians impacts plant development through the dynamic fas-mediated production of a cytokinin mix. Mol Plant Microbe Interact 23: 1164–1174 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Putnam ML, Miller ML (2007) Rhodococcus fascians in herbaceous perennials. Plant Dis 91: 1064–1076 [DOI] [PubMed] [Google Scholar]
- Qu L, Li S, Xing S (2010) Methylation of phytohormones by the SABATH methyltransferases. Chin Sci Bull 55: 2211–2218 [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JDG (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10: 372–379 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kasahara H, Ueda N, Kojima M, Takei K, Hishiyama S, Asami T, Okada K, Kamiya Y, Yamaya T, et al. (2005) Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc Natl Acad Sci USA 102: 9972–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stes E, Francis I, Pertry I, Dolzblasz A, Depuydt S, Vereecke D (2013) The leafy gall syndrome induced by Rhodococcus fascians. FEMS Microbiol Lett 342: 187–194 [DOI] [PubMed] [Google Scholar]
- Stes E, Vandeputte OM, El Jaziri M, Holsters M, Vereecke D (2011) A successful bacterial coup d’état: how Rhodococcus fascians redirects plant development. Annu Rev Phytopathol 49: 69–86 [DOI] [PubMed] [Google Scholar]
- Takei K, Dekishima Y, Eguchi T, Yamaya T, Sakakibara H (2003) A new method for enzymatic preparation of isopentenyladenine-type and trans-zeatin-type cytokinins with radioisotope-labeling. J Plant Res 116: 259–263 [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T (2001) Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem 276: 26405–26410 [DOI] [PubMed] [Google Scholar]
- Taravov VIK, Kolyachkina SV, Alexeev CS, Mikhailov SN (2011) N6-Acetyl-2′,3′,5′-tri-O-acetyladenosine; a convenient, ‘missed out’ substrate for regioselective N6-alkylations. Synthesis 15: 2483–2489 [Google Scholar]
- Temmerman W, Vereecke D, Dreesen R, Van Montagu M, Holsters M, Goethals K (2000) Leafy gall formation is controlled by fasR, an AraC-type regulatory gene in Rhodococcus fascians. J Bacteriol 182: 5832–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann KV, Sachs T (1966) The role of cytokinins in the “fasciation” disease caused by Corynebacterium fascians. Am J Bot 53: 731–739 [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters DR, McRoberts N, Fitt BDL (2008) Are green islands red herrings? Significance of green islands in plant interactions with pathogens and pests. Biol Rev Camb Philos Soc 83: 79–102 [DOI] [PubMed] [Google Scholar]
- Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T (2006) New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 8: 371–381 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.