Abstract
To explore the potential of tumor-specific DNA as a biomarker for head and neck squamous cell carcinomas (HNSCC), we queried DNA from saliva or plasma of 93 HNSCC patients. We searched for somatic mutations or human papillomavirus genes, collectively referred to as tumor DNA. When both plasma and saliva were tested, tumor DNA was detected in 96% of 47 patients. The fractions of patients with detectable tumor DNA in early- and late-stage disease were 100% (n = 10) and 95% (n = 37), respectively. When segregated by site, tumor DNA was detected in 100% (n = 15), 91% (n = 22), 100% (n = 7), and 100% (n = 3) of patients with tumors of the oral cavity, oropharynx, larynx, and hypopharynx, respectively. In saliva, tumor DNA was found in 100% of patients with oral cavity cancers and in 47 to 70% of patients with cancers of the other sites. In plasma, tumor DNA was found in 80% of patients with oral cavity cancers, and in 86 to 100% of patients with cancers of the other sites. Thus, saliva is preferentially enriched for tumor DNA from the oral cavity, whereas plasma is preferentially enriched for tumor DNA from the other sites. Tumor DNA in saliva was found postsurgically in three patients before clinical diagnosis of recurrence, but in none of the five patients without recurrence. Tumor DNA in the saliva and plasma appears to be a potentially valuable biomarker for detection of HNSCC.
INTRODUCTION
Head and neck squamous cell carcinomas (HNSCCs) are the seventh most common cancer worldwide, occurring in more than half a million new patients each year and in >50,000 patients in the United States alone (1, 2). The incidence of certain types of HNSCC appears to be increasing, especially among young people, in part due to the increasing prevalence of human papilloma virus (HPV) (3–7). HNSCCs are associated with a poor 5-year overall survival of only ~50% that has remained relatively unchanged, especially for patients with HPV-negative tumors (8). Only a few targeted therapies for this disease are available, in part because of the paucity of activating mutations in oncogenes that contribute to tumor development; most genetic alterations in HNSCCs inactivate tumor suppressor genes (9–12). There are also no available biomarkers for HNSCC to measure disease burden or response to therapy, further limiting progress in mitigating the impact of this often morbid and potentially lethal disease on human health.
Although HNSCC tumors are usually classified on the basis of histology, their biomedical properties, including demographics, risks factors, and clinical behavior, differ by anatomic site (Fig. 1) (13, 14). Anatomically, the tumors are categorized as squamous cell carcinomas (SCCs) of the oral cavity (including the oral tongue), oropharynx (including the base of the tongue), larynx, and hypopharynx. Oral cavity SCC, with the exception of those of the oral tongue, is declining in incidence in the United States because of the reduction in cigarette smoking (4). In contrast, there is an increasing incidence of oropharyngeal SCC involving the palatine and lingual (base of the tongue) tonsils, particularly in younger men. These tumors are often associated with HPV. The survival of these patients is better than for those whose tumors are un-associated with HPV (6, 15). Laryngeal SCC is declining in incidence and, unlike HNSCC at other sites, is generally associated with limited regional metastasis due to anatomic barriers (16). The hypopharynx is the least common site for HNSCC and has decreasing incidence but relatively poor prognosis (17, 18).
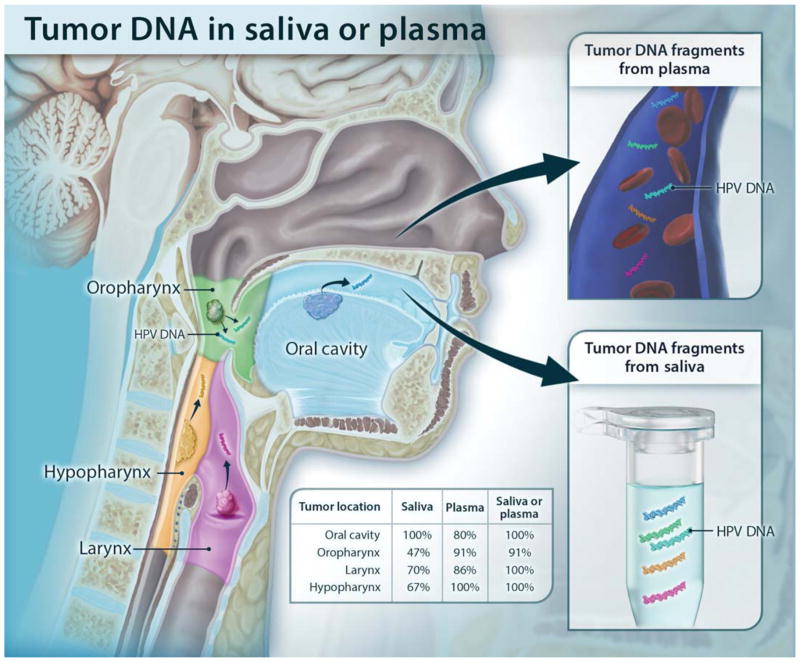
Fig. 1. Schematic showing the shedding of tumor DNA from head and neck cancers into the saliva or plasma.
Tumors from various anatomic locations shed DNA fragments containing tumor-specific mutations and HPV DNA into the saliva or the circulation. The detectability of tumor DNA in the saliva varied with anatomic location of the tumor, with the highest sensitivity for oral cavity cancers. The detectability in plasma varied much less in regard to the tumor’s anatomic location.
The idea that the genetic alterations present in tumors can be used as biomarkers for cancer was proposed more than two decades ago (19–22). The advantage of genetic alterations over conventional bio-markers such as carcinoembryonic antigen or prostate-specific antigen is that genetic changes are exquisitely specific for neoplastic cells. One challenge in exploiting genetic alterations for this purpose is that the concentration of mutant templates is often low in bodily fluids. Over the last several years, however, technological advances have made it possible to detect such mutations even when they are rare. These advances have facilitated the detection of altered DNA sequences in plasma, stool, Pap smear fluids, sputum, and urine (20, 21, 23–30).
In this proof-of-principle study, we determined whether genetically altered DNA could be detected in the saliva or plasma of HNSCC patients with tumors of various stages and anatomical sites. We chose these two bodily fluids for obvious reasons: plasma has been shown to harbor tumor DNA from many cancers, including HNSCC, though only a few HNSCCs, all of late stage, have been previously examined (23, 31, 32). Tumor DNA that is released from the basal side of HNSCC epithelial cells into the lymphatics or venous system should be detectable in this compartment. On the other hand, DNA that is released primarily on the apical side of HNSCC should be detectable in the saliva (23, 31, 32). The studies described below were performed to test these hypotheses.
RESULTS
Mutations in primary tumors
Ninety-three patients with HNSCC were enrolled in this study. Their average age was 60 and the majority (83%) were male, as is typical of HNSCC patients (table S1). Forty-six, 34, 10, and 3 samples were from the oral cavity, oropharynx, larynx, and hypopharynx, respectively. Twenty patients (22%) had early (stage I or II) disease, and the remaining 73 patients (78%) had advanced (stage III or IV) disease.
To begin this study, we attempted to identify at least one genetic alteration in each tumor. We first searched for the presence of either HPV type 16 (HPV16) or HPV18 sequences in tumor DNA. HPV is a well-established etiologic agent for a growing subset of HNSCCs, specifically oropharyngeal SCC (6, 15). With polymerase chain reaction (PCR) primer pairs specific for the E7 gene of the high-risk HPV types responsible for the overwhelming majority of HPV-associated HNSCCs, we identified 30 patients (32%) whose tumors contained HPV16 DNA and no patients with HPV18. The preponderance of HPV16 is not surprising given prior epidemiologic studies of this tumor type (33). In the other 63 patients (all of those without HPV), we searched for somatic mutations in genes or gene regions commonly altered in HNSCC, including TP53, PIK3CA, CDKN2A, FBXW7, HRAS, and NRAS, using multiplex PCR and massively parallel sequencing (primer sequences are listed in table S2) (9, 11, 12). This allowed us to identify a driver mutation in 58 of the 63 samples. In the remaining five samples, genome-wide sequencing was performed at low coverage with the goal of identifying one driver mutation or translocation as previously described (34). Ultimately, we identified and validated one genetic alteration in each of these 63 samples (table S3). The most commonly mutated gene was TP53 (86% of 63 patients). We also searched for mutations in the tumors of 25 of the patients with HPV and found mutations in 12 of those samples (table S3).
Mutations in saliva and plasma
Important characteristics of screening tests are that samples can be easily collected without discomfort and that the collection process is standardized. To achieve these goals, we used oral rinses, plasma, and commercially available kits to prepare DNA for conventional genotyping purposes. For saliva, we used the entire contents of the collection tube (including cells and cell debris) to prepare DNA. Of the 93 patients who donated saliva for this study before their surgery, 47 patients (51%) volunteered to donate plasma at the same time. DNA from plasma was purified as previously described (23). Digital PCR was used to query HPV sequences and translocations (35), whereas point mutations were assessed by Safe-SeqS (Safe-Sequencing System), a PCR-based technology for the detection of low-frequency mutations, as previously described (23, 25, 34–36).
Tumor DNA was identified in 76% (n = 93) and 87% (n = 47) of the saliva and plasma samples from these patients, respectively (Table 1 and tables S1 and S3). In the subset of patients with both plasma and saliva samples, 96% (n = 47) of patients had a tumor-specific alteration identified in at least one bodily fluid. Twenty-one of the 47 patients had HPV-positive tumors. Eighteen of the 21 patients (86%) had detectable HPV DNA in their plasma and/or saliva (Table 1 and tables S1and S3). Because HPV16 is rarely found in oral specimens of healthy individuals, we analyzed 10 saliva or plasma samples from patients whose tumors were not HPV-positive as controls (37, 38). As expected, no HPV was detected in any of these samples, confirming the specificity of the test. In all 26 patients without HPV-positive tumors, endogenous DNA mutations, mostly in the TP53 gene (92%), were identified in plasma or saliva (table S3). Thus, somatic mutations and HPV sequences were both useful as biomarkers for malignancy. The sensitivity of these biomarkers for detecting cancer was greatly improved when both plasma and saliva were examined, compared to testing saliva or plasma alone. There was no significant correlation between the amounts of tumor DNA in saliva versus plasma in the patients in whom both sample types were available (correlation coefficient of 0.074, P = 0.74), However, the use of versus plasma, and HPV versus somatic mutations, differed considerably with respect to the site of disease, as noted below.
Table 1. Detection of tumor-derived DNA in saliva and plasma.
The percentages of patients whose tumors were detectable through the examination of saliva, plasma, or both are shown, grouped by tumor site, stage, and HPV status.
| Saliva, % with mutations (95% confidence intervals) (total number studied) | Plasma, % with mutations (95% confidence intervals) (total number studied) | Saliva or plasma, % with mutations (95% confidence intervals) (total number studied)* | |
|---|---|---|---|
| Site | |||
| Oral cavity | 100 (92–100%) (46) | 80 (52–96%) (15) | 100 (78–100%) (15) |
| Oropharynx | 47 (30–65%) (34) | 91 (71–99%) (22) | 91 (71–99%) (22) |
| Larynx | 70 (35–93%) (10) | 86 (42–99%) (7) | 100 (59–100%) (7) |
| Hypopharynx | 67 (9.4–99%) (3) | 100 (29–100%) (3) | 100 (29–100%) (3) |
| Stage | |||
| Early (I and II) | 100 (83–100%) (20) | 70 (35–93%) (10) | 100 (69–100%) (10) |
| Late (III and IV) | 70 (58–80%) (73) | 92 (78–98%) (37) | 95 (82–99%) (37) |
| HPV | |||
| Positive | 40 (23–59%) (30) | 86 (64–97%) (21) | 86 (64–97%) (21) |
| Total | 76 (66–85%) (93) | 87 (74–95%) (47) | 96 (85–99%) (47) |
Includes only patients from whom both saliva and plasma were available.
Site
All (100%) of the 46 patients with oral cavity cancers harbored detectable tumor DNA in their saliva (Table 1). The sensitivities of detection in saliva of malignancy at sites not directly sampled by an oral rinse were lower: 47% (n = 34), 70% (n = 10), and 67% (n = 3) of patients with oropharyngeal cancers, laryngeal cancers, and hypopharyngeal cancers had detectable tumor DNA, respectively. The detection rate of tumor DNA in plasma varied less with site, as expected: 80% (n = 15), 91% (n = 22), 86% (n = 7), and 100% (n = 3) of tumors of the oral cavity, oropharynx, larynx, and hypopharynx, respectively, had detectable tumor DNA in plasma.
It is well known that HPV-associated tumors are most often found at specific sites, particularly the oropharynx. Twenty-nine of the 34 (85%) oropharyngeal cancers were HPV-positive. The remaining five oropharyngeal cancers were negative for HPV by PCR, were associated with tobacco use, and harbored TP53 mutations. In contrast, all but 1 of 59 samples from the oral cavity, larynx, and hypopharynx were HPV-negative. The finding that only 1 of the 46 oral cavity cancers tested was HPV-positive is consistent with recent evidence about the low prevalence of HPV-related cancers in the oral cavity (39, 40). For the HPV-associated cancers, which represent 30 (32%) of the total HNSCCs in our study, the presence of HPV DNA in bodily fluids represents a very convenient marker: HPV was detected in 40% (n = 30) of saliva samples and 86% (n = 21) of available plasma samples with a single primer pair specific for the E7 gene of HPV16 (Table 1).
Collectively, these data indicate that plasma rather than saliva is the optimal fluid for detecting tumor DNA in tumors of the oropharynx, larynx, and hypopharynx. Of the 32 patients with tumors from these sites in which both plasma and saliva were available, mutant DNA was detected in more plasma samples than saliva samples (29 versus 18, respectively). The amount of detectable mutant DNA alleles, expressed as a fraction of the total alleles assessed, was ~10-fold higher in the plasma compared with the saliva of these patients (median, 0.146% versus 0.015%; P = 0.005, Wilcoxon rank sum test; table S3). The higher fraction of alleles considerably simplifies the task of identifying such mutations. This pattern was not observed in the oral cavity: the fraction of patients harboring mutant DNA, as well as the mutant allele fraction, was similar in the saliva and plasma (median, 0.65% versus 0.54%; P = 0.14, Wilcoxon rank sum test; table S3).
Stage
Most HNSCC patients have advanced disease (stage III or IV) at diagnosis (2). Accordingly, only 22% of the 93 patients in our cohort presented with early-stage disease (table S1). Overall, tumor-specific DNA could be detected in the plasma or saliva of 100% (n = 20) and 86% (n = 73) of patients with early and advanced disease, respectively (P = 0.116, Fisher’s exact test). Saliva provided a more sensitive predictor of early-stage disease than plasma: 100% (15 of 15 oral cavity cancers, 3 of 3 oropharyngeal cancers, and 2 of 2 laryngeal cancers) versus 70% (5 of 7 oral cavity cancers, 2 of 2 oropharyngeal cancers, and 0 of 1 laryngeal cancers), respectively (P = 0.03, Fisher’s exact test; Table 1 and table S1). Contributing to the high sensitivity in saliva was the fact that 75% (15 of 20) of the early-stage cancers in the study were from the oral cavity, which are most readily detectable in saliva and are preferentially treated with surgery, explaining their enrichment in our study. As expected, plasma provided a more sensitive predictor than saliva in patients with advanced-stage disease: 92% (n = 37) versus 70% (n = 73), respectively (P = 0.008, Fisher’s exact test; Table 1). When segregated by nodal status, tumor-specific DNA could be detected in the plasma or saliva of 83% (n = 59) and 100% (n = 34) of patients with or without nodal metastasis, respectively. When both saliva and plasma were available, there was little difference between the detectability of cancers with respect to stage of disease (Table 1). An important caveat is that only five patients with early-stage disease of non-oral cavity sites were available; although tumor DNA was detectable in all of these patients (all had detectable tumor DNA in saliva; two of the three patients with available plasma also had detectable tumor DNA in their plasma), the amount of tumor DNA was considerably lower than that of late-stage patients (median, 0.007% versus 0.06%; P = 0.03, Wilcoxon rank sum test).
Human papilloma virus
Thirty patients harbored HPV16 DNA in their tumors when assessed by PCR, and none had HPV18. Of these 30 tumors, 29 (97%) were thought to be HPV-associated upon clinical presentation on the basis of in situ hybridization with high-risk HPV sequences or immunohistochemistry with antibodies to p16; in one case, the HPV status had not been determined in the clinic. Additionally, there were no patients who were considered to have HPV-associated tumors in the clinic and did not have HPV16 DNA identified in their tumors by PCR. This supports the specificity and sensitivity of our assays. As expected from the literature, all except 1 of the 30 tumors containing HPV DNA were found in the oropharynx (15, 39). As expected, plasma from HPV-associated tumors was more informative than saliva; HPV DNA was detectable in the plasma of 86% (n = 21) of the patients but in only 40% (n = 30) of the saliva from these patients (Table 1).
Surveillance
Although not the primary purpose of this study, it was of interest to determine whether tumor DNA could be found in the saliva or plasma of patients after surgical removal of their tumors. “Follow-up” samples were available in nine patients in whom tumor DNA could be identified before therapy. Three of these patients were found to have tumor DNA in their saliva or plasma after surgery, but before clinical evidence of disease recurrence (fig. S1). For example, patient HN 399, with cancer of the oral cavity, was found to have tumor DNA in his saliva and plasma 4 months after surgery, whereas the recurrence only became evident clinically 19 months later (23.6 months after surgery). Similarly, tumor DNA was found in the saliva and plasma of patient HN 402, with cancer of the oral cavity, at 8 months after surgery, 9 months before the recurrence was clinically evident. Patient HN 380 with cancer of the larynx was found to have tumor DNA in his saliva 7 months after surgery, before any clinical or radiologic evidence of disease recurrence; the patient died of recurrent disease soon thereafter. Tumor DNA was detectable in the saliva of patient HN 367 with cancer of the oropharynx 25 months after surgery; at the time of writing (36 months after surgery), no biopsy-proven disease is yet evident, but the clinical course has been complicated with suspicious imaging for locoregional and metastatic disease. No tumor DNA was detectable in the saliva and/or plasma of the other five patients in whom samples were available, all of whom have shown no clinical evidence of recurrence for a median follow-up of 12 months (fig. S2).
DISCUSSION
Current diagnostic methods for HNSCC make the detection of early disease, assessment of response to treatment, and differentiation between the adverse effects of treatment versus persistent or recurrent disease challenging. These issues collectively compromise clinical decision-making and impair patient management. Although it is now abundantly clear that all cancers, including HNSCC, are the result of genetic alterations, this knowledge is just beginning to be applied to meet diagnostic challenges such as those described above (10). In this proof-of-principle study, we show that tumor-derived DNA can be detected in the saliva of patients with HNSCC. We also show that the evaluation of plasma can complement that of saliva, together allowing detection of tumor-derived DNA in readily obtainable bodily fluids in >90% of the studied patients. Our findings lay the conceptual and practical foundation for clinical tests designed for the earlier detection of HNSCC, either for patients at high risk for the disease or for patients previously treated for HNSCC who are at risk for disease recurrence. Moreover, these results establish a paradigm for monitoring the response to treatment.
There were several notable findings in this study. The sensitivity for detection of tumor-derived DNA in the saliva was site-dependent and most efficient for tumors in the oral cavity. Not only was tumor DNA detectable in every one of the 46 patients with cancers of the oral cavity, but also the fraction of mutant DNA in the saliva was particularly high (median, 0.65%; interquartile range, 0.17 to 2.2%; mean, 3.46%). Moreover, early-stage oral cavity cancers were highly detectable; 75% of the patients with oral cavity cancer were at an early stage (stage I or II), and all were detectable. The high fraction of tumor DNA in the saliva of patients with oral cancers makes anatomical sense and demonstrates the advantage of examining local bodily fluids for optimal sensitivity in this type of assay.
HNSCCs distal to the oral cavity (oropharynx, larynx, and hypopharynx) were still often detectable through the examination of saliva, but the frequency of their detection (47, 70, and 67%, respectively) and the fraction of mutant alleles (median, 0.015%) were considerably lower than found in the oral cavity (0.65%). Anatomical locations likely explain this difference. Gargling might increase the detectability of tumor DNA in these distal compartments, but to test this idea, a reproducible procedure for gargling would have to be devised.
One striking aspect of this study is the increased sensitivity demonstrated when assays of two compartments are combined. This increased sensitivity is possible only because of the exquisite specificity of mutant DNA as biomarker; because no false-positives are expected, any number of assays can be combined, increasing sensitivity without compromising specificity. The combination of saliva and plasma allowed detection of 96% of the cancers when both fluids were available, higher than obtained with either saliva or plasma alone.
We emphasize that our study establishes the proof of principle for the use of saliva and plasma to reveal the presence of HNSCCs, but does not comprise a clinical test. In each patient, we first evaluated the tumor, then used an alteration (either the presence of HPV or a somatic mutation) to query the saliva or plasma. In an actual diagnostic test, a panel of genes would have to be used to assess each case. Fortunately, technologies are available for finding mutations, even those present at low frequencies (25, 35). On the basis of the results presented herein, as well as large studies of HNSCC genetics (9, 11, 12), a panel including HPV16 DNA sequences, TP53, PIK3CA, NOTCH1, and CDKN2A would be able to detect >95% of invasive HNSCCs. Another limitation of our study is that the number of early-stage cancers beyond the oral cavity was small, in part reflecting the unfortunate fact that most of these cancers are detected only when they are late-stage. Future larger studies should be able to determine how often early-stage cancers of the oropharynx, larynx, and hypopharynx can be detected using the approach described here. The fact that at least 70% of the oropharyngeal cancers in the United States are associated with HPV simplifies this task (6, 15).
One important application of the results described in this paper is in the diagnosis of clinically suspicious lesions. The often complex and highly specialized nature of current HNSCC diagnostic procedures can lead to delays in diagnosis and treatment, negatively impacting prognosis and survival (41–46). These delays could be prevented in many patients through the examination of saliva and plasma for tumor DNA. Such a test could potentially be incorporated into routine examinations to complement current diagnostic modalities and inform clinical decision-making. Another obvious application is in disease monitoring and surveillance. In nine patients with positive pre-treatment saliva and/or plasma, samples were collected at various times after surgery. The fact that no mutations were identified after surgery in the five patients whose tumors did not recur highlights the specificity of the mutation-based assay. It was also encouraging that we identified tumor DNA in the saliva of patients whose tumors were found to recur at the clinical level only months later, suggesting that these tests could provide a clinically meaningful lead time. The results presented here provide the setting for a larger study to explore whether the presence of tumor DNA in either saliva or plasma can be used to help manage patients who appear free of disease after definitive treatment by clinical criteria.
MATERIALS AND METHODS
Study design
This was a retrospective study with sample collection performed prospectively from 93 HNSCC patients donating saliva, 47 of whom are also donating plasma. Data analysis was performed in a blinded fashion, and all patient samples were de-identified.
Samples
All samples from the 93 patients in this study were collected using Institutional Review Board–approved protocols at Johns Hopkins University (JHU) and MD Anderson Medical Center. None of the patients in the current study were included in the previously published study from our groups, in which the genomic landscapes of HNSCC were described (9). Saliva samples were collected before definitive treatment for primary HNSCC (n = 71, 76% of 93 patients) and before salvage treatment for recurrent HNSCC (n = 22, 24% of 93 patients). In a subset of these patients (n = 9), posttreatment saliva was also collected for surveillance. Most patients (95% of the 93) underwent a biopsy of the primary tumor and/or metastatic lymph node, on average 44 days before the first sample collection (table S3). For the 22 patients with recurrent disease, previous treatment including an iteration of surgery, radiation, and/or chemotherapy was completed an average of 2.9 years before sample collection (table S3).
Whole blood was collected from 47 of the 93 patients before treatment. Four to 10 ml of plasma was used for DNA purification, with the average amount of plasma being 6 ml.
Saliva was collected using two different protocols. Under the JHU protocol, patients were asked to swish 15 to 20 ml of 0.9% sodium chloride in their mouths for 10 to 15 s before spitting into the collection tube. Under the MD Anderson protocol, patients were asked to allow saliva to collect in the floor of the mouth for 5 min without swallowing before spitting into the collection vial. There was no significant difference in the amounts of DNA purified, the fraction of mutant DNA, or the amount of HPV sequences found with the two protocols. Saliva was frozen at −80°C until DNA purification, and the entire volume of saliva, without centrifugation of cells, was used for DNA purification. The amount of saliva used averaged 15 ml (range, 10 to 20 ml).
When fresh tumor tissue from a surgical specimen of invasive SCC was available, it was immediately frozen at −80°C. When frozen tissue was not available, formalin-fixed, paraffin-embedded (FFPE) tissues were used for DNA purification. In either case (fresh-frozen or FFPE), tumors were macrodissected to ensure neoplastic cellularity exceeding 30%. DNA was purified from the white blood cell pellet (normal DNA), saliva, and tumor using an AllPrep Kit (Qiagen, catalog #80204), and from plasma using an QIAamp Circulating Nucleic Acid Kit (Qiagen, catalog #55114).
Tumor mutational profiling
A tiered approach was used to identify a somatic mutation within each tumor. Initially, the presence of HPV16 and HPV18 was assessed using the primers specific for the E7 oncogene of these variants (HPV16: TGTGACTCTACGCTTCGGTTG and GCCCATTAACAGGTCTTCCA; HPV18: GCATGGACCTAAGGCAACAT and GAAGGTCAACCGGAATTTCAT). When no HPV was present, multiplex PCRs containing primers amplifying regions of interest in TP53, PIK3CA, CDKN2A, FBXW7, HRAS, and NRAS were used to identify driver mutations in the tumors (table S2). If a mutation was not identified in the queried regions, paired-end libraries of DNA from the tumors and white blood cell pellets of each patient were prepared and used for low-coverage whole-genome sequencing or exomic sequencing as previously described (47). Massively parallel sequencing was carried out on an Illumina HiSeq instrument, either in the Goldman Sequencing Facility at Johns Hopkins Medical Institutions or at PGDx Inc. Point mutations were identified as previously described (9, 47, 48), using the following criteria: allele fraction >20%, >5 reads for point mutations, and >1 read for translocations. Genomic rearrangements were identified through an analysis of discordantly mapping paired-end reads. The discordantly mapping paired-end reads were grouped into 1-kb bins when at least five distinct tag pairs (with distinct start sites) spanned the same two 1-kb bins, and further annotated on the basis of the approximate breakpoint (34). One selected mutation was confirmed through an independent PCR and sequencing reaction, and then used to query the saliva or plasma. All oligonucleotides used in this study were synthesized by TriLink Biotechnologies.
Mutation detection in saliva and plasma
The same primers used to detect HPV16 in tumor DNA via PCR were used to detect HPV16 sequences in the DNA from saliva or plasma. Each saliva DNA or plasma DNA sample was assessed in at least three independent PCR assays, and all three assays had to be positive for the sample to be counted as positive. As an additional control for specificity, the PCR products were sequenced to ensure that they represented HPV16 sequences. To quantify the amount of HPV16 sequences present in saliva or plasma, we used digital PCR with the same primers (36). Digital PCR was also used to quantify the amount of sequences with translocation using primers spanning the breakpoints, as previously described (49). For evaluation of point mutations in saliva or plasma, we used Safe-SeqS, a PCR-based error reduction technology for detection of low-frequency mutations in reactions each containing up to 3 ng of input DNA (23, 25). High-quality sequence reads were selected on the basis of quality scores, which were generated by the sequencing instrument to indicate the probability a base was called in error (50). The template-specific portion of the reads was matched to reference sequences. Reads from a common template molecule were then grouped on the basis of the unique identifier sequences (UIDs) that were incorporated as molecular barcodes. Artifactual mutations introduced during the sample preparation or sequencing steps were reduced by requiring a mutation to be present in >90% of reads in each UID family (“supermutant”). Each PCR assay for each plasma or saliva sample was independently repeated at least three times, with the mutant allele fractions defined as the total number of supermutants divided by the total number of UIDs in all experiments. DNA from normal individuals was used as control, using at least five independent assays per queried mutation. Only saliva or plasma samples in which the mutant allele fractions significantly exceeded their frequencies in control DNA (P < 0.05) were scored as positive (table S3).
Statistical analysis
Sensitivity for the detection of tumor-specific mutations in the blood and saliva was calculated by tumor site, stage, and among HPV-associated tumors. Ability to detect tumor DNA in saliva and/or plasma was tested using Fisher’s exact tests, and Wilcoxon rank sum tests were used to compare amounts of tumor DNA in saliva versus plasma (51). For the comparison of mutant fractions in patients versus control in Safe-SeqS assays, P values were calculated using a two-sided χ2 test of equal proportions or Fisher’s exact test when conditions of the χ2 test are not met. The concordance between mutant fractions in saliva and plasma was calculated using Pearson’s product-moment correlation coefficient, a standard measure of linear dependence between two variables. All statistical analyses were performed using the R statistical package version 3.1.2.
Supplementary Material
Acknowledgments
We thank our patients for their courage and generosity. We thank J. Ptak, L. Dobbyn, C. Blair, K. Judge, M. Popoli, J. Bondy, M. Goldsmith, S. Greeley, and Z. Khan for technical assistance. We thank E. Cook for her artistic contribution.
Funding: This work was supported by the Virginia and D.K. Ludwig Fund for Cancer Research, the Conrad R. Hilton Foundation, the Banyan Gate Foundation, Swim Across America, the Sol Goldman Sequencing Facility at Johns Hopkins, and NIH grants CA43460, CA057345, CA152753, and NIH/National Institute of Dental and Craniofacial Research (NIDCR) Specialized Program of Research Excellence grant DE019032.
Footnotes
Author contributions: C.R.P., J.A.B., C.H.C., J.A.C., D.W.E., C.F., C.G.G., P.K.H., H.K., A.K., W.M.K., J.N.M., H.Q., J.D.R., D.S., R.P.T., W.H.W., B.V., and N.A. contributed to sample acquisition. Y.W., S.S., C.B., L.A.D., N.P., K.W.K., B.V., and N.A. contributed to the design of the study. N.S., J.S., J.A.B., and W.H.W. contributed to sample preparation. Y.W., S.S., N.P., K.W.K., B.V., and N.A. contributed to molecular genetic analysis. Y.W., S.S., C.L.M., M.S., N.J., E.M.R., T.G., N.P., K.W.K., B.V., and N.A. contributed to data analysis. Y.W., S.S., N.P., K.W.K., B.V., and N.A. wrote early drafts of the paper. All authors reviewed and approved the manuscript.
Competing interests: Under agreements between the JHU, Genzyme, Sysmex-Inostics, Qiagen, Invitrogen, and Personal Genome Diagnostics, L.D., N.P., B.V., and K.W.K. are entitled to a share of the royalties received by the University on sales of products related to genes and technologies described in this manuscript. L.D., N.P., B.V., and K.W.K. are co-founders of Personal Genome Diagnostics and PapGene Inc., are members of the Scientific Advisory Boards of Sysmex-Inostics, Personal Genome Diagnostics, and PapGene Inc., and own Personal Genome Diagnostics and PapGene Inc. stock, which is subject to certain restrictions under Johns Hopkins University policy. The terms of these arrangements are managed by the Johns Hopkins University in accordance with its conflict-of-interest policies. Additionally, B.V. and K.W.K. are on the Scientific Advisory Board of Morphotek Inc., and N.J. is affiliated with the Emmes Corporation.
www.sciencetranslationalmedicine.org/cgi/content/full/7/293/293ra104/DC1
Fig. S1. Tumor DNA is detectable in the saliva of patients before recurrence becomes clinically evident.
Fig. S2. Patients with undetectable tumor DNA after surgery have better disease-free survival.
Table S1. Patient demographics.
Table S2. Primer sequences used in multiplex assay for identification of driver mutations in tumors.
Table S3. Amounts of tumor-derived DNA in saliva and plasma.
REFERENCES AND NOTES
- 1.Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Simard EP, Ward EM, Siegel R, Jemal A. Cancers with increasing incidence trends in the United States: 1999 through 2008. CA Cancer J Clin. 2012;62:118–128. doi: 10.3322/caac.20141. [DOI] [PubMed] [Google Scholar]
- 4.Patel SC, Carpenter WR, Tyree S, Couch ME, Weissler M, Hackman T, Hayes DN, Shores C, Chera BS. Increasing incidence of oral tongue squamous cell carcinoma in young white women, age 18 to 44 years. J Clin Oncol. 2011;29:1488–1494. doi: 10.1200/JCO.2010.31.7883. [DOI] [PubMed] [Google Scholar]
- 5.Li R, Faden DL, Fakhry C, Langelier C, Jiao Y, Wang Y, Wilkerson MD, Pedamallu CS, Old M, Lang J, Loyo M, Ahn SM, Tan M, Gooi Z, Chan J, Richmon J, Wood LD, Hruban RH, Bishop J, Westra WH, Chung CH, Califano J, Gourin CG, Bettegowda C, Meyerson M, Papadopoulos N, Kinzler KW, Vogelstein B, DeRisi JL, Koch WM, Agrawal N. Clinical, genomic, and metagenomic characterization of oral tongue squamous cell carcinoma in patients who do not smoke. Head Neck. 2014 doi: 10.1002/hed.23807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26:612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 8.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975–2011. National Cancer Institute; Bethesda, MD: 2014. http://seer.cancer.gov/csr/1975_2011/ [Google Scholar]
- 14.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, Rosenberg PS, Bray F, Gillison ML. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31:4550–4559. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Sou za G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, Gillison WHML. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–1956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 16.Pfister DG, Spencer S, Brizel DM, Burtness B, Busse PM, Caudell JJ, Cmelak AJ, Colevas AD, Dunphy F, Eisele DW, Gilbert J, Gillison ML, Haddad RI, Haughey BH, Hicks WL, Jr, Hitchcock YJ, Jimeno A, Kies MS, Lydiatt WM, Maghami E, Martins R, McCaffrey T, Mell LK, Mittal BB, Pinto HA, Ridge JA, Rodriguez CP, Samant S, Schuller DE, Shah JP, Weber RS, Wolf GT, Worden F, Yom SS, McMillian NR, Hughes M. Head and neck cancers, version 2.2014. Clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2014;12:1454–1487. doi: 10.6004/jnccn.2014.0142. [DOI] [PubMed] [Google Scholar]
- 17.Gourin CG, Terris DJ. Carcinoma of the hypopharynx. Surg Oncol Clin N Am. 2004;13:81–98. doi: 10.1016/S1055-3207(03)00122-4. [DOI] [PubMed] [Google Scholar]
- 18.Kuo P, Chen MM, Decker RH, Yarbrough WG, Judson BL. Hypopharyngeal cancer incidence, treatment, and survival: Temporal trends in the United States. Laryngoscope. 2014;124:2064–2069. doi: 10.1002/lary.24651. [DOI] [PubMed] [Google Scholar]
- 19.Sidransky D. Nucleic acid-based methods for the detection of cancer. Science. 1997;278:1054–1059. doi: 10.1126/science.278.5340.1054. [DOI] [PubMed] [Google Scholar]
- 20.Sidransky D, Von Eschenbach A, Tsai YC, Jones P, Summerhayes I, Marshall F, Paul M, Green P, Hamilton SR, Frost P, Volgelstein B. Identification of p53 gene mutations in bladder cancers and urine samples. Science. 1991;252:706–709. doi: 10.1126/science.2024123. [DOI] [PubMed] [Google Scholar]
- 21.Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, Vogelstein B. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–105. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- 22.Boyle JO, Mao L, Brennan JA, Koch WM, Eisele DW, Saunders JR, Sidransky D. Gene mutations in saliva as molecular markers for head and neck squamous cell carcinomas. Am J Surg. 1994;168:429–432. doi: 10.1016/s0002-9610(05)80092-3. [DOI] [PubMed] [Google Scholar]
- 23.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SKN, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih IM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA., Jr Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diehl F, Schmidt K, Durkee KH, Moore KJ, Goodman SN, Shuber AP, Kinzler KW, Vogelstein B. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology. 2008;135:489–498. doi: 10.1053/j.gastro.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih IM, Kurman R, Dao F, Levine DA, Giuntoli R, Roden R, Eshleman JR, Carvalho JP, Marie SK, Papadopoulos N, Kinzler KW, Vogelstein B, Diaz LA., Jr Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013;5:167ra164. doi: 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 27.Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW, Jr, Alizadeh AA, Diehn M. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martignetti JA, Camacho-Vanegas O, Priedigkeit N, Camacho C, Pereira E, Lin L, Garnar-Wortzel L, Miller D, Losic B, Shah H, Liao J, Ma J, Lahiri P, Chee M, Schadt E, Dottino P. Personalized ovarian cancer disease surveillance and detection of candidate therapeutic drug target in circulating tumor DNA. Neoplasia. 2014;16:97–103. doi: 10.1593/neo.131900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ralla B, Stephan C, Meller S, Dietrich D, Kristiansen G, Jung K. Nucleic acid-based biomarkers in body fluids of patients with urologic malignancies. Crit Rev Clin Lab Sci. 2014;51:200–231. doi: 10.3109/10408363.2014.914888. [DOI] [PubMed] [Google Scholar]
- 30.Hubers AJ, Prinsen CF, Sozzi G, Witte BI, Thunnissen E. Molecular sputum analysis for the diagnosis of lung cancer. Br J Cancer. 2013;109:530–537. doi: 10.1038/bjc.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol. 2015;12:11–26. doi: 10.1038/nrclinonc.2014.192. [DOI] [PubMed] [Google Scholar]
- 32.Ahn SM, Chan JYK, Zhang Z, Wang H, Khan Z, Bishop JA, Westra W, Koch WM, Califano JA. Saliva and plasma quantitative polymerase chain reaction–based detection and surveillance of human papillomavirus–related head and neck cancer. JAMA Otolaryngol Head Neck Surg. 2014;140:846–854. doi: 10.1001/jamaoto.2014.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: A systematic review. Cancer Epidemiol Biomarkers Prevent. 2005;14:467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 34.Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA, Jr, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE, Hogarty MD. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2012;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreimer AR, Bhatia RK, Messeguer AL, González P, Herrero R, Giuliano AR. Oral human papillomavirus in healthy individuals: A systematic review of the literature. Sex Transm Dis. 2010;37:386–391. doi: 10.1097/OLQ.0b013e3181c94a3b. [DOI] [PubMed] [Google Scholar]
- 38.Gillison ML, Broutian T, Pickard RKL, Tong ZY, Xiao W, Kahle L, Graubard BI, Chaturvedi AK. Prevalence of oral HPV infection in the United States, 2009–2010. JAMA. 2012;307:693–703. doi: 10.1001/jama.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isayeva T, Li Y, Maswahu D, Brandwein-Gensler M. Human papillomavirus in non-oropharyngeal head and neck cancers: A systematic literature review. Head Neck Pathol. 2012;6(Suppl 1):S104–S120. doi: 10.1007/s12105-012-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lingen MW, Xiao W, Schmitt A, Jiang B, Pickard R, Kreinbrink P, Perez-Ordonez B, Jordan RC, Gillison ML. Low etiologic fraction for high-risk human papillomavirus in oral cavity squamous cell carcinomas. Oral Oncol. 2013;49:1–8. doi: 10.1016/j.oraloncology.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Kowalski LP, Carvalho AL. Influence of time delay and clinical upstaging in the prognosis of head and neck cancer. Oral Oncol. 2001;37:94–98. doi: 10.1016/s1368-8375(00)00066-x. [DOI] [PubMed] [Google Scholar]
- 42.Guggenheimer J, Verbin RS, Johnson JT, Horkowitz CA, Myers EN. Factors delaying the diagnosis of oral and oropharyngeal carcinomas. Cancer. 1989;64:932–935. doi: 10.1002/1097-0142(19890815)64:4<932::aid-cncr2820640428>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 43.Wildt J, Bundgaard T, Bentzen SM. Delay in the diagnosis of oral squamous cell carcinoma. Clin Otolaryngol Allied Sci. 1995;20:21–25. doi: 10.1111/j.1365-2273.1995.tb00006.x. [DOI] [PubMed] [Google Scholar]
- 44.Allison P, Franco E, Black M, Feine J. The role of professional diagnostic delays in the prognosis of upper aerodigestive tract carcinoma. Oral Oncol. 1998;34:147–153. doi: 10.1016/s1368-8375(97)00088-2. [DOI] [PubMed] [Google Scholar]
- 45.Carvalho AL, Pintos J, Schlecht NF, Oliveira BV, Fava AS, Curado MP, Kowalski LP, Franco EL. Predictive factors for diagnosis of advanced-stage squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 2002;128:313–318. doi: 10.1001/archotol.128.3.313. [DOI] [PubMed] [Google Scholar]
- 46.Koivunen P, Rantala N, Hyrynkangas K, Jokinen K, Alho OP. The impact of patient and professional diagnostic delays on survival in pharyngeal cancer. Cancer. 2001;92:2885–2891. doi: 10.1002/1097-0142(20011201)92:11<2885::aid-cncr10119>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 47.Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ, Wang LD, Wang L, Yang W, Velculescu VE, Vogelstein B, Papadopoulos N, Kinzler KW, Meltzer SJ. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, Rodriguez FJ, Cahill DP, McLendon R, Riggins G, Velculescu VE, Oba-Shinjo SM, Marie SKN, Vogelstein B, Bigner D, Yan H, Papadopoulos N, Kinzler KW. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA, Jr, Velculescu VE. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 51.Agresti A. Categorical Data Analysis. 2. John Wiley & Sons, Incorporated; New York: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.