Nitrate sensed by the NRT1.1/NPF6.3 nitrate transceptor activates a PLC activity which causes an increase in the concentration of cytoplasmic Ca2+ and stimulates expression of nitrate responsive genes.
Abstract
Understanding how plants sense and respond to changes in nitrogen availability is the first step toward developing strategies for biotechnological applications, such as improvement of nitrogen use efficiency. However, components involved in nitrogen signaling pathways remain poorly characterized. Calcium is a second messenger in signal transduction pathways in plants, and it has been indirectly implicated in nitrate responses. Using aequorin reporter plants, we show that nitrate treatments transiently increase cytoplasmic Ca2+ concentration. We found that nitrate also induces cytoplasmic concentration of inositol 1,4,5-trisphosphate. Increases in inositol 1,4,5-trisphosphate and cytoplasmic Ca2+ levels in response to nitrate treatments were blocked by U73122, a pharmacological inhibitor of phospholipase C, but not by the nonfunctional phospholipase C inhibitor analog U73343. In addition, increase in cytoplasmic Ca2+ levels in response to nitrate treatments was abolished in mutants of the nitrate transceptor NITRATE TRANSPORTER1.1/Arabidopsis (Arabidopsis thaliana) NITRATE TRANSPORTER1 PEPTIDE TRANSPORTER FAMILY6.3. Gene expression of nitrate-responsive genes was severely affected by pretreatments with Ca2+ channel blockers or phospholipase C inhibitors. These results indicate that Ca2+ acts as a second messenger in the nitrate signaling pathway of Arabidopsis. Our results suggest a model where NRT1.1/AtNPF6.3 and a phospholipase C activity mediate the increase of Ca2+ in response to nitrate required for changes in expression of prototypical nitrate-responsive genes.
Plants are sessile organisms that evolved sophisticated sensing and response mechanisms to adapt to changing environmental conditions. Calcium, a ubiquitous second messenger in all eukaryotes, has been implicated in plant signaling pathways (Harper et al., 2004; Hetherington and Brownlee, 2004; Reddy and Reddy, 2004; Hepler, 2005). Multiple abiotic and biotic cues elicit specific and distinct spatiotemporal patterns of change in the concentration of cytosolic Ca2+ ([Ca2+]cyt) in plants (Sanders et al., 2002; Hetherington and Brownlee, 2004; Reddy and Reddy, 2004; Hepler, 2005). Abscisic acid and heat shock treatments cause a rapid intracellular Ca2+ increase that is preceded by a transient increase in the level of inositol 1,4,5-trisphosphate (IP3; Sanchez and Chua, 2001; Zheng et al., 2012). Ca2+ signatures are detected, decoded, and transmitted to downstream responses by a set of Ca2+ binding proteins that functions as Ca2+ sensors (White and Broadley, 2003; Dodd et al., 2010).
Nitrate is the main source of N in agriculture and a potent signal that regulates the expression of hundreds of genes (Wang et al., 2004; Vidal and Gutiérrez, 2008; Ho and Tsay, 2010). Despite progress in identifying genome-wide responses, only a handful of molecular components involved in nitrate signaling has been identified. Several pieces of evidence indicate that NITRATE TRANSPORTER1.1 (NRT1.1)/Arabidopsis (Arabidopsis thaliana) NITRATE TRANSPORTER1 PEPTIDE TRANSPORTER FAMILY6.3 (AtNPF6.3) is a nitrate sensor in Arabidopsis (Ho et al., 2009; Gojon et al., 2011; Bouguyon et al., 2015). NRT1.1/AtNPF6.3 is required for normal expression of more than 100 genes in response to nitrate in Arabidopsis roots (Wang et al., 2009). Downstream of NRT1.1/AtNPF6.3, CALCINEURIN B-LIKE INTERACTING SER/THR-PROTEINE KINASE8 (CIPK8) is required for normal nitrate-induced expression of primary nitrate response genes, and the CIPK23 kinase is able to control the switch from low to high affinity of NRT1.1/AtNPF6.3 (Ho et al., 2009; Hu et al., 2009; Ho and Tsay, 2010; Castaings et al., 2011). CIPKs act in concert with CALCINEURIN B-LIKE proteins, plant-specific calcium binding proteins that activate CIPKs to phosphorylate downstream targets (Albrecht et al., 2001). Early experiments using maize (Zea mays) and barley (Hordeum vulgare) detached leaves showed that nitrate induction of two nitrate primary response genes was altered by pretreating leaves with the calcium chelator EGTA or the calcium channel blocker LaCl3 (Sakakibara et al., 1997; Sueyoshi et al., 1999), suggesting an interplay between nitrate response and calcium-related signaling pathways. However, the role of calcium as a second messenger in the nitrate signaling pathway has not been directly addressed.
We show that nitrate treatments cause a rapid increase of IP3 and [Ca2+]cyt levels and that blocking phospholipase C (PLC) activity inhibits both IP3 and [Ca2+]cyt increases after nitrate treatments. We provide evidence that NRT1.1/AtNPF6.3 is required for increasing both IP3 and [Ca2+]cyt in response to nitrate treatments. Altering [Ca2+]cyt or blocking PLC activity hinders regulation of gene expression of nitrate-responsive genes. Our results indicate that Ca2+ is a second messenger in the nitrate signaling pathway of Arabidopsis.
RESULTS
Nitrate Treatments Increase [Ca2+]cyt Rapidly and Transiently in Arabidopsis Roots
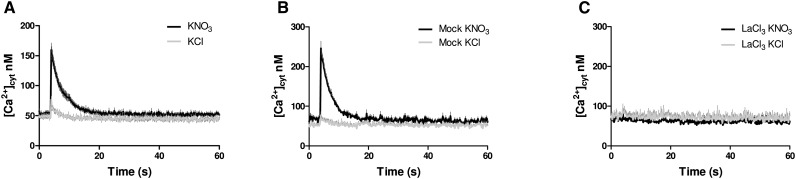
Calcium is an essential second messenger in plant signaling processes (Bush, 1995; Trewavas and Malhó, 1998). Increase in [Ca2+]cyt has been recorded in cellular responses to several stimuli (Sanders et al., 1999). As a first step to determine whether calcium acts as a second messenger in the nitrate signaling pathway, we measured [Ca2+]cyt in roots of Arabidopsis, where the transcriptomic and phenotypic responses to nitrate have been well documented (Wang et al., 2004; Gifford et al., 2008; Gutiérrez et al., 2008; Vidal et al., 2010, 2013a, 2013b, 2014; Alvarez et al., 2014). Plants expressing cytoplasmic aequorin (WT-AQ; Gao et al., 2004) were grown hydroponically for 2 weeks with ammonium as the only N source. Plant roots were excised, and luminescence was recorded every 0.2 s after treating the roots with 5 mm KNO3 or KCl as control. As shown in Figure 1A, nitrate treatment elicited a rapid and transient increase in [Ca2+]cyt in roots. KCl treatment also generated a rapid and transient peak; however, this calcium peak was considerably lower than the one obtained after nitrate treatments (Fig. 1A). After reaching a maximum, [Ca2+]cyt decreased to near basal levels (Fig. 1A).
Figure 1.
Nitrate treatments increase [Ca2+]cyt in Arabidopsis roots. Wild-type plants expressing WT-AQ were grown hydroponically for 2 weeks with 1 mm ammonium as the only N source. Aequorin was reconstituted by incubating plant roots in 2.5 µm coelenterazine (CTZ) overnight in the dark. [Ca2+]cyt levels were monitored in excised roots in response to 5 mm KNO3 or KCl treatment (A) without pretreatment (B) or with pretreatment with the Ca2+ channel blocker lanthanum chloride (LaCl3; C). Values plotted correspond to the means of at least three independent biological replicates of five plants per treatment ± sd.
It is known that abiotic and biotic cues, such as sugar, salt, and drought stress, cause transient [Ca2+]cyt in roots and leaves (Furuichi et al., 2001; Choi et al., 2014; Michal Johnson et al., 2014). This increase in [Ca2+]cyt can be partially abolished by the use of Ca2+ channel blockers, such as lanthanum chloride (Knight et al., 1996; Choi et al., 2014). Pretreatment of WT-AQ root and seedlings with 5 mm LaCl3 for 1 h inhibited the [Ca2+]cyt increase observed in response to nitrate treatment (Fig. 1B). These results indicate that nitrate treatments cause a specific increase in [Ca2+]cyt in Arabidopsis.
Phosphatidylinositol-PLC Activity Is Required for Changes in [Ca2+]cyt Levels in Response to Nitrate Treatments in Arabidopsis Roots
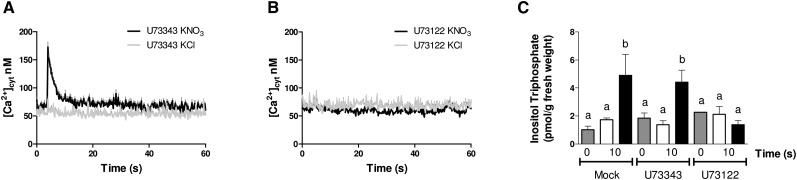
To identify components of the signal transduction pathway mediating changes in cytoplasmic calcium levels in response to nitrate, we first determined whether a PLC-dependent pathway was implicated in this [Ca2+]cyt increase. We evaluated the effect of a PLC inhibitor (U73122) and a nonfunctional PLC inhibitor analog (U73343) in WT-AQ lines in response to KNO3 or KCl treatments. WT-AQ plants were pretreated for 1 h with 10 µm U73122 or U73343, and luminescence of excised plant roots was recorded every 0.2 s after 5 mm KNO3 or KCl treatment. The presence of the PLC inhibitor (U73122) altered the [Ca2+]cyt increase in response to nitrate treatments (Fig. 2A). However, treatments with the nonfunctional analog (U73343) did not affect the [Ca2+]cyt increase in Arabidopsis roots (Fig. 2A). These results suggest that products of PLC enzyme activity or metabolites produced thereof trigger the [Ca2+]cyt increase in response to nitrate treatments. As an independent confirmation of PLC activity implicated in nitrate signaling, we measured IP3 content after nitrate treatments in Arabidopsis roots. Wild-type plants were grown and treated with KNO3 or KCl under the same experimental conditions described above, and roots were quickly collected and frozen in liquid nitrogen. Treatment with 5 mm KNO3 resulted in a 3-fold increase in IP3 levels compared with the KCl control 10 s after the treatment (Fig. 2B). Pretreatment of plants with U73122 (but not with U73343) completely blocked IP3 increase in response to nitrate (Fig. 2B). In addition, LaCl3 reduced IP3 levels in all tested conditions, suggesting that a calcium-dependent PLC activity is implicated (Supplemental Fig. S1).
Figure 2.
A PLC inhibitor blocks the increase in [Ca2+]cyt and IP3 levels in response to nitrate treatments in Arabidopsis roots. Wild-type plants expressing WT-AQ were grown hydroponically for 2 weeks with 1 mm ammonium as the only nitrogen source, and [Ca2+]cyt and IP3 levels were assayed as described in the text. [Ca2+]cyt levels were monitored in excised roots pretreated with U73343 (nonfunctional analog; A) or U73122 (PLC inhibitor; B) after we treated with KNO3 and KCl. C, Plants were pretreated with mock, U73122 (inhibitor of PLC), or U73343 (analogous no function), and we evaluated the IP3 content in Arabidopsis roots in response to 5 mm KNO3 or KCl. Values plotted correspond to the means of three independent biological replicates ± sd. Gray bars represent time zero (before treatment), white bars represent KCl treatment, and black bars represent KNO3 treatment. The letters indicate means that significantly differ between control and treatment conditions (P < 0.05).
These results indicate that a PLC activity is required for IP3 accumulation as well as increasing [Ca2+]cyt in response to nitrate treatments under our experimental conditions.
NRT1.1 Is a Positive Regulator of the [Ca2+]cyt Increase in Response to Nitrate Treatments in Arabidopsis Roots
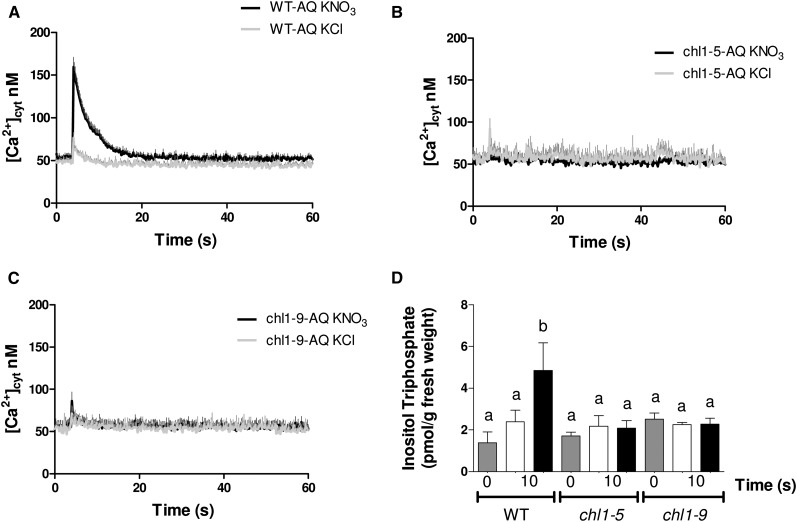
Several lines of evidence indicate that the nitrate transporter NRT1.1/AtNPF6.3 acts as a nitrate sensor in Arabidopsis plants (Ho et al., 2009; Wang et al., 2009; Gojon et al., 2011). To determine whether the increase in [Ca2+]cyt in response to nitrate requires a functional NRT1.1/AtNPF6.3, we generated a stable transgenic line that constitutively expresses aequorin in an nrt1.1 mutant background. Aequorin-expressing chlorate-resistant1-5 (chl1-5) line (chl1-5-AQ) was generated by crossing chl1-5 (Gao et al., 2004) with a transgenic line containing the 35S::aequorin construct (WT-AQ). We measured [Ca2+]cyt in chl1-5-AQ plant roots in response to nitrate using the same experimental strategy described above. As shown in Figure 3A, the increase in [Ca2+]cyt elicited by nitrate was significantly reduced in the chl1-5-AQ line compared with in the wild-type plants.
Figure 3.
NRT1.1/AtNPF6.3 is required for increases in [Ca2+]cyt and IP3 levels in response to nitrate treatments in Arabidopsis roots. Wild-type, chl1-5, and chl1-9 plants were grown hydroponically for 2 weeks with ammonium as the only nitrogen source, and [Ca2+]cyt and IP3 contents were assayed as described in the text. WT-AQ (A), chl1-5-AQ (B), and chl1-9-AQ (C) plants were reconstituted by incubating plant roots in 2.5 µm CTZ overnight in the dark. [Ca2+]cyt levels were monitored over time. D, Wild-type (WT), chl1-5, and chl1-9 plants were treated with 5 mm KNO3 and 5 mm KCl as control for 10 s, and then, we evaluated the IP3 content. Values plotted correspond to the means of at least three independent biological replicates ± sd. Gray bars represent time zero (before treatment), white bars represent KCl treatment, and black bars represent KNO3 treatment. The letters indicate means that significantly differ between control and treatment conditions (P < 0.05).
We also evaluated [Ca2+]cyt in response to nitrate treatments in aequorin reporter lines in the chl1-9 mutant background. chl1-9 has a P492L point mutation that has been shown to reduce NRT1.1/AtNPF6.3 nitrate uptake without affecting the signaling function of NRT1.1 over the NRT2.1 nitrate transporter (Ho et al., 2009). It was recently shown that this point mutation causes abnormal NRT1.1/AtNPF6.3 localization (Bouguyon et al., 2015). As shown in Figure 3B, [Ca2+]cyt is lower in chl1-9-AQ roots compared with the wild type in response to nitrate treatments and comparable with the results obtained for the chl1-5-AQ line. These results indicate that the increase in [Ca2+]cyt by nitrate depends on NRT1.1/AtNPF6.3.
To evaluate whether NRT1.1/AtNPF6.3 was part of the nitrate-PLC-Ca2+ pathway, we measured IP3 content in chl1-5 and chl1-9 mutant plant roots after nitrate treatments. chl1-5 and chl1-9 plants were grown for 15 d and treated with 5 mm KNO3 or KCl as control, and IP3 content was measured. In contrast to the increase in IP3 levels in wild-type roots, there was no significant increase in IP3 content in chl1-5 and chl1-9 mutant roots after nitrate treatments (Fig. 3C). This result indicates that accumulation of IP3 in Arabidopsis root in response to nitrate treatments also requires NRT1.1/AtNPF6.3 for activation of a PLC activity.
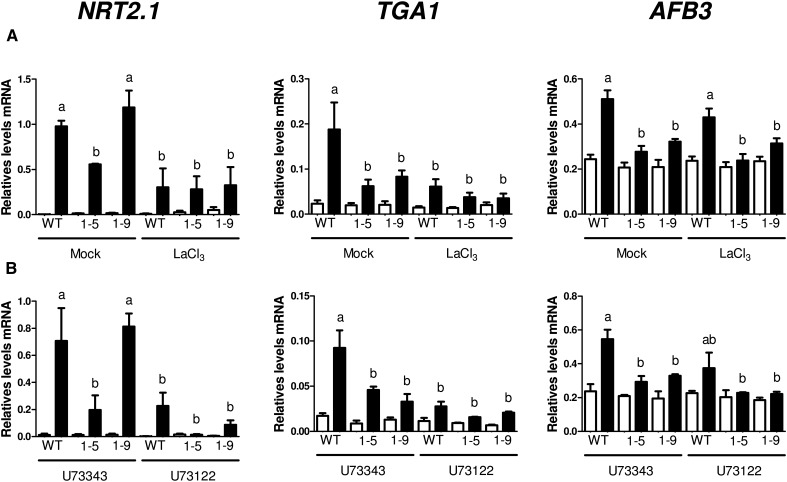
Nitrate-Induced Gene Expression Is Mediated by NRT1.1/NPF6.3, PLC, and Ca2+
To determine the impact of this signaling pathway on nitrate regulation of gene expression, we analyzed the expressions of nitrate-responsive genes, which have been shown to play important roles in nitrate-dependent root growth (Ho et al., 2009; Alvarez et al., 2014; Vidal et al., 2014) in wild-type, chl1-5, and chl1-9 plants treated with the calcium channel blocker LaCl3 or the PLC inhibitor U73122. Total RNA was isolated from roots, and mRNA levels were measured for selected genes using quantitative real-time reverse transcription (qRT)-PCR. As shown in Figure 4, NRT2.1, TGACG SEQUENCE-SPECIFIC BINDING PROTEIN1 (TGA1), and AUXIN SIGNALING F-BOX3 (AFB3) gene expressions are induced after KNO3 treatments. Consistent with previous reports (Ho et al., 2009; Alvarez et al., 2014; Vidal et al., 2014), nitrate regulation of gene expression of these genes was significantly altered in the chl1-5 and chl1-9 mutants under our experimental conditions. Similarly, nitrate inductions of NRT2.1 and TGA1 were significantly reduced in the presence of LaCl3 or U73122 but not mock or U73343 treatment (Fig. 4). Interestingly, induction of AFB3 by nitrate was not significantly affected in the presence of U73122 or LaCl3 (Fig. 4). In addition, NITRITE REDUCTASE (NIR) and NRT3.1 gene expressions behaved similarly to TGA1, with altered response to nitrate treatments in chl1-5 or chl1-9 mutant plants and in the presence of U73122 or LaCl3 (Supplemental Fig. S2). This indicates that NRT1.1/AtNPF6.3, a PLC activity, and an increase in [Ca2+]cyt levels are required for changes in gene expression in response to nitrate treatments in Arabidopsis. Moreover, these results suggest the existence of Ca2+-dependent and -independent pathways downstream of NRT1.1/AtNPF6.3 to control gene expression of nitrate-responsive genes (Fig. 5).
Figure 4.
Regulation of gene expression in response to nitrate treatments is mediated by NRT1.1/AtNPF6.3, a PLC activity, and Ca2+ in Arabidopsis roots. Col-0, chl1-5, and chl1-9 plants were grown for 15 d. Plants were pretreated with 5 mm LaCl3 (A) or 10 µm U73122 or U73343 (B) and then, treated with 5 mm KNO3 or KCl as control. Values plotted correspond to the means of three independent biological replicates ± sd. White bars represent KCl treatment, and black bars represent KNO3 treatment. The ADAPTOR PROTEIN4 μ-ADAPTIN gene (At4g24550) was used as a normalization reference (Aceituno et al., 2008). The letters indicate means that significantly differ between control and pharmacological treatment (P < 0.05). WT, Wild type.
Figure 5.
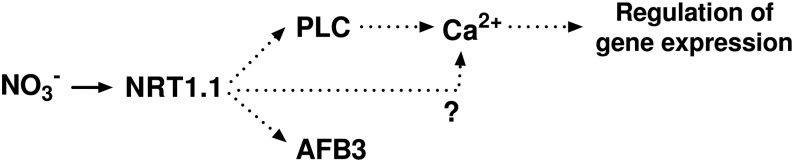
A simplified model of the NRT1.1/AtNPF6.3 calcium-dependent and -independent nitrate signaling pathway. Nitrate is sensed by NRT1.1/AtNPF6.3 and activates a PLC activity that increases [Ca2+]cyt. Increase in [Ca2+]cyt activates gene expression of nitrate-responsive genes.
DISCUSSION
Calcium is a second messenger implicated in various signaling pathways in plants (Sanders et al., 2002; Harper et al., 2004; Hetherington and Brownlee, 2004; Reddy and Reddy, 2004; Hepler, 2005; Dodd et al., 2010), and changes in [Ca2+]cyt are an important component of these calcium signaling networks. These changes can be induced by diverse environmental stimuli, including salt and oxidative stress, cold, light, hormones, and bacterial and fungal pathogens (Polisensky and Braam, 1996; Stoelzle et al., 2003; Chen and Kao, 2012; Choi et al., 2014; Gilroy et al., 2014). We found that nitrate is also able to trigger changes in [Ca2+]cyt. Moreover, we found that nitrate treatments increase IP3 levels, which correlate with an increase in [Ca2+]cyt. In animals, IP3 is generated by the cleavage of phosphatidylinositol (PI) 4,5-bisphosphate by PI-PLC enzymes (Alexandre et al., 1999; Hirose et al., 1999). This effect was abolished in chl1-5 and chl1-9 mutant plants, indicating that NRT1.1/AtNPF6.3 function is required for increased Ca2+ and IP3 in response to nitrate treatments. We found that gene expression in response to nitrate is affected by a PLC inhibitor and a Ca2+ channel blocker, suggesting existence of a signaling pathway for nitrate sensing and signal transduction involving a perception event at or downstream of NRT1.1/AtNPF6.3, activation of a PLC activity, and calcium as a second messenger to regulate gene expression.
Arabidopsis has nine actively transcribed PI-PLC genes. AtPLC2 is expressed constitutively, but expressions of the remaining eight PI-PLC genes have been shown to be regulated by salt, cold and dehydration stress, abscisic acid, and other perturbations (Tasma et al., 2008). Interestingly, the expression of AtPLC4 and AtPLC5 genes is regulated by nitrate in Arabidopsis roots (Wang et al., 2003, 2004; Vidal et al., 2013a, 2013b; Alvarez et al., 2014; Canales et al., 2014). Our results show that inhibition of PLC activity in plant roots blocks the increase in cytosolic IP3 and Ca2+ levels in response to nitrate treatments. In addition, LaCl3 also blocked the increase in IP3 and [Ca2+]cyt levels by nitrate treatments, suggesting a calcium-dependent PLC activity (Hunt et al., 2004). These results support the idea that one or more PLCs are implicated in Arabidopsis root nitrate signaling.
The mechanism by which PLC catalyzes the generation of diacylglycerol and IP3 in animals is well understood (Alexandre et al., 1999; Hirose et al., 1999). However, although accumulation of IP3 can be detected in plants in response to various stimuli and this increase in IP3 levels correlates with increases in cytoplasmic Ca2+ levels (Sanchez and Chua, 2001; Zheng et al., 2012), no homologs of animal IP3 receptors have been described in Arabidopsis (Nagata et al., 2004). IP3 can be further phosphorylated into inositol hexaphosphate (IP6; Laxalt and Munnik, 2002; Lemtiri-Chlieh et al., 2003; Meijer and Munnik, 2003; Munnik and Vermeer, 2010). Thus, IP3 levels may function directly or through its phosphorylated product IP6 in nitrate-mediated Ca2+ release. Similarly, DAG accumulation can lead to an increase in phosphatidic acid (PA), probably by action of a phospholipase D activity (Katagiri et al., 2001; Munnik, 2001; Sang et al., 2001). PA has been shown to act as second messenger in plant signaling pathways (Katagiri et al., 2001; Munnik, 2001; Sang et al., 2001), and previous work showed that phospholipase Dε and PA participate in N signaling during nitrogen deprivation in Arabidopsis (Hong et al., 2009). However, it is unclear whether PA has an effect over cytoplasmic calcium levels in Arabidopsis.
In Arabidopsis roots, the nitrate transporter NRT1.1/AtNPF6.3 is thought to be a nitrate sensor essential for regulation of gene expression in response to changes in external nitrate (Ho et al., 2009). Mutation of NRT1.1/AtNPF6.3 and U73122 treatments have a similar inhibitory effect over [Ca2+]cyt, which suggests that NRT1.1/AtNPF6.3 and PLC belong to the same signal transduction pathway to control cytoplasmic calcium levels in response to nitrate. We found that normal response to nitrate of NIR, NRT2.1, TGA1, and NRT3.1 depends on NRT1.1/AtNPF6.3, PLC activity, and Ca2+. However, we did not observe an additional effect of U73122 or LaCl3 on nitrate regulation of gene expression in chl1-5 or chl1-9 mutant backgrounds. Our results indicate existence of a PLC-dependent signaling pathway downstream of NRT1.1/AtNPF6.3.
Treatment of detached maize and barley leaves with protein kinase inhibitors has been shown to alter the nitrate regulation of nitrate-responsive genes (Sakakibara et al., 1997; Sueyoshi et al., 1999). Furthermore, nitrate treatments induce changes in phosphorylation levels of proteins (Engelsberger and Schulze, 2012; Wang et al., 2012). Transcriptomics analysis of the nitrate response has shown that several protein kinases and phosphatases are regulated by nitrate availability (Canales et al., 2014), and the Ca2+-dependent protein kinase CIPK8 controls the nitrate response of primary nitrate-responsive genes downstream of NRT1.1 (Hu et al., 2009). These studies are consistent with our results and suggest kinase targets of the nitrate-NRT1.1-Ca2+ pathway described here to control gene expression.
We have previously shown that regulatory factors AFB3 and TGA1 are downstream of NRT1.1/AtNPF6.3 function in the Arabidopsis root nitrate response (Alvarez et al., 2014; Vidal et al., 2014). As our results indicate, TGA1 and its target NRT2.1 would operate downstream of NRT1.1/AtNPF6.3 through a calcium-dependent signaling pathway, whereas AFB3 would operate downstream of NRT1.1/AtNPF6.3 through a calcium-independent signaling pathway. This observation is consistent with previous results that indicate that AFB3- and TGA1-mediated responses act independently to control root system architecture in response to nitrate (Alvarez et al., 2014; Vidal et al., 2014). More recently, using transcriptomics and phenotypic analysis of NRT1.1/NPF6.3 mutants, Bouguyon et al. (2015) showed that multiple signaling pathways act downstream of NRT1.1/NPF6.3. Our results are also consistent with these observations and show that at least one signaling pathway downstream of NRT1.1/NPF6.3 depends on PLC, IP3, and Ca2+.
Our combined cell biology and molecular genetics approach allowed us to identify steps in the nitrate signaling pathway that involve Ca2+ as second messenger in the regulation of prototypical nitrate-responsive genes. Mapping components in the nitrate signaling pathway contributes to our understanding of how plants sense and respond to changes in N availability and provides unique targets for improving N use efficiency in crops.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) was used for all experiments. The Arabidopsis line expressing WT-AQ (Gao et al., 2004) was obtained from Christoph Plieth. The chl1-5 and chl1-9 mutants were donated by Yi-Fang Tsay. Plants were grown in hydroponic culture under long-day (16-h light/8-h-dark cycle) conditions at 22°C (in Percival incubators) using Murashige and Skoog salt medium without N (M531; Phytotechnology Laboratories) supplemented with 0.5 mm ammonium succinate and 0.1% (w/v) Suc. Plants were treated for the indicated periods of time at the beginning of the light cycle on day 15 with 5 mm KNO3 or KCl as a control.
Chemical Treatment of Plants
U73122, U73343, and LaCl3 were purchased from Sigma-Aldrich. Before harvesting plant material for analysis of gene expression, Col-0 seedlings were pretreated in petri dishes for 1 h in the presence of 10 µm U73122, 10 µm U73343, or 5 mm LaCl3. Plants were then treated for the indicated periods of time with 5 mm KNO3 or KCl. For aequorin measurements, plant pretreatment with all pharmacological agents was done 1 h before the addition of KNO3 or KCl to excised roots. U73122 and U733343 were dissolved in 0.1% (v/v) dimethyl sulfoxide, and LaCl3 was dissolved in water.
In Vivo Reconstitution of Aequorin and Ca2+-Dependent Luminescence Measurements
Reconstitution of aequorin in vivo with CTZ was performed as described previously (Knight et al., 1996). Synthetic native CTZ was obtained from Sigma-Aldrich. Briefly, for each experiment, we incubated 14-d-old seedlings overnight in the dark with 2.5 µm CTZ. Plant were washed with water, and roots were excised and placed in a cuvette to measure luminescence immediately after treatments. Luminescence was recorded for the duration of the experiment every 0.2 s. To convert luminescence into Ca2+ concentrations, 1 m CaCl2 and 10% (v/v) ethanol were added to discharge the remaining aequorin. Calculations of Ca2+ concentrations were performed as previously mentioned (Knight et al., 1996). Luminescence measurements were performed using a Sirius Single-Tube Luminometer (Berthold Detection Systems).
IP3 Assays
IP3 was measured as described previously (Heilmann and Perera, 2013). Briefly, plants were treated with 5 mm KNO3 or KCl for 10 s, and roots were harvested and frozen immediately in liquid N2. Frozen tissue (approximately 0.1 g) was grounded to powder and incubated with 200 µL of 10% (v/v) perchloric acid on ice for 20 min. Samples were centrifuged to remove the precipitate, the supernatant was transferred to a new tube, and the pH was adjusted to 7.5 using 1.5 m KOH and 60 mm HEPES. IP3 was measured using the Inositol 1,4,5-Triphosphate (3H) Radioreceptor Assay Kit (Perkin Elmer) according to the instructions of the manufacturer.
RNA Isolation and qRT-PCR
RNA was isolated from whole roots with the PureLink RNA Mini Kit (12183020; Life Technologies) according to the instructions of the manufacturer. Complementary DNA synthesis was carried out using the Improm-II Reverse Transcriptase according to the instructions of the manufacturer (Promega). qRT-PCR was carried out using the Brilliant SYBR Green QPCR Reagents on a Stratagene MX3000P qPCR System. The RNA levels were normalized relative to ADAPTOR PROTEIN4 μ-ADAPTIN (At4g24550; Aceituno et al., 2008).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. LaCl3 reduced the IP3 levels in roots.
Supplemental Figure S2. NRT1.1/AtNPF6.3, PLC activity, and Ca2+ are required for the nitrate-dependent up-regulation of NRT3.1 and NIR.
Supplementary Material
Acknowledgments
We thank Dr. Christoph Plieth for providing the Arabidopsis line expressing WT-AQ and Dr. Yi-Fang Tsay for donating chl1-5 and chl1-9 mutants.
Glossary
- [Ca2+]cyt
concentration of cytosolic Ca2+
- Col-0
Columbia-0
- CTZ
coelenterazine
- IP3
inositol 1,4,5-trisphosphate
- PA
phosphatidic acid
- qRT
quantitative real-time reverse transcription
- WT-AQ
cytoplasmic aequorin
Footnotes
This work was supported by the Howard Hughes Medical Institute, the Fondo de Desarrollo de Areas Prioritarias Center for Genome Regulation (grant no. 15090007), the Millennium Nucleus Center for Plant Systems and Synthetic Biology (grant no. NC130030), the Comisión Nacional de Investigación Científica y Tecnológica (PhD fellowship no. AT–24121649 to E.R., postdoctoral scholarship no. 3140336 to J.M.A., and PSD–74 Academy Insertion Fellowship to E.A.V.), and the Fondo Nacional de Desarrollo Científico y Tecnológico (grant nos. 11121225 to E.A.V., 11110095 to A.V., and 1141097 to R.A.G.).
References
- Aceituno FF, Moseyko N, Rhee SY, Gutiérrez RA (2008) The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genomics 9: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J (2001) The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandra J, Lassalles JP, Kado RT (1999) Opening of Ca2+ channels in isolated red beet root vacuole membrane by inositol 1,4,5-trisphosphate. Nature 343: 567–570 [Google Scholar]
- Alvarez JM, Riveras E, Vidal EA, Gras DE, Contreras-López O, Tamayo KP, Aceituno F, Gómez I, Ruffel S, Lejay L, et al. (2014) Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J 80: 1–13 [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, Kubeš M, Pervent M, Leran S, Lacombe B, Krouk G, Guiderdoni E, Zažímalová E, et al. (2015) Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat Plants 1: 1–8 [DOI] [PubMed] [Google Scholar]
- Bush DS. (1995) Calcium regulation in plant cells and its role in signaling. Annu Rev Plant Physiol Plant Mol Biol 46: 95–122 [Google Scholar]
- Canales J, Moyano TC, Villarroel E, Gutiérrez RA (2014) Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front Plant Sci 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Marchive C, Meyer C, Krapp A (2011) Nitrogen signalling in Arabidopsis: how to obtain insights into a complex signalling network. J Exp Bot 62: 1391–1397 [DOI] [PubMed] [Google Scholar]
- Chen YH, Kao CH (2012) Calcium is involved in nitric oxide- and auxin-induced lateral root formation in rice. Protoplasma 249: 187–195 [DOI] [PubMed] [Google Scholar]
- Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S (2014) Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci USA 111: 6497–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D (2010) The language of calcium signaling. Annu Rev Plant Biol 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX (2012) Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J 69: 978–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi T, Mori IC, Takahashi K, Muto S (2001) Sugar-induced increase in cytosolic Ca(2+) in Arabidopsis thaliana whole plants. Plant Cell Physiol 42: 1149–1155 [DOI] [PubMed] [Google Scholar]
- Gao D, Knight MR, Trewavas AJ, Sattelmacher B, Plieth C (2004) Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol 134: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Suzuki N, Miller G, Choi WG, Toyota M, Devireddy AR, Mittler R (2014) A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci 19: 623–630 [DOI] [PubMed] [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E (2011) Nitrate transceptor(s) in plants. J Exp Bot 62: 2299–2308 [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A (2004) Decoding Ca(2+) signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Heilmann I, Perera IY (2013) Measurement of inositol (1,4,5) trisphosphate in plant tissues by a competitive receptor binding assay. Methods Mol Biol 1009: 33–41 [DOI] [PubMed] [Google Scholar]
- Hepler PK. (2005) Calcium: a central regulator of plant growth and development. Plant Cell 17: 2142–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Brownlee C (2004) The generation of Ca2+ signals in plants. Annu Rev Plant Biol 55: 401–427 [DOI] [PubMed] [Google Scholar]
- Hirose K, Kadowaki S, Tanabe M, Takeshima H, Iino M (1999) Spatiotemporal dynamics of inositol 1,4,5-trisphosphate that underlies complex Ca2+ mobilization patterns. Science 284: 1527–1530 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Ho CH, Tsay YF (2010) Nitrate, ammonium, and potassium sensing and signaling. Curr Opin Plant Biol 13: 604–610 [DOI] [PubMed] [Google Scholar]
- Hong Y, Devaiah SP, Bahn SC, Thamasandra BN, Li M, Welti R, Wang X (2009) Phospholipase D epsilon and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J 58: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF (2009) AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J 57: 264–278 [DOI] [PubMed] [Google Scholar]
- Hunt L, Otterhag L, Lee JC, Lasheen T, Hunt J, Seki M, Shinozaki K, Sommarin M, Gilmour DJ, Pical C, et al. (2004) Gene-specific expression and calcium activation of Arabidopsis thaliana phospholipase C isoforms. New Phytol 162: 643–654 [DOI] [PubMed] [Google Scholar]
- Katagiri T, Takahashi S, Shinozaki K (2001) Involvement of a novel Arabidopsis phospholipase D, AtPLDdelta, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J 26: 595–605 [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxalt AM, Munnik T (2002) Phospholipid signalling in plant defence. Curr Opin Plant Biol 5: 332–338 [DOI] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EAC, Webb AAR, Manison NF, Brownlee C, Skepper JN, Chen J, Prestwich GD, Brearley CA (2003) Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc Natl Acad Sci USA 100: 10091–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HJG, Munnik T (2003) Phospholipid-based signaling in plants. Annu Rev Plant Biol 54: 265–306 [DOI] [PubMed] [Google Scholar]
- Michal Johnson J, Reichelt M, Vadassery J, Gershenzon J, Oelmüller R (2014) An Arabidopsis mutant impaired in intracellular calcium elevation is sensitive to biotic and abiotic stress. BMC Plant Biol 14: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T. (2001) Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci 6: 227–233 [DOI] [PubMed] [Google Scholar]
- Munnik T, Vermeer JEM (2010) Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ 33: 655–669 [DOI] [PubMed] [Google Scholar]
- Nagata T, Iizumi S, Satoh K, Ooka H, Kawai J, Carninci P, Hayashizaki Y, Otomo Y, Murakami K, Matsubara K, et al. (2004) Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data. Mol Biol Evol 21: 1855–1870 [DOI] [PubMed] [Google Scholar]
- Polisensky DH, Braam J (1996) Cold-shock regulation of the Arabidopsis TCH genes and the effects of modulating intracellular calcium levels. Plant Physiol 111: 1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Reddy ASN (2004) Proteomics of calcium-signaling components in plants. Phytochemistry 65: 1745–1776 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kobayashi K, Deji A, Sugiyama T (1997) Partial characterization of the signaling pathway for the nitrate-dependent expression of genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant Cell Physiol 38: 837–843 [Google Scholar]
- Sanchez JP, Chua NH (2001) Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13: 1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Brownlee C, Harper JF (1999) Communicating with calcium. Plant Cell 11: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell (Suppl) 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Cui D, Wang X (2001) Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol 126: 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoelzle S, Kagawa T, Wada M, Hedrich R, Dietrich P (2003) Blue light activates calcium-permeable channels in Arabidopsis mesophyll cells via the phototropin signaling pathway. Proc Natl Acad Sci USA 100: 1456–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi K, Mitsuyama T, Sugimoto T, Kleinhofs A, Warner RL, Oji Y (1999) Effects of inhibitors for signaling components on the expression of the genes for nitrate reductase and nitrite reductase in excised barley leaves. Soil Sci Plant Nutr 45: 1015–1019 [Google Scholar]
- Tasma IM, Brendel V, Whitham SA, Bhattacharyya MK (2008) Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol Biochem 46: 627–637 [DOI] [PubMed] [Google Scholar]
- Trewavas AJ, Malhó R (1998) Ca2+ signalling in plant cells: the big network! Curr Opin Plant Biol 1: 428–433 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Álvarez JM, Gutiérrez RA (2014) Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signal Behav 9: e28501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA (2008) A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol 11: 521–529 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Krouk G, Katari MS, Tanurdzic M, McCombie WR, Coruzzi GM, Gutiérrez RA (2013a) Integrated RNA-seq and sRNA-seq analysis identifies novel nitrate-responsive genes in Arabidopsis thaliana roots. BMC Genomics 14: 701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA (2013b) Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc Natl Acad Sci USA 110: 12840–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM (2009) A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol 151: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Zou H, Sun SSM, He JX (2012) A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. J Proteome Res 11: 2301–2315 [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR (2003) Calcium in plants. Ann Bot (Lond) 92: 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SZ, Liu YL, Li B, Shang ZL, Zhou RG, Sun DY (2012) Phosphoinositide-specific phospholipase C9 is involved in the thermotolerance of Arabidopsis. Plant J 69: 689–700 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.