Abstract
Evidence for Trp-independent IAA synthesis is critically reevaluated in the light of tryptophan synthase proteome data, local IAA synthesis and Trp, indole-3-pyruvate, and IAA turnover.
Trp-independent synthesis of indole-3-acetic acid (IAA) was proposed back in the early 1990s based on observations from Trp auxotrophs in maize (Zea mays; Wright et al., 1991) and Arabidopsis (Arabidopsis thaliana; Normanly et al., 1993). Recently, Wang et al. (2015) published new data suggesting that a cytosolic indole synthase (INS) may catalyze the first step separating the Trp-dependent and Trp-independent pathways in Arabidopsis. If this is the case, it would be a major breakthrough; however, in this article, I critically evaluate both recent and older evidence for the Trp-independent route and suggest that the INS is more likely to participate in Trp-dependent IAA production.
The original work supporting Trp-independent IAA production was carried out prior to the availability of genome/proteome data and before the discovery that the final step of Trp-dependent IAA synthesis is carried out by a large number of YUCCA homologs operating in a highly localized manner (Zhao, 2008). I argue that experimental data supporting the Trp-independent route needs to be reconsidered in light of complete proteome data. Further, the evidence from feeding labeled compounds should be critically evaluated in light of recent data on the highly localized nature of IAA synthesis as well as older quantitative data on Trp, indole-3-pyruvic acid (IPA), and IAA turnover from my own laboratory (Cooney and Nonhebel, 1991). I conclude that evidence for the Trp-independent route is at best equivocal, and that it is not a conserved source of IAA in angiosperms.
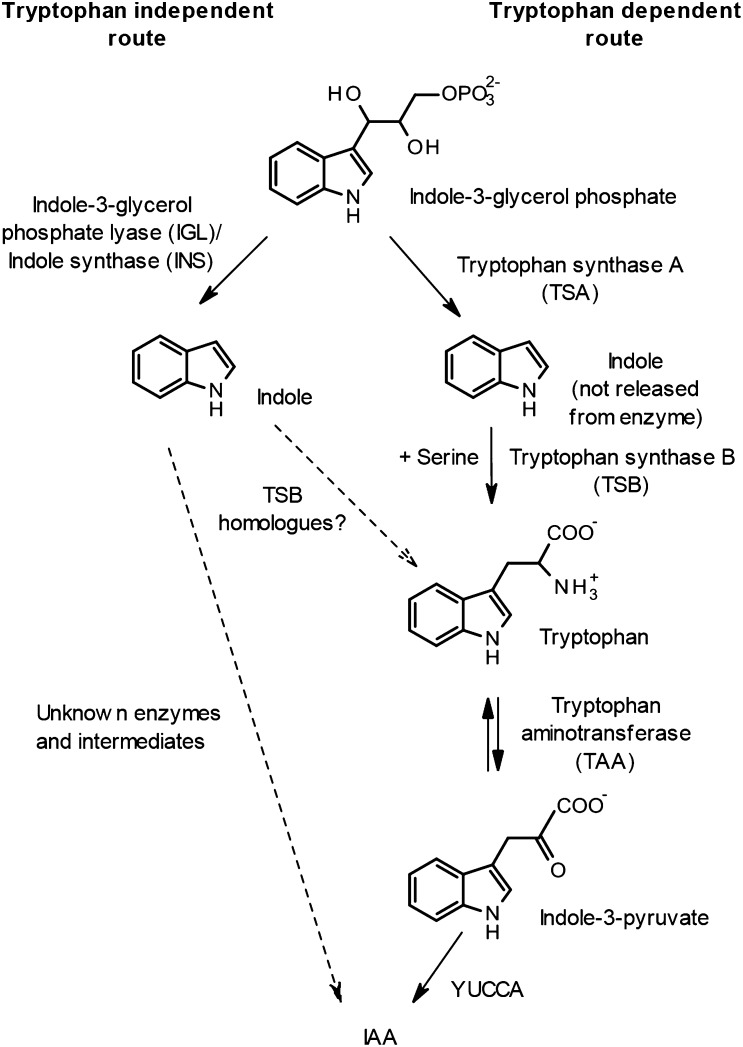
Figure 1 shows the major Trp-dependent route for IAA production whereby Trp, produced by the concerted action of Trp synthase α- and β-subunits, is converted to IAA in a further two steps catalyzed by Trp aminotransferase and the flavin monooxygenases commonly known as YUCCA (Mashiguchi et al., 2011; Won et al., 2011). This is compared with the Trp-independent route in which IAA may be produced from free indole by an unknown route (Ouyang et al., 2000; Wang et al., 2015).
Figure 1.
Outline of the major pathway for Trp-dependent IAA synthesis and the proposed Trp-independent route. The role proposed for Trp synthase beta (TSB) homologs discussed in the present paper is also shown. For clarity, reactions are simplified to show only the major compounds relevant to IAA synthesis.
The Trp-independent route was originally based on data from Trp auxotrophs that have mutations in genes encoding either the α- or β-subunits of Trp synthase. The α-subunit catalyzes the removal of the side chain from indole-3-glycerol phosphate, passing the indole product directly to the β-subunit where the Trp side chain is created from a Ser substrate (Pan et al., 1997). In plants, this is a chloroplast-localized enzyme. Elevated levels of IAA have been reported in Trp auxotrophs of both maize and Arabidopsis. However, the trp3-1 and trp2-1 mutants of Arabidopsis, deficient in the α- and β-subunits, respectively, only showed an increase in total IAA measured following conjugate hydrolysis. No difference in free IAA levels was found (Normanly et al., 1993). The orange pericarp (orp) maize mutant was reported to have 50 times more IAA than the wild type (Wright et al., 1991). However, this was also total IAA; no data on free IAA were published. Work by Müller and Weiler (2000) indicated that IAA measured following conjugate hydrolysis could have originated via the degradation of indole-3-glycerol phosphate that accumulates in trp3-1 mutants. Further doubt regarding the accuracy of IAA measurements following conjugate hydrolysis has recently been published. Yu et al. (2015) have shown that conjugate hydrolysis treatment substantially overestimates the actual conjugated IAA due to degradation of glucobrassicin and proteins. In addition, neither report (Wright et al., 1991; Normanly et al., 1993) described a high auxin phenotype for the Trp auxotrophs. This contrasts with the superroot1 (sur1) and sur2 mutants, where the accumulation of indole intermediates resulted in a high level of free IAA as well as a high auxin phenotype (Boerjan et al., 1995; Delarue et al., 1998). It is therefore doubtful that Trp auxotrophs actually accumulate more IAA than the wild-type plants.
In addition, proteome data have revealed new homologs of TSB in both Arabidopsis and maize that may contribute to Trp production in TSB mutants; these have not been considered in arguments supporting Trp-independent IAA synthesis. Maize orp has mutations in two TSB genes, resulting in a seedling lethal phenotype with high levels of accumulated indole. However, proteome sequence information now indicates that maize has three TSB genes. Plants and bacteria have divergent forms of TSB, type 1 and type 2 (Xie et al., 2001); the major TSB genes responsible for Trp synthase activity in maize and Arabidopsis are type 1. The third maize TSB gene, maize locus ID GRMZM2G054465, is a member of the TSB type 2 group. Its product is reported not to interact directly with a Trp synthase alpha (TSA) subunit but has experimentally demonstrated catalytic activity converting indole and Ser to Trp (Yin et al., 2010). This type 2 TSB may allow orp plants to make sufficient Trp for IAA production from the accumulated free indole.
When the original work on trp2 mutants of Arabidopsis was carried out, two TSB genes were known (Last et al., 1991). As the trp2 plants were deficient only in TSB1, they were able to make sufficient Trp to survive under low-light conditions. Full proteome data now indicate that Arabidopsis has four TSB-like genes; in addition to TSB1 and TSB2, there is a third type 1 TSB gene, Arabidopsis locus ID AT5G28237. The product of this gene has not been experimentally characterized. The fourth gene, AT5G38530, encodes a type 2 TSB with demonstrated catalytic activity similar to ZmTSB type 2 mentioned above (Yin et al., 2010). Thus, the trp2 plants may also make enough Trp for IAA production. It is even possible that one of the minor forms of TSB has a specific role in IAA production. Type 2 TSBs are conserved throughout the plant kingdom, and the biological role for this protein is not known (Xie et al., 2001).
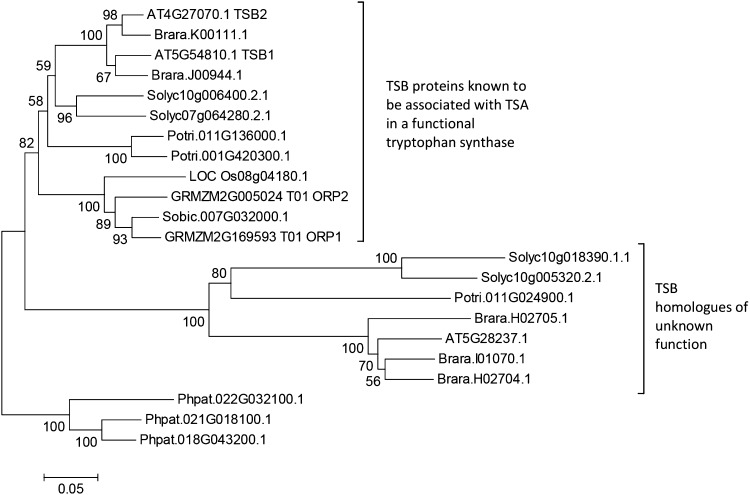
A phylogenetic analysis of type 1 TSBs is shown in Figure 2. This indicates that the product of AT5G28237 belongs to a eudicot-conserved TSB type 1-like clade, divergent from that containing major experimentally characterized TSBs. A multiple sequence alignment (not shown) reveals that members of this divergent clade have a shortened N terminus with respect to the major chloroplast-localized TSB proteins. A localization prediction carried out in CELLO (Yu et al., 2006) suggests a cytosolic location for these proteins. Examination of EST databases indicates that the genes encoding these proteins are expressed. It is possible that the product of AT5G28237 could interact with the cytosolic INS studied by Wang et al. (2015), or separately with its indole product, to produce Trp that is further converted to IAA.
Figure 2.
Phylogeny of TSB type 1 homologs from Oryza sativa (LOC_Os), Sorghum bicolor (Sobic), Z. mays (GRMZM), Arabidopsis (AT), Brassica rapa (Brara), Solanum lycopersicum (Solyc), Populus trichocarpa (Potri), and Physcomitrella patens (Phpat). Protein sequences were downloaded from Phytozome v10.2 (Goodstein et al., 2012). The phylogenetic analysis was conducted in MEGA6 (http://megasoftware.net; Tamura et al., 2013) with multiple sequence alignment by MUSCLE (Edgar, 2004) and evolutionary history inferred using the neighbor-joining method (Saitou and Nei, 1987). The optimal tree is shown; the percentage of replicate trees in which the associated sequences clustered together in the bootstrap test (500 replicates) is shown next to the branches (Felsenstein, 1985). The tree is drawn to scale; the scale bar indicates the number of amino acid substitutions per site. It is rooted with type 1 TSBs from the moss P. patens.
The second major line of evidence for Trp-independent IAA synthesis comes from isotopic labeling experiments. Wright et al. (1991) observed greater incorporation of 2H into IAA than Trp in orp seedlings grown on 2H2O. Normanly et al. (1993) reported higher enrichment of 15N in IAA than Trp in trp2-1 mutants of Arabidopsis grown on 15N anthranilate; very poor incorporation of deuterium from 2H-Trp into IAA was reported in the trp2-1 plants. A number of similar reports relating to other plants have been published showing differences in the incorporation of label from Trp into IAA depending on experimental tissue and environmental conditions (e.g. Michalczuk et al., 1992; Rapparini et al., 2002; Sztein et al., 2002). This evidence has been persuasive; however, it assumes a single pool of Trp to which 15N anthranilate and 2H-Trp contribute and from which IAA is made. If Trp is made at different rates in different parts of the plant, and/or exogenous 2H-Trp does not equilibrate with newly synthesized Trp, then the ratio of 15N to 2H in Trp will vary in different plant organs/tissues/cells. Trp turnover and thus incorporation of label from 15N anthranilate are likely to differ substantially throughout the plant, with the highest rates of labeling occurring in cells with high rates of protein synthesis. This would not be a problem for the experiment if IAA is made at equal rates in different parts of the plant, but we know it is not. The Trp aminotransferase/YUCCA pathway of IAA synthesis elegantly shown to be responsible for the bulk of IAA synthesis (Mashiguchi et al., 2011; Won et al., 2011) appears to be locally controlled in Arabidopsis via 11 different YUCCA-encoding genes that have highly localized expression (Zhao, 2008). Adding to the complication is the need for 15N anthranilate and 2H-Trp to move into and through the plant to regions of Trp and IAA synthesis, respectively. This is likely to occur at different rates due to differing transporter requirements.
Data from my own laboratory (Cooney and Nonhebel, 1991) is particularly relevant to this discussion. We monitored incorporation of 2H from deuterated water into IAA and Trp in tomato (S. lycopersicum) shoots. Unlike the other studies, we also measured the incorporation of label into IPA. Our data showed that IPA became labeled at a rate consistent with this compound acting as the major/sole precursor of IAA. Crucially, the proportion of labeled Trp was lower than 2H-IPA. Our interpretation of these data was that IPA and IAA were produced from newly synthesized Trp, and that Trp was not uniformly labeled throughout the shoot. At the time, we suggested different subcellular pools of Trp; this may be the case, but in light of new knowledge of localized IAA synthesis, it is most likely that substantial differences in Trp and IAA turnover in different cells/tissues may be the reason for these observations.
The arguments above cast doubt on the existence of the Trp-independent route; however, a recent publication by Wang et al. (2015) claims to provide new evidence for its importance. They present the interesting finding that Arabidopsis plants with a null mutation in INS, a cytosolic TSA homolog previously shown to have indole-3-glycerol phosphate lyase (IGL) activity (Zhang et al., 2008), had reduced levels of IAA. The mutation particularly affected early embryo development. I suggest that the INS may make a contribution to IAA synthesis, but the only specific evidence that it does so via a Trp-independent route is the observation that the ins-1 mutation has an additive effect with the weakly ethylene insensitive8-1 Trp aminotransferase mutation. This evidence is indicative rather than conclusive. The possibility that INS may act in concert with a minor TSB homolog, as suggested in Figure 1, needs to be considered.
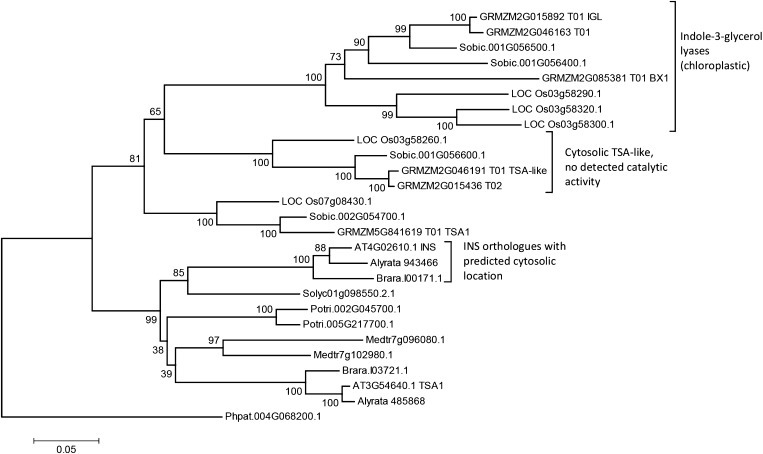
In addition, Wang et al. (2015) focus on Arabidopsis alone. If INS has a key role in IAA synthesis, then evolutionary theory predicts a conserved protein with wide taxonomic distribution. On the contrary, an exhaustive BLAST search (Altschul et al., 1997) of diverse taxa in Phytozome v10.2 (Goodstein et al., 2012) and GenBank (Benson et al., 2013) revealed that INS orthologs with cytosolic prediction and shortened N terminus occur only in members of the Brassicaceae (Eutrema salsugineum, Arabis alpina, Camelina sativa, Capsella rubella, Brassica napus, Boechera stricta, Arabidopsis lyrata, B. rapa) and in Tarenaya hassleriana from the Brassicaceae sister family, the Cleomaceae. The phylogenetic tree in Figure 3 shows relationships between INS and TSA homologs from several plant species and indicates the separate clade of cytosolic INS homologs in the Brassicaceae. In this diagram, the sequence most closely related to INS from another group is that from tomato. This protein is the only TSA found in tomato and has an unambiguous chloroplast signal peptide. P. trichocarpa and M. truncatula as well as other eudicots outside the Brassicaeae and Cleomaceae also lack cytosolic TSA homologs. Furthermore, INS and its orthologs are phylogenetically distinct from the other experimentally characterized indole-3-glycerol phosphate lyases benzoxazin1 and IGL (Frey et al., 2000) and their orthologs. The latter are restricted to the grasses where they are involved in the production of cyclic hydroxamic acid defense compounds (Frey et al., 2000). The grasses also have an additional separate clade of cytosolic TSA homologs, although work by Kriechbaumer et al. (2008) did not detect any catalytic activity for the product of GRMZM2G046191_T01. The phylogeny of INS and its orthologs would suggest the major role of these proteins may be the production of lineage-specific metabolites such as the indole-derived defense compounds produced in grasses; any role in IAA synthesis may be incidental and restricted to the Brassicaeae and Cleomaceae.
Figure 3.
Phylogeny of TSA homologs from O. sativa (LOC_Os), S. bicolor (Sobic), Z. mays (GRMZM), Arabidopsis (AT), A. lyrata (Alyrata), B. rapa (Brara), S. lycopersicum (Solyc), Medicago truncatula (Medtr), P. trichocarpa (Potri), and P. patens (Phpat). Protein sequences were downloaded from Phytozome v10.2 (Goodstein et al., 2012). The phylogenetic analysis was conducted in MEGA6 (Tamura et al., 2013) with multiple sequence alignment by MUSCLE (Edgar, 2004) and evolutionary history inferred using the neighbor-joining method (Saitou and Nei, 1987). The rooted optimal tree is shown; the percentage of replicate trees in which the associated sequences clustered together in the bootstrap test (500 replicates) is shown next to the branches (Felsenstein, 1985). The tree is drawn to scale; the scale bar indicates the number of amino acid substitutions per site. It is rooted with the TSA ortholog from the moss P. patens.
In conclusion, I contend that experimental data relating to IAA synthesis in Arabidopsis, including that suggesting the involvement of a cytosolic INS, can be explained by the Trp-dependent IAA synthesis pathway. I show that INS and its orthologs are not found outside the Brassicaceae and a closely related sister clade; any alternative IAA synthesis pathway in which they may be involved is likely to have similar limited taxonomic occurrence. Furthermore, Arabidopsis and its relatives contain two additional TSB homologs that could convert free indole into Trp. Curiously, both of these proteins have a wider taxonomic distribution. A priority for further experimental work should be testing the involvement of minor TSB homologs in IAA synthesis, including the highly conserved type 2 TSBs as well as a eudicot-specific clade of possibly cytosolic type 1 TSBs. Work would also have to establish whether free indole exists in plants other than the Brassicaceae and the grasses. Finally, I argue that isotope-labeling experiments do not provide strong support for the Trp-independent route, as IAA production is highly localized. Previously published data from my laboratory clearly show that the main Trp-dependent IAA precursor IPA becomes more highly labeled from 2H2O than Trp, even though the latter is produced from Trp in a single reaction. Thus, it cannot be argued that differences in isotope enrichment between Trp and IAA demonstrate the existence of a Trp-independent route.
Glossary
- IAA
indole-3-acetic acid
- INS
indole synthase
- IPA
indole-3-pyruvate acid
- TSA
Trp synthase alpha
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41: D36–D42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D (1995) superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney TP, Nonhebel HM (1991) Biosynthesis of indole-3-acetic acid in tomato shoots: measurement, mass-spectral identification and incorporation of (-2)H from (-2)H2O into indole-3-acetic acid, D- and L-tryptophan, indole-3-pyruvate and tryptamine. Planta 184: 368–376 [DOI] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C (1998) Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J 14: 603–611 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Frey M, Stettner C, Pare PW, Schmelz EA, Tumlinson JH, Gierl A (2000) An herbivore elicitor activates the gene for indole emission in maize. Proc Natl Acad Sci USA 97: 14801–14806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Weigang L, Fiesselmann A, Letzel T, Frey M, Gierl A, Glawischnig E (2008) Characterisation of the tryptophan synthase alpha subunit in maize. BMC Plant Biol 8: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Bissinger PH, Mahoney DJ, Radwanski ER, Fink GR (1991) Tryptophan mutants in Arabidopsis: the consequences of duplicated tryptophan synthase β genes. Plant Cell 3: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalczuk L, Ribnicky DM, Cooke TJ, Cohen JD (1992) Regulation of indole-3-acetic acid biosynthetic pathways in carrot cell cultures. Plant Physiol 100: 1346–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Weiler EW (2000) Indolic constituents and indole-3-acetic acid biosynthesis in the wild-type and a tryptophan auxotroph mutant of Arabidopsis thaliana. Planta 211: 855–863 [DOI] [PubMed] [Google Scholar]
- Normanly J, Cohen JD, Fink GR (1993) Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci USA 90: 10355–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Shao X, Li J (2000) Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J 24: 327–333 [DOI] [PubMed] [Google Scholar]
- Pan P, Woehl E, Dunn MF (1997) Protein architecture, dynamics and allostery in tryptophan synthase channeling. Trends Biochem Sci 22: 22–27 [DOI] [PubMed] [Google Scholar]
- Rapparini F, Tam YY, Cohen JD, Slovin JP (2002) Indole-3-acetic acid metabolism in Lemna gibba undergoes dynamic changes in response to growth temperature. Plant Physiol 128: 1410–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sztein AE, Ilic N, Cohen JD, Cooke TJ (2002) Indole-3-acetic acid biosynthesis in isolated axes from germinating bean seeds: the effect of wounding on the biosynthetic pathway. Plant Growth Regul 36: 201–207 [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, Li J (2015) Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci USA 112: 4821–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AD, Sampson MB, Neuffer MG, Michalczuk L, Slovin JP, Cohen JD (1991) Indole-3-acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 254: 998–1000 [DOI] [PubMed] [Google Scholar]
- Xie G, Forst C, Bonner C, Jensen RA (2001) Significance of two distinct types of tryptophan synthase beta chain in bacteria, archaea and higher plants. Genome Biol 3: RESEARCH0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Frey M, Gierl A, Glawischnig E (2010) Plants contain two distinct classes of functional tryptophan synthase beta proteins. Phytochemistry 71: 1667–1672 [DOI] [PubMed] [Google Scholar]
- Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64: 643–651 [DOI] [PubMed] [Google Scholar]
- Yu P, Lor P, Ludwig-Müller J, Hegeman AD, Cohen JD (2015) Quantitative evaluation of IAA conjugate pools in Arabidopsis thaliana. Planta 241: 539–548 [DOI] [PubMed] [Google Scholar]
- Zhang R, Wang B, Ouyang J, Li J, Wang Y (2008) Arabidopsis indole synthase, a homolog of tryptophan synthase alpha, is an enzyme involved in the Trp-independent indole-containing metabolite biosynthesis. J Integr Plant Biol 50: 1070–1077 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2008) The role of local biosynthesis of auxin and cytokinin in plant development. Curr Opin Plant Biol 11: 16–22 [DOI] [PubMed] [Google Scholar]