An actin nucleation protein affects water loss by regulating stomatal density.
Abstract
The global problem of drought threatens agricultural production and constrains the development of sustainable agricultural practices. In plants, excessive water loss causes drought stress and induces early senescence. In this study, we isolated a rice (Oryza sativa) mutant, designated as early senescence1 (es1), which exhibits early leaf senescence. The es1-1 leaves undergo water loss at the seedling stage (as reflected by whitening of the leaf margin and wilting) and display early senescence at the three-leaf stage. We used map-based cloning to identify ES1, which encodes a SCAR-LIKE PROTEIN2, a component of the suppressor of cAMP receptor/Wiskott-Aldrich syndrome protein family verprolin-homologous complex involved in actin polymerization and function. The es1-1 mutants exhibited significantly higher stomatal density. This resulted in excessive water loss and accelerated water flow in es1-1, also enhancing the water absorption capacity of the roots and the water transport capacity of the stems as well as promoting the in vivo enrichment of metal ions cotransported with water. The expression of ES1 is higher in the leaves and leaf sheaths than in other tissues, consistent with its role in controlling water loss from leaves. GREEN FLUORESCENT PROTEIN-ES1 fusion proteins were ubiquitously distributed in the cytoplasm of plant cells. Collectively, our data suggest that ES1 is important for regulating water loss in rice.
Rice (Oryza sativa) is a major worldwide food crop, but it consumes more water than most crops (Linquist et al., 2015), with water consumption for rice cultivation accounting for approximately 65% of agricultural water usage. Rice provides a staple food for about 3 billion people while using an estimated 24% to 30% of the world’s developed freshwater resources (Bouman et al., 2007). Severe water shortages restrict the expansion of rice production and hinder the irrigation of existing paddy fields (Zhu and Xiong, 2013). One effective way to overcome water shortages is to reduce water loss in rice plants, thus allowing the cultivation of this key crop in environments with less water (Nguyen et al., 1997).
In plants, the stomata on the leaf surface work as the main channels for the discharge of water and the entry of carbon dioxide, thus strongly affecting physiological processes such as transpiration and photosynthesis. Previous studies showed that mutations in some genes could affect stomatal density or differentiation, such as the Arabidopsis (Arabidopsis thaliana) genes TOO MANY MOUTHS (AtTMM; Yang and Sack, 1995), SCREAM2 (SCRM2; Kanaoka et al., 2008), STOMATA DENSITY AND DISTRIBUTION1 (AtSDD1; Von Groll et al., 2002), and AtYODA2, a mitogen-activated protein kinase kinase kinase (Bergmann et al., 2004). Stomata show a regular distribution on rice leaves (Huang et al., 2009) during plant growth and development, and various environmental factors affect the density and size of stomata as well as the chlorophyll contents of rice leaves. For example, rice leaves that develop under water stress show substantially fewer stomata compared with leaves that develop under well-watered conditions (Huang et al., 2009). Changes in stomatal density and morphology affect water loss (Boonrueng et al., 2013). In rice, SIMILAR TO RADICAL-INDUCED CELL DEATH1 enhances drought tolerance by regulating stomatal closure (You et al., 2013), while the zinc finger protein DROUGHT AND SALT TOLERANCE functions in drought and salt tolerance by adjusting stomatal aperture (Huang et al., 2009). In Arabidopsis, overexpression of the magnesium chelatase H subunit in guard cells confers drought tolerance by promoting stomatal closure (Tsuzuki et al., 2013). Thus, stomatal closure and low stomatal density enhance drought tolerance by reducing water loss in plants.
The epicuticular wax layer in plants acts as the first barrier to environmental conditions by reducing water loss due to transpiration and preventing plant damage due to excessively strong sunlight (Riederer and Schreiber, 2001). Leaf transpiration involves both stomatal and cuticular transpiration. Stomatal conductance controls stomatal transpiration, but the physicochemical properties of the leaf surface mainly control cuticular transpiration. For example, the composition, thickness, and microstructure of the cuticular wax affect water permeability and transport (Svenningsson, 1988; Xu et al., 1995; Buschhaus and Jetter, 2012). Rice DROUGHT-INDUCED WAX ACCUMULATION1, GLOSSY1, and WAX SYNTHESIS REGULATORY GENE1 affect drought tolerance by regulating the deposition or biosynthesis of cuticular wax (Islam et al., 2009; Wang et al., 2012; Zhou et al., 2013; Zhu and Xiong, 2013).
Besides, leaf trichomes can also affect water loss (Konrad et al., 2015) and leaf trichomes closely linked with the actin cytoskeleton. The involvement of the actin cytoskeleton in controlling directional cell expansion in trichomes has received much attention (Zhang et al., 2005). Generally, genes that affect cytoplasmic organization can be studied by screening leaf trichome mutants (Qiu et al., 2002). In Arabidopsis, a reproducible morphogenetic program directs the polarized development of trichome branches (Mathur et al., 1999; Szymanski et al., 1999; Le et al., 2006). Some of these genes affect the cytoskeleton and also affect the morphology of normal plant cells, especially epidermal cells. For example, mutation of SPIKE1 in Arabidopsis causes epidermal cells to show simple arrangements and morphologies (i.e. all cells dividing along a single axis; Qiu et al., 2002).
To study the molecular mechanisms underlying water loss in rice, we isolated and characterized the early senescence1-1 (es1-1) rice mutant, which showed excessive water loss and early senescence phenotypes. Map-based cloning data showed that ES1 encodes a SCAR-LIKE PROTEIN2, and its Arabidopsis homolog affects the polymerization of actin. The es1-1 mutants showed obvious changes in leaf trichomes, similar to Arabidopsis. However, few studies have reported a connection between the actin cytoskeleton and water loss in Arabidopsis. Our results demonstrated a critical role of the actin cytoskeleton in regulating water loss in rice.
RESULTS
Identification and Characterization of Early Senescence Mutants
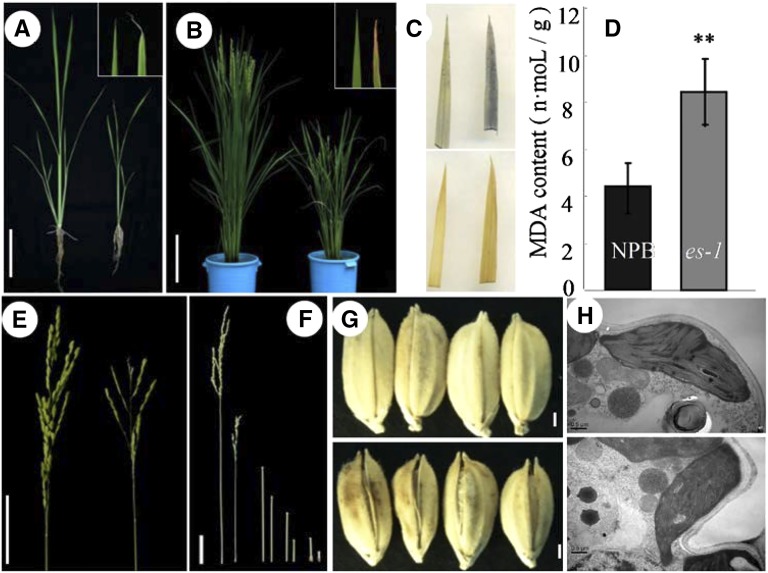
To study the mechanisms of senescence in rice, we screened a large pool of mutants generated in the japonica rice ‘Nipponbare’ (NPB) background by mutagenesis using ethyl methanesulfonate. From this pool, we identified two mutants with an early senescence phenotype. The two mutants showed similar phenotypes under the same growth conditions, and F1 hybrid individuals produced by the two parental mutants exhibited phenotypes like the parental line with the weaker phenotype (subsequently named es1-1; see below), indicating that the two mutants are allelic (Supplemental Table S1); therefore, we termed these mutants es1-1 and es1-2. Under normal growth conditions, es1-1 plants showed severely retarded development. At the seedling stage, the es1-1 mutants displayed whitish and yellowish leaf tips (Fig. 1A), a hallmark of early senescence (Li et al., 2014); this phenotype increased with increasing leaf age, becoming more severe at the heading stage (Fig. 1B). Newly developed leaves showed yellowing margins that rolled inward or formed a spiral, and old leaves showed spots with a rusty color and water deficiency phenotypes, including wilting (Fig. 1, A and B). Histochemical analysis showed that concentrations of senescence-related substances, including hydrogen peroxide, superoxide radical, and malondialdehyde, were higher in es1-1 leaves than in wild-type leaves (Fig. 1, C and D), indicating that the mutant does show senescence phenotypes (Moradi and Ismail, 2007). Moreover, we measured the expression of two senescence marker genes, STAYGREEN and Os185, in cv NPB and es1-1 (Supplemental Fig. S1, F and G). We found that the transcript levels of these two genes were substantially higher in es1-1 than in cv NPB, suggesting that es1-1 caused senescence (Lee et al., 2001; Park et al., 2007). We also measured plant height, finding that es1-1 plants were much shorter than wild-type plants from the seedling to the mature stages (Fig. 1, A and B; Supplemental Table S2). At the mature stage, es1-1 mutants showed degraded or white panicles with a low seed-setting rate (only 2.4%; Fig. 1E; Supplemental Table S2), short internodes (Fig. 1F; Supplemental Table S2), and brown, open glumes (Fig. 1G). Scanning transmission electron microscopy showed that es1-1 mutants had disordered thylakoids compared with the neat and well-ordered thylakoids observed in wild-type plants; moreover, es1-1 mutants had lower chlorophyll contents than wild-type cv NPB.
Figure 1.
Phenotypes of wild-type and es1-1 plants. A, Plants in the seedling stage. Wild-type (cv NPB; left) and es1-1 (right) plants are shown. Bar = 3 cm. B, Plants at the heading stage. Wild-type (cv NPB; left) and es1-1 (right) plants are shown. Bar = 10 cm. C, Nitroblue tetrazolium (NBT) staining (top: cv NPB, left; es1-1, right) and diaminobenzidine (DAB) staining (bottom: cv NPB, left; es1-1, right). D, Malondialdehyde (MDA) contents at the seedling stage (cv NPB, left; es1-1, right). E, Panicle rachis (cv NPB, left; es1-1, right). Bar = 5 cm. F, Leaf at different parts (cv NPB, left; es1-1, right): from left to right, internode lengths of the top, second from top, third from top, and fourth from top leaves. Bar = 3 cm. G, Grain size (cv NPB, top; es1-1, bottom). Bars = 1 mm. H, Chloroplasts observed by transmission electron microscopy (cv NPB, top; es1-1, bottom). Bars = 0.5 μm.
Compared with es1-1, es1-2 plants had more tillers (Supplemental Fig. S1, A and C), were taller (Supplemental Fig. S1, A and D), and showed a stronger early senescence phenotype in the leaves (Supplemental Fig. S1B). In this study, we mainly focused on es1-1 mutants, unless specified otherwise.
Map-Based Cloning of ES1
To clarify whether es1-1 phenotypes were caused by a single gene or multiple genes and dominant or recessive genes, we crossed the mutant plants directly and reciprocally with three wild-type cultivars. All the F1 plants showed no symptoms of early senescence. In the F2 segregating populations of these three crosses, normal and early senescent plants showed a typical segregation ratio of 3:1 (Supplemental Table S3). This result suggested that a single recessive nuclear gene controls the early senescence phenotype of es1-1.
For cloning ES1, we crossed the mutant es1-1 (japonica) with the typical indica cv Nanjing 6 (NJ6). Plants with the es1-1 phenotype were identified from the F2 generation and used as the initial mapping population. For primary chromosome mapping, we used 164 pairs of simple sequence repeat primers distributed on the 12 chromosomes of rice at a distance of 10 to 30 centimorgan (cM) from each other to detect differences between es1-1 and cv NJ6. A significant amplification difference was detected on chromosome 1 using the simple sequence repeat marker M1, showing linkage with a small population in the F2 generation. The M1 marker was further used to analyze 320 mutants in the F2 population, which detected 15 recombinants. Genetic linkage analysis mapped the M1 marker to a distance of 2.3 cM from ES1 (Fig. 2A).
Figure 2.
Map-based cloning of ES1, and phenotype of an es1-1 complementation transgenic line. A, Location of es1-1 on rice chromosome 1. B, Coarse linkage map of es1-1. C, es1-1 gene structure. D, Reverse transcription of es1-1. E, Schematic diagram of es1-1 and mutated proteins encoded by es1-1 and es1-2. F, Phenotype of the complementation transgenic line: es1-1 (left), the wild type (cv NPB; middle), and the transgenic line (right). Bar = 10 cm.
For fine-mapping of ES1, the mutation in ES1 must be flanked by two molecular markers. Thus, we designed multiple pairs of primers at positions relatively far from M1 (Supplemental Table S4). The marker M6 also showed polymorphisms between es1-1 and cv NJ6. Screening of the primary mapping population (320 mutants) identified 11 recombinants that differed from those identified using M1 (Fig. 2A). Linkage analysis mapped the M6 marker to a distance of 1.8 cM from ES1. Next, we used M1 and M6 to screen the entire population (2,850 plants), which identified 50 and 39 recombinants, respectively (Fig. 2A).
To further narrow the ES1 region, we developed a large number of sequence-tagged site and cleaved-amplified polymorphic sequence markers between M3 and M4. Primers C1 to C9 showed good polymorphisms between es1-1 and cv NJ6. Thus, we used these primers to analyze the recombinants obtained by preliminary screening and to construct fine genetic and physical maps of ES1 (Fig. 2B; Supplemental Table S4). The number of recombinants detected by C1 and C10 decreased to 28 and 35, respectively. Similarly, the number of recombinants screened by C2 and C9 decreased to 10 and 12, respectively, and those detected by C3 and C8 further decreased to three and four, respectively. The recombinants detected by C3 and C8 corresponded to different individual plants.
We also designed primers for the region between C3 and C8 to further screen the recombinants. Finally, these new primers detected only one recombinant; thus, these markers tightly cosegregate with ES1 (Fig. 2B). The physical distance between the end markers C5 and C6 was 13.1 kb, which included two predicted open reading frames.
Sequencing analysis detected a unique mutation in LOC-Os01g11040 within the 13.1-kb region in es1-1 compared with the wild-type sequence. This single-base mutation replaced G with A in the first nucleotide of the third intron (nucleotide 2,275; Fig. 2C). To test whether this mutation alters the normal splicing of the third intron, we generated es1-1 and wild-type genomic complementary DNAs (cDNAs) using reverse transcription. Sequencing showed that the es1-1 cDNA was 62 bases longer than the wild-type cDNA (Fig. 2D). This increase in length coincided with the length of the third intron, which contains a stop codon that results in the premature termination of protein translation (Fig. 2E). Similarly, sequencing analysis showed that the mutation in es1-2 was a single-base substitution (G to A) at nucleotide 4,660. This mutation changed the Trp codon into a stop codon, thus resulting in the premature termination of protein translation (Fig. 2E).
We then used genetic complementation to verify the identity of ES1. A 10.6-kb fragment containing ES1 (including the upstream 2-kb and downstream 1-kb sequences) was introduced into es1-1 and es1-2 mutants, and 52 and 36 transgenic plants, respectively, were recovered. Exogenous ES1 rescued the phenotypes of es1-1 and es1-2 (Fig. 2F), and the complementation reversed the mutant phenotype of es1-1. Taken together, these data demonstrate that LOC-Os01g11040 (Os01g0208600) is ES1.
ES1 Encodes a SCAR-Like Protein of the ACTIN-RELATED2/3-SCAR Pathway
We performed a BLAST search to identify ES1 homologs in more than 20 different plant species. ES1 showed high amino acid sequence similarity to proteins in other species, such as Oryza brachyantha (85%), Brachypodium distachyon (68%), maize (Zea mays; 60%), and Setaria italica (64%; Fig. 3E). ES1 also shares high amino acid sequence similarity to proteins in both monocots and dicots, suggesting that ES1 and its homologs may perform similar functions. Examination of ES1 homologs in Arabidopsis showed that ES1 (Os01g0208600) encodes a SCAR-LIKE PROTEIN2; its closest homolog in Arabidopsis forms part of the ACTIN-RELATED2/3 (ARP2/3) complex, which functions in regulating actin filament polymerization and has been examined in detail in Arabidopsis. ARP2/3 alone is inactive in Arabidopsis. A domain formed by the SCAR-like family proteins, and members of other protein families (e.g. SRA1 and ABSCISIC ACD-INSENSITIVE [ABI] families), are responsible for binding to and activation of the ARP2/3 complex (Zhang et al., 2005). In Arabidopsis, mutation of the gene encoding SCAR-LIKE PROTEIN2 leads to changes in leaf trichomes and stomata (Basu et al., 2005). In our study, we observed substantial changes in leaf trichomes of es1-1, mainly reflected by the serious degradation of the trichome tip (Fig. 3, A and B). The leaves of es1-1 had smooth leaf margins without a jagged pattern (Fig. 3A).
Figure 3.
ES1 is a SCAR homolog. A, Leaf margins (left, cv NPB; right, es1-1). Arrows indicate the trichome tip. B, Trichome phenotype (macro hairs; top, cv NPB; bottom, es1-1). C and D, F-actin organization was visualized with Alexa Fluor 488-phalloidin in root cells of the wild type (C) and es1 (D). Over 40 roots were used in this analysis. Arrows indicate the continuity of F-actin. Bars = 5 μm. E, Phylogenetic tree of SCAR-LIKE PROTEIN2 in several species.
To examine the possible role of ES1 in regulating actin filaments, we examined the organization of F-actin in the roots of the wild type and es1-1 mutants by Alexa Fluor 488-phalloidin staining. The actin filaments were well organized in the wild type (Fig. 3C). The actin filaments in the wild type were complete and orderly arranged, while es1-1 showed an irregular arrangement. Moreover, the actin filaments in es1-1 were shorter than those in the wild type (Fig. 3D, arrows), which indicated that ES1 likely functions in the ARP2/3-SCAR pathway in rice and that a mutation in ES1 affects the actin cytoskeleton in rice cells (Dyachok et al., 2011).
Expression of ES1 and Subcellular Localization of ES1
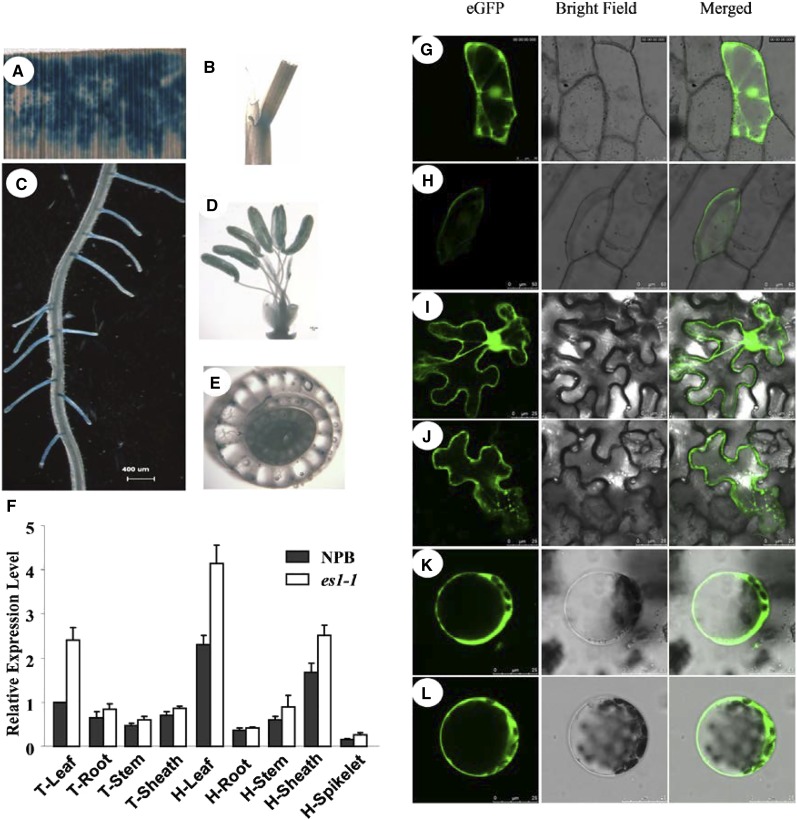
To monitor the tissue-specific expression of ES1, we generated an ES1:GUS reporter construct containing the 2-kb genomic region upstream of the start codon of ES1. We detected high GUS activity in leaves, leaf sheaths, and root tips and also detected lower GUS activity in stamens and vascular bundles (Fig. 4, A–E). Reverse transcription-quantitative PCR (RT-qPCR) assays showed that ES1 was ubiquitously expressed in various tissues at the tillering and heading stages, including roots, culms, leaves, sheaths, and spikelets. We observed the highest levels of ES1 expression in the leaf and leaf sheath (Fig. 4F). Consistent with the RT-qPCR data, ES1:GUS expression was higher in rice leaves and leaf sheaths than in other plant tissues.
Figure 4.
Expression of ES1 and subcellular localization of ES1. A to E, GUS staining of leaf (A), leaf sheath (B), fibrous root (C), stamen (D), and culm vascular bundles (E). F, Quantitative real-time PCR analysis of ES1 expression in different tissues at the tillering stage (T) and the heading stage (H). The results are given as means of three independent assays, and each error bar represents the percentage sd of the mean. G to L, GFP fluorescence visualized by confocal microscopy. G and H, Transient expression of eGFP (G) and eGFP-ES1 cDNA (H) in epidermal cells of onion. 35S::eGFP-ES1 cDNA and 35S::GFP were transiently expressed in onion epidermal cells. I to L, Transient expression of eGFP in epidermal cells (I) and protoplasts (K) and expression of eGFP-ES1 in epidermal cells (J) and protoplasts (L) of Nicotiana benthamiana leaves. 35S::eGFP-ES1 and 35S::GFP were transiently expressed in N. benthamiana leaves.
To investigate the subcellular localization of ES1, we constructed a construct to express enhanced green fluorescent protein (eGFP)-ES1 (35S::eGFP-ES1 DNA) and introduced it into onion (Allium cepa) and tobacco (Nicotiana tabacum) epidermal cells. Fluorescence signals were detected in both cell types, and the signals surrounded the cytoplasm, nucleus, and cell membrane (Fig. 4, G–J). Consistent results were obtained by examining protoplasts extracted from tobacco leaves after the expression of the fusion protein (Fig. 4, K and L). These results indicated that ES1 was ubiquitously distributed in the cytoplasm of plant cells.
ES1 Affects the Density of Leaf Stomata in Rice
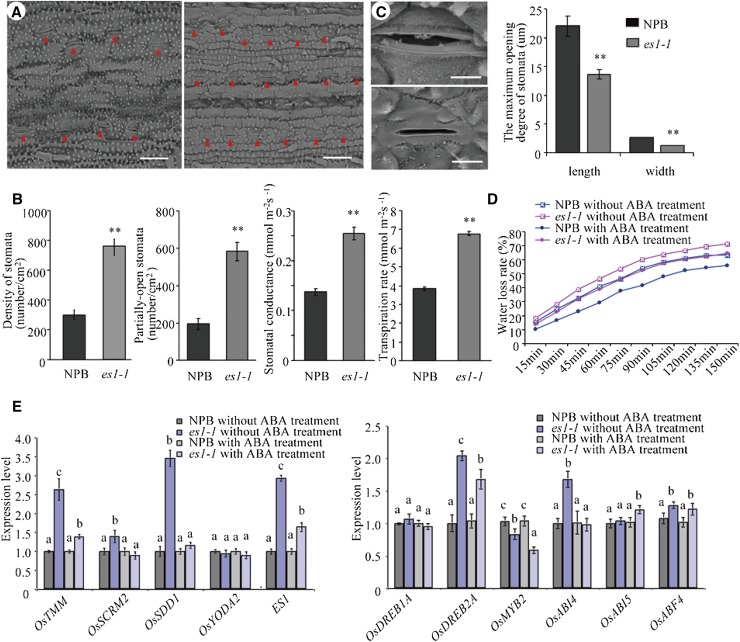
Scanning electron microscopy of es1-1 leaves showed that these leaves had smoother surfaces than the leaves of wild-type plants. In addition, these leaves showed substantially smaller siliceous protrusions at the seedling stage and substantially more stomata per unit of leaf area compared with wild-type leaves. The number of semiopen stomata in es1-1 was also larger than that in the wild type (Fig. 5, A and B). Moreover, the tips of trichomes and the waxy layer were severely degraded on the leaf surface of es1-1 mutants (Fig. 5A). Statistical data showed that, regardless of the position, stomatal density was significantly higher in es1-1 than in the wild type. While both stomatal conductance and transpiration rate were also detected, they were also significantly higher in es1-1; these data indirectly suggested that stomatal density in es1-1 was higher (Fig. 5B). The maximum and minimum stomatal apertures were greater in the wild type compared with es1-1 (Fig. 5C). At the seedling stage, the transcriptional levels of several genes that could affect the increase of stomatal density were analyzed by real-time PCR, including OsTMM, OsSCRM, OsSDD1, and OsYODA2 (Du et al., 2014). The expression levels of OsTMM, OsSCRM, and OsSDD1 were significantly higher in es1-1 than in the wild type, and the ES1 gene also showed a higher expression level in es1-1 (Fig. 5E).
Figure 5.
Observation of stomata between cv NPB and es1-1. A, Stomata density of the wild-type (left) and es1-1 (right) leaves. The red triangles indicate the positions of stomata. Bars = 30 µm. B, Statistical analysis of stomatal parameters, such as stomata density, partially open stomata, stomatal conductance, and transpiration rate. C, Morphological characteristics (left) and maximum opening size (right) of stomata between wild-type (top) and es1-1 (bottom) plants. Bars = 5 µm. D, Comparison of water loss rate between wild-type and es1-1 plants with and without 300 μm ABA treatment at the three-leaf-stage. E, RT-qPCR analysis of stomata density-related genes (OsTMM, OsSCRM, OsSDD1, and OsYODA2), ES1, and some ABA response-related genes (OsDREB1A, OsDREB2A, OsMYB2, OsABI4, OsABI5, and a basic leucine zipper transcription factor, OsABF4) in wild-type and es1-1 leaves before and after 300 μm ABA treatment at the three-leaf-stage. The values were normalized to the ACTIN1 gene. Asterisks indicate significance (Student’s t test) at P < 0.01. Different letters indicate significant differences at the 1% level (Duncan’s multiple range test).
Leaves of the wild type and es1-1 at the seedling stage were sprayed with 300 μmol L−1 abscisic acid (ABA), and after 3 h, quantitative analysis of OsTMM, OsSCRM, OsSDD1, and OsYODA2 was taken. In the wild type, the expression level of these genes almost did not change, but in es1-1, the majority of these genes showed a significantly decreased expression level; this indicated in es1-1 that those genes that could affect the increase of stomatal density were sensitive to ABA. We also detected the rate of in vitro water loss (RWL) of seedling leaves from the same position of the wild type and es1-1 before and after ABA treatment. The results showed that, no matter with or without ABA treatment, the RWL value was always higher in es1-1 than in the wild type. The RWL value in the wild type was reduced after ABA treatment, and the value in es1-1 was also reduced after ABA treatment, but it was not so obvious compared with the wild type (Fig. 5D). Scanning electron microscopy assay on the leaves of the wild type and es1-1 with ABA treatment showed that nearly all stomata of both wild-type and es1-1 plants were closed. Moreover, we also examined the expression levels of multiple ABA response-related genes (ABA signaling genes) in the wild type and es1-1 before and after ABA treatment (Shang et al., 2010) and discovered that, after ABA treatment, OsDREB2A (a transcription factor) was up-regulated while OsABI4 was down-regulated. These results suggest that both wild-type and es1-1 plants were sensitive to ABA (Fig. 5E).
The es1-1 Plants Showed Higher Water Loss Rates Than Wild-Type Plants
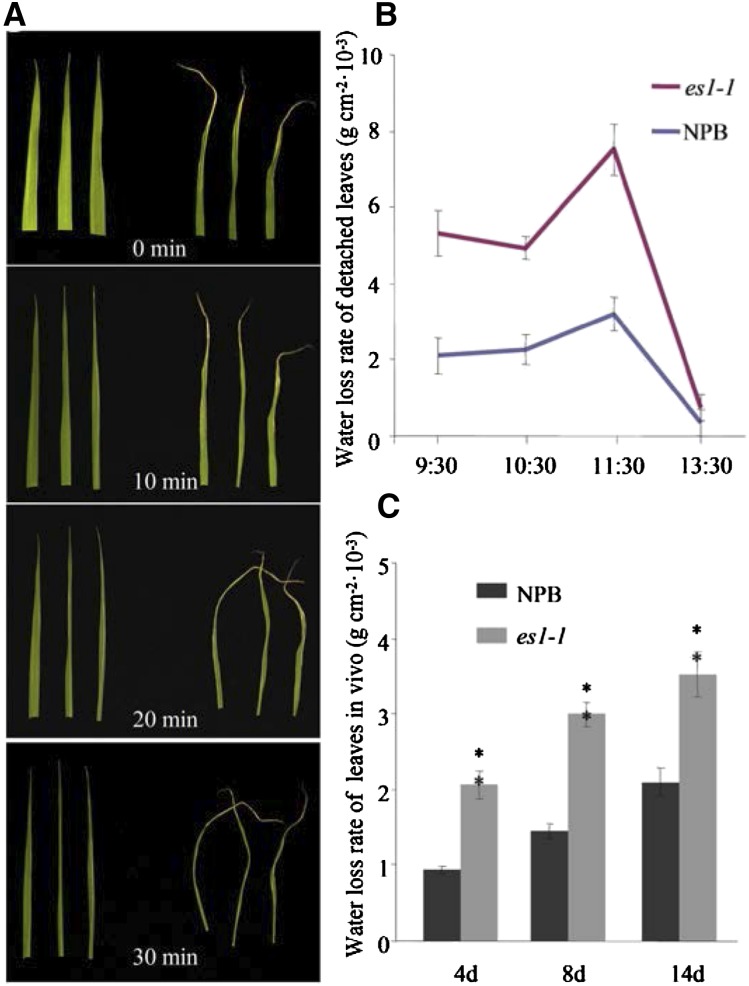
In vitro, the rate of water loss of es1-1 was higher than that of cv NPB at the seedling stage (Fig. 6A), as the detached leaves of es1-1 shrunk into a line at 30 min but the cv NPB leaves remained nearly normal in width (Fig. 6A). To confirm this phenomenon, we also measured water loss per unit of time (g cm−2 10−3) at the tillering stage in vivo and in vitro (Fig. 6, B and C), which gave similar results: the water loss per unit of time in vitro (detached leaves) in different periods was uniformly higher in es1-1 than in cv NPB (Fig. 6B). The water loss per unit of time in vivo was also uniformly higher in es1-1 than in cv NPB at 4, 8, and 14 d after tillering (Fig. 6C).
Figure 6.
Observation of water loss rate between wild-type and es1-1 plants. A, Water loss rate of detached leaves at the seedling stage (left, cv NPB; right, es1-1). B, Water loss rate of detached leaves at the tillering stage. C, Water loss rate per unit of time of living leaves between cv NPB and es1-1 plants at the tillering stage. Error bars indicate sd (n = 15). Asterisks indicate the significance of differences between cv NPB and es1-1 plants, as determined by Student’s t test: *, 0.01 ≤ P < 0.05 and **, P < 0.01.
To verify excessive water loss in es1-1, we performed a guttation experiment, where we looked for the exudation of drops of xylem sap on the leaf periphery in plants grown at different humidities. Hydroponic seedlings were cultured in the light, in an incubator at different humidities, from 30% to 90%, and es1-1 and wild-type plants were examined for guttation. At the seedling stage (two-leaf stage), both the wild-type and es1-1 plants were capable of guttation at 30% humidity. However, higher humidity was required for guttation with plant growth. Until the strong seedling stage (two-tiller stage), guttation was observed in wild-type plants but not in es1-1 (Supplemental Fig. S2). This result indicated that water loss occurred more rapidly in es1-1 mutants than in the wild-type plants.
Water Absorption and Transport Capacity Were Increased in es1-1 Mutants
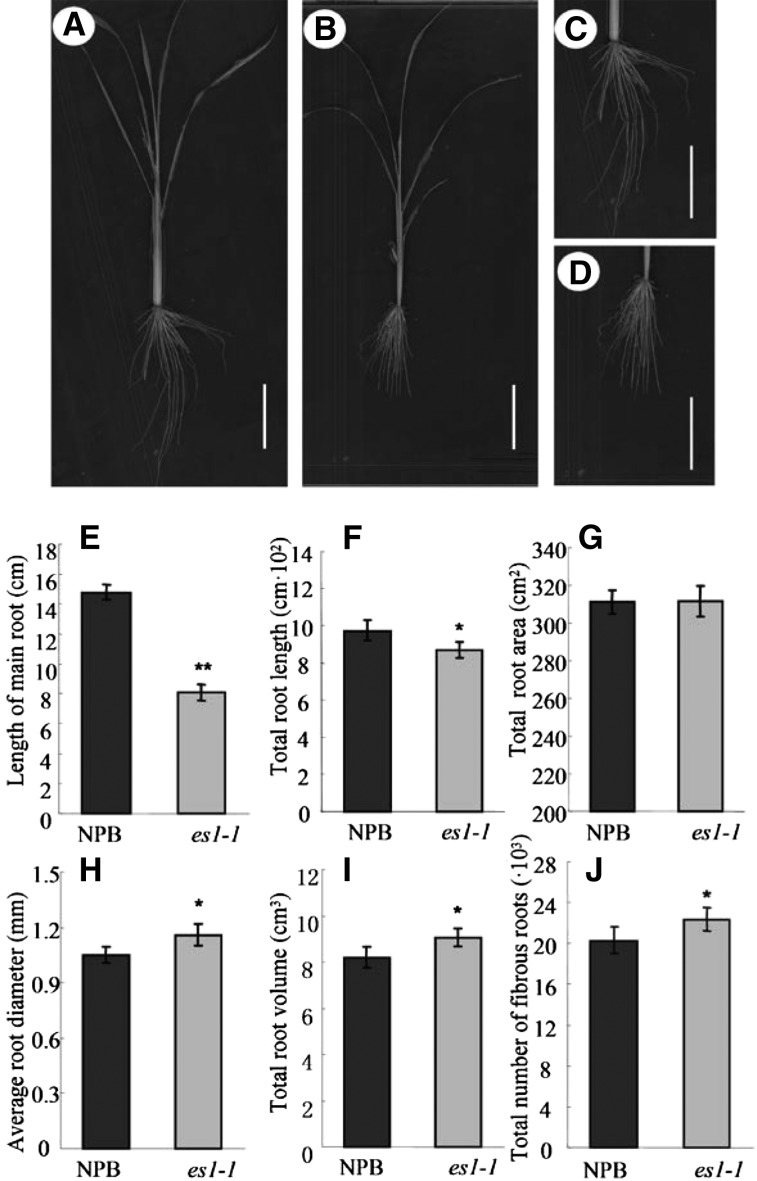
It is unknown whether excessive water loss through leaves affects water absorption in the root. To address this, we measured root traits in es1-1 and wild-type plants (Fig. 7, A–D). The es1-1 plants had smaller primary and total root lengths (Fig. 7, E and F) but higher average root diameters, total root volumes, and total numbers of fibrous roots compared with wild-type plants (Fig. 7, G–J). As fibrous roots are a major tissue for water adsorption in rice (Smith and De Smet, 2012), an increase in fibrous root number indicated an increase in the water absorption capacity of the roots in es1-1 mutants.
Figure 7.
Phenotype comparison of the roots between cv NPB and es1-1 plants. A, Root scanning of cv NPB plants. Bar = 5 cm. B, Root scanning of es1-1 plants. Bar = 5 cm. C, Enlarged image of roots in cv NPB plants. Bar = 5 cm. D, Enlarged image of roots in es1-1 plants. Bar = 5 cm. E to J, Statistical analysis of the main root length (E), total root length (F), total area of the root (G), average diameter of the root (H), total volume of the root (I), and total number of fibrous roots (J) of cv NPB and es1-1 plants. The results are given as means of three independent assays, and error bars indicate sd. Asterisks indicate the significance of differences between cv NPB and es1-1 plants, as determined by Student’s t test: *, 0.01 ≤ P < 0.05 and **, P < 0.01.
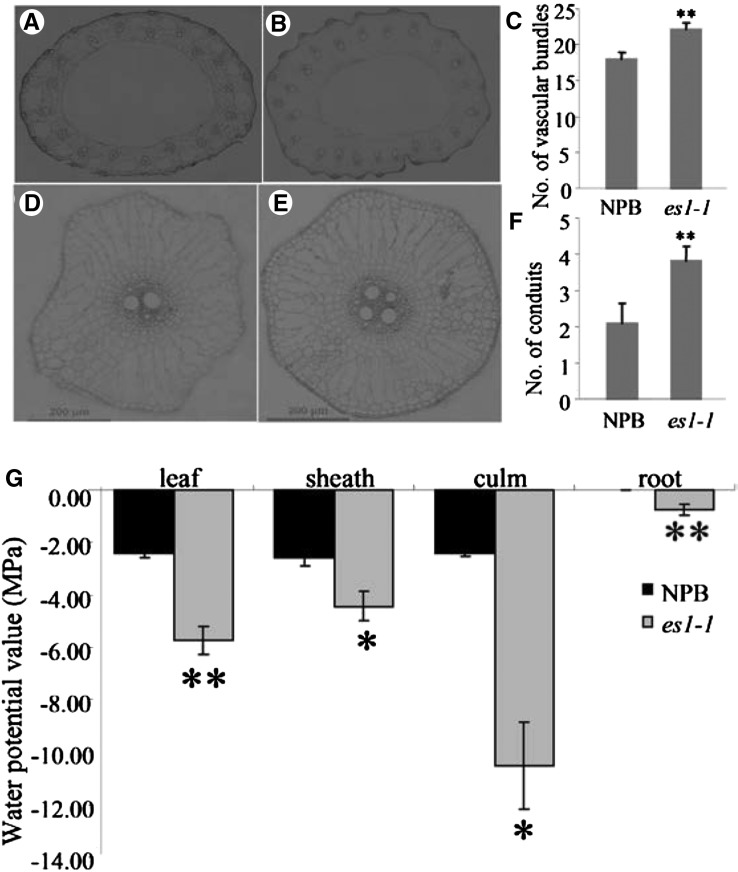
Examination of cross sections of the culm and primary root from es1-1 and cv NPB showed that the es1-1 stems had three to four additional vascular bundles (Fig. 8, A–C) and the es1-1 primary roots had two to three additional conduits compared with cv NPB (Fig. 8, D–F). Vascular bundles and conduits are necessary for water transport (Kim et al., 2014). Moreover, the water potential values were all higher in es1-1 than in the wild type in various tissues, including roots, stems, leaves, and sheath, which indicated that the ability to transport water was stronger in es1-1 than in the wild type (Fig. 8G; Da Silva et al., 2011). Therefore, our results indicate that es1-1 mutants have an enhanced ability to transport water.
Figure 8.
Number of vascular bundles and conduits in wild-type and es1-1 plants. A, Number of vascular bundles in cv NPB plants. B, Number of vascular bundles in es1-1 plants. C, Comparison of the number of vascular bundles between cv NPB and es1-1 plants. D, Number of conduits in cv NPB plants. E, Number of conduits in es1-1 plants. F, Comparison of the number of conduits between cv NPB and es1-1 plants. G, Detection of the water potential value in the wild type and es1-1. The relative water potential value in different tissues at the seedling stage of the wild type and es1-1 was analyzed with the WP4-T Dewpoint PotentiaMeter (Decagon Devices, Inc.). The results are given as means of three independent assays, and error bars indicate sd. Asterisks indicate significance at the level of 5% (*) and 1% (**).
Excessive Water Loss Causes in Situ Enrichment of Minerals in es1-1
In es1-1 mutants, excessive water loss through the leaves enhances the water absorption capacity of the roots and the water transportation capacity of the roots and culms. This, in turn, accelerates water circulation in the plant, thus leading to predicted in situ enrichment of metal ions cotransported with water (Kim et al., 2014). To test this hypothesis in this mutant, we determined the mineral concentrations of different tissues of es1-1 and wild-type plants at the mature stage. Consistent with the hypothesis, the concentrations of all four minerals tested (copper [Cu], iron [Fe], manganese [Mn], and zinc [Zn]) in the examined tissues were much higher in es1-1, especially in the roots and new leaves, than in wild-type plants (Table I). The Mn concentration in es1-1 leaves (719 mg kg−1) was seven times higher than that in wild-type plants (100 mg kg−1). The concentrations of the four minerals were also higher in es1-1 seeds than in cv NPB (Table I). These results suggested that mutation in ES1 promoted the accumulation of minerals in plants. The substantial accumulation of metal ions in es1-1 plants was likely due to excessive water loss and higher transpiration pull in the upper part of the plant.
Table I. Comparison of mineral contents between wild-type and es1-1 plants.
The results are given as means ± sd of three independent assays. Asterisks indicate significance at the level of 1%.
| Tissue | Plant | Cu | Fe | Mn | Zn |
|---|---|---|---|---|---|
| mg kg−1 | |||||
| Root | cv NPB | 23.5 ± 3.83 | 3,757.7 ± 122.55 | 17.75 ± 1.75 | 28.49 ± 1.12 |
| es1-1 | 446.3 ± 18.52** | 18,155.3 ± 326.27** | 58.97 ± 3.88** | 91.26 ± 5.88** | |
| Culm | cv NPB | 8.61 ± 1.21 | 35.52 ± 3.7 | 32.19 ± 1.3 | 28.75 ± 1.81 |
| es1-1 | 20.18 ± 4.7** | 46.05 ± 3.54** | 147.99 ± 10.06** | 85.83 ± 6.35** | |
| Sheath | cv NPB | 6.59 ± 1.15 | 136.2 ± 8.59 | 92.331 ± 5.9 | 42.52 ± 3.4 |
| es1-1 | 16.88 ± 2.43** | 240.2 ± 14.65** | 582.2 ± 13.22** | 65.36 ± 7.92** | |
| Old leaf | cv NPB | 9.32 ± 2.1 | 228.8 ± 15.1 | 100.01 ± 7.1 | 29.89 ± 2.56 |
| es1-1 | 18.27 ± 3.88** | 358.3 ± 21.94** | 719.05 ± 34.8** | 64.04 ± 7.5** | |
| Young leaf | cv NPB | 8.02 ± 1.16 | 102.1 ± 6.78 | 27.51 ± 3.32 | 30.48 ± 3.44 |
| es1-1 | 17.07 ± 2.91** | 223.1 ± 11.4** | 175.81 ± 12.24** | 62.22 ± 4.28** | |
| Seed | cv NPB | 4.26 ± 0.64 | 4.05 ± 0.35 | 31.62 ± 2.86 | 24.23 ± 2.6 |
| es1-1 | 5.91 ± 0.72** | 6.98 ± 1.06** | 50.36 ± 4.1** | 53.13 ± 8.12** | |
DISCUSSION
In this study, we characterized es1-1, an early senescence mutant that experienced excessive water loss. Map-based cloning showed that ES1 encodes a SCAR-LIKE PROTEIN2, a component of the suppressor of cAMP receptor/Wiskott-Aldrich syndrome protein family verprolin-homologous (SCAR/WAVE) complex that is involved in actin polymerization. This suggests that a mutation in ES1 affects the polymerization of actin and causes an abnormality in the cytoskeleton that promotes higher stomatal density (Huang et al., 2000; Yu et al., 2001; Xiao et al., 2003; Zhang et al., 2012), thus leading to excessive water loss.
Under stress conditions, plants develop a wax layer on the leaf surface or moderately roll their leaves to reduce transpiration, thereby protecting themselves from excessive water loss (Hu et al., 2010; Jäger et al., 2014). Although stomata account for only 1% to 2% of the leaf area, approximately 90% of water loss occurs through stomata (Buckley, 2005). Under drought conditions, rice cultivars that can maintain relatively high water potential and low transpiration rate in their leaves have strong drought resistance (Luo and Zhang, 2001). These drought-resistant cultivars commonly retain water in the plant body by closing their stomata. Small stomatal aperture and large diffusion resistance can jointly reduce water loss due to transpiration, thus avoiding the adverse effects of drought. Grill and Ziegler (1998) proposed that controlling the stomata is the most important way to improve the efficiency of water use in plants. In this study, scanning electron microscopy showed that the es1-1 mutants had substantially more stomata than wild-type plants. The expression levels of OsTMM, OsSCRM, and OsSDD1 were significantly higher in es1-1 than in the wild type (Fig. 5E), which was closely related to stomatal density. Stomatal density was significantly higher in es1-1 than in the wild type. While both stomatal conductance and transpiration were also significantly higher in es1-1 than in the wild type (Fig. 5B), these data suggested that es1-1 definitely loses water more quickly than the wild type. Nearly all stomata of both wild-type and es1-1 plants were closed after ABA treatment, and the RWL value in the wild type was reduced after ABA treatment; the value in es1-1 was also reduced after ABA treatment, but it was not so obvious compared with the wild type (Fig. 5D). Moreover, the wax layer on the leaf surface of es1-1 mutants was considerably degraded. Therefore, we thought the excessive water loss in es1-1 mainly due to increased stomatal density; meanwhile, the degradation of the waxy layer and leaf hairs may aggravate the rate of water loss.
Excessive water loss in the leaves inevitably depletes water from the body of es1-1 plants. Thus, more water needs to be absorbed through the roots to meet the metabolic requirements of es1-1 plants. In this study, we analyzed the roots of es1-1 and found that the total volume of the root and the number of fibrous roots increased compared with the wild type (Fig. 7, I and J), indicating that the water absorption capacity of the roots was enhanced in es1-1. Cross sections of the primary root and stems also showed that the number of vascular bundles in the stems increased by four (Fig. 8C) and those in the roots increased by two to three in es1-1 compared with wild-type plants (Fig. 8F). Moreover, the water potential values were all higher in es1-1 than in the wild type in various tissues (Fig. 8G). These observations confirmed that the water transport capacity of the roots was enhanced in es1-1.
All the above changes inevitably accelerate water circulation and cause minerals commonly cotransported with water to accumulate in the various tissues of es1-1 plants. Indeed, in different tissues, the concentrations of four minerals were higher in es1-1 than in cv NPB (Table I). For example, the Cu concentration in es1-1 (446.3 mg kg−1) was 18 times higher than in wild-type plants (23.5 mg kg−1), which could cause Cu overload in es1-1 plants. Enrichment of low-mobility metal elements (e.g. Cu, Fe, Mn, and Zn) in various tissues of es1-1 may, in turn, accelerate the onset of senescence. Moreover, the higher concentrations of mineral nutrients in seeds from the es1-1 mutants might produce more nutritious food; however, the accumulation of toxic elements such as arsenic might produce food that could adversely affect human health.
In our study, we found that actin filaments were shorter in es1-1, and the continuity was poor compared with the wild type (Fig. 3, C and D). Actin filaments function in many endomembrane processes, such as endocytosis of plasma membrane, vacuole formation, plasma membrane protrusion in crawling cells, and vesicle transport from the Golgi complex (Svitkina and Borisy, 1999; Eitzen et al., 2002; Kaksonen et al., 2003; Mathur et al., 2003; Chen et al., 2004). The ARP2/3 complex functions as a key regulator of actin filament nucleation in plants (Zhang et al., 2008). ARP2/3 is inactive by itself, as actin polymerization requires activation by the SCAR/WAVE complex, formed by proteins of the SCAR family. The SCAR/WAVE complex is highly conserved in plants. A deficiency of the SCAR/WAVE complex often leads to morphological changes in leaf trichomes and epidermal cells. This phenomenon has been reported in several studies in Arabidopsis (Li et al., 2003; Mathur et al., 2003; Saedler et al., 2004) but not in rice. In our study, es1-1 showed evident morphological changes in leaf trichomes (Figs. 3, A and B, and 5C), confirming that the function of the SCAR/WAVE complex is conserved in different species. Additional components of the SCAR/WAVE complex can be identified by screening for mutants that affect the leaf trichomes of the es1-1 mutant. A companion paper reveals that TUTOU1 (TUT1), identical to ES1, also plays a conserved role in trichome development (Bai et al., 2015).
Actin polymerization occurs in the cytoplasm. In this study, the fluorescent signal of eGFP-ES1 was detected in the cytoplasm and plasma membrane of tobacco and onion epidermal cells, a localization consistent with the predicted function of ES1 in affecting the actin cytoskeleton. After fusing the ES1 promoter with the GUS reporter gene, GUS staining was observed in various tissues, including the roots, root hairs, stems, leaves, leaf sheath, and vascular bundles, which are associated with water absorption, transportation, and loss. The ES1 expression level was relatively high in the leaves and leaf sheaths, the two major tissues involved in water loss. All these results are consistent with the proposed function of ES1.
The increased number of stomata would increase water loss and induce drought stress in es1-1 plants. Drought stress affects photosynthetic capacity, respiration, membrane permeability, osmotic adjustment, enzyme activity, and hormone levels in rice leaves, thus reducing leaf area, leaf area index, and dry weight. Drought stress also prevents the accumulation of dry matter and accelerates senescence (Hu et al., 2010; Ding et al., 2014; Sellammal et al., 2014). In this study, es1-1 plants showed early senescence characterized by yellowish leaf color and wilting of leaf margins at the seedling stage. Transmission electron microscopy showed that the volume percentage of chloroplasts in the es1-1 cells was lower than that in wild-type cells; however, the number of chloroplasts (irregular and fine prolate) was higher in es1-1 cells than in wild-type cells (Fig. 1H). This might have caused the decline in chlorophyll content in es1-1, causing the leaves to turn yellowish. In es1-1, the chloroplasts contained disorganized thylakoids (Fig. 1H) as opposed to the well-organized thylakoids in wild-type plants. The disordered thylakoids may have reduced the capacity to absorb light energy, which, in turn, would decrease the photosynthetic rate, thus causing early senescence in plant leaves.
Early senescence in rice (especially in functional leaves) is extremely unfavorable for rice growth and production because of its serious effects on yield and quality (Zhu et al., 2012). Therefore, rice breeders generally avoid lines showing early senescence. Similarly, the stay-green trait in rice also does not improve yield; although the stay-green trait promotes the ability of source tissues to fix carbon, it does not necessarily enhance the contents of sink tissues in plants. For example, a study on a stay-green mutant of rice (sgr[t]) reported that late photosynthesis and yield traits (seed-setting rate and thousand-grain weight) were not enhanced in sgr[t] mutants (Cha et al., 2002). In this study, the early-senescing mutant es1-1 showed other phenotypes, such as decreased number of tillers and substantially reduced seed-setting rate. Our data provide new insights into the molecular mechanisms underlying early senescence in plants, which will help genetically improve rice yield.
MATERIALS AND METHODS
Plant Growth and Sampling
The rice (Oryza sativa) es1-1 and es1-2 mutants were isolated from the japonica cv NPB that was subjected to ethyl methanesulfonate treatment. After multiple generations of inbreeding, the phenotypes of these two mutants were stably inherited, and homozygous mutant strains were obtained. The homozygous mutant es1-1 was directly and reciprocally crossed with two typical japonica cultivars (cv Zhonghua 11 and cv Chunjiang 06) and one typical indica rice cultivar (cv NJ6), providing a relatively clear genetic background to construct the genetic population. The obtained seeds of the F1 generation were sown and transplanted as individual plants to generate the F2 generation by inbreeding. The numbers of individuals showing wild-type and early senescence phenotypes were counted at the seedling stage. The segregation ratio was calculated using statistical methods. Genetic analysis was performed on es1-1, and recessive individuals were retrieved for preliminary mapping of the mutation. Plant materials were bred in an experimental field of the China National Rice Research Institute, in Hangzhou.
Detection of Reactive Oxygen Species and Malondialdehyde
Hydrogen peroxide and superoxide radical were detected using NBT and DAB, respectively, according to Li et al. (2010) and Wi et al. (2010), with some modifications. Leaves of 3-week-old plants that were grown in growth chambers were incubated in 0.05% (w/v) NBT (Duchefa) or 0.1% (w/v) DAB (Sigma) at room temperature in darkness with gentle shaking for 12 h Chlorophyll was cleared by treating with 90% (v/v) ethanol at 80°C. The concentration of malondialdehyde was measured according to the method described by Moradi and Ismail (2007) with minor modifications.
Determination of Water Potential
Ten seeds of cv NPB and es1-1 were selected, germinated for 3 d, and then cultured in an incubator at 28°C for 60 d. The WP4-T Dewpoint PotentiaMeter was preheated for 30 min until the temperature remained stable. Various tissues such as leaf, sheath, culm, and root of cv NPB and es1-1 were harvested from the same position on each plant and cut into short pieces. After making sure that the sample temperature was lower than the room temperature, the samples were put in the measuring apparatus, which showed the water potential value of the sample. Every sample measurement had six independent repeats, and the weight of the samples was equal.
Map-Based Cloning of ES1
The mutation in es1-1 was mapped in the F2 population of a cross between es1-1 and the indica cv NJ6. To finely map the ES1 locus, new molecular markers were developed using public databases (Supplemental Table S4). Furthermore, ES1 was sequenced using specific primers (Supplemental Table S4).
Fluorescence Microscopy of Actin Filaments
For F-actin observation, plants were grown in a culture chamber with a 16/8-h photoperiod at 30°C. Seedling roots were used to observe actin organization. F-actin was stained using the previously described method (Olyslaegers and Verbelen, 1998; Yang et al., 2011; Zhang et al., 2011). Seven-day-old roots were fixed in PEM buffer (100 mm PIPES, 10 mm EGTA, 0.3 m mannitol, and 5 mm MgSO4, pH 6.9) with 3% paraformaldehyde for 30 min and washed three times with PEM. After washing, roots were incubated in PEM buffer with 0.66 mm Alexa Fluor 488-phalloidin (Life Technologies). After overnight incubation at 4°C, samples were washed three times with PEM buffer and then observed in 50% glycerol with a confocal microscope (Olympus Fluoview FV1000).
Phylogenetic Analyses
A BLAST search of the SCAR-LIKE PROTEIN2 amino acid sequence was performed against the database (http://www.ebi.ac.uk/Tools/sss/ncbiblast/). The phylogenetic tree was constructed based on the amino acid sequences. Maximum likelihood phylogenetic analysis was performed by MEGA6.0 (Tamura et al., 2013). Maximum likelihood analysis of the SCAR-LIKE PROTEIN2 family was based on the amino acid sequences using the Jones-Taylor-Thornton model, which was chosen after model testing. Support for the maximum likelihood trees was evaluated by 1,000 bootstrap replicates.
Plasmid Construction and Plant Transformation
The cDNA sequence of ES1 was amplified using the primers cES1-F and cES1-R (Supplemental Table S3). This cDNA was fused in frame with eGFP in pCambia1300 (http://www.cambia.org) with minor modifications to generate the transient expression vector 35S::eGFP-cES1. The 35S::eGFP-cES1 plasmids were transferred into Agrobacterium tumefaciens strain EHA105 by electroporation; the cells of rice ES1, tobacco (Nicotiana tabacum), and onion (Allium cepa) were transformed according to a previously described method (Hiei et al., 1994; Lv, 2010).
For the molecular complementation assay, a 10.6-kb genomic fragment comprising the ES1 promoter region, coding sequence, and 3′ untranslated region was PCR amplified from the genomic DNA of wild-type plants using pES-F and pES-R primers (Supplemental Table S3). The PCR amplification product was cloned into pCambia1300 to generate the ES1:ES1 construct. Briefly, the pCambia1300::ES1 vector was first digested with the restriction enzymes PstI and XbaI. Next, the fragment containing ES1 was digested with restriction enzymes Sse83871 and NheI. All the restriction enzymes used above are isocaudomers. The pCambia1300::ES1 plasmids were transformed into A. tumefaciens strain EHA105 by electroporation. The es1-1 and es1-2 plants were transformed according to a published method (Hiei et al., 1994).
To generate the ES1:GUS fusion construct, a 2-kb upstream genomic region of the ES1 start codon was amplified with the primers ES1-gus-F and ES1-gus-R (Supplemental Table S3). The amplified fragment was linked with the reporter GUS gene and together inserted in the pCambia1305 vector (http://www.cambia.org).
GUS Staining
Samples (proES1::GUS) were stained with a solution containing 50 mm NaPO4 buffer, 1 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcA, 0.4 mm K3Fe(CN)6, 0.4 mm K4Fe(CN)6, and 0.1% (v/v) Triton X-100. The samples were then incubated at 37°C in the dark for 10 h. Chlorophyll was removed with 70% ethanol for observation.
RNA Extraction and RT-qPCR
Total RNA was extracted from the roots, culms, leaves, sheaths, and spikelets of wild-type and es1-1 plants at the seedling, tillering, and heading stages using the RNeasy Plant Mini Kit (Qiagen). Reverse transcription was handled with the ReverTra Ace qPCR-RT Kit (Toyobo); the primers are described in Supplemental Table S4. RT-qPCR analysis was conducted with the StepOnePlus System (Applied Biosystems) using Thunderbird qPCR Mix (Toyobo); the reaction procedure was 95°C for 30 s, 95°C for 5 s, 55°C for 10 s, and 72°C for 15 s for 40 cycles. OsACTIN was used as a control. The 2−ΔΔCt method was used for quantification (Livak and Schmittgen, 2001).
Microscopy
In different periods, especially at noon when the sunlight was strongest (and, thus, the stomata are likely to be closed), fresh leaf specimens were collected from the same position (one-fourth, one-half, and three-fourths of the leaf blade) in the mutant es1-1 and the control cv NPB from plants that showed consistent growth. The specimens were cut into approximately 1- × 2-mm pieces and processed as follows for microscopic examination.
For dual fixation, the specimens were fixed in 2.5% glutaraldehyde solution, vacuumed in a vacuum apparatus for approximately 20 to 30 min (which allowed the specimens to be immersed in the fixative as completely as possible), and kept at 4°C overnight. Thereafter, the fixative was discarded, and the specimens were rinsed with 0.1 m phosphate-buffered saline, pH 7. The specimens were rinsed three to four times (15 min each). The specimens were then fixed in 1% osmium tetroxide for at least 1 h. The fixative was discarded, and the specimens were rinsed with 0.1 m phosphate-buffered saline three times (15 min each) as described above.
For dehydration, the specimens were first dehydrated with a gradient of ethanol (50%, 70%, 80%, 90%, and 95%; 15–20 min at each gradient), processed in anhydrous ethanol for 20 min, and treated in pure acetone for 20 min.
After dehydration, the specimens were immersed in a mixture of ethanol and isoamyl acetate esters (volume ratio, 1:1) for 30 min and then treated with pure isoamyl acetate for 1 to 2 h. The specimens were dried, coated, and examined using the Hitachi TM-1000 scanning electron microscope.
For transmission electron microscopy of the leaves and cross-sectional examination of the leaves and roots, the specimens were pretreated as described above. After dehydration, the specimens were treated with a mixture of embedding agent and pure acetone at a volume ratio of 1:1 for 1 h, followed by treatment with a mixture of embedding agent and pure acetone at a volume ratio of 3:1 for 3 h and pure embedding agent overnight. The specimens were then embedded in small boxes and heated at 70°C for approximately 9 h. The obtained embedded samples were sliced into 70- to 90-nm sections using a Reichert ultramicrotome and stained with lead citrate solution and saturated uranyl acetate solution in 50% ethanol for 15 min each. The sections were examined and photographed using the Hitachi H-7650 transmission electron microscope.
Determination of Minerals
Wild-type and es1-1 plants were cultured in complete hydroponic nutrient solution up to the heading stage. Tissue specimens were taken from the roots, stems, old leaves, leaf sheaths, and new leaves. The weight of fresh specimens was more than 10 g for the different tissues. The specimens were oven dried at 80°C for 3 d to achieve a constant weight. After grinding, the dry samples (0.25 g for both es1-1 and cv NPB) were digested with a mixture of concentrated nitric acid and perchloric acid (volume ratio, 1:3). The volume of the digestion solution was adjusted to 25 mL with single-distilled water. The treated samples were then used for elemental analysis. Concentrations of elements were determined using full-spectrum direct reading inductively coupled plasma-atomic emission spectroscopy (IRIS Intrepid II XSP ICP-OES PE-2100; Thermo Electron; Shao et al., 2008).
Detection of the Rate of Water Loss
In Vitro Detection
The same leaf parts were selected from cv NPB and es1-1 plants at the seedling stage. These leaf parts were left at room temperature and observed and photographed every 10 min to examine their dehydration. At the tillering stage, the same leaf parts of cv NPB and es1-1 were quickly weighed (W1), and the total area (S) was measured. Moreover, the weights of the leaf blade (W2, W3, and W4) were also measured every 1 h. The formula (W1 − W2) S−1 was used to calculate the rate of water loss. Accordingly, the rate of water loss at different time points was obtained.
In Vivo Detection
After accelerating germination, wild-type and es1-1 seeds were cultured in fine sand and watered twice per day (in the morning and evening). At the one- to two-leaf stage, rice seedlings were washed off and wrapped in a long sponge, then placed in a closed bottle with clean water. At the three-leaf stage, clean water was replaced by complete nutrient solution. After the seedlings developed new roots, all the bottles were filled with a sufficient volume of nutrient solution, and the level was marked for subsequent measurement. The leaves were weighed on days 1 (W1), 4 (W2), 8 (W3), and 14 (W4). The corresponding leaf areas were also measured (S2, S3, and S4). Water loss per unit of time in 4 d was calculated as (W1 − W2) S2−1. Six plants were used for every time interval measurement (the results are presented as mean values). Analogously, (W1 − W3) S3−1 was the amount of water loss per unit of time in 8 d, and (W1 − W4) S4−1 was the amount of water loss per unit of time in 14 d.
Measurement of Root Traits
Hydroponic experiments were conducted using wild-type and es1-1 plants. The seeds were soaked for 2 d, during which time the water was changed once. Next, the seeds were transferred into a germination box to accelerate germination. Three days later, seeds with radicles breaking through the testa were transferred into fine sand for cultivation. After incubation for 7 d, the seedlings were washed carefully to avoid damage to the roots. The plants were transferred into a black bucket containing tap water. After the transition, the plants were supplied with complete nutrient solution. The seedlings entered the two-leaf stage after 3 d. At the five-leaf stage, seedlings of the mutant and wild-type plants (10 each) were taken out for root measurements. The hydroponic seedlings were rinsed and placed in a transparent rectangular box containing single-distilled water and loaded onto a scanner (ScanWizard Pro: ScanMarker 1000XL) for transparent scanning. The obtained images were loaded into Win-RHIZO Pro. V. 2002c for data analysis, and the results were exported to Excel 2011 (Microsoft) for analysis.
Sequence data from this article are available at the National Center for Biotechnology Information Web site (http:www.ncbi.nlm.nih.gov) with the following accession numbers: LOC_Os01g11040 (NP_001172227.1), OB01G1689 (XP_006643892.1), Bradi2g06610 (XP_003565325.1), rice indica (OsI_00845), S. italica (Si000056m.g), Hordeum vulgare var distichum (M0XEC2), Aegilops tauschii (F775_07293), Triticum aestivum (W5CZS7), maize (ZEAMMB73_679274), Triticum urartu (TRIUR3_31093), Musa acuminata ssp. malaccensis (M0UB73), Medicago truncatula (MTR_8g086300), Theobroma cacao (TCM_029698), Phaseolus vulgaris (PHAVU_002G238500g), Jatropha curcas (JCGZ_21566), Glycine max (I1K3Z5), Populus trichocarpa (POPTR_0004s05600g), Prunus persica (PRUPE_ppa000443mg), Citrus sinensis (CISIN_1g001053mg), Citrus clementina (CICLE_v10014081mg), Eucalyptus grandis (EUGRSUZ_D00489), Solanum lycopersicum (Solyc02g076840.2), Erythranthe guttata (MIMGU_mgv1a000504mg), Vitis vinifera (VIT_10s0003g01270), Ricinus communis (RCOM_0850090), Amborella trichopoda (AMTR_s00021p00061660), Morus notabilis (L484_002198), and Arabidopsis lyrata ssp. lyrata (ARALYDRAFT_903078).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypes of allelic mutants.
Supplemental Figure S2. Comparison of guttation between wild-type and es1-1 plants.
Supplemental Table S1. Allelic analysis of es1-1 and es1-2.
Supplemental Table S2. Phenotypic data of cv NPB and es1-1.
Supplemental Table S3. Reciprocal cross experiments between es1-1 and three crop strains (cv Zhonghua 11, cv Chunjiang 06, and cv NJ6).
Supplemental Table S4. Primers used for PCR in this study.
Acknowledgments
We thank Jianru Zuo (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for sharing unpublished data and Lizhong Xiong (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University) for critical reading and suggestions for our manuscript.
Glossary
- NPB
Nipponbare
- cM
centimorgan
- cDNA
complementary DNA
- RT-qPCR
reverse transcription-quantitative PCR
- ABA
abscisic acid
- RWL
rate of in vitro water loss
- Cu
copper
- Fe
iron
- Mn
manganese
- Zn
zinc
- NJ6
Nanjing 6
- NBT
nitroblue tetrazolium
- DAB
diaminobenzidine
- eGFP
enhanced green fluorescent protein
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31201183, 31221004, 31171531, 31171520, and 91435105), the State Key Basic Research Program (grant no. 2013CBA01403), the Ministry of Agriculture of China for Transgenic Research (grant no. 2014ZX08009003–001), and the China Postdoctoral Science Foundation (grant no. 2014M561108).
Articles can be viewed without a subscription.
References
- Bai J, Zhu X, Wang Q, Zhang J, Chen H, Dong G, Zhu L, Zheng H, Xie Q, Nian J, et al. (2015) Rice TUTOU1 Encodes a Suppressor of cAMP Receptor-Like Protein That Is Important for Actin Organization and Panicle Development. Plant Physiol 169: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Le J, El-Essal Sel-D, Huang S, Zhang C, Mallery EL, Koliantz G, Staiger CJ, Szymanski DB (2005) DISTORTED3/SCAR2 is a putative Arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17: 502–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Boonrueng N, Anuntalabhochai S, Jampeetong A (2013) Morphological and anatomical assessment of KDML 105 (Oryza sativa L. spp. indica) and its mutants induced by low-energy ion beam. Rice Sci 20: 213–219 [Google Scholar]
- Bouman B, Humphreys E, Tuong T, Barker R (2007) Rice and water. Adv Agron 92: 187–237 [Google Scholar]
- Buckley TN. (2005) The control of stomata by water balance. New Phytol 168: 275–292 [DOI] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R (2012) Composition and physiological function of the wax layers coating Arabidopsis leaves: β-amyrin negatively affects the intracuticular water barrier. Plant Physiol 160: 1120–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha KW, Lee YJ, Koh HJ, Lee BM, Nam YW, Paek NC (2002) Isolation, characterization, and mapping of the stay green mutant in rice. Theor Appl Genet 104: 526–532 [DOI] [PubMed] [Google Scholar]
- Chen JL, Lacomis L, Erdjument-Bromage H, Tempst P, Stamnes M (2004) Cytosol-derived proteins are sufficient for Arp2/3 recruitment and ARF/coatomer-dependent actin polymerization on Golgi membranes. FEBS Lett 566: 281–286 [DOI] [PubMed] [Google Scholar]
- Da Silva D, Favreau R, Auzmendi I, DeJong TM (2011) Linking water stress effects on carbon partitioning by introducing a xylem circuit into L-PEACH. Ann Bot (Lond) 108: 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Li Y, Li Y, Shen Q, Guo S (2014) Effects of drought stress on photosynthesis and water status of rice leaves. Chin J Rice Sci 28: 65–70 (in Chinese with English abstract) [Google Scholar]
- Du H, Chang Y, Huang F, Xiong L (January 30, 2015) GID1 modulates stomatal response and submergence tolerance involving abscisic acid and gibberellic acid signaling in rice. J Integr Plant Biol http://dx.doi.org/10.1111/jipb.12313 [DOI] [PubMed] [Google Scholar]
- Dyachok J, Zhu L, Liao F, He J, Huq E, Blancaflor EB (2011) SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes. Plant Cell 23: 3610–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen G, Wang L, Thorngren N, Wickner W (2002) Remodeling of organelle-bound actin is required for yeast vacuole fusion. J Cell Biol 158: 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Ziegler H (1998) A plant’s dilemma. Science 282: 252–253 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hu L, Wang Z, Huang B (2010) Diffusion limitations and metabolic factors associated with inhibition and recovery of photosynthesis from drought stress in a C perennial grass species. Physiol Plant 139: 93–106 [DOI] [PubMed] [Google Scholar]
- Huang RF, Wang XC, Lou CH (2000) Cytoskeletal inhibitors suppress the stomatal opening of Vicia faba L. induced by fusicoccin and IAA. Plant Sci 156: 65–71 [DOI] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MA, Du H, Ning J, Ye H, Xiong L (2009) Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol Biol 70: 443–456 [DOI] [PubMed] [Google Scholar]
- Jäger K, Fábián A, Eitel G, Szabó L, Deák C, Barnabás B, Papp I (2014) A morpho-physiological approach differentiates bread wheat cultivars of contrasting tolerance under cyclic water stress. J Plant Physiol 171: 1256–1266 [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG (2003) A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115: 475–487 [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Park J, Hwang I (2014) Investigating water transport through the xylem network in vascular plants. J Exp Bot 65: 1895–1904 [DOI] [PubMed] [Google Scholar]
- Konrad W, Burkhardt J, Ebner M, Roth-Nebelsick R (2015) Leaf pubescence as a possibility to increase water use efficiency by promoting condensation. Ecohydrology 8: 480–492 [Google Scholar]
- Le J, Mallery EL, Zhang C, Brankle S, Szymanski DB (2006) Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr Biol 16: 895–901 [DOI] [PubMed] [Google Scholar]
- Lee RH, Wang CH, Huang LT, Chen SC (2001) Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J Exp Bot 52: 1117–1121 [DOI] [PubMed] [Google Scholar]
- Li J, Pandeya D, Nath K, Zulfugarov IS, Yoo SC, Zhang H, Yoo JH, Cho SH, Koh HJ, Kim DS, et al. (2010) ZEBRA-NECROSIS, a thylakoid-bound protein, is critical for the photoprotection of developing chloroplasts during early leaf development. Plant J 62: 713–725 [DOI] [PubMed] [Google Scholar]
- Li S, Blanchoin L, Yang Z, Lord EM (2003) The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiol 132: 2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Liu L, Liu Q, Bi Z, Yu N, Cheng S, Cao L (2014) Fine mapping of the lesion mimic and early senescence 1 (lmes1) in rice (Oryza sativa). Plant Physiol Biochem 80: 300–307 [DOI] [PubMed] [Google Scholar]
- Linquist BA, Anders MM, Adviento-Borbe MA, Chaney RL, Nalley LL, da Rosa EF, van Kessel C (2015) Reducing greenhouse gas emissions, water use, and grain arsenic levels in rice systems. Glob Change Biol 21: 407–417 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luo L, Zhang Q (2001) The status and strategy on drought resistance of rice (Oryza sativa L.). Chin J Rice Sci 15: 209–214 (in Chinese with English abstract) [Google Scholar]
- Lv F. (2010) Optimization for Agrobacterium tumefaciens-mediated transformation system for exogenous genes of tobacco. J Anhui Agric Sci 38: 10065–10066 (in Chinese with English abstract) [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hülskamp M (2003) Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15: 1632–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Spielhofer P, Kost B, Chua N (1999) The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126: 5559–5568 [DOI] [PubMed] [Google Scholar]
- Moradi F, Ismail AM (2007) Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann Bot (Lond) 99: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Babu RC, Blum A (1997) Breeding for drought resistance in rice: physiology and molecular genetics considerations. Crop Sci 37: 1426–1434 [Google Scholar]
- Olyslaegers G, Verbelen JP (1998) Improved staining of F-actin and co-localization of mitochondria in plant cells. J Microsc 192: 73–77 [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14: 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M, Schreiber L (2001) Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot 52: 2023–2032 [DOI] [PubMed] [Google Scholar]
- Saedler R, Mathur N, Srinivas BP, Kernebeck B, Hülskamp M, Mathur J (2004) Actin control over microtubules suggested by DISTORTED2 encoding the Arabidopsis ARPC2 subunit homolog. Plant Cell Physiol 45: 813–822 [DOI] [PubMed] [Google Scholar]
- Sellammal R, Robin S, Raveendran M (2014) Association and heritability studies for drought resistance under varied moisture stress regimes in backcross inbred population of rice. Rice Sci 21: 150–161 [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Chen M, Wang D, Xu C, Mou R, Cao Z, Zhang X (2008) Iron manure regulation of cadmium accumulation in rice. Chin Sci Bull (C) Life Sci 38: 180–187 [DOI] [PubMed] [Google Scholar]
- Smith S, De Smet I (2012) Root system architecture: insights from Arabidopsis and cereal crops. Philos Trans R Soc Lond B Biol Sci 367: 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsson M. (1988) Epi- and intracuticular lipids and cuticular transpiration rates of primary leaves of eight barley (Hordeum vulgare) cultivars. Plant Physiol 73: 512–517 [Google Scholar]
- Svitkina TM, Borisy GG (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol 145: 1009–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM (1999) Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11: 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Takahashi K, Tomiyama M, Inoue S, Kinoshita T (2013) Overexpression of the Mg-chelatase H subunit in guard cells confers drought tolerance via promotion of stomatal closure in Arabidopsis thaliana. Front Plant Sci 4: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Groll U, Berger D, Altmann T (2002) The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14: 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wan L, Zhang L, Zhang Z, Zhang H, Quan R, Zhou S, Huang R (2012) An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol Biol 78: 275–288 [DOI] [PubMed] [Google Scholar]
- Wi SJ, Jang SJ, Park KY (2010) Inhibition of biphasic ethylene production enhances tolerance to abiotic stress by reducing the accumulation of reactive oxygen species in Nicotiana tabacum. Mol Cells 30: 37–49 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Chen Y, Wang X (2003) Filaments skeleton in guard cell. Chin Bull Bot 20: 489–494 [Google Scholar]
- Xu H, Gauthier L, Gossenlin A (1995) Stomatal and cuticular transpiration of greenhouse tomato plants in response to high solution electrical conductivity and low soil water content. J Am Soc Hortic Sci 120: 417–422 [Google Scholar]
- Yang M, Sack FD (1995) The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7: 2227–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Ren S, Zhang X, Gao M, Ye S, Qi Y, Zheng Y, Wang J, Zeng L, Li Q, et al. (2011) BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23: 661–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Zong W, Li X, Ning J, Hu H, Li X, Xiao J, Xiong L (2013) The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J Exp Bot 64: 569–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Huang RF, Wang XC, Yuan M (2001) Microtubule dynamics are involved in stomatal movement of Vicia faba L. Protoplasma 216: 113–118 [DOI] [PubMed] [Google Scholar]
- Zhang C, Mallery EL, Schlueter J, Huang S, Fan Y, Brankle S, Staiger CJ, Szymanski DB (2008) Arabidopsis SCARs function interchangeably to meet actin-related protein 2/3 activation thresholds during morphogenesis. Plant Cell 20: 995–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Lin F, Mao T, Nie J, Yan M, Yuan M, Zhang W (2012) Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24: 4555–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dyachok J, Krishnakumar S, Smith LG, Oppenheimer DG (2005) IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17: 2314–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang Y, Tan H, Wang Y, Li G, Liang W, Yuan Z, Hu J, Ren H, Zhang D (2011) RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23: 681–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Ni E, Yang J, Zhou H, Liang H, Li J, Jiang D, Wang Z, Liu Z, Zhuang C (2013) Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS ONE 8: e65139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LF, Yu SM, Jin QY (2012) Effects of aerated irrigation on leaf senescence at late growth stage and grain yield of rice. Rice Sci 19: 44–48 [Google Scholar]
- Zhu X, Xiong L (2013) Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc Natl Acad Sci USA 110: 17790–17795 [DOI] [PMC free article] [PubMed] [Google Scholar]