Phosphorylation of the presequence of an RNA editing factor by cytosolic kinases and its dephosphorylation by an outer mitochondrial membrane phosphatase contribute to mitochondrial import of the factor protein.
Abstract
The nucleus-encoded mitochondria-targeted proteins, multiple organellar RNA editing factors (MORF3, MORF5, and MORF6), interact with Arabidopsis (Arabidopsis thaliana) PURPLE ACID PHOSPHATASE2 (AtPAP2) located on the chloroplast and mitochondrial outer membranes in a presequence-dependent manner. Phosphorylation of the presequence of the precursor MORF3 (pMORF3) by endogenous kinases in wheat germ translation lysate, leaf extracts, or STY kinases, but not in rabbit reticulocyte translation lysate, resulted in the inhibition of protein import into mitochondria. This inhibition of import could be overcome by altering threonine/serine residues to alanine on the presequence, thus preventing phosphorylation. Phosphorylated pMORF3, but not the phosphorylation-deficient pMORF3, can form a complex with 14-3-3 proteins and HEAT SHOCK PROTEIN70. The phosphorylation-deficient mutant of pMORF3 also displayed faster rates of import when translated in wheat germ lysates. Mitochondria isolated from plants with altered amounts of AtPAP2 displayed altered protein import kinetics. The import rate of pMORF3 synthesized in wheat germ translation lysate into pap2 mitochondria was slower than that into wild-type mitochondria, and this rate disparity was not seen for pMORF3 synthesized in rabbit reticulocyte translation lysate, the latter translation lysate largely deficient in kinase activity. Taken together, these results support a role for the phosphorylation and dephosphorylation of pMORF3 during the import into plant mitochondria. These results suggest that kinases, possibly STY kinases, and AtPAP2 are involved in the import of protein into both mitochondria and chloroplasts and provide a mechanism by which the import of proteins into both organelles may be coordinated.
Chloroplasts and mitochondria are endosymbiotic organelles that are intimately involved in energy metabolism in plants (Araújo et al., 2014). The majority of proteins located in chloroplasts and mitochondria are encoded in the nucleus, translated in the cytosol, and imported into the organelles (Murcha et al., 2014). For both chloroplasts and mitochondria, over 1,000 different proteins are required to be specifically imported into each organelle. Furthermore, while most proteins are imported specifically into one organelle, a significant number of proteins are dually targeted to both via ambiguous targeting signals (Carrie et al., 2009a; Ye et al., 2012, 2015). The sorting of proteins to chloroplast and mitochondria is achieved through the inclusion of targeting signals in newly synthesized proteins that act in combination with receptor domains present in outer membrane multisubunit protein complexes to specifically direct proteins to their destination compartments (Jarvis, 2008; Shi and Theg, 2013; Murcha et al., 2014). Chloroplast targeting signals (transit peptides) and mitochondrial targeting signals (presequences) are recognized by receptors on the translocase of the outer chloroplast envelope (TOC) and the translocase of the outer mitochondria membrane (TOM), respectively (Ye et al., 2015). In addition to the targeting signals and protein receptors, it has been proposed that cytosolic chaperone proteins may also play a role in maintaining precursor proteins in an import-competent state and may play a role in determining targeting specificity. However, in plants, the role of cytosolic chaperones has only been characterized to some extent for protein import into chloroplasts (Jarvis, 2008; Fellerer et al., 2011; Flores-Pérez and Jarvis, 2013; Lee et al., 2013; Schweiger et al., 2013) but not into mitochondria.
In addition to cytosolic chaperone factors, three STY kinases are also involved in phosphorylating the transit peptides of several chloroplast precursor proteins, such as the precursor for the small subunit of Rubisco (pSSU) and the precursor for HIGH CHLOROPHYLL FLUORESCENCE136 (pHCF136), but not the presequence of the tobacco (Nicotiana tabacum) precursor for the β-subunit of the mitochondria ATP synthase (pF1β; Martin et al., 2006; Lamberti et al., 2011a, 2011b). In addition, a pea (Pisum sativum) 14-3-3 protein has also been reported to bind to the phosphorylated Ser on the transit peptide of pSSU but not to an S→A mutant. The formation of the pSSU/14-3-3/HEAT SHOCK PROTEIN70 (HSP70) complex enhances the kinetics of the import of pSSU into chloroplasts, as free pSSU is imported relatively slowly (May and Soll, 2000). Phosphatase inhibitors, NaF and NaMoO4, inhibit pSSU import into plastids in a reversible manner, suggesting that the dephosphorylation of the phosphorylated transit peptide of pSSU is required for import (Flügge and Hinz, 1986; Waegemann and Soll, 1996). A model for pSSU recognition and TOC translocation has been proposed in which the transit peptide of pSSU is phosphorylated at Ser-34 and the transit peptide binds to Toc33 and Toc159 to form a trimeric complex (Becker et al., 2004; Oreb et al., 2011). Hydrolysis of GTP at Toc33 dissociates it from the complex, and an as yet unknown phosphatase dephosphorylates pSer-34 on pSSU, allowing import to proceed through the combined action of Toc159 and Toc75 (Becker et al., 2004; Oreb et al., 2011). Phosphorylation seems to affect import, as phosphomimicking transit peptides of pSSU and pHCF136 reduced their import rates into chloroplasts (Lamberti et al., 2011a; Nickel et al., 2015).
Unlike chloroplast import, there is no evidence that the phosphorylation and dephosphorylation of plant mitochondrial presequences are required for efficient protein import. In yeast (Saccharomyces cerevisiae), Tom22 functions as a cytosolic facing receptor and transfers precursor proteins to the Tom40 channel. It can be phosphorylated by Casein Kinase2 (CK2) and the mitochondria-bound CK1 to stimulate the activity and assembly of the TOM complex (Harbauer et al., 2014). Protein Kinase A (PKA) can phosphorylate Tom22, impairing its import rate. Thus, PKA, CK1, and CK2 act antagonistically. It has also been demonstrated that the cyclin-dependent kinase CDK1 stimulated assembly of the TOM complex by the phosphorylation of Tom6, an accessory subunit of the TOM complex, enhancing its import into mitochondria (Gerbeth et al., 2013). Thus, in yeast mitochondria, the phosphorylation of the protein import machinery itself appears to play a role in import.
While the overall theme of mitochondrial protein targeting, machinery, and pathways utilized is conserved between different systems, significant variations have been observed in plants. First, the plant outer mitochondrial protein receptor Tom20 is not an ortholog to the yeast or mammalian receptors (Perry et al., 2006). Structural studies on the plant Tom20 import receptor suggest a discontinuous bidentate hydrophobic binding mechanism, somewhat different from that observed in other systems (Rimmer et al., 2011). Furthermore, plant mitochondria are required to distinguish between chloroplast and mitochondrial proteins. Transit peptides and presequences display some similarities in that both are enriched in positively charged residues. However, while mitochondrial presequences are proposed to form α-amphiphilic structures, chloroplast transit peptides do not and, instead, are predicted to form β-sheet secondary structures (Zhang and Glaser, 2002). The identification of a plant-specific outer membrane receptor, Toc64, involved in the import of specific precursor proteins (Chew et al., 2004) highlights the possibility of additional and varied mechanisms between plant mitochondrial protein targeting and other systems.
Previously, we identified an outer mitochondrial membrane protein, called PURPLE ACID PHOSPHATASE2 (AtPAP2), that is also dually targeted to the outer envelope in chloroplasts (Sun et al., 2012a, 2012b). Similar to Toc33/34 and Tom20, AtPAP2 is anchored on the outer membranes by a C-terminal transmembrane motif. Overexpression of AtPAP2 resulted in an altered growth phenotype with elevated ATP levels (Sun et al., 2012b); thus, the function of this protein was investigated. Here, we show that the phosphorylation and dephosphorylation of the presequence of the precursor MULTIPLE ORGANELLAR RNA EDITING FACTOR3 (pMORF3) alter the kinetics of its import in Arabidopsis (Arabidopsis thaliana). Furthermore, it was shown that the phosphorylation of pMORF3, when translated in wheat germ lysate (WGL), resulted in relatively slow import kinetics, and the inhibition of phosphorylation by site-directed mutagenesis resulted in faster import kinetics. Together, these results show that the phosphorylation and dephosphorylation of a mitochondrial precursor protein play a role in its rate of import. The roles of the outer membrane AtPAP2 protein and STY kinases were also investigated, supporting a role for phosphorylation and dephosphorylation in the import of some precursor proteins into plant mitochondria.
RESULTS
AtPAP2 Interacts with the Presequences of MORF Proteins
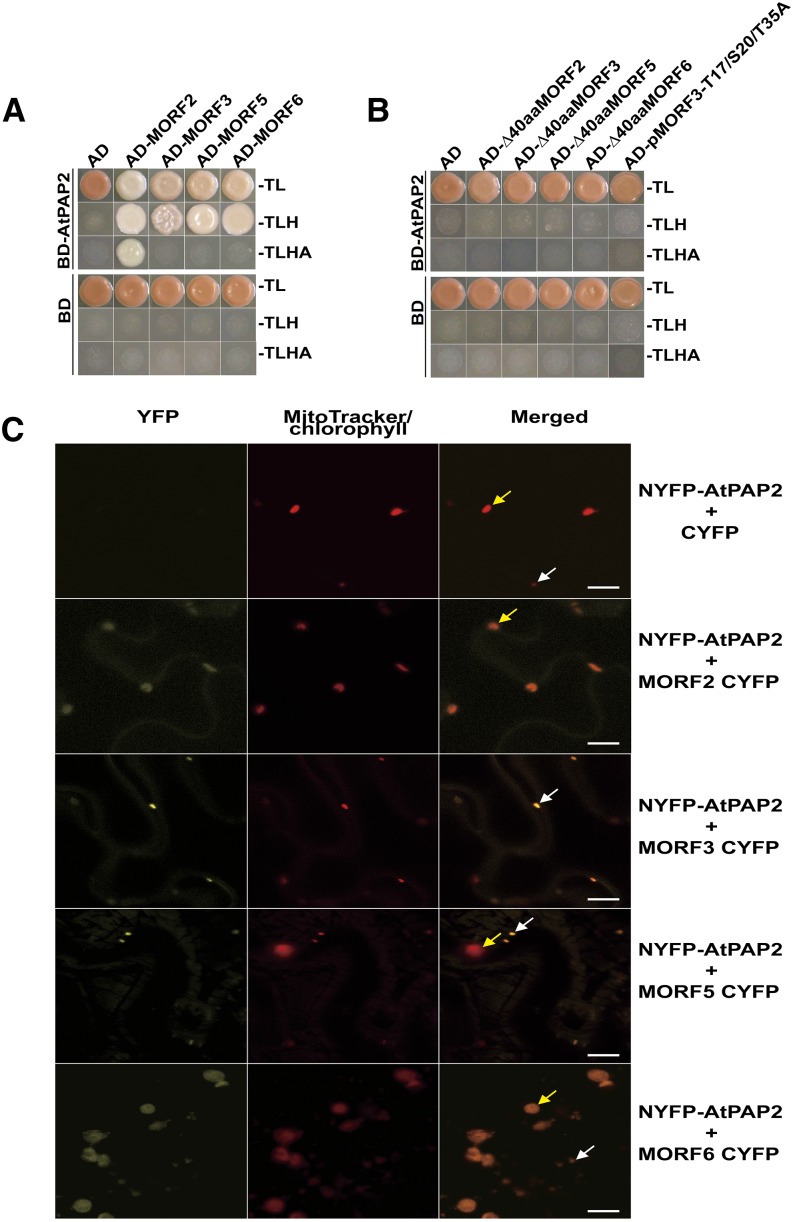
Using AtPAP2 as the bait in a yeast two-hybrid screen, four MORF family proteins, MORF2/3/5/6, were identified (Fig. 1A). When the predicted mitochondrial targeting presequences of MORF2 (∆40aaMORF2) and MORF3/5/6 (∆50aaMORF3/5/6) were removed, their interactions with AtPAP2 were completely abolished (Fig. 1B). Previously, a large-scale protein phosphorylation study in Arabidopsis revealed three experimentally identified phosphorylation sites (Thr-17, Ser-20, and Thr-35) on the presequence of MORF3 (Wang et al., 2013). Mutations were introduced within the predicted phosphorylation sites of the MORF3 presequence, pMORF3-T17/S20/T35A, and it was found that the mutated prey construct did not interact with AtPAP2 (Fig. 1B).
Figure 1.
Interactions of AtPAP2 with MORF proteins as demonstrated by yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays. A, Yeast two-hybrid assay using MORF2/3/5/6 proteins and AtPAP2 in the double dropout medium without Trp and Leu (-TL) and in the triple dropout medium without Trp, Leu, and His (-TLH). MORF2 also shows interactions with quadruple dropout medium without Trp, Leu, His, or adenine (-TLHA). AD, Activation domain; BD, binding domain. B, The interaction is abolished when the presequences were removed (∆40aaMORF2 and ∆50aaMORF3/5/6) or when three Thr residues on the presequence were changed to Ala (AD-pMORF3-T17/S20/T35A mutant). The pGBKT7 and pGADT7 vectors were used as negative controls. C, BiFC visualization of interactions between AtPAP2 and MORFs in Agrobacterium tumefaciens-infiltrated Nicotiana benthamiana leaves. The coexpression of NYFP-AtPAP2 and MORF2-CYFP reconstituted a functional YFP signal in the chloroplasts. The coexpression of NYFP-AtPAP2 and MORF3/5-CYFP reconstituted a functional YFP signal in the mitochondria. The coexpression of NYFP-AtPAP2 and MORF6-CYFP reconstituted functional YFP signals in both mitochondria and chloroplasts. The yellow arrows represent chloroplasts, and the white arrows represent mitochondria. Bars = 10 µm.
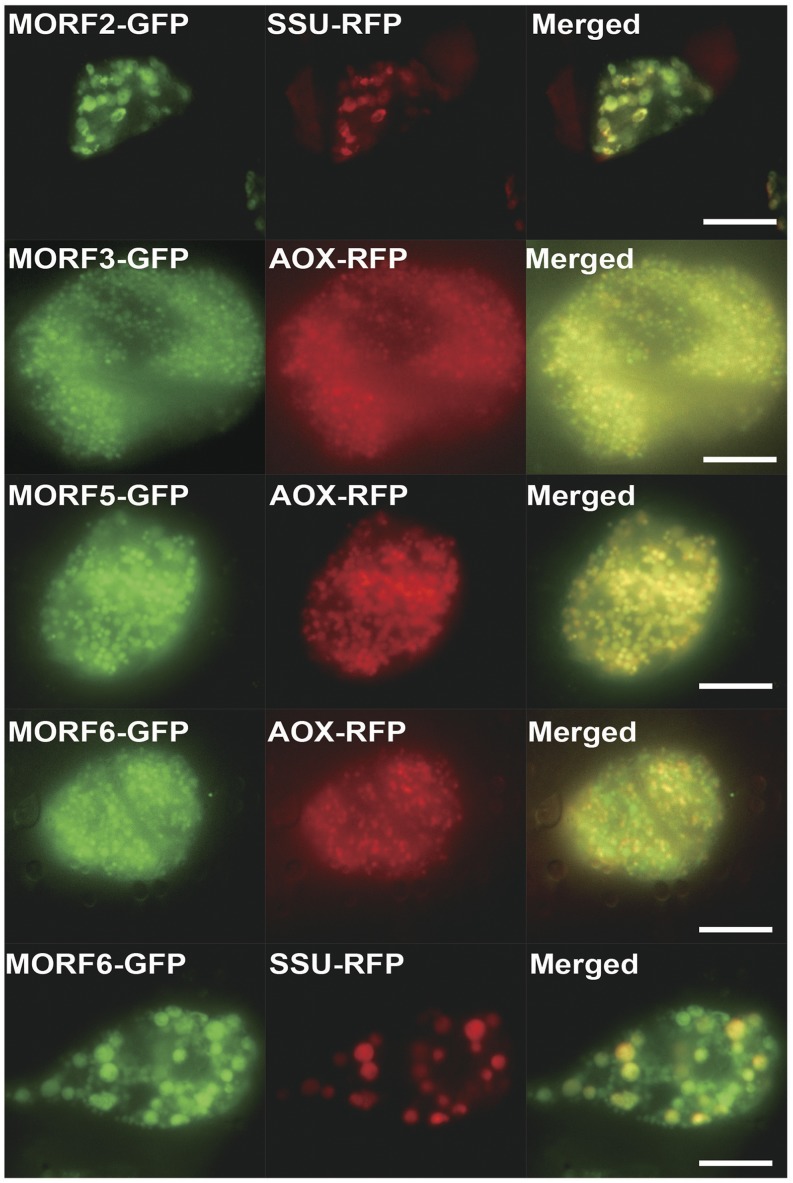
To confirm these interaction assays, the localization of MORF2/3/5/6 was tested by biolistic transformation with GFP fusion constructs in combination with the specific organelle markers alternative oxidase-red fluorescent protein (RFP; mitochondria) and SSU-RFP (chloroplasts). The MORF2-GFP fusion protein was observed to be targeted to plastids, the MORF3-GFP and MORF5-GFP fusion proteins were observed to be targeted to mitochondria, and the MORF6-GFP fusion protein was observed to be targeted to both plastid and mitochondria (Fig. 2; Supplemental Fig. S1). A BiFC assay was used to verify in vivo interactions between AtPAP2 and MORF2/3/5/6. In the BiFC assay, the coexpression of NYFP-AtPAP2 and MORF2-CYFP reconstituted a functional YFP signal in the chloroplasts, the coexpression of NYFP-AtPAP2 and MORF3/5-CYFP reconstituted a functional YFP signal in the mitochondria, and the coexpression of NYFP-AtPAP2 and MORF6-CYFP generated functional YFP signals in both the chloroplasts and mitochondria (Fig. 1C).
Figure 2.
Subcellular localization of MORFs in Arabidopsis cell suspension. GFP was fused to the C termini of Arabidopsis MORF2/3/5/6 (MORF2/3/5/6-GFP), while RFP was fused to the C termini of soybean (Glycine max) mitochondrial alternative oxidase (AOX-RFP; Carrie et al., 2009b) and pea small subunit of Rubisco (SSU-RFP; Carrie et al., 2009b). Bars = 10 µm.
STY8 and STY17 Phosphorylate the Presequences of Nucleus-Encoded Mitochondrial Proteins
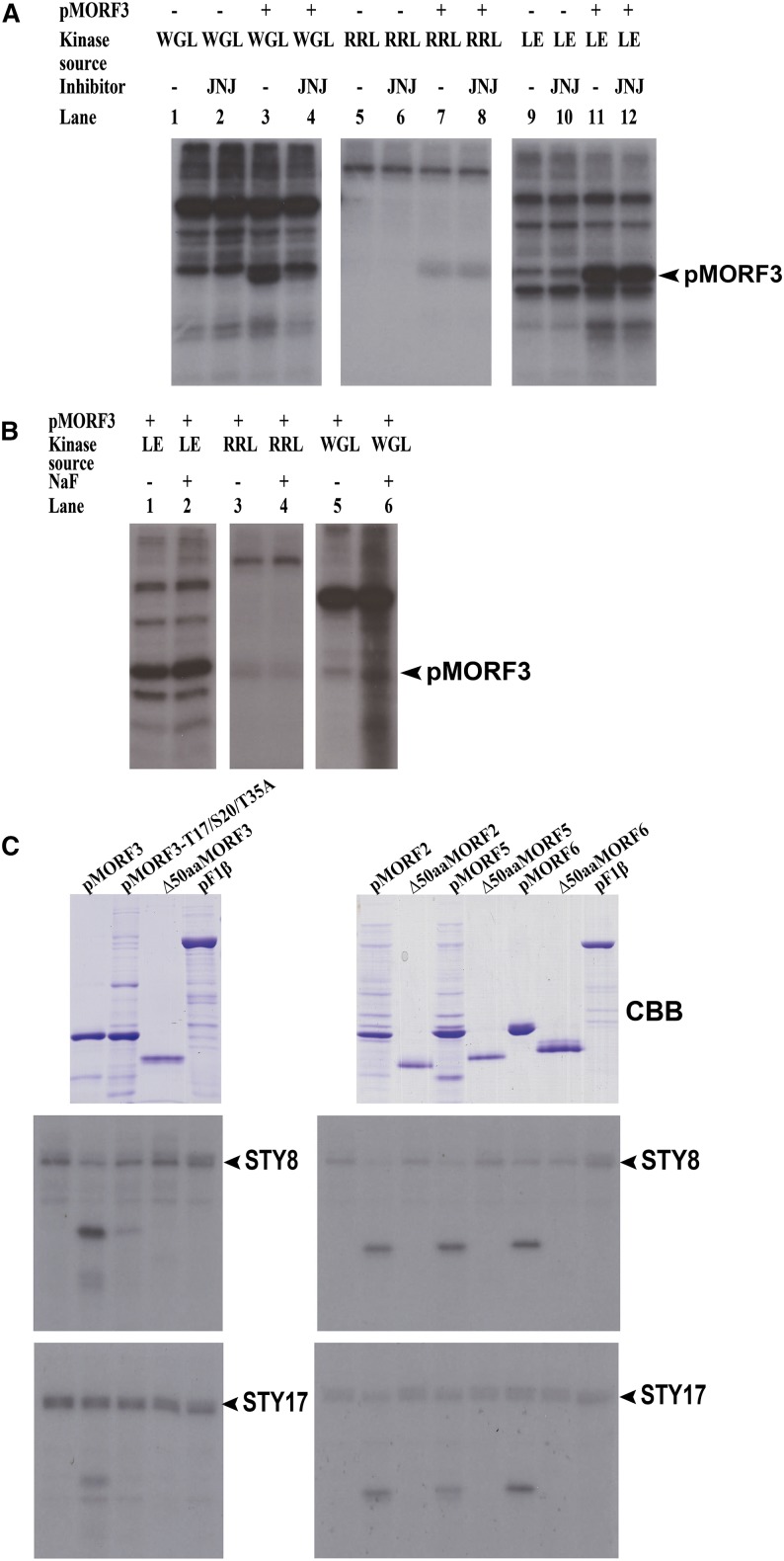
It was found that rabbit reticulocyte lysate (RRL), WGL, and leaf extract (LE) contain kinases that phosphorylate pMORF3, but the pMORF3-phosphorylating kinase activity of RRL is much weaker than that of WGL and LE (Fig. 3A). The specific kinase inhibitor, JNJ-101198409 (JNJ), inhibits STY kinase autophosphorylation and chloroplast substrate phosphorylation, and the nonspecific phosphatase inhibitor, NaF, inhibits chloroplast substrate dephosphorylation (Waegemann and Soll, 1996; Lamberti et al., 2011b). An in vitro phosphorylation assay indicated that JNJ can specifically inhibit the phosphorylation of pMORF3 by an intrinsic kinase in WGL, but the phosphorylation of intrinsic proteins in WGL was not affected (Fig. 3A, lane 3 versus lane 4). However, the phosphorylation of pMORF3 by an intrinsic kinase in LE was not inhibited by JNJ (Fig. 3A, lane 12 versus lane 11), suggesting that the pMORF3-phosphorylating kinases in WGL and LE have different properties. The weak pMORF3-phosphorylating activity in RRL was not affected by JNJ (Fig. 3A, lane 8 versus lane 7). Compared with WGL (Fig. 3A, lane 1) and LE (Fig. 3A, lane 9), RRL had very weak kinase activity, not only to pMORF3 (Fig. 3A, lane 7) but also toward intrinsic proteins (Fig. 3A, lane 5). The results of the in vitro kinase assay reflect a balance of intrinsic kinase and phosphatase activities. Therefore, the activities of phosphatases in WGL, RRL, and LE were determined (Fig. 3B) by including a phosphatase inhibitor (NaF) in the kinase assays. Our results show that WGL not only has rich kinase activities but is also rich in phosphatase activities that could counteract the kinase activities in the WGL (Fig. 3B, lane 6 versus lane 5). LE also contained substantial phosphatase activities (Fig. 3B, lane 2 versus lane 1). For RRL, NaF had little effect on its kinase activities, suggesting that the lack of phosphorylation of pMORF3 is because RRL does not have rich intrinsic kinase activities and is not due to the presence of high intrinsic phosphatase activities.
Figure 3.
Phosphorylation of recombinant pMORF3 protein by various kinase sources and the effects of inhibitors. A, The ability of RRL (200 µg), WGL (35 µg), and LE (10 µg) to phosphorylate recombinant pMORF3 protein in the presence or absence of the kinase inhibitor JNJ was examined. B, The ability of RRL, WGL, and LE to dephosphorylate the phosphorylated recombinant pMORF3 protein was analyzed in the presence or absence of the phosphatase inhibitor NaF. C, Recombinant pMORFs, the corresponding mature proteins (mMORFs), and a substitution mutant of pMORF3 (pMORF3-T17/S20/T35A) were produced in Escherichia coli and purified. The recombinant proteins were phosphorylated by recombinant STY8 and STY17 kinases. pF1β was used as a control. CBB, Coomassie Brilliant Blue.
The STY8 and STY17 kinases phosphorylate transit peptides for some nucleus-encoded chloroplast proteins (Martin et al., 2006; Lamberti et al., 2011b). The precursor protein (pMORF3) but not ∆50aaMORF3 was phosphorylated in the in vitro phosphorylation assay with STY8 or STY17 (Fig. 3C). The phosphorylation-deficient mutant of pMORF3-T17/S20/T35A was not phosphorylated by STY8 or STY17, indicating that one or all of Thr-17, Ser-20, and Thr-35 are the target phosphorylation sites of STY8 and STY17 (Fig. 3C). Additionally, pMORF2/5/6 were all phosphorylated by STY8 and STY17, but upon removal of the presequences (∆40aaMORF2 and ∆50aaMORF3/5/6), no phosphorylation by STY8 or STY17 could be observed (Fig. 3C).
Dephosphorylation of the Presequence of pMORF3 Is Required for Import into Mitochondria
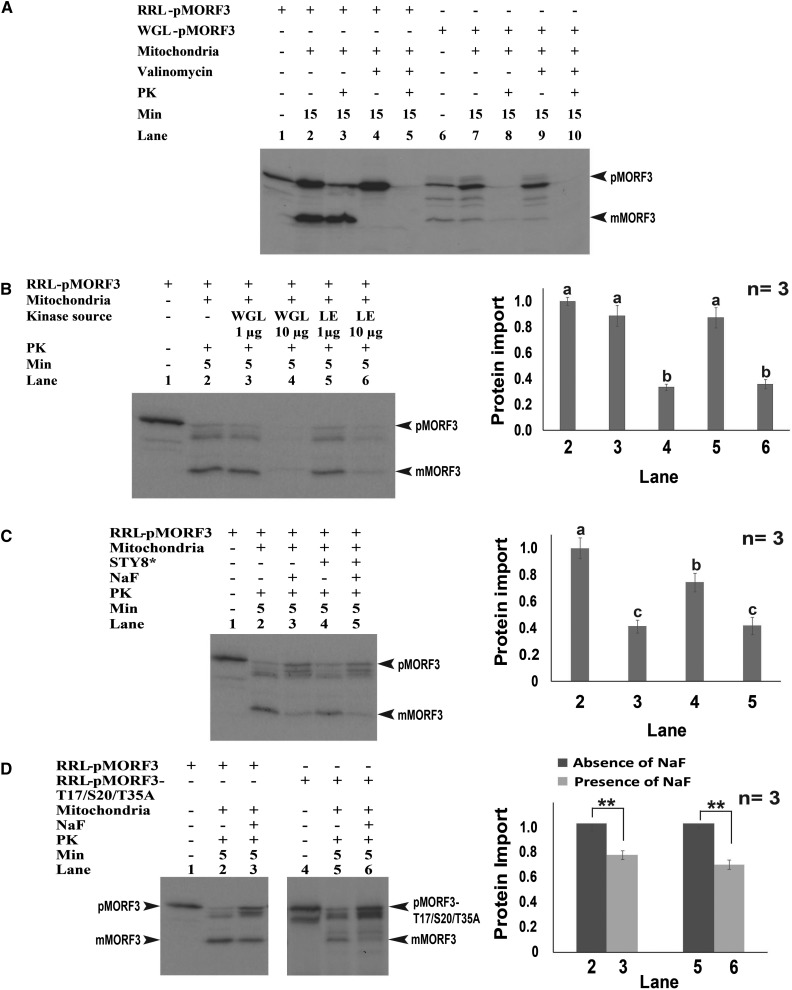
RRL and WGL have both been used to transcribe and translate precursor proteins for in vitro chloroplast and mitochondria protein import studies, although it has been observed that WGL-synthesized precursors are generally import incompetent into mitochondria (Dessi et al., 2003; Biswas and Getz, 2004). Here, we examined whether pMORF3 synthesized in RRL (RRL-pMORF3) and WGL (WGL-pMORF3) are import competent into isolated Arabidopsis mitochondria. In the mitochondrial import assay, RRL-pMORF3 was imported and protease protected when incubated with mitochondria, as evidenced by the generation of the mature form of MORF3 (mMORF3; Fig. 4A, lanes 2 and 3). As the generation of mMORF3 was sensitive to the addition of valinomycin, an ionophore that dissipates the membrane potential (Fig. 4A, lanes 4 and 5), the generation of mMORF3 is thus a direct result of correct import and processing. Interestingly, the transcription and translation of WGL-pMORF3 results in a mature-like product of the same apparent Mr as mMORF3 prior to the addition of mitochondria (Fig. 4A, lane 6). This has been reported previously for other mitochondrial precursor proteins synthesized in WGL and is likely generated by a mitochondrial peptidase/protease within the WGL, as wheat germ is a rich source of mitochondrial proteins (Peiffer et al., 1990). Nevertheless, the import and processing of WGL-pMORF3, as determined by the generation of a protease-protected band (Fig. 4A, lane 8), were very weak compared with RRL-pMORF3 (Fig. 4A, lane 3). Furthermore, the truncated precursor protein of pMORF3 translated in RRL (RRL-∆50aaMORF3) could not be imported into mitochondria, indicating that the presequence of pMORF3 is essential for import (Supplemental Fig. S2).
Figure 4.
Mitochondrial import efficiency of RRL-pMORF3 and WGL-pMORF3 under various conditions. A, RRL-pMORF3 (lanes 2 and 3) and WGL-pMORF3 (lanes 7 and 8) were successfully imported into mitochondria. In the presence of valinomycin, the import of RRL-pMORF3 (lanes 4 and 5) and WGL-pMORF3 (lanes 9 and 10) was inhibited. Proteinase K (PK) was used to digest nonimported pMORF3 (lanes 3, 5, 8, and 10). B, Preincubation of RRL-pMORF3 with WGL (lanes 4 and 5) and LE (lane 7) reduced the mitochondrial import efficiency. Import experiments were repeated three times, and statistical differences were based on one-way ANOVA followed by Tukey’s honestly significant difference test. The amount of imported mMORF3 without the addition of WGL or LE (lane 2) was taken as 1. Values marked with different letters are significantly different (P < 0.01). C, Preincubation of RRL-pMORF3 with NaF (lane 3), STY8 (lane 4), or both (lane 5) reduced the mitochondrial import efficiency. The asterisk represents an additional 10 min of incubation with kinase assay buffer before the in vitro kinase assay. Import experiments were repeated three times, and statistical differences were based on one-way ANOVA followed by Tukey’s honestly significant difference test. The amount of imported mMORF3 without STY8 or NaF treatment (lane 2) was taken as 1. Values marked with different letters are significantly different (P < 0.01). D, RRL-pMORF3-T17/S20/T35A (lanes 4 and 5) was imported normally into mitochondria. The amount of imported mMORF3 without NaF treatment (lanes 2 and 5) was taken as 1. In the presence of NaF, the import rate was reduced. Statistical differences at P < 0.01 (**) were based on Student’s t test.
We have shown that WGL and LE contain kinases that could phosphorylate pMORF3 in the in vitro kinase assay (Fig. 3A), and as it had been shown previously that the addition of WGL to RRL-synthesized precursor proteins reduces the rate of import into mitochondria (Dessi et al., 2003), we further tested if this was also the case for pMORF3. The import rates of both LE-treated and WGL-treated RRL-pMORF3 were reduced when treated prior to import into isolated mitochondria, suggesting that LE and WGL both contain unknown inhibitor(s) that can reduce the import ability of RRL-pMORF3 into mitochondria (Fig. 4B). As pMORF3 can be phosphorylated by intrinsic kinases in WGL and LE, but not RRL (Fig. 3A), it was tested if the phosphorylation of pMORF3 in WGL and LE affected the rate of import uptake into isolated mitochondria. Although RRL contains an unknown inhibitor of the STY-dependent phosphorylation of pMORF3 (Supplemental Fig. S3), the treatment of RRL-pMORF3 by recombinant STY8 could increase the amount of phosphorylated RRL-pMORF3 (Supplemental Fig. S3). RRL-pMORF3 preincubated with STY8 kinase to increase the amount of the phosphorylated pMORF3 prior to mitochondrial import displayed reduced amounts of protein import into mitochondria compared with untreated RRL-pMORF3 (Fig. 4C). In the presence of the phosphatase inhibitor NaF, the import rate of STY8-treated RRL-pMORF3 exhibited additive inhibitory effect (Fig. 4C, lane 5). These results suggest that STY8 phosphorylation of the presequence of pMORF3 impedes its import into mitochondria and that its dephosphorylation is required prior to translocation.
Next, we examined whether the phosphorylation of the presequence is necessary for the import of pMORF3 into mitochondria. As shown in Figure 4D, the RRL-synthesized phosphorylation-deficient mutant pMORF3-T17/S20/T35A was imported normally, similar to wild-type RRL-pMORF3, indicating that the phosphorylation of the presequence of pMORF3 is not required for import. Taken together, unlike in the import of pSSU into chloroplasts, the phosphorylation of pMORF3 by STY kinases does not enhance, but rather impedes, import into mitochondria. Furthermore, the addition of NaF reduced the import rate of RRL-pMORF3-T17/S20/T35A. Therefore, the slower import rates of pMORF3 (Fig. 4D, lane 3) and pMORF3-T17/S20/T35A (Fig. 4D, lane 6) in the presence of the phosphatase inhibitor NaF may be due to its effects on the phosphorylation status of the TOM and TIM complexes, in addition to the phosphorylation status of the presequence of pMORF3 (Sugiyama et al., 2008; Nakagami et al., 2010; Schmidt et al., 2011; Havelund et al., 2013).
HSP70 and 14-3-3 Interact with pMORF3 But Not with pMORF3-T17/S20/T35A
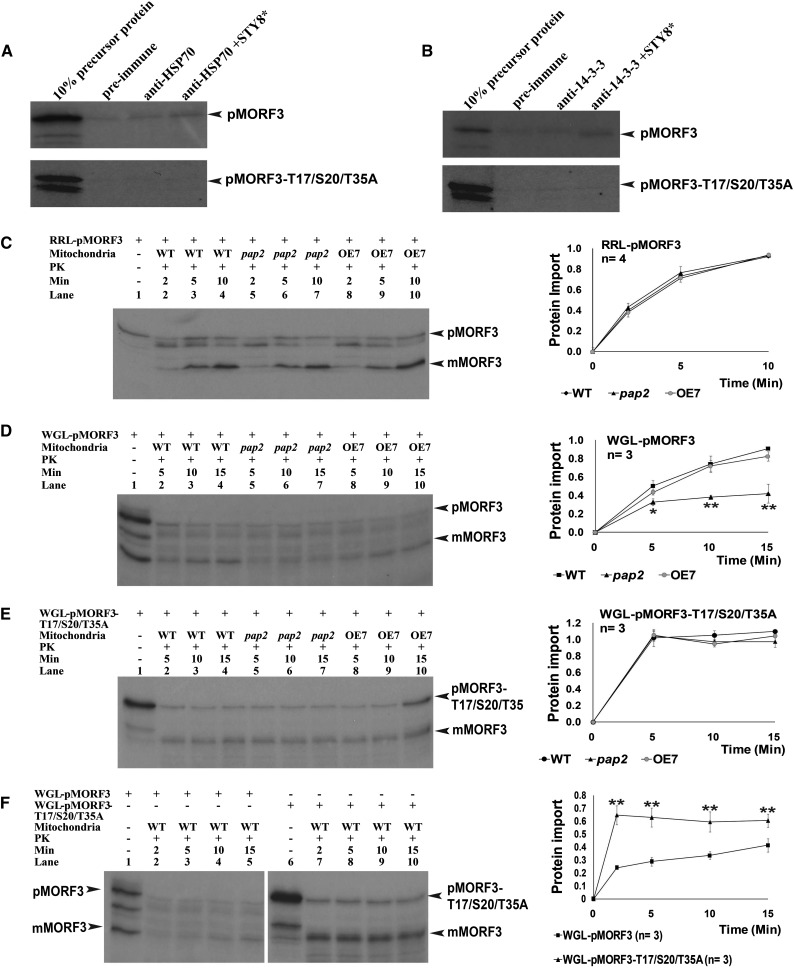
HSP70 interacts with precursor proteins during translocation into mitochondria, chloroplasts, and endoplasmic reticulum (Deshaies et al., 1988; Zimmermann et al., 1988; Beckmann et al., 1990), whereas 14-3-3 proteins recognize transit peptides of chloroplast precursors containing a phosphopeptide-binding motif and then form a guidance complex together with HSP70 (Muslin et al., 1996). It was investigated if pMORF3 also has the ability to interact with 14-3-3 and HSP70 using a coimmunoprecipitation assay. STY8-treated RRL-pMORF3 interacted with HSP70 and 14-3-3, whereas the STY8-treated RRL-pMORF3-T17/S20/T35A mutant could not interact with HSP70 or 14-3-3 (Fig. 5, A and B). This result suggests that the phosphorylation of the presequence of pMORF3 by STY8 kinase enables it to bind to both HSP70 and 14-3-3 proteins in vitro.
Figure 5.
Comparative coimmunoprecipitation and mitochondrial import assays between pMORF3 and pMORF3-T17/S20/T35A. A and B, RRL-pMORF3 and pMORF3-T17/S20/T35A, with or without 15 min of treatment with STY8 kinase, were immunoprecipitated with anti-HSP70 (A) and anti-14-3-3 (B) antibodies. Without STY8 treatment, pMORF3 showed a weak interaction with HSP70. In contrast, STY8 treatment of RRL-pMORF3 enhanced pMORF3’s interaction with 14-3-3 and HSP70 but had no effect on pMORF3-T17/S20/T35A. The asterisk represents an additional 10 min of incubation in kinase assay buffer before the in vitro kinase assay. C, Import efficiencies of RRL-pMORF3 into wild-type (WT; lanes 2–4), OE7 (lanes 5–7), and pap2 (lanes 8–10) mitochondria. Import experiments were repeated three times, and the import rates were not significantly different between mitochondria. D and E, Import efficiencies of WGL-pMORF3 (D) and WGL-pMORF3-T17/S20/T35A (E) into wild-type, pap2, and OE7 mitochondria. Import experiments were repeated three times. The import rate of WGL-pMORF3 into pap2 (lanes 5–7) mitochondria was significantly reduced, with P < 0.05 (^) and P < 0.01 (**) using Student’s t test, but the import rates of WGL-pMORF3-T17/S20/T35A into different mitochondria were not significantly different. F, The import rate of WGL-pMORF3-T17/S20/T35A into wild-type mitochondria was significantly faster than that of WGL-pMORF3, with P < 0.01 (**) using Student’s t test. Import experiments were repeated three times.
The Import Rate of WGL-pMORF3 into pap2 Transfer DNA Mitochondria Is Slower
The C-terminal transmembrane domain of PAP2 anchors AtPAP2 to the outer membrane of chloroplasts and mitochondria (Sun et al., 2012a). AtPAP2 was highly expressed in mitochondria of an Arabidopsis overexpressing AtPAP2 line (OE7) but was not expressed in the pap2 transfer DNA (T-DNA) line (Supplemental Fig. S4). We compared the import rates of RRL-pMORF3 and WGL-pMORF3 into wild-type (Columbia-0 [Col-0]), OE7, and pap2 mitochondria. The import rates of largely nonphosphorylated RRL-pMORF3 were significantly faster than the import rates of more extensively phosphorylated WGL-pMORF3 into the mitochondria of all three lines (Fig. 5, C and D). As the presequence of pMORF3 translated in WGL is phosphorylated (Fig. 3A), the presequence must be dephosphorylated before import. By contrast, the import rate of WGL-pMORF3 into mitochondria isolated from pap2 was lower when compared with the wild type (Col-0; Fig. 5D, lanes 5–7), and this decrease in import was not observed for mitochondria isolated from OE7. The import rate of WGL-pMORF3-T17/S20/T35A was also tested into mitochondria isolated from the wild type (Col-0), OE7, and pap2, and no disparity in import ability was observed (Fig. 5E). In addition, the import rate of WGL-pMORF3 and WGL-pMORF3-T17/S20/T35A was tested into mitochondria isolated from the wild type (Col-0), and it was determined that the import of WGL-pMORF3-T17/S20/T35A reached maximum ability within the first 2 min of the import assay, unlike WGL-pMORF3, which reached maximum import uptake at 15 min, suggesting that the import kinetics between the two precursors are altered (Fig. 5F).
DISCUSSION
The outer mitochondrial membrane preprotein receptors of plants are not orthologous to those in fungal or mammalian systems (Chew et al., 2004; Perry et al., 2006). This is proposed to be due to the unique cellular environments of plant cells, which contain plastids, and thus the requirement to sort proteins between these organelles, a situation that does not exist in animals or fungi. The in vivo and in vitro import of precursor proteins into mitochondria from plants (Boutry et al., 1987; Whelan et al., 1988) has been known for some time. While in vivo methods are generally considered to reflect the in planta situation with respect to targeting specificity, in vitro approaches allow the dissection of the molecular mechanisms of protein import, from synthesis of the precursor protein in the cytosol to assembly into a functional unit in mitochondria (Murcha et al., 2014). For in vitro studies, precursor proteins are synthesized in cell-free translation lysates, most typically the RRL. These approaches have been very successful in elucidating the molecular mechanisms required to achieve mitochondrial protein import.
A noticeable and puzzling feature of in vitro protein import into plant mitochondria is that while it is readily supported by import from RRLs, wheat germ translation lysates do not generally support mitochondrial protein import (Dessi et al., 2003). This is puzzling given the efficient import of chloroplast precursor protein from wheat germ translation lysates (Waegemann and Soll, 1996; May and Soll, 2000), although some inhibitory effects on protein import into chloroplasts can also be attributed to proteinaceous factors in wheat germ translation lysates (Schleiff et al., 2002). Detailed studies with wheat germ translation lysates show that, not only can mitochondrial import not be supported, but, in fact, the addition of WGL to otherwise mitochondrial import-competent precursor protein inhibits import (Dessi et al., 2003). Thus, it has been proposed that there are inhibitory factors present in the WGL, although the identity of these factors remains unidentified.
Here, it is presented that the phosphorylation of at least one mitochondrial precursor protein, pMORF3, results in a large decrease in the amount of import into mitochondria. Several lines of evidence show this decrease associated with phosphorylation. First, mutation of Thr as well as Ser residues to Ala, which cannot be phosphorylated, results in much greater import from a wheat germ translation lysate. Second, the addition of WGL or LE, shown to phosphorylate pMORF3, reduces the amount of import into mitochondria, and the addition of NaF, a general inhibitor of phosphatases, results in a reduction in the amount of protein import into mitochondria. Third, the Arabidopsis line pap2, which has the outer membrane phosphatase inactivated, imports WGL-pMORF3 slower than the wild type or lines that have it overexpressed. Finally, it can be shown that the presequence was the site of action, as it was required to interact with AtPAP2, and that purified STY7 and STY8 can phosphorylate the presequence of pMORF3. Combined, these findings show a role for phosphorylation and dephosphorylation in the import of pMORF3 into mitochondria on the presequence of pMORF3.
STY kinases are present in the cytosol, and AtPAP2 is present on the outer membranes of both organelles (Martin et al., 2006; Sun et al., 2012a). In vivo BiFC assays showed that pMORF3/5 interacted with AtPAP2 on mitochondria but not with AtPAP2 on chloroplasts (Fig. 1C). In contrast, in vivo BiFC assays showed that the chloroplast-targeted precursor protein pMORF2 interacted with AtPAP2 on chloroplasts but not with AtPAP2 on the mitochondria (Fig. 1C). These data indicate that the interactions of these nucleus-encoded proteins with AtPAP2 are not the sole determining factors of their targeting to these two organelles. Instead, other receptors on TOC (Toc34, Toc64, and Toc159) and TOM (Tom20 and OM64) complexes are responsible for cargo specificity (Ye et al., 2012, 2015).
What is the purpose of the phosphorylation and dephosphorylation of transit peptides and presequences of precursor proteins when both processes are energy consuming? The localizations of at least 751 mitochondrial proteins (Duncan et al., 2011) and 1,323 chloroplastic proteins (Smith et al., 2004) have been confirmed. Both mitochondria- and chloroplast-targeting signals are located at their N termini. Although these sequences are enriched in basic and hydroxylated amino acids and are deficient in acidic residues (Perry et al., 2008), their sequences are versatile. The process of STY kinase phosphorylation, 14-3-3 binding, and interaction with TOC/TOM can greatly reduce the variety of sequences of transit peptides. Complex formation also enhances the import efficiency of the chloroplast precursor proteins into chloroplasts and impedes the import efficiency of mitochondrial precursor proteins into mitochondria.
This design may serve to coordinate protein import into chloroplasts and mitochondria and may have evolved early in the green lineage. A single STY-like gene and a single AtPAP2-like gene are present in the genome of Ostreococcus tauri (Supplemental Fig. S5; Supplemental Table S1), a unicellular photosynthetic alga that contains a single chloroplast and a single mitochondrion per cell. Both STY-like kinases and AtPAP2-like phosphatases can be found in green plants (Supplemental Fig. S5; Supplemental Table S1), suggesting that the genes may have coevolved in the plant kingdom (Martin et al., 2006; Lamberti et al., 2011b; Sun et al., 2012b). Conversely, MORF family members only evolved in flowering plants (Supplemental Fig. S5) and work together with RNA sequence-specific pentatricopeptide repeat family members in RNA editing in mitochondria and chloroplasts (Takenaka et al., 2012). Bioinformatics analysis of the first 66 amino acids of 334 MORFs from flowering plants showed that Ser is the dominant residue at positions 17 to 29 (Supplemental Fig. S6), implying that phosphorylation at the Ser residues at the presequences of MORFs may be of biological significance.
Modulation of the mitochondrial import process or import apparatus was shown to have various effects on the physiology of plants. For example, the limiting step for the import of precursor protein appears to be the amount of the inner membrane translocase TIM17:23 (Wang et al., 2012). Increasing the amount of AtTim23 resulted in increased protein import but significantly reduced growth, while significantly reducing the rate of protein import into mitochondria had no detectable effect on the growth rate in Arabidopsis (Wang et al., 2012). By contrast, Arabidopsis plants with all three Tom20 receptor isoforms inactivated show only some reduced growth rate (Lister et al., 2007). In the case of AtPAP2, while overexpression of AtPAP2 results in enhanced growth and higher seed yield (Sun et al., 2012a, 2012b), sole targeting of AtPAP2 to mitochondria results in early senescence and lower seed yield (Supplemental Fig. S7). This shows the in vivo biological effects of the overexpression of AtPAP2 in mitochondria on the physiology of plants.
In conclusion the observation that at least some mitochondrial precursor protein can be phosphorylated, and that this impedes import into mitochondria, uncovers a novel step in the import of precursor protein into mitochondria and may have implications for protein sorting and the specificity of import.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) ecotype Col-0, Arabidopsis overexpressing AtPAP2 (OE7), and a T-DNA insertion mutant (pap2 [Salk_013567]; Sun et al., 2012b) were used in this study. All of the plants were grown under long-day conditions (16 h of light/8 h of dark) in a controlled-environment chamber. Arabidopsis seeds were sterilized with 20% (v/v) bleach and grown for 14 d on Murashige and Skoog (MS) medium that was supplemented with 3% (w/v) Suc.
Plasmid Construction
First-strand complementary DNA (cDNA) was reverse transcribed by the Reverse Transcription System (Promega) according to the manufacturer’s instructions. Full-length coding sequences of MORF2/3/5/6, AtPAP2, pF1β, STY8, and STY17, coding sequence of MORF2 with deletion of the first 120 nucleotides following the start codon (∆40aaMORF2), and coding sequences of MORF3/5/6 with deletion of the first 150 nucleotides following the start codon (∆50aaMORF3/5/6) were amplified by Platinum Pfx DNA Polymerase (Life Technologies) using the primers listed in Supplemental Table S2. For site-directed mutagenesis, the phosphorylation-deficient mutant MORF3-T17/S20/T35A was amplified by primers containing mutated bases as described by Heckman and Pease (2007).
To produce transgenic lines that only overexpressed AtPAP2 on mitochondria, cDNA encoding amino acids 1 to 588 of AtPAP2 and amino acids 163 to 202 of Tom20-3 was amplified separately, and then both purified PCR products were combined and a chimeric PCR product was amplified by a forward primer of AtPAP2 and a reverse primer of Tom20-3. The PCR product was cloned into the pCXSN vector (Chen et al., 2009) to generate pCXSN-P2Tom.
For the yeast two-hybrid study, the PCR product of AtPAP2 was digested with NdeI and SalI and cloned into pGBKT7 vector (Clontech). The PCR products of MORF2/3/5/6, ∆40aaMORF2, ∆50aaMORF3/5/6, and MORF3-T17/S20/T35A were digested with NdeI and XhoI, and the PCR product of STY8 was digested with NdeI and SacI and ligated into the pGADT7 vector (Clontech). For the BiFC assay, the cDNA of AtPAP2 was amplified and digested with SpeI and cloned into the pSPYNE vector, while the cDNAs of MORF2/3/5/6 were amplified and cut by XbaI and SalI and cloned into the pSPYCE vector (Walter et al., 2004). For the overproduction of recombinant proteins, the PCR products of MORF2/3/5/6, ∆40aaMORF2, ∆50aaMORF3/5/6, MORF3-T17/S20/T35A, and pF1β were digested with NdeI and SalI and cloned into the pET21a vector (Novagen), and the PCR products of STY8 and STY17 were digested with NcoI and SacI and cloned into pET28a vector (Novagen). For GFP analysis, the PCR products of MORF2/3/5/6 containing attB sites were transferred into the pDONR201 vector (Life Technologies) by Gateway BP reaction (Life Technologies) according to the manufacturer’s instructions. The pDONR201 vector containing the fragment of interest was transferred into the C-terminal GFP vector (Life Technologies) by Gateway LR reaction (Life Technologies) according to the manufacturer’s instructions.
Expression of Recombinant Proteins
The coding sequences of STY8 and STY17 were subcloned into the pET28a vector (Novagen), and the coding sequences of pMORF2/3/5/6, mMORF2/3/5/6, and pMORF3-T17/S20/T35A were subcloned into the pET21a vector (Novagen). The overexpression and purification of STY8 and STY17 were performed as described by Lamberti et al. (2011b). The other recombinant proteins were purified as described by Inoue and Akita (2008).
Mitochondria Isolation
Fourteen-day-old seedlings that were grown on MS agar were used for mitochondria isolation as described by Day et al. (1985) and Lister et al. (2007). For immunoblot analysis, wash buffer (300 mm Suc and 10 mm TES) without bovine serum albumin was used for the final mitochondria wash.
In Vitro Mitochondrial Protein Import
Rabbit reticulocyte TNT in vitro transcription/translation lysate (RRL; Promega) and wheat germ TNT in vitro transcription/translation lysate (WGL; Promega) were used to synthesize [35S]Met-labeled precursor proteins as described previously (Dessi et al., 2003). The in vitro mitochondrial protein import assay was performed as described by Lister et al. (2007).
In Vitro Phosphorylation Assay
This assay was performed as described (Martin et al., 2006). The recombinant substrate proteins and recombinant STY8 and STY17 kinases were expressed in the Escherichia coli BL21 (PLysS) strain and purified. Two micrograms of recombinant substrate proteins was incubated with 2 µg of recombinant STY8/17, 35 µg of WGL, 200 µg of RRL, and 10 µg of LE in the reaction buffer (20 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 0.5 mm MnCl2, and 2.5 µm ATP) containing 5 µCi of [γ-32P]ATP. The reaction was incubated at 30°C for 15 min and terminated by adding 20 µL of 5× SDS loading buffer. The reaction products were analyzed on a 15% (v/v) SDS-PAGE gel and exposed to Hyperfilm MP (GE Healthcare).
Yeast Two-Hybrid Interaction and BiFC Assays
The Clontech Matchmaker two-hybrid system was used according to the manufacturer’s instructions. Positive interactions were verified on triple dropout medium without Trp, Leu, and His. BiFC assays were performed as described (Walter et al., 2004). The pSPYNE and pSPYCE vectors containing genes of interest were cotransformed into the leaf cells of Nicotiana benthamiana by Agrobacterium tumefaciens infiltration. Biofluorescence was detected with an LSM710 confocal laser scanning microscope (Zeiss).
Immunoblot Assay
Fourteen-day-old wild-type Arabidopsis Col-0, Arabidopsis overexpressing AtPAP2 (OE7), and the pap2 T-DNA insertion mutant (Salk_013567) were used for mitochondrial isolation. Mitochondrial proteins were separated on a 10% (w/v) SDS-PAGE gel and transferred to Hybond-C nitrocellulose membranes (GE Healthcare). The membranes containing mitochondrial proteins were immunodetected by anti-AtPAP2 antibodies as described by Sun et al. (2012b).
Coimmunoprecipitation Assay
[35S]Met-labeled pMORF3 and [35S]Met-labeled pMORF3-T17/S20/T35A synthesized by RRL (Promega), with or without a 15-min treatment of STY8, were incubated with 2 μm JNJ and 10 µg of WGL (Promega), which was used as a source of 14-3-3 and HSP70 protein. The mixture were then mixed with anti-14-3-3 (a gift from Carol MacKintosh) and anti-HSP70 (Agrisera) antibodies for 1 h, and the bound proteins were pulled down using Sepharose-Protein A (Amersham) and detected by autoradiography (Dessi et al., 2003).
GFP Subcellular Localization Studies
Full-length MORF2/3/5/6 was cloned into the Gateway vector pDONR201 and then transferred into a vector containing GFP according to the manufacturer’s instructions (Invitrogen). MORF2/3/5/6 fused to GFP was cotransformed with a positive control containing RFP into Arabidopsis cell suspensions by the PDS-1000/He biolistic transformation system (Bio-Rad) as described by Xu et al. (2013).
Plant Transformation
The pCXSN-P2Tom vector was transformed into A. tumefaciens strain GV3101 by the freeze/thaw method as described by Weigel and Glazebrook (2006). After that, A. tumefaciens containing pCXSN-P2Tom was introduced into the wild type by floral dip as described previously (Clough and Bent, 1998). Homozygous CaMV35S:P2-Tom overexpression lines were selected on MS plates containing antibiotics as described previously by Chen et al. (2009). Multiple independent expression lines were verified by immunoblot assay using anti-AtPAP2 antibody.
Phylogenetic Analysis
For phylogenetic analysis of PAP2, two steps were conducted to identify PAPs, including AtPAP2/9-like proteins across 64 species ranging from algae to higher plants to human, with genome sequences available. First, BLASTP (e ≤ 1e-5) was used to search homologous protein using 42 seed PAP proteins, including PAPs with a unique C-terminal hydrophobic motif, against nonredundant protein sequences of plants (taxid: 3193) in which all PAP candidate sequences with signals of the five conserved motifs (DxG, GDISY, GNHE, QGHR, and GHVH) within the proteins were identified. Second, an HMM matrix was built for the C-terminal hydrophobic motif of PAP proteins using HMM build, and the candidate PAP sequences in step 1 were used to identify PAP proteins with a C-terminal hydrophobic motif using HMM search, in which the AtPAP2/9-like proteins were identified. For phylogenetic analysis of STY proteins, a similar strategy was implemented. First, BLASTP (e ≤ 1e-5) was used to search homologous proteins using 20 seed STYs (each with an ACT motif and a kinase activate segment motif) against the 64 species protein database, in which STY candidate sequences were identified. Second, to make sure that each candidate STY contains the ACT and activate segment motifs, an HMM matrix was built for the ACT and kinase activate segment motifs, respectively, and the candidate STYs identified in step 1 were searched using HMM search, in which the STY protein set for each species was determined. For phylogenetic analysis of MORF, a similar strategy to that used for PAP and STY was implemented. First, BLASTP (e ≤ 1e-5) was used to search homologous proteins using nine seed MORF proteins from Arabidopsis against the 64 species protein database in which the MORF candidate sequences were identified. Second, an HMM matrix was used to screen the identified orthologs for a conserved GCT motif.
Sequence data from this work can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT1G13900 (AtPAP2), AT2G33430 (MORF2), AT3G06790 (MORF3), AT1G32580 (MORF5), AT2G35240 (MORF6), AT2G17700 (STY8), and AT4G35780 (STY17).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Negative controls of subcellular localizations of MORFs in Arabidopsis cell suspension.
Supplemental Figure S2. Mitochondrial import efficiency of RRL-pMORF3 and RRL-∆50aaMORF3.
Supplemental Figure S3. The effects of RRL and WGL on the activities of STY8 kinase.
Supplemental Figure S4. The expression level of AtPAP2 in mitochondrial protein of wild-type, OE7, and pap2 T-DNA line.
Supplemental Figure S5. Distribution of STY, PAP, and MORF genes.
Supplemental Figure S6. Ser is the dominant amino acid at the presequences of MORF family proteins.
Supplemental Figure S7. Growth phenotypes of P2Tom OE lines.
Supplemental Table S1. Summary of STYs, MORFs, and AtPAP2 in different plant sources.
Supplemental Table S2. List of primer sequences used for overproduction of recombinant proteins, site-directed mutagenesis, yeast-two hybrid, GFP, and BiFC studies.
Supplementary Material
Acknowledgments
We thank Dr. Clive Lo (University of Hong Kong) for generous gifts of BiFC vectors.
Glossary
- BiFC
bimolecular fluorescence complementation
- RRL
rabbit reticulocyte lysate
- WGL
wheat germ lysate
- LE
leaf extract
- T-DNA
transfer DNA
- Col-0
Columbia-0
- MS
Murashige and Skoog
- cDNA
complementary DNA
- JNJ
JNJ-101198409
Footnotes
This work was supported by the Seed Funding Program for Basic Research (grant no. 201311159043) and the Strategic Research Theme on Clean Energy of the University of Hong Kong, by the General Research Fund (grant nos. HKU772710M and HKU772012M), by the Innovation and Technology Fund of the Hong Kong Special Administrative Region, China, and by the University of Hong Kong (PhD scholarships to Y.-S.L., R.Z., S.C., and X.G.).
Articles can be viewed without a subscription.
References
- Araújo WL, Nunes-Nesi A, Fernie AR (2014) On the role of plant mitochondrial metabolism and its impact on photosynthesis in both optimal and sub-optimal growth conditions. Photosynth Res 119: 141–156 [DOI] [PubMed] [Google Scholar]
- Becker T, Jelic M, Vojta A, Radunz A, Soll J, Schleiff E (2004) Preprotein recognition by the Toc complex. EMBO J 23: 520–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann RP, Mizzen LE, Welch WJ (1990) Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science 248: 850–854 [DOI] [PubMed] [Google Scholar]
- Biswas TK, Getz GS (2004) Requirement of different mitochondrial targeting sequences of the yeast mitochondrial transcription factor Mtf1p when synthesized in alternative translation systems. Biochem J 383: 383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutry M, Nagy F, Poulsen C, Aoyagi K, Chua NH (1987) Targeting of bacterial chloramphenicol acetyltransferase to mitochondria in transgenic plants. Nature 328: 340–342 [DOI] [PubMed] [Google Scholar]
- Carrie C, Giraud E, Whelan J (2009a) Protein transport in organelles: dual targeting of proteins to mitochondria and chloroplasts. FEBS J 276: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Carrie C, Kühn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J (2009b) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew O, Lister R, Qbadou S, Heazlewood JL, Soll J, Schleiff E, Millar AH, Whelan J (2004) A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett 557: 109–114 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Day D, Neuburger M, Douce R (1985) Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Funct Plant Biol 12: 219–228 [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R (1988) A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332: 800–805 [DOI] [PubMed] [Google Scholar]
- Dessi P, Pavlov PF, Wållberg F, Rudhe C, Brack S, Whelan J, Glaser E (2003) Investigations on the in vitro import ability of mitochondrial precursor proteins synthesized in wheat germ transcription-translation extract. Plant Mol Biol 52: 259–271 [DOI] [PubMed] [Google Scholar]
- Duncan O, Taylor NL, Carrie C, Eubel H, Kubiszewski-Jakubiak S, Zhang B, Narsai R, Millar AH, Whelan J (2011) Multiple lines of evidence localize signaling, morphology, and lipid biosynthesis machinery to the mitochondrial outer membrane of Arabidopsis. Plant Physiol 157: 1093–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellerer C, Schweiger R, Schöngruber K, Soll J, Schwenkert S (2011) Cytosolic HSP90 cochaperones HOP and FKBP interact with freshly synthesized chloroplast preproteins of Arabidopsis. Mol Plant 4: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú, Jarvis P (2013) Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta 1833: 332–340 [DOI] [PubMed] [Google Scholar]
- Flügge UI, Hinz G (1986) Energy dependence of protein translocation into chloroplasts. Eur J Biochem 160: 563–570 [DOI] [PubMed] [Google Scholar]
- Gerbeth C, Schmidt O, Rao S, Harbauer AB, Mikropoulou D, Opalińska M, Guiard B, Pfanner N, Meisinger C (2013) Glucose-induced regulation of protein import receptor Tom22 by cytosolic and mitochondria-bound kinases. Cell Metab 18: 578–587 [DOI] [PubMed] [Google Scholar]
- Harbauer AB, Opalińska M, Gerbeth C, Herman JS, Rao S, Schönfisch B, Guiard B, Schmidt O, Pfanner N, Meisinger C (2014) Mitochondria: cell cycle-dependent regulation of mitochondrial preprotein translocase. Science 346: 1109–1113 [DOI] [PubMed] [Google Scholar]
- Havelund JF, Thelen JJ, Møller IM (2013) Biochemistry, proteomics, and phosphoproteomics of plant mitochondria from non-photosynthetic cells. Front Plant Sci 4: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2: 924–932 [DOI] [PubMed] [Google Scholar]
- Inoue H, Akita M (2008) The transition of early translocation intermediates in chloroplasts is accompanied by the movement of the targeting signal on the precursor protein. Arch Biochem Biophys 477: 232–238 [DOI] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Lamberti G, Drurey C, Soll J, Schwenkert S (2011a) The phosphorylation state of chloroplast transit peptides regulates preprotein import. Plant Signal Behav 6: 1918–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti G, Gügel IL, Meurer J, Soll J, Schwenkert S (2011b) The cytosolic kinases STY8, STY17, and STY46 are involved in chloroplast differentiation in Arabidopsis. Plant Physiol 157: 70–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Jung C, Hwang I (2013) Cytosolic events involved in chloroplast protein targeting. Biochim Biophys Acta 1833: 245–252 [DOI] [PubMed] [Google Scholar]
- Lister R, Carrie C, Duncan O, Ho LHM, Howell KA, Murcha MW, Whelan J (2007) Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19: 3739–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Sharma R, Sippel C, Waegemann K, Soll J, Vothknecht UC (2006) A protein kinase family in Arabidopsis phosphorylates chloroplast precursor proteins. J Biol Chem 281: 40216–40223 [DOI] [PubMed] [Google Scholar]
- May T, Soll J (2000) 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Kmiec B, Kubiszewski-Jakubiak S, Teixeira PF, Glaser E, Whelan J (2014) Protein import into plant mitochondria: signals, machinery, processing, and regulation. J Exp Bot 65: 6301–6335 [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84: 889–897 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, Shirasu K (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol 153: 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel C, Soll J, Schwenkert S (2015) Phosphomimicking within the transit peptide of pHCF136 leads to reduced photosystem II accumulation in vivo. FEBS Lett 589: 1301–1307 [DOI] [PubMed] [Google Scholar]
- Oreb M, Höfle A, Koenig P, Sommer MS, Sinning I, Wang F, Tews I, Schnell DJ, Schleiff E (2011) Substrate binding disrupts dimerization and induces nucleotide exchange of the chloroplast GTPase Toc33. Biochem J 436: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer WE, Ingle RT, Ferguson-Miller S (1990) Structurally unique plant cytochrome c oxidase isolated from wheat germ, a rich source of plant mitochondrial enzymes. Biochemistry 29: 8696–8701 [DOI] [PubMed] [Google Scholar]
- Perry AJ, Hulett JM, Likić VA, Lithgow T, Gooley PR (2006) Convergent evolution of receptors for protein import into mitochondria. Curr Biol 16: 221–229 [DOI] [PubMed] [Google Scholar]
- Perry AJ, Rimmer KA, Mertens HDT, Waller RF, Mulhern TD, Lithgow T, Gooley PR (2008) Structure, topology and function of the translocase of the outer membrane of mitochondria. Plant Physiol Biochem 46: 265–274 [DOI] [PubMed] [Google Scholar]
- Rimmer KA, Foo JH, Ng A, Petrie EJ, Shilling PJ, Perry AJ, Mertens HD, Lithgow T, Mulhern TD, Gooley PR (2011) Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J Mol Biol 405: 804–818 [DOI] [PubMed] [Google Scholar]
- Schleiff E, Motzkus M, Soll J (2002) Chloroplast protein import inhibition by a soluble factor from wheat germ lysate. Plant Mol Biol 50: 177–185 [DOI] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, et al. (2011) Regulation of mitochondrial protein import by cytosolic kinases. Cell 144: 227–239 [DOI] [PubMed] [Google Scholar]
- Schweiger R, Soll J, Jung K, Heermann R, Schwenkert S (2013) Quantification of interaction strengths between chaperones and tetratricopeptide repeat domain-containing membrane proteins. J Biol Chem 288: 30614–30625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2013) The chloroplast protein import system: from algae to trees. Biochim Biophys Acta 1833: 314–331 [DOI] [PubMed] [Google Scholar]
- Smith MD, Rounds CM, Wang F, Chen K, Afitlhile M, Schnell DJ (2004) atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J Cell Biol 165: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Carrie C, Law S, Murcha MW, Zhang R, Law YS, Suen PK, Whelan J, Lim BL (2012a) AtPAP2 is a tail-anchored protein in the outer membrane of chloroplasts and mitochondria. Plant Signal Behav 7: 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Suen PK, Zhang Y, Liang C, Carrie C, Whelan J, Ward JL, Hawkins ND, Jiang L, Lim BL (2012b) A dual-targeted purple acid phosphatase in Arabidopsis thaliana moderates carbon metabolism and its overexpression leads to faster plant growth and higher seed yield. New Phytol 194: 206–219 [DOI] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Kugelmann M, Härtel B, Brennicke A (2012) Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc Natl Acad Sci USA 109: 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K, Soll J (1996) Phosphorylation of the transit sequence of chloroplast precursor proteins. J Biol Chem 271: 6545–6554 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Gu LF, Ye M, Zou H, Sun SSM, He JX (2013) A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J Proteomics 78: 486–498 [DOI] [PubMed] [Google Scholar]
- Wang Y, Carrie C, Giraud E, Elhafez D, Narsai R, Duncan O, Whelan J, Murcha MW (2012) Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2006) Transformation of Agrobacterium using the freeze-thaw method. CSH Protoc 2006: pdb.prot4666. [DOI] [PubMed] [Google Scholar]
- Whelan J, Dolan L, Harney MA (1988) Import of precursor proteins into Vicia faba mitochondria. FEBS Lett 236: 217–220 [Google Scholar]
- Xu L, Carrie C, Law SR, Murcha MW, Whelan J (2013) Acquisition, conservation, and loss of dual-targeted proteins in land plants. Plant Physiol 161: 644–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Spånning E, Glaser E, Mäler L (2015) Interaction of the dual targeting peptide of Thr-tRNA synthetase with the chloroplastic receptor Toc34 in Arabidopsis thaliana. FEBS Open Bio 5: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Spånning E, Unnerståle S, Gotthold D, Glaser E, Mäler L (2012) NMR investigations of the dual targeting peptide of Thr-tRNA synthetase and its interaction with the mitochondrial Tom20 receptor in Arabidopsis thaliana. FEBS J 279: 3738–3748 [DOI] [PubMed] [Google Scholar]
- Zhang XP, Glaser E (2002) Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci 7: 14–21 [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Sagstetter M, Lewis MJ, Pelham HR (1988) Seventy-kilodalton heat shock proteins and an additional component from reticulocyte lysate stimulate import of M13 procoat protein into microsomes. EMBO J 7: 2875–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.