Abstract
Kidney fibrosis is marked by an epithelial–to–mesenchymal transition (EMT) by tubular epithelial cells (TECs). Here we find that during renal fibrosis TECs acquire a partial EMT program during which they remain associated with their basement membrane and express markers of both epithelial and mesenchymal cells. The functional consequence of EMT program during fibrotic injury is an arrest in the G2 phase of the cell cycle and lower expression of several transporters in TECs. We also found that transgenic expression of Twist or Snai1 expression is sufficient to promote prolonged TGF-β1–induced G2 arrest of TECs, limiting their potential for repair and regeneration. Also, in mouse models of experimentally-induced renal fibrosis, conditional deletion of Twist1 or Snai1 in proximal TECs resulted in inhibition of the EMT program and the maintenance of TEC integrity, while restoring proliferation, de–differentiation–associated repair and regeneration of the kidney parenchyma and attenuating interstitial fibrosis. Thus, inhibition of EMT program in TECs during chronic renal injury represents a potential anti–fibrosis therapy
Kidney fibrosis is associated with a reduction in the functional renal parenchyma, leading to compromised kidney functions and eventual organ death1. There is no effective treatment for renal fibrosis and its occurrence is on the rise2. But understanding the complex mechanisms and cellular mediators of kidney fibrosis could offer new therapeutic avenues3–5. To this end, protecting the integrity of the functioning parenchyma is critical for preserving overall tissue function in organ fibrosis. Importantly, a feature of kidney fibrosis includes the transition of tubular epithelial cells (TECs) into cells with mesenchymal features4,6–18, a so-called epithelial-mesenchymal transition (EMT). The EMT of TECs has also been observed in human fibrotic kidneys and enrichment in transcription factors associated with EMT correlate with disease progression19–21. However, the precise contribution of EMT to the progression of kidney fibrosis remains a subject of debate22–24 and only a minor population of αSMA+ myofibroblasts are derived from TECs4. Induction of expression of the transcriptional regulator of EMT, Snail, in the renal epithelial cells of adult mice is sufficient to provoke expression of mesenchymal features in TECs and results in a marked deposition of interstitial collagen deposition20, yet the functional role of EMT of injured TECs during renal fibrosis remains unknown. In particular, while repair of injured TECs involves the proliferation of these cells to repopulate the renal tubules with functional epithelium25,26, it remains unclear whether EMT is directly associated with cell cycle arrest of these cells, and whether gain of mesenchymal features by these cells are linked to loss of normal TEC function in the kidney.
RESULTS
Genetic targeting of EMT improves tubular health
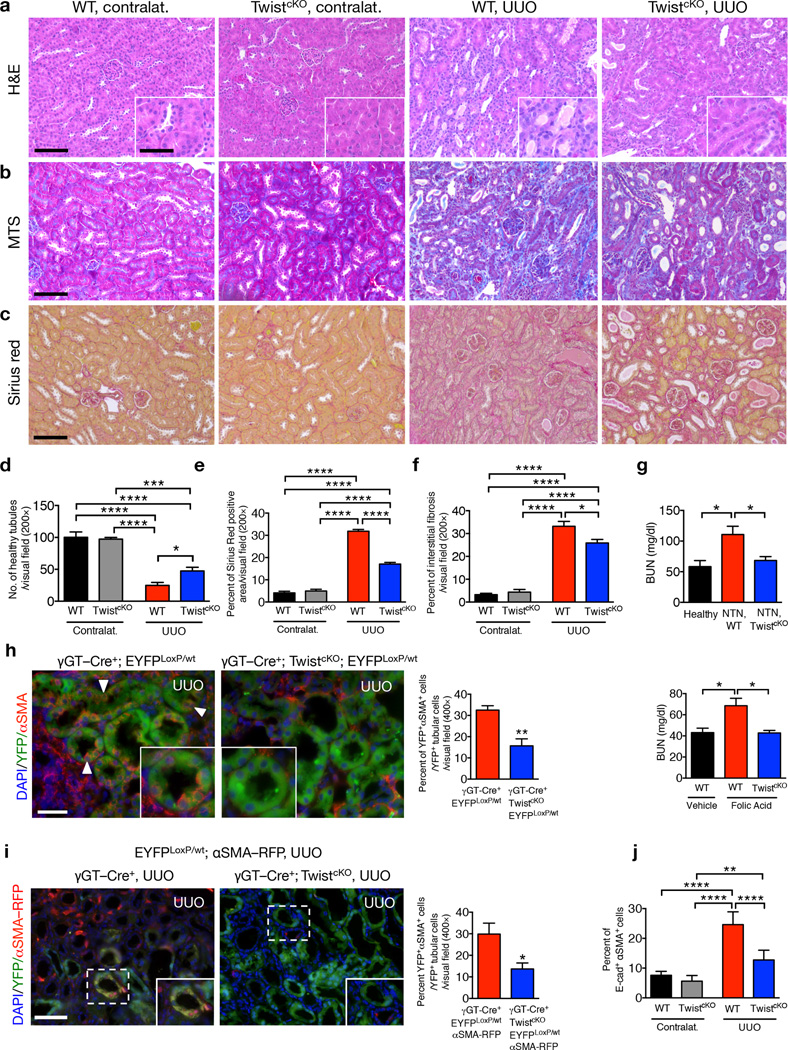
To determine the functional significance of the EMT program in renal fibrosis we crossed mice harboring a γGT–Cre locus, which allows for deletion of genes specifically in proximal TECs3, to mice with floxed alleles of Twist1 (encoding for Twist) or Snai1 (encoding for Snail) (Supplementary Fig. 1a), two transcriptional regulators of EMT (reviewed in9) and known to participate in kidney fibrosis16,20. The resulting progeny (γGT–Cre+; TwistLoxP/LoxP – hereafter referred to as TwistcKO, and γGT–Cre+; SnailLoxP/LoxP – hereafter referred to as SnailcKO) were born in expected Mendelian ratio. They were fertile and could generate offspring at a normal frequency, reached adulthood without any phenotypic abnormalities compared to litter mates and presented no obvert histopathological changes in the kidney, as well as other organs (Supplementary Fig. 1b–c). These mice, together with litter mate controls (γGT–Cre−; TwistLoxP/LoxP, and γGT–Cre−; SnailLoxP/LoxP – hereafter referred to as wild–type (WT) or control mice) were subjected to unilateral ureter obstruction (UUO), nephrotoxic serum–induced nephritis (NTN) and folic acid (FA)-induced nephropathy to induce a damaging insult to the renal parenchyma and renal fibrosis3,27 In the UUO model, histopathological analyses revealed no obvert changes in heart, lung, liver, pancreas and intestine (Supplementary Fig. 1d), whereas analyses of fibrotic kidneys following H&E, MTS (Masson Trichrome staining) and Sirius red staining revealed improved tubular health and a lower degree of interstitial fibrosis in TwistcKO compared to WT, on day 5 and day 10 after UUO (Fig. 1a–f and Supplementary Fig. 1e). Interstitial collagen deposition, measured by hydroxyproline release assay, was lower in TwistcKO UUO compared to WT UUO kidneys (Supplementary Fig. 1f). The lower degree of renal fibrosis in TwistcKO kidneys was associated with a significantly lower presence of αSMA+ myofibroblasts (Supplementary Fig. 1g). Twist genetic deletion also correlated with better renal function (Fig. 1g) and tubular health, and a lower degree of interstitial fibrosis in mice challenged with NTN (Supplementary Fig. 2a) or FA (Supplementary Fig. 2b) compared to their respective control littermates.
Figure 1.
Genetic targeting of EMT reduces renal fibrosis and improves tubular health. (a–c) Representative images (8 visual fields for each tissue analyzed) of H&E (a), MTS (b) and Sirius Red (c) staining of kidneys from the indicated experimental groups. Scale bar, 100 ~m; insert, 25 ~m. (d) Number of healthy tubules. WT contralat., n = 4; TwistcKO contralat., n = 3; WT UUO, n = 9; TwistcKO UUO, n = 9. (e–f) Interstitial fibrosis, based on MTS (e) and Sirius Red (f) staining. WT contralat., n = 4; TwistcKO contralat., n = 4; WT UUO, n = 9; TwistcKO UUO, n = 9. (g) Blood urea nitrogen (BUN) levels. Healthy, n = 3; WT NTN, n = 7; TwistcKO NTN; n = 7. WT vehicle, n = 4; WT FA, n = 4; TwistcKO FA, n = 4. (h) Representative images (8 visual fields for each tissue analyzed) of immunolabeling for αSMA and YFP (left) and quantification of the percent YFP+αSMA+ cells per total number of YFP+ tubular epithelial cells (right). WT UUO, n = 3; TwistcKO UUO, n = 4. Scale bar, 20 ~m. White arrowheads, YFP+/αSMA+ cells. (i) Representative images (3 visual fields for each tissue analyzed) of YFP+ proximal tubules and αSMA–RFP+ myofibroblasts in UUO-treated mice (left) and quantification of the percent of YFP+RFP+ cells (right). γGT–Cre; LSL–EYFPLoxP/+; αSMA–RFP, n = 3; γGT–Cre; LSL–EYFPLoxP/+; αSMA–RFP; TwistcKO, n = 4. Scale bar, 20 ~m. (j) Percent E–cadherin+αSMA+ cells (as measured by flow cytometry) in the indicated experimental groups (n = 5). Data is represented as mean ± SEM. One–way ANOVA with Tukey post–hoc analysis. h and i, unpaired two–tailed t–test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Contralat.: contralateral kidney.
SnailcKO mice (Supplementary Fig. 1a) challenged with UUO or FA also showed better tubular health and renal function, less interstitial fibrosis and a lower number of αSMA+ myofibroblasts (Supplementary Fig. 2c–g). As anticipated, deletion of Twist1 and Snai1 was correlated with a lower frequency of EMT compared to control mice with wild type Twist and Snail alleles, observed with lineage tracing of proximal TECs (γGT–Cre+; TwistcKO; EYFPLoxP/+, γGT–Cre+; SnailcKO; EYFPLoxP/+ and control γGT–Cre+; EYFPLoxP/+) and immunolabeling for the mesenchymal marker αSMA (Fig. 1h and Supplementary Fig. 3a–b) or direct visualization of αSMA–RFP transgene expression (γGT–Cre+; TwistcKO; EYFPLoxP/+; αSMA–RFP and control γGT–Cre+; EYFPLoxP/+; αSMA–RFP mice) (Fig. 1i and Supplementary Fig. 3e). Analyses of EMT frequency assayed by flow cytometry (E–cadherin+αSMA+ cells) showed a 17.33–24.6% of EMT in fibrotic kidney compared to 12.74% (TwistcKO) and 6.177% (SnailcKO), or overall 48.2 to 64.3% decrease in EMT in mice lacking proximal TEC expression of Twist or Snail, respectively (Fig. 1j and Supplementary Fig. 3f–h). Altogether these results indicate that EMT suppression in injured TECs suppressed kidney damage and promoted tubular health.
EMT inhibition prevents loss of TEC transporters
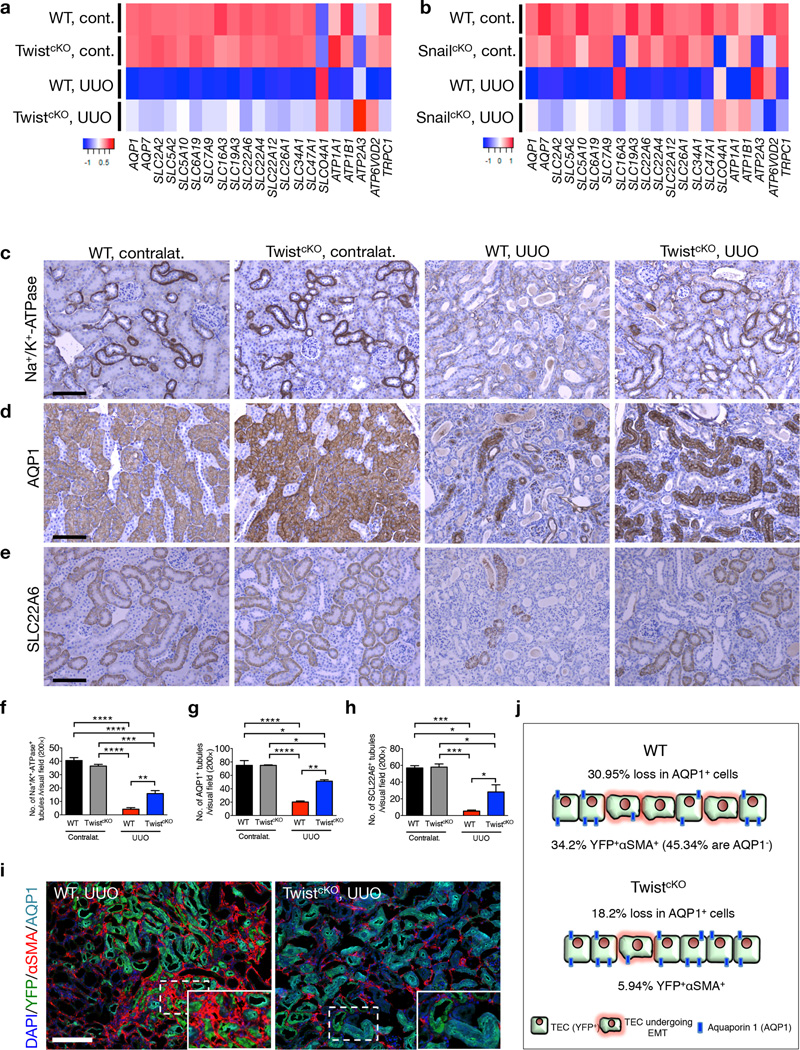
In light of the histopathological renoprotective effect noted when suppressing Twist1 and Snai1 in TECs of the injured kidney (Fig. 1 and Supplementary Fig. 1–3), global gene expression profiling was performed to assay transcriptomic changes in kidneys from TwistcKO and SnailcKO and WT mice from the UUO group and compared them to contralateral kidneys, as well as healthy controls of various genotypes (Fig. 2a–b and Supplementary Fig. 4). While contralateral kidneys from WT mice and healthy kidneys from TwistcKO and SnailcKO mice showed a similar transcriptomic signature compared to healthy kidneys from WT mice (γGT–Cre− and γGT–Cre+ WT) (Supplementary Fig. 4a,c), a specific decrease in expression of genes associated with tubular cell functions was noted when comparing contralateral and diseased kidneys (Fig. 2a – b and Supplementary Fig. 4b,d). Such decrease was attenuated in the fibrotic kidneys of TwistcKO and SnailcKO mice (Fig. 2a – b and Supplementary Fig. 4b,d), supporting the observed improved tubular cell function (Fig. 1 and Supplementary Fig. 2). Gene expression analyses and gene set enrichment analysis (GSEA) also confirmed the enrichment for an EMT signature in fibrotic kidneys from WT UUO-treated mice compared to contralateral kidney, and an attenuation of this signature in fibrotic kidneys of TwistcKO and SnailcKO UUO-treated mice (Supplementary Fig. 4f–h). In support of the better tubular function in fibrotic kidneys of TwistcKO and SnailcKO UUO-treated mice, GSEA confirmed a strong enrichment for kidneys specific genes associated with solute transport (Supplementary Fig. 4e), as well as fatty acid metabolism and β–oxidation (Supplementary Figure 4i), which were recently demonstrated to be defective and associated with cell death and dedifferentiation in tubular epithelial cells in fibrotic kidneys28.
Figure 2.
Inhibition of EMT prevents loss of TEC associated solute and solvent transporters. (a–b) Heatmaps representing averaged intensity of expression of genes associated with TEC function in kidneys of the indicated TwistcKO (a) and SnailcKO (b) experimental groups. WT cont., n = 3 ; TwistcKO cont., n = 3; WT UUO, n = 3; TwistcKO UUO, n = 4. WT cont., n = 3 ; SnailcKO cont., n = 3; WT UUO, n = 3; SnailcKO UUO, n = 3. (c–e) Representative images (8 visual fields for each tissue analyzed) of immunolabeling for Na+/K+ ATPase (c), AQP1 (d), and SLC22A6 (e) in the indicated experimental groups. Scale bar, 100 ~m. (f–h) Number of Na+/K+ ATPase+ (f), AQP1+ (g), and SLC22A6+ (h) tubules per visual field (200×). WT contralat., n = 3 ; TwistcKO contralat., n = 3; WT UUO, n = 3; TwistcKO UUO, n = 3. (i) Representative images (4 visual fields for each tissue analyzed) of immunolabeling for YFP, AQP1 and αSMA in WT UUO and TwistcKO UUO. (j) Schematic representation of the relative changes percentages of AQP1 and αSMA in YFP+ tubular epithelial cells, in kidneys from WT and TwistcKO UUO mice. WT UUO, n = 6; TwistcKO UUO, n = 3. Scale bar, 100 ~m. Data is represented as mean ± SEM. One–way ANOVA with Tukey post–hoc analysis was used. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. cont. or contralat.: contralateral kidney, EMT: epithelial to mesenchymal transition as defined by YFP+αSMA+ cells.
Loss of Na+/K+–ATPase solute transporter in TECs is associated with renal fibrosis29, and the loss of organic anion transporter SLC22A6 has been proposed to promote toxic accumulation of metabolites and uremic toxin30. Healthy kidneys from TwistcKO and SnailcKO mice showed a similar Na+/K+–ATPase, SLC22A6, and AQP1 expression pattern compared to healthy kidneys from WT (γGT–Cre− and γGT–Cre+) mice (Supplementary Fig. 5a). Genetic targeting of Twist1 and Snai1 prevented the dramatic loss in Na+/K+–ATPase, AQP1 and SLC22A6 expression compared to respective fibrotic kidneys from WT mice in all fibrotic models evaluated, including UUO (day 5 and day 10 post UUO, Fig. 2c–h, Supplementary Fig. 5b–c and Supplementary Fig. 6a–c), NTN (Supplementary Fig. 5d and Supplementary Fig. 6d), and FA (Supplementary Fig. 6e–f) models. Transcriptomic and protein expression analyses also corroborated the preserved Na+/K+–ATPase, AQP1 and SLC22A6 expression in fibrotic kidneys from TwistcKO and SnailcKO UUO-treated mice (Supplementary Fig. 6a–d and Supplementary Fig. 7). Immunological stainings showed that TECs in EMT transition (YFP+αSMA+) partake in the overall loss of AQP1 expression (with 45.34% of YFP+αSMA+ cells losing AQP1 expression), and suppression of EMT in TwistcKO UUO kidneys reduces the 30.95% loss of AQP1 in YFP+ TECs of fibrotic kidneys from WT UUO-treated mice down to 18.2% in fibrotic kidneys from TwistcKO UUO-treated mice (Fig. 2i–j and Supplementary Fig. 8). Similar findings were noted in fibrotic kidneys from SnailcKO compared to fibrotic kidneys from WT UUO-treated mice (Supplementary Fig. 9). These data support the notions that suppressing TECs from undergoing EMT preserve tubular AQP1 expression.
EMT reduces TEC transporters in subjects with renal fibrosis
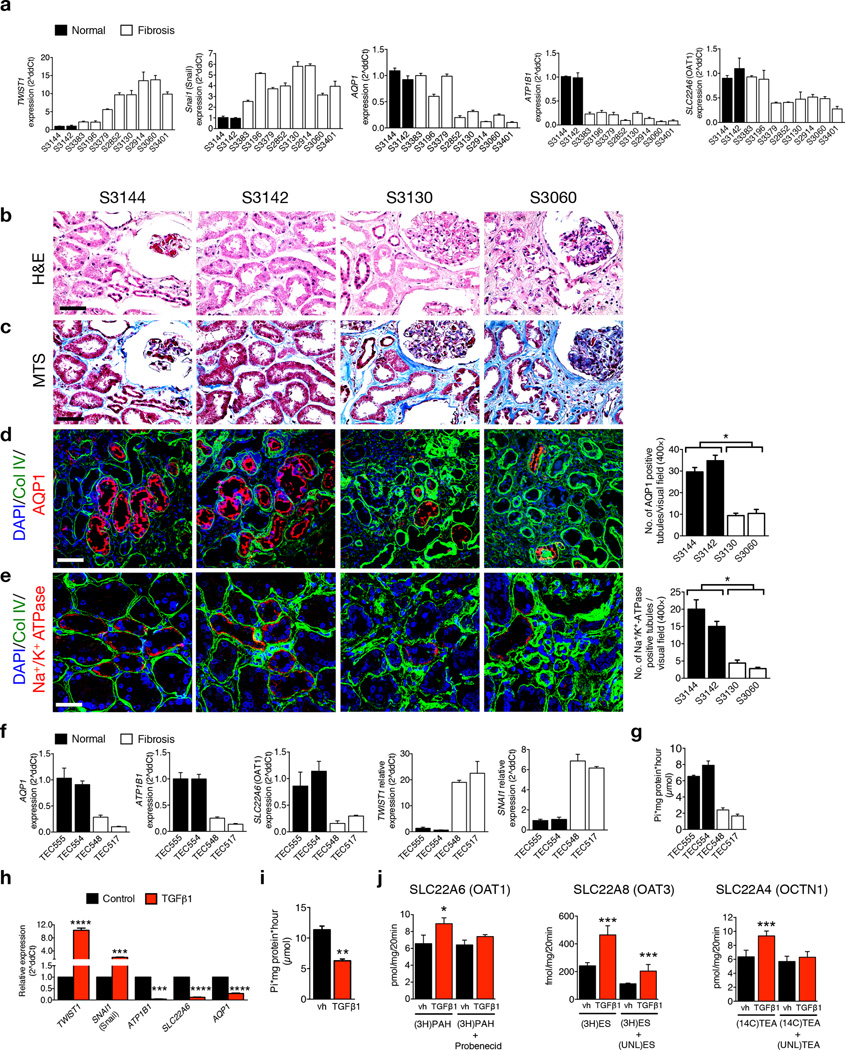
We next explored the link between an EMT program and expression of solute and solvent transporters in TECs during progression of chronic kidney disease in humans. We probed the expression of EMT–related genes and TEC transporters in gene expression datasets (www.nephromine.org) from kidney biopsies from subjects with kidney fibrosis and TGF-β1–induced EMT of cultured human HK2 TECs (GSE20247, Ref. 31), and performed additional gene expression and functional analyses on human–derived biopsies, human–derived cultured TECs (obtained from subjects with diverse fibrotic etiologies, Supplementary Fig. 10a) and HK2 cells. The EMT signature, including specific expression of TWIST1 and SNAI1, in a cohort of biopsy samples from normal and fibrotic kidneys and in the primary TECs isolated from these biopsies, was positively associated with tubulointerstitial fibrosis (Fig. 3a and Supplementary Fig. 10b).
Figure 3.
EMT program is associated with deregulated expression and functionality of TEC transporter in subjects with kidney disease. (a) Relative expression of the indicated genes in renal biopsies from healthy individuals and subjects with renal fibrosis. (b–c) Representative images of H&E (b) and MTS (c) staining of kidneys from human biopsies. (d–e) Representative images (3 visual fields for each tissue analyzed) of co–immunolabeling for Collagen IV and AQP1 (d), and Collagen IV and Na+/K+ ATPase (e) and associated quantification. Scale bar, 50 ~m. (f) Relative expression of the indicated genes in TECs isolated from renal biopsies from healthy individuals and subjects with renal fibrosis. (g) Na+/K+ ATPase activity assay, Pi: inorganic phosphate. (h) Relative expression of the indicated genes in HK2 cells with and without TGF-β1 treatment, n = 3. (i) Na+/K+ ATPase activity assay of HK2 cells treated with vehicle or TGF-β1. (j) Transporter uptake assays of vehicle or TGF-β1–treated HK2 cells cultured with radio–labeled substrates in the presence or absence of unlabeled inhibitors. Data is presented as mean ± SEM. Vehicle (vh) and TGF-β1 treatment were conducted for 72 hours, n = 3. a and f, error bars denote variation in technical replicates, all other error bars depict variation in biological replicates. d,e and h,i, unpaired two–tailed t–test was used. *P < 0.05, **P < 0.01, **P < 0.001, ****P < 0.0001. (3H)PAH: 3H–p–aminohippuric acid, (3H)ES: 3H–estrone sulfate, (14C)TEA: 14C–tetraethylammonium, UNL(TEA): unlabeled tetraethylammonium.
Previous studies demonstrated that decreased expression of organic anion transporters OAT1 and OAT3 was linked to accelerated oxidative stress and inflammation30. We assayed expression of OAT1 and OAT3 as well as amino acid transporter SLC7A9 and sodium–dependent phosphate transporter SLC34A1, as these transporters were among 16 candidates associated with a decreased glomerular filtration rate identified via metanalysis of genome-wide association study data of cohorts from the Heart and Ageing in Genomic Epidemiology and CKDGen consortia32 and subsequent clustering using known transcriptional networks33. Our analyses showed that transcript and protein levels of selected TEC transporters were deregulated in kidney biopsies (Fig. 3a–e and Supplementary Fig. 10c–f) from fibrotic kidneys of individuals when compared to normal kidneys, including AQP1, ATPB1, SLC22A6 (Fig. 3a), and ATP1A1, SLC22A4, SLC22A8, SLC16A3, SLC22A1, SLC22A2, SLC22A3, SLC7A9, SLC34A1, SLCO4C1, SLC22A7, while SLC22A11, SLC16A9, SLC16A12, and SLC22A13 expression was unchanged (Supplementary Fig. 10c–d). ATP1B1, which encodes the β1 essential subunit of the Na+/K+–ATPase pump, and AQP1 are predominantly expressed in TECs, and their relative expression is significantly downregulated in human biopsies with various histology and renal fibrosis (Fig. 3a–e). Gene expression analyses also indicate a deregulation of transporter expression in TECs from fibrotic kidneys (TEC548 and TEC517) compared to healthy kidneys (TEC555 and TEC554), and a specific deregulation in AQP1, ATP1B1, SLC22A6 expression (Fig. 3f), as well as other transporters (Supplementary Fig. 10g). SLCL7A9, SLCO4C1 and OCTN1 protein levels were also specifically deregulated in fibrotic TECs (Supplementary Fig. 10h). Supporting the decreased in ATP1B1 transcript levels, the function of the Na+/K+–ATPase pump was reduced in TECs from fibrotic kidneys compared to TECs from healthy kidneys (Fig. 3g). Fibrotic TEC changes in transporter transcript, protein, and activity levels, were associated with an EMT program (Fig. 3f and Supplementary Fig. 10i). Similar patterns of gene expression were also noted in the TEC cell line HK2, in which the treatment with TGF-β1 results a robust induction of an EMT program (assayed by gene expression changes (Fig. 3h and Supplementary Fig. 11a), morphological changes, and β–catenin nuclear translocation (Supplementary Fig. 11b), with concomitant deregulation in the expression of TEC transporters (Fig. 3h and Supplementary Fig. 11c – d) and a significant decrease in Na+/K+–ATPase activity (Fig. 3i). TGF-β1–induced EMT in HK2 cells was associated with intracellular accumulation of transporter–specific substrates as a result of decreased efflux via OCT1 and OCT3 and increased influx via OCTN1 (Fig. 3j), supporting that an EMT program induces the loss of TEC transporter functions (Supplementary Fig. 11f).
Among the upregulated transporters, MCT4 expression was increased in fibrotic kidney biopsies (Supplementary Fig. 10c,e) consistent with previous reports in which MCT4 was found upregulated in several human kidney diseases34–37. MCT4 mediates lactic acid efflux and is associated with lactate accumulation and acid microenvironment, both pro–fibrotic stimuli38,39. The up–regulation of MCT transporters (Supplementary Fig. 11c) in TGF-β1–stimulated HK2 cells was associated with an increase in lactate levels and decreased pH in cell culture media (Supplementary Fig. 11e). In addition, organic cation transporter–novel 1 (OCTN1) is associated with chronic inflammation and is directly induced by inflammatory cytokines40. A recent study demonstrated that over–expression of SLCO4C1 in transgenic rats reduced accumulation of uremic toxins, ameliorating renal inflammation and interstitial fibrosis41. We show that an EMT program in TGF-β1–exposed HK2 cells was associated with decreased SLCO4C1 expression (Supplementary Fig. 11c).
Inhibition of EMT decreases immune infiltration
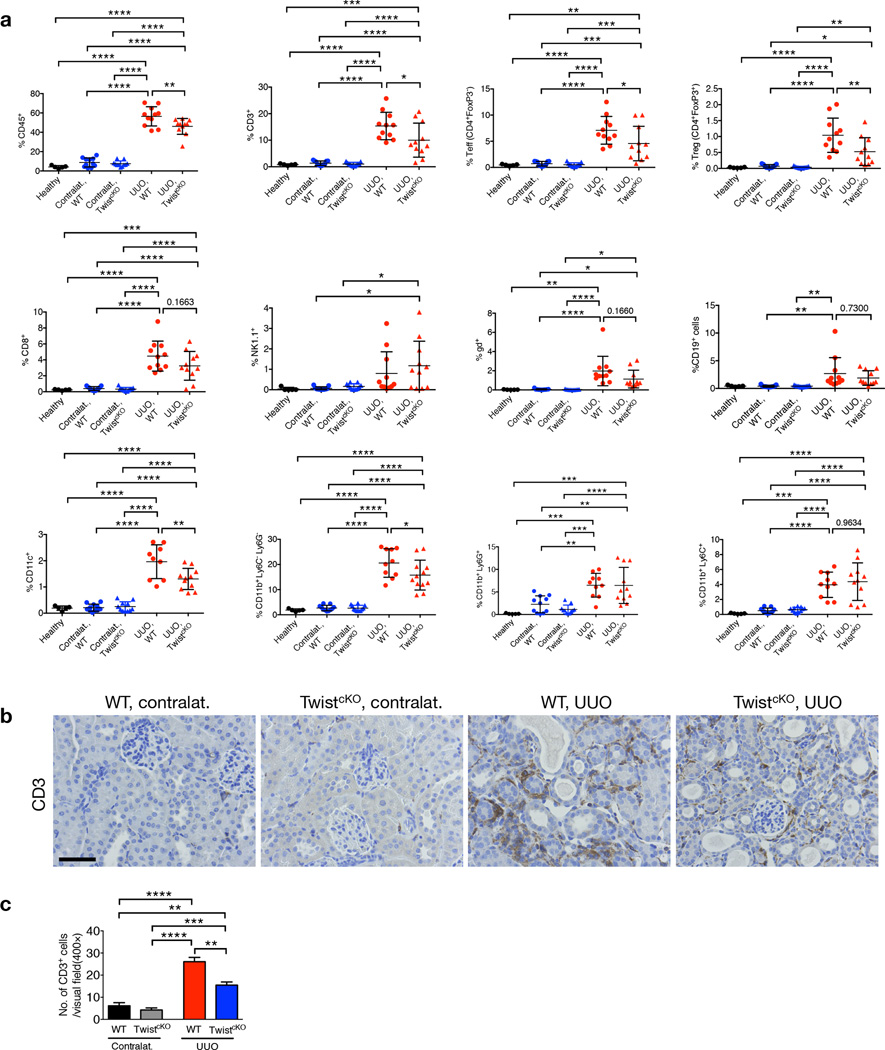
Inflammation has been widely described to be a key driver of kidney fibrosis42. Initiated as a protective response to injury, the immune infiltrate evolves and may contribute to fibrotic scarring, leading to the loss of functional parenchyma. We assayed by flow cytometry and immunostaining the differential composition of the immune infiltrate in WT and TwistcKO UUO kidneys compared to contralateral and to kidneys from healthy mice. While contralateral and healthy kidneys showed similar immune infiltrate (Fig. 4a–c), UUO kidneys showed a significant increase in CD45+ leukocyte infiltration (Fig. 4a). The percent CD3+ T cells, both effector (CD4+Foxp3−) and regulator (CD4+Foxp3+) T cells, cytotoxic CD8+ T cells, natural killer cells (NK1.1+), gamma delta (γδ+) T cells, CD11c+ dentritic cells, and CD19+ B cells were significantly increased in fibrotic kidneys compared to contralateral kidneys in WT mice (Fig. 4a).
Figure 4.
Inhibition of EMT reduces immune infiltration in kidney fibrosis. (a) Percentages of CD45+, CD3+, CD4+FoxP3− Teff, CD4+FoxP3+ Treg, CD8+, NK cells, CD11c+, γδ+, CD19+, CD11b+Ly6C−Ly6G−, CD11b+Ly6G+, CD11b+Ly6C+ in the indicated experimental groups. Healthy, n = 5; WT contralat., n = 11; TwistcKO contralat., n = 11; WT UUO, n = 11; TwistcKO UUO, n = 11. (b) Representative images (8 visual fields for each tissue analyzed) of immunolabelling for CD3 in the indicated experimental groups. Scale bar, 50 ~m. (c) Quantification of the number of CD3+ cells per visual field in the indicated experimental groups. WT contralat., n = 4 ; TwistcKO contralat., n = 4; WT UUO, n = 4; TwistcKO UUO, n = 4. For panel a data is presented as mean ± SD; for c data is presented as mean ± SEM. One–way ANOVA with Tukey post–hoc analysis was used. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. contralat.: contralateral kidney.
In agreement with histopathological improvement noted in fibrotic kidneys from TwistcKO UUO-treated mice, the higher degree of CD45+ leukocyte infiltration was suppressed when compared to kidneys from WT UUO-treated mice (Fig. 4a). Specifically, fibrotic kidneys from TwistcKO UUO-treated mice showed a significantly lower degree of T cell infiltration (CD4+Foxp3−, CD4+Foxp3+, and cytotoxic CD8+ T cells, except γδ+ T cells) and CD11c+ dendritic cells compared to fibrotic kidneys from WT UUO-treated mice, while NK and B cells were not changed. Kidney fibrosis was also associated with an influx of macrophages (CD11b+Ly6C−Ly6C−), and CD11b+Ly6C+ and CD11b+Ly6G+ myeloid–derived suppressor cells, and fibrotic kidneys from TwistcKO UUO-treated mice showed suppressed macrophage infiltration compared to fibrotic kidneys from WT UUO mice (Fig. 4a). The improved kidney histopathology and renal functions in TwistcKO and SnailcKO mice challenged with UUO was thus associated with a suppressed recruitment of pro-inflammatory cells. Expression of pro–inflammatory cytokines was higher in fibrotic kidneys from WT UUO-treated mice compared to contralateral kidneys, and the expression of cytokines transcripts was lower in fibrotic kidneys from TwistcKO and SnailcKO UUO-treated mice compared to fibrotic WT UUO-treated mice (Supplementary Fig. 11g). Immunolabeling for interstitial CD3+ T cells and F4/80+ macrophages also supports a decrease in T cells when suppressing EMT in kidney fibrosis in both TwistcKO and SnailcKO UUO-treated mice (Fig. 4b–c and Supplementary Fig. 11h–i). Altogether these results support that a specific decrease in EMT in TECs of fibrotic kidneys results in improved histopathology associated with suppressed immune infiltration.
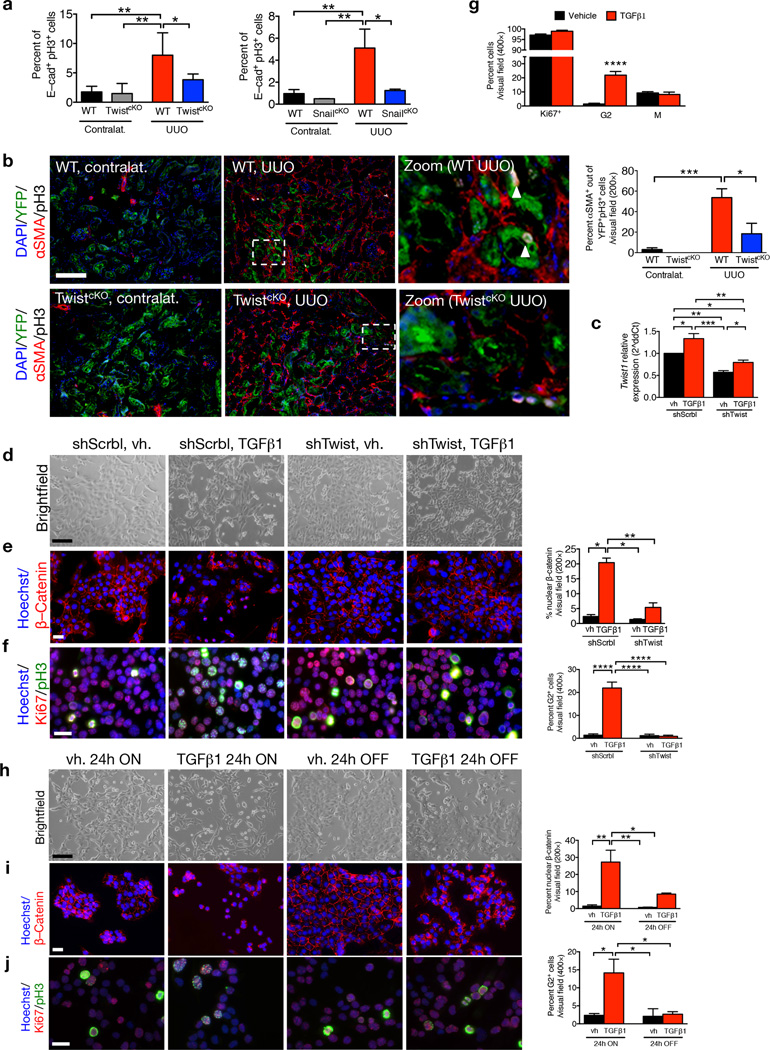
EMT arrests tubular epithelial cells in the G2 phase
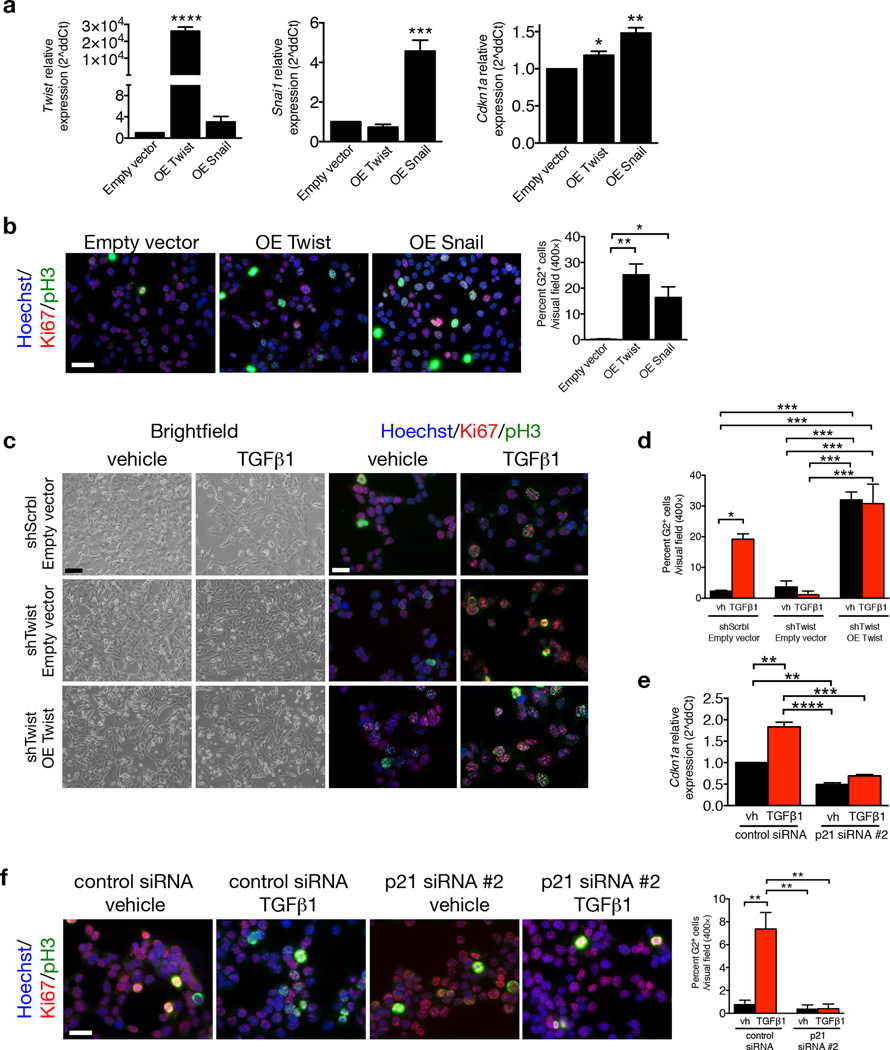
Chronic kidney disease is associated with inadequate cell cycle progression of the tubular epithelial cells, which arrest in the G2/M phase25,26,43,44. TGF-β1 stimulation of mouse TECs also results in a cell cycle arrest at G2/M phase45. We probed whether acquisition of an EMT program in the injured nephron may contribute to G2/M arrest, limiting their regenerative potential. Fibrotic kidneys from WT UUO-treated mice exhibit greater phospho–histone H3 (pH3) expression in TECs compared to fibrotic kidneys of TwistcKO and SnailcKO UUO-treated mice, as evaluated by flow cytometry (Fig. 5a and Supplementary Fig. 12a–b) and immunohistochemistry analyses (Fig. 5b and Supplementary Fig. 12c–f and Supplementary Fig. 13), indicative of a prolonged G2/M phase of the cell cycle. The overall number of proliferating TECs (those that are Ki67+) and cells in the S–phase of the cell cycle (those that are BrdU+) were unchanged in WT, TwistcKO and SnailcKO mice (Supplementary Fig. 12e–f). These results suggest that the greater number of pH3+ TECs in fibrotic kidneys WT vs. TwistcKO and SnailcKO UUO-treated mice likely reflected a prolonged arrest of TECs in G2/M phase, and suppressing the EMT program relieves this brake. We noted that αSMA expression is enriched in pH3+ TECs (Fig. 5b and Supplementary Fig. 13), indicating that cells undergoing EMT are delayed in G2/M phase. These data support the notion that proliferation of epithelial cells is, in part, mutually exclusive from their acquisition of mesenchymal features via an EMT program. Similar findings were noted in vitro, employing a mouse proximal tubular epithelial cell line (MCT46). EMT induction via TGF-β1 stimulation, as assayed by gene expression analyses (Fig. 5c and Supplementary Fig. 14a), morphological changes (Fig. 5d) and β–catenin nuclear accumulation (Fig. 5e), induced a G2–specific arrest with a characteristic punctate nuclear staining of pH3+Ki67+ cells (Fig. 5f–g and Supplementary Fig. 14b–e). The EMT-induced G2 arrest was reversible when EMT induction by TGF-β1 was relieved (24h OFF TGF-β1 after a 24h ON TGF-β1 stimulation (Fig. 5h–j). RNAi–mediated targeting of Twist1 and Snai1 expression (shTwist and shSnail) prevented acquisition of TGF-β1–induced EMT features (Fig. 5d–e and Supplementary Fig. 15a) and G2 arrest (Fig. 5f–g and Supplementary Fig. 15a). Over–expression of Twist and Snail was sufficient to induce p21 expression (Fig. 6a) and G2 arrest (Fig 6b). The re–expression (rescue) of Twist and Snail in MCT shTwist and MCT shSnail, respectively, re–induced a G2 block, supporting that the TGF-β1–induced G2 arrest is indeed mediated by Twist or Snail (Fig. 6c–d and Supplementary Fig. 15b–c). Insignificant changes in the proliferation rate of MCT cells – with or without Twist1 suppression – by TGF-β1 stimulation were detected (Supplementary Fig. 15d). DNA damage was not observed as analyzed by comet assay (Supplementary Fig. 15e) and TUNEL labeling of kidneys from WT and TwistcKO UUO mice (Supplementary Fig. 15f).
Figure 5.
EMT program G2 cell cycle arrest. (a) Percent E–cadherin+pH3+ cells in kidneys from the indicated experimental groups. WT contralat., n = 4; TwistcKO contralat., n = 5; WT UUO, n = 4; TwistcKO UUO, n = 5. WT contralat., n = 3; SnailcKO contralat., n = 3; WT UUO, n = 4; SnailcKO UUO, n = 3. (b) Representative images (5 visual fields for each tissue analyzed) of immunolabeling for YFP, AQP1 and pH3 and percentage of αSMA+ cells out of the YFP+pH3+ TECs (contralat., n = 6; UUO n = 3). Scale bar, 100 ~m. (c) Relative Twist1 expression in indicated cells. (d) Representative images of brightfield imaging in indicated cells. (e) Representative images (3 visual fields for each tissue analyzed) of immunolabeling for β–catenin and percent nuclear accumulation. Scale bar, 50 ~m. (f,g) Representative images (3 visual fields for each tissue analyzed) of immunolabeling for Ki67 and pH3 (f) and percent cells in G2 (g). Scale bar, 50 ~m. (h) Representative images of brightfield imaging in indicated cells. (i) Representative images (3 visual fields for each tissue analyzed) of immunolabeling for β–catenin and percent nuclear accumulation. Scale bar, 50 ~m. (j) Representative images (3 visual fields for each tissue analyzed) of immunolabeling for Ki67 and pH3 (f) and percent cells in G2 (g). Scale bar, 50 ~m. Vehicle (vh) and TGF-β1 treatment were conducted for 24 hours, followed by vehicle or TGF-β1 withdrawal for 24 hours, n = 3. Data is represented as mean ± SEM. Hoechst: nucleus. One–way ANOVA with Tukey post–hoc analysis. b, unpaired one–tailed t–test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure 6.
p21 controls EMT program G2 cell cycle arrest. (a) Relative expression of Twist1 Snai1 and Cdkn1a (p21) in MCT cells transfected with empty vector, Twist or Snail overexpression vectors, 24 hours post transfection, n = 3. (b) Representative images (3 visual fields for each tissue analyzed) of vehicle or MCT cells transfected with empty vector, Twist or Snail overexpression vectors, 24 hours post transfection, immunolabeled for Ki67 and pH3 and respective quantification of the percentage of cells in G2 phase, n = 3. Scale bar: 50 ~m. (c) Phase contrast light microscopy and immunolabeling for Ki67 and pH3 of MCT shScrbl and MCT shTwist cells transfected with empty or Twist overexpression (OE Twist) plasmids and treated with vehicle or TGF-β1. (d) Quantification of the percentage of empty or Twist OE transfected MCT shScrbl and MCT shTwist cells in G2 phase of the cell cycle comparing cells treated with vehicle or TGF-β1, n = 3. (e) Relative expression of Cdkn1a (p21). (f) Representative images (3 visual fields for each tissue analyzed) of immunolabeling for Ki67 and pH3 of control or p21 siRNA–transfected MCT cells and percent of cells in G2 phase. Scale bar, 50 ~m. Data is represented as mean ± SEM. Hoechst: nucleus. One–way ANOVA with Tukey post–hoc analysis. b and e, unpaired one–tailed t–test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Over–expression of Snai1 in cultured canine kidney epithelial cells (MDCK) induced a cell cycle arrest involving enhanced p21WAF1/CIP1 expression47. Over–expression of Twist and Snail induced p21 expression and G2 arrest in MCT cells (Fig. 6a). Absence of p21WAF1/CIP1 (using Cdkn1a knockout mice) improved progression of kidney disease in the remnant kidney model in association with increased TEC proliferation48. Here, TGF-β1–treated MCT cells arrest in G2 is concomitant with a higher expression of p21 expression (Fig. 6e–f), and RNAi–mediated silencing of p21 prevented TGF-β1–induced G2 arrest (Fig. 6f), supporting that p21 plays a regulatory role in the EMT–induced cell cycle arrest of TECs. Collectively, these results indicate that TGF-β1–mediated induction of an EMT program is associated with a p21–mediated G2 phase cell cycle arrest that requires, at least in part, Twist1 and Snai1, but is independent of DNA damage–associated cell cycle check point activation.
DISCUSSION
These results provide an opportunity to consider a working model that incorporates acquired EMT program as a component of the early initiating damage to the functional renal parenchyma (Supplementary Fig. 15g). It is conceivable that EMT program observed during early embryogenesis and organ development results in the generation of fully functional mesenchymal cells, but in diseases such as organ fibrosis and cancer, a complete phenotypic conversion might be rare with injured epithelial cells exhibiting a more hybrid phenotype with partial EMT program18,49. Kidneys can encounter many different types of injuries that can damage their TECs. In response to injury, TECs release growth factors, cytokines, chemokines and matrix metalloproteinases (MMPs) to initiate host repair and regenerative response. Such a response results in vasodilation, basement membrane remodeling, and initial influx of immune cells, such as macrophages30,33. Damaged TECs are vulnerable and experience apoptotic pressure and other stressors such as macrophage-generated TGF-β1, among other cytokines30. Injury to TECs results in loss of functional parenchyma and also evasive survival mechanisms such as an initiation of an EMT program mediated via TGF-β1–induced expression of Twist1 and Snai19,30. But the EMT program further damages TECs, leading to a compromise of their functional capabilities, generation of a pathological secretome and induction of cell cycle arrest. These events further augment the host injury response, resulting in robust immune reaction and recruitment of myofibroblasts. Apoptosis and an EMT program of TECs lead to a vicious cycle of damage and host response, leading to chronic fibrosis. Prevention of the acquisition of an EMT program in injured TECs results in restoration of functional TECs, decreased pathological secretome and alleviation from cell cycle arrest, facilitating improved organ function26,50. Subsequently, a decrease in myofibroblasts and ECM accumulation also occurs.
Here we show that a fibrotic injury–induced EMT program in TECs leads to a p21–mediated G2 cell cycle arrest, depletion in several TEC solute and solvent transporter genes such as Na+/K+–ATPase pump, AQP1, SLC22A6 and anion and cation transporters, among others. TECs undergoing EMT thus emerged with a pronounced G2 cell cycle arrest and deregulated transporter activities. Induced Twist or Snai1 expression in TECs is sufficient to induce p21–mediated G2 arrest, and promotes prolonged TGF-β1–induced cell cycle arrest in G2 phase, limiting their potential for repair and regeneration and exacerbating chronic fibrosis. Mice with the conditional deletion of EMT–inducing transcription factors, Twist1 or Snai1, in proximal TECs were subjected to kidney fibrosis. Inhibition of an EMT program led to protection of TEC integrity, restored proliferation and de–differentiation–associated repair and regeneration, attenuated myofibroblast accumulation, fibrosis, and immune infiltration. Inhibition of an EMT program in TECs protects functional kidney parenchyma by facilitating cell cycle–dependent repair, regeneration of the fibrotic kidney and informs potential anti–fibrosis therapies.
Our results support the studies in liver fibrosis where specific deletion of Snai1 in hepatocytes and subsequent induction of CCL4–induced liver injury led to attenuation of liver fibrosis27. In summary, our experiments offer evidence for the functional relevance and importance of an EMT program in the progression of chronic kidney injury and support the notion of targeting the EMT program as a viable therapeutic strategy for protecting functional parenchyma in kidney fibrosis.
ONLINE METHODS
Animal studies
γGT–Cre mice were previously described3,6. SnailLoxP/LoxP mice were kindly provided by S.J. Weiss, University of Michigan, Ann Arbor. TwistLoxP/LoxP mice were kindly provided by R. R. Behringer (UT MDACC, Houston, TX via the Mutant Mouse Regional Resource Center (MMRRC) repository. R26R–LoxP–Stop–LoxP–EYFP (Rosa26–LSL–EYFP) were purchased from Jackson Laboratories. αSMA–RFP mice were described in51. TwistLoxP/LoxP and SnailLoxP/LoxP mice were bred with γGT–Cre mice to generate mice with a deletion of Twist or Snail transcription factors in the proximal tubular cells. γGT–Cre+;TwistLoxP/wt showed a wild–type like phenotype and were included with γGT–Cre−;TwistLoxP/LoxP mice and compared to γGT–Cre+;TwistLoxP/LoxP mice. γGT–Cre−;SnailLoxP/LoxP mice were compared to γGT–Cre+;SnailLoxP/LoxP mice. All mice were on a mixed 129/Sv, C57BL/6 and Balb/c genetic background and littermates with similar genetic background were analyzed. 10–14 weeks male and female mice were used for all experiments. The sample size was chosen based on our previous experience and for each experiment it is indicated in the figure legend. UUO and NTN renal fibrosis models were performed as previously described3. UUO mice were euthanized at 5 or 10 days post surgery and NTN mice were euthanized 9 weeks after the injection of nephrotoxic serum (kindly provided by Dr. Salant, Boston University, Boston, MA). Folic acid induced nephropathy was performed by daily i.p. injection of 150 mg/kg BW folic acid (Sigma–Aldrich) dissolved in 150 mM sodium bicarbonate, for three weeks. One hour prior to sacrifice mice were injected i.p. with 40 mg/Kg BrdU (Sigma). BUN measurement was performed on mouse serum samples collected at the time of the sacrifice using the Urea Assay Kit (Cell Biolabs), according to manufacturer’s instructions. All animal procedures were reviewed and approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee. Investigators were not blinded for group allocation but were blinded for the assessment of the phenotypic outcome assessed by histological analyses. No randomization method was used and no animals were excluded from the analysis.
Histology and Histopathology
Kidneys were fixed in 10% neutral buffered formalin and embedded in paraffin. 5 ~m sections were cut for hematoxylin and eosin (H&E) and Masson’s trichrome (MTS) stainings (Leica). Picrosirius red staining for collagen was performed using 0.1% picrosirius red (Direct Red 80, Sigma) and counterstained with Weigert’s hematoxylin. The extent of renal injury was estimated by morphometric assessment of tubular damage and interstitial fibrosis. To estimate the protection from tubular damage in UUO, NTN and folic acid mice, eight 200× visual fields were randomly selected for each slide and the number of healthy tubules was manually counted using the count tool of Adobe Photoshop. Tubules were defined as healthy when the dimension, structure, relative nucleus–cytoplasm disposition, integrity of the brush border and of the basal membrane resemble those of normal tubules from healthy kidneys. For the analysis of the interstitial fibrosis in UUO, NTN and folic acid mice, eight 200× visual fields were also randomly selected for each MTS and picrosirius red stained kidney section and interstitial fibrosis was manually evaluated by a grid intersection analysis using Adobe Photoshop. Representative images were acquired with Leica DM 1000 LED microscope and the MC120 HD Microscope Camera with Las V4.4 Software (Leica).
Immunohistochemistry
Formalin–fixed paraffin–embedded kidney sections 5 ~m thick were deparaffinized, rehydrated and antigen retrieved at 98 °C for 10 minutes in 10 mM citrate buffer pH 6. The tissue sections were incubated with 4% CWFS gelatin (Aurion) in TBS or PBS for 1 hour prior to the overnight incubation with the primary antibody. The following primary antibodies were used: anti–rabbit Ki67 (Thermo Scientific, RM–9106–S, 1:400), anti–mouse phospho–histone H3 (Abcam, ab14955, 1:3,000), anti–mouse BrdU (BD, 347580, 1:100), anti–mouse αSMA (Sigma, A5228, 1:400), anti–rabbit Na+K+ ATPase (Abcam, ab58475, 1:100), anti–rabbit AQP1 (Alpha Diagnostic, AQP11–A, 1:300), anti–rabbit SLC22A6 (Abcam, ab183086, 1:100), anti–rabbit CD3 (Dako, A0452, 1:100), and anti–rat F4/80 (Abcam, ab6640, 1:200). Staining for αSMA, BrdU and phospho–histone H3 were processed using the M.O.M. Kit (Vector Laboratories) according to the manufacturer’s recommendations. CD3 staining was processed using anti–rabbit HRP–polymer (Biocare, MACH4). For all other stainings, sections were incubated with biotinylated goat anti–rabbit and streptavidin HRP (Biocare Medical), each for 10 minutes. For all stainings, counterstaining with haematoxylin was performed and DAB positivity was analyzed in eight visual fields at 200× magnification. Ki67, BrdU and phospho–histone H3 (pH3) stainings were quantified by counting the number of tubular epithelial cells displaying positive nuclei, per visual field (400×). Na+K+ ATPase, AQP1 and SLC22A6 stainings were quantified by counting the number of positive tubules, per visual field (200×). αSMA staining was quantified with ImageJ. TUNEL assay was performed on paraffin sections using the In situ cell death detection kit, TMR Red (Roche) according to manufacturer’s instructions. For multiplexed EMT and AQP1 or pH3 staining, FFPE slides were deparaffinized and antigens were retrieved using a heated citric acid buffer for 15 min. Slides were serial stained with anti–mouse αSMA (DAKO, M0851, 1:2,000; LifeTechnologies SuperPicture–HRP polymer; PerkinElmer TSA–Cy5), anti–YFP (Abcam, ab13970, 1:3,000; Life Technologies chicken–HRP 1:1,000; PerkinElmer TSA–Cy3), anti–rabbit AQP1 (Alpha Diagnostic, AQP11–A, 1:2,000; LifeTechnologies SuperPicture–HRP polymer; PerkinElmer TSA–Fluorescein), anti–mouse phospho–histone H3 (Abcam, ab14955, 1:6,000; LifeTechnologies SuperPicture–HRP polymer; PerkinElmer TSA–Fluorescein) and DAPI (LifeTechnologies, 1:20,000). Slides were scanned at 200× magnification, using the PerkinElmer Vectra multispectral slide scanning system. Five random fields for each mouse were selected and the number of YFP+ tubule cells were scored for the presence or absence of αSMA and/or AQP1 co–staining or αSMA and/or pH3 co–staining. Quantification was performed using PerkinElmer InForm Analysis Software. Pseudocoloring and non–linear adjustment (gamma changes) were applied in order for the printed images to accurately represent the quality of the high resolution screens. For Na+K+ ATPase and Col IV immunolabelling of human biopsies, formalin–fixed, HOPE–embedded kidney sections (5µm thickness) were deparaffinized, rehydrated and steam–boiled for 40 minutes in 10 mM citrate buffer pH 6. The tissue sections were incubated with 1% bovine serum albumin (Sigma Aldrich) in PBS for 1 hour prior to the overnight incubation with the primary antibody anti–rabbit Na+K+ ATPase (Abcam, ab58475, 1:100) at 4 °C. The day after, an AlexaFluor568–labeled secondary antibody (Invitrogen, 1:200) was used and slides were incubated for 45 minutes at ambient temperature. An AlexaFluor488–labeled antibody raised against type IV Collagen (SouthernBiotech, 1340–30, 1:10) was used for 3 hours at ambient temperature to visualize the basal membrane and slides were mounted with Vectashield Mounting Medium with DAPI (Vectashield) and a glass coverslip. Na+K+ ATPase staining was quantified by counting the number of positive tubules per 5 visual fields (400×) and representative confocal pictures are shown in 400× magnification.
YFP and RFP visualization and immunofluorescence
Mouse kidneys were fixed in 4% paraformaldehyde overnight at 4°C and equilibrated in 30% sucrose overnight at 4°C. Kidneys were then embedded in OCT compound, and 5 ~m–thick frozen sections were blocked for one hour with 5% normal goat serum and immunostained overnight with anti–αSMA–Cy3 (Sigma–Aldrich, C6198, 1:200). Slides were then mounted with Vectashield Mounting Medium with DAPI (Vectashield) and a glass coverslip and visualized using a YFP and RFP fluorescent filter. The number of double YFP and αSMA and of single YFP positive tubular cells per visual field (400×) was counted in three fields of view. For cell cycle analysis, MCT cells adhered to coverslips were fixed with ice cold acetone for 10 minutes at room temperature and then blocked with 4% CWFS gelatin (Aurion) in PBS for 1 hour prior to the incubation with anti–phospho histone H3 (Abcam, ab14955, 1:1000) and anti–Ki67 eFluor570 (eBioscience, 41–5698, 1:500) for 1 hour. Cells were then incubated with anti AlexaFluor488 anti–mouse (Invitrogen, 1:800) and with Hoechst (Invitrogen, 1:10,000 in PBS) for nuclear staining. For cell cycle profiling, four fields of view (400×) were counted and quantified as follows: percentage of Ki67+ cells was calculated as ratio between number of Ki67+ cells and total number of cells stained with the nuclear dye Hoechst, per field of view; percentage of G2+ cells was calculated as ratio between number of cells in the G2 phase, identified for the characteristic phospho–histone H3 punctate nuclear staining (see Supplementary Fig. 14b), and number of Ki67+ cells per field of view; percentage of cells in M phase was calculated as the ratio between the sum of cells in prophase (P), metaphase (M), anaphase (A) and telophase (T) (each phase identified by the unique pattern of phospho–histone H3 staining, see Supplementary Fig. 14b) and number of Ki67+ cells per field of view. For the analysis of β–Catenin localization, MCT cells adhered to coverslips were fixed with 100% Methanol 5 minutes at –20 °C and then blocked in blocking buffer (0.1% Tween, 1%BSA, 10% normal goat serum, 0.3M glycine in PBS) for one hour at room temperature prior to incubation with anti–β–Catenin (Abcam, ab16051, 1 ~g/ml diluted in 0.1% Tween, 1% BSA PBS) overnight at 4 °C. the following day cells were incubated with anti AlexaFluor594 anti–rabbit (Invitrogen, 1:250) and with Hoechst for nuclear staining. Representative images were acquired with Demo Axio Observer.Z1 motorized inverted microscope with Axiocam 506 monochrome camera and ZEN software (Zeiss). In order for the printed images to accurately represent the quality of the high resolution screens, the color brightness was modified by using Adobe Photoshop and was equally applied across the entire image and equally increased in all the acquired images.
Flow cytometry
For the characterization of immune infiltration, one quarter of contralateral and one quarter of UUO kidneys were minced and allowed to digest in a 2 ml mixture of collagenase (400 U type II collagenase; Worthington) in DMEM media at 37 °C for 25 minutes. The tissue lysate was filtered through a 100 nm mesh prior to immunostaining. The resulting single–cell suspension was stained with fixable viability dye eFluor 780, anti–CD45.2 Pacific Blue, anti–CD3 PE–Cy7, anti–CD3 Alexa Fluor 700, anti–FoxP3 Alexa Fluor 700, anti–CD11c eFluor 615, and anti–NK1.1 PE (all from eBioscience); anti–Granzyme B APC and anti–CD4 Qdot605 (Life Technologies); anti CD8 Brilliant Violet 650, anti–CD11b BrilliantViolet 570, anti–CD19 Brilliant Violet 650 (all from BioLegend); and anti–Ly6C APC, anti–Ly6G PE–Cy7, and anti–Ki–67 FITC (BD Biosciences). The CD45+ cells were analyzed for the expression of the various markers using FlowJo. Doublets were gated out using forward–scatter width/height and side–scatter width/height event characteristics.
For EMT and P–H3 analysis, one quarter of contralateral and one quarter of UUO kidneys were minced and allowed to digest in a 2 ml mixture of collagenase (400 U type II collagenase; Worthington) in DMEM media at 37 °C for 25 minutes. The tissue lysate was filtered through a 100 mm mesh prior to immunostaining. One fifth of the resulting single–cell suspension was incubated with anti–Ecadherin eFluor660 (eBioscience, 50–3249, 1:100 in PBS–1%BSA) overnight at 4 °C. The following day cells were washed twice with PBS–1%BSA and then fixed and permeabilized using the FoxP3 staining buffer set (BD Bioscience, 00–5523), according to manufacturer’s instructions. Half of the cells were incubated with anti–αSMA–Cy3 (Sigma–Aldrich, C6198, 1:100 in permeabilization buffer) or with anti–pH3–PE (Cell Signaling, 5764, 1:50 in permeabilization buffer) on ice for one hour. Cells were finally washed twice with permeabilization buffer and resuspended in FACS staining buffer (1% FBS, 0.5 mM EDTA PBS).
Protein extraction and Western blot analysis
Kidneys were homogenized in RIPA lysis buffer (150 mM NaCl, 50 mM Tris–HCl pH 8, 1% Igepal, 0.5% sodiumdeoxycholate, 0.1% SDS) added with a protease inhibitor cocktail (Complete™, Roche). Total protein amount was measured with BCA assay (Thermo Scientific) and 20 ~g of total lysate for each samples were denatured at 95 °C for 10 minutes in Laemli buffer containing β–mercaptoethanol and then separated in 4–12% Bis–Tris gels (Bolt®, Life Technology). Primary antibodies used were: anti–rabbit AQP1 (Alpha Diagnostic, AQP11–A, 1:500), anti–rabbit Na+K+ ATPase (Cell Signaling, 3010, 1:1,000), anti–rabbit SLC22A6 (Abcam, ab183086, 1:1,000), anti–goat GAPDH (Abcam, ab9483, 1:800), and anti–rabbit vinculin (Abcam, ab18058, 1:10,000).
Hydroxyproline assay
Analysis of collagen content was performed in contralateral and UUO kidneys of WT, TwistcKO and SnailcKO mice by using the hydroxyproline assay kit (Sigma–Aldrich), according to manufacturer instructions. Measurements were normalized to total protein amount.
Gene expression profiling and quantitative real–time PCR analysis
Kidneys were homogenized in TRIzol reagent (Invitrogen) and total RNA was extracted according to the manufacter’s directions, from UUO kidneys and contralateral healthy kidneys in γGT–Cre−;TwistLoxP/LoxP (WT) and γGT–Cre+;TwistLoxP/LoxP (TwistcKO) and in γGT–Cre−;SnailLoxP/LoxP (WT) and γGT–Cre+;SnailLoxP/LoxP (SnailcKO) mice and submitted to the Microarray Core Facility at MD Anderson Cancer Center. Gene expression analysis was performed using Mouse Ref6 Gene Expression Bead Chip (Illumina). The Limma package from R Bioconductor52 was used to do quantile normalization of expression arrays and analyze differentially expressed genes between any two sample groups (e.g., TwistcKO_UUO vs WT_UUO) (p ≤ 0.05 and fold change ≥ 1.5). Gene set enrichment analysis (GSEA) was used for the analysis of gene pathways/datasets. Gene expression microarray data was deposited in GEO (GSE60685). For real–time PCR analysis in WT, TwistcKO and SnailcKO kidneys total RNA and in MCT and HK2 cells, cDNA synthesis was performed using the Applied Biosystems cDNA synthesis kit according to the manufacter’s directions. Quantitative PCR was performed to analyze the gene expression profiles of the listed genes using SYBR Green PCR Master Mix in a 7300 Sequence Detector System (Applied Biosystems), and measurements were standardized to the expression of the GAPDH housekeeping gene. For Twist1 expression levels measurements in MCT cells, RNA from vehicle or TGF-β1–treated MCT shScrbl and shTwist cells was extracted with Pure Link RNA Mini Kit (Ambion) and subjected to the one–step RT–qPCR reaction using SuperScript III Platinum Sybr Green One–Step qRT–PCR kit (Invitrogen), according to manufacturer’s instructions. The expression level of Twist1 was normalized to the expression of Gapdh housekeeping gene. The expression data is presented as 1/dCt or fold change (2^ddCt) with control group normalized to a fold value of 1. dCt were used to measure statistical significance in observed changes. Genes and primer sequences are listed below (F: forward, R: reverse primer).
| Gene (human) | Sequence |
|---|---|
| ATP1A1 F | ACAGACTTGAGCCGGGGATTA |
| ATP1A1 R | TCCATTCAGGAGTAGTGGGAG |
| ATP1B1 F | CCGGTGGCAGTTGGTTTAAGA |
| ATP1B1 R | GCATCACTTGGATGGTTCCGA |
| MCT4 F | TACATGTAGACGTGGGTCGC |
| MCT4 R | CACAAGTTCTCCAGTGCCATT |
| MCT9 F | CCGTCAGGCGACTTTTTAAG |
| MCT9 R | CCGGAGGCTTCACAATCTAT |
| MCT12 F | GGAATGCTGCAGTGTAGCTG |
| MCT12 R | ATCAGCGCTTGGTGTGGTAT |
| OAT1 F | GGCAGTCATGCTCACCAGT |
| OAT1 R | CTGTATCCCACAATGATCCG |
| OAT2 F | CACACTCCATCCAGCAAGG |
| OAT2 R | TTGTACCCTACGGTGCTCAG |
| OAT3 F | GCCATGAAGATAGACTGGGC |
| OAT3 R | CTGGGTCTACAACAGCACCA |
| OAT4 F | GAAGATGGTGCTGGTGCC |
| OAT4 R | CTTTATCTGGGGCCTCCTCT |
| OAT10 F | GTGGTGTCCTTCAGGTGCTT |
| OAT10 R | TATGGGCTGGGAATATCCTG |
| SLC7A9 F | CTTTGAGCATGTGACCCTCC |
| SLC7A9 R | TTTTGTGGCATTTTCAACCA |
| SLC34A1 F | CCACAAAGATGTGGTTGCAT |
| SLC34A1 R | GCCACCCAGACTCCTTACAG |
| OCTN1 F | TACGAAGAACAGGGAGGTGG |
| OCTN1 R | GTTCAGCCAGGACGTCTACC |
| SLCO4C1 F | CAACTGGTGCTATTCCTTGTTG |
| SLCO4C1 R | TTGGACTGGGAGCACTTGTA |
| COL1A2 F | GGTGAAGTGGGTCTTCCAGG |
| COL1A2 R | TAAGGCCGTTTGCTCCAGG |
| VIMENTIN F | ATTCCACTTTGCGTTCAAGG |
| VIMENTIN R | CTTCAGAGAGAGGAAGCCGA |
| S100A4 F | TCTTTCTTGGTTTGATCCTGACT |
| S100A4 R | AGTTCTGACTTGTTGAGCTTGA |
| ACTA2 F | AAGCACAGAGCAAAAGAGGAAT |
| ACTA2 R | ATGTCGTCCCAGTTGGTGAT |
| E–CADHERIN F | CATGAGTGTCCCCCGGTATC |
| E–CADHERIN R | CAGTATCAGCCGCTTTCAGA |
| TWIST F | CTCAAGAGGTCGTGCCAATC |
| TWIST R | CCCAGTATTTTTATTTCTAAAGGTGTT |
| SNAIL F | GGCAATTTAACAATGTCTGAAAAGG |
| SNAIL R | GAATAGTTCTGGGAGACACATCG |
| SLUG F | ACTCCGAAGCCAAATGACAA |
| SLUG R | CTCTCTCTGTGGGTGTGTGT |
| GAPDH F | undisclosed |
| GAPDH R | undisclosed |
| Gene (mouse) | Sequence |
|---|---|
| TWIST F | CTGCCCTCGGACAAGCTGAG |
| TWIST R | CTAGTGGGACGCGGACATGG |
| SNAIL F | CACACGCTGCCTTGTGTCT |
| SNAIL R | GGTCAGCAAAAGCACGGTT |
| GAPDH F | AGGTCGGTGTGAACGGATTTG |
| GAPDH R | TGTAGACCATGTAGTTGAGGTCA |
| CDKN1A F | GTGGCCTTGTCGCTGTCTT |
| CDKN1A R | GCGCTTGGAGTGATAGAAATCTG |
| AQP1 F | AGGCTTCAATTACCCACTGGA |
| AQP1 R | GTGAGCACCGCTGATGTGA |
| ATP1B1 F | TCGGGACCATCCAAGTAA |
| ATP1B1 R | TGATGTTTAGCACGTAGGC |
| OAT1 F | CTGATGGCTTCCCACAACAC |
| OAT1 R | GTCCTTGCTTGTCCAGGGG |
| ACTA2 F | GTCCCAGACATCAGGGAGTAA |
| ACTA2 R | TCGGATACTTCAGCGTCAGGA |
| FIBRONECTIN F | GCTCAGCAAATCGTGCAGC |
| FIBRONECTIN R | CTAGGTAGGTCCGTTCCCACT |
| VIMENTIN F | CTTGAACGGAAAGTGGAATCCT |
| VIMENTIN R | GTCAGGCTTGGAAACGTCC |
| COL1A1 F | CTCCTCTTAGGGGCCACT |
| COL1A1 R | CCACGTCTCACCATTGGGG |
| CDH16 F | TTTGCTCTCGGTCCCAAT |
| CDH16 R | AATGCCAAGGTGAGGTAG |
Cell culture, constructs and treatments
MCT (mouse) and HK2 (human) proximal tubular epithelial cells were cultured in DMEM medium supplemented with 10% heat–inactivated fetal bovine serum and penicillin/streptomycin (100 ~g/ml) at 37 °C and 5% CO2. The cells were routinely tested and found free from mycoplasma. For the induction of EMT, MCT cells were treated with 5 ng/ml human recombinant TGF-β1 (R&D Systems) for 24 hours and HK2 cells were treated with 10 ng/ml TGF-β1 for 72 hours in serum free medium. For MCT experiments, control for TGF-β1 treatment employed vehicle (4 mM HCl in H2O with 1 mg/ml BSA). MCT shScrbl (scrambled shRNA) and shTwist were generated by transducing MCT cells with a lentivirus encoding for a modified version of a short harping sequence against GFP (control, Addgene plasmid 10900) or Twist1 (Addgene plasmid 1784)53, respectively. MCT shScrbl and shSnail were generated by transducing MCT cells with a lentivirus encoding a scramble short hairpin sequence or a sequence against Snai1 (pLKO Mission® shRNA, SHC016 and TRCN0000218784, respectively; Sigma–Aldrich). Transduced cells were selected with 0.75 ~g/ml puromycin (Sigma–Aldrich). For Twist and Snail overexpression, MCT cells were transfected with pcDNA3–Twist (kindly provided by Dr. Roberta Maestro, CRO National Cancer Institute, Italy) or pCMV6–Snail (Origene) using Lipofectamine® 2000 (Life Technology) according to manufacturer’s instructions and harvested 24 hours post–transfection. For p21 silencing, MCT cells were transfected with Negative Control siRNA (Qiagen, 1027310) or with Cdkn1a_2 siRNA (Qiagen, SI02652503) using Lipofectamine ®2000 (Life Technology) and harvested 24 hours post–transfection.
MTT assay
MCT cells were seeded in a 96 well plate (3,500 cells/well), treated with vehicle or TGF-β1 for 24 hours and then incubated with 3–(4,5–dimethylthiazol–2–yl)–2,5–diphenyltetrazolium bromide (MTT, Sigma–Aldrich) according to manufacturer’s instructions.
COMET assay
Comet assay was performed as previously described54. Briefly, a single cells suspension of 20,000, vehicle or TGF-β1–treated, MCT shGFP or shTwist cells were included in low–melting agarose and run on a pre–coated agarose–covered slide. After agarose has gelled, slides were incubated with neutral lysis buffer and run at 0.6 V/cm for 30 minutes. Slides were then stained with 2.5 ~g/ml propidium iodide and analyzed under the fluorescent microscope. UV exposure (30 min) was used as positive control using the same number of cells.
Lactate Assay
Lactate concentrations were determined using Lactate Assay Kit (Sigma–Aldrich, St. Louis, USA) according to the manufacturer’s protocol. Briefly, 50 µl of medium was mixed with 46 µl Lactate Assay Buffer, 2 µl Lactate Enzyme Mix and 2 µl Lactate Probe. Colorimetric measurements were performed using a spectrophotometric multiwall plate reader.
ELISA assay
Transporter protein levels were quantified in total kidney lysates and HK2 cells after EMT induction using ELISA assay kits against OCTN1, SLC7A9 (Cloud–Clone, Houston, USA) and SLCO4C1 (Cusabio Biotech, Wuhan, China) according to the manufacturer’s protocol, measurements were done in triplicate.
Na+/K+–ATPase activity assay
ATPase activity was determined by ATPase Colorimetric Assay Kit (Novus Biologicals). Briefly, 100 µg cell lysates were mixed in the reaction buffer which consisted of 50 mmol/L Tris, 2.5 mmol/L MgCl2 and 0.5 mmol/L ATP and total ATPase activity was assayed by measurement of liberated inorganic phosphate (Pi) compared to a standard curve. The resting ATPase activity, except Na+/K+–ATPase, was assayed in the presence of an additional 1 mmol/L ouabain, a specific inhibitor of Na+/K+–ATPase. The Na+/K+–ATPase activity was calculated as the difference between the amounts of inorganic phosphate liberated in the absence of ouabain minus that liberated in the presence of 1 mmol/L ouabain. Activity was expressed as µmol of Pi liberated per mg of protein per hour, after subtraction of a blank run.
Transporter uptake assay
HK2 cells were seeded in collagen–coated 24–well plates at a density of 20,000 cells per well. After 24 hours, cells were treated with 10 ng/ml TGF-β1 in DMEM without additives to induce EMT, HK2 cells in DMEM alone served as controls. TGF-β1 was renewed after 48 hours and transporter measurements were performed after 72 hours. Radio–labeled substrates were dissolved in lactated Ringer’s solution in the absence or presence of unlabeled specific inhibitors. In detail, SLC22A1 (OCT1) uptake was measured using [3H]PAH, SLC22A3 (OCT3) uptake using [3H]ES and SLC22A4 (OCTN1) uptake was measured using [14C]TEA. Inhibition was performed by adding Probenecid (OCT1), excess unlabeled ES (OCT3) or excess unlabeled TEA (OCTN1). Radio–uptake measurements were performed after supernatant removal measuring intra–cellular [3H] (OCT1 and OCT3) or [14C] emission (OCTN1) and done in triplicate. Measurements were normalized to total protein amounts in each well.
Ethical research conduct
The use of parts of kidney biopsies and primary cell cultures for research purposes was approved by the Ethics Committee of the University Medical Center Göttingen, and written consent was obtained from all subjects before kidney biopsy.
Equipment and settings
Brightfield images: representative images at 200× magnification were acquired with Axiovert 200 and Axiocam HRc camera (Zeiss). The brightness was modified by using Adobe Photoshop and was equally applied across the entire image and equally increased in all the acquired images. All images were acquired at 300 dpi (or higher) resolution. Immunohistochemistry: representative images at 200× and 400× magnification were acquired with Leica DM 1000 LED microscope and the MC120 HD Microscope Camera with Las V4.4 Software (Leica). All images were acquired at 300 dpi (or higher) resolution. Immunofluorescence: representative images at 200× and 400× magnification were acquired with Demo Axio Observer.Z1 motorized inverted microscope with Axiocam 506 monochrome camera and ZEN software (Zeiss). Color brightness was modified using Adobe Photoshop and modification was equally applied across the entire image and equally increased in all the acquired images. All images were acquired at 300 dpi (or higher) resolution. Immunofluorescence (PerkinElmer TSA technology): slides were scanned at 200× magnification, using the PerkinElmer Vectra multispectral slide scanning system and quantification was performed using PerkinElmer InForm Analysis Software. Pseudocoloring and non-linear adjustment (gamma changes) were applied to accurately represent the quality of the high resolution screens. All images were acquired at 300 dpi (or higher) resolution.
Statistical analysis
Statistical analyses of immunohistochemical/immunofluorescence quantifications and of qPCR analysis were performed by using one–way ANOVA or unpaired one– or two–tailed Student’s t–test with Welch’s correction with GraphPad Prism (GraphPad Software). Statistical significance was defined as P < 0.05.
Supplementary Material
ACKNOWLEDGMENTS
This work was primarily supported with funds from MD Anderson Cancer Center (MDACC) and partially by the Cancer Prevention and Research Institute of Texas. R.K. is also supported by the US National Institutes of Health Grants CA–155370, CA–151925, DK–081576, DK–55001 and Metastasis Research Center at the MD Anderson Cancer Center (P30CA016672). V.S.L. is supported by the US National Institutes of Health under the award number P30CA016672 and the Khalifa Bin Zayed Al Nahya Foundation. This research was performed in the Flow Cytometry & Cellular Imaging Facility at UT MDACC, which is supported in part by the US National Institutes of Health through MDACC Support Grant CA–016672. This work was in part supported by the Deutsche Forschungsgemeinschaft (equipment grant INST1525/16–1 FUGG). SnailL/L mice were kindly provided by S.J. Weiss, University of Michigan, Ann Arbor, and TwistL/L mice were kindly provided by R. R. Behringer, UT MDACC, Houston, TX via the Mutant Mouse Regional Resource Center (MMRRC) repository. pcDNA3–Twist plasmid was kindly provided by R. Maestro, CRO National Cancer Institute, Italy. We thank E. Lawson for technical help with immunostaining and L. Gibson for the help with breeding and genotyping mice.
Footnotes
ACCESSION NUMBER
The NCBI GEO accession number for the gene expression data reported in this paper is GSE60685.
AUTHOR CONTRIBUTIONS
R.K. conceptually designed the strategy for this study, participated in discussions, provided intellectual input, supervised the studies and wrote the manuscript. V.S.L. designed the study, provided intellectual input, supervised the studies, designed and performed experiments, generated the figures and wrote the manuscript. S.L. designed and performed experiments, collected the data, generated the figures and participated in the writing the manuscript. S.L., V.S.L., B.T., H.S., K.V., J.L.C., C.–C.W., Y.H., B.C.B., T.H., and H.N. performed some experiments and collected data. The data was analyzed by S.L., V.S.L., B.T., J.L.C., C.–C.W. and T.H. J.P.A. and M.Z. participated in discussions, provided intellectual input, supervised the studies and edited the manuscript.
COMPETING FINANCIAL INTERESTS
Dr. James P. Allison is an Inventor of intellectual property owned by the University of California, Berkeley, and licensed to Bristol Meyers–Squibb.
References
- 1.Zeisberg M, Kalluri R. Cellular mechanisms of tissue fibrosis. 1. Common and organ-specific mechanisms associated with tissue fibrosis. Am. J Physiol. Cell Physiol. 2013;304:216–225. doi: 10.1152/ajpcell.00328.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grams ME, et al. Lifetime incidence of CKD stages 3–5 in the United States. Am. J Kidney Dis. 2013;62:245–252. doi: 10.1053/j.ajkd.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugimoto H, et al. Activin-like kinase 3 is important for kidney regeneration and reversal of fibrosis. Nat. Med. 2012;18:396–404. doi: 10.1038/nm.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeBleu VS, et al. Origin and function of myofibroblasts in kidney fibrosis. Nat. Med. 2013;19:1047–1053. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bechtel W, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat. Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwano M, et al. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin. Invest. 2002;110:341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeisberg M, Kalluri R. Fibroblasts emerge via epithelial-mesenchymal transition in chronic kidney fibrosis. Front. Biosci. 2008;13:6991–6998. doi: 10.2741/3204. [DOI] [PubMed] [Google Scholar]
- 8.Zeisberg EM, et al. Fibroblasts in kidney fibrosis emerge via endothelial-to-mesenchymal transition. J Am. Soc. Nephrol. 2008;19:2282–2287. doi: 10.1681/ASN.2008050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K, Nelson CM. New insights into the regulation of epithelial-mesenchymal transition and tissue fibrosis. Int. Rev. Cell Mol. Biol. 2012;294:171–221. doi: 10.1016/B978-0-12-394305-7.00004-5. [DOI] [PubMed] [Google Scholar]
- 11.Zeisberg M, et al. Renal fibrosis: collagen composition and assembly regulates epithelial-mesenchymal transdifferentiation. Am. J Pathol. 2001;159:1313–1321. doi: 10.1016/S0002-9440(10)62518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeisberg M, et al. Renal fibrosis Extracellular matrix microenvironment regulates migratory behavior of activated tubular epithelial cells. Am. J Pathol. 2002;160:2001–2008. doi: 10.1016/S0002-9440(10)61150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeisberg M, Kalluri R. The role of epithelial-to-mesenchymal transition in renal fibrosis. J Mol. Med. (Berl) 2004;82:175–181. doi: 10.1007/s00109-003-0517-9. [DOI] [PubMed] [Google Scholar]
- 14.Burns WC, Kantharidis P, Thomas MC. The role of tubular epithelial-mesenchymal transition in progressive kidney disease. Cells Tissues Organs. 2007;185:222–231. doi: 10.1159/000101323. [DOI] [PubMed] [Google Scholar]
- 15.Teng Y, Zeisberg M, Kalluri R. Transcriptional regulation of epithelial-mesenchymal transition. J Clin. Invest. 2007;117:304–306. doi: 10.1172/JCI31200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kida Y, et al. Twist relates to tubular epithelial-mesenchymal transition and interstitial fibrogenesis in the obstructed kidney. J Histochem. Cytochem. 2007;55:661–673. doi: 10.1369/jhc.6A7157.2007. [DOI] [PubMed] [Google Scholar]
- 17.Strutz F, et al. Role of basic fibroblast growth factor-2 in epithelial-mesenchymal transformation. Kidney Int. 2002;61:1714–1728. doi: 10.1046/j.1523-1755.2002.00333.x. [DOI] [PubMed] [Google Scholar]
- 18.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hertig A, et al. Early epithelial phenotypic changes predict graft fibrosis. J Am. Soc. Nephrol. 2008;19:1584–1591. doi: 10.1681/ASN.2007101160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boutet A, et al. Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO J. 2006;25:5603–5613. doi: 10.1038/sj.emboj.7601421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rastaldi MP. Epithelial-mesenchymal transition and its implications for the development of renal tubulointerstitial fibrosis. J Nephrol. 2006;19:407–412. [PubMed] [Google Scholar]
- 22.Kriz W, Kaissling B, Le Hir M. Epithelial-mesenchymal transition (EMT) in kidney fibrosis: fact or fantasy? J Clin. Invest. 2011;121:468–474. doi: 10.1172/JCI44595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeisberg M, Duffield JS. Resolved: EMT produces fibroblasts in the kidney. J Am. Soc. Nephrol. 2010;21:1247–1253. doi: 10.1681/ASN.2010060616. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y. New insights into epithelial-mesenchymal transition in kidney fibrosis. J Am. Soc. Nephrol. 2010;21:212–122. doi: 10.1681/ASN.2008121226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, et al. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat. Med. 2010;16:535–543. doi: 10.1038/nm.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canaud G, Bonventre JV. Cell cycle arrest and the evolution of chronic kidney disease from acute kidney injury. Nephrol. Dial. Transplant. 2015;30:575–583. doi: 10.1093/ndt/gfu230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowe RG, et al. Hepatocyte-derived Snail1 propagates liver fibrosis progression. Mol. Cell. Biol. 2011;31:2392–2403. doi: 10.1128/MCB.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang HM, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajasekaran SA, et al. Na,K-ATPase subunits as markers for epithelial-mesenchymal transition in cancer and fibrosis. Mol. Cancer Ther. 2010;9:1515–1524. doi: 10.1158/1535-7163.MCT-09-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poesen R, et al. Renal clearance and intestinal generation of p-cresyl sulfate and indoxyl sulfate in CKD. Clin. J Am. Soc. Nephrol. 2013;8:1508–1514. doi: 10.2215/CJN.00300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hills CE, Willars GB, Brunskill NJ. Proinsulin C-peptide antagonizes the profibrotic effects of TGF-beta1 via up-regulation of retinoic acid and HGF-related signaling pathways. Mol. Endocrino. 2010;24:822–831. doi: 10.1210/me.2009-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kottgen A, et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martini S, et al. Integrative Biology Identifies Shared Transcriptional Networks in CKD. J Am. Soc. Nephrol. 2014;25:2559–2572. doi: 10.1681/ASN.2013080906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reich HN, et al. A molecular signature of proteinuria in glomerulonephritis. PLoS One. 2010;5:13451. doi: 10.1371/journal.pone.0013451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid H, et al. Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes. 2006;55:2993–3003. doi: 10.2337/db06-0477. [DOI] [PubMed] [Google Scholar]
- 36.Neusser MA, et al. Human nephrosclerosis triggers a hypoxia-related glomerulopathy. Am. J Pathol. 2010;176:594–607. doi: 10.2353/ajpath.2010.090268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodgin JB, et al. A molecular profile of focal segmental glomerulosclerosis from formalin-fixed, paraffin-embedded tissue. Am. J Pathol. 2010;177:1674–1686. doi: 10.2353/ajpath.2010.090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin. Invest. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stern R, et al. Lactate stimulates fibroblast expression of hyaluronan and CD44: the Warburg effect revisited. Exp. Cell Res. 2002;276:24–31. doi: 10.1006/excr.2002.5508. [DOI] [PubMed] [Google Scholar]
- 40.Maeda T, et al. Mechanism of the regulation of organic cation/carnitine transporter 1 (SLC22A4) by rheumatoid arthritis-associated transcriptional factor RUNX1 and inflammatory cytokines. Drug Metab. Dispos. 2007;35:394–401. doi: 10.1124/dmd.106.012112. [DOI] [PubMed] [Google Scholar]
- 41.Toyohara T, et al. SLCO4C1 transporter eliminates uremic toxins and attenuates hypertension and renal inflammation. J Am. Soc. Nephrol. 2009;20:2546–2555. doi: 10.1681/ASN.2009070696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witzgall R, et al. Localization of proliferating cell nuclear antigen vimentin, c-Fos clusterin in the postischemic kidney Evidence for a heterogenous genetic response among nephron segments, and a large pool of mitotically active and dedifferentiated cells. J Clin. Invest. 1994;93:2175–2188. doi: 10.1172/JCI117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duffield JS, et al. Restoration of tubular epithelial cells during repair of the postischemic kidney occurs independently of bone marrow-derived stem cells. J Clin. Invest. 2005;115:1743–1755. doi: 10.1172/JCI22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu CF, et al. Transforming growth factor beta-1 stimulates profibrotic epithelial signaling to activate pericyte-myofibroblast transition in obstructive kidney fibrosis. Am. J Pathol. 2013;182:118–131. doi: 10.1016/j.ajpath.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haverty TP, et al. Characterization of a renal tubular epithelial cell line which secretes the autologous target antigen of autoimmune experimental interstitial nephritis. J Cell Biol. 1988;107:1359–1368. doi: 10.1083/jcb.107.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vega S, et al. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Megyesi J, et al. The lack of a functional p21(WAF1/CIP1) gene ameliorates progression to chronic renal failure. Proc. Natl. Acad. Sci. U S A. 1999;196:10830–10835. doi: 10.1073/pnas.96.19.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooke VG, et al. Pericyte depletion results in hypoxia-associated epithelial-to-mesenchymal transition and metastasis mediated by met signaling pathway. Cancer cell. 2012;21:66–81. doi: 10.1016/j.ccr.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wynn TA. Fibrosis under arrest. Nature medicine. 2010;16:523–525. doi: 10.1038/nm0510-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online method references
- 51.LeBleu VS, et al. Identification of human epididymis protein-4 as a fibroblast-derived mediator of fibrosis. Nature medicine. 2013;19:227–231. doi: 10.1038/nm.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smyth GK. Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions using R and Bioconductor. 2005 [Google Scholar]
- 53.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nature protocols. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.