Abstract
In solid tumors, resistance to therapy inevitably develops upon treatment with cytotoxic drugs or molecularly targeted therapies. Here, we describe a system that enables pooled shRNA screening directly in mouse hepatocellular carcinomas (HCC) in vivo to identify genes likely to be involved in therapy resistance. Using a focused shRNA library targeting genes located within focal genomic amplifications of human HCC, we screened for genes whose inhibition increased the therapeutic efficacy of the multikinase inhibitor sorafenib. Both shRNA-mediated and pharmacological silencing of Mapk14 (p38α) were found to sensitize mouse HCC to sorafenib therapy and prolong survival by abrogating Mapk14-dependent activation of Mek-Erk and Atf2 signaling. Elevated Mapk14-Atf2 signaling predicted poor response to sorafenib therapy in human HCC, and sorafenib resistance of p-Mapk14-expressing HCC cells could be reverted by silencing Mapk14. Our results suggest that a combination of sorafenib and Mapk14 blockade is a promising approach to overcoming therapy resistance of human HCC.
Cancer genomes are heterogeneous and complex, and distinguishing oncogenic drivers from bystander lesions that occur as a result of genomic instability remains a major challenge. In contrast to some hematopoietic malignancies, for which molecular therapies can induce long-lasting tumor remissions, clinical experiences over the past couple of years have revealed that in the most common types of solid tumors, acquired therapy resistance against molecular therapies is inevitable 1–3. Hepatocellular carcinoma can be seen as a prototypical therapy-resistant tumor, and it represents a major health problem, causing more than 700,000 deaths annually worldwide4. HCC shows intrinsic resistance to cytotoxics5,6, and although the multikinase inhibitor sorafenib was recently approved as the first systemic treatment for patients with advanced HCC, the survival advantage conferred to these patients from sorafenib therapy averages only 2.8 months7. Sorafenib targets wild-type Raf1, mutant and wild-type Braf, and vascular endothelial growth factor receptors 2 and 3 (Vegfr2, Vegfr3) 8, and it is currently unclear how sorafenib resistance occurs at the molecular level.
Taking advantage of a recently developed system for transposon-mediated in vivo delivery of miRNA-based short hairpin RNAs (shRNAs)9,10, we developed a platform that can be used to conduct negative-selection shRNA screens directly in mouse liver carcinomas in vivo. A pooled shRNA screen conducted to identify target genes whose inhibition increases the therapeutic efficacy of sorafenib identified Mapk14-dependent activation of Mek-Erk and Atf2 signaling as a key mechanism of sorafenib resistance in mouse and human liver cancer. Combined sorafenib treatment and Mapk14 inhibition decreased proliferation of HCCs in vivo and significantly prolonged survival of tumor-bearing mice. Our results establish a tractable system for functional and direct in vivo identification of treatment-response modifiers in HCC and suggest that Mapk14 inhibition is a promising strategy to increase the therapeutic efficacy of sorafenib.
RESULTS
Generation of therapy-resistant mouse HCCs using a transposon-based mouse model
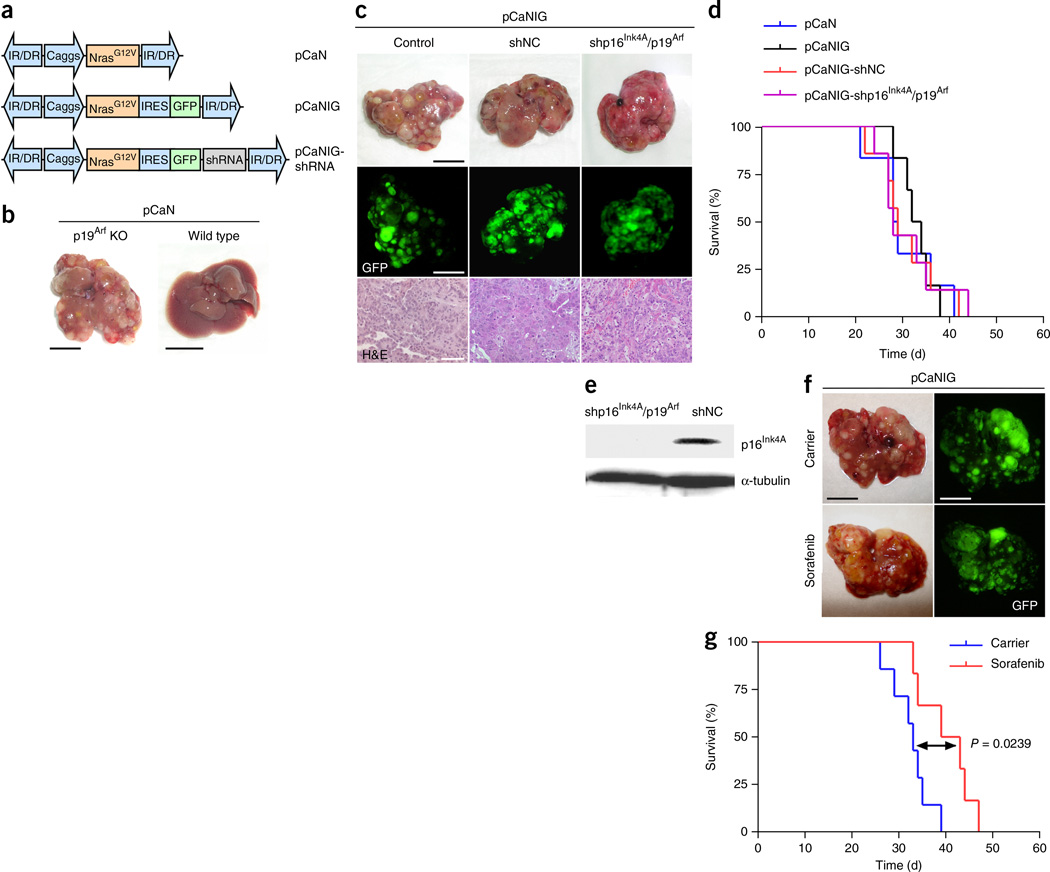
To model hepatocellular carcinoma in mice, we took advantage of a well-established mouse model in which transposable elements are stably delivered into the liver via hydrodynamic tail-vein injection10,11 (Supplementary Fig. 1). Stable delivery of oncogenic NrasG12V (using the pCaN vector; Fig. 1a) into the livers of p19Arf-deficient mice triggers the growth of aggressive multifocal HCCs, whereas, as also reported recently10, no tumor growth is observed when NrasG12V was delivered into C57BL/6 wild-type livers (Fig. 1b). To facilitate imaging and quantification of HCCs, we generated a transposon vector for coexpression of NrasG12V and green fluorescent protein (GFP) (pCaNIG; Fig. 1a) and found that GFP expression did not affect either the tumor burden or the survival of tumor-bearing mice (Fig. 1c,d).
Figure 1.
A transposon-based mouse model of liver cancer shows therapy resistance resembling that of human HCC. (a) Schematic representation of transposable elements encoding oncogenic NrasG12V, GFP and miR30- based shRNAs. Caggs, CAGGS promoter; IR/DR, inverted repeats and direct repeats; IRES, internal ribosome entry site. (b) Representative photographs of intrahepatic tumor burden 5 weeks after delivery of NrasG12V (pCaN) into p19Arf-deficient or wild-type mice (n = 6 for each condition; scale bars, 1 cm). (c) Representative photographs (top), GFP imaging (middle) and H&E staining (bottom) 5 weeks after delivery of pCaNIG (Control), pCaNIG-shNC (containing a noncoding shRNA) and pCaNIG-shp16Ink4A/p19Arf into p19Arf-deficient mice (n = 6 for each condition; scale bars: 1 cm (top and middle), 50 µm (bottom)). (d) Survival analyses (Kaplan-Meier format) of the same p19Arf-deficient mice after delivery of pCaN (n = 6), pCaNIG (n = 6), pCaNIG-shNC (n = 7) and pCaNIG-shp16Ink4A/p19Arf (n = 7) transposons (no statistical significant difference between the different groups could be found by log-rank test). (e) In vivo knockdown test of transposon-encoded shRNAs by p16Ink4A western blot analysis from tumors triggered by pCaNIG-shp16Ink4A/p19Arf or pCaNIG-shNC delivery into p19Arf-deficient mice (n = 3 for each condition). (f) Intrahepatic tumor burden of p19Arf-deficient mice 5 weeks after injection of pCaNIG and treatment with sorafenib (100 mg/kg, n = 5) or carrier (n = 6) (representative photographs and GFP images; scale bars, 1 cm). (g) Survival analyses (Kaplan-Meier format) of p19Arf-deficient mice after injection of pCaNIG and treatment with sorafenib (n = 6) or carrier (n = 7) (treatment was started 1 week after injection; statistical significance was calculated using a log-rank test).
We recently showed that miRNA-based shRNAs (hereafter referred to as shRNAs) can be expressed efficiently from transposons to generate stable knockdown phenotypes in mouse livers9. To explore whether oncogene-encoding transposable elements can be used to engineer mouse HCCs with stable knockdown of target genes, we generated transposons (pCaNIG-shRNA; Fig. 1a) encoding NrasG12V, GFP, and either different noncoding shRNAs (shNC) or a previously described shRNA targeting Cdkn2a that encodes the tumor suppressors p16Ink4A and p19Arf (ref. 12) (shp16Ink4A/p19Arf). Transposon vectors were stably delivered into the livers of p19Arf-deficient mice, where they triggered HCCs with latency comparable to those of vectors without shRNA expression (Fig. 1c,d). Tumors stably expressing pCaNIG-shp16Ink4A/p19Arf showed efficient intratumoral p16Ink4A knockdown; however, p16Ink4A silencing did not further accelerate tumor growth (Fig. 1d,e). Histopathological analyses revealed that engineered mouse HCCs resemble moderately to poorly differentiated human HCC in their histopathology (Fig. 1c). In summary, these data show that multifocal HCCs can be triggered by direct and stable intrahepatic delivery of transposons encoding oncogenes, marker genes and shRNAs and that intratumoral shRNA expression results in high knockdown efficiency.
Currently more than 90% of all new oncology drugs entering early clinical testing fail in later phases of clinical development13,14, indicating that higher bars are needed in preclinical drug testing. To assess whether our transposon-based mouse model of liver cancer recapitulates the therapy resistance of human HCC, we treated mice harboring NrasG12V-driven HCCs with sorafenib. Strikingly, sorafenib barely reduced intrahepatic tumor burden (Fig. 1f) and resulted in a very limited, although statistically significant, survival advantage averaging 8 d (P = 0.0239) (Fig. 1g), which, taking into account the different lifespans of mice and humans, reflects the limited efficiency of sorafenib in human HCC7.
Direct in vivo RNAi screening for sorafenib sensitizer genes
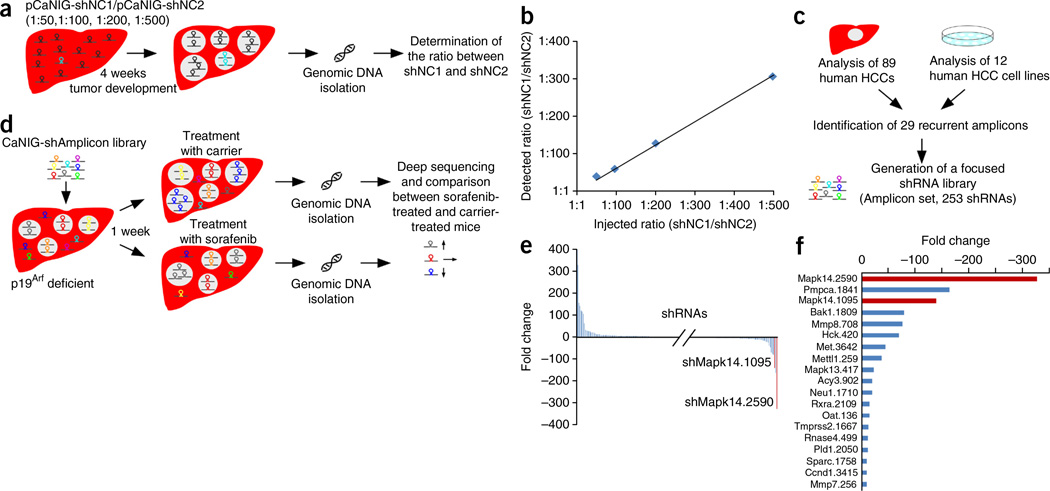
We reasoned that the established mouse model would be ideally suited for direct in vivo negative-selection RNA interference (RNAi) screens to identify gene products that mediate resistance to sorafenib. To determine the complexity of shRNA pools that could be screened in our model, we stably delivered two different noncoding shRNAs (shNC1 and shNC2) into mouse livers (pCaNIG vector; Fig. 1a) at different ratios. After 4 weeks, we harvested genomic DNA from tumor-bearing livers and used it for PCR amplification of shRNA cassettes (Fig. 2a). We performed deep sequencing of PCR products to quantify the abundance of each shRNA and calculated the ratios between them. Given that both shRNAs could be safely recovered from tumor-bearing livers, even when shRNAs were injected at a 1:500 ratio (Fig. 2b), we reasoned that pools with a complexity of up to 500 shRNAs could be used for shRNA screening in our mouse model.
Figure 2.
Outline and results of an in vivo RNAi screen to identify sorafenib treatment response modifiers. (a) Schematic outline of a dilution experiment to determine the maximum number of shRNAs that can be recovered from HCC-bearing mouse livers. (b) Results of the dilution experiment outlined in a. Plotted is the detected ratio of shNC1 to shNC2 in p19Arf-deficient livers 4 weeks after delivery of pCaNIG-shNC1 and pCaNIG-shNC2 (y axis), in relation to the ratio injected (x axis) (values represent mean; n = 2). (c) Schematic outline of the workflow for compilation of a focused shRNA library (Amplicon set). (d) Scheme of the in vivo RNAi screen. (e) Enrichment or depletion of individual shRNAs that were screened as described in d. The representation of each shRNA after sorafenib treatment (n = 9) was compared to its representation after treatment with carrier alone (n = 9). (f) Overview of shRNAs that showed at least 10-fold depletion in sorafenib-treated mice compared to carrier-treated mice. Plotted are the fold changes of shRNA abundance in the sorafenib-treated group compared to carrier-treated group. Top-scoring shRNAs targeting Mapk14 are marked in red.
Oncogenic drivers of human tumors are often found in focal chromosomal amplifications and might also be involved in mediating therapy resistance. We therefore compiled a focused shRNA library targeting genes located within focal genomic amplifications of approximately 100 human HCCs15 (Amplicon set, on average 3.4 shRNAs per gene) (Fig. 2c).
We generated pCaNIG-shRNA transposons encoding the amplicon shRNA set and delivered them into p19Arf-deficient mice. We treated half of the mice with sorafenib and the other half with carrier (Fig. 2d). After 5 weeks, we extracted genomic DNA from tumor-bearing livers of both groups and determined the abundance of each shRNA in each liver by deep sequencing (Fig. 2d). We calculated average values for the total number of sequence reads to determine the proportion of each shRNA within the whole population, and later used these data to calculate the fold enrichment or depletion of each shRNA between sorafenib- and carrier-treated mice (Fig. 2e,f and Supplementary Table 1). As expected, the majority of shRNAs had no impact on sorafenib sensitivity, and therefore their frequencies did not differ significantly between sorafenib- and carrier-treated mice (Fig. 2e,f). However, 28 shRNAs were enriched more than 10-fold, and 19 shRNAs were depleted more than 10-fold, in sorafenib- compared to carrier-treated mice (Fig. 2f, Supplementary Table 1). For follow-up studies, only gene targets that were represented by two independent shRNAs that each showed at least 10-fold depletion were included. Only one candidate gene, the mitogen-activated protein kinase 14 (Mapk14 or p38α) gene, fulfilled these criteria, with shMapk14.1095 being depleted 138-fold and shMapk14.2590 being depleted 324-fold.
shRNA-mediated Mapk14 knockdown sensitizes liver carcinomas for sorafenib
Mapk14, a member of the MAP kinase family, can be activated by various environmental stresses and proinflammatory cytokines. It represents a dual-function protein, as it can inhibit cell cycle progression or even induce cellular senescence or apoptosis in normal cells but increase proliferation in cancer cells16,17.
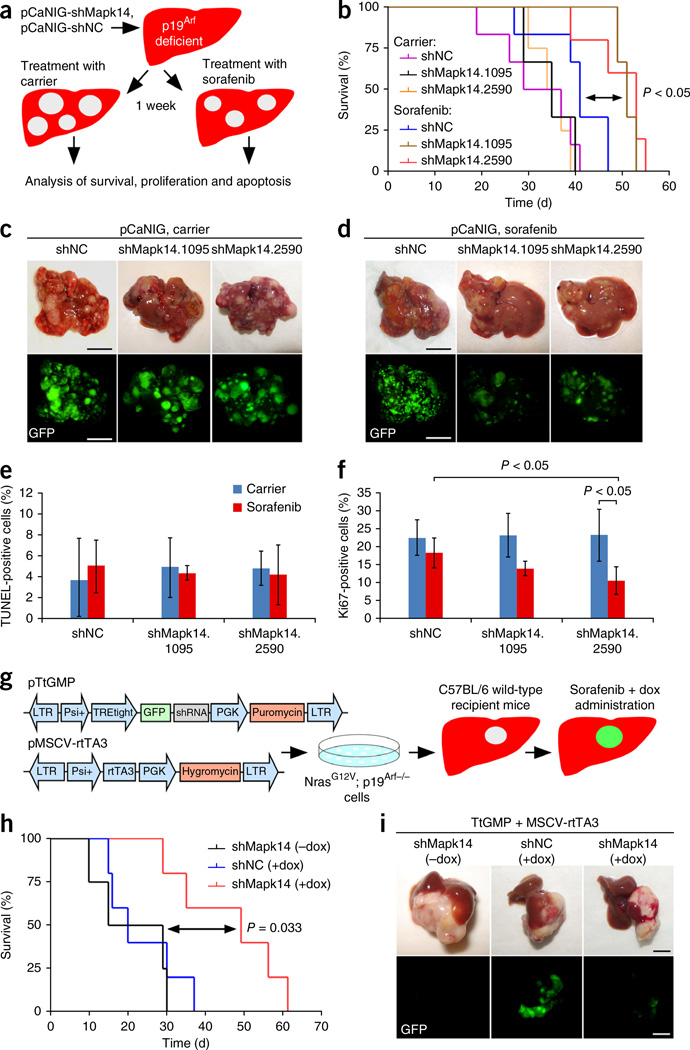
Both Mapk14 shRNAs identified in the screen yielded good knockdown efficiencies (Supplementary Fig. 2). We tested them individually using the experimental setup employed in the screen by hydrodynamically injecting p19Arf-deficient mice with pCaNIG-shMapk14.1095, pCaNIG-shMapk14.2590 or pCaNIG-shNC and treating them with sorafenib or carrier (Fig. 3a). Stable knockdown of Mapk14 per se had no impact on intrahepatic tumor burden or overall mouse survival (Fig. 3b,c), but intratumoral knockdown of Mapk14 sensitized the mice to sorafenib treatment and resulted in lower tumor burden and significantly longer survival than was observed for mice bearing shNC-expressing tumors treated with sorafenib (P < 0.05) (Fig. 3b,d). TdT-mediated dUTP-biotin nick end-labeling (TUNEL) showed no greater apoptosis of sorafenib-treated tumors as a result of Mapk14 knockdown (Fig. 3e and Supplementary Fig. 3a); however, Ki67 staining revealed markedly lower proliferation of sorafenib-treated shMapk14-expressing tumors compared to shNC-expressing tumors (Fig. 3f and Supplementary Fig. 3b), thus suggesting that sorafenib sensitization through Mapk14 inhibition is due mainly to reduced tumor cell proliferation.
Figure 3.
Functional genetic validation of Mapk14 as a sorafenib sensitizer gene. (a) Schematic outline of functional genetic testing of Mapk14 shRNAs in NrasG12V; p19Arf-deficient mouse HCCs. (b) Survival analyses (Kaplan-Meier format) of p19Arf-deficient mice injected with pCaNIG-shMapk14 or pCaNIG-shNC transposons treated with sorafenib or carrier starting 1 week after injection (carrier: shNC, n = 6; shMapk14.1095, n = 3; shMapk14.2590, n = 4; sorafenib: shNC, n = 6; shMapk14.1095, n = 3; shMapk14.2590, n = 5; statistical significance was calculated using a log-rank test). (c,d) Intrahepatic tumor burden of p19Arf-deficient mice 5 weeks after injection of pCaNIG-shMapk14 or pCaNIG-shNC transposons and administration of carrier or sorafenib (carrier: shNC, n = 6; shMapk14.1095, n = 5; shMapk14.2590, n = 5; sorafenib: shNC, n = 6; shMapk14.1095, n = 5; shMapk14.2590, n = 5; representative photographs and GFP images; scale bars, 1 cm). (e,f) Quantification of TUNEL- (e) and Ki67-positive (f) cells in p19Arf-deficient tumor nodules 5 weeks after injection of pCaNIG-shMapk14 or pCaNIG-shNC transposons and administration of carrier or sorafenib (carrier: shNC, n = 3; shMapk14.1095, n = 3; shMapk14.2590, n = 3; sorafenib: shNC, n = 4; shMapk14.1095, n = 3; shMapk14.2590, n = 3; represent mean ± s.d.; statistical significance calculated using two-tailed Student’s t-test). (g) Schematic outline for the generation of NrasG12V-driven liver tumors with inducible shRNA expression. (h) Survival analyses (Kaplan-Meier format) of wild-type mice after in situ transplantation of NrasG12V; p19Arf-deficient liver tumor cells stably expressing MSCV-rtTA3 and TtGMP-shMapk14 or TtGMP-shNC, with doxycycline (dox) and sorafenib administered starting 1 week after injection (shMapk14 - doxycycline, n = 4; shNC + doxycycline, n = 5; shMapk14 + doxycycline, n = 5; statistical significance was calculated using a log-rank test). (i) Representative pictures of tumor burden 6 weeks after in situ transplantation of NrasG12V; p19Arf-deficient cells expressing TtGMP-shMapk14 or TtGMP-shNC and treatment as described in i (scale bars, 1 cm; n = 5 for each condition).
To explore whether the combined use of sorafenib and Mapk14 inhibition shows efficacy against advanced HCC, we next took advantage of a mouse model enabling conditional RNA interference in progressed HCC. We injected mouse NrasG12V; p19Arf-deficient HCC cells stably expressing doxycycline-responsive Mapk14 or control shRNAs (pTtGMP and rtTA3 vectors; Fig. 3g) under the liver capsules of immunocompetent C57BL/6 mice, where orthotopic HCCs developed. Doxycycline was administered 1 week after cell injection, and treatment with sorafenib or carrier was started simultaneously (Fig. 3g). Strikingly, we found marked therapeutic efficacy even in this intervention treatment setup of already advanced HCCs. Mice with induced shMapk14 (+doxycycline) lived significantly longer than mice without Mapk14 knockdown under sorafenib treatment (P = 0.033 shNC versus shMapk14, +doxycycline) and also showed a strongly reduced tumor burden (Fig. 3h,i). Notably, tumors that arose in doxycycline-treated shMapk14 mice showed weak to no GFP expression (Fig. 3i), which, as GFP and shRNA expression are linked in our system, suggests strong selection against shRNA expression. Therefore, the data we obtained may even underestimate the real therapeutic efficacy of combined sorafenib and Mapk14 inhibition treatment.
Pharmacological inhibition of Mapk14 increases therapeutic efficacy of sorafenib
We next set out to explore whether our genetic findings could be translated into a pharmacological combination therapy. Several Mapk14 inhibitors are available18,19, and we confirmed efficient Mapk14 inhibition in HCC cells upon exposure to the inhibitors BIRB796 and SB202190 by detection of reduced levels of phosphorylated Hsp27, an established downstream target of Mapk14 (Supplementary Fig. 4).
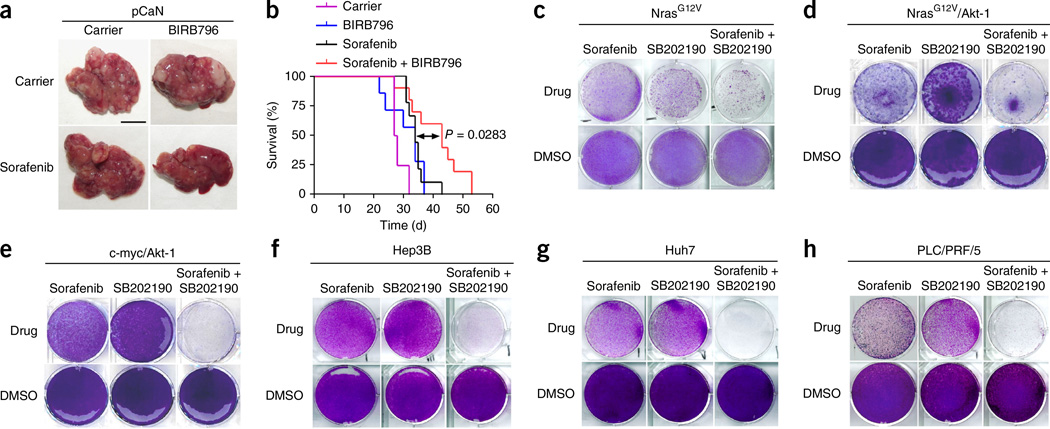
To test the efficacy of pharmacological combination therapy of sorafenib and Mapk14 inhibition, we next treated mice with sorafenib and BIRB796 after orthotopic and autochthonous HCCs had been triggered by oncogenic NrasG12V in the livers of p19Arf-deficient mice. We treated tumor-bearing mice with either carrier, sorafenib or BIRB796 alone or in combinations, again starting 1 week after induction of tumor growth. Strikingly, mice receiving BIRB796 combined with sorafenib showed markedly lower intrahepatic tumor burden and significantly longer survival than mice given monotherapy (Fig. 4a,b).
Figure 4.
Pharmacological inhibition of Mapk14 sensitizes to sorafenib therapy. (a) Intrahepatic tumor burden of p19Arf-deficient mice 4 weeks after stable delivery of NrasG12V (pCaN) and treatment with sorafenib, BIRB796, sorafenib and BIRB796 combination and corresponding carriers (representative photographs; scale bar, 1 cm; n = 5 for each condition). (b) Survival analyses (Kaplan-Meier format) of p19Arf-deficient mice after injection of NrasG12V and indicated treatment, as described in a (carrier, n = 4; BIRB796, n = 7; sorafenib, n = 9; sorafenib + BIRB796, n = 10; statistical significance was calculated using a log-rank test). (c) Crystal violet staining of NrasG12V; p19Arf-deficient cells after 4 d of treatment with sorafenib, SB202190, a combination thereof or corresponding DMSO concentrations (representative photographs, n = 3 for each condition). (d,e) Colony formation (crystal violet staining) of p19Arf-deficient NrasG12V/Akt-1 and c-myc/Akt-1 cells after 4 d of treatment with sorafenib, SB202190, a combination thereof or corresponding DMSO concentrations (representative photographs, n = 3 for each condition). (f–h) Colony formation of Hep3B, Huh7 and PLC/PRF/5 cells after 4 d of the indicated treatment. Cells were treated with sorafenib, SB202190, a combination thereof or corresponding DMSO concentrations (representative photographs, n = 3 for each condition).
To confirm our results with a second, independent Mapk14 inhibitor, we incubated cells from an NrasG12V-driven mouse liver tumor with SB202190. In line with our previous data, we found only a subtle reduction in colony formation upon sorafenib or SB202190 monotherapy; however, colony formation was strongly suppressed when sorafenib and SB202190 were combined (Fig. 4c).
We next extended our studies to mouse liver cancer cells of other genotypes to address whether combined sorafenib and Mapk14 inhibitor therapy is applicable to liver carcinomas with different driving oncogenes. Strikingly, we observed similar activity of our combination therapy both in NrasG12V-driven liver carcinomas, which additionally harbor a constitutively active form of Akt-1 (NrasG12V; Akt-1; p19Arf-deficient cells) (Fig. 4d), and in tumor cells outgrown from c-myc/ Akt-1–driven liver carcinomas (Fig. 4e). We also found pronounced therapeutic efficacy when we combined sorafenib and different Mapk14 inhibitors to treat several different human HCC cells, namely Hep3B, Huh7 and PLC/PRF/5 cells (Fig. 4f–h and Supplementary Fig. 5). Taken together, our data suggest that sorafenib and Mapk14 inhibition therapy shows robust therapeutic activity in mouse and human liver carcinomas of different genetic backgrounds.
Second-generation Mapk14 inhibitors combine high target selectivity and therapeutic efficacy without adverse drug effects in vivo
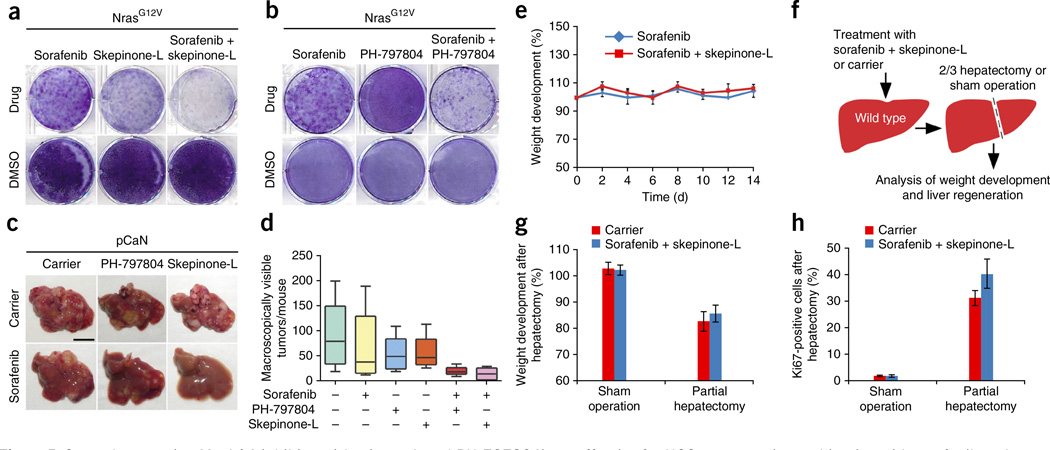
To set the basis for a rapid translation of our results into clinical trials, we next explored the therapeutic efficacy of highly selective and potent second-generation Mapk14 inhibitors (skepinone-L and PH-797804), which, in contrast to first-generation Mapk14 inhibitors20, show no significant side effects when used preclinically and in clinical trials21–24. Although skepinone-L is still undergoing pre-clinical testing, PH-797804 is currently in a phase 2 clinical trial for the treatment of patients with chronic obstructive lung disease (COPD) and rheumatoid arthritis, and interim analyses have yielded no signs of significant adverse side effects24. Both second-generation Mapk14 inhibitors showed pronounced therapeutic efficacy when combined with sorafenib for the treatment of mouse and human HCCs in vitro (Fig. 5a,b and Supplementary Fig. 6a) and in vivo (Fig. 5c,d), and skepinone-L by trend showed superior efficacy (Fig. 5a–d and Supplementary Fig. 6a).
Figure 5.
Second-generation Mapk14 inhibitors (skepinone-L and PH-797804) are effective for HCC treatment in combination with sorafenib and exert no adverse effects. (a) Colony formation (crystal violet staining) of NrasG12V; p19Arf-deficient HCC cells after a 4-d treatment with sorafenib, skepinone-L, a combination thereof or corresponding DMSO concentrations (representative photographs, n = 3 for each condition). (b) Crystal violet staining of NrasG12V; p19Arf-deficient cells after a 4-d treatment with sorafenib, PH-797804, a combination thereof or corresponding DMSO concentrations (representative pictures, n = 3 for each condition). (c) Intrahepatic tumor burden of p19Arf-deficient mice 4 weeks after NrasG12V injection (pCaN) and treatment with sorafenib, skepinone-L or PH-797804, combinations thereof and carrier (scale bar, 1 cm; n = 5 (carrier, PH-797804, skepinone-L, sorafenib + PH-797804, sorafenib + skepinone-L) or n = 6 (sorafenib)). (d) Quantification of macroscopically visible tumors 4 weeks after NrasG12V injection and treatments as described in c. (e) Weight changes in sorafenib-treated and sorafenib- and skepinone-L-treated p19Arf-deficient mice after NrasG12V delivery (mean ± s.d., n = 3 for each condition). (f) Testing of liver regeneration in sorafenib- and skepinone-L-treated mice after partial (two-third) hepatectomy. (g) Body weight development of mice treated with sorafenib and skepinone-L or carrier 42 h after two-third hepatectomy or sham surgery (mean ± s.d., n = 3 (carrier: sham operation, carrier: partial hepatectomy, treated: sham operation) or n = 4 (treated: partial hepatectomy)). (h) Percentage of Ki67-positive cells in livers, 42 h after two-third hepatectomy or sham surgery in sorafenib and skepinone-L or carrier mice (mean ± s.d., n = 3 (carrier: sham operation, carrier: partial hepatectomy, treated: sham operation) or n = 4 (treated: partial hepatectomy)).
As an indicator of potential adverse drug effects, we followed up weight development of mice treated with sorafenib + skepinone-L combination therapy, but we did not detect any decrease in body weight over time (Fig. 5e). As liver carcinomas mostly arise in chronically damaged livers with impaired function and regenerative capacity, it is of utmost importance that antitumorigenic treatments of HCC do not negatively impact the regenerative capacity of hepatocytes. To address whether our combination therapy affects liver regeneration, we performed two-third hepatectomies on mice that were on combined sorafenib + Mapk14 blockade treatment (Fig. 5f). Importantly, mice on combination therapy showed normal gain of body weight and even slightly better liver regeneration upon partial hepatectomy compared to carrier-treated mice (Fig. 5g,h and Supplementary Fig. 6b). These data show that a combinatorial treatment with sorafenib and second-generation Mapk14 inhibitors is well tolerated and has no negative impact on liver regeneration.
Mapk14 inhibition attenuates Mek-Erk signaling in sorafenib-treated HCC
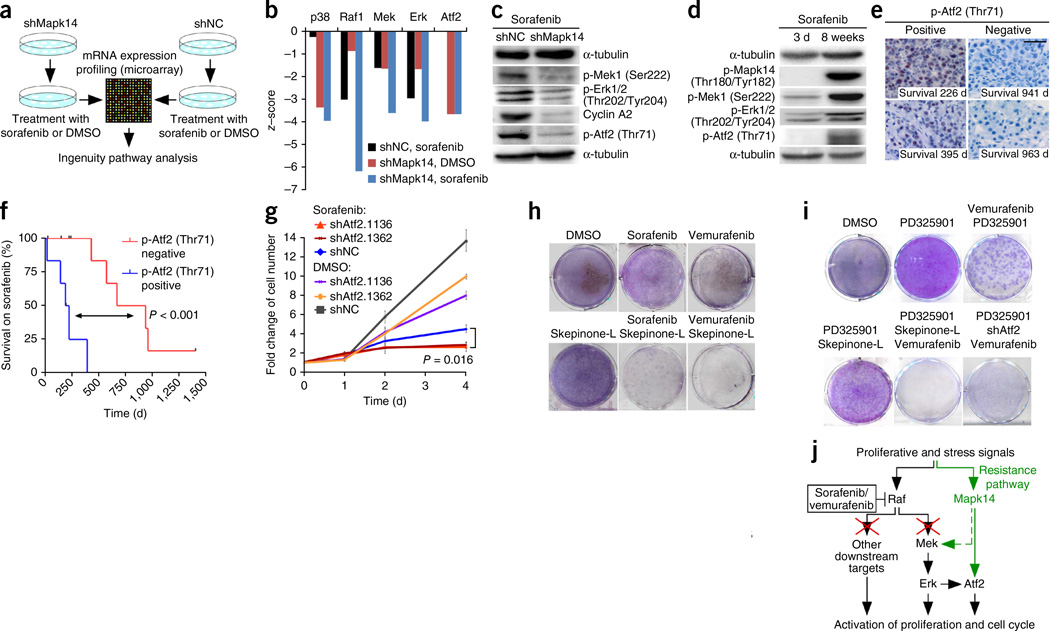
To gain mechanistic insights into how Mapk14 inhibition sensitizes HCCs to sorafenib, we subjected shMapk14-expressing and shNC-expressing mouse HCC cells to sorafenib treatment and conducted mRNA expression profiling and Ingenuity pathway analyses (Fig. 6a and Supplementary Fig. 7). These analyses revealed significantly downregulated Mapk14 (p38α)-dependent signaling in shMapk14-expressing cells compared to shNC-expressing cells, thus confirming the validity of our approach (P < 0.05) (Fig. 6b). In line with sorafenib’s high affinity for Raf proteins8, we also found a much lower number of Raf1-, Mek- and Erk-dependent signaling events in shNC-expressing cells treated with sorafenib than in those treated with DMSO. Unexpectedly, however, we also found that Raf1-, Mek- and Erk-dependent signaling was further attenuated upon shRNA-mediated silencing of Mapk14 (Fig. 6b), suggesting that there is cross-talk between Mapk14 signaling and the Raf-Mek-Erk cascade in liver cancer cells. Western blot analyses of sorafenib-treated shNC or shMapk14 cells confirmed that Mapk14 silencing resulted in a marked reduction of phospho- (p-) Mek1 and p-Erk1/2 using phospho-specific antibodies, which was accompanied by reduced cell proliferation, as indicated by strongly decreased expression of cyclin A (Fig. 6c).
Figure 6.
Cross-talk of Mapk14 and Mek-Erk signaling in HCC and its role in sorafenib resistance. (a) Outline of gene expression and Ingenuity pathway analysis of mouse hepatoma cells. (b) Ingenuity analysis of mRNA expression data of NrasG12V; p19Arf-deficient hepatoma cells expressing shNC or shMapk14 after treatment with sorafenib or DMSO. Data were normalized against those for DMSO-treated NrasG12V-shNC cells. (c) Sorafenib-treated cells as described in b were analyzed by western blotting using the indicated antibodies (n = 3 for each condition; representative images from at least two blots). (d) Western blot of NrasG12V; p19Arf-deficient mouse HCC cells upon 3 d or 8 weeks of sorafenib treatment (n = 3 for each condition; representative images from at least two blots) using the indicated antibodies. (e) p-Atf2 immunostaining of human HCC sections from biopsies taken before sorafenib therapy. Numbers indicate survival time under treatment. Images are representative of 16 patients. (f) Kaplan-Meier survival analyses of sorafenib-treated HCC patients with high (positive, n = 6) versus low (negative, n = 10) p-Atf2 expression in biopsies taken before sorafenib therapy (log-rank test). (g) Cell doubling rates of NrasG12V-shNC and NrasG12V-shAtf2 cells treated with sorafenib or DMSO for 4 d (mean ± s.d., n = 4; two-tailed Student’s t-test). (h) Colony formation (crystal violet staining) of NrasG12V; p19Arf-deficient HCC cells after 4 d of the indicated treatment (representative pictures, n = 3 for each condition). (i) Colony formation of NrasG12V; p19Arf-deficient HCC cells after 4 d of treatment with the indicated inhibitors (representative pictures, n = 3 for each condition). (j) Scheme indicating the roles of Mapk14 and Raf-Mek-Erk signaling in sorafenib sensitivity and resistance.
Mek-Erk signaling is reported to be upregulated in more than 50% of all human HCCs25,26 and represents a major oncogenic driver in liver cancer25,27. Our data thus far suggested that sorafenib-mediated suppression of the Mek-Erk cascade (via Raf inhibition) is not fully efficient, and that suppression can be further enhanced through Mapk14 silencing. Given the important role of Mek-Erk signaling in controlling the proliferation of liver cancer cells, we hypothesized that Mapk14-dependent cross-signaling into the Mek-Erk cascade might be involved in mediating sorafenib resistance, as upregulation of Mapk14 signaling would allow Mek-Erk signaling to be maintained even when Raf was blocked by sorafenib. To address this hypothesis experimentally, we subjected mouse and human liver cancer cells to short- and long-term sorafenib treatment and analyzed p-Mapk14 as well as p-Mek and p-Erk levels. Indeed, long-term exposure of mouse or human hepatoma cells to sorafenib resulted in strong induction of p-Mapk14 accompanied by a marked increase in p-Mek and p-Erk (Fig. 6d and Supplementary Fig. 8a). HCC cells with p-Mapk14 induction and reactivated Mek-Erk signaling were no longer responsive to sorafenib treatment and regained high proliferation rates (Supplementary Fig. 8b). Detailed time-course analyses in panels of mouse and human hepatoma cells and NrasG12V-driven tumors in vivo revealed that the timing of p-Mapk14 induction under sorafenib treatment varied between different types of HCC (Supplementary Fig. 9). Furthermore, expression profiling and Ingenuity pathway analyses suggested that Tgfbr1-mediated signaling may be involved in Mapk14 induction and development of sorafenib resistance (Supplementary Fig. 10).
In further support of the hypothesis that induction of Mapk14 signaling may be a key mechanism of resistance to sorafenib treatment, we found marked clonal expression of p-Mapk14 in HCC tissue obtained from biopsies of two patients receiving sorafenib therapy (Supplementary Fig. 11a). For ethical reasons tumor biopsies are not routinely taken from patients undergoing sorafenib therapy, and we therefore also aimed to assess whether the pretherapeutic Mapk14 activation level can predict the outcome of sorafenib therapy in patients with HCC. Interestingly, high pretherapeutic staining of both p-Mapk14 and its downstream target p-Atf2 correlated with a shorter survival on sorafenib therapy (Fig. 6e,f and Supplementary Fig. 11b,c), whereas p-Atf2 staining more strongly predicted response to sorafenib therapy than p-Mapk14 (P < 0.001). This appears to be an example of the common phenomenon of signal amplification of downstream factors in a signaling pathway and highlights p-Atf2 as a robust biomarker for predicting response to sorafenib therapy. In further support of the idea that pretherapeutic differences in Mapk14 activity exist between sorafenib-sensitive and sorafenib-resistant tumors, studies on different mouse and human hepatoma cells revealed that pretherapeutic Mapk14 activation status correlated with response to sorafenib therapy (Supplementary Fig. 12).
Because mRNA expression profiling and western blot analyses also revealed pronounced downregulation of Atf2-dependent signaling as a result of Mapk14 silencing (Fig. 6b,c), we sought to explore whether Atf2 reduction upon Mapk14 silencing per se contributes to sorafenib sensitization or whether the increased therapeutic efficacy of sorafenib is solely due to reduced Mek-Erk signaling in Mapk14-silenced cells. We generated mouse liver cancer cells with stable knockdown of Atf2 (Supplementary Fig. 13a), subjected them to sorafenib or DMSO treatment and quantified cell proliferation (Fig. 6g and Supplementary Fig. 13b). Although in the absence of sorafenib Atf2 knockdown reduced cell proliferation only slightly, the combination of sorafenib treatment and stable Atf2 suppression reduced cell proliferation below the level seen with sorafenib therapy alone. Therefore, our data suggest that decreased Atf2-dependent transcription may contribute to sorafenib sensitization upon Mapk14 blockade.
Recent reports suggest that, in addition to inducing Mek-Erk activation, Raf proteins can signal to other downstream targets in cancer cells28,29. To explore the significance of such additional Raf downstream targets in liver cancer, we compared the therapeutic efficacies of combinations comprising Raf inhibition (vemurafenib) plus Mapk14 blockade or Mek1/2 blockade (PD325901) plus Mapk14 blockade. Strikingly, whereas combined vemurafenib and Mapk14 blockade phenocopied the results obtained with sorafenib and Mapk14 blockade (Fig. 6h), blockade of both Mek and Mapk14 produced only a marginal antiproliferative effect (Fig. 6i). However, as expected, full therapeutic efficacy was observed when Raf inhibition, Mek blockade and Mapk14 inhibition were combined (Fig. 6i). Likewise, full therapeutic efficacy was observed when liver cancer cells with shRNA-mediated Atf2 knockdown were treated with a combination of Mek and Raf inhibitors, whereas combined Raf and Mek inhibition without additional Atf2 silencing did not yield the full antiproliferative effect (Fig. 6i). In summary, these data suggest a working model whereby full therapeutic efficacy of sorafenib and Mapk14 blockade therapy relies on three modules: (i) Mek-Erk inhibition, (ii) inhibition of non–Mek-Erk downstream targets of Raf and (iii) Atf2 inhibition (Fig. 6j).
Importantly, particularly in regard to clinical trials to test sorafenib and Mapk14 inhibition in patients with HCC, we found that sorafenib plus Mapk14 inhibition is effective against liver cancer cells that have developed sorafenib resistance (Supplementary Fig. 14). Accordingly, clinical trials testing sorafenib and Mapk14 inhibition can be designed so that patients can first undergo standard therapy (sorafenib) and then participate in a trial that explores the combination treatment.
DISCUSSION
Recent work has shown that RNAi-based functional genetic screening is a powerful approach to characterize molecular mechanisms of therapy resistance30–33. However, such screens were mostly conducted in vitro and therefore cannot survey gene activities that are related to the complex three-dimensional tumor architecture and microenvironment.
Here we present a unique platform that for the first time (to our knowledge) makes it possible to conduct shRNA drop-out screens in tumors directly in vivo—that is, in a setting where the tumor cells to be screened have never been removed from their natural three-dimensional environment. We reasoned that our system would therefore be ideally suited to dissect molecular mechanisms of therapy resistance in liver cancer and to pinpoint new target genes whose inhibition can increase the therapeutic efficacy of the multikinase inhibitor sorafenib, the only systemic treatment for HCC currently approved by the US Food and Drug Administration. Our screening platform turned out to be highly robust and functionally identified Mapk14 as a target whose inhibition strongly sensitizes tumors to sorafenib therapy. Against the background of its promiscuous cellular effects, it is unlikely that Mapk14 would have been selected as a candidate gene for sorafenib sensitization based on hypothesis-driven approaches. Therefore, our data emphasize the power of unbiased functional genetic screens to identify new gene targets in different biological systems.
Our mouse model is highly flexible and can also be used for screening other focused shRNA libraries, either as straight lethality screens to directly pinpoint new vulnerabilities or, in conjunction with other targeted therapies or cytotoxics, as a means to identify new drug sensitizer or resistance genes. Notably, as our data show that pools of up to 500 shRNAs can be screened per mouse with our approach, it is even conceivable to conduct genome-scale screens, assuming that a genome-scale library is split into sets of maximum 500 shRNAs and screened in a sufficient number of mice. We believe that the results we present herein have high and direct translational potential, as combination therapies of sorafenib and second-generation Mapk14 inhibitors should be practicable for patients with HCC without imposing significant side effects. Although first-generation p38 inhibitors lacked specificity for Mapk14 and therefore exerted toxicity in clinical trials20, highly specific and efficient Mapk14 inhibitors are now available that show no or very benign toxicity profiles21,24. For example, PH-797804 and ARRY-371797 have completed phase 1 or phase 2 clinical trials for different non-cancer-related diseases (for example, NCT00729209, NCT01366014, NCT00383188 and NCT00559910). The lack of toxicity, especially liver toxicity, is of particular importance for the treatment of patients with liver cancer as the majority of hepatocellular carcinomas arise in cirrhotic livers with impaired liver function. Regarding the design of clinical trials to test the efficacy of Mapk14 inhibition in combination with sorafenib in human HCC, it is noteworthy that Mapk14 blockade can restore sorafenib sensitivity in tumors that have become resistant upon long-term sorafenib treatment. This finding might facilitate the initiation of clinical trials as patients could first undergo standard therapy (sorafenib) and then participate in a trial exploring the combinatorial use of sorafenib and Mapk14 blockade. As our data suggest the Mapk14 downstream factor Atf2 as a robust biomarker to predict poor responses toward sorafenib therapy, it should be explored whether patients with higher pretherapeutic p-Atf2 signals are candidates to start combination therapy directly.
ONLINE METHODS
Vector and shRNA design
Sleeping beauty transposase (SB13) and NrasG12V transposon plasmids have been described recently9,10. The NrasG12V sequence in the transposon plasmid was first replaced by a polylinker. Next NrasG12V, c-myc and Akt-1 were inserted by PCR cloning using primers with AscI, MluI and NotI or AgeI restriction sites. GFP was added into transposon plasmids via PCR cloning using primers with NotI and AgeI restriction sites. The miR30 5′ sequence was inserted using AgeI and NheI. The shRNA against p16Ink4A/p19Arf and the noncoding shRNAs have been described before12,34. The Amplicon shRNA library was generated against genes that were initially identified based on ROMA analysis of genomic DNA of 89 human HCCs and 12 human HCC cell lines as described earlier15. The shRNA library and new single hairpins were designed using Biopred algorithms35 and new single shRNAs were ordered as 97-mer DNA oligos from MWG (Ebersberg, Germany). The shRNAs were PCR cloned into MSCV plasmids using XhoI and EcoRI and shuttled into transposon plasmids using XhoI, MluI and AscI fragments. The shRNA library was subcloned maintaining a 1,000-fold over-representation of colonies/number of shRNAs.
Animal strains and methods
All animal experiments were approved by the German or American legal authorities, and mice were kept under pathogen-free conditions in accordance with the institutional guidelines of the Helmholtz Centre for Infection Research (Braunschweig), the Cold Spring Harbor Laboratory (Cold Spring Harbor, New York, USA) and the University of Tuebingen (Tuebingen, Germany). For all animal experiments, only mice in a C57BL/6 background were used (equal gender distribution in randomized groups). C57BL/6 wild-type mice were purchased from Harlan (Rossdorf, Germany) or Janvier (Saint Berthevin, France). No statistical method was used to predetermine sample size. p19Arf-deficient mice were generated by C. Sherr (St. Jude Children’s Research Hospital, Memphis, Tennessee, USA) and were obtained in a C57BL/6 background from S.W. Lowe (Memorial Sloan Kettering Cancer Center, New York, New York, USA). DNA for hydrodynamic tail vein injection was prepared using Qiagen EndoFreeMaxi Kit and dissolved in 0.9% NaCl solution to a final volume of 10% of body weight. Animals (6 weeks old) were injected within 10 s with 25 µg transposon plasmids and 5 µg transposase (mice that were not efficiently injected were excluded from the experiment). For orthotopic transplantation of tumor cells, mice (10 weeks old) were anesthetized with rompun and ketamine and subjected to laparotomy, followed by injection of 106 cells into the left liver lobe. To avoid seeding of tumor cells into the abdominal cavity, the peritoneum was washed with distilled water. Doxycycline hyclate (Sigma-Aldrich) was added to the drinking water to a final concentration of 2 mg ml−1 supplemented with saccharose (1 g 100 ml−1 water). For drug administration, mice were treated with 100 mg kg−1 body weight sorafenib (Nexavar) and 50 mg kg−1 BIRB796, 40 mg kg−1 skepinone-L or 40 mg kg−1 PH-797804. Sorafenib and skepinone-L were dissolved in a 4× cremophor EL/95% ethanol solution (50:50). BIRB796 was dissolved in ethanol/water solution (25:75). Treatment was performed by oral gavage every second day. Two-third hepatectomy was done after mice (10 weeks old) were anesthetized with rompun and ketamine as described earlier36. Livers were photographed and sampled, and GFP imaging of mice and livers was done using a Hamamatsu imaging system. Counting of tumors was done in a blinded fashion.
Histopathology and immunohistochemistry
Haematoxylin and eosin (H&E) staining was performed on paraffin-embedded sections and evaluated by experienced pathologists (T.L. and P.S.). Sections of snap-frozen tissues were subjected to Ki67 (Abcam, ab15580, 1:200) and TUNEL (terminal deoxynucleotidyl transferase dUTP nick end-labeling, Roche) staining. Ten independent optical fields were counted in a blinded fashion (400× magnification).
Pseudonymized human HCC samples were provided by the tissue bank of the National Center for Tumor Diseases Heidelberg after approval by the ethics committee (project ID: 1359). Immunohistochemistry was performed on human HCC samples using an automated immunostainer (Ventana BenchMark XT, Ventana Medical Systems, Tucson, Arizona, USA) and standard protocols as supplied by the manufacturer. Citrate buffer (pH 6), for antigen retrieval, and the following antibodies were used: p-p38 (Thr180/Tyr182) (Cell Signaling, #4511 (D3F9), 1:1,000) and p-Atf2 (Thr71) (Cell Signaling, #9221, 1:50). Staining was assessed qualitatively and semiquantitatively using the immunoreactive score as described previously37.
Cell culture
Cells were cultured in DMEM medium supplemented with 10% FCS at 37 °C and 7% CO2. Generation of cell lines from mouse tumor nodules was accomplished under sterile conditions via dispase (1,000 U ml−1) and collagenase (0.1 U ml−1) (Roche) digestion in DMEM medium containing HEPES buffer for 30 min at 37 °C with gentle shaking. The cells were passed through nylon mesh filter (100 µm), centrifuged at 80 × g for 10 min and washed twice with medium. The cells were placed on a 0.1% gelatin-coated culture dish and maintained in culture. Huh7, Hep3B, PLC/PRF/5 and HepG2 cells were obtained from ATCC and authenticated. Mycoplasma contamination was excluded via a PCR-based method. Production of retroviral particles was accomplished using Phoenix packaging cells. The Phoenix cells were transfected with retroviral DNA via calcium phosphate-mediated transfection. The viral supernatant was applied directly on target cells and polybrene was added (1–10 µg ml−1) to enhance infection efficiency. Target cells were selected using puromycin (1–10 µg ml−1) or hygromycin (300–1,000 µg ml−1). For drug testing, cells were treated either with sorafenib (8 µM for mouse cells, 2 µM for Hep3B and 4 µM for Huh7 and PLC/PRF/5 cells), with SB202190 (50 µM for mouse cells, 12 µM for Hep3B and 20 µM for Huh7 and PLC/PRF/5 cells), with BIRB796 (12 µM for Hep3B and 20 µM for Huh7 and PLC/PRF/5 cells), with skepinone-L (20 µM for mouse cells and 15 µM for Hep3B cells), with PH797804 (20 µM), with vemurafenib (16 µM), with PD325901 (10 µM) or with corresponding combinations. The compounds were dissolved in DMSO and treated 1 d after plating. Cell proliferation rates were quantified using the cell doubling assay on a Guava flow cytometer. The doubling time was calculated as fold changes in comparison to the initial point. For clonogenic assays 104 cells were plated and cells were stained with 0.07% crystal violet solution. Visualization of drug- and DMSO-treated cells was done with crystal violet staining according to standard protocols.
Immunoblot analysis
Cells were washed in PBS and then lysed in NP40 lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP40, and protease and phosphatase inhibitors). 50–80 µg of protein were separated by SDS-polyacrylamide electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes (Amersham Hybond ECL) using a semidry blotting system. The blots were incubated with antibodies specific to α-tubulin (Cell Signaling 2125, (11H10) 1:3,000), p16Ink4A (Santa Cruz Biotechnology sc-1207, 1:500), p-Hsp27 (Ser82) (Cell Signaling 2401, 1:1,000), p-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (Cell Signaling 4370 (D13.14.4E), 1:1,000), p-Mek1 (Ser222) (Santa Cruz Biotechnology sc-293106, 1:200), p-p38 (Thr180/Tyr182) (Cell Signaling 4511 (D3F9), 1:1,000), cyclin A (Santa Cruz Biotechnology sc-596, 1:200), p38α (Cell Signaling 9218, 1:1,000), p-Tgfbr1 (Ser165) (Abcam ab112095, 1:500) and p-Atf2 (Thr71) (Cell Signaling 9221, 1:1,000), followed by peroxidase-conjugated secondary antibodies. Images were detected by ChemiDoc MP Imaging System (Bio-Rad).
mRNA expression, Ingenuity pathway and quantitative PCR analysis
mRNA was isolated from whole cells via TRIZOL (Invitrogen) and the RNeasy Mini Kit (Qiagen). mRNA gene expression microarrays were performed using Affymetrix GeneChips MOE430 2.0 and analyzed using GeneSpring 11.5.1 software (Agilent Technologies). Based on these data, the activity of signaling pathways was identified by Ingenuity pathway analysis software (IPA, Ingenuity Systems, Inc., http://www.ingenuity.com/). The data were analyzed using upstream regulator analysis (P ≤ 0.05) including only interactions that were experimentally observed or predicted with high confidence (Ingenuity Knowledge Base). cDNA synthesis was done with TaqMan Kit (Applied Biosystems) using random hexamer primers. Quantitative qPCR was performed with SYBRgreen Master Mix (Applied Biosystems). Values were normalized toward beta-actin quantification.
ShRNA recovery, identification and quantification
For deep sequencing, DNA was isolated from whole liver tissues or cells. Hairpin sequences were amplified using primers harboring the Illumina adaptor sequence38 and a sample identifier (3-base barcode). Deep sequencing analyses were performed in the Genome Analytics Group at the Helmholtz Centre for Infection Research (R.G.) using an Illumina GA IIx with a 46-bp single end run. Image analysis, base calling and first quality filtering steps of the sequencing results were done by the Illumina Pipeline Vers. 1.8., producing a short-read container file in FASTQ format. Data analysis and barcode attribution was performed by using a Perl-Script. The sequencing results were blasted against hairpin sequences of shRNA library database (only 100% matches). We performed Laplace correction and calculated fold changes by quotients of mean values of the corresponding groups.
Statistics
In mouse experiments, survival was calculated from the date of hydrodynamic tail-vein injection to death. It was assumed that treatment would prolong the survival of treated mice. The association between survival and the investigated treatment was represented using Kaplan-Meier plots and assessed by log-rank tests. For human patients survival was calculated from initiation of sorafenib treatment and survival time was censored for patients who did not experience the investigated event, i.e. were alive at last contact. p-Mapk14 (Thr180/Tyr182) and p-Atf2 (Thr71) expression were dichotomized based on the median. The association between survival and expression was represented using Kaplan-Meier plots and compared by log-rank tests. Statistical analyses were implemented using Graph Pad Prism 4.03. P < 0.05 was considered statistically significant.
Other statistical analyses were performed using two-tailed Student’s t-test (using Graph Pad Prism 4.03 software, if not otherwise stated). P-values <0.05 were considered to indicate statistical significance.
ACKNOWLEDGMENTS
We thank A. Rinkel, N. Struever, C. Hermann, V. Geissler, U. Koppenhoefer, H. Riedesel, M. Jarek, M. Scharfe and the HZI Genome Analytics Group team for technical assistance. We thank the tissue bank of the National Center for Tumor Diseases Heidelberg for providing human HCC tissues. This work was supported by the German Research Foundation, DFG (Emmy Noether Programme ZE 545/2-1 to L.Z., the “Rebirth” Cluster of Excellence, project “Liver regeneration”, SFB/TRR77 and SFB685), the Helmholtz Association of German Research Centers (VH-NG-424 to L.Z.), the European Commission (project ‘Heptromic’) and the Wilhelm Sander Stiftung.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
The study was designed by L.Z. Research was conducted by R.R., D.D., T.L., K.M., T.W., T.-W.K., A.H., M.P., J.L., A.v.T., P.S., J.Z., K.-H.W., S.P., N.P.M., M.E., B.S., S.W.L., R.G., S.L. and L.Z. The manuscript was written by L.Z., R.R. and D.D.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Poulikakos PI, Rosen N. Mutant BRAF melanomas—dependence and resistance. Cancer Cell. 2011;19:11–15. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Hartsough E, Shao Y, Aplin AE. Resistance to RAF inhibitors revisited. J. Invest. Dermatol. 2014;134:319–325. doi: 10.1038/jid.2013.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berasain C. Hepatocellular carcinoma and sorafenib: too many resistance mechanisms? Gut. 2013;62:1674–1675. doi: 10.1136/gutjnl-2013-304564. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yau T, Chan P, Epstein R, Poon RT. Evolution of systemic therapy of advanced hepatocellular carcinoma. World J. Gastroenterol. 2008;14:6437–6441. doi: 10.3748/wjg.14.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lord R, Suddle A, Ross PJ. Emerging strategies in the treatment of advanced hepatocellular carcinoma: the role of targeted therapies. Int. J. Clin. Pract. 2011;65:182–188. doi: 10.1111/j.1742-1241.2010.02545.x. [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 8.Wilhelm S, et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 9.Wuestefeld T, et al. A direct in vivo RNAi screen identifies MKK4 as a key regulator of liver regeneration. Cell. 2013;153:389–401. doi: 10.1016/j.cell.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 10.Kang TW, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 11.Carlson CM, Frandsen JL, Kirchhof N, McIvor RS, Largaespada DA. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc. Natl. Acad. Sci. USA. 2005;102:17059–17064. doi: 10.1073/pnas.0502974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braumüller H, et al. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013;494:361–365. doi: 10.1038/nature11824. [DOI] [PubMed] [Google Scholar]
- 13.Holohan C, Van SS, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 14.Margutti S, Laufer SA. Are MAP kinases drug targets? Yes, but difficult ones. ChemMedChem. 2007;2:1116–1140. doi: 10.1002/cmdc.200600271. [DOI] [PubMed] [Google Scholar]
- 15.Sawey ET, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 17.Nebreda AR, Porras A. p38 MAP kinases: beyond the stress response. Trends Biochem. Sci. 2000;25:257–260. doi: 10.1016/s0968-0004(00)01595-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Shen B, Lin A. Novel strategies for inhibition of the p38 MAPK pathway. Trends Pharmacol. Sci. 2007;28:286–295. doi: 10.1016/j.tips.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein DM, Kuglstatter A, Lou Y, Soth MJ. Selective p38alpha inhibitors clinically evaluated for the treatment of chronic inflammatory disorders. J. Med. Chem. 2010;53:2345–2353. doi: 10.1021/jm9012906. [DOI] [PubMed] [Google Scholar]
- 20.Dominguez C, Powers DA, Tamayo N. p38 MAP kinase inhibitors: many are made, but few are chosen. Curr. Opin. Drug Discov. Devel. 2005;8:421–430. [PubMed] [Google Scholar]
- 21.Koeberle SC, et al. Skepinone-L is a selective p38 mitogen-activated protein kinase inhibitor. Nat. Chem. Biol. 2012;8:141–143. doi: 10.1038/nchembio.761. [DOI] [PubMed] [Google Scholar]
- 22.Hope HR, et al. Anti-inflammatory properties of a novel N-phenyl pyridinone inhibitor of p38 mitogen-activated protein kinase: preclinical-to-clinical translation. J. Pharmacol. Exp. Ther. 2009;331:882–895. doi: 10.1124/jpet.109.158329. [DOI] [PubMed] [Google Scholar]
- 23.Fischer S, et al. Dibenzosuberones as p38 mitogen-activated protein kinase inhibitors with low ATP competitiveness and outstanding whole blood activity. J. Med. Chem. 2013;56:241–253. doi: 10.1021/jm301539x. [DOI] [PubMed] [Google Scholar]
- 24.MacNee W, Allan RJ, Jones I, De Salvo MC, Tan LF. Efficacy and safety of the oral p38 inhibitor PH-797804 in chronic obstructive pulmonary disease: a randomised clinical trial. Thorax. 2013;68:738–745. doi: 10.1136/thoraxjnl-2012-202744. [DOI] [PubMed] [Google Scholar]
- 25.Calvisi DF, et al. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J. Hepatol. 2011;54:311–319. doi: 10.1016/j.jhep.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvisi DF, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Guichard C, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steelman LS, et al. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 29.Hindley A, Kolch W. Extracellular signal regulated kinase (ERK)/mitogen activated protein kinase (MAPK)-independent functions of Raf kinases. J. Cell Sci. 2002;115:1575–1581. doi: 10.1242/jcs.115.8.1575. [DOI] [PubMed] [Google Scholar]
- 30.Turner NC, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008;27:1368–1377. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanton C, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Giroux V, Iovanna J, Dagorn JC. Probing the human kinome for kinases involved in pancreatic cancer cell survival and gemcitabine resistance. FASEB J. 2006;20:1982–1991. doi: 10.1096/fj.06-6239com. [DOI] [PubMed] [Google Scholar]
- 33.Huang S, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgess DJ, et al. Topoisomerase levels determine chemotherapy response in vitro and in vivo . Proc. Natl. Acad. Sci. USA. 2008;105:9053–9058. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huesken D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 37.Schlaeger C, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511–520. doi: 10.1002/hep.22033. [DOI] [PubMed] [Google Scholar]
- 38.Dow LE, et al. A pipeline for the generation of shRNA transgenic mice. Nat. Protoc. 2012;7:374–393. doi: 10.1038/nprot.2011.446. [DOI] [PMC free article] [PubMed] [Google Scholar]