Abstract
Reanalysis of published mass spectrometry data on Tyr-phosphorylated chloroplast proteins indicates that the majority of peptide spectrum matches reporting Tyr phosphorylation are ambiguous.
Tyr phosphorylation is a controversial issue in plant phosphoproteomics, ever since early analyses reported up to 5% Tyr phosphorylation in Arabidopsis (Arabidopsis thaliana), despite the lack of a classical Tyr kinase in the Arabidopsis genome (Sugiyama et al., 2008; de la Fuente van Bentem and Hirt, 2009). The same controversy extends to the phosphorylation of chloroplast proteins. In the past 20 years, several indications for Tyr phosphorylation in chloroplasts were reported, and Rubisco is annotated as Tyr phosphorylated protein (www.arabidopsis.org). Initially, Tullberg et al. (1998) found the protein Tyr kinase inhibitor genistein to inhibit the phosphorylation of thylakoid membrane proteins. Supported by the observed stability of some thylakoid phosphoproteins against acid and base hydrolysis, a characteristic property of phospho-Tyr, the authors argue that Tyr phosphorylation of thylakoid membrane proteins is vital for short-term acclimation responses. Similar biochemical properties were observed for autophosphorylation of the chloroplast sensor kinase CSK (Puthiyaveetil et al., 2008). Support for Tyr phosphorylation came from the cross-reactivity of thylakoid membrane proteins and Calvin cycle enzymes (e.g. Rubisco) with phospho-Tyr-specific antibodies (Forsberg and Allen, 2001; Fedina et al., 2008; Ghelis et al., 2008). With the same set of methods, no Tyr phosphorylation was observed in mitochondrial proteins (Forsberg and Allen, 2001).
The above reported data are indirect hints for Tyr phosphorylation, and none of the applied methods is sufficiently specific to serve as solid evidence. For example, all phospho-Tyr-specific antibodies have significant cross reactivity with phospho-Ser and phospho-Thr when these have an aromatic amino acid in the +1 position (Zerweck et al., 2009). Using phospho-Tyr-specific antibodies, Forsberg and Allen (2001) found genistein inhibition of light-harvesting complex II phosphorylation with a 50% inhibition of initial activity of around 15 µm. Surprisingly, the same inhibition kinetics were observed with phospho-Thr-specific antibodies, suggesting a lack of specificity of either genistein or the phospho-amino acid antibodies, or both. So far, direct proof for Tyr phosphorylation in chloroplasts by phospho-amino acid analyses is missing. However, mass spectrometry-based phosphoproteomics experiments with plant cell extracts reported phospho-Tyr-containing peptides in chloroplasts but surprisingly not in abundant thylakoid membrane proteins or Calvin cycle enzymes. A recent meta-analysis collated data from 27 published studies and several internal data sets, resulting in a cumulative data set with 5% Tyr-phosphorylated peptides in the entire data set and 12% to 19% in the mitochondria (van Wijk et al., 2014). In this data set, almost 30% of the plastid phosphoproteins are flagged as Tyr phosphorylated (90 proteins from around 300; see supplemental table 5B in van Wijk et al., 2014), standing in stark contrast to dedicated plastid phosphoproteome analyses that identified less than 1% Tyr phosphorylation in the cellular phosphoproteome and none in chloroplast proteins (Reiland et al., 2009).
Many of the phospho-Tyr-containing peptides were identified in analyses that applied multistage activation to elevate fragment ion intensity in spectra dominated by the neutral loss of phosphoric acid from phospho-Ser and/or phospho-Thr, sometimes in combination with searches for the phospho-Tyr-specific immonium ion at mass-to-charge ratio 216.0426 (see table 1 in van Wijk et al., 2014). In one instance, phospho-Tyr-specific antibodies were used to enrich Tyr phosphorylated proteins from Arabidopsis full cell extracts (Mithoe et al., 2012). Remarkably, there is almost no overlap in phospho-Tyr peptide identification between the different studies (Mithoe et al., 2012; Wu et al., 2013; van Wijk et al., 2014). Although this could be the result of diverse acquisition methods, enrichment strategies, and data interpretation software in different analyses (Bodenmiller et al., 2007), the low reproducibility and the discrepancies in phospho-Tyr detection among different analyses require further attention, because both are characteristic for incorrect peptide spectrum matches. This is a specific problem here, because false discovery rates (FDRs) accumulate in cumulated data sets.
Therefore, we decided to assess the quality of matches to Tyr phosphorylated peptides by a dedicated reanalysis of the original data and benchmarked the robustness of peptide identification by using different software tools for spectra interpretation. Different tools use different scoring schemes to calculate identification probabilities from the fragment ion spectrum; however, they all use basic rules for spectrum matching, such as consecutive b- or y-ion series, matching of the highest intensity peaks to peptide fragments, and identification of matches to plausible derivatives of the major fragments such as losses of ammonia or water. Because of the differences in scoring the identified fragment ions, software tools may interpret spectra differently. However, since the basic rules for peptide matching apply to all identifications, it is clear that robust and reliable identifications are made by different tools that agree on the same interpretation for a spectrum. A specific aspect in the interpretation of phosphopeptide spectra is the assignment of the exact modification site. Common database matching software is usually unsuitable to distinguish modifications at closely spaced amino acids, and it is insufficiently explicit when spectra do not allow distinguishing between alternatives. To circumvent this problem, specialized software tools were developed that score spectra for alternative phosphorylation sites within the peptide sequence by searching for specific fragment ions supporting one or another phosphorylation site (MacLean et al., 2008; Martin et al., 2010).
We extracted from the different data sets Tyr-phosphorylated chloroplast proteins and extracted the spectrum information in the form of a MASCOT generic file (mgf) from either PhosphAT (van Wijk et al., 2014) or PRIDE (Mithoe et al., 2012). This resulted in 139 spectra identifying 53 unique peptides representing putative Tyr phosphorylation sites in 53 chloroplast proteins (Supplemental Table S1A; Supplemental Data Set S1). This set of spectra was reanalyzed with MASCOT to assess the significance of the identifications and two alternative software tools established for database searches: PEAKS, a database matching software with a de novo sequencing option (Ma et al., 2003); and Proteome Discoverer with the search engine SEQUEST (Thermo Scientific). With the original search parameters of dynamic phosphorylation of Ser, Thr, and Tyr, dynamic oxidation of Met, fixed carbamidomethylation of Cys, and maximum of two missed cleavages at mass tolerances for precursor and fragment ion matching of 20 ppm/0.5 D (Wu et al., 2013), 50 ppm/0.8 D (internal data sets in van Wijk et al., 2014), and 10 ppm/0.9 D (Mithoe et al., 2012), 11 out of 53 unique peptides were identified with the reported amino acid sequence above the MASCOT significance threshold of P < 0.05, while 42 mgf matchings were reported as insignificant or gave rise to an unrelated peptide identification (www.matrixscience.com; Supplemental Table S1B). The lack of significance correlates with the relaxed search parameters and the many degrees of freedom allowed for peptide matching. With a variation of the above mass tolerance settings, PEAKS identified five (9%) and Proteome Discoverer identified 11 (21%) out of 53 peptides from the data set at a fully relaxed FDR, of which three (6%) identifications by PEAKS and one (2%) by Proteome Discoverer were significant (Table I, asterisks). Two peptides were identified by both tools (6%), one of them, GLAYDTSDDQQDITR, with significant scores (Table I).
Table I. Search results obtained with alternative software for 139 spectra resulting in 53 reported unique Tyr phosphorylated peptides (van Wijk et al., 2014).
Presented are those peptides that were identified at least once with one of the alternative tools at one of the indicated mass tolerance settings: precursor tolerance/tandem mass spectrometry/ion match tolerance 50 ppm/0.8 D, 20 ppm/0.5 D, or 10 ppm/0.9 D. We reported all identifications irrespective of the score. Identifications considered significant are labeled with asterisks. Provided is the FDR at which the identification was made. Proteome Discoverer has two FDR settings: below 1% (stringent) or below 5% (relaxed). All matches above a 5% FDR threshold are considered insignificant. The PEAKS FDR is calculated individually for every peptide. Dashes indicate that the peptide was not identified.
| Peptide | PEAKS |
Proteome Discoverer |
||||
|---|---|---|---|---|---|---|
| 50 ppm/0.8 D | 20 ppm/0.5 D | 10 ppm/0.9 D | 50 ppm/0.8 D | 20 ppm/0.5 D | 10 ppm/0.9 D | |
| VIYELIDDVR | 0%* | – | – | – | – | – |
| SLKPFDLYTIGNSVK | – | – | – | – | – | >5% |
| RSSVLYPASLK | – | – | – | – | – | >5% |
| RSFNVYYEDK | – | – | – | – | >5% | – |
| RRSMEPSNVYVASNSTEMEIGSHDIVK | – | – | – | >5% | – | >5% |
| LDESTGIVDYDMLEK | 0%* | 0%* | 0%* | – | – | – |
| IMESISVGGEAGGAGGAYSYNALKR | – | – | – | >5% | – | – |
| GTFYGKTEEKEPSK | – | – | – | >5% | >5% | >5% |
| GSRYVPAAFLTGLLDPVSSR | – | – | – | >5% | – | – |
| GLAYDTSDDQQDITR | 0%* | – | – | <1%* | <1%* | <1%* |
| ETYQEEQLK | – | – | – | >5% | – | – |
| EAYLDLVKKIR | – | 100% | – | – | – | – |
| YKIMGGVPVSHFNIYK | 19.20% | – | 68.80% | >5% | <1%* | <1%* |
| YIDWEVLK | – | – | – | – | >5% | >5% |
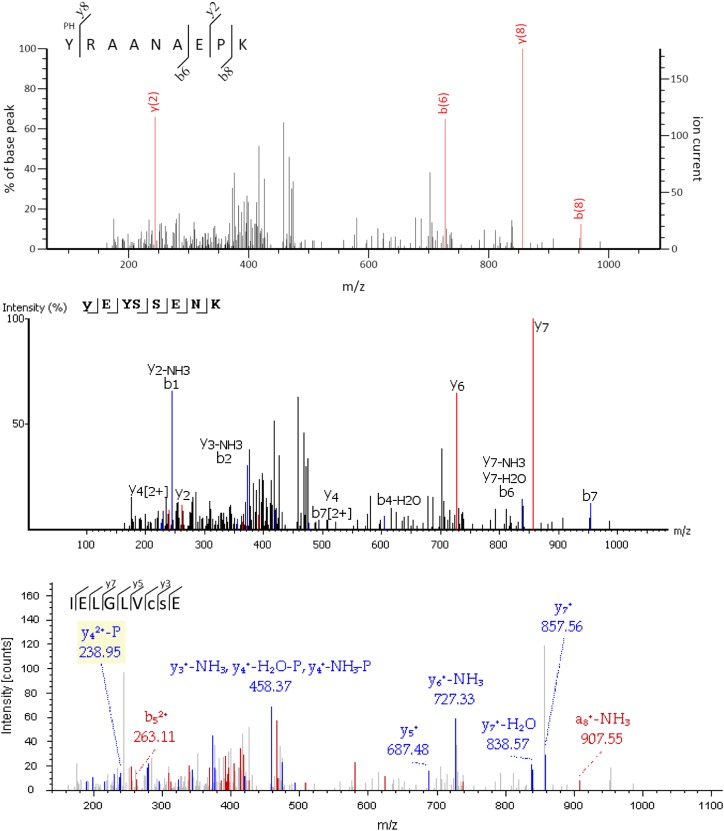
The small overlap in the identification of phosphopeptides between different software tools from the same spectra is uncommon (Kapp et al., 2005) and specific for the data set assembled here. This is illustrated by two control sets comprising either 114 randomly chosen phosphopeptides from PhosphAT (set A) or 295 mgf files from Wu et al. (2013; set B, without acetylated and pY-containing peptides). PEAKS identified 42 (37%) peptides from set A and 158 (54%) peptides from set B with the reported amino acid sequence. Proteome Discoverer identified 36 (32%) peptides from set A and 81 (27%) peptides from set B, while 27 (24%) peptides in set A and 67 (23%) peptides in set B were identified by both software tools (Supplemental Table S2). This suggests that there is no major identification problem of different software tools with the mgf compressed files, except for a small detection bias of Proteome Discoverer (see below). However, since we cannot exclude that some spectra were incorrectly matched because of compression artifacts, we next assessed the detection rate of Tyr phosphorylated peptides with uncompressed files. To this end, we downloaded the original raw files that resulted in the reporting of 27 unique Tyr phosphorylated peptides in 27 chloroplast proteins (van Wijk et al., 2014; Supplemental Table S1A). At FDRs of 2% and 5%, respectively, neither PEAKS nor Proteome Discoverer identified any of the 27 phospho-Tyr-containing peptides in chloroplast proteins, suggesting that the detection problem highlighted above is a property of poor spectrum quality (i.e. a small number of fragment ions and a weak signal-to-noise separation). Under these circumstances, ambiguous matches are reported as exemplified in Figure 1 for the spectrum that gave rise to the reported sequence pYRAANAEPK (http://phosphat.uni-hohenheim.de). In this example, all three software tools rated the match as not significant, because the quality of the spectrum is insufficient for an unambiguous match, suggesting that the original assignment was ambiguous.
Figure 1.
Different interpretations for the spectrum that gave rise to the reported sequence pYRAANAEPK. The reported sequence was retrieved from MASCOT (top). PEAKS also identifies a phosphorylated Tyr within the sequence but assigns the spectrum to a different peptide (YEYSSENK) in a nonchloroplast protein (middle; At4g24430). The best Proteome Discoverer match identified carbamidomethylated Cys and phosphorylated Ser within the sequence IELGLVCSE (bottom). There is a greater diversity of possible assignments in large search spaces (many degrees of freedom; see text); thus, care must be taken in the definition of search parameters and in the significance settings of the different identification softwares. Note that none of the identifications shown here is considered significant by the software used for the matching.
Fourteen peptides from the original data set were identified as Tyr phosphorylated with at least one alternative software tool, but the identification scores for 10 of these are connected with high FDRs (Table I) because the fragment ion matching does not sufficiently comply with the basic rules for reliable peptide identification (see above and the selection of spectra in Supplemental Fig. S1). For example, the fragment ion spectrum of RRSMEPSNVYVASNSTEMEIGSHDIVK contains few matches, unassigned high peaks, and no consecutive row of b- or y-ions, and the phosphorylation site is assigned to Ser-13 instead of Tyr-10 by PhosCalc (MacLean et al., 2008; Supplemental Fig. S1; Supplemental Table S3). Similarly, the spectrum quality for ETYQEEQLK is poor by the above standards (Supplemental Fig. S1), and PhosCalc is unable to distinguish between phosphorylation at Tyr-3 or Thr-2 (Supplemental Table S3). The same ambiguity exists for the singly phosphorylated peptide GLAYDTSDDQQDITR and the amino acids Tyr-4 and Thr-6 (Supplemental Table S3). This peptide from Rubisco activase was previously identified as Ser/Thr phosphorylated by the characteristic dominant neutral loss peak of phosphoric acid in the fragment spectrum generated by collision-induced dissociation (Reiland et al., 2009; Thingholm et al., 2009). The only significant PEAKS and PhosCalc match was obtained for the phosphorylation of LDESTGIVDYDMLEK at Tyr-10 and with relaxed PhosCalc parameters for VIYELIDDVR at Tyr-3 (Table I; Supplemental Table S3; serine hydroxymethyltransferase3 [SHM3; AT4G32520] and translation initiation factor-2 [IF-2; AT1G17220]). The mgf files for both spectra were not recognized by Proteome Discoverer because they are highly compressed and contain only matching peaks (Supplemental Fig. S1).
We started the analysis here with the goal to identify high-confidence peptide spectrum matches to phospho-Tyr-containing peptides in chloroplast proteins. However, after critical scrutiny with different software tools, de novo sequencing, and cross comparison with information in the literature, we have to conclude that the analyzed 139 spectra do not unambiguously identify phospho-Tyr in chloroplast proteins, with the possible exception of LDESTGIVDYDMLEK in SHM3 and VIYELIDDVR in IF-2. It is clear that our analysis is not suitable to prove individual reported peptide spectrum matches wrong, because spectrum assignment is often a matter of interpretation (for an example, see Fig. 1). However, our analysis illustrates that the evidence for Tyr phosphorylation in chloroplasts is weak and that the identifications of Tyr phosphorylated chloroplast proteins are uncertain, as illustrated by insignificant and contradicting peptide spectrum matches obtained with three established software tools. This shows that Tyr phosphorylation remains a rare posttranslational modification in this organelle, which is supported by low reproducibility of phospho-Tyr detection between different laboratories. From the collated data sets reporting chloroplast Tyr phosphorylated proteins (see above; Supplemental Table S1A), 77 out of 79 unique peptides were identified exclusively in one laboratory, and only two peptides (i.e. MGLVNESDSEDSSEHDKDVDDEKYWSE and YAGTEVEFNDVK) were identified by different laboratories (http://phosphat.uni-hohenheim.de).
Although we were unsuccessful in unambiguously identifying phospho-Tyr in chloroplast proteins, we do not claim by any means that it does not occur. In fact, there is no reason why chloroplasts should not use the phosphorylation of Tyr residues in signaling and why a Tyr-specific protein kinase should be absent from this organelle. Recent years uncovered that even bacterial systems utilize Tyr phosphorylation as an important part of their signaling, and Rubisco is clearly Tyr phosphorylated in Rhodomicrobium vannielii (Mann and Turner, 1988). In prokaryotes, Tyr phosphorylation is catalyzed by different kinases that have no homologs in eukaryotes (the bacterial tyrosine kinases and the odd Tyr kinases) but also by Hanks-type kinases that resemble eukaryotic dual-specificity kinases (Chao et al., 2014). Similarly, Tyr phosphorylation was also reported for cyanobacteria (Warner and Bullerjahn, 1994), and a dual-specificity kinase was identified in tobacco (Nicotiana tabacum) chloroplasts (Cho et al., 2001). Thus, there are several reasons why it is possible or even likely that chloroplasts use Tyr phosphorylation in their signaling; however, our search for clear-cut evidence for Arabidopsis chloroplast proteins was unsuccessful, and the putative targets for Tyr phosphorylation remain elusive.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Selection of spectra.
Supplemental Table S1. Phospho-Tyr containing peptides and MASCOT search results.
Supplemental Table S2. Results of searches with alternative tools.
Supplemental Table S3. Phosphorylation site assignment based on PhosCalc.
Supplemental Data Set S1. Combined mgf files.
Supplementary Material
Acknowledgments
We thank Waltraud Schulze for providing original raw data and MASCOT search results.
Glossary
- FDR
false discovery rate
- mgf
mascot generic file
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. Ba1902/2–2 and grant no. W21004490, Land Sachsen-Anhalt).
References
- Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R (2007) Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods 4: 231–237 [DOI] [PubMed] [Google Scholar]
- Chao JD, Wong D, Av-Gay Y (2014) Microbial protein-tyrosine kinases. J Biol Chem 289: 9463–9472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HS, Yoon GM, Lee SS, Kim YA, Hwang I, Choi D, Pai HS (2001) A novel dual-specificity protein kinase targeted to the chloroplast in tobacco. FEBS Lett 497: 124–130 [DOI] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Hirt H (2009) Protein tyrosine phosphorylation in plants: more abundant than expected? Trends Plant Sci 14: 71–76 [DOI] [PubMed] [Google Scholar]
- Fedina EO, Karimova FG, Tarchevsky IA, Toropygin IY, Khripach VA (2008) Effect of epibrassinolide on tyrosine phosphorylation of the Calvin cycle enzymes. Russ J Plant Physiol 55: 193–200 [Google Scholar]
- Forsberg J, Allen JF (2001) Protein tyrosine phosphorylation in the transition to light state 2 of chloroplast thylakoids. Photosynth Res 68: 71–79 [DOI] [PubMed] [Google Scholar]
- Ghelis T, Bolbach G, Clodic G, Habricot Y, Miginiac E, Sotta B, Jeannette E (2008) Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol 148: 1668–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp EA, Schütz F, Connolly LM, Chakel JA, Meza JE, Miller CA, Fenyo D, Eng JK, Adkins JN, Omenn GS, et al. (2005) An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics 5: 3475–3490 [DOI] [PubMed] [Google Scholar]
- Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom 17: 2337–2342 [DOI] [PubMed] [Google Scholar]
- MacLean D, Burrell MA, Studholme DJ, Jones AM (2008) PhosCalc: a tool for evaluating the sites of peptide phosphorylation from mass spectrometer data. BMC Res Notes 1: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann NH, Turner AM (1988) Covalent modification of ribulose 1,5-bisphosphate carboxylase/oxygenase in Rhodomicrobium vannielii. Mol Microbiol 2: 427–432 [DOI] [PubMed] [Google Scholar]
- Martin DM, Nett IR, Vandermoere F, Barber JD, Morrice NA, Ferguson MA (2010) Prophossi: automating expert validation of phosphopeptide-spectrum matches from tandem mass spectrometry. Bioinformatics 26: 2153–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithoe SC, Boersema PJ, Berke L, Snel B, Heck AJ, Menke FL (2012) Targeted quantitative phosphoproteomics approach for the detection of phospho-tyrosine signaling in plants. J Proteome Res 11: 438–448 [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S, Kavanagh TA, Cain P, Sullivan JA, Newell CA, Gray JC, Robinson C, van der Giezen M, Rogers MB, Allen JF (2008) The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc Natl Acad Sci USA 105: 10061–10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingholm TE, Jensen ON, Larsen MR (2009) Analytical strategies for phosphoproteomics. Proteomics 9: 1451–1468 [DOI] [PubMed] [Google Scholar]
- Tullberg A, Håkansson G, Race HL (1998) A protein tyrosine kinase of chloroplast thylakoid membranes phosphorylates light harvesting complex II proteins. Biochem Biophys Res Commun 250: 617–622 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ, Friso G, Walther D, Schulze WX (2014) Meta-analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell 26: 2367–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner KM, Bullerjahn GS (1994) Light-dependent tyrosine phosphorylation in the cyanobacterium Prochlorothrix hollandica. Plant Physiol 105: 629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XN, Sanchez Rodriguez C, Pertl-Obermeyer H, Obermeyer G, Schulze WX (2013) Sucrose-induced receptor kinase SIRK1 regulates a plasma membrane aquaporin in Arabidopsis. Mol Cell Proteomics 12: 2856–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerweck J, Masch A, Schutkowski M (2009) Peptide microarrays for profiling of modification state-specific antibodies. Methods Mol Biol 524: 169–180 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.