A salt-related transcription factor regulates abscisic acid synthesis and signaling genes in germinating Arabidopsis seeds under saline conditions.
Abstract
Abiotic stresses, such as salinity, cause global yield loss of all major crop plants. Factors and mechanisms that can aid in plant breeding for salt stress tolerance are therefore of great importance for food and feed production. Here, we identified a MYB-like transcription factor, Salt-Related MYB1 (SRM1), that negatively affects Arabidopsis (Arabidopsis thaliana) seed germination under saline conditions by regulating the levels of the stress hormone abscisic acid (ABA). Accordingly, several ABA biosynthesis and signaling genes act directly downstream of SRM1, including SALT TOLERANT1/NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3, RESPONSIVE TO DESICCATION26, and Arabidopsis NAC DOMAIN CONTAINING PROTEIN19. Furthermore, SRM1 impacts vegetative growth and leaf shape. We show that SRM1 is an important transcriptional regulator that directly targets ABA biosynthesis and signaling-related genes and therefore may be regarded as an important regulator of ABA-mediated salt stress tolerance.
In plants, the phytohormone abscisic acid (ABA) regulates seed dormancy, seedling development, and responses to abiotic, including drought and salt, and biotic stresses (Cutler et al., 2010). Under drought and salt stress conditions, ABA is de novo synthesized and activates downstream signal transduction pathways, which leads to physiological and cellular responses aimed at countering the stress. The biosynthetic route of ABA is well established in Arabidopsis (Arabidopsis thaliana; Schwartz et al., 2003; Nambara and Marion-Poll, 2005; Finkelstein, 2013), where genes encoding the enzymes involved in its synthesis have been identified and characterized (Marin et al., 1996; Schwartz et al., 1997, 2003; Iuchi et al., 2000; Seo et al., 2000; Bittner et al., 2001; Cheng et al., 2002; González-Guzmán et al., 2002; North et al., 2007). The key regulatory enzymatic step in ABA biosynthesis is mediated by 9-cis-epoxycarotenoid dioxygenases (NCEDs) that cleave the C40 epoxycarotenoids to produce xanthoxin (Schwartz et al., 2003; Tan et al., 2003). Among the nine NCED family members in Arabidopsis, NCED3/SALT TOLERANT1 (STO1) is the major NCED and affects both constitutive and stress-induced ABA levels in shoot tissues (Iuchi et al., 2001). Moreover, NCED3/STO1 is induced by salt and external ABA applications (Barrero et al., 2006), and overexpression of NCED3/STO1 results in increased endogenous ABA levels. However, although the loss of NCED3/STO1 confers salt-resistant germination (Ruggiero et al., 2004), the mutant displays hypersensitivity to drought stress (Iuchi et al., 2001; Wan and Li, 2006). These observations indicate that improving abiotic stress tolerance in plants by directly altering the levels of NCED3/STO1 is unlikely to be rewarding.
The central role of NCED3/STO1 in stress-responsive ABA production, and the pleiotropic phenotypes that have resulted from direct manipulation of the enzyme, have led to efforts to delineate the transcriptional regulation of NCED3/STO1. Interestingly, NCED3/STO1 can be regulated via epigenetic means (Chinnusamy and Zhu, 2009). More specifically, the ARABIDOPSIS HOMOLOG of TRITHORAX1 factor can bind to the NCED3/STO1 locus and thereby change the binding affinity of RNA-polymerase II to the locus (Ding et al., 2011). Recently, the two transcription factors (TFs) ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR1 (ATAF1) and WRKY57, involved in drought tolerance, were shown to associate with the NCED3/STO1 promoter (Jiang et al., 2012; Jensen et al., 2013). Overexpression of the ATAF1 caused higher levels of NCED3/STO1 and ABA levels (Jensen et al., 2013). However, the biological meaning of this finding is unclear, as it has been reported that the loss of ATAF1 can both cause tolerance as well as hypersensitivity to drought (Lu et al., 2007; Wu et al., 2009). The TF WRKY57 is expressed ubiquitously at low levels in Arabidopsis and conferred drought tolerance to Arabidopsis plants by increasing ABA biosynthesis through alterations in NCED3/STO1 expression (Jiang et al., 2012). However, the binding of WRKY57 to the NCED3/STO1 promoter only occurred during drought conditions. While these studies have begun to shed light on the transcriptional regulation and coordination of the genes involved in drought-responsive ABA biosynthesis and signaling, much remains to be identified. This gap in knowledge is certainly also evident for regulatory aspects of ABA production during abiotic stresses apart from drought.
Drought and salt stress trigger the activation of numerous stress-responsive genes, including TFs, chaperones, osmotins, water channels, and signal transduction-related genes (Shinozaki and Yamaguchi-Shinozaki, 2007; Nakashima et al., 2009). The transcriptional response during stress is in part ABA dependent, as drought, salt, and ABA cause overlapping changes in gene expression (Fujita et al., 2011). A substantial fraction of the induced TFs belong to the basic Leu Zipper Domain, MYB, and the plant-specific NAC (NO APICAL MERISTEM [NAM], ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR [ATAF], CUP-SHAPED COTYLEDON [CUC]) domain TFs. Three closely related NAC TFs are regarded as important stress response integrators, Arabidopsis NAC DOMAIN CONTAINING PROTEIN19 (ANAC019), ANAC055, and ANAC072/RESPONSIVE TO DESICCATION26 (RD26; Fujita et al., 2004; Tran et al., 2004). Overexpression of these TFs activates a range of abiotic stress-related genes and the transgenic plants displayed improved drought tolerance (Tran et al., 2004). Yeast one-hybrid assays have begun to unravel potential upstream regulators of these TFs (Hickman et al., 2013). In addition, the bHLH TF MYC2 can bind and activate the promoters of ANAC019, ANAC055, and RD26 in response to coronatine, a toxin produced by Pseudomonas syringae (Zheng et al., 2012). A recent study further revealed that RD26 and some other NAC TFs can be antagonistically regulated by ABA and brassinosteroids (Chung et al., 2014). Nevertheless, abiotic stress-related regulators of these NAC TFs are not clearly defined.
Here, we report that a MYB-like R2R3 TF, which we refer to as Salt-Related MYB1 (SRM1), can directly activate the expression of both the key ABA biosynthetic NCED3/STO1 gene and the two prominent stress integrators RD26 and ANAC019. We conclude that SRM1 regulates ABA synthesis and signaling during seed germination and seedling development on saline conditions and that it influences vegetative growth in Arabidopsis.
RESULTS
SRM1 Affects Salt Tolerance in Arabidopsis
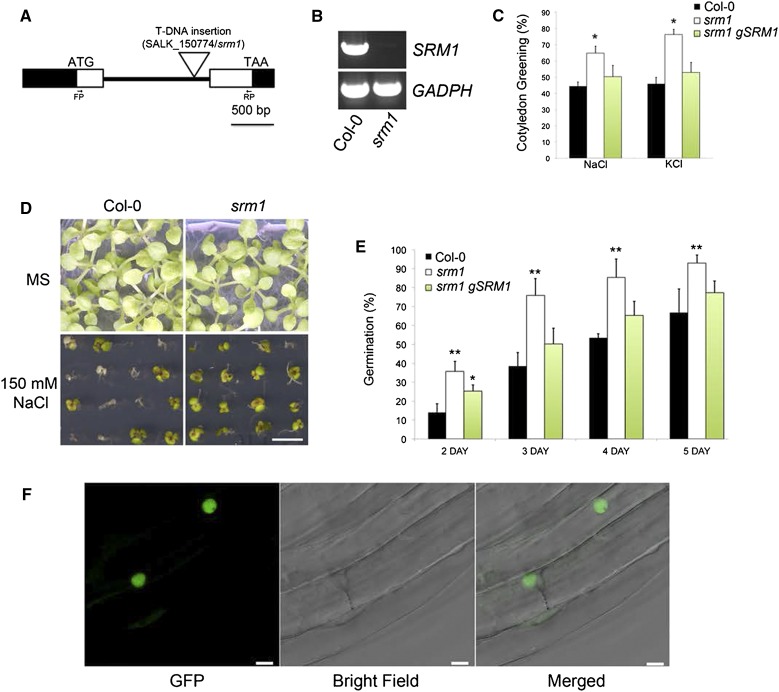
To identify stress tolerance regulators in Arabidopsis, we performed a survival-based screen on Murashige and Skoog (MS) medium supplemented with high concentration of salt (150 mm NaCl) on a homozygous transfer DNA (T-DNA) insertion line collection (Alonso et al., 2003). One T-DNA line (SALK_150774), with an insertion in the intron region of At5g08520 (Fig. 1A), survived better on the salt-containing plates than the wild type (Fig. 1, C and D). The location of the T-DNA insert was validated by sequencing, and the absence of the full-length transcript in the mutant was confirmed by reverse transcription (RT)-PCR (Fig. 1B). Apart from seedling survival, the germination of mutant seeds on NaCl (125 mm)-supplemented MS medium was also improved (Fig. 1E). At5g08520 is annotated as a MYB-related TF, which we confirmed using BLAST searches, and we therefore referred to the protein as SRM1. SRM1 is part of a clade of R2R3 MYB TFs that appears to have diversified in dicot species and that have one close homolog in rice (Oryza sativa; Supplemental Fig. S1A). To assess whether the enhanced survival of srm1 seedlings was specific to NaCl, we also performed survival tests of srm1 on MS medium supplemented with 150 mm KCl or 275 mm mannitol, or 300 mm sorbitol that has comparable osmolarity to the NaCl medium used in our screen. We found that the srm1 seedlings survived better than wild-type seedlings on both NaCl- and KCl-supplemented medium (Fig. 1C) but that seedling survival was similar to, or even worse than, the wild type on the mannitol- or sorbitol-containing medium (Supplemental Fig. S1, B and C). To confirm that the observed phenotype is due to the T-DNA insertion in the SRM1 locus, we complemented the line using a genomic SRM1 (gSRM1) construct, which restored the salt germination and seedling survival phenotypes (Fig. 1, C and E). Moreover, introduction of a 35S promoter-driven SRM1-GFP construct into the srm1 mutant also complemented the salt-related phenotypes (Supplemental Fig. S1D). Microscopic observations revealed that the SRM1-GFP protein was localized to the nucleus (Fig. 1F), consistent with a role of SRM1 as a TF. Importantly, the SRM1-GFP expression in the srm1 mutant was only moderately increased compared with that of the endogenous SRM1 gene (Fig. 2A), and the line did not show any phenotypic changes compared with wild-type plants. Taken together, our data suggest that the nuclear-localized SRM1 TF is a negative regulator of seed germination and seedling survival during salt stress.
Figure 1.
SRM1 impacts germination on medium with high salt levels. A, T-DNA insertion site in the SRM1 gene. Black boxes indicate untranslated regions, white boxes indicate exons, the black thick line denotes intron, and ATG and TAA indicate start and stop codons, respectively. FP (Forward Primer) and RP (Reverse Primer) indicate primers used for SRM1 RT-PCR. B, RT-PCR of SRM1 transcript in the wild type and srm1 (3-week-old soil-grown seedlings). Glyceraldehyde 3-phosphate dehydrogenase (GADPH) was used as mRNA control. C, Quantification of seedling greening on NaCl and KCl plates. D, Cotyledon greening of wild-type and srm1 seedlings on MS medium or MS medium supplemented with 150 mm NaCl. E, Germination time courses of srm1, Col-0, and T3 homozygous transgenic plants carrying the SRM1 genomic fragment (gSRM1) in the srm1 mutant background in the presence of 125 mm NaCl. All germination and cotyledon greening experiments were performed in triplicate, and more than 100 seeds and seedlings were used for each replicate. F, A functional 35S-driven SRM1-GFP in srm1 mutant background (35S:SRM1-GFP srm1) localizes to the nucleus in Arabidopsis seedling root cells. Error bars indicate sd. *, P < 0.05; and **, P < 0.01 (Student’s t test). Bars = 5 mm (D) and 5 µm (F).
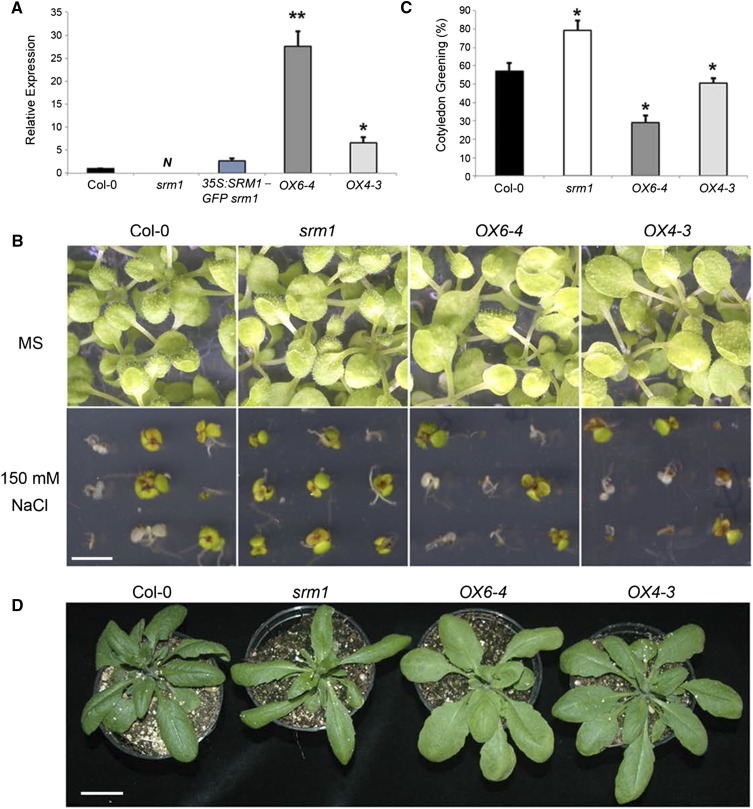
Figure 2.
SRM1 overexpressing plants display opposing phenotypes to srm1 mutant. A, qRT-PCR of SRM1 in the wild type, srm1, complementation line 35S:SRM1-GFP in srm1 mutant background (35S:SRM1-GFP srm1), and two SRM1 overexpression lines (OX6-4 and OX4-3). Three-week-old soil-grown plants were used for mRNA samples. Data are from one out of three experiments with similar profiles. B, Cotyledon greening of the wild type, srm1, and the two SRM1 overexpressing lines (OX6-4 and OX4-3) on MS medium or MS medium supplemented with 150 mm NaCl. C, Quantification of seedling greening on plates such as those in B. Three independent experiments were performed. D, Four-week-old rosettes of soil-grown wild type, srm1, and the two SRM1 overexpressing lines under long-day conditions. Error bars indicate sd. *, P < 0.05; and **, P < 0.01 (Student’s t test). Bars = 5 mm (B) and 2 cm (D).
SRM1 Is Mainly Expressed in Vegetative Tissues of Arabidopsis
To investigate the expression pattern of SRM1, we generated plants expressing a reporter gene fusion of the SRM1 promoter (ProSRM1) and a GUS reporter gene. We detected strong GUS activity in young seedlings (Supplemental Fig. S2, A–C), which is in agreement with the germination and survival phenotype of the srm1 mutant. In addition, the ProSRM1 showed activity in rosette leaves, especially in expanding and maturing leaves (Supplemental Fig. S2D). Whereas the first four true leaves, i.e. the oldest leaves, displayed weak GUS staining, the fifth and sixth leaf displayed relatively strong GUS staining (Supplemental Fig. S2D). These expression patterns were confirmed by quantitative RT (qRT)-PCR (Supplemental Fig. S2G). Apart from the leaves, also other tissues, including sepals and trichomes, displayed a strong GUS signal (Supplemental Fig. S2, E and F). These data are in close agreement with publicly available microarray data experiments (eFP browser; Winter et al., 2007).
srm1 Has Altered Leaf Morphology
The expression pattern of SRM1 in leaves prompted us to investigate whether the gene has a role in vegetative development. Interestingly, we observed differences in both rosette leaf size and shape in srm1 compared with control plants (Fig. 2D; Supplemental Fig. S3A). We sampled and aligned individual rosette leaves and found differences from the third leaf onwards of soil-grown plants (Supplemental Fig. S3B). To quantify the differences, we analyzed shape-related parameters of the third leaves from both the wild type and srm1 using the program SHAPE (Iwata and Ukai, 2002). Supplemental Figure S3C shows the major separations of the three first principal component scores of leaf shape variation, of which the first principal component accounts for 75% of the total variation. Based on the loadings of these scores, we conclude that the srm1 plants hold leaves with narrow leaf shapes compared with wild-type leaves. These data are consistent with the strong expression of SRM1 in expanding and maturing rosette leaves.
Overexpression of SRM1 Impairs Salt Tolerance But Promotes Vegetative Growth
To further investigate the impact of SRM1 on salt tolerance and plant growth, we generated SRM1 overexpressing lines. We selected two lines, transgenic lines OVEREXPRESSING LINE6-4 (OX6-4) and OX4-3, which had a 27.5-fold and 3.6-fold increase in SRM1 transcript levels compared with the wild type, respectively, for further testing (Fig. 2A). We first assessed the salt tolerance of these lines by performing a survival assay. We therefore germinated wild-type, srm1, OX4-3, and OX6-4 seeds on MS plates supplemented with 150 mm NaCl and monitored seedling survival. Figure 2, B and C, shows that the two SRM1 overexpression lines survived less well and that the greening of the cotyledons was reduced compared with both srm1 and wild-type seedlings on the salt-supplemented medium.
Furthermore, the overexpressing lines developed larger rosette leaves compared with wild-type and srm1 plants when grown on soil (Fig. 2D). While these phenotypes were largely opposite to that of the srm1 plants, it is worth noting that the increased rosette size was attributed both to changes in length and width (Supplemental Fig. S3D). More detailed measurements revealed that the first pair of rosettes leaves was not significantly different between the mutant and the two overexpression lines. Instead, the size differences started from the second pair of leaves, in which OX6-4 and OX4-3 held leaves with both increased length and width (Supplemental Fig. S3D).
These results suggest that SRM1 regulates both vegetative plant growth and salt stress tolerance of seedlings in Arabidopsis.
SRM1 Affects the Expression of Genes Related to ABA Biosynthesis and Signaling
To determine the effect of loss of SRM1 function on global gene expression, we performed microarray experiments in which we compared rosette leaves of 3-week-old soil-grown srm1 mutant plants with those of the wild type. The transcript profiling revealed 39 differentially expressed genes (with a 2-fold significant difference threshold) in the srm1 mutant compared with the wild type (Supplemental Table S1), of which 20 were down-regulated and 19 were up-regulated in the mutant (Supplemental Table S1).
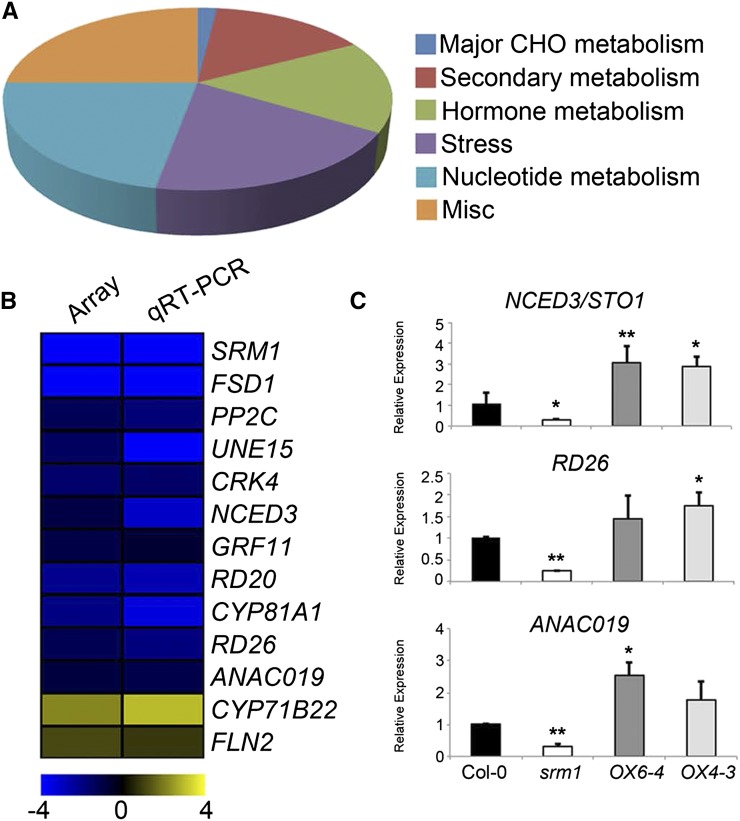
The online tool PageMan (Usadel et al., 2006) was used to analyze the overrepresentation of gene ontology (MapMan) terms of the differentially expressed genes. Apart from miscellaneous protein functions, the bins Hormone metabolism, Nucleotide metabolism, and Stress were the main processes changed in srm1 compared with the wild type (Fig. 3A; Supplemental Table S1). More specifically, the genes NCED3/STO1, RD26, RD20, and CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASE4 (CRK4; At3g05640) were significantly down-regulated in the mutant (Fig. 3B; Supplemental Table S1). These genes are linked to hyperosmotic responses and ABA biosynthesis and signaling pathways (Iuchi et al., 2000; Fujita et al., 2004; Aubert et al., 2010; Wrzaczek et al., 2010). Genes with potential ABA glucosyltransferase activity, i.e. UDP-GLUCOSYLTRANSFERASE75B1 (UGT/UGT75B1) and UGT73B1 (Lim et al., 2005), together with EARLY RESPONSIVE TO DEHYDRATION5 (ERD5), on the other hand, showed increased transcript levels in the srm1 mutant compared with the wild type (Fig. 3B; Supplemental Table S1). Taken together, several differentially expressed genes in srm1 are involved in ABA biosynthesis and signaling, indicating a role of SRM1 in ABA-related processes.
Figure 3.
Microarray and qRT-PCR analysis of altered gene expression in srm1 mutant. A, Pie chart of overrepresented PageMan ontology terms associated with genes up- (log2FC > 1, P < 0.05) or down-regulated (log2FC < –1, P < 0.05) in the srm1 compared with the wild type. B, Confirmation of microarray data using qRT-PCR. Blue and yellow colors indicate decrease and increase of gene expression, respectively. Scale bar shows log2 fold changes. C, qRT-PCR of three genes (NCED3/STO1, RD26, and ANAC019) differentially expressed from the microarray experiments in the wild type, srm1, and two SRM1 overexpression lines. Data represent three biological replicates. Error bars indicate sd. *, P < 0.05; and **, P < 0.01 (Student’s t test). CHO, CARBOHYDRATE; CYP83A1, CYTOCHROME P450 FAMILY83-SUBFAMILY A-POLYPEPTIDE1; FLN2, FRUCTOKINASE-LIKE PROTEIN2; FSD1, FE-SUPEROXIDE DISMUTASE1; GRF11, GENERAL REGULATORY FACTOR11; PP2C, PROTEIN PHOSPHATASE 2C; UNE15, UNFERTILIZED EMBRYO SAC15.
We confirmed the microarray results using qRT-PCR assays for selected genes (Fig. 3B; Supplemental Table S1). Of these genes, seven are involved in ABA and stress response signaling pathways, i.e. RD26, NCED3/STO1, PROTEIN PHOSPHATASE2C (At3g16800), RD20, CRK4, and ANAC019 (all down-regulated in srm1) and ERD5 (up-regulated in srm1). Here, it is worth noting that some of the genes picked for qRT-PCR were not significantly changed in the srm1 mutant according to the microarray analysis. However, in the context of changes in expression of other genes, we deemed some of them as interesting for qRT-PCR investigation. These included COLD REGULATED78, ARABIDOPSIS RAB GTPASE HOMOLOG B18, and ANAC019 (Supplemental Table S1). All of the genes showed significant changes in the qRT-PCR experiments in the same direction as obtained in the microarray analysis (Fig. 3B; Supplemental Table S1). In addition, the expression level of three down-regulated genes in srm1, NCED3/STO1, RD26, and ANAC019 were found to be up-regulated in the OX6-4 and OX4-3 plants (Fig. 3C; Supplemental Fig. S4). Furthermore, the expression level of NCED3/STO1, RD26, and RD20 were significantly reduced in 7-d-old srm1 seedlings (Supplemental Fig. S4B), which might explain the altered salt tolerance of srm1 seed germination and seedling survival.
SRM1 Affects ABA Responses during Seed Germination
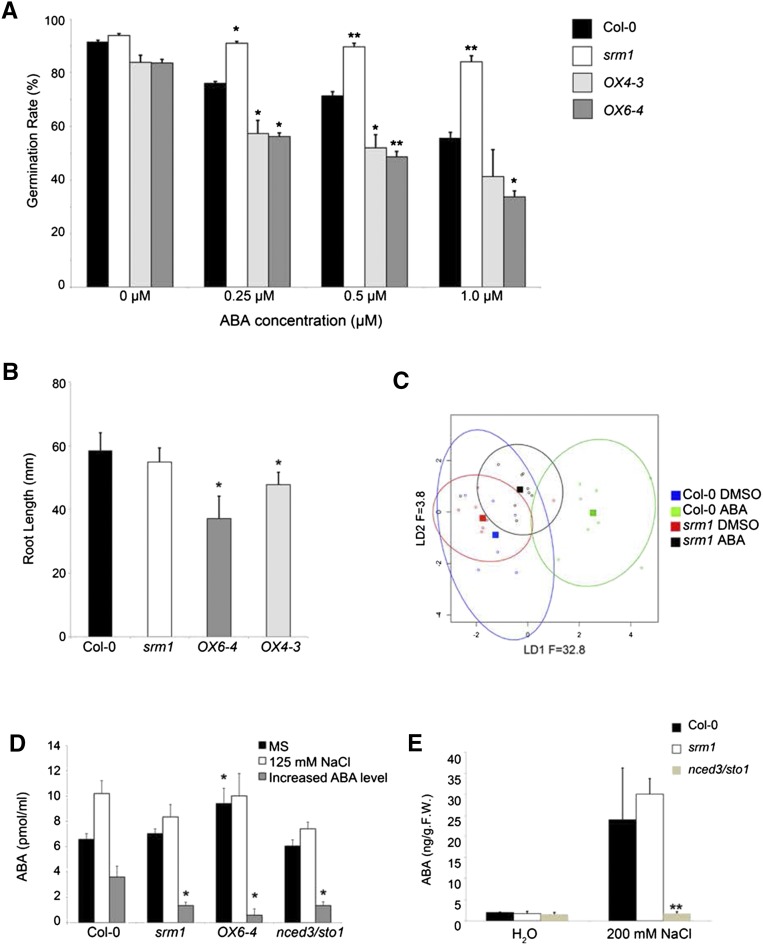
Given that the expression of key ABA-related genes was affected in plants with altered SRM1 transcript levels, we next analyzed the response of the transgenic lines to exogenous ABA. ABA mainly affects two developmental stages: seed dormancy and vegetative stage propagation (Cutler et al., 2010; Finkelstein, 2013). We therefore first investigated whether seed germination was affected by ABA in the transgenic lines. Figure 4A shows that the srm1 mutant was less sensitive to the inhibitory effect of ABA on seed germination compared with wild-type and overexpressing lines. On 1 µm ABA, only around 56% of the wild-type seeds germinated, whereas the germination of srm1 seeds was around 84%. Furthermore, the two overexpressing lines (OX6-4 and OX4-3) showed increased sensitivity to ABA, displaying germination rates of around 34% and 41%, respectively, on ABA-containing plates (Fig. 4A).
Figure 4.
SRM1 affects plant responses to ABA and impacts ABA production. A, Germination assay of the wild type, srm1, and two SRM1 overexpressing lines on MS medium or on medium supplemented with different concentrations of ABA. B, Root growth assay of the wild type, srm1, and two SRM1 overexpressing lines. Seeds for the different genotypes were germinated on MS medium and grown for 7 d under 16-h-light/8-h-dark conditions. Seedlings were then transferred to MS medium supplemented with 50 µm ABA or to plates with control medium. Root growth was measured after an additional 10 d on the new medium. C, LDA plot of leaf shape analysis of srm1 and the wild type with or without ABA treatment. Seven-day-old MS-grown seedlings were transferred to DMSO-containing and ABA-containing (2 µm) MS medium for another 2 weeks for growing. D and E, ABA levels in germinating seedlings (3-d-old seedlings and seeds grown in control plates and 5-d-old seedlings and seeds in 125 mm NaCl-containing plates) and in 3-week-old rosette leaves (without or with 2-h 200 mm NaCl treatment). All experiments were performed independently three times. Error bars indicate sd. *, P < 0.05; and **, P < 0.01 (Student’s t test). F.W., fresh weight.
We next tested whether transgenic seedling root growth was affected by exogenous application of ABA. We therefore grew seedlings for 7 d in the light on normal MS medium and then transferred them to medium supplemented with 50 µm ABA. While we did not observe any differences in root growth for the srm1 mutant, the roots of the two SRM1 overexpressing lines grew significantly less than the wild type and srm1 mutant (Fig. 4B). We also investigated whether the leaf shape phenotypes observed in the srm1 mutant were associated with ABA. To this end, we grew wild-type and srm1 seedlings for 7 d on normal MS medium and then transferred them to medium supplied with 2 μm ABA, where they were maintained for another 2 weeks. Figure 4C shows the separation achieved using linear discriminant analysis (LDA) and plots of the average shape for each genotype and treatment. Leaf shape was significantly affected by both genotype (P = 0.008) and ABA treatment (P < 0.001). However, the interaction between genotype and treatment was not significant (P = 0.07), indicating that the ABA treatment had the same effect on the wild type and srm1. Interestingly, ABA had a dramatic effect on the shape of leaf 5 in wild-type plants (P < 0.001), making the leaves broader, particularly close to the leaf base. This was manifested on the first linear discriminant axis (LD1; Fig. 4C), where the distance between the average shapes is an indication of the strength of the ABA response. The srm1 exhibited a qualitatively similar response to ABA, with a broadening of the leaf, which was highly significant (P = 0.008); however, the response was not as strong as in the wild type (61% lower separation along LD1), indicating that the mutant leaf shape is affected differently by ABA. Nevertheless, ABA did affect the leaf width, and we therefore conclude that at least part of the narrow leaf phenotype in srm1 may be attributed to changes in ABA levels or signaling.
SRM1 Affects Endogenous ABA Levels during Seed Germination under Salt Stress
Because germination of the SRM1 transgenic lines on saline medium differed from that of the wild type, we speculated that ABA levels could be changed. To assess this, we germinated srm1, one overexpressing line (OX6-4), and wild-type seeds on either MS medium or medium supplemented with 125 mm NaCl for 3 or 5 d, respectively, and measured the endogenous ABA levels. As mutations in NCED3/STO1 result in deficiency in stress-induced ABA production (Wan and Li, 2006), we used a T-DNA knockout insertion line for NCED3/STO1 (GK_129B08) as control. Not surprisingly, in srm1, nced3/sto1, and wild-type seeds germinated on MS medium, we detected very low levels of ABA content (Fig. 4D). By contrast, germinating seeds of the overexpression line OX6-4 contained significantly higher levels of ABA (Fig. 4D), indicating that overproduction of SRM1 influences ABA levels. On MS medium supplemented with 125 mm NaCl, the ABA level was significantly increased in wild-type seeds (Fig. 4D); however, this increase was considerably attenuated in the nced3/sto1 and srm1 mutants and also only modestly changed in the OX6-4 to levels similar to those in the wild type. Interestingly, the increase in ABA was less than 25% in germinating srm1 seeds compared with the wild type (Fig. 4D). These data are very similar to the reduced ABA induction in sto1/nced3 (Fig. 4D) and indicate that the salt-resistant germination phenotype of srm1 may be explained by lower endogenous ABA levels.
We also tested whether the ABA levels in nced3/sto1 and srm1 rosette leaves were changed after salt treatment compared with the wild type. However, we did not find any significant changes in the ABA content of srm1 compared with that of the wild type after the treatment (Fig. 4E). In agreement with this, we did not see any drought-sensitive phenotype in srm1 rosette leaves (Supplemental Fig. S5), which clearly was observed in nced3/sto1 (Wan and Li, 2006). This suggests that the srm1-related leaf shape phenotype may be a result of subtle changes in the basal ABA levels during vegetative growth.
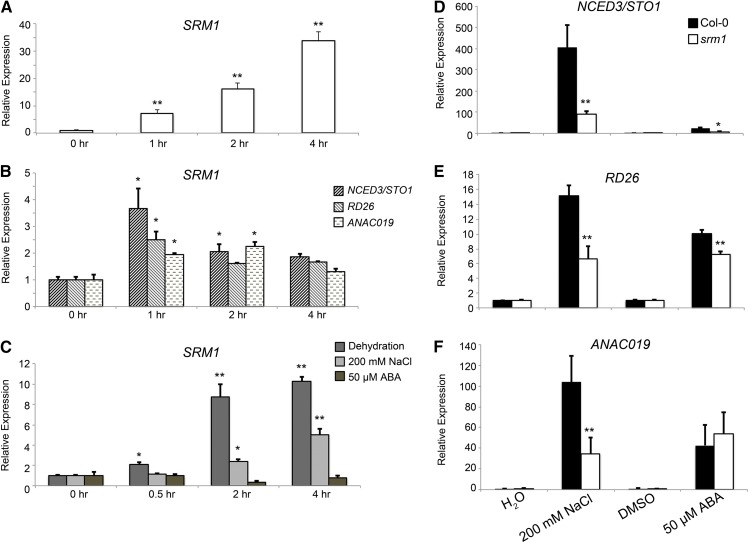
Induction of SRM1 Activates NCED3/STO1, RD26, and ANAC019
Our data indicated that SRM1 regulates genes involved in ABA-related stress responses. To find out the temporal induction scheme of these genes by SRM1, we generated an estradiol-inducible (Zuo et al., 2000) expression construct for SRM1 and transformed this into the srm1 mutant. Induction of SRM1 was confirmed via qRT-PCR (Fig. 5A). We then monitored the induction of genes from the microarray analyses above. From the genes tested, only three genes, i.e. NCED3/STO1, RD26, and ANAC019, showed an increase in expression after short-term SRM1 induction (Fig. 5B). The response of these genes was the strongest at initial time points (1 and 2 h) after SRM1 induction, at which time we observed a 2- to 4-fold increase in the expression of the three genes (Fig. 5B). It is possible that SRM1 needs further coactivators not present in sufficient amounts to sustain expression of the tested genes or that SRM1 can additionally activate other processes that to some degree counteract the inducing effect. In any case, we conclude that induction of SRM1 significantly induced NCED3/STO1, RD26, and ANAC019 expression.
Figure 5.
SRM1 is induced by stress but not by ABA and is necessary for induction of NCED3/STO1, RD26, and ANAC019 during salt stress. A, qRT-PCR of an estradiol-inducible SRM1 line after 1, 2, and 4 h of induction. B, Expression of NCED3/STO1, RD26, and ANAC019 after induction of SRM1 using estradiol. ACTIN2 was used for normalization of expression. Seven-day-old light-grown seedlings were used for mRNA extraction. C, Induction of SRM1 after treatment with 50 µm ABA, dehydration, or 200 mm NaCl in light-grown 7-d-old seedlings. Expression was normalized against ACTIN2. D to F, Differential mRNA accumulation of NCED3/STO1, RD26, or ANAC019 in srm1 mutant under stress condition. Light-grown 7-d-old wild-type and srm1 seedlings were treated with 200 mm NaCl and 50 µm ABA for 2 h, respectively. All experiments were performed independently three times. Error bars indicate sd. *, P < 0.05; and **, P < 0.01 (Student’s t test).
Because SRM1 affects several stress-responsive genes, it may also be assumed that SRM1 is induced by exogenous stress treatment. To test this, we exposed wild-type seedlings to stress treatments, i.e. salt (200 mm NaCl) or dehydration for 30 min, 2 h, and 4 h, and assessed changes in SRM1 expression using qRT-PCR. Figure 5C shows that SRM1 is induced by salt and dehydration treatment. NCED3/STO1, ANAC019, and RD26 have been shown to be induced by a variety of stresses, including salt, dehydration, and ABA (Fujita et al., 2004; Barrero et al., 2006; Jiang et al., 2009). To investigate whether SRM1 is required for the stress-induced transcriptional response of these genes, we subjected 7-d-old srm1 and wild-type seedlings to 200 mm NaCl for 2 h and measured the transcript levels of NCED3/STO1, ANAC019, and RD26 using qRT-PCR. Interestingly, the response of all three genes was attenuated after the salt treatment in srm1 compared with wild-type seedlings (Fig. 5, D–F). These data show that SRM1 is required for full induction of the genes in response to salt in Arabidopsis seedlings. In addition, the salt stress-responsive induction of ANAC019 and RD26 was also attenuated in the nced3/sto1 mutant (Supplemental Fig. S6, A and B), indicating that the salt stress-related induction of ANAC019 and RD26 is partly dependent on ABA synthesis.
SRM1 Is Induced Independently of ABA
To assess whether SRM1 expression is also induced by ABA, we subjected 7-d-old seedlings to ABA (50 µm). Interestingly, SRM1 expression was not induced by the treatment (Fig. 5C). We further confirmed this observation by treating nced3/sto1 mutants with salt (200 mm) while monitoring SRM1 expression. The salt-induced expression of SRM1 occurred independently of NCED3/STO1 (Supplemental Fig. S6C), supporting a role for SRM1 upstream of NCED3/STO1. Conversely, ABA-induced expression of NCED3/STO1 was significantly decreased in the srm1 mutant compared with that observed in wild-type seedlings (Fig. 5D). These data suggest that although SRM1 is not induced at the expression level by ABA, it contributes to an ABA-mediated response via NCED3/STO1. By contrast, while RD26 showed modest changes in its ABA induction, the induction of ANAC019 expression was unchanged in the srm1 mutant (Fig. 5, E and F). These results suggest that although SRM1 is an activator of the three genes under certain stress conditions, other unknown processes may work in parallel to SRM1, at least for the induction of RD26 and ANAC019, in response to ABA.
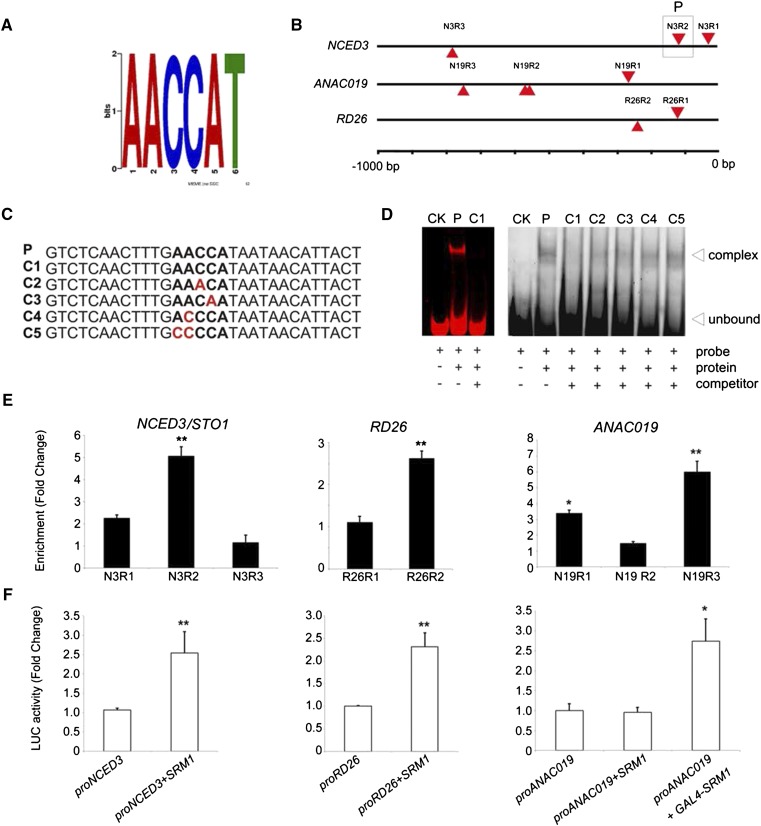
SRM1 Binds to an MYB-Related ABA Promoter Motif in Vitro
Our data suggest that SRM1 may be a positive regulator of a salt stress-mediated ABA response. TFs typically recognize specific DNA motifs in the promoter regions of target genes to regulate their expression. For members of the MYB TF family, specific DNA-binding motifs have been reported (Abe et al., 2003; Borg et al., 2011). To see whether some of these cis-elements were present in the promoters of the genes obtained in the expression analyses, we used the online motif analysis tools PLACE and MEME (Higo et al., 1999; Bailey et al., 2009). We analyzed the promoters of the ABA- and stress-related genes down-regulated in the srm1 mutant from the microarray data and found that they contained, among others, the common motif (A/T)AACCAT (Fig. 6A), which is similar to the regulatory segment AACCA, involved in the ABA signaling pathway via AtMYB2 (Abe et al., 2003). To investigate whether SRM1 can interact with this motif, we performed electrophoretic mobility shift assays (EMSAs) using a DNA fragment containing the (A/T)AACCAT motif found in the NCED3/STO1 promoter (segment P; Fig. 6, B and C). In the absence of SRM1 protein, the Cyanine5 (Cy5)-labeled probe migrated as a single band corresponding to the unbound DNA (Fig. 6D). However, when the probe was incubated together with in vitro synthesized SRM1 protein, we observed a clear retardation of a fraction of the DNA in accordance with binding of the protein to the probe (Fig. 6D). This band disappeared when including excess unlabeled probe, corroborating that the SRM1 binds to regions containing the (A/T)AACCAT motif.
Figure 6.
SRM1 binds to specific elements in the promoters of NCED3/STO1, RD26, and ANAC019 and activates them in vitro and in vivo. A and B, PLACE and MEME analyses identified an AACCAT motif in the promoters of NCED3/STO1, RD26, and ANAC019 (A) at sites indicated in B. Numbers indicate base pairs from the ATG start site of the respective genes. C, Probes used for EMSAs in D. Sequence P was the probe designed from the NCED3/STO1 promoter (N3R2), C1 was the competitor with the same sequence as P, and C2, C3, and C4 were the competitors with a mutated binding motif in their sequences. D, EMSA analyses to test binding of SRM1 to the DNA fragment harboring the AACCAT motif. A segment corresponding to the second AACCAT motif in the NCED3/STO1 promoter (segment P in B) was used for the analysis. SRM1 interacted with this segment probe as indicated by the band shift in D (left section), which could be reversed if an excess of unlabeled probe was included in the assay. The right section shows competitive EMSA analyses with mutated AACCAT motifs in the probe segments. Note that only the nonmutated probe can completely outcompete the labeled probe. CK indicates the probe alone without protein or competitor addition. E, ChIP-qPCR analyses of immune precipitates of a functional SRM1-GFP. Different regions of the promoters that contained AACCAT related motifs were used as templates for the qPCR analysis (see outline in B). GFP control plants were used as normalization for the qPCR. Experiments were performed independently three times. F, Transactivation assays for the NCED3/STO1, RD26, and ANAC019 promoters driving luciferase genes by the SRM1. Three-week-old rosette leaves from the srm1 mutant were used for protoplast isolation, and the relative luciferase activity was normalized by Renilla luciferase activity and represented as fold change. Error bars indicate sd. *, P < 0.05; and **, P < 0.01 (Student’s t test).
To provide further evidence that SRM1 recognizes this motif, we performed a competition assay using the Cy5-labeled (A/T)AACCAT DNA probe (segment P) and mutated competitor probes (Fig. 6D). As can be seen in Figure 6D, the unlabeled probe without mutations readily outcompeted the labeled probe. However, mutated unlabeled probes clearly did this less efficiently (Fig. 6D). These data corroborate that SRM1 binds to the (A/T)AACCAT motif in vitro.
SRM1 Binds and Activates NCED3/STO1, RD26, and ANAC019 Promoters in Vivo
To determine whether SRM1 recognizes promoters containing the (A/T)AACCAT motif in vivo, we performed chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR) assays using plants expressing the functional SRM1-GFP fusion protein in the srm1 background (Fig. 1E). For the ChIP assay, we tested the promoter regions of NCED3/STO1, RD26, and ANAC019. Enrichment tests were used to verify that the sequences corresponding to these genes were truly occupied in vivo by SRM1. As the promoters contained multiple (A/T)AACCAT-related motifs (Fig. 6B), we performed multiple ChIP-qPCRs per promoter (Fig. 6E). For all tested promoters, we observed clear enrichment for at least one region per promoter, corroborating that SRM1 directly binds to the NCED3/STO1, RD26, and ANAC019 promoters in planta.
While the inducible expression of SRM1 supports an activation of NCED3/STO1, RD26, and ANAC019 in vivo, we also explored whether we could activate the promoter by using a transactivation system. We therefore performed transactivation assays using Arabidopsis srm1 leaf protoplasts. We tested 1-kb promoter regions of RD26 and ANAC019, which contain the (A/T)AACCAT motif that was tested in the ChIP-qPCR assay. For NCED3/STO1, we used a 3-kb promoter segment upstream of the transcription start site of the gene, as it has been shown that this region is required for stress-related induction of the gene (Behnam et al., 2013). The protoplasts were cotransfected with a firefly LUCIFERASE (LUC) reporter gene behind the NCED3/STO1, RD26, or ANAC019 promoter segments and an effector plasmid containing a 35S promoter driving SRM1 expression. Both the RD26 and NCED3/STO1 promoters gave strong and reproducible induction signals using this setup (Fig. 6F). However, we were unable to induce the ANAC019 promoter (Fig. 6F). To test whether SRM1 could interact with the ANAC019 promoter while requiring additional proteins for activation, we fused SRM1 with a GAL4 activation domain via its N terminus and then redid the assay. This construct readily activated the ANAC019 promoter (Fig. 6F), thus indicating that SRM1 binds and can activate the three promoters in planta.
DISCUSSION
Abiotic stress is detrimental to plant growth, development, and yield. ABA is a key molecule in abiotic stress response, and understanding regulatory aspects of ABA synthesis and signaling is therefore of great importance. While the biosynthetic machinery of ABA synthesis and many downstream elements of its signaling are well established, the coordination of abiotic stress with ABA production and signaling remains poorly defined. Here, we report on a MYB-like TF (SRM1) that regulates ABA biosynthesis- and signaling-related genes and that influences plant growth during salt stress.
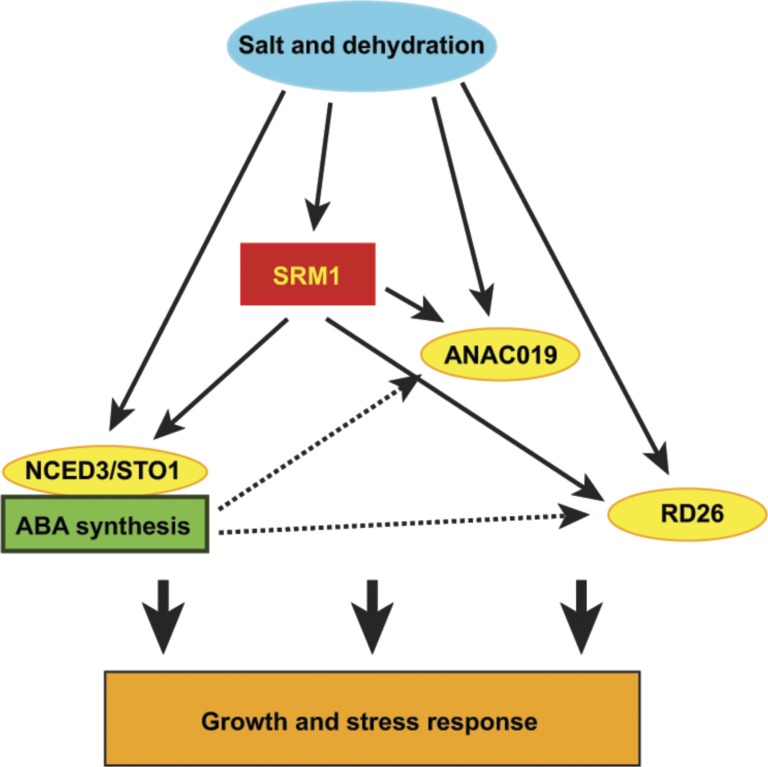
As the largest clade of the MYB TFs in plants, the R2R3 MYBs are likely to have diverse functions during plant growth and development (Dubos et al., 2010). Our data support a model in which the R2R3 MYB SRM1 coordinates the expression of NCED3/STO1, ANAC019, and RD26 by direct binding to an (A/T)AACCAT motif in their promoters. This binding is supported by a recent large-scale screening effort of TF-promoter interactions (Weirauch et al., 2014). These data, together with salt-related phenotypes and reduced ABA levels after salt treatment, solidly place SRM1 as a transcriptional regulator of ABA synthesis and signaling in response to salt stress during seed germination and seedling development (Fig. 7). In addition, SRM1 expression was induced by salt and drought, but not by ABA, which induced both RD26 and ANAC019 expression also in the absence of SRM1 (Fig. 7).
Figure 7.
Schematic model for SRM1 function during salt stress. SRM1 is induced in response to salt and dehydration but not to ABA. SRM1 controls expression of NCED3/STO1, RD26, and ANAC019 both during normal growth conditions and under salt stress conditions. However, ABA can, by a currently unknown mechanism, induce RD26 and ANAC019 in the absence of SRM1, indicating a complex regulatory principle for RD26 and ANAC019. The activation of the ABA synthesis and signaling components by SRM1 impacts the growth and development of the plant and its ability to withstand external stress.
While the role of SRM1 during seed and seedling development therefore appears to be clarified, the function of the TF during vegetative growth is less clear. SRM1 is highly expressed during leaf growth, supporting the leaf shape phenotypes observed in the SRM1 transgenic lines. Here, it is plausible that subtle changes in the ABA levels might be caused by perturbations in the expression of SRM1 and that these changes might influence leaf shape development. This notion is supported by the fact that the leaf shape phenotypes were ABA dependent. Nevertheless, it is also plausible that SRM1 is associated with processes that go beyond ABA synthesis and signaling and that part of the vegetative phenotypes observed for the SRM1 transgenic plants could be due to such effects. In addition, the phenotypic differences between, on the one hand, seeds and seedlings and, on the other hand, vegetative stages could be achieved by different SRM1 interactors or perhaps via SRM1 posttranslation modifications that could fine-tune the regulation of downstream targets. Of note, SRM1 was reported to undergo phosphorylation in an ABA-dependent manner in a large-scale phosphoproteomics study (Wang et al., 2013) Hence, although ABA does not induce the expression of SRM1, it still could influence the activity of the SRM1 protein that, in turn, could impact its function.
Abiotic stress activates expression of genes that synthesize ABA, which, in turn, activates downstream ABA signaling networks that allow for the plant to adapt to the environmental stress. ANAC019 and RD26/ANAC072 mRNA accumulate to different extents in response to dehydration, salt, ABA, hydrogen peroxide, and methyl jasmonate (MeJA; Bu et al., 2008; Jiang et al., 2009). We show that SRM1 can directly activate RD26 and binds to the promoter of ANAC019 but that it also requires additional coactivators to induce its expression. It is plausible that SRM1 is required for the initial induction of these genes during salt stress. However, subsequent increase in ABA levels (Schmidt et al., 2013) may then suffice to further enhance the induction. We show here that induction of SRM1 results only in a transient activation of these genes in the absence of stress (Fig. 5B). This could lead to activation schemes of the two genes that are independent of SRM1. Interestingly, leaf growth phenotypes in srm1 mimicked those of RD26 dominant-negative transgenic lines, which produced longer petioles and smaller leaves when grown on MS medium (Fujita et al., 2004). While we did not detect changes in endogenous ABA levels in srm1 leaves, the leaf shape was clearly altered, and this phenotype could be restored by application of exogenous ABA. It is therefore possible that the srm1 leaf phenotype is related to changes in RD26 activity.
In summary, we show that SRM1 acts as a positive regulator of the key ABA biosynthetic gene NCED3/STO1 and the two ABA- and stress-related transcriptional activators RD26 and ANAC019 to modulate the trade-off between growth and salt tolerance. Our data position SRM1 as an important integrator of ABA synthesis and signaling during seed germination and seedling development on saline medium in Arabidopsis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The srm1 mutant (At5g08520, SALK_150774) is a T-DNA insertion line obtained from the Nottingham Arabidopsis Stock Centre (http://www.arabidopsis.info; Alonso et al., 2003). Primers used for PCR to obtain homozygous insertion lines are listed in Supplemental Table S2. Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) was used as the wild type. Plants were grown in soil under a 16-h-light (120–150 μE)/8-h-dark (22°C/18°C) regime. For in vitro culture, sterilized seeds were stratified in the dark at 4°C for 2 d and sowed on plates containing one-half-strength MS medium with 0.1% (w/v) MES, 0.8% (w/v) agar, and 1% (w/v) Suc. Plates were sealed and incubated at 22°C/16°C with a 16-h-light/8-h-dark photoperiod.
Seed Germination Assay and Stress Treatment
To propagate seeds for germination assay, plants were grown on soil at 22°C under long days (16 h of light/8 h of dark). Care was taken to always analyze seed germination from batches of seeds that had been grown and harvested together under the same conditions. Harvested mature seeds were stored at a dehumidifier cabinet for at least 2 months before the seed germination test was performed. The quality of the different batch of seeds was tested by germination percentage (radicle emergence), and the seed batches with similar germination rates were used for germination assay. Three independently grown seed batches were used to measure percentage of seed germination, and more than 100 seeds were used for each replicate. Sterilized seeds were subsequently plated on MS medium (Sigma-Aldrich) containing 1% (w/v) Suc supplemented with ABA or NaCl with different concentration, respectively. Stock solution of ABA and MeJA (mixed isomers; Sigma-Aldrich) were dissolved in dimethyl sulfoxide (DMSO). Control plates contained equal amounts of the corresponding solvents. Plates were kept at 4°C in darkness for 2 d for stratification and then transferred to a tissue culture room set at 22°C with a 16-h-light/8-h-dark photoperiod. Plants grown on different plates for 2 weeks were used for the cotyledon greening assessments. For the root growth upon ABA treatment, 7-d-old seedlings were transferred from one-half-strength MS plates to the new MS plates with or without 50 μm ABA (or 50 μm MeJA) for another 10 d. Plates with DMSO were used as control. For short-time stress treatment, 7-d-old seedlings grown in one-half-strength MS plates with 1% (w/v) Suc were transferred to the liquid system with 200 mm NaCl (water as control), 50 μm ABA, or MeJA (DMSO as control) for different time points.
Isolation of RNA and Analysis of RNA Expression
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) and treated with DNase I (Ambion) following the manufacturer’s instructions. First-strand complementary DNA (cDNA) was synthesized from the DNA-free RNA samples using the Superscript III Reverse Transcriptase (Invitrogen) and oligo(dT) primer. First-strand cDNA of total RNA (1.5 µg) from leaves of 3-week-old plants was used for real-time PCR amplification. For real-time PCR, the resulting cDNA was directly used as DNA templates for PCR reaction. For quantitative real-time PCR, the resulting cDNA was diluted 1:3 with DNAse-free water. A mixture of 1 µL of cDNA, 5 µL of SYBR Green Master Mix (Applied Biosystems), and 4 µL of primer mix was run on a real-time PCR machine (Gene AMP 7900 Sequence Detector, Applied Biosystems). Data were normalized to reference gene ACTIN2 (At3g18780): ΔCT = CT (gene) – CT (ACTIN), where CT stands for cycle threshold. Sequences of oligonucleotides are given in Supplemental Table S2.
Construction of Plasmids and Plant Transformation
Oligonucleotides were ordered from Eurofins MWG Operon or Eurogentec and are listed in Supplemental Table S2. Primer sequences for the genotyping of the T-DNA lines were obtained from the iSct-Primer tool of the SIGnAL Web site (http://signal.salk.edu/tdnaprimers.2.html). For cloning procedures, the subcloning vectors CR II TOPO or CR2.1 TOPO (Invitrogen) and the binary destination vectors pCAMBIA1305.1 (CAMBIA), pER8 inducible expression vector (Zuo et al., 2000), and pEZR-H-LN and the pF3A WG (BYDV) Flexi Vector (Promega) for in vitro translation were used.
For complementation of srm1 (SALK_150774), the 35S:SRM1-GFP fusion construct was used. The SRM1 coding sequence (CDS) without stop codon was amplified from seedling cDNA by PCR and then cloned into the pEZRH-LN containing a C-terminal GFP using the restriction sites BamHI and NcoI. Another complementation construct, a 4.4-kb SRM1 genomic fragment (gSRM1) including the 1.9-kb upstream sequence and 2.5-kb full coding region plus introns, was amplified and cloned to pCAMBIA1305.1 using the restriction sites EcoRI and PmlI. For the overexpression construct, the SRM1 coding sequence (CDS) was amplified from seedling cDNA by PCR and then cloned into pCAMBIA1305.1 using the restriction sites BglII and PmlI. For the ProSRM1:GUS construct, a 1.9-kb fragment upstream of the SRM1 start codon was amplified and cloned into the pCAMBIA1305.1 using the restriction sites EcoRI and NcoI. For the inducible expression construct, SRM1 coding sequence was inserted to pER8 inducible vector with restriction sites XhoI and ApaI. For EMSA assay, the SRM1 CDS was ligated between the SgfI and PmeI restriction sites of the destination vector. Furthermore, the NucleoSpin Extract II kit and Nucleo Spin plasmid kit (Macherey-Nagel) were used for DNA purification and plasmid isolation, respectively.
The resulting constructs were introduced into Agrobacterium tumefaciens strain GV3101, and subsequently, the 35S:SRM1-GFP and pER8-SRM1 constructs were transformed into srm1 mutant background and the 35S:SRM1 and proSRM1:GUS constructs were transformed into wild-type background. Transformation of Arabidopsis was carried out by dipping the inflorescence into an A. tumefaciens solution as previously described (Clough and Bent, 1998). The obtained seeds were grown on plates containing the construct-specific antibiotic to select for positive transformants.
Promoter GUS Analysis of SRM1
Different plant tissues carrying the proSRM1:GUS construct were analyzed with GUS-staining as previously described (Sánchez-Rodríguez et al., 2012). In brief, flowers were transferred to GUS staining solution (1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 0.1 m sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, 1 mm K3[Fe(CN)6], and 1 mm K4[Fe(CN)6]) and shortly vacuum infiltrated. After incubation at 37°C overnight, the tissue was cleared by washing several times with 70% (v/v) ethanol for a total of 4 h.
Light and Confocal Imaging
GUS staining was examined using a stereomicroscope (Leica) with the Leica Application Suite software. Distribution of SRM1-GFP fusion protein was analyzed by confocal fluorescence microscopy using a Leica SP5 microscope (Leica). Seven-day-old seedlings grown on the one-half-strength MS medium were analyzed with appropriate filter settings.
Leaf Shape Analyses
Leaf shape was quantified using elliptic Fourier analysis, a method that encodes two-dimensional shape outlines in the form of high-dimensional vectors, which contain information about the entire shape. On the basis of the quantitative shape data, an average shape can be calculated and drawn. The encoded shapes were then separated using multiclass LDA, a method that seeks to find the best separation among a set of groups. To assess and separate the contributions of genotype, treatment, and their interaction to the shape variation, a two-way multivariate ANOVA was performed. Pairwise comparisons between each group were performed by testing the separation along the first discriminant axis using the nonparametric Kolmogorov-Smirnov test, and 10 to 12 plant leaves from each genotype were used for analysis. For leaf width and length analysis, 3-week-old soil-grown rosette leaves were sampled for imaging and size measurement, with 10 leaves for each replicate.
Microarray Analysis
Three micrograms of quality-checked total RNA from 3-week-old rosette leaves (soil-grown plants) from Col-0 and srm1 mutant was processed for use in Affymetrix Gene Chip hybridizations representing approximately 22,000 Arabidopsis genes. Labeling, hybridization, washing, staining, and scanning procedures were performed by Affymetrix Authorized Service Provider (ATLAS Biolabs) as described in the Affymetrix technical manual. Raw data (CEL files) obtained from RNA hybridization experiments were normalized with the Robin (Lohse et al., 2010), and the online tool PageMan was used for overrepresentation analysis. Three biological repeats were used for the array experiments.
ABA Extraction and Measurement
For the ABA extraction and content quantification in seeds and seedlings, 3-d-old seedlings under control growth condition and 5-d-old seedlings under 125 mm NaCl condition were harvested and ground in liquid nitrogen, respectively. ABA was extracted as described previously (Lin et al., 2007) from 50 mg of materials, and ABA content was quantified by the ELISA method (Phytodetek ABA Kit; Agdia) according to the manufacturer’s protocol.
For vegetative-stage ABA quantification, samples were harvested from 3-week-old rosette leaves treated without or with 200 mm NaCl for 2 h, respectively. Frozen plant material (1 g) was ground and extracted by 10 mL of 80% (v/v) acetonitrile containing 1% (v/v) acetic acid and 6d-ABA (1 ng mL–1; Olchemin) overnight with slow shaking at 4°C. Supernatant was obtained after centrifugation at 4,000 rpm at 4°C for 10 min and evaporated without heating until less than 7 mL. The pellet was resuspended by mixing with the same extraction buffer (5 mL) and reextracted for 3 h with slow shaking at 4°C. Second supernatant was obtained after centrifugation at 4,000 rpm at 4°C for 10 min. The combined extract fractions were evaporated until less than 7 mL, and 10 mL of ULC-mass spectrometry-grade water containing 1% (v/v) aqueous acetic acid was added. After centrifugation, supernatant was loaded onto a Strata-X 33u Polymeric Reversed Phase column (60 mg, 3 mL; Phenomenex) and prewashed/equilibrated consecutively with 3 mL of methanol and 1% (v/v) acetic acid. After discarding the passed-through elute fraction following sample loading, the column was washed with 1% (v/v) aqueous acetic acid (1 mL). ABA fraction was eluted with 80% (v/v) methanol containing 1% (v/v) acetic acid (2 × 500 µL) and evaporated without heating. The dried elute was dissolved in 150 µL of 80% (v/v) MeOH with 1% (v/v) acetic acid. After centrifugation, supernatant was subjected to liquid chromatography-tandem mass spectrometry in negative ion detection mode using the HPLC Surveyor System coupled to the LTQ Linear Ion Trap Electrospray Ionization-Mass Spectrometry system (Thermo Finnigan). HPLC separation was performed with a Luna C18 column (2.0 × 150 mm, 3-μm particle size; Phenomenex) at a flow rate of 200 μL min–1 of 0.1% (v/v) aqueous formic acid as solvent A and 0.1% (v/v) acetic acid-acetonitrile as solvent B with the following linear gradient: from 10% B to 30% B for 1 min and to 50% B for 9 min. The initial condition was restored and allowed to wash (100% B) and equilibrate (10% B) for 2 min, respectively. The ABA-specific mass fragment peak (152.3–162.3 mass-to-charge ratio) derived from tandem mass spectrometry fragmentation (collision energy, 70 eV) of the molecular parental ion peak ranging from 262.3 to 272.3 mass-to-charge ratio was profiled. An obtained chromatogram was processed and picked peak area using the Quan Browser of the Xcalibur software (Thermo Finnigan). Absolute concentration of endogenous ABA was estimated by the ratio to concentration of internal 6d-ABA.
EMSA and ChIP-qPCR
To obtain protein for EMSA assays, the SRM1 CDS was subcloned into the pF3A WG Flexi vector (Promega). In vitro expression and EMSA assays were done as previously described (Schmidt et al., 2013). In short, the TNT SP6 High-Yield Wheat Germ Mastermix containing 1 µL of FluoroTec Green Lys (Promega) was used to generate SRM1 protein. For gel shift assays, Cy5-labeled probes were generated based on the NCED3/STO1 promoter containing the SRM1 binding site (see Supplemental Table S2). Subsequent binding reactions were performed using the LightShift chemiluminescent assay kit (Pierce) according to the manufacturer’s instructions. Protein-DNA complexes were separated on a 5% (v/v) native polyacrylamide gel, after which the Cy5 signal was imaged using a Typhoon Scanner (GE Healthcare).
35S:SRM1-GFP transgenic lines in the srm1 mutant background (35S:SRM1-GFP srm1) used for ChIP-qPCR were grown on one-half-strength MS plates (1% [w/v] Suc) for 12 d. Whole seedlings were cross linked with 1% (v/v) formaldehyde for 10 min. For the immunopurification of the SRM1-GFP:DNA complex, a mouse anti-GFP antibody (Roche Applied Science) was used. ChIP analysis with the EpiQuik Plant ChIP kit (Epigentek Group) was essentially performed as previously described (Schmidt et al., 2013; Lu et al., 2014). Primers used are listed in Supplemental Table S2 and were designed with the Primer3 software tool (Rozen and Skaletsky, 2000).
Transactivation Assay
Arabidopsis rosette leaf protoplasts from the srm1 mutant were isolated and transformed according to Wu et al. (2009). Constructs for transactivation assay were generated by amplifying 1-kb upstream promoter sequences of RD26 and ANAC019 and a 3-kb upstream promoter sequence of NCED3/STO1 by PCR (Supplemental Table S2). After cloning into pENTR/D-TOPO (Invitrogen), the promoter sequences were recombined with the p2GWL7.0 vector (Licausi et al., 2011). To place them upstream of the firefly LUC gene, the wild-type SRM1 CDSs were cloned from pENTR/D-TOPO into the vector p2GW7.0 containing the Cauliflower mosaic virus 35S promoter. Protoplasts were cotransformed with the recombinant p2GWL7.0 vectors, the normalization vector containing 35S:RLUC, and recombinant p2GW7.0 vectors using 5 mg of each plasmid. Dual luciferase reporter assays (Promega) were performed as previously described (Licausi et al., 2011; Schmidt et al., 2013). Luminescence was measured using a GloMax 20/20 luminometer (Promega).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SRM1, At5g08520; NCED3/STO1, At3g14440; RD26, At4g27410; ANAC019, At1g52890; FSD1, At4g25100; PP2C, At3g16800; RD20, At2g33380; UNE15, At4g13560; CRK4, At3g45860; GRF11, At1g34760; CYP83A1/REF2, At4g13770; CYP71B22, At3g26200; FLN2, At1g69200; COR47, At1g20440; COR15A, At2g42540; COR15B, At2g42530; ERD5, At3g30775; RAB18, At1g43890; and COR78, At5g52310.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. SRM1 does not impact seed germination on mannitol-containing medium.
Supplemental Figure S2. Expression pattern of SRM1.
Supplemental Figure S3. Assessments of rosette leaf shape and size in srm1 and SRM1 overexpressing lines.
Supplemental Figure S4. qRT-PCR of selected genes from the wild-type versus srm1 microarray experiment.
Supplemental Figure S5. Drought stress treatment of wild-type, srm1, SRM1 overexpressor OX6-4, and nced3/sto1 plants.
Supplemental Figure S6. Induction of expression of RD26, ANAC019, and SRM1 in nced3/sto1 mutant after salt stress.
Supplemental Table S1. Genes with altered expression in srm1.
Supplemental Table S2. Primer sequences used in the study.
Acknowledgments
We thank Meryem Oral, Norma Funke, and Drs. Dandan Lu, Nadine Tiller, and Guozhang Wu for technical aid.
Glossary
- ABA
abscisic acid
- TF
transcription factor
- MS
Murashige and Skoog
- T-DNA
transfer DNA
- RT
reverse transcription
- qRT
quantitative reverse transcription
- LDA
linear discriminant analysis
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative PCR
- Col-0
Columbia-0
- DMSO
dimethyl sulfoxide
- cDNA
complementary DNA
- LD1
first linear discriminant axis
- MeJA
methyl jasmonate
Footnotes
This work was supported by the Max-Planck Gesellschaft (to T.W., T.T., A.I., A.R.F., M.M., and S.P.) and Australian Research Council Discovery (grant no. DP150103495 to S.P.).
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T, Vavasseur A, Galaud JP (2010) RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol 51: 1975–1987 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Rodríguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29: 2000–2008 [DOI] [PubMed] [Google Scholar]
- Behnam B, Iuchi S, Fujita M, Fujita Y, Takasaki H, Osakabe Y, Yamaguchi-Shinozaki K, Kobayashi M, Shinozaki K (2013) Characterization of the promoter region of an Arabidopsis gene for 9-cis-epoxycarotenoid dioxygenase involved in dehydration-inducible transcription. DNA Res 20: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner F, Oreb M, Mendel RR (2001) ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem 276: 40381–40384 [DOI] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Khatab H, Sidorova A, Lingaya M, Twell D (2011) The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell 23: 534–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18: 756–767 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Kwon SI, Choe S (2014) Antagonistic regulation of Arabidopsis growth by brassinosteroids and abiotic stresses. Mol Cells 37: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Ding Y, Avramova Z, Fromm M (2011) The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J 66: 735–744 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. (2013) Abscisic acid synthesis and response. The Arabidopsis Book 11: e0166, doi/10.1199/tab.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124: 509–525 [DOI] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman R, Hill C, Penfold CA, Breeze E, Bowden L, Moore JD, Zhang P, Jackson A, Cooke E, Bewicke-Copley F, et al. (2013) A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J 75: 26–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2000) A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol 123: 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Ukai Y (2002) SHAPE: a computer program package for quantitative evaluation of biological shapes based on elliptic Fourier descriptors. J Hered 93: 384–385 [DOI] [PubMed] [Google Scholar]
- Jensen MK, Lindemose S, de Masi F, Reimer JJ, Nielsen M, Perera V, Workman CT, Turck F, Grant MR, Mundy J, et al. (2013) ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 3: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Li H, Bu Q, Li C (2009) The RHA2a-interacting proteins ANAC019 and ANAC055 may play a dual role in regulating ABA response and jasmonate response. Plant Signal Behav 4: 464–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yu D (2012) Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol Plant 5: 1375–1388 [DOI] [PubMed] [Google Scholar]
- Licausi F, Weits DA, Pant BD, Scheible WR, Geigenberger P, van Dongen JT (2011) Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol 190: 442–456 [DOI] [PubMed] [Google Scholar]
- Lim EK, Doucet CJ, Hou B, Jackson RG, Abrams SR, Bowles DJ (2005) Resolution of (+)-abscisic acid using an Arabidopsis glycosyltransferase. Tetrahedron Asymmetry 16: 143–147 [Google Scholar]
- Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH (2007) Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol 143: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Nunes-Nesi A, Krüger P, Nagel A, Hannemann J, Giorgi FM, Childs L, Osorio S, Walther D, Selbig J, et al. (2010) Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol 153: 642–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wang T, Persson S, Mueller-Roeber B, Schippers JH (2014) Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat Commun 5: 3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63: 289–305 [DOI] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15: 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- North HM, De Almeida A, Boutin JP, Frey A, To A, Botran L, Sotta B, Marion-Poll A (2007) The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J 50: 810–824 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Ruggiero B, Koiwa H, Manabe Y, Quist TM, Inan G, Saccardo F, Joly RJ, Hasegawa PM, Bressan RA, Maggio A (2004) Uncoupling the effects of abscisic acid on plant growth and water relations: analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol 136: 3134–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Bauer S, Hématy K, Saxe F, Ibáñez AB, Vodermaier V, Konlechner C, Sampathkumar A, Rüggeberg M, Aichinger E, et al. (2012) Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24: 589–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Mieulet D, Hubberten HM, Obata T, Hoefgen R, Fernie AR, Fisahn J, San Segundo B, Guiderdoni E, Schippers JH, et al. (2013) Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25: 2115–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Léon-Kloosterziel KM, Koornneef M, Zeevaart JA (1997) Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol 114: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JA (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Peeters AJ, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JA, Koornneef M, Kamiya Y, Koshiba T (2000) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97: 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XR, Li L (2006) Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem Biophys Res Commun 347: 1030–1038 [DOI] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci USA 110: 11205–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpiński S, Karpińska B, Kangasjärvi J (2010) Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]